Abstract
ADNP and POGZ are two top-ranking risk factors for autism spectrum disorder and intellectual disability, but how they are linked to these neurodevelopmental disorders is largely unknown. Both ADNP and POGZ are chromatin regulators, which could profoundly affect gene transcription and cellular function in the brain. Using post-mortem tissue from patients with autism spectrum disorder, we found diminished expression of ADNP and POGZ in the prefrontal cortex, a region highly implicated in neurodevelopmental disorders. To understand the functional role of these neurodevelopmental disorder risk factors, we used viral-based gene transfer to investigate how Adnp or Pogz deficiency in mouse prefrontal cortex affects behavioural, transcriptomic and synaptic function. Mice with prefrontal cortex deficiency of Adnp or Pogz exhibited specific impairment of cognitive task performance. RNA-sequencing revealed that Adnp or Pogz deficiency induced prominent upregulation of overlapping genes enriched in neuroinflammation, similar to the elevation of pro-inflammatory genes in humans with neurodevelopmental disorders. Concomitantly, Adnp or Pogz deficiency led to the significant increase of pro-phagocytic microglial activation in prefrontal cortex, as well as the significant decrease of glutamatergic transmission and postsynaptic protein expression. These findings have uncovered the convergent functions of two top risk factors for autism spectrum disorder and intellectual disability in prefrontal cortex, providing a mechanism linking chromatin, transcriptional and synaptic dysregulation to cognitive deficits associated with neurodevelopmental disorders.
Keywords: neurodevelopmental disorders, ADNP, POGZ, microglia, synaptic deficits
ADNP and POGZ encode chromatin regulators and are top-ranking risk genes for autism and intellectual disability. Conrow-Graham et al. show that PFC deficiency of Adnp or Pogz in mice causes pro-inflammatory gene upregulation, leading to pro-phagocytic microglial activation and synaptic deficits, and resulting in cognitive impairment.
Introduction
The neurodevelopmental disorders (NDD) autism spectrum disorder (ASD) and intellectual disability (ID) co-occur at a rate of 30–70%. Converging evidence from whole-exome sequencing suggests the causal link of relatively few key genes to ASD/ID.1–4 Two identified top-ranking ASD/ID risk genes with the highest confidence are ADNP (activity-dependent neuroprotective protein) and POGZ (pogo transposable element with zinc finger domain).1,3,4 ADNP and POGZ are among the genes exhibiting the most frequent de novo loss-of-function mutations in ASD/ID.2,3,5 The ADNP syndrome, also called Helsmoortel-van der Aa syndrome, is characterized by moderate to severe ID and developmental delay, with ASD present in the majority of cases.6,7 POGZ syndrome, or White-Sutton syndrome, is similarly marked by a strong preponderance of ID and developmental delays, with comorbid ASD in about half of cases.8,9 At the molecular level, these ASD/ID risk genes share further similarities. Both are chromatin regulators/transcription factors involved in gene repression, probably through interaction with the chromatin architectural protein HP1 (heterochromatin protein 1).10–13 Recent evidence suggests that ADNP and POGZ are direct binding partners, acting together to regulate transcription.14 ADNP is also involved in the regulation of microtubules, autophagy, apoptosis and DNA stability, all of which are essential for neuronal homeostasis,15–18 while POGZ has additional functions in mitotic progression and DNA repair.10,19
The parallels of ADNP and POGZ in phenotypic expression and molecular function prompted us to investigate whether a commonly dysregulated pathway links these two risk factors to NDD. Similar to human syndromes, Adnp- or Pogz-deficient mice exhibit behavioural abnormalities and developmental delays,20–22 as well as impaired neural differentiation and neurogenesis during early development.21–23 Reduced dendritic spine density and altered synaptic gene expression have also been reported in Adnp-deficient mice.20,24 A recently generated mouse model expressing the most common ADNP syndrome mutation, p.Tyr718*, similarly shows developmental delays and reduced synaptic density.25 Yet how Adnp or Pogz deficiency is linked to synaptic and behavioural deficits in post-natal stages remains largely unknown.
One key region involved in ASD/ID is prefrontal cortex (PFC), a high-level cognitive centre controlling working memory, social functioning, self-regulatory and goal-directed behaviours.26 Clinical studies demonstrate severely impaired glutamatergic transmission and synaptic organization in PFC of ASD and ID.27,28 Restoring PFC synaptic function ameliorates behavioural deficits in ASD/ID mouse models.29–32 The PFC is the last brain region to mature, with much of the key synaptic development and circuitry refinement occurring during late childhood to early adulthood in humans, analogous to the juvenile to adolescent period (3–7 weeks) in rodents. Revealing the biological function of ADNP and POGZ in PFC during this critical period is vital for explaining pathophysiological mechanisms and potential treatment strategies for ASD/ID. Here we have used viral-based knockdown (KD) of Adnp or Pogz in PFC of juvenile mice to determine the mechanism linking chromatin, transcriptional and synaptic dysregulation to cognitive deficits associated with NDD.
Materials and methods
Animals and human post-mortem tissue
All animal studies were performed with the approval of the Institutional Animal Care and Use Committee of the State University of New York at Buffalo, USA. C57BL6/J mice, both male and female, were used for all experiments. Animals underwent stereotaxic viral injection at 4 weeks of age and were allowed at least 10 days for recovery and viral expression before being used for experiments. Frozen human post-mortem tissues (Brodmann’s area 9) were provided by the NIH Neuro BioBank (Supplementary Table 1). Human samples with RNA integrity number (RIN) <5.0 were excluded from analysis. The two groups (control and ASD) do not differ significantly in average RIN, post-mortem interval or age.
Virus generation
Short-hairpin RNA (shRNA) against mouse Adnp or Pogz were designed using the RNAi Consortium shRNA Library at the Broad Institute. To test the KD effect, we transfected the shRNA plasmids into 80–90% confluent mouse N2a cells using the Lipofectamine 2000 method. A plasmid containing a scrambled shRNA sequence was used as control. After 48 h of transfection, RNA was extracted for quantitative (q)PCR experiments. These verified shRNA plasmids were cloned into green fluorescent protein (GFP)-tagged adeno-associated virus (AAV) (serotype 9) vectors (Addgene) under the control of the U6 promotor. Viral particles were produced by the viral core centre of Emory University.
Animal surgeries
Four-week-old mice received bilateral stereotaxic injection of AAV (0.5 μl per hemisphere, 1.0 μl total volume) into medial PFC as described previously.31–34 In brief, mice were anaesthetized and placed on a stereotaxic apparatus (David Kopf Instruments). The injection was performed with a Hamilton syringe (gauge 31) at a speed of 0.1 μl/min, and the needle was kept in place for an additional 5 min. The virus was delivered to the target area using the following coordinates: 2.0-mm anterior to bregma; 0.25-mm lateral and 2.25-mm dorsal to ventral. The viral expression was observed throughout medial PFC (cingulate cortex, prelimbic and infralimbic). All experiments were conducted 10–20 days after surgery.
Real-time qPCR
Quantitative PCR was performed as previously described.32,33 Details are included in the Supplementary material.
Western blotting
Details are included in the Supplementary material.
Immunohistochemistry
Details are included in the Supplementary material.
Behavioural testing
Behavioural tests, including Barnes maze, Temporal order recognition, Novel object recognition, Three-chamber social preference, Social approach, RotaRod, Locomotion, Self-grooming and startle response/pre-pulse inhibition were carried out as previously described.35–39 Details are included in the Supplementary material.
Electrophysiological recording
Details are included in the Supplementary material.
RNA-sequencing and analysis
Total RNA was isolated from mouse PFC punches containing GFP fluorescence using the RNAeasy Mini kit coupled to an RNase-free DNase step (Qiagen). The dissected brain tissues from three mice were pooled as a single sample. RNA-seq libraries were constructed by TruSeq stranded total RNA plus Ribo-Zero kits (Illumina). Sequencing was carried out with the HiSeq 2500 platform (Illumina) at the Genomics and Bioinformatics Core of the State University of New York at Buffalo. Raw fastq paired-end sequencing reads were aligned to the mouse reference genome mm10 using RNA STAR (Galaxy v.2.7.5b). Gene expression of mapped reads was then measured using featureCounts (Galaxy v.1.6.4 + galaxy1), producing compatible gene expression matrices for downstream analyses. We then used DEseq2 (Galaxy v.2.11.40.2) for differential gene expression analysis, with featureCounts data as the input, under default settings. Differentially expressed genes (DEGs) were selected on the basis of the criteria: P < 0.05, fold change >1.2 and average count per million >300. Genes without a canonical name, as well as ribosomal and mitochondrial RNA products, were excluded. Gene ontology (GO) analysis was then performed using DAVID Functional Annotation Bioinformatics Microarray Analysis.40 Functional protein classification analyses were undertaken using Panther and Enrichr. The combined score generated by Enrichr was used to identify top biological process enrichment for each dataset. Hub genes were identified and interactome networks were generated using the GeneMANIA tool in Cytoscape software, v.3.8.0. Network edges are annotated according to GeneMANIA classification. Genes functionally relevant to microglia, astrocytes and neurons were identified using AmiGO searches for ‘Annotations’ between GO terms associated with each cell type. The results were compared to our identified DEGs.
