Abstract
Characterization of transcriptional networks is one of the main strategies used to understand how bacteria interact with their environment. To reveal novel regulatory elements in the human pathogen Staphylococcus aureus , we adapted a traditional transduction protocol to be used in a high-throughput format in combination with the publicly available S. aureus Nebraska Transposon Mutant Library. Specifically, plasmid transductions are performed in 96-well format, so that a single plasmid can be simultaneously transferred into numerous recipient strains. When used in conjunction with bioluminescent reporter constructs, this strategy enables parallel and continuous monitoring of downstream transcriptional effects of hundreds of defined mutations. Here, we use this workflow in a proof-of-concept study to identify novel regulators of the staphylococcal metalloprotease aureolysin. Importantly, this strategy can be utilized with any other bacterium where plasmid transduction is possible, making it a versatile and efficient tool to probe transcriptional regulatory connections.
Keywords: Staphylococcus aureus, transduction, reporter plasmid, screening, S. aureus Nebraska Transposon Mutant Library, NTML
Introduction
Staphylococcus aureus is a problematic human pathogen that encodes an arsenal of virulence determinants. While these factors enable the bacterium to infect nearly every niche within the human host, their expression is tightly controlled to maintain energy homeostasis [1, 2]. This regulation often occurs on a transcriptional level and is one of the main strategies for adaptation to the host environment during infection [1, 3]. The governing transcriptional networks are highly complex, incorporate many internal and external stimuli, and, consequently, allow the bacterium to adjust to, and survive, in a variety of niches within the host [1, 4–6].
Several strategies exist for the discovery and characterization of regulatory networks, though few approaches can investigate these connections in a high-throughput format. Historically, simple phenotypical readouts, including plating on agar containing substrates, e.g. for staphylococcal secreted virulence factors such as nucleases [7], proteases [8] and haemolysins [9, 10], have been used to characterize the effects of specific (regulatory) mutations on a gene of interest (GOI). While the need to isolate or create individual mutants for such screens was a significant bottleneck for staphylococcal research in the past, this limitation has been surmounted by the generation of the S. aureus Nebraska Transposon Mutant Library (NTML) [11]. This annotated array of ~2000 mutants of all non-essential genes in the S. aureus JE2 background is a rich resource to investigate staphylococcal physiology. In its 96-well format, the library has been the basis for several phenotypic screens exploring the effects of individual mutations on various aspects of staphylococcal biology, including, but not limited to, S. aureus pathogenesis [12, 13], adaptation to and survival of the host environment and corresponding stresses [14, 15], and bacterial ability to withstand antimicrobials [16–19].
However, these experimental strategies are limited by the availability of existing assays to probe the activity of the protein of interest. Furthermore, such approaches often only assess the endpoint of an experiment and do not take temporal aspects, such as growth phase-dependent effects, into account. The same is true for more current methodologies, including RNA seq approaches, that probe downstream transcriptional effects of individual mutations at only a specific time point, therefore merely providing a snapshot of the transcriptional landscape.
Due to these challenges, one of the most effective ways to probe regulatory links is through the use of transcriptional reporters, preferably those that can be easily measured and do not require the addition of external substrates. Bioluminescent reporters are a prime example, as reporter activity can be monitored in a plate reader over the course of the experiment. Nevertheless, once a reporter plasmid is generated, the transfer of reporter constructs to numerous genetic backgrounds (i.e. mutants of regulatory genes) can be labour intensive and therefore prevents high-throughput screens. To overcome these challenges, various studies have combined reporter plasmids with the use of transposon mutagenesis [20–22]. Specifically, mutant generation is frequently performed in the parental strain harbouring the reporter plasmid, presenting technical challenges during library preparation, and necessitating sequencing of strains identified in the screen.
