Abstract
The lipid phosphatase PTEN (phosphatase and tensin homologue on chromosome 10) is a key tumour suppressor gene and an important regulator of neuronal signalling. PTEN mutations have been identified in patients with autism spectrum disorders, characterized by macrocephaly, impaired social interactions and communication, repetitive behaviour, intellectual disability, and epilepsy. PTEN enzymatic activity is regulated by a cluster of phosphorylation sites at the C-terminus of the protein. Here, we focused on the role of PTEN T366 phosphorylation and generated a knock-in mouse line in which Pten T366 was substituted with alanine (PtenT366A/T366A). We identify that phosphorylation of PTEN at T366 controls neuron size and connectivity of brain circuits involved in sensory processing. We show in behavioural tests that PtenT366/T366A mice exhibit cognitive deficits and selective sensory impairments, with significant differences in male individuals. We identify restricted cellular overgrowth of cortical neurons in PtenT366A/T366A brains, linked to increases in both dendritic arborization and soma size. In a combinatorial approach of anterograde and retrograde monosynaptic tracing using rabies virus, we characterize differences in connectivity to the primary somatosensory cortex of PtenT366A/T366A brains, with imbalances in long-range cortico-cortical input to neurons. We conclude that phosphorylation of PTEN at T366 controls neuron size and connectivity of brain circuits involved in sensory processing and propose that PTEN T366 signalling may account for a subset of autism-related functions of PTEN.
Keywords: PTEN, developmental disorders, cortical connectivity, cognitive behaviour, neuronal morphology
PTEN is a key regulator of neuronal signalling, and PTEN mutations have been identified in patients with autism spectrum disorders. Using a newly generated knock-in mouse, Ledderose et al. show that phosphorylation of PTEN at threonine 366 controls neuron size and connectivity of brain circuits involved in sensory processing.
See Binder and Bordey (https://doi.org/10.1093/brain/awac350) for a scientific commentary on this article.
See Binder and Bordey (https://doi.org/10.1093/brain/awac350) for a scientific commentary on this article.
Introduction
The lipid phosphatase PTEN (phosphatase and tensin homologue on chromosome 10) was first described as a tumour suppressor1–3 in a few sporadic and heritable tumours in humans.4,5 More recently, PTEN mutations have also been known to occur in various forms of autism spectrum disorders (ASD).6–10 PTEN is an important regulator in the PI3K/AKT/mTor signalling pathway.11,12 By dephosphorylating phosphatidylinositol-3,4,5-triphosphate to phosphatidylinositol-4,5-bisphosphate, PTEN is involved in growth-dependent processes such as proliferation, cell survival or differentiation.13–17 In PTEN conditional knock-out mouse models—homozygous Pten mice are lethal18—different effects of PTEN on neuron size were found.19,20 In Ptenfl/fl/Nse-Cre knockout (KO) mice, loss of PTEN leads to enlarged somata of cells in the dentate gyrus, enlargement of dendritic trees and greater spine density in hippocampal neurons.21,22 In cortical neurons, when Pten was deleted in Camk2a-cre+/–; PtenloxP/loxP; Thy1-gfp mice, a compartmentalized effect was observed in pyramidal cells of cortical layer (L) 2/3.23 Further, these PTEN-deficient neurons show changes in their connectivity.24
In relation to ASD, 35 missense mutations have been identified in the Pten gene, most of which share a macrocephaly phenotype.25,26 This macrocephaly phenotype correlates with findings from loss-of-function mouse models.16,21,27 Thus, in a Pten mouse model for autism—germline haploinsufficient Pten+/– mice—increased brain volume, altered neuronal morphology and altered social behaviour were observed.28–30 Brain volume in those mice also scales differently during development.27 Similar characteristics are found in PTEN ASD patients, who show altered neuronal morphology and anatomy, increased white matter and enlargement of the perivascular space.31–33 Cognitive defects in these patients, such as limited ability to socialize or to communicate,34 could relate to altered connections between sensory areas.
The activity and stability of the PTEN protein is crucially regulated by phosphorylation of a serine and threonine cluster at the C-terminus of the gene. Phosphorylation at threonine 366 occurs by polo-like-kinase (Plk3) and glycogen synthase kinase 3 (GSK3). Phosphorylation at T366 shows different effects in different cell types; PTEN is stabilized by phosphorylation at T366 in glioma cells,35,36 but destabilized in epithelial cells.37–39 This property is interesting with regard to the wide range of symptoms associated with Pten mutations and to diverse possibilities of regulatory mechanisms through PTEN.
To investigate the phosphorylation of PTEN at T366 in vivo and to understand its mechanistic significance, we generated a knock-in mouse in which threonine at 366 is replaced by alanine (PtenT366A/T366A). We show that these mice have altered neuronal morphology: soma and dendrites are enlarged and cortical connectivity is altered. In addition, these mice show sensory abnormalities in exploration, freezing and in their response to vibrissal stimuli, which may relate to their altered neuronal morphology. We conclude that intact phosphorylation at PTEN T366 is essential for the establishment of proper cortical connectivity and for normal sensory processing. Our work identifies that T366 phosphorylation of PTEN might serve as an important signalling regulator in ASD.
Materials and methods
Ethics statement and handling of mice
Experiments were conducted under the licences T0143/11, G0621/12 and G0189/14 in accordance with the guidelines of the Universitätsmedizin Charité, Berlin and the Landesamt für Gesundheit und Soziales. Behavioural studies were conducted in the Animal Models Core at The Scripps Research Institute and approved by The Scripps Research Institute’s Institutional Animal Care and Use Committee, and met the NIH guidelines detailed in the ‘Guide for the Care and Use of Laboratory Animals’. For morphology experiments, PtenT366A/T366A mice and wild-type (wt) mice were in FVB/N background; for behaviour in C57BL6/J background. Mice were kept in groups of two to three individuals under standard conditions in a 12-h day–night cycle with water and food ad libitum.
Generation of PtenT366A/T366A knock-in mice
A BAC construct containing the mouse Pten gene was used as a genomic source to create a targeting vector. Briefly, a 5′ homology arm of exon 8 followed by exon 9 containing a threonine 366 to alanine mutation and a right homology arm containing intron 9 were inserted into targeting vector pKO V915 (Dana Farber Cancer Center) flanking a LoxP-pGK-Neo-LoxP selection cassette. The linearized targeting vector was electroporated into 129S6/SV EV Tac embryonic stem cells and clones were screened by southern blot analysis for correct targeting into the Pten locus. Two correctly targeted ES cell clones were injected into blastocysts to generate chimeric mice in C57BL/6J background. Mice with greater than 80% chimerism, as determined by agouti coat colour, were bred with C57BL/6J mice and progeny were screened by southern blot to ascertain germline transmission. Heterozygous mice were bred with E2a-cre mice (Jackson Lab, #003314) to excise the pGK-Neo selection cassette and progeny were screened by PCR to confirm successful cre-mediated excision. Heterozygous mice lacking pGK-Neo were bred with C57BL/6J mice to generate PtenT366A/T366A mice.
DNA isolation, genotyping and sequencing
Genomic DNA from PtenT366A/T366A tail or ear tissue was incubated in 80% Proteinase-K in TE buffer for 6 h. DNA was purified (DNA purification kit, Thermo Fisher Scientific) and analysed with standard PCR conditions using primers targeting the Pten gene location at threonine 366 (forward 5′-AGCAGTGCCCTTCAGAATTC-3′, reverse 5′-TCAGCCACTTCAGCTGGTGAC-3′). The genotype of PtenT366A/T366A mice was determined by sequencing the PCR product of 600 bp with the forward primer.
Primary cell culture
Hippocampi and cortices were dissected from wild-type and PtenT366A/T366A mice at embryonic stage (E) 16.5 and cells dissociated in papain (Worthington) according to the manufacturer’s protocol. Twenty-four-well dishes (MatTek Corporation) were coated with 30 ng/µl poly-D-lysine and cells were plated in growth medium (neurobasal medium supplemented with 1% Glutamax, 1% B27). After 2, 4, 8 and 14 days in vitro (DIV), cells were fixed in 4% paraformaldehyde (PFA) and processed for immunohistochemistry.
Protein lysate preparation and western blotting
Forebrain homogenates from postnatal Days (P) 7 and P14 were weighed and homogenized with a glass pistil in 4× the volume in RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP40, 0.1% sodium dodecyl sulphate) supplemented with protease inhibitors (Calbiochem set III) and phosphatase inhibitors (1 mM Na2MO4, 1 mM NaF, 20 mM β-glycerophosphate, 1 mM Na3VO4, 500 nM cantharidin). Homogenates were centrifuged at 20 000g and the supernatant collected for protein quantification using BCA Thermo Scientific Pierce™ Protein Assay. The cells were washed once with cold phosphate-buffered saline (PBS) and lysed in cold RIPA buffer. Cell lysates were centrifuged at 20 000g and the supernatant transferred to Roti load I sample buffer. Protein lysates in Roti-Load sample buffer (5–10 µg total protein) were loaded on 8% sodium dodecyl sulphate gels, stacked at 60 V for 30 min and separated at 120 V for 60 min. Proteins were transferred to a nitrocellulose membrane for 2 h in a wet blot tank system (Bio-Rad) and membranes were blocked with 5% skimmed milk for 30 min at room temperature (RT) before incubating with primary antibody. Primary antibodies were prepared in 5% skimmed milk and incubated overnight at 4°C. Following incubation, membranes were washed three times with TBS-T at RT for 5 min. Secondary antibodies were prepared in 5% skimmed milk, and membranes incubated for 30 min at RT, washed three times with TBS-T and imaged using the Fusion SL system from Vilber Lourmat. Quantification of band densities was performed by Fiji/ImageJ as previously described.40 The area of the band and the mean grey values were measured to obtain a relative density. For relative quantifications, measurements were normalized to the loading control.