Statistical analyses
Most statistical analyses were performed with GraphPad Prism. Hypergeometric analysis was performed with the Graeber laboratory online calculator. Experiments with two groups were analysed using two-tailed unpaired t-tests. Experiments with more than two groups were subjected to one-way or two-way ANOVA with Bonferroni correction for multiple post hoc comparisons. Data were tested for normality before parametric analysis. All data points represent distinct samples. All data are presented as the mean ± SEM. Data points identified as statistically significant outliers (determined by the Grubb’s test, P < 0.05) were removed from the analyses. Sample sizes were determined on the basis of power analyses and were similar to those reported in previous works.31–36,41,42
Data availability
The RNA-seq data generated in this study have been deposited in the GEO public repository under accession code GSE188865. Source data are provided with this paper.
Results
ADNP and POGZ are diminished in ASD, and ADNP or POGZ deficiency in PFC impairs cognitive behaviours
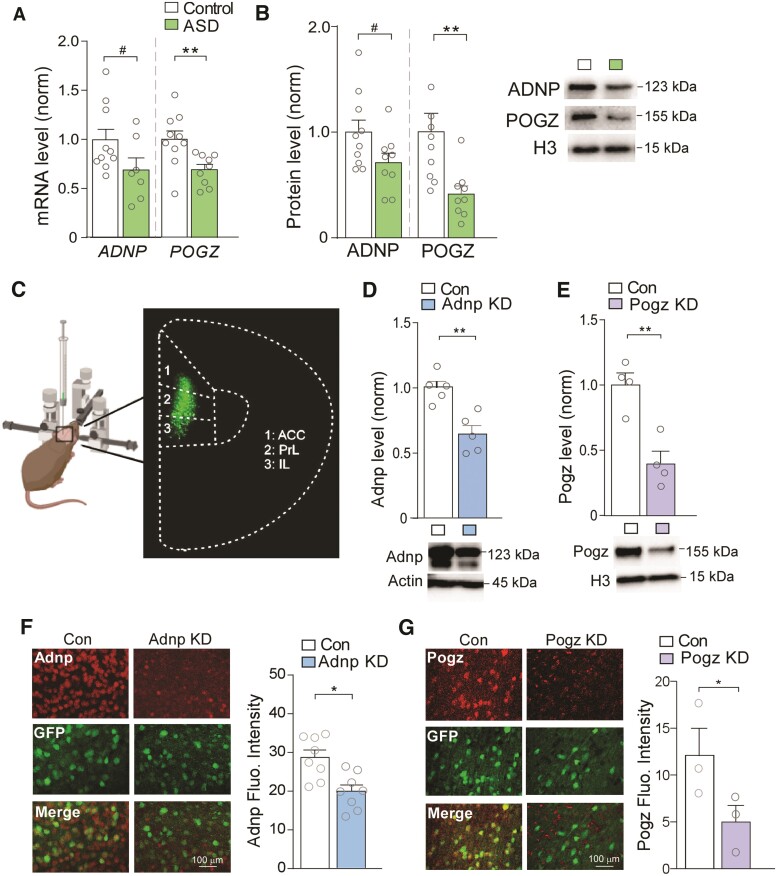
Because of the genetic link of ADNP and POGZ to ASD/ID, we assessed the expression of these risk factors in PFC, a central cognitive region that is impaired in NDD.27 Post-mortem tissues (Brodmann’s area 9) from human patients with ASD and control subjects were compared. The ages of the subjects ranged from 4 to 19 years old, encompassing the critical period of PFC synaptic refinement.43 PFC samples from humans with ASD showed reduction of ADNP and POGZ mRNA (Fig. 1A), as well as lower levels of ADNP and POGZ protein in the nuclear fraction (Fig. 1B). It is unlikely that these ASD subjects carry ADNP or POGZ mutations, as such mutations are each estimated to account for <0.2% of ASD/ID cases.6,44 Therefore, these findings indicate that diminished expression of ADNP and POGZ in PFC extends beyond the relatively rare mutation syndromes to affect patients with idiopathic ASD as well.
Figure 1.
ADNP and POGZ are diminished in PFC of ASD patients, which is simulated by viral-based KD of these NDD risk factors in mouse PFC. (A) Bar graphs showing mRNA levels of ADNP and POGZ in human post-mortem PFC tissue from control (n = 10) and ASD subjects (ADNP: t = 1.86, P = 0.085, n = 7; POGZ: t = 3.05, P = 0.0082, n = 9; unpaired t-tests). (B) Bar graphs showing protein levels of ADNP and POGZ in nuclear fractions isolated from human control (n = 10) and ASD (n = 9) subjects (ADNP: t = 2.00, P = 0.062; POGZ: I = 3.08, P = 0.0093; unpaired t-tests). Inset: representative western blot images. (C) Schematic representation of stereotaxic injection of GFP-tagged shRNA AAV into mouse medial PFC. Inset: representative image of GFP signal in viral-infected PFC. (D and E) Bar graphs showing Adnp (D) or Pogz (E) protein levels in PFC from mice with stereotaxic injection of a control shRNA versus Adnp shRNA or Pogz shRNA AAV (Adnp: n = 5/group, t = 4.31, P = 0.0030, Pogz: n = 4/group, t = 4.57, P = 0.0038, unpaired t-tests). Inset: representative western blot images. (F and G) Immunostaining images and quantification of Adnp (red, F) or Pogz (red, G) in PFC slices from mice injected with a control shRNA versus Adnp shRNA (F) or Pogz shRNA (G) AAV (F: n = 8 images/group, t = 3.47, P = 0.0038, G: n = 3 images/group, t = 2.15, P = 0.049, unpaired t-test). Green, AAV-linked GFP signal. All data are presented as mean ± SEM. #P < 0.1, *P < 0.05, **P < 0.01.
To determine the impact of deficiency in each of these NDD risk factors on PFC function, we used viral-based gene KD. The Adnp or Pogz shRNA, which effectively diminished the corresponding gene expression in mouse N2A cell lines (Supplementary Fig. 1), was cloned into a GFP-tagged AAV9 vector, and stereotaxically injected bilaterally into mouse medial PFC including prelimbic and infralimbic areas (Fig. 1C). Western blotting showed significantly lower expression of Adnp (Fig. 1D) or Pogz (Fig. 1E) in PFC from mice injected with Adnp shRNA (Adnp KD) or Pogz shRNA (Pogz KD) respectively, demonstrating the efficacy of gene KD in vivo. Immunohistochemical staining also confirmed the significant reduction of Adnp (Fig. 1F) and Pogz (Fig. 1G) signals in PFC cells from Adnp KD and Pogz KD mice.
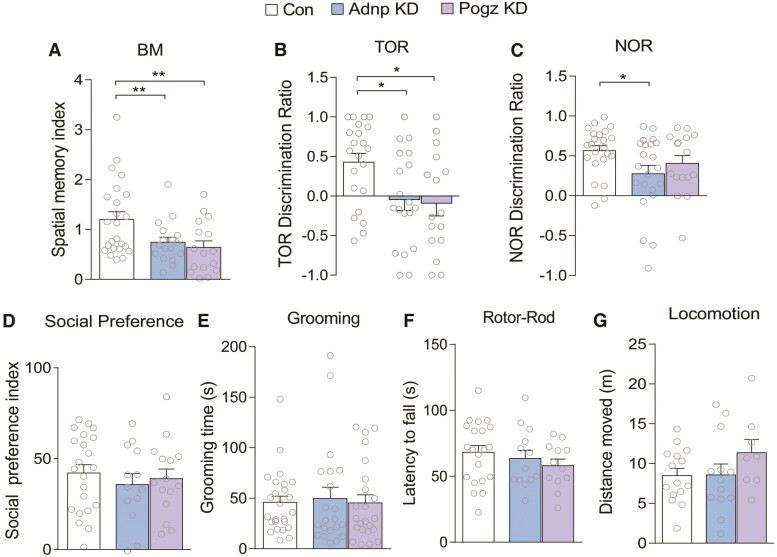
Next, we performed a series of behavioural assays to examine the effect of Adnp or Pogz deficiency. Given the link of ADNP and POGZ to ID, we first examined cognitive behaviours mediated by PFC and interacting regions45,46 We found significantly decreased performance of Adnp or Pogz KD mice in Barnes maze (BM) tests of spatial memory, in which the animal is trained to use visual cues to identify an escape hole from several incorrect holes on a circular platform. Adnp KD and Pogz KD mice demonstrated a lower spatial memory index on this task, indicating impaired memory for the correct position compared to control animals (Fig. 2A). Both KD groups also demonstrated impairment on the temporal order recognition task (TOR), a PFC-dependent cognitive task testing the animal’s ability to discriminate between two objects presented at different timepoints.45 Adnp KD and Pogz KD mice exhibited lower discrimination ratios between the novel (less recent) object and the familiar object (Fig. 2B). For the novel object recognition task (NOR), Adnp KD mice exhibited significantly lower discrimination ratios, compared to control animals (Fig. 2C). Detailed interaction times are shown in Supplementary Fig. 2A–C.