In this study, we take advantage of the versatility of transcriptional reporter plasmids as well as the availability of the NTML to develop a simple workflow that allows for the parallel creation of hundreds of mutant reporter strains within a short amount of time. Through transduction of a reporter plasmid in a 96-well format, the effects of a large number of mutations on the expression of a GOI can be assessed within a matter of days. The method was used in a proof-of-concept study to test the effects of mutations on the promoter activity of the aur gene, which encodes the important staphylococcal metalloprotease aureolysin [23, 24]. This virulence factor is essential for immune evasion [25–27] and is known to be under the control of various regulatory proteins [28]. The results of the reporter screen were validated by plating mutants on agar containing the aureolysin substrate casein, highlighting the robustness and reproducibility of the approach. In conclusion, the method described here will be a valuable tool for the identification of new regulatory connections in S. aureus as well as in other tractable bacterial backgrounds, where a collection of mutants is available, and transduction is genetically feasible.
Methodology and discussion
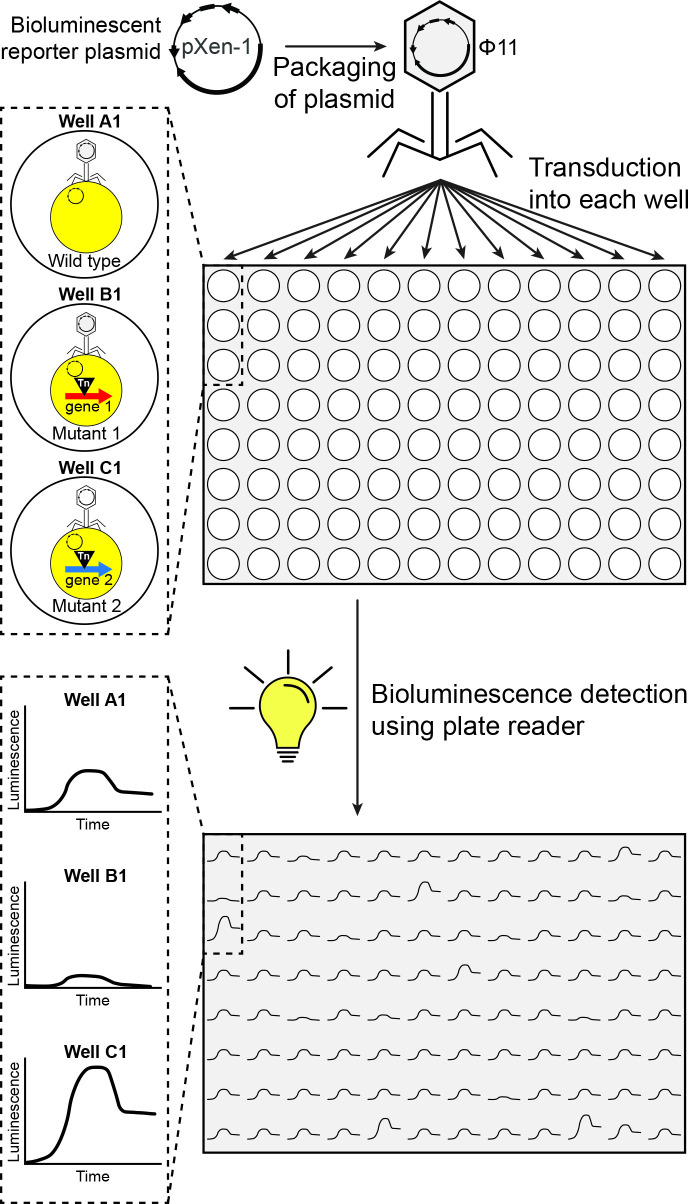
Transfer of genetic material by phage transduction has been a widely applied tool in bacteriology in general, and in staphylococcal research specifically. To increase the throughput of this strategy, a traditional phage transduction protocol was adapted to be applied to a 96-well plate layout and used in concert with the NTML that is often maintained in the same format (see supporting information for a detailed protocol). We reasoned that such a strategy would allow the simultaneous transduction of a bioluminescent reporter construct for a GOI into each of the mutants in the library. The resultant reporter activity could then be assessed continuously throughout the duration of an experiment using a luminescence plate reader (Fig. 1).
Fig. 1.