Antibodies
Western blotting
Primary antibodies: anti-PTEN rabbit (Cell Signaling, #9559, 1:1000), anti-pThr308AKT rabbit (Cell Signaling, #9275, 1:1000), anti-pSer473AKT rabbit (Cell Signaling, #4060, 1:1000), anti-pS6 rabbit (Cell Signaling #2211, 1:1000) and anti-tubulin mouse (Sigma #PT6199,1:5000). Secondary antibodies: (1:3000) horseradish peroxidase (HRP)-conjugated anti-rabbit, anti-mouse (Vector Labs, #PI1000, Cat no. PI2000).
Immunohistochemistry
Primary antibodies: anti-MAP2 mouse (1:500, Sigma M9942), anti-MAP2 guinea pig (1:500, Synaptic System, 188 004), anti-GFP chicken (1:5000, Abcam ab13970), anti-BrdU rat (1:250, Biorad, OBT0030), anti-Cre rabbit (1:1000, Abcam ab41104), anti-FOXP2 (1:1000, Abcam ab16046), anti-NeuN mouse (1:1000, Millipore MAB377), anti-Cux1 rabbit (1:1000, Santa Cruz, sc13024), anti-CTIP2 rat (1:1000, Abcam ab18465) and anti-pS6 rabbit (1:500, Cell Signaling #2211). Secondary antibodies (all 1:500): anti-mouse Alexa Fluor 488 (Dianova, 015-540-003), anti-rabbit 488 (Dianova 711-545-152), anti-rabbit Cy3 (Dianova 711-167-003), anti-mouse Cy5 (Dianova, 715-175-151), anti-chicken Alexa Fluor 488 (Dianova, 703-546-155) and HOECHST (1:5000, Sigma Aldrich 14533).
Production of recombinant adenovirus and rabies virus
Viruses were produced in the Viral Core facility of the Universitätsmedizin Charité, Berlin (https://vcf.charite.de/)41–43 using standard protocols.44 For morphological analysis, an adenovirus, tagged to a nuclear green fluorescent protein (AAV9-CAG-GFP, Addgene plasmid #37825) was used. For rabies virus production, B7GG cells were transfected with rabies genomic vectors SADdeltaG-F3-mcherry (Addgene plasmid #32634), pcDNA-SADB19N (Addgene plasmid #32630), pcDNA-SADB19P (Addgene plasmid #32631), pcDNA-SADB19L (Addgene plasmid #32632) and pcDNA-SADB19G (Addgene plasmid #32633) and cells were transfected with Lipofectamine 3000 (ThermoFisher Scientific). The rabies was pseudotyped with envelope protein EnvA of the Avian Sarcoma and Leukosis virus and collected, filtered and concentrated by ultracentrifugation after 3–5 days.
Rabies virus injection scheme
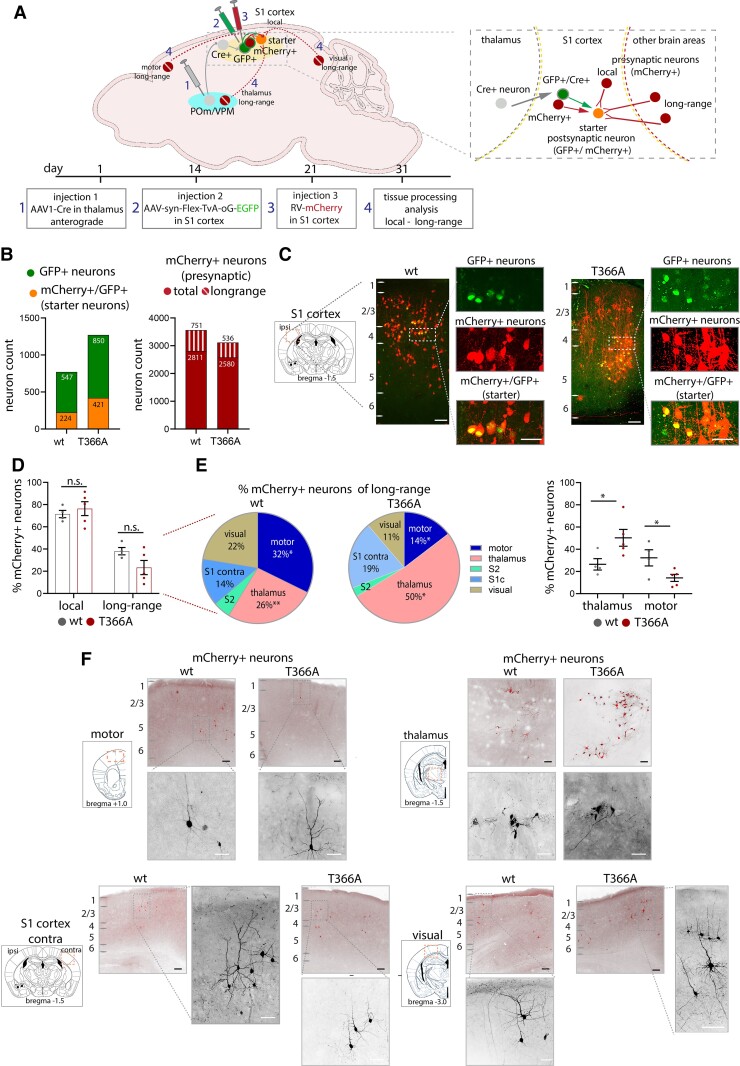
To examine the effect of PTEN on thalamocortical circuit connectivity, a dual transynaptic strategy combining retrograde tracing with monosynaptic rabies virus43,45 and AAV1-Cre46 was used in both wild-type and PtenT366A/T366A mice (Fig. 5). To target cortical neurons, postsynaptic to thalamic inputs in somatosensory (S1) cortex, AAV1.hSyn.Cre.WPRE.hGH (Addgene, #105553) was injected into the ventrobasal thalamus. This step puts Cre trans-synaptically into S1 cortical neurons receiving input from the posterior medial thalamus (POm) and ventral posteriomedial thalamus (VPm). After allowing Cre expression for 2 weeks, a second injection of a helper virus for rabies, a Cre-dependent AAV coupled to nuclear GFP linked to an EnvA interacting cognate avian viral receptor TVA receptor and to a rabies G protein (AAV-FLeX-EnvA-TvA-GFP), was injected in S1. This virus expressed TVA receptor and G protein that can be targeted by the rabies virus. The rabies virus coupled to mCherry and a deleted rabies glycoprotein (RABB-SADB19dG-mCherry)43 and was injected 2 weeks later into S1 cortex. Starter neurons in S1 expressed this construct (and turned yellow, expressing both mCherry and GFP) and neurons presynaptic to starter neurons expressed mCherry. Brains were removed and placed in fixative 10 days after the third injection. This strategy provides enough time for rabies virus to express in local and long-range neurons that are presynaptic to the S1 neurons.
Figure 5.
Presynaptic input to S1 cortex in PtenT366A/T366A mice. (A) Schematic showing injection procedure of three viruses (also see the ‘Materials and methods' section). AAV1-Cre was injected (injection 1) into POm and VPm thalamus. Two weeks later, the trans-synaptically labelled S1 cortical neurons expressing Cre, i.e. receiving thalamic input, were targeted with a flex AAV to express the TVA receptor (injection 2). Two weeks after injection 2, the TVA-expressing neurons (GFP+ S1 neurons) were targeted with rabies virus (injection 3). The Cre+, TVA-expressing (green) and mCherry+ expressing (red) neurons were the starter neurons (yellow). One week after injection of rabies, brains were harvested, and local and long-range retrograde-labelled neurons could be observed (red presynaptic neurons). (B) Left graph showing number of (Cre+)/GFP+/mCherrry+ (starter) neurons, right graph showing all presynaptic mCherry+ labelled neurons. (C) Examples with enlarged images showing expression of GPF+ neurons, GFP+/mCherry+ (starter) neurons and presynaptic mCherry+ neurons in wild-type and PtenT366A/T366A S1 cortices. (D) Graph showing the percentage of presynaptic neurons local in S1 cortex and long-range neurons from other cortical and thalamic areas in PtenT366A/T366A and wild-type brains. (E) Pie charts showing percentages of dissected presynaptic inputs from long-range areas to S1 in PtenT366A/T366A and wild-type brains. Graph showing input to motor cortices and thalamus in PtenT366A/T366A and wild-type brains. (F) Example images of presynaptic neurons in motor cortices, thalamus, S1 cortex contralateral and visual cortices in PtenT366A/T366A and wild-type brains. Higher magnification of single neurons. Statistical analysis with unpaired t-test, *P < 0.05. Analysis details are provided in Supplementary Table 8. Scale bars = 100 µm, 50 µm in enlarged images.
