Abstract
Purpose:
On the basis of preclinical evidence of epigenetic contribution to sensitivity and resistance to immune checkpoint inhibitors (ICI), we hypothesized that guadecitabine (hypomethylating agent) and atezolizumab [anti–programmed cell death ligand 1 (PD-L1)] together would potentiate a clinical response in patients with metastatic urothelial carcinoma (UC) unresponsive to initial immune checkpoint blockade therapy.
Patients and Methods:
We designed a single arm phase II study (NCT03179943) with a safety run-in to identify the recommended phase II dose of the combination therapy of guadecitabine and atezolizumab. Patients with recurrent/advanced UC who had previously progressed on ICI therapy with programmed cell death protein 1 or PD-L1 targeting agents were eligible. Preplanned correlative analysis was performed to characterize peripheral immune dynamics and global DNA methylation, transcriptome, and immune infiltration dynamics of patient tumors.
Results:
Safety run-in enrolled 6 patients and phase II enrolled 15 patients before the trial was closed for futility. No dose-limiting toxicity was observed. Four patients, with best response of stable disease (SD), exhibited extended tumor control (8–11 months) and survival (>14 months). Correlative analysis revealed lack of DNA demethylation in tumors after 2 cycles of treatment. Increased peripheral immune activation and immune infiltration in tumors after treatment correlated with progression-free survival and SD. Furthermore, high IL6 and IL8 levels in the patients’ plasma was associated with short survival.
Conclusions:
No RECIST responses were observed after combination therapy in this trial. Although we could not detect the anticipated tumor-intrinsic effects of guadecitabine, the addition of hypomethylating agent to ICI therapy induced immune activation in a few patients, which associated with longer patient survival.
Translational Relevance.
We designed a phase II clinical trial testing the combination of guadecitabine (DNA methyltransferase inhibitor) and atezolizumab (anti–programmed cell death ligand 1) with the goal to restimulate the immune-mediated anticancer effects of immune checkpoint inhibitors (ICI) in patients with metastatic urothelial carcinoma who were resistant to prior ICI treatment. No RECIST criteria responses were observed, however, patients could be separated into those with either progressive or stable disease outcomes. Analysis of the tumors did not show substantial DNA hypomethylation in the tumor or the upregulation of viral mimicry after two cycles of combination treatment. We did, however, observe that the therapy associated with markers of immune activation in circulating immune cells and abundance of tumor-infiltrating CD8 T cells within the tumor, which was correlated with a longer survival outcome. Our results suggest that further studies to target the reprogramming of T cells with alternate methodologies may be of benefit in this disease setting.
Introduction
Urothelial carcinoma (UC) of the bladder is the 6th most common cancer (4th most common in men) with an estimated 80,000 cases diagnosed annually, and close to 17,000 deaths per year attributed to the disease. The overall survival (OS) for patients with advanced or metastatic bladder cancer is just 15 months using standard first-line therapies (1–4). Once metastatic, UC is considered incurable and treatments are aimed at controlling the disease while optimizing quality of life.
Immunotherapy agents designed to block immune checkpoints, such as the programmed cell death protein 1 (PD-1)/programmed cell death ligand 1 (PD-L1) axis, lead to robust antitumor responses in multiple solid tumors (5). Clinical trials of atezolizumab in metastatic UC (3, 6) showed a response rate of 15% to 24%, and the drug is FDA-approved for metastatic UC for patients who are ineligible for cisplatin therapy (7, 8). Despite the excitement over the efficacy and acceptable toxicity of checkpoint inhibitors in advanced UC, the majority of treated patients show primary resistance to the drug and, critically, most responders eventually lose their response through unclear mechanisms (9–11), thus atezolizumab has recently been withdrawn from UC clinical trials. Sensitivity to immune checkpoint inhibitor (ICI) therapy depends in part on the genetic and epigenetic makeup of the cancer cells (12–15) and the CD8 T-lymphocytes that mediate immune responses (16, 17). Preclinical data suggest that epigenetic reprogramming of CD8 T-lymphocytes can sensitize to and/or reverse resistance to ICI (PD-1/PD-L1) therapy (16). Extensive work has shown that epigenetic modifiers can dramatically affect the tumor immune microenvironment and promote an “inflamed phenotype” in tumors that can synergize with immunotherapies (18, 19). In particular, DNA methyltransferase inhibitors (DNMTis) can upregulate the cytosolic double-strand RNA (dsRNA)-sensing viral defense pathway in cancer cells (18, 19), termed “viral mimicry”, through the transcriptional reactivation of transposable elements (TE). Viral mimicry upregulates immunostimulatory cytokines, antigen-presentation pathways, and interferon responses in tumors (19–22), which consequently attracts T and natural killer (NK) cells to the tumor environment and contributes to the activation of CD8 T cells (14, 15). Indeed, DNMTi has been shown to upregulate interferon genes in T24 bladder cancer cell line, suggesting potential benefit of DNMTi therapy in bladder cancer (23, 24). Furthermore, high expression of viral defense genes correlates with improved clinical outcome with immuno-oncology treatment (18).
UC provides an opportunity to study mechanisms of resistance and resensitization to ICI therapy because of the large population of patients who have primary or secondary resistance to this approach (ICI-resistant; ref. 7). Here, we report the results of a phase II clinical trial of guadecitabine (a second-generation DNMTi) in combination with atezolizumab (anti–PD-L1) in patients with advanced UC with progression during or after initial treatment with a checkpoint inhibitor. This single arm phase II study included a safety run-in to identify the recommended phase II dose of guadecitabine in combination with atezolizumab. Patients with recurrent/advanced metastatic UC (stage IV) who had previously progressed on ICI therapy with PD-1 or PD-L1 targeting agents were eligible for this study. The primary endpoint was overall response rate (ORR).
Patients and Methods
Trial design and patient eligibility
The investigator-initiated clinical trial enrolled patients with metastatic UC who had experienced primary or secondary progression on a checkpoint inhibitor prior to study entry, and were either not suitable for or had already received platinum-based chemotherapy. There was no other restriction placed on the number of prior therapies. Patients with severe immune-mediated adverse events or active autoimmune disease were excluded for safety concerns. Eastern Cooperative Oncology Group performance status 0–2 was required. The enrolling sites were Fox Chase Cancer Center, Johns Hopkins University, and University of Southern California. All participating institutions followed protocols approved by the Institutional Review Board and written informed consent was collected from each patient enrolled in the study. All studies were conducted in accordance with Good Clinical Practice and the Belmont Report; more information can be found on clinicaltrials.gov (NCT03179943).
The study was designed with a safety run-in phase during which 6 patients were treated at the target doses of guadecitabine 45 mg/m2 daily days 1 to 5 concurrent with atezolizumab 1,200 mg on days 1 and 22 of a 6-week cycle. A total of four cycles of combination therapy were planned, after which single-agent atezolizumab could be continued as standard of care at patient/physician discretion. Imaging for RECIST response assessment was performed after two treatment cycles, which equated to every 12 weeks. If safe, the study would expand to a total accrual of 43 patients (inclusive of the first six).
The primary endpoint of the trial was ORR employing the Simon's optimal two-stage design with a null hypothesis of 0.10 and an alternative hypothesis of 0.25. This design has 4.8% type I error (one-sided) and 80% power. Secondary endpoints were progression-free survival (PFS) and OS. A formal assessment of safety and efficacy was designed to occur after the 18th patient (Simon's Stage I) completed study treatment. Clinical trial will be stopped for futility if two or fewer objective responses are observed among the first 18 evaluable patients in Simon's Stage I. However, to avoid disruption in recruitment, after the 18 Simon's Stage I patients have been treated, Simon's Stage II patients may be enrolled while the remaining Stage I patients are followed for response.