Schematic overview of the high-throughput transduction and screening strategy.
For a proof-of-concept study using this method, the impact of 93 different NTML mutations on the expression of the S. aureus aur gene was determined. To do so, the aur promoter was amplified using primers OL5271 (CGCGAATTCAGGCATCTGGTTTGTCGTTG) and OL5272 (GGGGATCCTCTAGGCTAAATCCACCGTC). The resulting PCR product was purified and digested with EcoRI and BamHI. The fragment was ligated into the pXen-1 vector coding for the promoterless luxABCDE operon [29] (Fig. S1, available in the online version of this article), and transformed into chemically competent Escherichia coli DH5α. Following plasmid confirmation by Sanger sequencing, constructs were transformed into electrocompetent S. aureus RN4220 [30]. RN4220 carrying the pXen-1-P aur-luxABCDE plasmid was used to generate ϕ11 phage lysates by using a broth-based method. Of note, several protocols are amendable to generate staphylococcal phage lysates to be used in this protocol, including those described by Novick [31] and Krausz et al. [32]. Further details pertaining generating phage lysates as well as determining phage titer can be found in the detailed protocol (supporting information).
For the initial experiment, we aimed to transduce the aur reporter plasmid into a subsection of the NTML (Table S1), including a sarA mutant strain. SarA is a known regulator of aur, and therefore served as a control to validate the method. Of note, while the NTML was constructed in the JE2 background, the general transduction protocol performs with equal efficiency in other S. aureus strains (e.g. Newman or USA300 LAC/Houston). Therefore, we anticipate the approach to be also amendable to other S. aureus strains and arrayed mutant libraries in these backgrounds.
First, each well of a fresh 96-well plate was filled with 150 µl Tryptic Soy Broth (TSB) supplemented with erythromycin at 5 µg ml−1 (the antibiotic marker carried by the NTML strains). Using a 12-channel pipette, the 96-well plate was directly inoculated from a single NTML 96-well freezer stock plate. Three of the 96 mutants of the original NTML plate were omitted to allow for the growth of three replicates of JE2 wild-type to serve as a reference. After incubation overnight (37 °C, shaking, 180 r.p.m.), 25 µl of each well was transferred into a fresh 96-well plate containing (i) 175 µl TSB, (ii) 20 µl phage and (iii) 2.2 µl of 1 M CaCl2. Following a 30 min incubation (37 °C, static), 6.7 µl of 1 M sodium citrate was added to each well to chelate Ca2+ and inhibit phage adsorption. Subsequently, bacteria were pelleted by centrifugation (4000 g , 10 min), resuspended in TSB supplemented with 5 mM sodium citrate and incubated for 1 h (37 °C, shaking, 180 r.p.m.). After incubation, using a 12-channel pipette, 10 µl of each well was plated on a 150 mm TSA plate supplemented with 5 mM sodium citrate and 10 µg chloramphenicol ml−1 to select for bacteria carrying the reporter plasmid. The plate was incubated overnight at 37 °C (static). The following day, single colonies were transferred onto fresh TSA plates containing 10 µg chloramphenicol ml−1 and 5 mM sodium citrate. After overnight incubation at 37 °C, bacterial colonies were used to seed a 96-well plate containing TSB with 10 µg chloramphenicol ml−1 using a 12-channel pipette. The plate was grown for 12 h (37 °C, shaking, 180 r.p.m.) before bacteria were pelleted and resuspend in TSB with 20 % (v/v) glycerol. The plate was sealed and stored at −80 °C.
The resulting reporter strains were then used to inoculate a fresh 96-well plate containing TSB supplemented with 10 µg chloramphenicol ml−1. Inoculation of fresh media was performed directly from 96-well freezer stock plates using a 12-channel pipette. After overnight growth (37 °C, shaking, 180 r.p.m.), cultures were diluted 1 : 100 into 200 µl of fresh TSB and grown for 3 h (37 °C, shaking, 180 r.p.m.). Synchronous cultures were standardized to an OD600 of 0.05 in a black clear-bottom 96-well plate with a final volume of 200 µl. This plate was incubated in a BioTek Cytation 5 plate reader for 7 h (37 °C, continuous double orbital shaking) with assessment of bioluminescent signal as well as culture density through measurement of OD600 at 15 min intervals.