Injections in new-born mice
Injections into new-born mice were performed as described.47 In short, new-born pups (P0) were removed from their mother and briefly anaesthetized with isoflurane. AAV9-CAG-GFP virus was injected (1 µl, 125 nl/s) into both lateral ventricles using a 10 µl NANOfil syringe and a 34-gauge needle and a UltraMicroPump (World Precision Instruments). After injection, pups were allowed to recover on a heating pad at 37°C, and then placed in the home cage with their mother.
Stereotaxic injections in adult mice
Before surgery, mice were anaesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) and head-fixed on a stereotaxic frame with non-puncture ear bars and a nose clamp (Kopf Stereotaxic device, California, USA, Inc.). The extent of the anaesthetic was confirmed by a toe pinch. For analgesics, carpofen (5 mg/kg) intraperitoneal and lidocaine under the skull was injected. The skull was opened with scissors and a craniotomy (∼1 mm) was made on S1 cortex. Coordinates for S1 cortex were (S1, AP −1.5, lateral: 2.3–2.7), for POm/VPm thalamus (AP, −1.5, lateral, 1.5, ventral 3.0). A fine glass pipette was tip-filled with negative pressure and 200 nl of virus (20 nl/min) was injected under constant positive pressure (QSI, Quintessential Stereotaxic Injector). During surgery, the brain tissue was kept moist by applying sterile PBS and the eyes were lubricated with Bepanthen eye cream (Bayer). Following stereotaxic injection, the craniotomy was cleaned, the skull sutured and the mouse put on a heating pad at 37°C for waking up and returned to its home cage for full recovery. Caprofen (5 mg/kg) was administered for 2 days after surgery to reduce postoperative pain.
BrdU proliferation experiments
Birth dating experiments with BrdU were carried out as described previously.48 Time-pregnant mice were injected intraperitoneally with 50 mg/kg 5-bromo-2′-deoxyuridine (BrdU; Accurate Chemical & Scientific Corporation) at E11.5, E13.5 and E15.5. BrdU was allowed to express until P1 and P8. The brains were fixed and the number of BrdU+ neurons was analysed in S1 cortices.
Tissue processing and histological methods
Neuronal cultures
Neuronal cultures were fixed in 4% PFA for 30 min. Neurons were permeabilized in a cytoskeleton-stabilizing PHEM buffer (PIPES, HEPES, EDTA and MgCl2, pH 7.4) with 0.1% Triton for 10 min and blocked in PHEM buffer with 4% goat serum. Primary antibodies were incubated for 1 h followed by PBS washes and incubation of secondary antibodies for 1 h.
Brain tissue
Mice were deeply anaesthetized with isoflurane and trans-cardially perfused with PBS and 4% PFA and brains placed in 4% PFA overnight. For microtome processing, brains were soaked in 30% sucrose overnight and cut in 70–100 µm sections. Brain sections were stored in PBS for immediate immunohistochemistry or in cryo-protection medium (ethylene ethanol 30%, glycerol 30% in PBS) for long-term storage.
Immunohistochemistry
Brain sections were placed in blocking solution (5% normal goat serum), 0.1% Triton X-100 in PBS) for 2 h and incubated in primary antibodies overnight at RT. On the following day, sections were washed in PBS and incubated in secondary antibodies for 2 h. Following washes in PBS, brain sections were counterstained with HOECHST, washed with PBS and mounted in 80% glycerol (2.5% DAPCO in PBS).
Nissl staining
Brain sections were mounted on Super Frost Plus Slides (Thermo Fisher Scientific, 10149870). Sections were incubated in Nissl solution for 40 s to 1 min, then washed three times in water and dehydrated in ethanol (50–70–80–95%, 5 min each), followed by incubation in Xylol. Slices were mounted in Eukitt quick-hardening mounting medium (Fluka) and stored at 4°C.
Behavioural experiments
A full description of the behavioural tests (Y-maze, hanging wire, rotarod, open field, von Frey, vibrissae-stimulated reflex, Barnes maze, Morris water maze, conditioned fear) can be found in the Supplementary material.
Confocal imaging
Images were taken on confocal laser scanning microscopes (Leica SP5, Leica SP8 Nikon A1Rsi+) with 10× air, 20× or 63× oil immersion objectives (Leica: HCX PL APO 20×/0.7, HCX PL APO 63×/1.20 W motCORR CS; Nikon 20× Plan Apo, Air immersion, 0.8 NA, WD 1.000, DIC N2 VC, 40× Plan Fluor, oil immersion 1.3 NA, 200 WD, DIC N2 H). A 405 nm laser was used for detection of HOECHST, a 561 nm laser for detection of mCherry and a 488 nm laser for detection of GFP (Leica, 425/70, 515/25, 590/70; Nikon, 450/50, 525/50, 595/50). Images were taken at a resolution of 1024 × 1024 with a step size of 2 µm (10×, 20×) and with a step size of 1 µm (40×).
Data analysis
Cell counting
Cells were counted manually from confocal images in Fiji/ImageJ with the cell counter plugin. In cases of clear distinguishable cells, z-stacks were used. In less clear images, cells were counted while inspecting each confocal plane.
Soma size
For the analysis of soma size, maximal intensity projections of the confocal images were used, and soma size was measured with ImageJ/Fiji. The maximal projection was converted to 8-bit black and white images, and the threshold was adjusted manually.
pS6 level
For the analysis of pS6 level, Cy3 or Alexa 488 and HOECHST fluorescence of neurons was measured in single planes of 40× confocal images. The ratio of Cy3 (Alexa 488) to HOECHST was used to define the final fluorescence level.
Dendritic arborization
For the analysis of dendritic arborization of L2/3, L4 neurons and granule neurons in the dentate gyrus, dendrites were reconstructed from confocal images (2 µm, about 30 stacks per image of 70-µm thick brain sections) with the imaging analysis software IMARIS. The reconstructed neurons were displayed in maximal intensity projections in 3D and the complexity of the dendrites was analysed with Sholl analysis.49 L2/3 pyramidal neurons were defined by a triangular soma shape, by one apical dendrite reaching L1 and basal and apical dendrites. L4 interneurons were defined by a local arborization and somata with a round shape.
Defining cortical layers and brain areas
Cortical areas and cortical layers were defined according to the Allen brain reference atlas (https://mouse.brain-map.org/) and to the Paxinos and Franklin’s mouse brain coordinate atlas.50 Cortical areas are defined as: motor cortices (+1.0 to 2.5 from bregma, 1.0–1.5 lateral), S1 cortex (−1.2 to −1.8 from bregma, 2.3–2.7 lateral), S2 cortex (−1.2 to −1.8 from bregma, 3.0–3.5 lateral), visual cortices (−3.0 to −3.5 from bregma, 1.0–3.0 lateral). Cortical layers in S1 cortex were defined as, from pia: L1, <100 µm, L2/3, 100–300 µm; L4: 300–400 µm; L5, 400–700 µm; L6, 700–950 µm or >1000 µm from white matter.
Analysis of input to S1 cortex
The retrograde-labelled neurons with the rabies approach could be characterized as local (at the injection site) or long-range. The presynaptic input was obtained by counting mCherry+ (presynaptic) neurons in every second brain section (four brains wild-type, five brains PtenT366A/T366A). Local input was defined as 500 µm around the injection site in S1 cortex (bregma −1.2 to −1.7, lateral 2.3–2.7). Cells found in other areas of the brain were defined as long-range. We focused on five long-range areas that provided input to S1: M1, S2, visual, contralateral S1 and thalamus.
Statistical analysis
Data were statistically analysed with Graphpad/Prism software. Statistics for behaviour, morphology experiments and connectivity were calculated by Students t-test or one-way ANOVA with Bonferroni post hoc test. Sholl analysis for dendrite arborization was analysed by two-way ANOVA with Bonferroni post hoc test. Data are shown as mean ± SEM, P-values are indicated as: P > 0.05 ns, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.
Results
Generation of PTEN T366-deficient (PtenT366A/T366A) mice
To test the contribution of PTEN T366 to brain development and function, we analysed mice in which PTEN T366 phosphorylation was inactivated by the introduction of a threonine (T) to alanine (A) amino acid substitution, leading to constitutive inactive phosphorylation at PTEN T366 (thereafter abbreviated as T366A). Homozygous PtenT366A/T366A mice are viable and fertile. Embryos were born at expected Mendelian ratios and did not display overt phenotypes. We confirmed the point mutation in the Pten gene in PtenT366A/T366A mice by PCR and subsequent sequencing (Supplementary Fig. 1A and B). Forebrain homogenates from PtenT366A/T366A mice showed normal expression levels of PTEN protein from P7 to P14 compared to wild-type samples. Similarly, prominent signalling effectors of PTEN activity such as pAkt and pS6 did not show any differences in forebrain homogenates (Supplementary Fig. 1C and Supplementary Table 1). Immunolabelling against pS6 in PtenT366A/T366A and wild-type brain slices at P8 and P14 and quantification of fluorescence level in relation to HOECHST counterstain revealed increased fluorescence level in single L2/3 and L4 but not in L5 in PtenT366A/T366A neurons (Supplementary Fig. 1D and Supplementary Table 2). These results show that systemic loss of PTEN T366 phosphorylation does not overtly affect normal embryonal development, PTEN protein stability or PTEN signalling pathways when looking at the whole brain. On a single cell level, however, it suggests that phosphorylation of PTEN at T366 affects specifically cortical neuron morphology.