Figure 2.
Adnp or Pogz deficiency in mouse PFC induces cognitive deficits, but not social impairment, repetitive behaviours or motor problems. (A) Bar graphs of spatial memory index in Barnes maze (BM) tests of mice with PFC injection of a control shRNA, Adnp shRNA or Pogz shRNA AAV [n = 24 (control), 21 (Adnp), 16 (Pogz); F(2,58) = 6.21, P = 0.0036, one-way ANOVA]. (B) Bar graphs of discrimination ratio in temporal order recognition (TOR) tests for each group [n = 24 (control), 22 (Adnp), 16 (Pogz); F(2,59) = 4.30, P = 0.018, one-way ANOVA]. (C) Bar graphs of discrimination ratio in novel object recognition (NOR) tests for each group [n = 23 (control), 22 (Adnp), 16 (Pogz); F(2,58) = 2.99, P = 0.058, one-way ANOVA]. (D) Bar graphs of social preference index in three-chamber sociability tests for each group [n = 22 (control), 13 (Adnp), 15 (Pogz); F(2,47) = 0.20, P = 0.82, one-way ANOVA]. (E) Bar graphs of self-grooming time over a 10-minute period for each group [n = 34 (control), 27 (Adnp), 19 (Pogz); F(2,77) = 0.43, P = 0.65, one-way ANOVA]. (F) Bar graphs of latency to fall in rotor-rod tests for each group [n = 24 (control), 13 (Adnp), 12 (Pogz); F(2,46) = 1.14, P = 0.33, one-way ANOVA]. (G) Bar graphs of total distance travelled over a 1-h period for each group [n = 15 (control), 13 (Adnp), 16 (Pogz); F(2,33) = 1.35, P = 0.27, one-way ANOVA]. All data are presented as mean ± SEM. *P < 0.05, **P < 0.01.
As ADNP and POGZ are also linked to ASD, we further investigated social deficits and repetitive behaviour, two core phenotypes associated with ASD. In the three-chamber social preference test, a paradigm measuring the animal’s preferred interaction with a social stimulus (an age- and sex-matched mouse) over a non-social stimulus (a wooden block), no significant difference on the social preference index was observed in either Adnp KD or Pogz KD mice, compared with control animals (Fig. 2D). Similarly, Adnp or Pogz KD mice showed no alteration on the time spent interacting with a social stimulus in the social approach test (Supplementary Fig. 2D). Self-grooming, a marker of repetitive behaviour in mouse models of ASD, was also unchanged overall in Adnp KD and Pogz KD mice (Fig. 2E).
In addition, we examined motor function in mice with Adnp or Pogz deficiency in PFC. No significant alteration was observed in rotor-rod tests of motor coordination (Fig. 2F) or locomotor activity (Fig. 2G). Finally, we tested sensorimotor gating by measuring the pre-pulse inhibition and startle response, which is altered in patients and animal models of schizophrenia, as well as in some cases of ASD.47,48 No change in pre-pulse inhibition or startle response was found in Adnp KD or Pogz KD mice (Supplementary Fig. 2E and F).
To find out whether the behavioural phenotypes are sex specific, we further analysed the behavioural data in each sex. We found that females with PFC deficiency of Adnp or Pogz exhibited a significant impairment in two cognitive tasks (Barnes maze and temporal order recognition), while males with PFC deficiency of Adnp or Pogz only showed a trend of cognitive deficits (Supplementary Fig. 3).
Deficiency in Adnp or Pogz induces convergent genetic alterations that are shared in human ASD
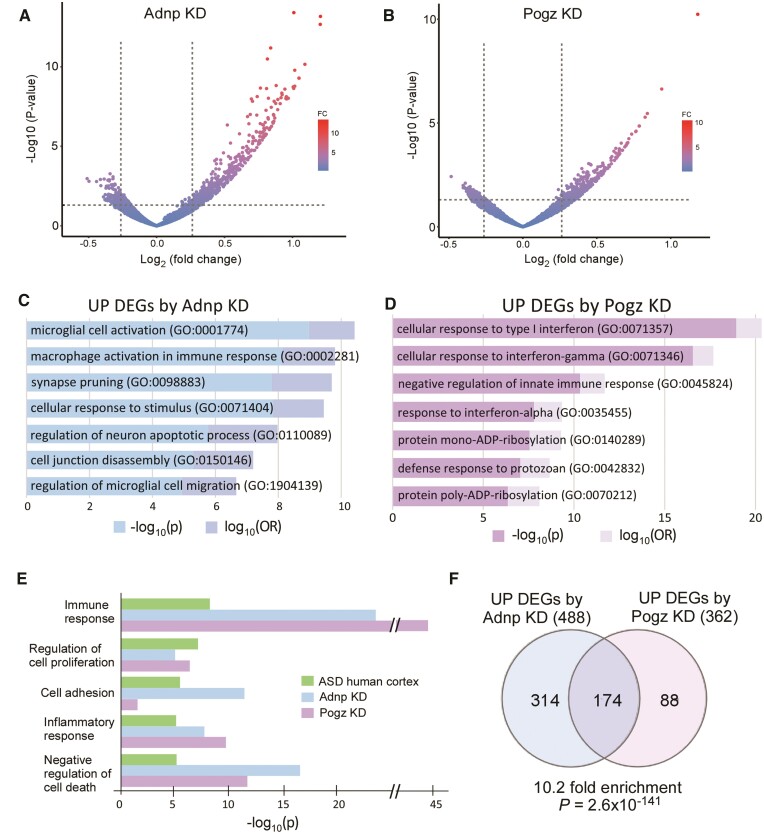
ADNP and POGZ are chromatin regulators controlling gene expression,12,21 so we examined genome-wide transcriptional changes induced by Adnp or Pogz deficiency using next-generation RNA-seq analyses of PFC tissue. We found prominent upregulation of gene transcription following Adnp or Pogz KD (Fig. 3A and B), consistent with their known roles as gene repressors. Many more upregulated genes were identified (488 in Adnp KD, and 362 in Pogz KD, Supplementary Table 2) than downregulated genes, so we focused most of our analyses on these upregulated DEGs.
Figure 3.
Adnp or Pogz deficiency in mouse PFC induces convergent transcriptomic alterations that are shared in human ASD. (A and B) Volcano plots showing DEGs in mice with PFC KD of Adnp (A) or Pogz (B), compared to controls. (C and D) Bar graphs showing top-ranking GO pathways of the upregulated genes from Adnp KD (C) and Pogz KD (D). (E) Bar graphs comparing GO pathways of upregulated genes in post-mortem cortical tissue from ASD patients,49 and mice with Adnp or Pogz KD in PFC. (F) Venn diagram comparing DEGs induced by Adnp KD or Pogz KD. Note the significant overlap of DEGs (P = 2.6 × 10−141, hypergeometric test) in the two groups.
We first examined the functional roles of the upregulated DEGs using GO pathway analysis to uncover the top biological processes affected by Adnp or Pogz deficiency. We found that many of the GO categories revolved around similar themes of immune response, microglial activation and stimulus response in mice with Adnp KD (Fig. 3C) or Pogz KD (Fig. 3D). Comparing these GO results with transcriptomic data from ASD humans, we found that, among the top five upregulated GO pathways previously identified in frontal cortex of autistic patients,49 both Adnp KD and Pogz KD similarly exhibited significant changes in these categories (Fig. 3E). This suggests that the transcriptional elevation of gene sets in mice with Adnp or Pogz deficiency is highly correlated with humans with NDD.
In addition to similarly enriched GO pathways in mice with Adnp or Pogz deficiency, a significant overlap between specific DEGs was observed (1/3 of Adnp KD DEGs overlapping with 1/2 of Pogz KD DEGs, Fig. 3F). This represented a 10-fold enrichment in overlapping genes, demonstrating that KD of either of these two ASD/ID risk factors leads to similar genetic alterations.