DNA whole-exome sequencing
Whole-exome sequencing (WES) of cancerous formalin-fixed, paraffin-embedded (FFPE) tissue obtained pre- and posttreatment (prior to Cycle 3 Day 1, or 12 weeks after start of treatment) by on-study core biopsy of a metastatic or primary site was performed on subjects of the clinical trial. Matched normal tissue was not available for these patients. A subset of patients had samples collected both pre- and posttreatment. VAI Genomics core extracted DNA from tumor samples using AllPrep DNA/RNA FFPE kit (Qiagen) and generated WES libraries with Sure Select Human All Exon v6 (Agilent) and the Sure Select XT2 library prep kits (Agilent). FASTQ reads were preprocessed using Trimmomatic (25) and aligned to hg38 using the Burrows–Wheeler Aligner (26). Somatic single-nucleotide variant and indel calling was performed using Mutect2 from GATK (27) in Tumor-Only mode. A somatic panel of normal tissue and Genome Aggregation Database (ref. 28; gnomAD, https://gnomad.broadinstitute.org/) germline resource was downloaded from the gatk-best-practices google cloud server. Mutations were filtered on the basis of the variants present in these two files to remove known genetic variants present across different human populations. VCF files produced by Mutect2 were annotated using ANNOVAR (29). Only exonic variants were kept for downstream analysis.
Remaining exonic variants were filtered on the basis of a minimum read-depth threshold of 10. Only variants with a variant-allele frequency of 0.1 < x < 0.9 were included in the final dataset. Tumor mutation burden was calculated as the total number of nonsynonymous single nucleotide variants and indels identified by Mutect2. Mutations listed in Supplementary Fig. S2 are identified in genes listed by the Catalogue of Somatic Mutations in Cancer (COSMIC; ref. 30) oncogene database. Moreover, we only included missense mutations that were documented in the Database of Curated Mutations database (31). This additional curation was performed to maximize the removal of artifacts.
DNA methylation analysis
DNA methylation was evaluated using the Illumina Human MethylationEPIC (EPIC) array (Illumina, CA). The EPIC platform analyzes the DNA methylation status of up to 863,904 CpG loci and 2,932 non-CpG cytosines, spanning gene-associated CpGs as well as a large number of enhancer/regulatory CpGs in intergenic regions (32). Briefly, DNA was quantified by Qubit fluorimetry (Life Technologies) and 500 ng of DNA from each sample was bisulfite-converted using the Zymo EZ DNA Methylation Kit (Zymo Research, Irvine, CA) following the manufacturer's protocol using the specified modifications for the Illumina Infinium Methylation Assay.
The signal intensities corresponding to methylated (M) and unmethylated (U) alleles were extracted from the IDAT files by the readIDATpair function in the R package SeSAMe (https://github.com/zwdzwd/sesame). A detection P value for each probe was calculated using pOOBAH (P value with Out-Of-Band probes for Array Hybridization), which is based on the empirical cumulative distribution function of the out-of-band signal from all Type I probes (33). The signal intensities were further processed with background correction and dye-bias correction. The background correction is based on the noob method (34). The dye-bias was corrected using a nonlinear quantile interpolation-based method using the dyeBiasCorrTypeINorm function (33). β values, defined as SM /(SM+SU) for each locus where SM and SU represent signal intensities for methylated and unmethylated alleles, were computed using the getBetas function. β values range from zero to one, with scores of zero indicating no DNA methylation and scores of one indicating complete DNA methylation. Probes with a detection P value greater than 0.05 in a given sample were masked as not available (NA). Additional experiment-independent masking of probes subject to cross-hybridization and genetic polymorphism (N = 105,454) was implemented according to the probe manifest (release 20180909) downloaded from http://zwdzwd.github.io/InfiniumAnnotation. Further information on the EPIC array, including detailed annotation of transcription association for each probe, was obtained from the same source. For copy-number variation (CNV) detection, we used “cnSegmentation” and “visualizeSegments” functions from the SeSAMe tool. We performed cell deconvolution with Epigenetic Dissection of Intra-Sample-Heterogeneity [EpiDISH (35, 36)] through the use of their EpiDISH web server (37) with bladder tissue as reference.
Peripheral blood mononuclear cell flow cytometry and plasma cytokine analysis
Blood was drawn from patients into two BD Vacutainer Sodium Citrate Mononuclear Cell Preparation Tubes (CPT), which were centrifuged onsite within 2 hours at 1,500 g × 20 minutes to isolate peripheral blood mononuclear cells (PBMC) and plasma. Plasma was centrifuged at 4,500 rpm × 20 minutes to remove residual cells and frozen at −70°C, and PBMC were washed 3 times in PBS + 10% FBS by centrifuging at 400 g × 10 minutes prior to freezing at −70°C in FBS containing 10% DMSO at 6 to 15 million cells/mL. Samples were stored until analysis in liquid nitrogen. Batches of cryopreserved PBMC were thawed in a 37°C water bath, transferred to warm RPMI1640 (without biotin or phenol red) + 10% FBS medium and centrifuged at 400 g × 10 minutes and 5×1 million cell aliquots were stained for 20 minutes on ice with up to 14 fluorophore-conjugated antibodies or viability dye as listed in Supplementary Table S5, washed twice with ice-cold wash buffer (HBSS + 10% FBS + 0.1% Na azide), with the final wash buffer containing propidium iodide viability dye (100 ng/mL). “Fix and Perm” tubes in Supplementary Table S5 were first treated with buffers from the eBiosciences FoxP3/transcription factor fixation/permeabilization buffers for 30 minutes on ice according to the manufacturer's instructions prior to staining with antibodies. Stained PBMC were then analyzed on a BD FACSAria II flow cytometer with four lasers (365 nm, 405 nm, 488 nm, and 633 nm). Between 100,000 and 400,000 events were acquired from each sample. The flow cytometer was calibrated daily with fluorescent BD CompBeads and compensation and PMT voltages were optimized at the beginning of the study and kept consistent for all samples. Data were collected with BD FACS Diva software (v6) and analyzed with FlowJo (BD Life Sciences; v10.3 or later), Microsoft Excel (2016), Prism (GraphPad Software, San Diego), and Matlab R2016b (The Mathworks). Gating for positive staining was determined in comparison to control tubes lacking these stains. Biomarker expression level was quantified as geometric mean fluorescence intensity (GMFI).
Ultrahigh sensitivity quantification of TNFα, IL8, and IL6 levels in plasma samples was performed using Simoa Service by Myriad RBM (Austin, TX).
Cox proportional hazards regression analysis (R v 4.2.1, survival package, https://cran.r-project.org/web/packages/survival/index.html) was used to screen individual markers for correlations between their pretreatment or mid-cycle measurements, to PFS. The percent change in biomarkers between these two timepoints was also individually correlated to PFS via Cox models. No PFS events occurred prior to the mid-cycle timepoint. Differential expression of markers across distinct timepoints, as well as differential expression of markers between progressive disease (PD) and stable disease (SD) were assessed using cumulative-probit mixed-effects ordinal regressions (R ordinal package, https://cran.r-project.org/web/packages/ordinal/index.html) fit with a Laplace approximation. Fixed effects included: timepoint (pretreatment, mid-cycle, and end of the first treatment), status (PD or SD), and their interaction timepoint x status. A random intercept for each patient was included to account for repeated measures. Timepoint differences were averaged over mean PD and SD effects to estimate changes irrespective of status. For all statistical comparisons, second-generation P values (SGPV) were used to control the false positive rate and determine if there was significant evidence of differences greater than ± 5% in regression estimates (38).