PtenT366A/T366A mice show selective sensory impairments
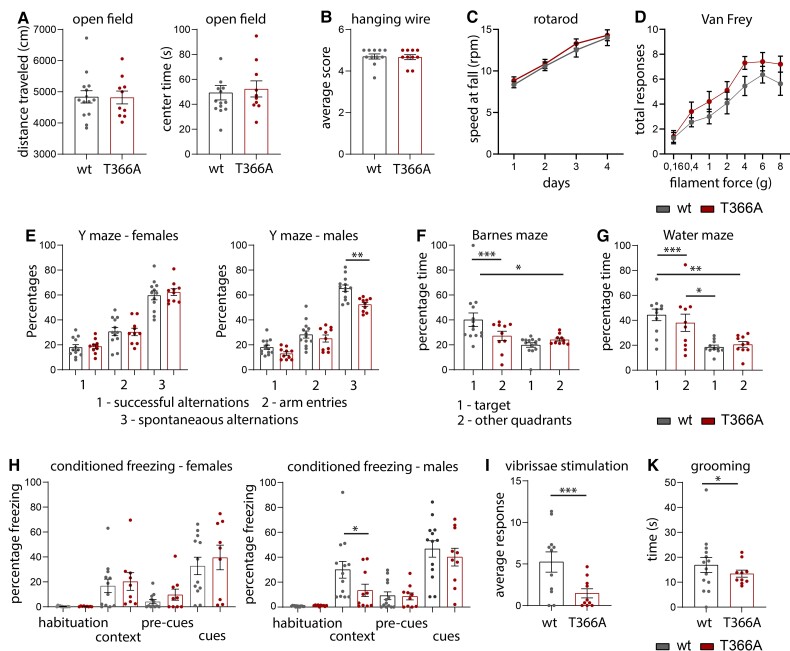
Social learning and social interaction is reduced in Pten neuron-specific enolase (Nse-Cre) mice,21 while heterozygous Pten+/– mice show increased anxiety and hyperactivity under stressful environments and decreased attention,28,30 and a depression-like-phenotype in Pten+/– male mice.29 To characterize whether the loss of PTEN phosphorylation at T366 leads to changes in levels of anxiety and other behaviours like grooming and changes in sensory motor behaviours, we performed standard behavioural tests in PtenT366A/T366A mice (Fig. 1 and Supplementary Table 3). Exploratory behaviour in the open field was similar in PtenT366A/T366A mice compared to their wild-type littermates, in terms of both the distance travelled and time spent in the centre of the field (Fig. 1A). Motor performance and strength as determined using the hanging wire test (Fig. 1B) and the rotarod (Fig. 1C) were similar in PtenT366A/T366A mice and their wild-type siblings. Response to mechanical stimulation in the von Frey test did not differ between genotypes (Fig. 1D).
Figure 1.
Behaviour in PtenT366A/T366A mice. (A) PtenT366A/T366A and wild-type (wt) siblings tested for general exploratory behaviour in the open field. (B and C) Motor skills as determined in the hanging wire test (B) and in the rotarod (C). (D) Percentage of total responses to mechanical stimulation in the von Frey test in PtenT366A/T366A and wild-type siblings. (E) Percentage of spontaneous alterations in the Y-maze in female and in male PtenT366A/T366A and wild-type siblings. (F) Percentage of time that PtenT366A/T366A and wild-type mice spent in the target area and other quadrants of the Barnes maze. (G) Percentage of time that PtenT366A/T366A and wild-type mice spent in the target area and other quadrants of the Morris water maze. (H) Percentage freezing in the conditioned fear-induced freezing in female and in male PtenT366A/T366A and wild-type siblings. (I) The average number of responses in the vibrissae-stimulated reflex test of PtenT366A/T366A and wild-type siblings. (K) Grooming time in PtenT366A/T366A and wild-type mice. Statistical analysis with one-way ANOVA (Bonferroni post hoc test), unpaired t-test, Wilcoxon test (grooming), *P < 0.05. For analysis details see Supplementary Table 3.
There were, however, differences in behavioural testing for cognitive abilities in PtenT366A/T366A mice. In the Y-maze, male PtenT366A/T366A mice showed a decrease in spontaneous alternation behaviour relative to wild-type siblings (Fig. 1E). The performance of PtenT366A/T366A mice, when both sexes were evaluated in the Barnes maze (Fig. 1F), and in the water maze was diminished (Fig. 1G). PtenT366A/T366A mice also performed poorly (compared to wild-type mice) in the probe tests, suggesting that these mice have spatial memory deficits. In the conditioned fear test, male PtenT366A/T366A mice showed less freezing in the context task compared to their wild-type siblings (Fig. 1H), supporting that decreased hippocampus-mediated contextual learning and memory occurs in these mice.
PtenT366A/T366A mice also demonstrated a decreased number of responses to vibrissae stimulation (Fig. 1I) and grooming time in the open field test was reduced (Fig. 1K). Overall, these phenotypes suggest differences in cognitive and sensory processing in PtenT366A/T366A mice, which could be based on differences of cortical morphology or connectivity of cortical neurons.
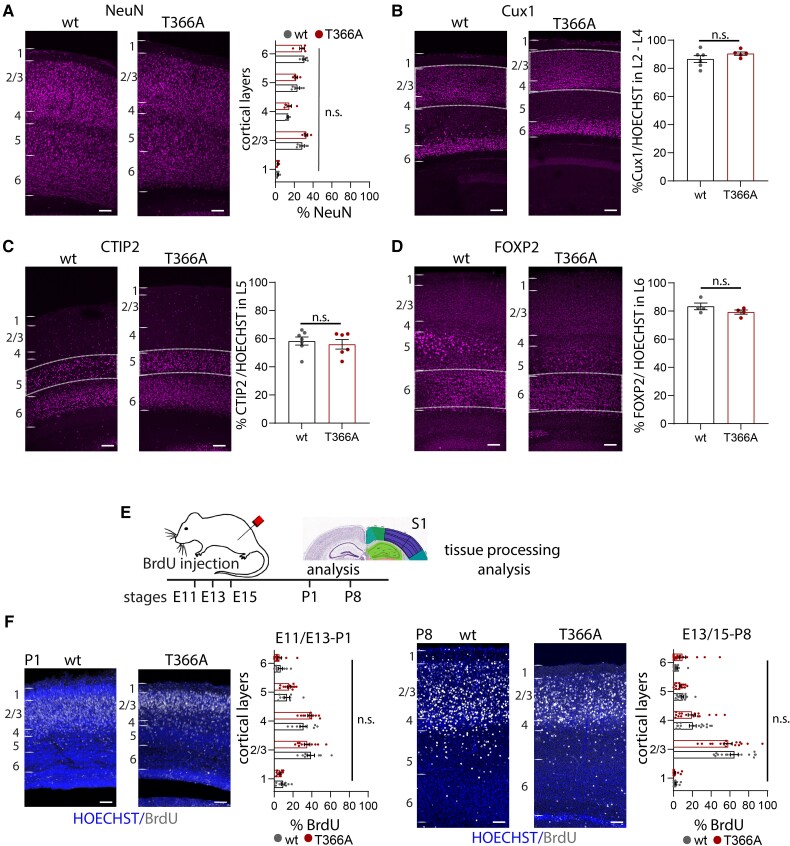
Layer formation and cortical proliferation are normal in PtenT366A/T366A brains
Germline PTEN mutations show disrupted cortical proliferation51 and affected scaling across brain areas during development.27 To characterize whether loss of PTEN T366 phosphorylation alters cortical layering, we performed immunolabelling, exploiting different layer-specific markers. We used immunolabelling against NeuN for all cortical neurons; Cux1 for L2/3 to L4; CTIP2 for L5; and FOXP2 for L6 (Fig. 2 and Supplementary Table 4). The overall percentage of NeuN+ neurons in the single layers did not show significant differences in PtenT366A/T366A mice compared to wild-type at P14 (Fig. 2A). Neither the percentage of Cux1+ neurons in L2/3 (Fig. 2B), nor the percentage of CTIP2+ neurons in L5 (Fig. 2C) or the percentage of FOXP2+ neurons in L6 (Fig. 2D) differed significantly between the genotypes when analysed in relation to HOECHST counterstain, suggesting that cortical layers develop normally in PtenT366A/T366A mice.
Figure 2.