GO pathways of downregulated DEGs in mice with Adnp or Pogz deficiency are divergent (Supplementary Fig. 4). Top pathways downregulated by Adnp KD were primarily related to metabolic biosynthesis, while Pogz KD mainly resulted in disruption of pathways for action potential and membrane depolarization. Given the role of Adnp and Pogz primarily in gene repression, the downregulated DEGs could be indirectly affected by their deficiency.
Genes commonly upregulated by Adnp or Pogz deficiency are enriched in glial regulation
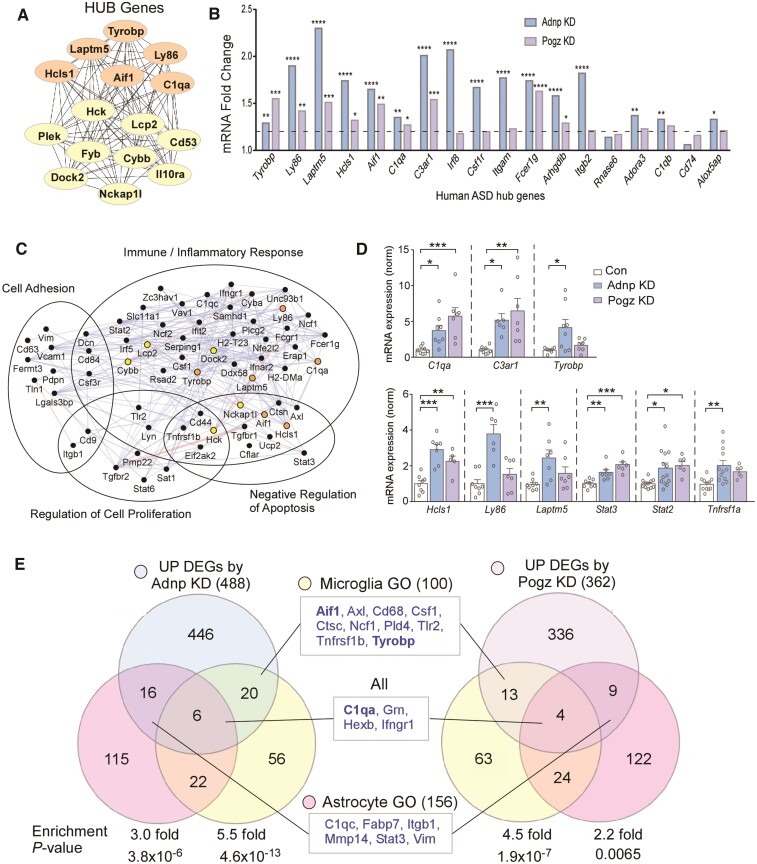
Recent transcriptomic profiling of several major neuropsychiatric disorders identified genes related to microglia as a unique and significant transcriptomic signature of ASD.50 From this profiling, the top 20 hub genes driving the altered microglia-related transcription were identified. To determine whether the transcriptional alteration by Adnp or Pogz deficiency might exhibit a similar pattern, we performed hub gene analysis on the 174 overlapping genes in Adnp or Pogz KD mice. Of these top hub genes common to Adnp or Pogz deficiency in PFC, 40% overlapped with the identified ASD human hub genes (Fig. 4A). Examining our RNA-seq data from Adnp or Pogz KD mice, we found that 16 out of the 18 orthologous ASD human hub genes were significantly upregulated (nine in both KD conditions and seven in Adnp KD only, Fig. 4B).
Figure 4.
Commonly upregulated genes by Adnp or Pogz deficiency are enriched in glial regulation. (A) Network of top 15 hub genes identified from the overlapping DEGs induced by Adnp KD or Pogz KD. Some of these genes (orange) are also hub genes identified in human ASD.50 (B) Bar graphs representing fold change of top hub genes identified in human ASD in RNA-seq data from mice with Adnp KD or Pogz KD in PFC. Dotted line signifies cutoff at 1.2-fold increase over control. (C) Network of top overlapping DEGs induced by Adnp KD or Pogz KD, grouped by GO categories enriched in human ASD. Hub genes identified in Fig. 4A are marked with their respective colours. Blue lines represent colocalization of gene expression, while pink lines represent physical interactions between gene products. (D) Bar graphs of qPCR data showing mRNA levels of top DEGs in mice with Adnp KD or Pogz KD in PFC (n = 5–13/group, one-way ANOVA). (E) Venn diagrams showing the overlap of DEGs induced by Adnp KD or Pogz KD with GO terms related to microglia (yellow), astrocytes (pink) or both (centre). Genes commonly found in both KD conditions are listed in the centre boxes. Identified HUB genes are demarcated in bold font. Enrichment significance calculated by hypergeometric tests. All data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The functional network of overlapping upregulated genes by Adnp or Pogz deficiency (Fig. 4C) demonstrates interactions between hub genes and other upregulated DEGs, which are clustered into the top GO categories identified in ASD: immune/inflammatory response, regulation of cell proliferation, cell adhesion and negative regulation of apoptosis. We next performed qPCR assays to verify the RNA-seq data for top DEGs identified either through hub gene or network analysis. We found that mRNA levels of most of these genes were significantly elevated in mice with Adnp KD or Pogz KD, including the complement genes C3ar1 and C1qa, immune signalling adaptor Hcls1 and transcription factors Stat2 and Stat3 (Fig. 4D).
We further compared our RNA-seq data to previously generated datasets from Adnp- or Pogz-deficient models: Adnp+/− heterozygous mouse hippocampus (GSE72664)51 and Pogz brain-specific conditional knockout (cKO) mouse hippocampus.21 We found 314 upregulated DEGs from Adnp+/− (P < 0.05 and fold change > 1.2) and 586 upregulated DEGs from Pogz cKO (FDR < 0.05). We then compared GO categories that were enriched in at least three of the four datasets (Adnp KD, Pogz KD, Adnp+/− and Pogz cKO). Interestingly, the commonly enriched GO categories included several pathways related to immune response, including ‘macrophage activation’ and ‘positive regulation of inflammatory response’ (Supplementary Fig. 5).
Since our transcriptomic data indicated an increase of pro-inflammatory transcription by Adnp or Pogz deficiency, we compared DEGs from Adnp KD or Pogz KD to genes functionally related to microglia and astrocytes, two major inflammation-involved glial types. We found significant enrichment in both DEG sets with 100 microglia genes and 156 astrocyte genes (Fig. 4E, Supplementary Table 3). Microglia genes upregulated in both Adnp KD and Pogz KD included Aif1, Axl, Cd68, Csf1, Ctsc, Ncf1, Pld4, Tlr2, Tnfrsf1b and Tyrobp. Aif1 was also upregulated in RNA-seq data from Adnp+/− mice. Astrocyte genes upregulated in both Adnp KD and Pogz KD included C1qc, Fabp7, Itgb1, Mmp14, Stat3 and Vim. Several genes common to microglia and astrocytes, including C1qa, Grn, Hexb and Ifngr1, were upregulated in both KD conditions. Greater enrichment was seen in microglia than in astrocytes with Adnp or Pogz KD.
In accordance with many previous findings in ASD/ID,32,34,49 the upregulated DEGs by Adnp or Pogz deficiency showed significant under-enrichment of neuronal genes (Supplementary Fig. 6A and B, Supplementary Table 4). On the other hand, downregulated DEGs by Adnp or Pogz deficiency were over-enriched in this category (Supplementary Fig. 6C and D), indicating the decreased transcription of some neuron-related genes by deficiency of these NDD risk factors. This is also in agreement with the recent discovery that Pogz may promote transcription of clustered synaptic genes in early development.14 No significant overlap was seen between the downregulated DEGs of Adnp KD or Pogz KD with either glial subtype (data not shown).