IHC
UC tumor needle biopsies were fixed uniformly in 10% NBF and histology processed and sectioned with hematoxylin and eosin (H&E) staining by the histology department in Pathology Biorepository Core at VAI. H&E review by study pathologist assessed biopsy adequacy, tumor content, and extent of stroma/residual lymph node. Study biopsies were cut at 5-μm thickness onto standard charged slides. Tissue adequacy was assessed by H&E staining and minimum criteria of >25 tumor cells required for analyses. IHC data analysis included PD-L1 clone 22C3, Dako, 1:50 (RRID:AB_2833074); CD8 clone 4B11, Bio-Rad, 1:35 (RRID:AB_322868); MDA5 clone 33H12L34, ThermoFisher Scientific, 1:1,600 (RRID: AB_2532316); and CCL5 AF278, R&D Systems, 1:40 (RRID:AB_354440). Automated immunostaining was performed by Autostainer Link 48, Dako, Inc. Heat-induced epitope retrieval (HIER) at low pH for PD-L1 and CCL5 and HIER at high pH for CD8 and MDA5. Singly stained slides were scanned at 20× by digital imaging Aperio scanner (Leica, Inc.) and scored using modified H-Score method comprised two components of stain intensity (0–3) multiplied by stain prevalence (0–6) with total H-score of range 0–18. Also, PD-L1, MDA5, and CCL5 were scored by tumor and stroma compartment separately. CD8 has a relatively uniform intensity score and received a stromal score only. Intensity score was modified 1 to 3 to capture the pattern of CD8 infiltration: lack of directionality tumor or stroma (1), aggregating at tumor edge (2), and actively infiltrating tumor (3). Clinical trial samples were reviewed and scored by the pathologist in a double-blind manner and were analyzed at separate institutions. Independent pathology review of separate tumor set of same IHC single-stained markers and this modified H-Score method showed > 90% concordance.
IHC data is presented two ways, one with jittered data at both pretreatment and posttreatment timepoints colored by outcome with means ± Gini mean difference (a nonparametric measure of dispersion/spread) calculated using R 4.2.1 Hmisc package (https://cran.r-project.org/package=Hmisc) and the second where we connect the individuals who had both timepoint measures. At pretreatment and posttreatment timepoints, PD and SD tumor samples were tested one of two ways: (i) outcomes with fewer than 6 possible values (i.e., scores ranged from 0–3) were analyzed using cumulative-probit mixed-effects ordinal regression with a Laplace approximation and random intercepts for each patient; this random intercept was removed from the model in outcomes where the random effect had estimated variance equal to 0, and (ii) all other metrics were analyzed via natural log transformed robust linear mixed-effects models with an unstructured covariance structure (39, 40). All models included fixed effects: timepoint (pretreatment, mid-cycle, and end of the first treatment), status (PD or SD), and their interaction timepoint x status. Normality assumptions for the linear mixed-effects models were assessed using qq-plots; no concerning deviations were noted. For all statistical comparisons, SGPVs were used to determine if there was significant evidence of a difference where the null interval was set at ± 5% difference in regression estimates.
RNA sequencing and analysis
Total RNA was extracted by the VAI Genomics core using RNeasy kit (Qiagen) followed by on-column DNase I digestion according to the manufacturer's protocol. Sequencing libraries were prepared using the KAPA RNA HyperPrep Kit with RiboErase (HMR; Roche) and sequenced on an Illumina NovaSeq 6000 as 51 bp paired-end (PE) reads. FASTQ reads from total RNA sequencing (RNA-seq) libraries were trimmed and aligned to the hg38 genome using HISAT2 (41). Then the aligned reads were processed with StringTie (42) to quantify the gene expression using GENCODE v37 annotation. Gene count files were generated using StringTie prepDE3.py script, and then further processed with DESeq2 (43) to identify differentially expressed genes (counts ≥ 10, fold-change > 1, adj. P value < 0.05). Next, we identified gene set enrichment analysis (GSEA) Hallmark pathway enrichment of differentially expressed genes using the fgsea package in R (44). To quantify intergenic TE expression, we used featureCounts (45) to identify how many non-duplicated reads overlap TE copies annotated by TEtranscripts (46). Multimapped reads were assigned to their primary location by specifying “-primary” in featureCounts. To minimize potential biases from duplicate read percentage or library size, we normalized each sample by dividing each TE read count by the total number of non-duplicated uniquely and multi-mapped reads to generate Counts Per Million (CPM) values. Next, we performed filtering of > 1 CPM to identify adequately expressed or induced TEs across DAC treatment timepoints. Immune deconvolution using quanTIseq was performed following the author's instructions (47). In R, statistical significance for immune fraction differences was calculated by using mixed-effects beta regression (glmmTMB package; ref. 48) followed by Likelihood Ratio Test with unstructured covariance. P values were corrected for multiple comparisons with Benjamini–Hochberg method.
Data availability
Sequencing data from tumor RNA-seq, WES, and DNA methylome analysis is available on Gene Expression Omnibus repository (GSE222934). H-scores from IHC analysis and data collected from flow cytometry analysis are available in the Supplementary Tables.
Results
Patient characteristics, enrollment, and outcome
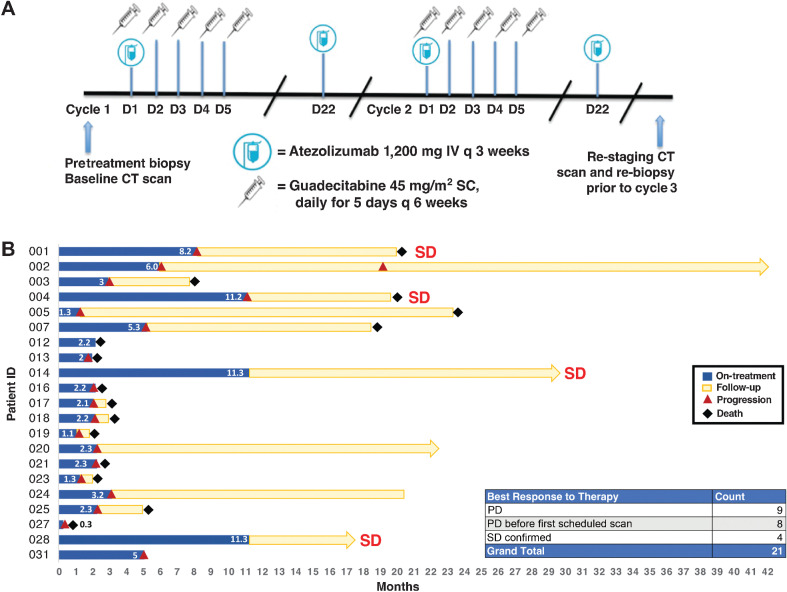
The study enrolled 6 patients to the safety run-in. Prespecified safety analysis determined the target dose combination to be safe. The study enrolled a total of 21 patients (Supplementary Table S1) to receive a combination of guadecitabine (45 mg/m2 subcutaneously, first 5 days of each cycle) and atezolizumab (1,200 mg intravenously, at Day 1 and Day 22) in a 6-week treatment cycle (Fig. 1A). No dose-limiting toxicities were observed. The trial was closed for futility given that no RECIST responses were seen. Median OS was 8.6 months [95% confidence interval (CI), 3.4–21.5 months) and median PFS was 3.0 months (95% CI, 1.9–5.5 months). Eight patients experienced clinical progression before their first scheduled scan. One patient died of cardiopulmonary failure during cycle 1 possibly related to treatment. Fourteen additional patients experienced grade 3–4 adverse events as defined by Common Terminology Criteria for Adverse Events (CTCAE), predominantly cytopenias related to guadecitabine, which have been commonly observed in past hypomethylating agent therapies. For the purposes of correlative analysis, tumors were categorized as PD (n = 17) or SD (n = 4) based on the best RECIST response (Fig. 1B). We did not detect any observable association between previous immunotherapy treatment nor past best RECIST response and the clinical responses documented in this combination treatment trial (Table 1). To elucidate the probable reasons for the lack of clinical response and identify potential prognostic biomarkers that associate with longer patient survival and disease stability, we performed comprehensive preplanned correlative analysis of various molecular assays generated from peripheral blood samples or tumor samples harvested by needle biopsies from all patients who passed initial screening (Table 2; Supplementary Fig. S1).