Cortical layers and proliferation in PtenT366A/T366A brains. (A–D) Immunolabelling for cortical layer markers and the percentages of neurons expressing in PtenT366A/T366A and wild-type (wt) mice. (A–D) The percentage of (A) NeuN+ neurons in cortical layers 1 to 6. (B–E) The percentage of (B) Cux1+ neurons in cortical layers 2–4, (C) CTIP2+ neurons in L5, (D) FOXP2+ neurons in L6 in relation to all neurons counterstained with HOECHST. (E) Proliferation of cortical progenitor cells in PtenT366A/T366A and wild-type siblings. Schematic for experimental set-up, BrdU injections were performed in time-pregnant heterozygous females at E11.5, E13.5 and E15.5. Analysis was undertaken at P1 and P8. (F) Example images of BrdU labelling and HOECHST counterstain at P1 and P8. Graphs showing distribution of BrdU+ neurons in PtenT366A/T366A and wild-type mice at P1 and P8, respectively. Each dot in graphs accounts for one brain section. Statistical analysis with unpaired t test and two-way ANOVA, Bonferroni post hoc test. For analysis details see Supplementary Table 4. Scale bars = 100 µm.
Because the PTEN behavioural phenotype is established early during development,52,53 we tested whether proliferation of cortical progenitors was affected in PtenT366A/T366A mice. We performed a BrDU proliferation assay in timed-pregnant PtenT366A/T366A and wild-type mice.48 We injected BrdU intraperitoneally in pregnant females at embryonic stages E11.5, E13.5 and E15.5, and processed brains of offspring for immunohistochemistry of BrdU at P1 and P8 (Fig. 2E and F and Supplementary Table 4). Cortical layer formation in the mouse cortex starts at E10.5 with formation of the preplate and projection neurons develop in a tightly controlled temporal and spatial order from E11.5 to E17.5 with L6 and L5, continuing to L4, and L2/3 at stages E15/E1654,55. In PtenT366A/T366A mice, we found BrdU+ neurons in upper cortical L4 to L1, which did not differ across layers. We conclude that cortical layers form appropriately in PtenT366A/T366A mice.
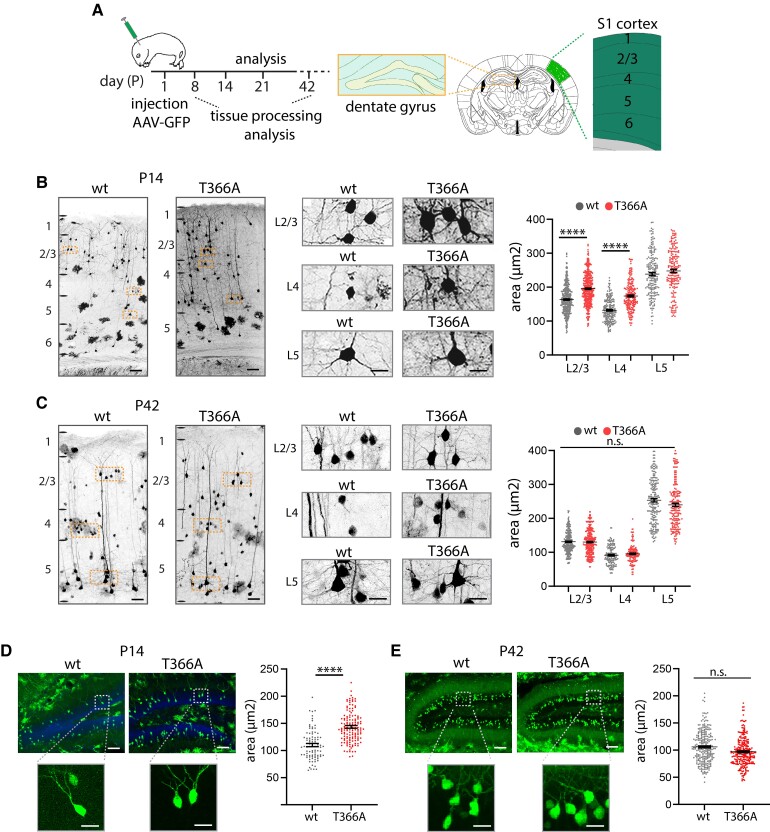
Soma size in cortical upper layers is affected in PtenT366A/T366A brains
Loss of PTEN leads to a developmental state-dependent increase in soma size and dendritic arborization.21,23 To determine whether PTEN T366 affects neuronal growth, we analysed neuronal soma size in PtenT366A/T366A cortices at various stages of development. We injected GFP-tagged AAV in new-born mice at P0 and analysed GFP+ neurons at P8, P14, P21 and P42. GFP+ neurons were present in all cortical layers and in the hippocampus (schematic, Fig. 3A). We defined cortical layers according to the Allen brain atlas and with HOECHST counterstain. We concentrated our analysis on S1 cortex and on granule neurons of the dentate gyrus of the hippocampus. In S1 cortex, PtenT366A/T366A mice in comparison to wild-type showed increased neuronal soma size in pyramidal neurons in L2/3 and in L4 interneurons by P8 and P14 (Fig. 3A and Supplementary Fig. 2A). In contrast, by P21 and P42, pyramidal neurons in L2/3 and L5 and neurons in L4 in PtenT366A/T366A did not show any differences in soma size when compared to wild-type (Fig. 3C, Supplementary Fig. 2B and Supplementary Tables 5 and 6).
Figure 3.
Soma size in PtenT366A/T366A cortical and dentate gyrus neurons. (A) Schematic of AAV-GFP injection at P0 and expression of GFP in adult S1 cortex and dentate gyrus. Soma size was analysed at P14 and P42. Staining with HOECHST and the Allen brain mouse reference atlas were used to define cortical layers (also see the ‘Materials and methods' section). (B and C) GFP-injected neurons at P14 (B) and P42 (C) for PtenT366A/T366A and wild-type (wt) mice. Enlarged images show somata from pyramidal neurons in L2/3 and L5 and neurons in L4. Graphs for soma size in L2/3 to L5 PtenT366A/T366A and wild-type neurons at P14 and P42, respectively. (D and E) Analysis of soma size in dentate gyrus neurons at P14 (D) and at P42 (E) in PtenT366A/T366A and wild-type mice. Each dot in graphs accounts for one cell. Data shown as average ± SEM. Data from three brains each genotype, six sections per brain. Statistical analysis with one-way ANOVA, unpaired t-test for dentate gyrus, ***P < 0.001. Analysis details are provided in Supplementary Table 5. Scale bars = 100 µm, enlarged images in B 20 µm, in C–E, 50 µm.
We obtained similar results for the dentate gyrus. Labelled granule neurons in dentate gyrus of PtenT366A/T366A mice at P14 were significantly larger than somata in wild-type mice (Fig. 3D), while by P42 the soma size was similar in both genotypes (Fig. 3E). Statistics indicate that soma size in PtenT366A/T366A mice is increased significantly between postnatal stage P8 and P21 with a peak at P14.
Control measurements in primary neuronal cultures for cortical and hippocampal neurons plated at E16.5 and analysed at DIV2–4, 8 and 14 by immunolabelling with MAP2 showed no differences in soma size (Supplementary Fig. 3 and Supplementary Table 6). These results combined argue for layer- and neuron-specific difference in somata size in PtenT366A/T366A mice.
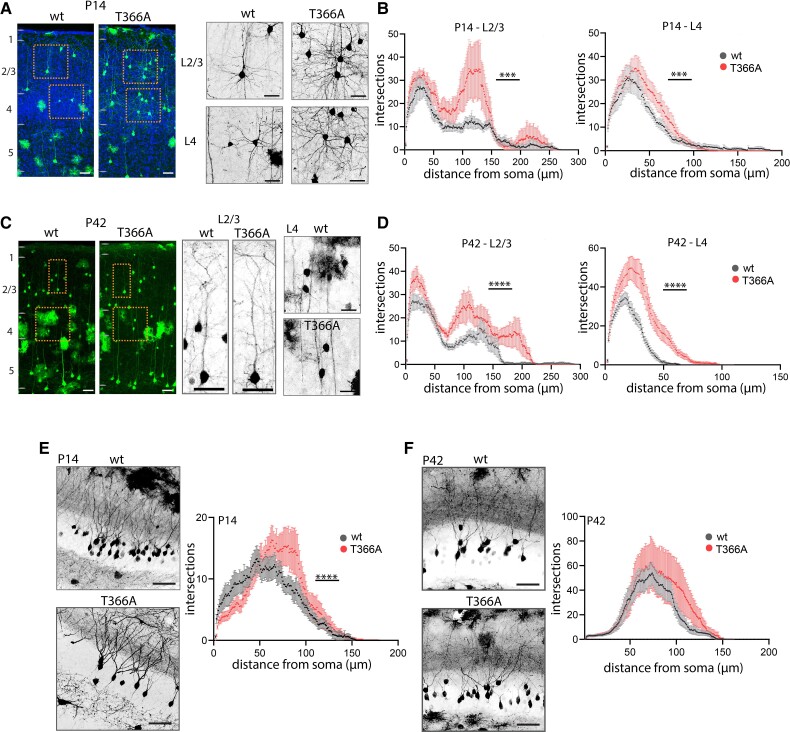
Dendritic arborization is increased in PtenT366A/T366A neurons
We next examined whether dendritic arborization of PtenT366A/T366A neurons was modified. We analysed dendritic complexity and dendritic length of pyramidal L2/3 and L4 interneurons by Sholl analysis49 (Fig. 4, Supplementary Fig. 4 and Supplementary Table 7). Analysis of dendritic arbours of pyramidal neurons in L2/3 and of interneurons in L4 in S1 cortex revealed an increase in dendritic arborization in PtenT366A/T366A brains from P8 onwards.