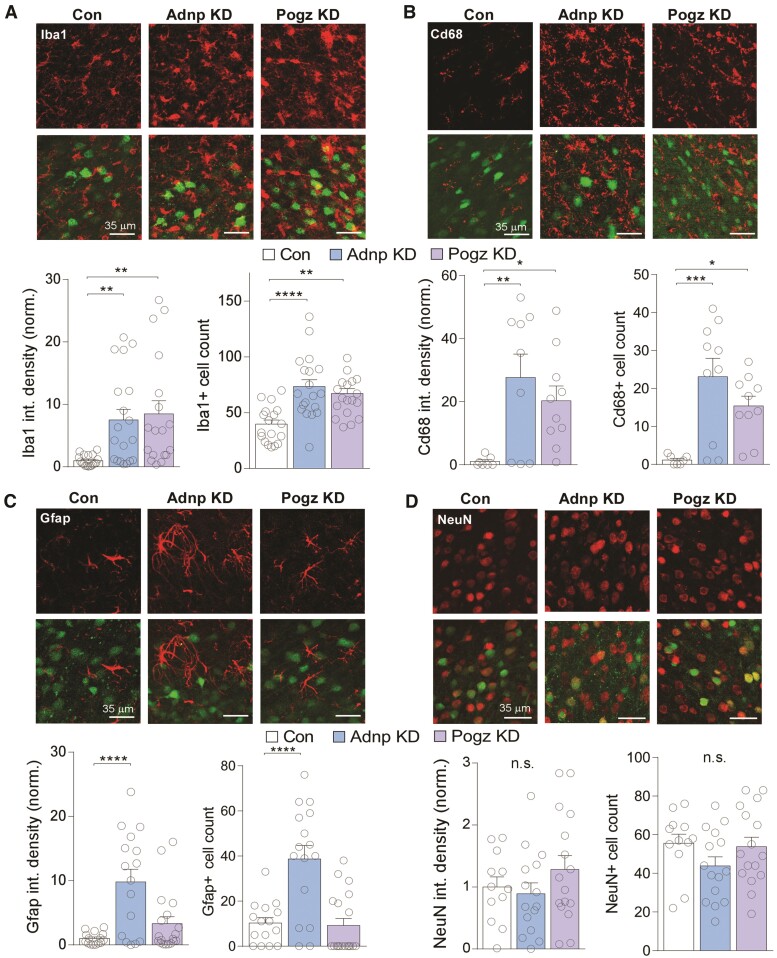
Adnp or Pogz deficiency in PFC induces a prominent increase of microglia and astrocytes.
The rise in immune- and inflammatory-related transcription by Adnp or Pogz KD prompted us to examine whether gliosis and neuroinflammation were occurring alongside these transcriptional changes. We performed immunostaining with the microglial marker Iba1 (gene name Aif1, one of our identified overlapping hub genes). We found that both the Iba1 protein level and the number of Iba1+ cells were significantly increased in PFC with Adnp KD and Pogz KD (Fig. 5A). While increase in Iba1 expression has been used as a marker of the pro-inflammatory phenotype in microglia, we further examined specifically pro-phagocytic ‘activated’ microglia, which could damage surrounding neurons. We found that Cd68 (macrosialin), a scavenger receptor upregulated in pro-phagocytic microglia, was significantly increased in PFC with Adnp KD or Pogz KD (Fig. 5B), confirming microglia activation with the deficiency of these NDD risk factors.
Figure 5.
Adnp or Pogz deficiency in mouse PFC increases microglia and astrocytes without changing neuron numbers. (A–D) Top: representative images of microglial marker Iba1 (red, A), activated microglial marker Cd68 (red, B), astrocyte marker Gfap (red, C) or neuronal marker NeuN (red, D) in PFC slices from mice with the injection of a control shRNA, Adnp shRNA or Pogz shRNA AAV. Green, AAV-linked GFP signal. Bottom: bar graphs of Iba1 (A), Cd68 (B), Gfap (C) or NeuN (D) integrated density (mean fluorescence intensity × area of fluorescence) and average number of Iba1+ (A), Cd68+ (B), Gfap+ (C) or NeuN+ (D) cells per image area (0.085 mm2) in each group (Iba1, n = 3 animals/18 images (control); 4 animals/19 images (Adnp); 4 animals/18 images (Pogz); density: F(2,52) = 6.71, P = 0.0026, number: F(2,52) = 11.96, P < 0.0001; Cd68, n = 2 animals/7 images (control); 3 animals/9 images (Adnp); 3 animals/10 images (Pogz); density: F(2,23) = 5.62, P = 0.010, number: F(2,24) = 8.84, P = 0.0013; Gfap, n = 3 animals/15 images (control); 4 animals/16 images (Adnp); 4 animals/19 images (Pogz); density: F(2,47) = 11.57, P < 0.0001, number: F(2,47) = 16.41, P < 0.0001; NeuN, n = 3 animals/12 images (control); 4 animals/15 images (Adnp); 4 animals/16 images (Pogz); density: F(2,40) = 1.18, P = 0.32, number: F(2,40) = 1.67, P = 0.20, one-way ANOVA). All data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Since astrocyte-related genes were also increased in Adnp KD and Pogz KD mice, we performed immunostaining with the astrocyte marker Gfap. We found that Gfap expression and Gfap+ cell numbers were significantly increased in PFC with Adnp KD, but not Pogz KD (Fig. 5C), indicating astrocyte activation specifically by Adnp deficiency.
Examining the overlap of GFP from control animals infected with Adnp or Pogz shRNA AAV (GFP-tagged) with each cell-type marker (Iba1, Gfap and NeuN), we found that most of the overlap was found in NeuN+ cells (Supplementary Fig. 7), suggesting that the viral KD of Adnp or Pogz occurs primarily in neurons. Finally, we tested whether the inflammatory activation by Adnp or Pogz deficiency could be due to an increase in neuronal cell death, triggering cleanup of cellular debris. Immunostaining with the neuronal marker NeuN found no change in neuron numbers in PFC with Adnp KD or Pogz KD (Fig. 5D), suggesting that glial upregulation is not occurring because of neuronal death.
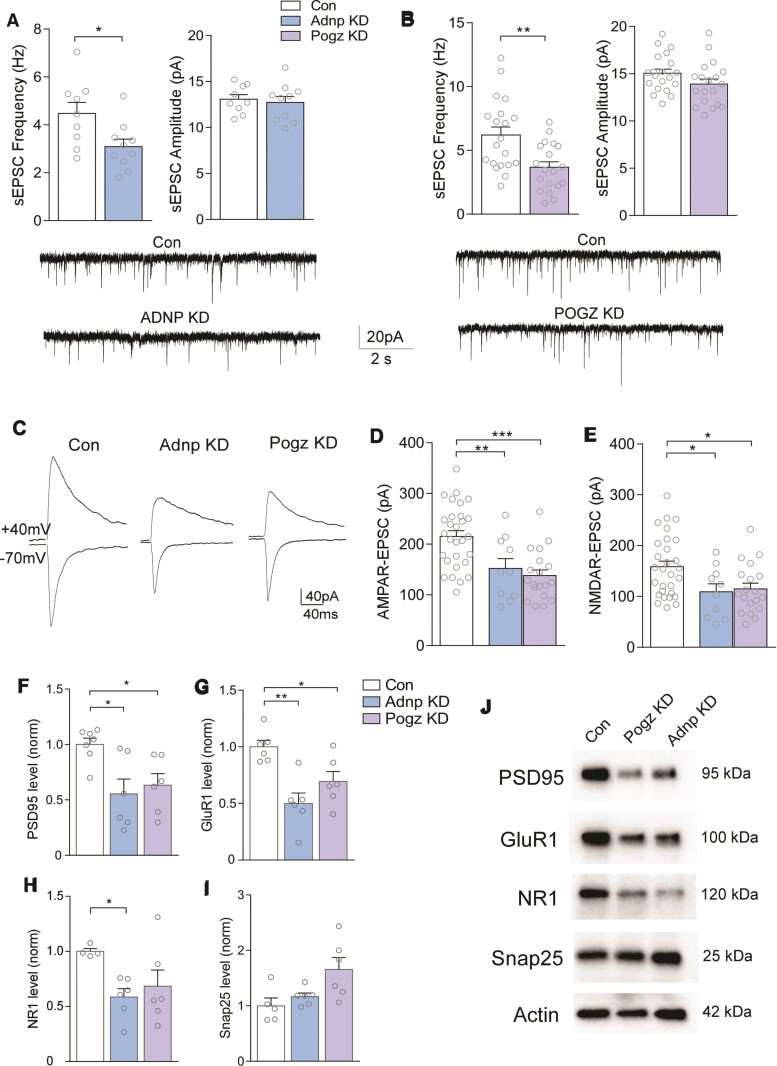
Adnp or Pogz deficiency in PFC induces similar synaptic deficits
Microglia have profound effects on synaptic function, particularly in disease conditions.52 The pro-phagocytic microglial upregulation in Adnp KD and Pogz KD could cause synaptic deficits. To test this possibility, we performed whole-cell patch-clamp recordings of synaptic currents in PFC pyramidal neurons with Adnp or Pogz deficiency. Significantly reduced frequencies of spontaneous excitatory postsynaptic currents (sEPSC) were found in PFC neurons with Adnp KD or Pogz KD (Fig. 6A and 6B). Next, we examined evoked EPSC mediated by AMPA and NMDA receptors. As shown in Fig. 6C–E, AMPAR-EPSC and NMDAR-EPSC were both significantly reduced in PFC neurons with Adnp KD or Pogz KD, indicating the diminished glutamatergic synaptic function by deficiency of these NDD risk factors.
Figure 6.