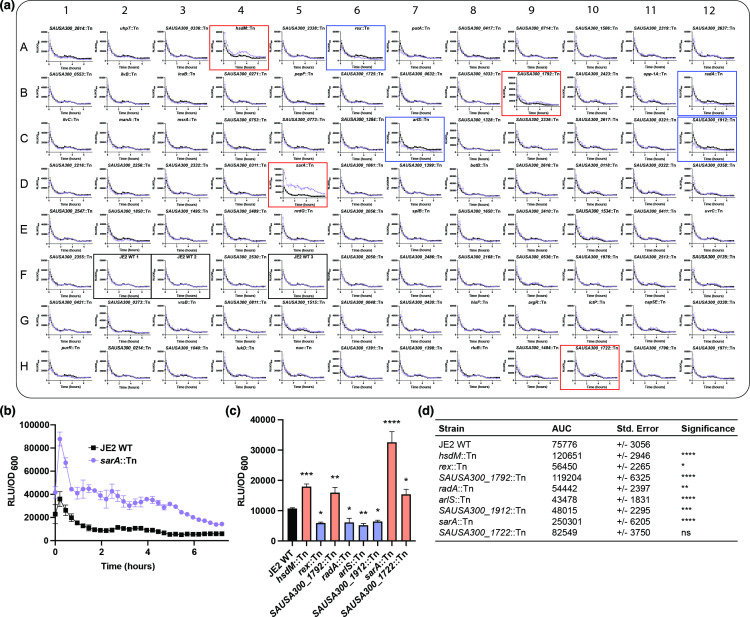
Several of the 93 transposon mutants included in this screen exhibited different levels of bioluminescence compared to JE2 wild-type (Fig. 2a, b). Comparisons were based either on OD600-normalized bioluminescence during the mid- to late exponential phase [3.5 h of growth, Fig. 2(c); raw relative light units (RLU) values are depicted in Fig. S2] or on cumulative bioluminescence throughout the entirety of the experiment, as determined by comparisons of the area under the curve (Fig. 2d). Four mutants, sarA::Tn, hsdM::Tn, SAUSA300_1722::Tn and SAUSA300_1792::Tn, showed increased luciferase expression, while four mutants, rex::Tn, radA::Tn, arlS::Tn and SAUSA300_1912::Tn, displayed lower luciferase activity. These data demonstrate that the method is suited to effectively assess positive and negative regulatory connections. In agreement with the known role of SarA as a repressor of aureolysin transcription [28, 33, 34], we observed a large increase in bioluminescence in the sarA::Tn strain (Fig. 2a–d, well D5), thereby validating our method. For a mutant of the redox-sensing transcriptional repressor, rex, a decrease in bioluminescence was observed (Fig. 2a, c and d, well A6). Notably, it has been reported that in a USA300 Houston rex::Tn mutant there is no change in aureolysin transcript levels during the post-exponential phase [28]. This could suggest that Rex functions in the regulation of aureolysin only prior to post-exponential growth or that strain-specific effects on aur regulation are detected. To our knowledge, for the remaining six transposon mutants showing differential expression of the luciferase operon (arlS::Tn, hsdM::Tn, radA::Tn, SAUSA300_1722::Tn, SAUSA300_1792::Tn and SAUSA300_1912::Tn), no roles for regulation of the aureolysin promoter have been assigned. This finding presents the exciting possibility of the discovery of novel regulatory connections for the proteolytic activity of S. aureus . Notably, the SAUSA300_1722::Tn mutant only showed significant differential expression at the 3.5 h timepoint (Fig. 2c), but not cumulatively over the course of the experiment (Fig. 2d). While determining the biological significance of this finding is beyond the scope of this methods paper, the observation highlights a strength of the approach. The ability to delineate growth phase-dependent effects thus offers a distinct advantage over strategies that only allow measurements at experimental endpoints and provide only cumulative data.