Figure 1.
Clinical trial overview and patient survival. A, Schematic describing the guadecitabine and atezolizumab dosing and timing strategy for this clinical trial. Each treatment cycle was 6 weeks. B, Swimmer plot of patients with UC treated with combination therapy in this trial. The length of treatment duration and patient follow-up durations are depicted.
Table 1.
Previous immunotherapy and best RECIST response documented of the patients recruited into this clinical trial.
| Previous IO therapy and best RECIST response documented | ||
|---|---|---|
| Best RECIST response (this clinical trial): | SD (N = 4) | PD (N = 17) |
| Pembrolizumab | Durvalumab+Tremelimumab | |
| PD (2 patients) | SD (1 patient) | |
| Pembrolizumab+Epacadostat | Pembrolizumab+Epacadostat | |
| Partial response (1 patient) | Partial response (1 patient) | |
| Atezolizumab | Pembrolizumab | |
| SD (1 patient) | Partial response (4 patients) | |
| SD (2 patients) | ||
| PD (6 patients) | ||
| Atezolizumab | ||
| SD (1 patient) | ||
| PD (2 patients) | ||
Table 2.
Metastatic location and tumor biopsy site.
| Best RECIST response (this clinical trial): | Metastatic site | # of patients | Tumor biopsy site | # of patients |
|---|---|---|---|---|
| PD | Lymph node | 4 | Lymph node | 3 |
| Lung | 5 | Lung | 4 | |
| Liver | 3 | Liver | 4 | |
| Multiple locations | 5 | Primary | 4 | |
| Other | 2 | |||
| SD confirmed | Lymph node | 4 | Lymph node | 2 |
| Primary | 2 |
Viral mimicry activation is not associated with clinical response to combination therapy
To evaluate the effect of guadecitabine in reducing DNA methylation in tumors, we extracted genomic DNA from available bulk tumor samples and measured the DNA methylation levels of CpGs with the Illumina Human MethylationEPIC array. We generated DNA methylome libraries of tumors from pretreatment and posttreatment [Cycle 3 Day 1 (C3D1) or End of Treatment, which were collected 12 weeks after start of treatment] timepoints. Due to aggressive nature of metastatic UC, only 7 posttreatment samples were available for DNA methylation profiling. Therefore, to evaluate decitabine's impact on tumor DNA methylomes, we focused the analysis on the global DNA methylation change in 7 tumor pairs with patient-matched timepoints, which ranged from −5% to +5% mean (−11% to +10% median) DNA methylation change (Supplementary Fig. S2A and S2B). Two of the four PD tumors showed a gain (4.3%–4.8%) of average DNA methylation after therapy, while all three SD tumors showed modest losses (0.7%–3.6%) of DNA methylation. Considering that tumor heterogeneity and cellularity can collectively contribute to changes in global DNA methylation levels (49), we queried whether the global methylation changes in tumors after treatment could be attributed to tumor progression or differences in the extent of immune infiltration in bulk tumors. This can be detected through leukocyte-specific DNA methylation patterns (50), as estimated by SeSAMe (33), and is inversely related to tumor purity (51). First, we identified the top 1,000 bladder tumor–specific hypermethylated CpG probes from The Cancer Genome Atlas (TCGA) BLCA methylomes (Supplementary Fig. S2C), which is representative of tumor methylation levels, and then performed CNV detection (Supplementary Fig. S3) and cell type deconvolution from the bulk tumor DNA methylomes. In the two bladder tumors (patients 3 and 7) that showed increases in global DNA methylation levels after treatment, we also observed dilution of DNA methylation signals in BLCA-hypermethylated probes, and accordingly decrease in epithelial cell fractions (bladder cancer cells often originate from the urothelium or transitional epithelium) and increases in the immune cell fractions estimated by EpiDISH (Supplementary Fig. S1C). This suggests the apparent increase in DNA methylation is likely due to a shift in cell populations, as normal cells have a lower degree of global hypomethylation than tumor cells (52). Consistent with this, tumors with global losses of DNA methylation after treatment showed increases in CNVs and a gain of epithelial cell fractions (proxy for tumor purity). Therefore, we speculate that the global differences in DNA methylation between tumors at pretreatment and posttreatment timepoints are reflecting tumor cellularity rather than the direct result of guadecitabine treatment.
Next, we asked whether the combination therapy was sufficient to trigger the induction of TE expression and viral mimicry activation in the bladder tumors. For gene and TE expression analysis, we generated total RNA-seq from available tumors, of which we had seven tumor pairs across pre- and posttreatment timepoints. We focused on transcripts derived from intergenic long terminal repeats (LTR) or long interspersed nuclear elements (LINE), as these have been canonically associated with viral mimicry activation (14, 18, 19), and quantified TE expression across all available tumor samples. We then calculated TE expression fold changes across treatment timepoints (Methods; Supplementary Fig. S4A). We detected no appreciable difference in TE abundance between PD or SD tumors at either treatment timepoints. Furthermore, we observed no correlation between global DNA methylation loss and TE expression fold-change induced by combination therapy and most tumors showed down-regulation of viral mimicry-related genes after three treatment cycles (Supplementary Fig. S4B and S4C).
Genetic and transcriptional signatures associated with tumor progression
Considering the lack of association between viral mimicry activation and clinical outcome, we were motivated to explore additional molecular features or biological mechanisms that could serve as potential biomarkers. First, we queried the mutation profiles of the tumors by performing WES to identify if certain mutation signatures might confer resistance or sensitivity to the combination therapy. We observed no statistically significant correlation between tumor mutation burden and clinical response (using Welch t-test) or PFS (using Cox proportional hazard; Supplementary Fig. S5; Supplementary Tables S2 and S3). Because of the lack of patient-matched controls, we prioritized analysis of known cancer-associated mutations identified by TCGA (53, 54) and COSMIC (ref. 30; Supplementary Table S4). Mutations in TP53 were most frequently found in ICI-resistant UC tumors followed by mutations in critical epigenetic regulators, such as ARID1A, KMT2C, and KMT2D (Supplementary Fig. S5). Although limited by low sample size, we identified recurrent mutated genes that were uniquely detected in SD tumors (RNF213, STAG1, ASXL1, PABPC1) and PD tumors (ARID1A, FGFR3, ATP2B3, WT1).