Adnp or Pogz deficiency in mouse PFC leads to diminished glutamatergic transmission and postsynaptic protein expression. (A and B) Bar graphs of spontaneous excitatory postsynaptic current (sEPSC) frequency and amplitude in PFC pyramidal neurons from mice with injection of a control shRNA, Adnp shRNA (A) or Pogz shRNA (B) AAV [A: n = 3 animals/group, cells: n = 9 (control), n = 10 (Adnp); t = 2.47, P = 0.027 (frequency); t = 0.408, P = 0.689 (amplitude); B: n = 4 animals/group, cells: n = 20 (control), n = 19 (Pogz); t = 3.28, P = 0.0021 (frequency); t = 1.65, P = 0.108 (amplitude), unpaired t-test]. Inset: representative sEPSC recordings from each group. (C) Representative AMPAR-EPSC (recorded at −70 mV) and NMDAR-EPSC (recorded at +40 mV) evoked on the same PFC pyramidal neurons from each group. (D and E) Bar graphs of AMPAR-EPSC amplitudes (D) and NMDAR-EPSC amplitudes (E, detected at 40 ms after current onset) in each group (n = 3–4 animals/10–29 cells/group, AMPA: F(2,55) = 11.43, P < 0.0001, NMDA: F(2,55) = 5.01, P = 0.01, one-way ANOVA). (F–I) Bar graphs comparing expression levels of synaptic proteins PSD95, GluR1, NR1 and Snap25 in PFC synaptic fraction from mice with injection of a control shRNA, Adnp shRNA or Pogz shRNA [n = 5–7 animals/protein, control; n = 6 (Adnp); n = 6 (Pogz); one-way ANOVA]. All proteins are normalized to actin. (J) Representative western blot images from each group. All data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate the potential reason for the reduction of glutamatergic transmission in Adnp KD or Pogz KD mice, we examined the expression of synaptic markers in the synaptic fraction of PFC. A significant reduction of the glutamatergic postsynaptic density protein PSD-95, as well as the AMPAR subunit GluR1, were found in PFC with Adnp or Pogz deficiency (Fig. 6F and G). The NMDAR subunit NR1 was also significantly decreased by Adnp KD (Fig. 6H), while the presynaptic protein Snap25 was not reduced in either condition (Fig. 6I). Representative blots are shown in Fig. 6J. These data indicate that spine synapses may be reduced in mice with Adnp or Pogz deficiency, leading to the compromised glutamatergic transmission.
Discussion
While the genetic heterogeneity of ASD and ID has complicated the quest for pathophysiological mechanisms, a wealth of evidence proposes that a convergent feature of ASD/ID is synaptic dysfunction.28,29,53,54 Some ASD/ID monogenic variants contribute directly to synapse formation or function32,41,55; however, many others, including ADNP and POGZ, are involved in the regulation of gene expression.4,33,34 How the transcriptomic changes eventually lead to the synaptic dysfunction in ASD/ID remains relatively unknown. Furthermore, many cases of ASD/ID cannot be traced to a single genetic mutation. It is possible that these idiopathic cases involve dysregulation of a network containing identified high-risk gene products, which could explain the phenotypic similarity despite the genotypic divergence.
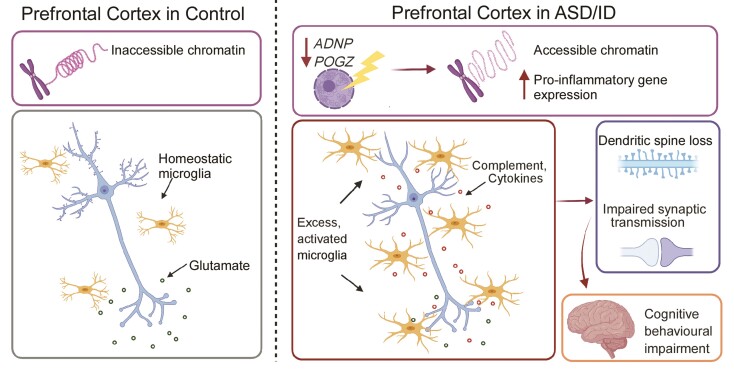
Here, we have identified functional commonalities between two high-confidence monogenic risk factors for ASD/ID. We found that both ADNP and POGZ were downregulated in PFC of humans with idiopathic ASD. Thus, the biological functions disrupted by haploinsufficiency of these NDD risk factors could represent dysregulated pathways in broad populations of ASD/ID. Using viral-based gene KD in PFC of juvenile mice, we find that deficiency of the transcriptional regulator Adnp or Pogz in PFC triggers the upregulation of immune response genes, such as complement, cytokines and other signalling molecules. We propose that such upregulation may be due to the reduced chromatin accessibility, as both Adnp and Pogz have been shown to act locally together with HP1 to maintain regions of chromatin in an inaccessible state.12,14,21 This recruits and activates glia, especially microglia, to a pathologically pro-phagocytic state, which significantly hampers glutamatergic synaptic transmission presumably by eliminating synapses, leading to the cognitive impairment associated with ASD/ID (Fig. 7).
Figure 7.
A schematic model illustrating a potential mechanism underlying the involvement of Adnp and Pogz in NDD. Deficiency of the chromatin regulator Adnp or Pogz causes pro-inflammatory gene upregulation, leading to the activation of microglia and astrocytes. This induces synaptic deficits via synapse modulation and/or elimination, resulting in cognitive impairment.
We have found significant deficits in spatial memory and recognition memory in mice with Adnp KD or Pogz KD in PFC. Consistently, heterozygous Adnp+/− mice demonstrate cognitive impairments in spatial memory,56 object recognition memory and social memory.57 Brain-specific Pogz+/− heterozygosity impairs spatial memory and learning,21 and whole-body Pogz haploinsufficiency impairs contextual fear learning.19 No changes were present in sociability, motor function or sensory gating process, indicating that Adnp or Pogz deficiency in PFC is primarily linked to cognitive impairment. While motor function was found to be altered in Adnp+/− mice20 and Pogz+/− mice,21 gene deficiency in these models encompasses the entire brain. Thus, the lack of motor deficits in our study is probably due to the PFC-specific deficiency of Adnp or Pogz.
In the Adnp+/− mouse model, sex differences were observed in some behavioural tests, including impaired object recognition only in males and impaired social preference only in females.20,57 Sex-specific changes in gene expression were also found in Adnp+/− mice.51,57 In the current study, we found that female Adnp KD and Pogz KD mice showed significant impairment in the Barnes maze and temporal order cognitive tasks, while males with PFC deficiency of Adnp or Pogz only showed a trend of cognitive deficits. This suggests that the two NDD risk factors may induce more pronounced phenotypes in females. Considering that ADNP expression differs by sex57 and regulates its own expression,58 it is possible that the developmental timepoint at which deficiency occurs affects sex-specific behavioural modulation. More studies are needed to determine whether the functions of these NDD risk factors are distinct in a sex-specific manner.
To reveal potential reasons underlying behavioural abnormalities induced by Adnp or Pogz deficiency, we examined transcriptomic changes using RNA-seq analyses. ADNP and POGZ share the binding partner HP1 and play similar roles in gene regulation.10–13 ADNP is thought to be essential for neuronal lineage specification through binding to HP1 to repress aberrant gene transcription.12 POGZ has been found to act as either a repressor or activator of gene expression, depending on the developmental and tissue-specific context.21,59 Here, we found predominant transcriptional upregulation with deficiency of either Adnp or Pogz in PFC. There was 30–50% overlap of the DEGs in Adnp KD and Pogz KD, underscoring their similar function. This is in line with the recent finding that POGZ and ADNP co-occupy a significant proportion of genetic loci, indicating shared targets of regulation.14 GO analysis revealed that many of these upregulated genes are related to immune function, inflammation and microglial function. Enrichment for immune-related biological processes was also found in RNA-seq datasets from Adnp- and Pogz-deficient mouse hippocampal tissue.21,51 Previous studies in non-neuronal cells suggested that ADNP has potential immunomodulatory functions.60,61 Interestingly, immune function has also been identified as the top upregulated biological process in PFC of ASD/ID patients,49 and the top five upregulated GO pathways identified in ASD/ID humans were all significantly increased in mice with Adnp or Pogz deficiency in PFC.
Although the role of the immune system in CNS is nebulous, it is generally agreed that microglia are the most relevant immune cells in the brain. Across a range of neuropsychiatric disorders, including ASD, schizophrenia, bipolar disorder and major depressive disorder, ASD is uniquely associated with altered microglial transcriptomic signatures.50 We also found that genes upregulated by Adnp or Pogz KD were significantly over-enriched for microglia-related genes. Out of the top 18 orthologous microglial hub genes found in human ASD,50 16 genes were upregulated in Adnp or Pogz KD mice. Additionally, of the hub genes identified from RNA-seq data of Adnp or Pogz KD mice, about half overlapped with the ASD human hub genes. Taken together, these findings indicate a highly significant effect on the microglial transcriptome by Adnp or Pogz deficiency in PFC.