Fig. 2.
Examining P aur activity in numerous transposon mutants. (a) JE2 wild-type (WT) and transposon mutants carrying pXen-1-P aur-luxABCDE were grown for 7 h and bioluminescence and OD600 were assessed every 15 min. The experiment was repeated three times and data for each well were combined. Purple lines show data generated from specific wells (e.g. individual mutants), while black lines represent the average of three biological replicates as well as three technical replicates for JE2 WT. Black boxes denote the parental strain. Red and blue boxes indicate strains with increased and decreased bioluminescence, respectively. Data are represented as relative light units (RLU) normalized by optical density (OD600) to account for potential growth differences among transposon mutants. (b) Enlarged representative graph of (a) showing reporter activity for the sarA::Tn mutant (purple) and JE2 WT (black). (c) Comparison of bioluminescent signals from selected mutant strains and JE2 WT at 3.5 h of growth. (d) Curves from (a) were utilized to determine and compare the area under the curve (AUC) for the parental strain and selected mutants. Data in (b) and (c) were analysed by one-way ANOVA comparing each transposon mutant to the JE2 parental strain. Significance is denoted as: *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, ns – not significant.
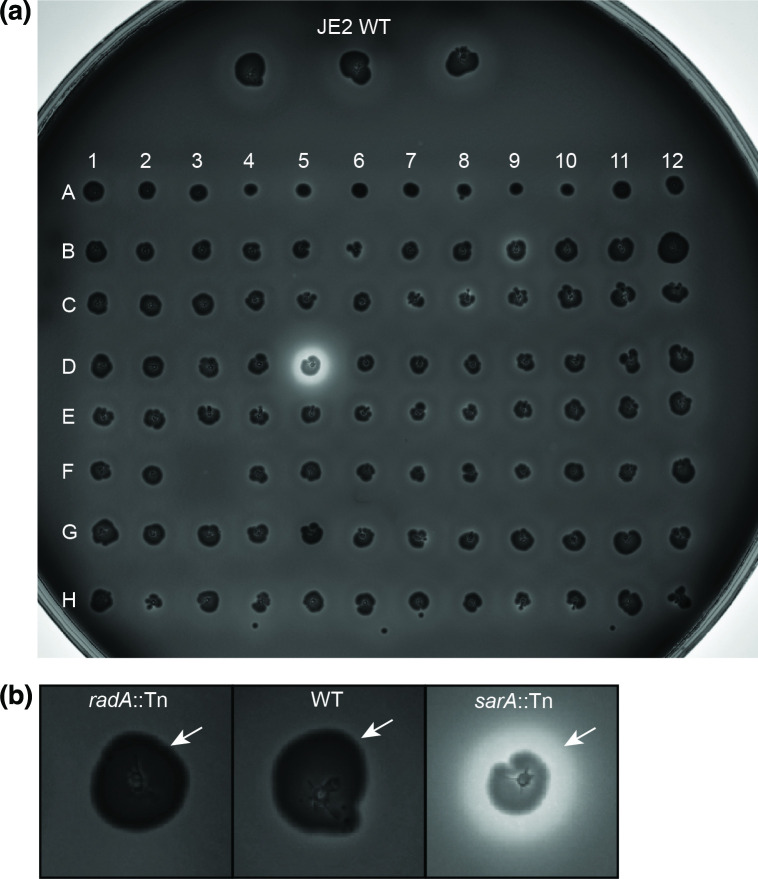
To confirm the results of the initial bioluminescence-based screen, we performed a separate assessment of aureolysin activity for each of the screened transposon mutants. We used the original plate containing the NTML mutants (without the reporter plasmid) alongside their parental strain to inoculate a casein agar plate and test aureolysin activity, as described previously [8]. Briefly, a casein agar plate composed of TSA supplemented with 5 % (w/v) dry milk was inoculated directly from glycerol stocks of the transposon mutants by lightly puncturing the surface of the agar using a multichannel pipette. The plate was incubated for 18 h at 37 °C (static) followed by 24 h at 4 °C (static) before being imaged using a Bio-Rad ChemiDoc MP Imaging System. The proteolytic cleavage of casein was assessed by observing clearing around the colonies on the plate (seen as the bright white clearing immediately surrounding the dark colonies, Fig. 3a, b). Our results were largely consistent with the findings obtained from the initial screen (Fig. 2). Most notably, we observed dramatically increased casein proteolysis by the sarA::Tn mutant (Fig. 3, well D5), mirroring the findings of the bioluminescent assay. The increased expression from the aureolysin promoter in the SAUSA300_1792::Tn mutant identified in the initial screen was also confirmed on casein agar (Fig. 3, well B9). These results suggest that SAUSA300_1792 may be a newly identified negative regulator of the aureolysin locus. In line with the results of the initial screen, we also observed a visible decrease in proteolysis for rex::Tn, radA::Tn and, to a slightly lesser extent, SAUSA300_1912::Tn (Fig. 3, wells A6, B12 and C12, respectively). Combined, these results confirm the robustness of our method. Nevertheless, some of the findings from the transcriptional screen were not recapitulated by the plate-based assay (including those for arlS::Tn and hsdM::Tn), highlighting the need for careful interpretation and evaluation of the generated data.