Figure 4.
Dendritic arborization in PtenT366A/T366A cortical and dentate gyrus neurons. (A–D) Dendritic arborization for L2/3 pyramidal neurons and L4 neurons at P14 (A and B) and P42 (C and D). Enlarged images showing L2/3 and L4 neurons, graphs showing quantification with Sholl analysis. (E and F) Dendritic arborization in hippocampal dentate gyrus neurons at P14 (E) and at P42 (F) with enlarged images, and quantification with Sholl analysis in graphs. Data shown as average ± SEM. Statistical analysis with two-way ANOVA, Bonferroni post hoc test, ****P < 0.0001. Analysis details are provided in Supplementary Table 7. Scale bars = 100 µm, 50 µm in enlarged images.
At P14, the dendritic arborization of PtenT366A/T366A L2/3 pyramidal neurons and PtenT366A/T366A L4 interneurons were significantly increased (Fig. 4A and B). In PtenT366A/T366A L2/3 neurons the extent of arborization differed from wild-type in the region of the oblique dendrites (70–150 µm from the soma) and in the region of the apical dendrites (200–250 µm from the soma). PtenT366A/T366A L4 neurons showed a significant increase in the number of proximal branches at 30–70 µm from the soma. At P42, dendritic arborization in PtenT366A/T366A neurons was significantly increased in L2/3 and L4 (Fig. 4C and D). In PtenT366A/T366A L2/3 neurons the extent of arborization differed from wild-type in the region of the oblique dendrites and in the region of the apical dendrites similarly to P14. PtenT366A/T366A L4 neurons showed a significant increase in the number of proximal branches at 10–50 µm from the soma. In contrast to soma size, the differences in dendritic arborization persisted from P8 to P42. In addition, the total dendritic lengths were significantly increased in PtenT366A/T366A L2/3 pyramidal neurons and L4 interneurons at P14. However, no changes were observed at P42 (Supplementary Fig. 4A–D).
A similar analysis of hippocampal dentate gyrus granule neurons showed the same effect as in cortical neurons (Fig. 4E and F). Dendritic arbours in PtenT366A/T366A mice were larger at both stages P14 and P42 compared to wild-type. Sholl analysis of all order of branches for hippocampal dentate gyrus granule neurons at P14 showed increased arborization of PtenT366A/T366A neurons from 50 to 100 µm distance from the soma. By comparison, wild-type dentate gyrus dendrites were normally distributed with a peak at 50 µm from soma (Fig. 4E). At P42, dendritic arbours in PtenT366A/T366A neurons showed more complexity between 100 and 140 µm from the soma (Fig. 4F). The total dendritic lengths, however, were not different in PtenT366A/T366A and wild-type dentate gyrus neurons at P14 and at P42 (Supplementary Fig. 4E and F). Taken together, these results demonstrate that morphology of cortical dendrites is affected in PtenT366A/T366A mice.
Disruption of the thalamocortical/corticocortical balance in PtenT366A/T366A mice
Earlier work using post-mortem tissue56,57 and magnetic resonance tomography connectivity mapping58,59 suggests that cortical connectivity might be affected in ASD. In humans, PTEN germline variations lead to imbalances of cortical connections.53,60 Similarly, in a Pten mouse model for autism (Pten+/–), connectivity between prefrontal cortex and amygdala is disturbed.30,61
Monosynaptic rabies virus tracing has been used in multiple studies to track presynaptic input.43,45,62 Here we used the rabies approach to test the hypothesis that in ASD/Pten mice the balance between corticocortical and thalamocortical connectivity is modified.59,63,64 To examine this in the PtenT366A/T366A mice, we combined anterograde trans-synaptic tracing with retrograde monosynaptic rabies tracing.65 We first injected an anterograde AAV1-Cre in the ventrobasal thalamus. AAV1-Cre is expressed in axon terminals of infected neurons (L5a and L1, POm thalamus; L4 and L2/3, VPm thalamus) and in post-synaptic target neurons in S1 cortex.66,67 We then used a second injection in S1 cortex to target those Cre+ neurons with a helper Flex virus (AAV1-Syn-Flex-nGToG-WPRE3) expressing a TVA receptor that can be recognized by the rabies virus.68 Finally, RABV-SADB19dG-mCherry, which targets the TVA receptor expressing neurons, was injected in S1 cortex. We analysed the expression of presynaptic neurons 10 days after the last injection in every second brain section (schematic Fig. 5A and Supplementary Fig. 5A and B also see ‘Materials and methods' section). Immunolabelling for Cre+ neurons in VPm/POm thalamus confirmed the presence of label at the ventrobasal injection site of the AAV1-Cre virus. The presence of Cre+ neurons in S1 cortex targeted by the AAV1-Syn-Flex-nGToG-WPRE3 additionally indicated that AAV1-Cre was functioning trans-synaptically from thalamocortical axons (Supplementary Fig. 5C).
At the injection site in S1 cortex, we counted 224 postsynaptic, GFP+/mCherry+ (yellow) starter neurons out of 547 GFP+ neurons or wild-type, and 421 starter neurons out of 850 GFP+ neurons for PtenT366A/T366A brains (Fig. 5B and Supplementary Table 8). These starter cells formed the cohort for which monosynaptic connections were examined. Note that total number of starter neurons was variable (Supplementary Fig. 5D). Earlier work has suggested that the total number of starter neurons could be an overestimate and depends on the conditions of the experiment.62,69 For statistical comparison, we therefore considered the percentage of input to each cortical and thalamic area in relation to total input. In total, we found 2811 presynaptic neurons in the wild-type brains and a total of 2580 presynaptic neurons in the PtenT366A/T366A brains (Fig. 5B). Example images of S1 cortex of wild-type and PtenT366A/T366A brains show specific label of GFP+/mCherry+ starter neurons in L2/3, L4, and L5 in S1 cortex (Fig. 5C).
We examined local and long-range input to S1 cortex by counting mCherry+, presynaptic neurons in S1 at the injection site and in other areas of neocortex and thalamus. We found local presynaptic neurons in S1 cortex in all layers in wild-type and PtenT366A/T366A brains. In wild-type, the local presynaptic axonal input to the starter cells constituted 72% of the total input to these cells, while in PtenT366A/T366A brains the local input accounted for 76% of total input to the starter neurons. When counting mCherry+ neurons from other cortical areas, we found that the long-range presynaptic input to S1 cortex from other cortical areas was 38% in wild-type compared to 23% in PtenT366A/T366A brains (Fig. 5D). Thus, the balance between local and long-range cortical connectivity was similar in PtenT366A/T366A and wild-type cortical neurons synaptically connected to the corticothalamic neurons.
However, when we examined the long-range input to the cortical neurons more closely, we observed a shift in the long-range connectivity, with a significant increase in thalamic inputs and a decrease in motor cortical inputs. For both wild-type and PtenT366A/T366A mice, the bulk of the long-range input arose from four main areas in the neocortex: motor, S1 contra, S2 and visual cortices (Fig. 5E). The input from visual cortex was 22% for wild-type and 11% for PtenT366A/T366A, input from S2 cortex was 5% for wild-type and 3% for PtenT366A/T366A, while the input from contralateral S1 cortex accounted for 14% in wild-type and 19% in PtenT366A/T366A. Inputs from motor cortices were significantly different for wild-type and PtenT366A/T366A brains: 32% for wild-type and 14% in PtenT366A/T366A. Also inputs from thalamic areas, 26% in wild-type and 50% in PtenT366A/T366A, were significantly different between the genotypes (Fig. 5E and F).
The significant difference in the input from thalamus to S1 cortex and from motor cortices to S1 cortex between the wild-type and PtenT366A/T366A mice suggests that the cortical connectivity of these neurons receiving input from thalamus was modified in PtenT366A/T366A brains. It further suggests that aspects of the cortical and the thalamocortical circuit organization were modified in PtenT366A/T366A mice. Note, however, that when we use rabies virus tracing45 in PtenT366A/T366A and wild-type mice without first using the trans-synaptic AAV1-Cre targeting neurons thalamus, there was no significant difference in the mixture of local and long-range inputs to S1 cortex (data not shown). These experiments suggest that the difference in cortical circuit organization in PtenT366A/T366A brains arises from modification in a few aspects of cortical and thalamocortical connectivity.
Discussion
We describe a new aspect of the phosphorylation site at T366 in the Pten gene in a newly generated knock-in mouse. PtenT366A/T366A mice are viable and reproduce according to Mendelian ratios; thus, they provide an excellent model for studying this phosphorylation site in vivo.