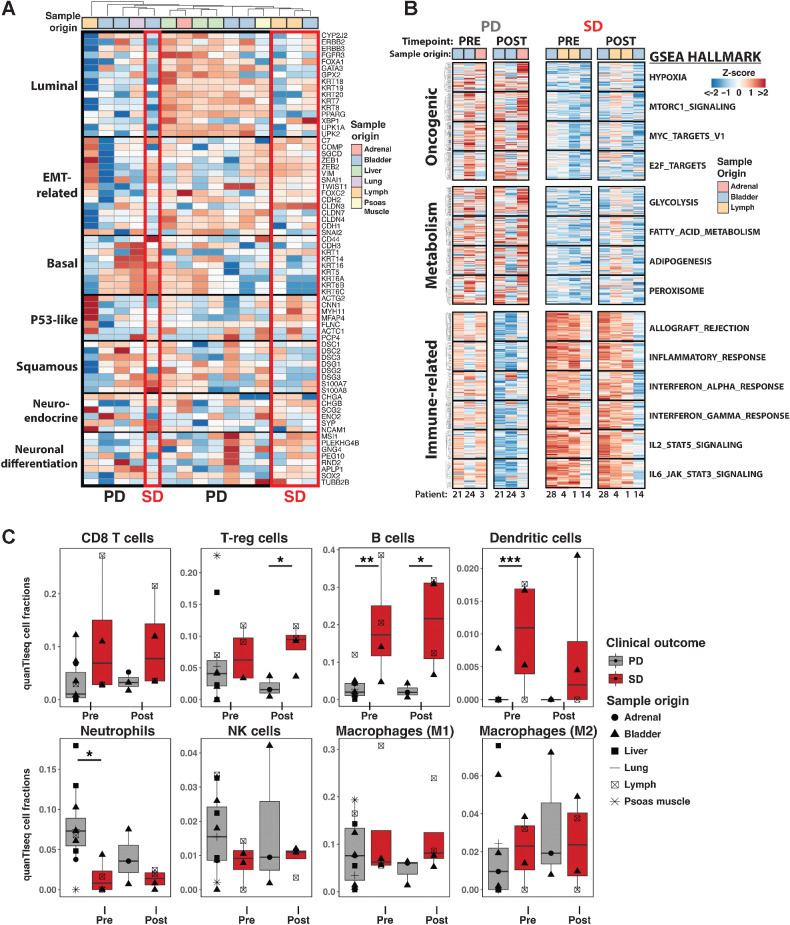
In addition to genetic-based molecular subtyping, UC tumors can be further categorized into distinct transcriptomic subtypes with variable clinical prognosis and clinically actionable targets (54–56). To investigate if particular UC tumor subtypes were enriched in PD versus SD tumors, we obtained a published list of marker genes that denote particular subsets of UC tumors (57). Using gene expression profiles from tumors before enrollment, we identified that luminal-subtype genes were enriched in 7 of 11 PD tumors while the other PD subset expressed basal-subtype and squamous-subtype genes (Fig. 2A). The majority of the SD tumors expressed EMT-related and Neuroendocrine/Neuronal differentiation signatures. Next, we performed pairwise comparisons of GSEA (44, 58) on PD tumors and SD tumors to identify particular gene pathways that might serve as prognostic markers for treatment efficacy (Supplementary Fig. S6A). Regardless of treatment timepoint, when compared with SD tumors, PD tumors had preferential expression of oncogenic pathways, such as “Hypoxia”, “Myc_targets_v1”, and “Mtorc1_signaling”, and metabolism-related pathways, such as “Glycolysis”, “Fatty_acid_metabolism”, and “Adipogenesis”, but lower expression of inflammatory gene pathways, including IL6, IL2, and interferon responses (Fig. 2B; Supplementary Fig. S6B). Interestingly, comparison of pretreatment and posttreatment PD or SD tumors revealed shared loss of interferon responses, suggesting that cancer progression despite combination therapy is associated with immunosuppression (Supplementary Fig. S6A). However, the expression of interferon response, IL6, and IL2 pathway genes were markedly reduced between pre- and posttreatment in PD tumors, whereas the higher levels of these were essentially maintained in SD tumors (Fig. 2B). In summary, transcriptomic analysis revealed that PD tumors enrich for luminal or basal/squamous subtypes and show increased metabolic demand and oncogenic programming while the strongest molecular signatures of SD tumors were elevated expression of immune-related gene pathways.
Figure 2.
Transcriptional features that associate with PD or SD status after combination therapy in UC tumors. A, Expression profile (Z-score) of bladder tumor subtype marker genes in pretreatment UC tumors. Tissue origin of tumors are provided. B, Gene expression of GSEA Hallmark pathways comparing available PD and SD tumors at pretreatment and posttreatment timepoints. C, Deconvolution of immune cell type abundance in tumors from bulk tumor RNA-seq using quanTIseq. Statistical significance was calculated by using mixed-effect beta regression followed by Likelihood Ratio Test. P values were corrected for multiple comparisons with Benjamini–Hochberg method. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Immune infiltration in tumors is associated with durable tumor control
The abundance and composition of tumor-infiltrating immune cells are strongly linked to favorable clinical prognosis after ICI therapy in various cancer types (8, 55, 59). Considering the strong immune-related signatures enriched in SD tumors, we asked whether differential immune cell infiltration of tumors could be contributing to the superior tumor control after combination therapy. It is important to note that although every SD patient had lymph node metastases, only half of the SD tumor samples were taken from the lymph node while the other half were harvested from primary tumors or other metastatic sites (Table 2). Therefore, the differences in immune signatures are not solely influenced by tumor biopsy location. We performed immune deconvolution analysis of bulk tumor transcriptome data to identify immune composition (quanTIseq; ref. 47) in our UC tumor samples. QuanTIseq quantifies and normalizes marker gene expression for ten different immune cell types from bulk RNA-seq data to estimate what fraction of the sample transcriptome is derived from particular immune cell types. This analysis revealed higher abundance of regulatory CD4 T cells (∼1.5-fold) at posttreatment timepoint in SD tumors compared with PD tumors and higher abundance of B cells (∼6-fold) fractions at both pretreatment and posttreatment timepoints (Fig. 2C). It is also important to note that, although not statistically significant, SD tumors had higher abundance of CD8 T cells (∼3-fold) than PD tumors. Altogether, these results were consistent with the immune fraction estimate from DNA methylome data by EpiDISH (Supplementary Fig. S6C). Indeed, rather than the tumor-intrinsic upregulation of immune-related genes, the increased abundance of these leukocytes could be the main contributor to the higher immune-related gene signatures in SD tumors (Fig. 2B). Tumor-associated macrophages are critical for modulating the tumor microenvironment (TME; refs. 60, 61), but no significant differences in macrophage fractions was detected between SD and PD tumors. Meanwhile, the presence of dendritic cells was detected in most SD tumors while the majority of PD tumors lacked these antigen-presenting cells (Fig. 2C). Interestingly, we noticed that neutrophil and NK fractions were elevated in some PD tumors at pretreatment timepoints. Of note, immune infiltration landscape remained largely unchanged in respect to combination therapy. Thus, the treatment did not significantly change the global immune repertoire in the tumors, but the SD patients had a greater presence of CD8 T cells, regulatory T cells, B cells, and dendritic cells in tumors at study onset. These immune deconvolution methods revealed pretreatment immune infiltration and composition differences that segregated the tumor samples based on disease progression after therapy.
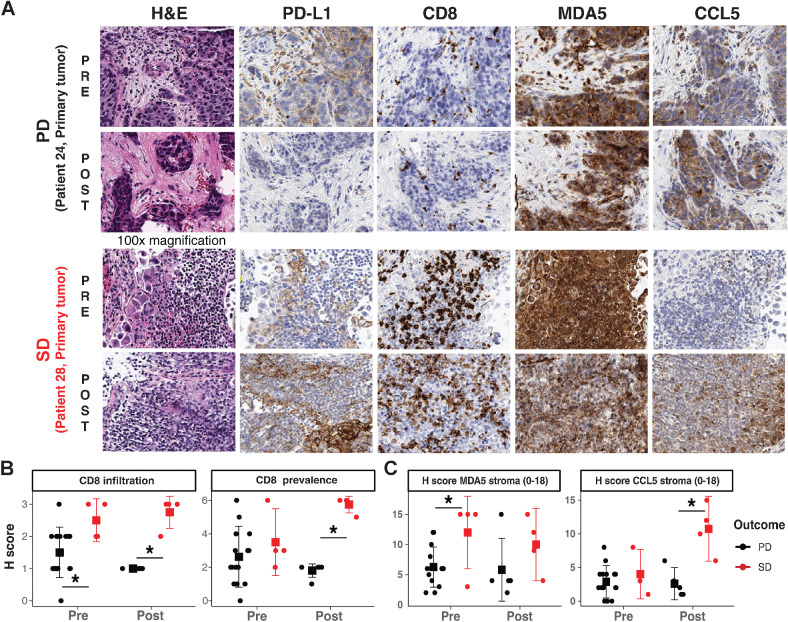
Considering the caveats of normalization and lack of spatial resolution in bulk immune deconvolution methods, we next performed quantitative IHC on available FFPE UC tumor samples to refine the spatial resolution of protein expression in stromal or epithelial sections of the TME (Fig. 3A; Supplementary Fig. S7). The tumors were reviewed and scored by two pathologists in a double-blind manner; the IHC staining of PD-L1 (immune checkpoint molecule), MDA5 (refs. 18, 19; dsRNA sensor), and CCL5 (ref. 62; immunostimulatory cytokine) were quantified using a modified H-Score method, which was calculated by stain intensity (0–3) multiplied by stain prevalence (0–6) for a total H-Score range between 0 and 18. CD8 infiltration was scored in tumors by substituting stain intensity (0–3) with patterns of CD8 infiltration: primarily present in stroma (1), aggregating at tumor edge (2), and actively infiltrating tumor (3) multiplied by CD8 abundance (0–6) for H-Score ranges of 0–18 (Methods; Supplementary Table S5). Past immunotherapy trials in muscle-invasive UCs suggest that higher PD-L1 levels might serve as a prognostic marker to enrich for patients with clinical response to ICI therapy (63). In this study of ICI-resistant metastatic UC tumors, we detected no statistically significant correlation between PD-L1 levels in tumor stroma or epithelial compartments and tumor progression (Supplementary Fig. S8A–S8C). However, we observed a modest increase in epithelial PD-L1 expression in all SD tumors after treatment suggesting that the SD tumors could be reactively compensating or transformed by the hypomethylating agent and immune checkpoint inhibition treatments (Supplementary Fig. S8C).