Fig. 3.
Evaluation of casein proteolysis using the transposon mutants. The proteolytic capacity of these transposon mutants was visually compared to that of the parental JE2 wild-type (WT) to confirm the results of the initial screen. The strains located at F2, F3 and F5 were not included in the initial screen to allow space for the JE2 WT. (b) Colonies of radA::Tn (B12), WT and sarA::Tn (D5) on the casein plate were enlarged and an arrow highlights the clearing of the casein plate around each strain.
Limitations and future perspectives
While in our hands the parallel transfer of reporter plasmids into numerous strains of the NTML was highly effective (success rate of 95–100 %), both, the application of phage-based methods to transfer genetic material between strains as well as the use of transposon mutants generally, carry inherent risks. Specifically, as with most phage-based methods, there is the possibility of phage integration into the genome and, subsequently, unwanted downstream effects. Analogously, when using transposon mutants, polar effects of the transposon insertions as well as the introduction of secondary mutations during library generation should be anticipated. Therefore, it is important to view approaches like the one described here as an initial screen that requires further validation of its results. Such confirmatory experiments could include the recapitulation of findings in strains with a deletion of a GOI, rather than through insertion as in a transposon mutant. Alternatively, specific transposon mutations for which regulatory effects were identified using our method could be transduced from the library into a new background (e.g. lab stock of WT S. aureus ) to recapitulate findings in a ‘clean’ background. Furthermore, validation of identified transcriptional connections should be performed by alternative methods, e.g. through RT-qPCR. Lastly, transcriptional effects do not necessarily result in alterations in protein abundance (e.g. as for arlS::Tn and hsdM::Tn in our screen) since additional (post-)translational regulatory mechanisms might be in place. Therefore, effects on protein abundance or functional assessments on the protein level should be conducted. Such experiments should also consider that plate-based assays (i) do not easily account for differences in growth rate between individual mutants and the parental strain (i.e. effects due to different colony sizes), and (ii) do not allow us to delineate results if a mutation only affects specific growth phases rather than the entire life cycle.
Conclusions
To our knowledge, this is the first description of transduction in S. aureus scaled up to high-throughput format and used in combination with the NTML and bacterial bioluminescent transcriptional reporter constructs. This method presents the possibility of genome-wide probing of transcriptional connections at low cost in a short amount of time. Consequently, this approach can assess expression of a GOI dependent on (i) (a set of) specific mutations like those included in the NTML, (ii) the growth-phase and (iii) the context of environmental stimuli such as chemical stressors, immune components or alterations in temperature. We believe that this flexible high-throughput approach will be a valuable tool to investigate transcriptional connections in S. aureus .