These microglial gene alterations could translate to changes in glial numbers or activation. Indeed, we found an increase in microglial numbers and phagocytic activation in PFC of mice with Adnp KD or Pogz KD. Consistently, in human subjects with ASD, increased microglial numbers,62 activation63 and density64 have also been observed. In addition, increased astrocyte numbers were found in Adnp KD mice, in agreement with the association of astrocytic activation with ASD50 and with astrocytic deficiencies in Adnp+/− mice.56 The lack of changes in neuronal numbers by Adnp or Pogz deficiency suggest that behavioural deficits are not due to loss of neurons.
Data from the Human Protein Atlas show that both ADNP and POGZ are highly expressed across all major cell types in the CNS (excitatory and inhibitory neurons, astrocytes, oligodendrocytes and microglia).65 ADNP was first isolated from astrocytes66 and has been localized to astrocytes and microglia following head trauma in mice.67 Thus, identifying the functional role of these NDD risk factors in various cell types is important to understanding NDD pathophysiology. Here, we have used an AAV serotype (AAV9) that shows marked preference for neurons over glia under the U6 universal promoter.68 Therefore, the current study provides a framework to understand the impact of ADNP or POGZ deficiency in neurons.
The overlap of GFP signal with NeuN, but not Iba1 or Gfap, indicates that the viral-based gene KD is mainly targeted to neurons. How does neuron-specific Adnp or Pogz deficiency lead to profound effects on glial activation? On the basis of the role of Adnp and Pogz in transcriptional repression, loss of these genes in PFC neurons probably allows for aberrantly upregulated transcription of factors like complement and other pro-inflammatory signalling molecules. Once released into the extracellular space, microglia (and potentially astrocytes) are quickly recruited and transformed to an ‘activated’ state, which could actively engulf synaptic material, critically affecting synaptic pruning and synaptic maturation during development.69,70
To test this, we evaluated the effects of Adnp KD and Pogz KD on synaptic transmission. Cell-type-specific expression of known ASD risk genes is found to be especially enriched in glutamatergic neurons.3 Deep layer PFC glutamatergic neurons show the most prominent deficits in humans with ASD.28 Thus, we investigated whether deficiency of Adnp or Pogz induces synaptic dysfunction in mouse layer V PFC pyramidal neurons. Significantly diminished AMPAR- and NMDAR-mediated synaptic currents, along with significantly reduced expression of postsynaptic proteins, were found in Adnp or Pogz KD mice, suggesting the loss of glutamatergic synapses. This is in line with previous findings of glutamatergic dysfunction in Adnp+/− mice, including reduced dendritic spine density20 and vesicular glutamate transporter (VGLUT1) expression.24 Interestingly, a Pogz mouse model with a patient-derived mutation near the C terminal (Q1038R) exhibits increased pyramidal neuron activity in the cortex.22 This discrepancy could be due to the nature of the deficiency (acute KD as opposed to constitutive mutation).
In summary, we have proposed a convergent mechanism underlying the action of two top-ranking NDD risk factors. Intriguingly, neuron-specific knockout of another ASD/ID risk factor, Ctcf, a key player in 3D chromatin organization, led to very similar findings of behavioural dysfunction, increased cytokine transcription, altered microglial phenotype and spine loss.71 The ADNP complex competes with CTCF at a subset of DNA binding sites, acting as a local modulator of CTCF-mediated chromatin looping.72 This relationship may underlie the converging phenotypes by Adnp or Ctcf deficiency. Moreover, our data dovetail with the findings of increased microglial activation and density in PFC of humans with ASD/ID, further lending credence to the convergent nature of this mechanism.
Finally, it is important to note that this study does not diminish the importance of ADNP/POGZ deficiency on embryonic and early post-natal development. Constitutive Adnp or Pogz haploinsufficiency in mice has demonstrated behavioural and neuronal deficits at very early stages.20,21 The current study shows that these two genes remain essential during the period of post-natal synaptic refinement. The contributions of ADNP/POGZ deficiency to ASD pathology are probably multifaceted and compounded as the CNS undergoes multiple stages of maturation. Thus, the immune dysregulation and synaptic deficits shown here in the post-natal period represent one piece of the larger puzzle of ASD/ID pathology across the lifespan.
Supplementary Material
Acknowledgements
We thank Xiaoqing Chen, Kaijie Ma and Freddy Zhang for excellent technical support and Dr Kelcie Schatz for proofreading the manuscript.
Abbreviations
- AAV =
adeno-associated virus
- ASD =
autism spectrum disorder
- EPSC =
excitatory postsynaptic currents
- ID =
intellectual disability
- KD =
knockdown
- NDD =
neurodevelopmental disorders
- PFC =
prefrontal cortex
Contributor Information
Megan Conrow-Graham, Department of Physiology and Biophysics, State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
Jamal B Williams, Department of Physiology and Biophysics, State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
Jennifer Martin, Department of Physiology and Biophysics, State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
Ping Zhong, Department of Physiology and Biophysics, State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
Qing Cao, Department of Physiology and Biophysics, State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
Benjamin Rein, Department of Physiology and Biophysics, State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
Zhen Yan, Department of Physiology and Biophysics, State University of New York at Buffalo, Jacobs School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
Funding
This work was supported by a grant from the Nancy Lurie Marks Family Foundation to Z.Y. and an NIH F30 fellowship to M.C.G. (MH127800). We also thank E.F. Trachtman and the Varanasi family for their kind donations.
Competing interests
The authors report no competing financial or other interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stessman HAF, Xiong B, Coe BP, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coe BP, Stessman HAF, Sulovari A, et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet. 2019;51:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Satterstrom FK, Kosmicki JA, Wang J, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kosmicki JA, Samocha KE, Howrigan DP, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Helsmoortel C, Vulto-van Silfhout AT, Coe BP, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Dijck A, Vulto-van Silfhout AT, Cappuyns E, et al. Clinical presentation of a complex neurodevelopmental disorder caused by mutations in ADNP. Biol Psychiatry. 2019;85:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stessman Holly AF, Willemsen Marjolein H, Fenckova M, et al. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet. 2016;98:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. White J, Beck CR, Harel T, et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nozawa RS, Nagao K, Masuda HT, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010;12:719–727. [DOI] [PubMed] [Google Scholar]
- 11. Mandel S, Rechavi G, Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev Biol. 2007;303:814–824. [DOI] [PubMed] [Google Scholar]
- 12. Ostapcuk V, Mohn F, Carl SH, et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature. 2018;557:739–743. [DOI] [PubMed] [Google Scholar]
- 13. Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. [DOI] [PubMed] [Google Scholar]
- 14. Markenscoff-Papadimitriou E, Binyameen F, Whalen S, et al. Autism risk gene POGZ promotes chromatin accessibility and expression of clustered synaptic genes. Cell Rep. 2021;37:110089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oz S, Kapitansky O, Ivashco-Pachima Y, et al. The NAP motif of activity-dependent neuroprotective protein (ADNP) regulates dendritic spines through microtubule end binding proteins. Mol Psychiatry. 2014;19:1115–1124. [DOI] [PubMed] [Google Scholar]
- 16. Merenlender-Wagner A, Malishkevich A, Shemer Z, et al. Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatry. 2015;20:126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gozes I, Shazman S. STOP Codon mutations at sites of natural caspase cleavage are implicated in autism and Alzheimer’s Disease: The Case of ADNP. Front Endocrinol. 2022;13:867442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan Q, Wulfridge P, Doherty J, et al. Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat Commun. 2022;13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heath J, Cheyou ES, Findlay S, et al. POGZ promotes homology-directed DNA repair in an HP1-dependent manner. EMBO Rep. 2022;23:e51041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hacohen-Kleiman G, Sragovich S, Karmon G, et al. Activity-dependent neuroprotective protein deficiency models synaptic and developmental phenotypes of autism-like syndrome. J Clin Invest. 2018;128:4956–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suliman-Lavie R, Title B, Cohen Y, et al. Pogz deficiency leads to transcription dysregulation and impaired cerebellar activity underlying autism-like behavior in mice. Nat Commun. 2020;11:5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsumura K, Seiriki K, Okada S, et al. Pathogenic POGZ mutation causes impaired cortical development and reversible autism-like phenotypes. Nat Commun. 2020;11:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun X, Peng X, Cao Y, Zhou Y, Sun Y. ADNP promotes neural differentiation by modulating Wnt/β-catenin signaling. Nat Commun. 2020;11:2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sragovich S, Malishkevich A, Piontkewitz Y, et al. The autism/neuroprotection-linked ADNP/NAP regulate the excitatory glutamatergic synapse. Transl Psychiatry. 2019;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karmon G, Sragovich S, Hacohen-Kleiman G, et al. Novel ADNP syndrome mice reveal dramatic sex-specific peripheral gene expression with brain synaptic and Tau pathologies. Biol Psychiatry. 2021;128:4956–4969. [DOI] [PubMed] [Google Scholar]