Previous work has shown that Pten Nse-Cre mutant mice show deficits in social activity, reaction to sensory stimuli and increased anxiety-like behaviours.16,21,70 In the Pten+/– mouse, a model for autism, social behaviour was impaired and associated with increased brain size.28,29 In PtenT366A/T366A mice, exploratory behaviour in the open field and motor skills in the hanging wire and rotarod tests were similar to that observed in wild-type littermates, suggesting that general exploratory and motor function are intact in these mice. Deficits in spatial working memory (Y-maze), spatial navigation (Barnes maze and Morris water maze), contextual fear conditioning and vibrissae-stimulated reflexes in PtenT366A/T366A mice could support the idea that sensory integration with hippocampal and entorhinal circuitries involved in spatial cognition71 is impaired in PtenT366A/T366A mice. Grooming was reduced in PtenT366A/T366A mice, which could argue for an ASD-related phenotype. It is known that ASD is characterized by a strong sexual dimorphism—male individuals are more often affected than females.72,73 This observed behavioural bias in the PtenT366A/T366A male mice is another evidence of an ASD phenotype associated with this phosphorylation site.
Pten mutations affect neuronal morphology, soma size and dendritic arborization.16,21,30,74 Our findings that soma size is transiently regulated during postnatal development from P8 to P14 in PtenT366A/T366A mice, and cell-type specific to cortical neurons in upper layers, argues in favour for the idea of the stabilizing and destabilizing effects of the phosphorylation site depending on the cell type.35,36 Because cortical proliferation and layer formation were not affected in PtenT366A/T366A mice, we argue that the increase of soma size and dendritic arborization follows an intrinsic developmental programme initiated after birth.
Finally, connectivity tracing with rabies virus showed an imbalance of long-range cortical input to S1 cortex in PtenT366A/T366A mice, which could be due to the altered morphology of L2/3 pyramidal neurons in PtenT366A/T366A mice. Neurons in S1 cortex in PtenT366A/T366A mice had a smaller number of presynaptic inputs from motor cortices. Along with this modification in cortical connectivity, the thalamocortical pathway was disrupted in PtenT366A/T366A mice, meaning that neurons in the cortex that received thalamic input were better connected in PtenT366A/T366A mice than in wild-type mice. Note two potential caveats of our method. First, efficacy of synaptic uptake could be affected when three viruses are combined46,75 and an efficacy of rabies could be synapse-specific.76 Our results suggest that the behavioural changes seen in PtenT366A/T366A mice could arise from changes in long-range connectivity, which in turn could result in alterations in sensory processing. Imbalance of cortical connections have also been described in ASD patients.59,63,64 However, to further dissect the circuit properties in PtenT366A/T366A mice that underlie PTEN T366 function, a functional analysis of dendritic activity by two-photon Ca2+ imaging or of input by optogenetic approaches could prove useful.41,77
It remains to mention that an ASD patient has been identified who shows a decrease in phosphorylation at PTEN T366,78 thus mimicking the PTEN T366A phenotype. Several human PTEN ASD mutations affect protein stability and activity.26 Testing the phosphorylation site at PTEN T366 for protein stability and activity could provide an entry point for understanding the effect of phosphorylation at PTEN T366 on soma size or neuron size in general. Further, it could help in understanding the involvement of PTEN T366 phosphorylation in the context of ASD.
In summary, our analyses of the PtenT366A/T366A mice show that phosphorylation of PTEN at T366 impacts on cortical neuronal morphology and connectivity, which may account for changes in behavioural characteristics. Altogether, our analyses of T366 contribute to the understanding of PTEN function in sensory processing with a potential link to ASD.
Supplementary Material
Acknowledgements
We thank Dr Thorsten Trimbuch and the Viral Core facility for their support and the generation of the viruses used in this study. We thank Dr Jan Schmoranzer and the AMBIO facility for their support and the use of the microscope facility. We would like to thank Zara Khan for helping with collecting data for the analysis of dendrite arborization. We thank Dr Timothy Zolnik for helpful discussion on the manuscript. Finally, we thank Kristin Lehmann, Kerstin Schlawe and Beate Dietmar for excellent technical assistance.
Abbreviations
- ASD
autism spectrum disorders
- S1
primary somatosensory cortex
Contributor Information
Julia M T Ledderose, Institute for Biochemistry, Charité Universitätsmedizin Berlin, Berlin, Germany; Department of Biology, Humboldt Universität zu Berlin, 10117 Berlin, Germany.
Jorge A Benitez, Bristol Myers Squibb, San Diego, CA 92121, USA.
Amanda J Roberts, The Scripps Research Institute, Animal Models Core, La Jolla, CA 92037, USA.
Rachel Reed, Department of Medicine, Ludwig Cancer Institute, San Diego, USA.
Willem Bintig, Institute for Biochemistry, Charité Universitätsmedizin Berlin, Berlin, Germany.
Matthew E Larkum, Department of Biology, Humboldt Universität zu Berlin, 10117 Berlin, Germany; Neurocure Center for Excellence, Charité Universitätsmedizin Berlin, Germany.
Robert N S Sachdev, Department of Biology, Humboldt Universität zu Berlin, 10117 Berlin, Germany.
Frank Furnari, Department of Medicine, Ludwig Cancer Institute, San Diego, USA; Department of Medicine, University of California San Diego, La Jolla, USA.
Britta J Eickholt, Institute for Biochemistry, Charité Universitätsmedizin Berlin, Berlin, Germany; Neurocure Center for Excellence, Charité Universitätsmedizin Berlin, Germany.
Funding
Deutsche Forschungsgemeinschaft (DFG-SFB 665 A11, 12959370) to B.J.E. Charité fellowhip to J.L. Deutsche Forschungsgemeinschaft (DFG), Grant Nos. 246731133, 250048060 and 267823436 to M.L. DFG Project number 327654276 – SFB 1315 to M.L. European Commission Horizon 2020 Research and Innovation Program and Euratom Research and Training Program 2014–2018 (under grant agreement No. 670118 to M.L.). Human Brain Project, EU Commission Grant 720270 (SGA1), 785907 (SGA2) and 945539 (SGA3) to M.L. R01NS080939 (F.F.), and the Defeat GBM Research Collaborative, a subsidiary of the National Brain Tumor Society (F.F.).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 2. Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. [DOI] [PubMed] [Google Scholar]
- 3. Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. [DOI] [PubMed] [Google Scholar]
- 4. Marsh DJ, Dahia PL, Zheng Z, et al. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet. 1997;16:333–334. [DOI] [PubMed] [Google Scholar]
- 5. Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. [DOI] [PubMed] [Google Scholar]
- 6. Butler MG, Dazouki MJ, Zhou XP, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buxbaum JD, Cai G, Chaste P, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen MM. The AKT genes and their roles in various disorders. Am J Med Genet Part A. 2013;161:2931–2937. [DOI] [PubMed] [Google Scholar]
- 9. Rodríguez-Escudero I, Oliver MD, Andrés-Pons A, Molina M, Cid VJ, Pulido R. A comprehensive functional analysis of PTEN mutations: Implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20:4132–4142. [DOI] [PubMed] [Google Scholar]
- 10. Lv JW, Cheng TL, Qiu ZL, Zhou WH. Role of the PTEN signaling pathway in autism spectrum disorder. Neurosci Bull. 2013;29:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. [DOI] [PubMed] [Google Scholar]
- 12. Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. [DOI] [PubMed] [Google Scholar]
- 13. Tilot AK, Frazier TW II, Eng C. Balancing proliferation and connectivity in PTEN-associated autism spectrum disorder. Neurotherapeutics. 2015;12:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Diepen MT, Eickholt BJ. Function of PTEN during the formation and maintenance of neuronal circuits in the brain. Dev Neurosci. 2008;30:59–64. [DOI] [PubMed] [Google Scholar]
- 15. Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amiri A, Cho W, Zhou J, et al. Pten deletion in adult hippocampal neural stem/progenitor cells causes cellular abnormalities and alters neurogenesis. J Neurosci. 2012;32:5880–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groszer M, Erickson R, Scripture-Adams DD, et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. [DOI] [PubMed] [Google Scholar]
- 18. Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. [DOI] [PubMed] [Google Scholar]
- 19. van Diepen MT, Parsons M, Downes CP, Leslie NR, Hindges R, Eickholt BJ. MyosinV controls PTEN function and neuronal cell size. Nat Cell Biol. 2009;11:1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kwon C-H, Zhu X, Zhang J, Baker SJ. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci U S A. 2003;100:12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwon CH, Luikart BW, Powell CM, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwon CH, Zhu X, Zhang J, et al. Pten regulates neuronal soma size: a mouse model of Lhermitte–Duclos disease. Nat Genet. 2001;29:404–411. [DOI] [PubMed] [Google Scholar]
- 23. Chow DK, Groszer M, Pribadi M, et al. Laminar and compartmental regulation of dendritic growth in mature cortex. Nat Neurosci. 2009;12:116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia-Junco-Clemente P, Chow DK, Tring E, Lazaro MT, Trachtenberg JT, Golshani P. Overexpression of calcium-activated potassium channels underlies cortical dysfunction in a model of PTEN-associated autism. Proc Natl Acad Sci U S A. 2013;110:18297–18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rademacher S, Eickholt BJ. PTEN in autism and neurodevelopmental disorders. Cold Spring Harb Perspect Med. 2019;9:a036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spinelli L, Black FM, Berg JN, Eickholt BJ, Leslie NR. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J Med Genet. 2015;52:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clipperton-Allen AE, Cohen OS, Aceti M, et al. Pten haploinsufficiency disrupts scaling across brain areas during development in mice. Transl Psychiatry. 2019;9:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Page DT, Kuti OJ, Prestia C, Sur M. Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proc Natl Acad Sci U S A. 2009;106:1989–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clipperton-Allen AE, Page DT. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum Mol Genet. 2014;23:3490–3505. [DOI] [PubMed] [Google Scholar]