Figure 3.
IHC reveals immune recruitment in tumors is associated with SD. A, Representative IHC staining of PD and SD tumors in pretreatment and posttreatment timepoint. B, IHC H-scores of CD8 infiltration status and prevalence in PD and SD tumors. C, IHC histology scores (H scores) of stromal MDA5 levels, and stromal CCL5 levels in PD and SD tumors. Statistical significance was tested with log-transformed robust linear mixed-effects models; SGPVs were used to determine if there was significant evidence of a difference where the null interval was set at ± 5% difference in regression estimates. *SGPV = 0; there were no SGPVs = 1 (i.e., no evidence of approximate equivalence). Error bars represent means ± Gini mean difference (a nonparametric measure of dispersion/spread).
The most striking prognostic IHC marker in this clinical trial was CD8 expression with the higher preexisting CD8 abundance in tumors and the increase of CD8 infiltration after combination therapy in SD tumors compared with PD tumors (Fig. 3B; Supplementary Fig. S9A and S9B). Furthermore, the preexisting higher levels of MDA5 and strong augmentation of CCL5 after treatment in the stromal TME compartment were more prominent in SD tumors (Fig. 3C; Supplementary Fig. S9C). Considering that MDA5 and CCL5 are markers for interferon response and immune recruitment, respectively, we postulate that the combination therapy could have reshaped the TME to become more immunostimulatory in SD tumors, but failed to do so in PD tumors. Finally, we did not detect any statistically significant differences in MDA5 and CCL5 levels in the epithelial compartment of the UC TME (Supplementary Fig. S9D and S9E), suggesting that the combination therapy primarily impacted immune-mediated dynamics rather than tumor-intrinsic changes to influence tumor control.
Peripheral immune activation and dynamics associate with patient survival
Exhaustion of CD8 T-cell function, and potentially other immune cells, is mediated by DNA methylation, which can be reversed by DNMTi treatment (14–16, 64, 65). If an immune cell reaches terminal exhaustion, ICI therapy alone might not be sufficient to overcome the restrictive epigenetic barrier to rescue the effector function of these T cells (65). Therefore, we hypothesized that guadecitabine treatment would complement ICI therapy by epigenetically reversing the exhaustion state and ultimately rescuing immune effector function to target and control tumor growth. Bulk transcriptomics and IHC analysis revealed that tumor behavior is intricately tied to the increased immune expression signatures and CD8 T-cell infiltration status in the tumor (Fig. 2C and 3B). Furthermore, IHC analysis revealed that significant differences in the stroma of the TME, which is typically rich with immune cells, distinguished SD tumors from PD tumors especially after combination therapy (Fig. 3C). To characterize the global impact of the combination therapy on immune effector function in patients, we profiled patients’ PBMCs with multidimensional flow cytometry using T and NK cell–specific panels (Supplementary Table S6). The flow cytometry panel distinguished naïve, central memory, effector memory, and effector subtypes of CD8 and CD4 T cells, in addition to the CD56dim and CD56bright NK cells. Preliminary time-course profiling of PBMCs after combination therapy revealed reduction of lymphocyte and monocyte counts at 22 days after the start of the first treatment cycle, and this pattern persisted through subsequent three treatment cycles (Supplementary Fig. S10). The immune cell counts quickly rebounded by the end of each treatment cycle. Therefore, we compared patients’ PBMCs at the start of therapy (pretreatment), mid-cycle (Cycle 1 Day 22, C1D22), and end of the first treatment cycle (Cycle 2 Day 1, C2D1) to capture the preexisting immune state, acute effect of combination treatment, and preferential expansion of immune subtypes after treatment, respectively (Supplementary Table S7).
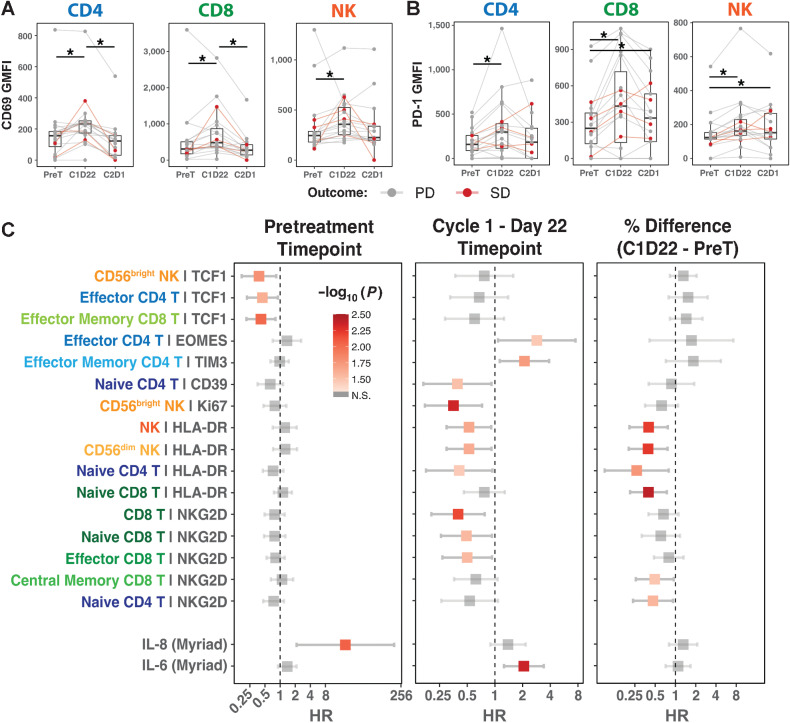
First, we investigated whether the combination therapy activated peripheral immune cells by querying established immune activation markers, CD69 (66) and PD-1 (67, 68). CD69 is an early marker of lymphocyte activation and was upregulated in all T- and NK-cell subtypes after treatment at the C1D22 timepoint and regressed back to basal levels by end of the treatment cycle (Fig. 4A; Supplementary Fig. S11A). Similarly, PD-1 levels were increased in all immune subtypes at C1D22 (Fig. 4B; Supplementary Fig. S11B), which substantiates that combination therapy activated the T and NK cells in the patients’ peripheral blood. Next, we explored whether particular immune features or ligands in specific immune subtypes at certain treatment timepoints might provide prognostic value in predicting survival time in patients. Cox proportional hazard regression analysis (Methods) revealed that greater expression of TCF1 at pretreatment timepoint in CD56bright NK cells, effector CD4 T cells, and effector memory CD8 T cells (HR range: 0.380–0.443; SGPV = 0) was associated with longer survival (Fig. 4C). Interestingly, we identified that increased induction of HLA-DR (HR range: 0.266–0.523, SGPV = 0) in naïve T and NK cells, and NKG2D (HR range: 0.398–0.499, SGPV = 0) in various CD8 T-cell subtypes and naïve CD4 T cells after the combination therapy treatment were also associated with longer survival (Supplementary Fig. S12A and S12B). Also, the abundance of NKG2D, a tumor-responsive activating receptor, was significantly higher on CD8 T cells in PBMCs from SD patients than PD patients at the C1D22 timepoint (Supplementary Fig. S12B). Finally, using the Myriad RBM Simoa immunoassay platform, we profiled TNFα, IL6, and IL8 cytokine abundance in patient plasma samples from pretreatment, C1D22, and C2D1 timepoints. With the exception of TIM3 expression on effector memory CD4 T cells that weakly correlated with inferior PFS, elevated baseline plasma IL8 levels at the pretreatment timepoint and increased levels of IL6 at C1D22 were strong predictors for short time to progression or death (Fig. 4C; Supplementary Fig. S12C). IL6 and IL8 are inflammatory cytokines, which are often associated with worse disease prognosis (69, 70), immunosuppressive myeloid-derived suppressor cells (71, 72), and reduced efficacy of ICI therapies (73–77). Therefore, elevated levels of IL6 and IL8 in patients’ plasma could serve as biomarkers for cancer progression after ICI therapy in metastatic UCs. In conclusion, flow cytometry analysis of patients’ PBMCs established that the combination therapy activated peripheral T and NK cells. Furthermore, higher expression of HLA-DR and NKG2D on T and NK cells associated with longer survival while IL6 and IL8 cytokine presence in patients’ plasma were strong risk factors for short survival.
