To the Editor—Darkoh et al [1] recently described high-level vancomycin resistance (≥32 μg/mL) in Clostridioides difficile isolates that were recovered from Houston, Texas, and Nairobi, Kenya. The authors concluded that C. difficile isolates with a reduced susceptibility to vancomycin are circulating within a broad population. They further concluded that the increasing presence of these isolates with reduced vancomycin susceptibility may result in therapeutic failures, thus calling for increased screening for C. difficile vancomycin resistance [1]. The authors report that 38% of the 536 stool samples acquired from patients with acute diarrhea had C. difficile growth on a screening medium containing 4-μg/mL vancomycin. These results are counter to previously reported vancomycin minimum inhibitory concentration (MIC) data from clinically relevant C. difficile isolates [2].
The investigators used a screening media that included vancomycin to select for isolates with an elevated vancomycin MIC (≥4 μg/mL) [1]. These methods have been well described for the selection of vancomycin-resistant enterococci, but there is a lack of evidence for identifying resistant C. difficile [3]. In addition, while Slanetz and Bartley medium was used to control for enterococcal contamination, there is no indication as to Darkoh et al controlled for additional vancomycin-resistant anaerobic organisms [1].
To replicate the findings in their report, and as part of an ongoing epidemiologic study, we cultured 10 stool specimens from patients that were positive for C. difficile by real-time polymerase chain reaction (PCR) to taurocholate-cefoxitin-cycloserine-fructose agar (TCCFA) and to TCCFA plus 4 μg/mL vancomycin (TCCFA-V). These stool specimens were collected from patients in the Chicago area between 1 September and 7 October 2022. All 10 specimens were positive for C. difficile using TCCFA, which was confirmed via matrix-assisted laser desorption/ionization (MALDI) or restriction endonuclease analysis. However, there was no growth of C. difficile using the TCCFA-V medium. Instead, we identified Clostridium innocuum from 4 of the 10 stool specimens, using the TCCFA-V medium confirmed with MALDI.
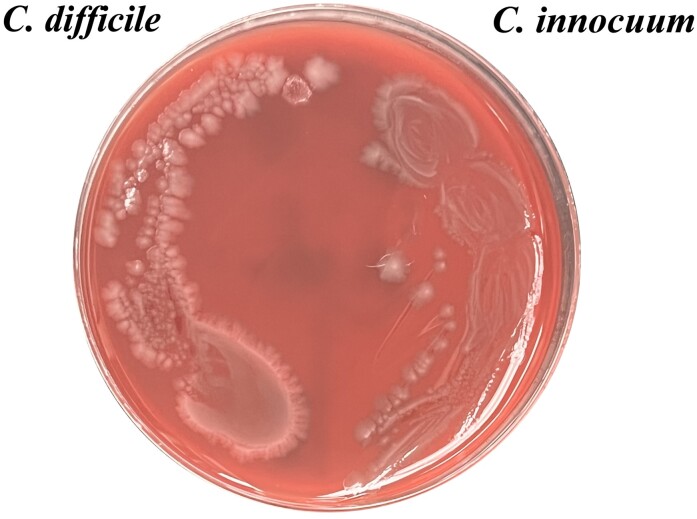
We subsequently determined the vancomycin MIC against C. innocuum, using the Clinical and Laboratory Standards Institute–recommended agar dilution method [4]. The vancomycin MIC against all 4 of these C. innocuum was ≥32 μg/mL. C. innocuum is an emerging stool pathogen that has been associated with diarrheal illnesses and is intrinsically resistant to vancomycin owing to the chromosomal genes racemase and ddlc. innocuum [5, 6]. In addition, the phenotypic appearance of C. innocuum is similar to that of C. difficile on a blood agar plate, which could lead to misidentification (Figure 1).
Figure 1.
Culture of Clostridioides difficile (left) and Clostridium innocuum (right) on blood agar plate.
Our results confirm previous findings that stool specimens can be coinfected by C. difficile and C. innoccum [7]. While Darkoh et al reported that PCR was performed to confirm the identification of C. difficile, it is unclear whether the isolates that underwent PCR were retrieved from the vancomycin screening plates or the standard C. difficile culture medium used [1]. While C. difficile isolates with moderately elevated MICs (4–8 µg/mL) have been reported, particularly in the epidemic BI/027/NAP1/ST1 strain, there are no corroborated data to suggest high-level resistance (eg, MICs ≥32 µg/mL) [8]. We caution against the use of unvalidated screening methods and bring to attention to the possibility of misidentifying other vancomycin-resistant organisms, such as C. innoccum.
Contributor Information
Andrew M Skinner, Department of Research and Medicine, Edward Hines Jr Veterans Administration Hospital, Hines, Illinois, USA; Loyola University Medical Center, Department of Medicine, Maywood, Illinois, USA.
Laurica Petrella, Department of Research and Medicine, Edward Hines Jr Veterans Administration Hospital, Hines, Illinois, USA.
Stacey Spandoni, Department of Research and Medicine, Edward Hines Jr Veterans Administration Hospital, Hines, Illinois, USA.
Fidel Serna-Perez, Department of Research and Medicine, Edward Hines Jr Veterans Administration Hospital, Hines, Illinois, USA.
Stuart Johnson, Department of Research and Medicine, Edward Hines Jr Veterans Administration Hospital, Hines, Illinois, USA; Loyola University Medical Center, Department of Medicine, Maywood, Illinois, USA.
Notes
Disclaimer . The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the US Department of Veteran Affairs.
Financial support . This work was supported by the National Center for Advancing Translational Sciences, National Institutes of Health (grant 5KL2TR002387-05; support to A. M. S. and the Institute for Translational Medicine) and the US Department of Veterans Affairs Research Service (support to S. J.).
References
- 1. Darkoh C, Keita K, Odo C, et al. Emergence of clinical Clostridioides difficile isolates with decreased susceptibility to vancomycin. Clin Infect Dis 2022; 74:120–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Snydman DR, McDermott LA, Thorpe CM, et al. Antimicrobial susceptibility and ribotypes of Clostridium difficile isolates from a phase 2 clinical trial of ridinilazole (SMT19969) and vancomycin. J Antimicrob Chemother 2018; 73:2078–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suwantarat N, Roberts A, Prestridge J, et al. Comparison of five chromogenic media for recovery of vancomycin-resistant enterococci from fecal samples. J Clin Microbiol 2014; 52:4039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard—ninth edition. Wayne, PA: Clinical and Laboratory Standards Institute, 2018. [Google Scholar]
- 5. Chia JH, Wu TS, Wu TL, et al. Clostridium innocuum is a vancomycin-resistant pathogen that may cause antibiotic-associated diarrhoea. Clin Microbiol Infect 2018; 24:1195–9. [DOI] [PubMed] [Google Scholar]
- 6. Cherny KE, Ozer EA, Kochan TJ, Kociolek LK. Complete genome sequence of Clostridium innocuum strain ATCC 14501. Microbiol Resour Announc 2020; 9:e00452-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cherny KE, Balaji A, Mukherjee J, et al. Identification of Clostridium innocuum hypothetical protein that is cross-reactive with C. difficile anti-toxin antibodies. Anaerobe 2022; 102555. doi: 10.1016/j.anaerobe.2022.102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thorpe CM, McDermott LA, Tran MK, et al. U.S.-based national surveillance for fidaxomicin susceptibility of Clostridioides difficile-associated diarrheal isolates from 2013 to 2016. Antimicrob Agents Chemother 2019; 63:e00391-19. [DOI] [PMC free article] [PubMed] [Google Scholar]