While we screened a single plate of the NTML, the simplicity of the method allows us to process multiple 96-well plates at a time. Given the scalability of the approach, it is possible to screen ~20 plates containing the entire library within a matter of weeks. Additionally, the setup can be adapted towards a specific research question by combining selected NTML mutants on individual 96-well plates (e.g. all known regulatory factors, individual two-component systems or strains deficient in specific metabolic pathways) to further streamline the screening process. The experimental approach outlined here can also serve as a blueprint for similar strategies in other bacteria for which mutant libraries are available and reporter plasmids (e.g. bioluminescent or short-lived fluorescent reporters) are a viable experimental tool.
Supplementary Data
Funding information
This work was supported by grants AI101171, R01AI073843 and R01AI069233 (E.P.S.) and AI124458 (L.N.S.) from the National Institute of Allergy and Infectious Diseases, as well as the Ernest W. Goodpasture Chair in Pathology (E.P.S.). A.W. was supported by the American Heart Association (18POST33990262), the NIH National Institute of Environmental Health Sciences (T32ES007028), and the NIH National Institute of Allergy and Infectious Diseases (F32AI157215).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: GOI, gene of interest; NTML, Nebraska Transposon Mutant Library; TSB, Tryptic Soy Broth.
Two supplementary figures, one supplementary table, and a detailed protocol are available with the online version of this article.
References
- 1.Jenul C, Horswill AR. Regulation of Staphylococcus aureus virulence. Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.GPP3-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim HK, Thammavongsa V, Schneewind O, Missiakas D. Recurrent infections and immune evasion strategies of Staphylococcus aureus . Curr Opin Microbiol. 2012;15:92–99. doi: 10.1016/j.mib.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mäder U, Nicolas P, Depke M, Pané-Farré J, Debarbouille M, et al. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet. 2016;12:e1005962. doi: 10.1371/journal.pgen.1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian D, Harper L, Shopsin B, Torres VJ. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis. 2017;75 doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibarra JA, Pérez-Rueda E, Carroll RK, Shaw LN. Global analysis of transcriptional regulators in Staphylococcus aureus . BMC Genomics. 2013;14:126. doi: 10.1186/1471-2164-14-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapun-Araiz B, Haag AF, De Cesare V, Gil C, Dorado-Morales P, et al. Systematic reconstruction of the complete two-component sensorial network in Staphylococcus aureus . mSystems. 2020;5:e00511-20. doi: 10.1128/mSystems.00511-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rydén AC, Lindberg M, Philipson L. Isolation and characterization of two protease-producing mutants from Staphylococcus aureus . J Bacteriol. 1973;116:25–32. doi: 10.1128/jb.116.1.25-32.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlsson A, Arvidson S. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect Immun. 2002;70:4239–4246. doi: 10.1128/IAI.70.8.4239-4246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, et al. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 10.Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus . J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4:e00537–12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang D, Ho YX, Cowell LM, Jilani I, Foster SJ, et al. A genome-wide screen identifies factors involved in S. aureus-induced human neutrophil cell death and pathogenesis. Front Immunol. 2019;10:45. doi: 10.3389/fimmu.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly J, Boldock E, Prince LR, Renshaw SA, Whyte MK, et al. Identification of Staphylococcus aureus factors required for pathogenicity and growth in human blood. Infect Immun. 2017;85:11. doi: 10.1128/IAI.00337-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golla RM, Mishra B, Dang X, Lakshmaiah Narayana J, Li A, et al. Resistome of Staphylococcus aureus in response to human cathelicidin LL-37 and its engineered antimicrobial peptides. ACS Infect Dis. 2020;6:1866–1881. doi: 10.1021/acsinfecdis.0c00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yee R, Feng J, Wang J, Chen J, Zhang Y. Identification of genes regulating cell death in Staphylococcus aureus . Front Microbiol. 