Figure 4.
Peripheral immune features and cytokine expression associated with PFS. A and B, GMFI distribution of CD69 (A) and PD-1 (B) in all immune subtypes detected in patient blood across various treatment timepoints. Statistical significance tested with cumulative probit ordinal mixed-effects models. C, Cox proportional hazard regression results comparing PFS and various immune features of peripheral blood immune subtypes in pretreatment (n = 20) and C1D22 (n = 19) timepoints. Third column presents whether the GMFI change from pretreatment to C1D22 associates with PFS (n = 19). HR with 95% CI are presented with error bars. For all statistical comparisons, SGPVs were used to determine if there was significant evidence of a difference where the null interval was set at ± 5% difference in regression estimates. *SGPV = 0; there were no SGPVs = 1.
Discussion
One hallmark of cancer is a tumor's ability to evade immune detection and elimination, often through the increased expression of immune checkpoint molecules (78). On the basis of demonstrated efficacy, ICI therapy has earned a valuable position in the clinic based on its ability to restore antitumor function in dysfunctional immune cells leading to measurable antitumor activity. However, there is high motivation to improve upon ICI monotherapy in UC given the observed ORRs is 20% to 30% in most trials (63). Specifically, there is a need to identify alternative strategies to treat the significant fraction of patients who become resistant or refractory to ICI therapy, which was the main goal of our study. Here, we present a phase II clinical trial to combine hypomethylating agent (guadecitabine) and ICI (atezolizumab) hoping to overcome ICI resistance in the context of metastatic UC. The trial was closed for futility following predefined early stopping rules after no RECIST-defined responses were noted among the first 18 patients. The stopping rule allowed for continued accrual while the 18th patient awaited their first scan. An additional 3 patients enrolled during this time, and non-RECIST responses were noted among them also. However, in a subset of ICI-resistant patients, the combination treatment induced durable tumor control that persisted for 8 to 11 months and extended patient survival to > 14 months. Although we recognize the inherent limitations of small sample size, low statistical power, and weak clinical endpoint, we performed various molecular profiling of tumor and peripheral blood samples in hopes of gleaning biological insights that might help extend this survival benefit to additional patients in the future.
First, we hypothesized that guadecitabine would reduce DNA methylation levels in tumors and consequential viral mimicry response. We observed only nominal gains or losses of global DNA methylation and, surprisingly, a loss of viral mimicry-associated gene expression in tumors after two treatment cycles (Supplementary Fig. S4B). One explanation for the lack of DNA demethylation could be that guadecitabine is simply not effective at inducing hypomethylation in UC tumors. However, it is also important to note that the two comparative timepoints were 12 weeks apart (after two cycles). Furthermore, the tumors in the posttreatment timepoints are collected at 6 weeks after the initial dose of guadecitabine treatment. The long interval between guadecitabine treatment and sample collection might provide ample time for tumors to recover and rebound from treatment-induced DNA demethylation effects, as was evident in the recovery of peripheral leukocytes at the same timepoint (Supplementary Fig. S10).
UCs show complex heterogeneity of transcriptomic profiles, which have been associated with patient outcome to conventional therapy. Using the bulk tumor transcriptome data, we measured the expression of marker genes known to be preferentially expressed in particular subtypes of UC. This analysis revealed that luminal and basal-subtype signatures, oncogenic pathways, and fatty acid metabolism were enriched in PD tumors (Fig. 2A and B). In contrast, SD tumors enriched for neuroendocrine/neuronal subtype signatures and immune-related gene pathways, such as inflammation and interferon response. Interestingly, correlative studies from IMvigor210 trial (atezolizumab monotherapy in metastatic UC) identified that luminal papillary tumors were found to be resistant (6), while neuronal subtypes showed 100% response rate with increased survival (79). In the ABACUS trial, atezolizumab was evaluated as a neoadjuvant in 95 patients with muscle-invasive UC (80). Biomarker studies from the ABACUS trial revealed that preexisting activated T cells was better correlated with clinical outcome than PD-L1 levels or tumor mutation burden. Indeed, IHC profiling of the UC tumors from our trial also revealed that high levels of CD8 infiltration before and after therapy was more pronounced in SD tumors (Fig. 3B). Furthermore, immune deconvolution analysis of the UC tumors corroborated the IHC findings, as we observed greater infiltration of CD8 T cells, B cells, and dendritic cells in SD tumors compared with PD tumors (Fig. 2C). Finally, we profiled peripheral blood immune cells and identified that higher baseline levels of TCF1 and higher levels of HLA-DR and NKG2D at C1D22 on T cells or NK cells positively correlate with PFS (Fig. 4). Considering that NKG2D expression is often associated with circulating antitumor CD8 T cells (81, 82), we speculate that NKG2D expression in peripheral immune cells could serve as prognostic marker for therapy efficacy. The molecular mechanism and insight in how these immune features provide therapeutic benefit warrants further investigation to improve immunotherapy-related precision oncology.
Strikingly, deconvolution of RNA-seq data revealed that neutrophils were highly abundant in PD tumors at baseline. Cytokine profiling from patients’ plasma across various treatment timepoints uncovered that elevated IL8 and IL6 levels associated with worse PFS (Supplementary Fig. S12D). These results support prior work demonstrating that systemic and tumor-associated IL8 levels correlate with worse response in metastatic bladder or kidney cancer patients treated with atezolizumab (77). Furthermore, higher baseline levels of IL6 was associated with shorter PFS in patients with metastatic melanoma treated with ICI therapy (75, 76). Considering that IL8 and IL6 cytokines are abundantly produced by neutrophils, our correlative insights imply that neutrophils might play an underappreciated role in tumor progression and resistance to ICI therapy. Indeed, large-scale retrospective analysis has shown that IL8 levels are associated with neutrophil abundance and reduced benefit from ICI therapy (73, 74). Finally, GSEA analysis revealed that molecular pathways related to glycolysis, adipogenesis, and fatty acid metabolism were upregulated in PD tumors. Whether this metabolic reprogramming in PD tumors contributes to the reshaping of the tumor-immune dynamics, potentially neutrophil recruitment, and the immunosuppressive TME is of great interest.