- 30. Huang W-C, Chen Y, Page DT, et al. Hyperconnectivity of prefrontal cortex to amygdala projections in a mouse model of macrocephaly/autism syndrome. Nat Commun. 2016;7:13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balci TB, Davila J, Lewis D, et al. Broad spectrum of neuropsychiatric phenotypes associated with white matter disease in PTEN hamartoma tumor syndrome. Am J Med Genet Part B, Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet. 2018;177:101–109. [DOI] [PubMed] [Google Scholar]
- 32. Vanderver A, Tonduti D, Kahn I, et al. Characteristic brain magnetic resonance imaging pattern in patients with macrocephaly and PTEN mutations. Am J Med Genet A. 2014;164A:627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hansen-Kiss E, Beinkampen S, Adler B, et al. A retrospective chart review of the features of PTEN hamartoma tumour syndrome in children. J Med Genet. 2017;54:471–478. [DOI] [PubMed] [Google Scholar]
- 34. Busch RM, Srivastava S, Hogue O, et al. Neurobehavioral phenotype of autism spectrum disorder associated with germline heterozygous mutations in PTEN. Transl Psychiatry. 2019;9:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu D, Yao Y, Jiang X, Lu L, Dai W. Regulation of PTEN stability and activity by Plk3. J Biol Chem. 2010;285:39935–39942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Al-khouri AM, Ma Y, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3β. J Biol Chem. 2005;280:35195–35202. [DOI] [PubMed] [Google Scholar]
- 37. Tibarewal P, Zilidis G, Spinelli L, et al. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Sci Signal. 2012;5:ra18. [DOI] [PubMed] [Google Scholar]
- 38. Maccario H, Perera NM, Davidson L, Downes CP, Leslie NR. PTEN is destabilized by phosphorylation on Thr366. Biochem J. 2007;405:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okumura K, Zhao M, Depinho RA, Furnari FB, Cavenee WK. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proc Natl Acad Sci U S A. 2005;102:2703–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schrötter S, Leondaritis G, Eickholt BJ. Capillary isoelectric focusing of Akt isoforms identifies highly dynamic phosphorylation in neuronal cells and brain. J Biol Chem. 2016;291:10239–10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zolnik TA, Ledderose J, Toumazou M, et al. Layer 6b is driven by intracortical long-range projection neurons. Cell Rep. 2020;30:3492–3505.e5. [DOI] [PubMed] [Google Scholar]
- 42. Sun Y, Nguyen AQ, Nguyen JP, et al. Cell-type-specific circuit connectivity of hippocampal CA1 revealed through Cre-dependent rabies tracing. Cell Rep. 2014;7:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim EJ, Jacobs MW, Ito-Cole T, Callaway EM. Improved monosynaptic neural circuit tracing using engineered rabies virus glycoproteins. Cell Rep. 2016;15:692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wickersham IR, Lyon DC, Barnard RJO, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zingg B, Chou X-L, Zhang Z-G, et al. AAV-mediated anterograde transsynaptic tagging: Mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron. 2017;93:33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pilpel N, Landeck N, Klugmann M, Seeburg PH, Schwarz MK. Rapid, reproducible transduction of select forebrain regions by targeted recombinant virus injection into the neonatal mouse brain. J Neurosci Methods. 2009;182:55–63. [DOI] [PubMed] [Google Scholar]
- 48. Inta D, Alfonso J, von Engelhardt J, et al. Neurogenesis and widespread forebrain migration of distinct GABAergic neurons from the postnatal subventricular zone. Proc Natl Acad Sci U S A. 2008;105:20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uylings HBM, van Pelt J. Measures for quantifying dendritic arborizations. Network. 2002;13:397–414. [PubMed] [Google Scholar]
- 50. Paxinos G, Franklin K. Paxinos and Franklin's the mouse brain in stereotaxic coordinates. 5th Edition. Elsevier; 2019. [Google Scholar]
- 51. Tilot AK, Gaugler MK, Yu Q, et al. Germline disruption of Pten localization causes enhanced sex-dependent social motivation and increased glial production. Hum Mol Genet. 2014;23:3212–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hobert JA, Embacher R, Mester JL, Frazier TW 2nd, Eng C. Biochemical screening and PTEN mutation analysis in individuals with autism spectrum disorders and macrocephaly. Eur J Hum Genet. 2014;22:273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Frazier TW. Autism spectrum disorder associated with germline heterozygous PTEN mutations. Cold Spring Harb Perspect Med. 2019;9:a037002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. [DOI] [PubMed] [Google Scholar]
- 55. Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. [DOI] [PubMed] [Google Scholar]
- 56. Wegiel J, Flory M, Kuchna I, et al. Stereological study of the neuronal number and volume of 38 brain subdivisions of subjects diagnosed with autism reveals significant alterations restricted to the striatum, amygdala and cerebellum. Acta Neuropathol Commun. 2014;2:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ecker C, Ronan L, Feng Y, et al. Intrinsic gray-matter connectivity of the brain in adults with autism spectrum disorder. Proc Natl Acad Sci. 2013;110:13222–13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Acevedo B, Aron E, Pospos S, Jessen D. The functional highly sensitive brain: A review of the brain circuits underlying sensory processing sensitivity and seemingly related disorders. Philos Trans R Soc Lond B Biol Sci. 2018;373:20170161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tomasi D, Volkow ND. Reduced local and increased long-range functional connectivity of the thalamus in autism spectrum disorder. Cereb Cortex. 2019;29:573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mighell TL, Thacker S, Fombonne E, Eng C, O’Roak BJ. An integrated deep-mutational-scanning approach provides clinical insights on PTEN genotype–phenotype relationships. Am J Hum Genet. 2020;106:818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barrows CM, McCabe MP, Chen H, Swann JW, Weston MC. PTEN loss increases the connectivity of fast synaptic motifs and functional connectivity in a developing hippocampal network. J Neurosci. 2017;37:8595–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hafner G, Witte M, Guy J, et al. Mapping brain-wide afferent inputs of parvalbumin-expressing GABAergic neurons in barrel cortex reveals local and long-range circuit motifs. Cell Rep. 2019;28:3450–3461.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nair A, Treiber JM, Shukla DK, Shih P, Müller R-A. Impaired thalamocortical connectivity in autism spectrum disorder: A study of functional and anatomical connectivity. Brain. 2013;136(Pt 6):1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Woodward ND, Giraldo-Chica M, Rogers B, Cascio CJ. Thalamocortical dysconnectivity in autism spectrum disorder: An analysis of the Autism Brain Imaging Data Exchange. Biol Psychiatry Cogn Neurosci Neuroimaging. 2017;2:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zingg B, Dong H-W, Tao HW, Zhang LI. Input–output organization of the mouse claustrum. J Comp Neurol. 2018;526:2428–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wimmer VC, Bruno RM, de Kock CPJ, Kuner T, Sakmann B. Dimensions of a projection column and architecture of VPM and POm axons in rat vibrissal cortex. Cereb Cortex. 2010;20:2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Petersen CCH. The functional organization of the barrel cortex. Neuron. 2007;56:339–355. [DOI] [PubMed] [Google Scholar]
- 68. Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim EJ, Juavinett AL, Kyubwa EM, Jacobs MW, Callaway EM. Three types of cortical layer 5 neurons that differ in brain-wide connectivity and function. Neuron. 2015;88:1253–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ogawa S, Kwon C-H, Zhou J, Koovakkattu D, Parada LF, Sinton CM. A seizure-prone phenotype is associated with altered free-running rhythm in Pten mutant mice. Brain Res. 2007;1168:112–123. [DOI] [PubMed] [Google Scholar]
- 71. Vorhees CV, Williams MT. Assessing spatial learning and memory in rodents. ILAR J. 2014;55:310–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ferri SL, Abel T, Brodkin ES. Sex differences in autism spectrum disorder: A review. Curr Psychiatry Rep. 2018;20:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang Y, Li N, Li C, et al. Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Transl Psychiatry. 2020;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Holtmaat A, Bonhoeffer T, Chow DK, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zingg B, Dong H-W, Tao HW, Zhang LI. Application of AAV1 for anterograde transsynaptic circuit mapping and input-dependent neuronal cataloging. Curr Protoc. 2022;2(1):e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rogers A, Beier KT. Can transsynaptic viral strategies be used to reveal functional aspects of neural circuitry? J Neurosci Methods. 2021;348:109005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ebner C, Ledderose J, Zolnik TA, et al. Optically induced calcium-dependent gene activation and labeling of active neurons using CaMPARI and Cal-Light. Front Synaptic Neurosci. 2019;11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wong CW, Or PMY, Wang Y, et al. Identification of a PTEN mutation with reduced protein stability, phosphatase activity, and nuclear localization in Hong Kong patients with autistic features, neurodevelopmental delays, and macrocephaly. Autism Res. 2018;11:1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.