2019;10:2199. doi: 10.3389/fmicb.2019.02199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vestergaard M, Leng B, Haaber J, Bojer MS, Vegge CS, et al. Genome-wide identification of antimicrobial intrinsic resistance determinants in Staphylococcus aureus . Front Microbiol. 2016;7:2018. doi: 10.3389/fmicb.2016.02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yee R, Cui P, Shi W, Feng J, Zhang Y. Genetic screen reveals the role of purine metabolism in Staphylococcus aureus persistence to rifampicin. Antibiotics (Basel) 2015;4:627–642. doi: 10.3390/antibiotics4040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Ingmer H, Vestergaard M. Genome-wide identification of resveratrol intrinsic resistance determinants in Staphylococcus aureus . Antibiotics (Basel) 2021;10:82. doi: 10.3390/antibiotics10010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wassmann CS, Rolsted AP, Lyngsie MC, Torres-Puig S, Kronborg T, et al. The menaquinone pathway is important for susceptibility of Staphylococcus aureus to the antibiotic adjuvant, cannabidiol. Microbiol Res. 2022;257:126974. doi: 10.1016/j.micres.2022.126974. [DOI] [PubMed] [Google Scholar]
- 20.Gongerowska-Jac M, Szafran MJ, Jakimowicz D. Combining transposon mutagenesis and reporter genes to identify novel regulators of the topA promoter in Streptomyces . Microb Cell Fact. 2021;20:99. doi: 10.1186/s12934-021-01590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolk CP, Cai Y, Panoff JM. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci U S A. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flentie K, Kocher B, Gammon ST, Novack DV, McKinney JS, et al. A bioluminescent transposon reporter-trap identifies tumor-specific microenvironment-induced promoters in Salmonella for conditional bacterial-based tumor therapy. Cancer Discov. 2012;2:624–637. doi: 10.1158/2159-8290.CD-11-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arvidson S. Studies on extracellular proteolytic enzymes from Staphylococcus aureus. II. Isolation and characterization of an EDTA-sensitive protease. Biochim Biophys Acta. 1973;302:149–157. doi: 10.1016/0005-2744(73)90017-x. [DOI] [PubMed] [Google Scholar]
- 24.Saheb SA. Purification and characterization of an extracellular protease from Staphylococcus aureus inhibited by EDTA. Biochimie. 1976;58:793–804. doi: 10.1016/s0300-9084(76)80310-0. [DOI] [PubMed] [Google Scholar]
- 25.Laarman AJ, Ruyken M, Malone CL, van Strijp JAG, Horswill AR, et al. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol. 2011;186:6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 26.Jusko M, Potempa J, Kantyka T, Bielecka E, Miller HK, et al. Staphylococcal proteases aid in evasion of the human complement system. J Innate Immun. 2014;6:31–46. doi: 10.1159/000351458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elmwall J, Kwiecinski J, Na M, Ali AA, Osla V, et al. Galectin-3 is a target for proteases involved in the virulence of Staphylococcus aureus . Infect Immun. 2017;85:e00177-17. doi: 10.1128/IAI.00177-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gimza BD, Larias MI, Budny BG, Shaw LN. Mapping the global network of extracellular protease regulation in Staphylococcus aureus . mSphere. 2019;4:e00676-19. doi: 10.1128/mSphere.00676-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francis KP, Joh D, Bellinger-Kawahara C, Hawkinson MJ, Purchio TF, et al. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect Immun. 2000;68:3594–3600. doi: 10.1128/IAI.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis . mBio. 2012;3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novick RP. Genetic systems in Staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 32.Krausz KL, Bose JL. Bacteriophage transduction in Staphylococcus aureus: broth-based method. Methods Mol Biol. 2016;1373:63–68. doi: 10.1007/7651_2014_185. [DOI] [PubMed] [Google Scholar]
- 33.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, et al. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol. 2012;86:1183–1196. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez AM, Beenken KE, Byrum SD, Tackett AJ, Shaw LN, et al. SarA plays a predominant role in controlling the production of extracellular proteases in the diverse clinical isolates of Staphylococcus aureus LAC and UAMS-1. Virulence. 2020;11:1738–1762. doi: 10.1080/21505594.2020.1855923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.