- 26. Amodio DM, Frith CD. Meeting of minds: the medial frontal cortex and social cognition. Nat Rev Neurosci. 2006;7:268–277. [DOI] [PubMed] [Google Scholar]
- 27. Schubert D, Martens GJM, Kolk SM. Molecular underpinnings of prefrontal cortex development in rodents provide insights into the etiology of neurodevelopmental disorders. Mol Psychiatry. 2015;20:795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stoner R, Chow ML, Boyle MP, et al. Patches of disorganization in the neocortex of children with autism. N Eng J Med. 2014;370:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan Z, Rein B. Mechanisms of synaptic transmission dysregulation in the prefrontal cortex: Pathophysiological implications. Mol Psychiatry. 2021;27:445–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sacai H, Sakoori K, Konno K, et al. Autism spectrum disorder-like behavior caused by reduced excitatory synaptic transmission in pyramidal neurons of mouse prefrontal cortex. Nat Commun. 2020;11:5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duffney LJ, Zhong P, Wei J, et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 2015;11:1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qin L, Ma K, Wang Z-J, et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci. 2018;21:564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qin L, Williams JB, Tan T, et al. Deficiency of autism risk factor ASH1L in prefrontal cortex induces epigenetic aberrations and seizures. Nat Commun. 2021;12:6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang ZJ, Rein B, Zhong P, et al. Autism risk gene KMT5B deficiency in prefrontal cortex induces synaptic dysfunction and social deficits via alterations of DNA repair and gene transcription. Neuropsychopharmacology. 2021;46:1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao Q, Wang W, Williams JB, Yang F, Wang ZJ, Yan Z. Targeting histone K4 trimethylation for treatment of cognitive and synaptic deficits in mouse models of Alzheimer's disease. Sci Adv. 2020;6:eabc8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rein B, Tan T, Yang F, et al. Reversal of synaptic and behavioral deficits in a 16p11.2 duplication mouse model via restoration of the GABA synapse regulator Npas4. Mol Psychiatry. 2021;26:1967–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng Y, Liu A, Wang ZJ, et al. Inhibition of EHMT1/2 rescues synaptic and cognitive functions for Alzheimer's disease. Brain. 2019;142:787–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuen EY, Wei J, Liu W, Zhong P, Li X, Yan Z. Repeated stress causes cognitive impairment by suppressing glutamate receptor expression and function in prefrontal cortex. Neuron. 2012;73:962–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rein B, Ma K, Yan Z. A standardized social preference protocol for measuring social deficits in mouse models of autism. Nat Protoc. 2020;15:3464–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 41. Rapanelli M, Tan T, Wang W, et al. Behavioral, circuitry, and molecular aberrations by region-specific deficiency of the high-risk autism gene Cul3. Mol Psychiatry. 2021;26:1491–1504. [DOI] [PubMed] [Google Scholar]
- 42. Wang ZJ, Zhong P, Ma K, et al. Amelioration of autism-like social deficits by targeting histone methyltransferases EHMT1/2 in Shank3-deficient mice. Mol Psychiatry. 2020;25:2517–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol. 2013;106–107:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Assia Batzir N, Posey JE, Song X, et al. Phenotypic expansion of POGZ-related intellectual disability syndrome (White-Sutton syndrome). Am J Med Genet A. 2020;182:38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barker GR, Bird F, Alexander V, Warburton EC. Recognition memory for objects, place, and temporal order:Aa disconnection analysis of the role of the medial prefrontal cortex and perirhinal cortex. J Neurosci. 2007;27:2948–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Binder S, Mölle M, Lippert M, et al. Monosynaptic hippocampal-prefrontal projections contribute to spatial memory consolidation in mice. J Neurosci. 2019;39:6978–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Braff DL, Geyer MA. Sensorimotor gating and schizophrenia. Human and animal model studies. Arch Gen Psychiatry. 1990;47:181–188. [DOI] [PubMed] [Google Scholar]
- 48. Madsen GF, Bilenberg N, Jepsen JR, Glenthøj B, Cantio C, Oranje B. Normal P50 gating in children with autism, yet attenuated P50 amplitude in the Asperger subcategory. Autism Res. 2015;8:371–378. [DOI] [PubMed] [Google Scholar]
- 49. Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gandal MJ, Haney JR, Parikshak NN, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Amram N, Hacohen-Kleiman G, Sragovich S, et al. Sexual divergence in microtubule function: The novel intranasal microtubule targeting SKIP normalizes axonal transport and enhances memory. Mol Psychiatry. 2016;21:1467–1476. [DOI] [PubMed] [Google Scholar]
- 52. Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med. 2017;23:1018–1027. [DOI] [PubMed] [Google Scholar]
- 53. Pinto D, Delaby E, Merico D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zoghbi HY. Postnatal neurodevelopmental disorders: Meeting at the synapse? Science. 2003;302:826–830.. [DOI] [PubMed] [Google Scholar]
- 55. Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vulih-Shultzman I, Pinhasov A, Mandel S, et al. Activity-dependent neuroprotective protein snippet NAP reduces tau hyperphosphorylation and enhances learning in a novel transgenic mouse model. J Pharmacol Exp Ther. 2007;323:438–449. [DOI] [PubMed] [Google Scholar]
- 57. Malishkevich A, Amram N, Hacohen-Kleiman G, Magen I, Giladi E, Gozes I. Activity-dependent neuroprotective protein (ADNP) exhibits striking sexual dichotomy impacting on autistic and Alzheimer's pathologies. Transl Psychiatry. 2015;5:e501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aboonq MS, Vasiliou SA, Haddley K, Quinn JP, Bubb VJ. Activity-dependent neuroprotective protein modulates its own gene expression. J Mol Neurosci. 2012;46:33–39. [DOI] [PubMed] [Google Scholar]
- 59. Gudmundsdottir B, Gudmundsson KO, Klarmann KD, et al. POGZ is required for silencing mouse embryonic β-like hemoglobin and human fetal hemoglobin expression. Cell Rep. 2018;23:3236–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Escher U, Giladi E, Dunay IR, Bereswill S, Gozes I, Heimesaat MM. Anti-inflammatory effects of the octapeptide NAP in human microbiota-associated mice suffering from subacute ileitis. Eur J Microbiol Immunol. 2018;8:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Braitch M, Kawabe K, Nyirenda M, et al. Expression of activity-dependent neuroprotective protein in the immune system: possible functions and relevance to multiple sclerosis. Neuroimmunomodulation. 2010;17:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tetreault NA, Hakeem AY, Jiang S, et al. Microglia in the cerebral cortex in autism. J Autism Dev Disord. 2012;42:2569–2584. [DOI] [PubMed] [Google Scholar]
- 63. Suzuki K, Sugihara G, Ouchi Y, et al. Microglial activation in young adults with autism spectrum disorder. JAMA Psychiatry. 2013;70:49–58. [DOI] [PubMed] [Google Scholar]
- 64. Morgan JT, Chana G, Pardo CA, et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. 2010;68:368–376. [DOI] [PubMed] [Google Scholar]
- 65. Karlsson M, Zhang C, Méar L, et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7:eabh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bassan M, Zamostiano R, Davidson A, et al. Complete sequence of a novel protein containing a femtomolar-activity-dependent neuroprotective peptide. J Neurochem. 1999;72:1283–1293. [DOI] [PubMed] [Google Scholar]
- 67. Gozes I, Zaltzman R, Hauser J, Brenneman DE, Shohami E, Hill JM. The expression of activity-dependent neuroprotective protein (ADNP) is regulated by brain damage and treatment of mice with the ADNP derived peptide, NAP, reduces the severity of traumatic head injury. Curr Alzheimer Res. 2005;2:149–153. [DOI] [PubMed] [Google Scholar]
- 68. Torregrosa T, Lehman S, Hana S, et al. Use of CRISPR/Cas9-mediated disruption of CNS cell type genes to profile transduction of AAV by neonatal intracerebroventricular delivery in mice. Gene Therapy. 2021;28:456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schafer DP, Lehrman EK, Kautzman AG, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Paolicelli RC, Bolasco G, Pagani F, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. [DOI] [PubMed] [Google Scholar]
- 71. McGill BE, Barve RA, Maloney SE, et al. Abnormal microglia and enhanced inflammation-related gene transcription in mice with conditional deletion of Ctcf in Camk2a-Cre-expressing neurons. J Neurosci. 2018;38:200–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kaaij LJT, Mohn F, van der Weide RH, de Wit E, Bühler M. The ChAHP complex counteracts chromatin looping at CTCF sites that emerged from SINE expansions in mouse. Cell. 2019;178:1437–1451.e14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study have been deposited in the GEO public repository under accession code GSE188865. Source data are provided with this paper.