In conclusion, our phase II trial combining guadecitabine and atezolizumab in patients with metastatic UC progression after initial ICI therapy (Supplementary Table S8) fell short of anticipated promise. Due to the long interval between treatment and sample collections, we could not confidently detect tumor DNA demethylation after combination therapy, which might reflect the lack of viral mimicry activation posttreatment in the UC tumors. However, the combination therapy was successful in stimulating immune cells, especially peripheral T and NK cells, and enhancing T cells to better infiltrate the tumor in a subset of patients. The pharmacologic reinvigoration of immune cells correlated with stabilization of tumor growth and PFS. Our findings introduce the translational implications of immune reprogramming induced by epigenetic therapy in a small subset of ICI-resistant patients in a clinical setting. There is an increased interest in exploring the synergistic potential of epigenetic therapy in combination with ICI therapy in the solid tumor arena. Therefore, it is critical to disseminate any and all clinical trial outcomes that provide correlative insights, which can improve future clinical trial design (e.g., the targeted patient recruitment and drug delivery methods) and maximize the efficacy of combination therapy in patients. Future work on improving the pharmacokinetics of the hypomethylating agents in solid tumors and dissecting the pharmacologic activation of select immune cells will help better achieve the therapeutic potential of combination epigenetic and immunotherapy.
Supplementary Material
Supplementary TablesSupplementary Table 1: Patient and tumor sample informationSupplementary Table 2: Tumor mutation burdenSupplementary Table 3: Unfiltered whole exome sequencing mutation analysisSupplementary Table 4: Filtered whole exome sequencing mutation analysisSupplementary Table 5: Immunohistochemistry scores of tumorsSupplementary Table 6: FACS panel for PBMC analysisSupplementary Table 7: Geometric mean fluorescence intensity (GMFI) from PBMC FACSSupplementary Table 8: Representativeness of Study Participants
Supplementary Figure 1: Patient recruitment and sample collection overview.
Supplementary Figure 2: Modest treatment-induced DNA methylation changes in UC tumors.
Supplementary Figure 3: Copy number variation detected by SeSAMe from DNA methylation data.
Supplementary Figure 4: Weak correlation of TE expression induction with DNA methylation change and viral mimicry activation.
Supplementary Figure 5: Overall tumor mutation burden and frequency of mutations in cancer-associated genes in UC tumors.
Supplementary Figure 6: Gene Set Enrichment Analysis reveals immune-related pathways discriminate PD and SD tumors.
Supplementary Figure 7: Representative IHC staining of additional PD and SD tumors in pre-treatment and post-treatment timepoint from additional patients.
Supplementary Figure 8: PD-L1 levels in tumor or tumor microenvironment does not associate with tumor progression or control.
Supplementary Figure 9: CD8 infiltration in tumor is associated with survival.
Supplementary Figure 10: Peripheral immune cell dynamics induced by combination therapy.
Supplementary Figure 11: Peripheral immune cells are activated mid-treatment cycle.
Supplementary Figure 12: HLA-DR and NKG2D abundance on peripheral lymphocytes associate with longer progression free survival.
Acknowledgments
This research is supported by a Stand Up To Cancer – Genentech Catalyst Research Team (grant number: SU2C -AACR -CT08–17). Stand up to cancer (SU2C) is a division of the Entertainment Industry Foundation. This SU2C grant is administered by the American Association for Cancer Research, the scientific partner of SU2C. We thank the Van Andel Institutes’ Genomics Core, Pathology and Biorepository Core, and Biostatistics & Bioinformatics Core for their valuable help with processing the tumor and peripheral blood samples for molecular analysis. Also, we are grateful for the technical assistance with flow cytometry from Ms. Bryn Queeley and Ms. Irina Shchaveleva and support from the following FCCC Core Research Facilities: Immune Monitoring, Cell Sorting, Cell Culture, and Biostatistics & Bioinformatics, as well as support from NCI CCSG CA06927 (Chernoff; FCCC). Research reported in this publication was supported by the NCI of the NIH under Award Number P30CA006927.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 2013
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
H.J. Jang reports grants from AACR during the conduct of the study. Z. Madaj reports grants from AACR during the conduct of the study. E.A. Ross reports grants from NIH during the conduct of the study, as well as grants from Varian outside the submitted work. M.R. Zibelman reports other support from Bristol Myers Squibb and Exelixis outside the submitted work. N.M. Hahn reports grants and personal fees from AstraZeneca, Merck, Genentech, Seattle Genetics, Incyte, EMD Serono, Astellas, Bristol Myers Squibb, Inovio, and Incyte; personal fees from GlaxoSmithKline, Ferring, Champions Oncology, Health Advances, Keyquest Health, Guidepoint Global, Mirati, TransMed, CicloMed, Janssen, Pfizer, Boehringer Ingelheim, Verity Pharmaceuticals, Protara Therapeutics, Creative Educational Concepts, Large Urology Group Practice Association, ImmunityBio, and Tyra Biosciences; and grants from HTG Diagnostics, Principia Biopharm, and Astex during the conduct of the study. In addition, N.M. Hahn has a patent for PCT/US2022/019757 pending. D.I. Quinn reports personal fees from AbbVie, Merck, EMD Serono, and Pfizer outside the submitted work. J.-P.J. Issa reports personal fees from Astex during the conduct of the study; in addition, J.-P.J. Issa is founder and stock owner of EpigenOnco. K.S. Campbell reports grants from Janssen R&D outside the submitted work. P.A. Jones reports grants from NCI during the conduct of the study, as well as personal fees from Zymo Corporation outside the submitted work. E.R. Plimack reports grants from Genentech during the conduct of the study, as well as personal fees from Astellas, AstraZeneca, Aveo, BMS, Calithera, EMD Serono, Exelixis, Genentech, IMV, Janssen, MEI, Merck, Pfizer, Regeneron, Seagen, Signatera, and Infinity Pharma outside the submitted work. No disclosures were reported by the other authors.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Authors' Contributions
H.J. Jang: Resources, data curation, software, formal analysis, validation, investigation, visualization, writing–original draft, writing–review and editing. G. Hostetter: Data curation, formal analysis, validation, investigation, visualization. A.W. MacFarlane: Data curation, formal analysis, validation, investigation. Z. Madaj: Data curation, formal analysis, validation, investigation, visualization. E.A. Ross: Supervision, project administration. T. Hinoue: Data curation, software, formal analysis, validation, investigation, visualization. J.R. Kulchycki: Software, formal analysis, validation, investigation, visualization. R.S. Burgos: Conceptualization, resources, data curation, funding acquisition, project administration. M. Tafseer: Data curation, project administration. R.K. Alpaugh: Data curation, project administration. C.L. Schwebel: Data curation, project administration. R. Kokate: Data curation, project administration. D.M. Geynisman: Data curation, project administration. M.R. Zibelman: Data curation, project administration. P. Ghatalia: Data curation, project administration. P.W. Nichols: Data curation, validation, investigation. W. Chung: Data curation, project administration. J. Madzo: Data curation, project administration. N.M. Hahn: Conceptualization, data curation, supervision, funding acquisition, project administration. D.I. Quinn: Conceptualization, data curation, supervision, funding acquisition, project administration. J.-P.J. Issa: Conceptualization, data curation, supervision, funding acquisition, project administration. M.J. Topper: Software, investigation. S.B. Baylin: Conceptualization, supervision, funding acquisition, project administration. H. Shen: Conceptualization, resources, supervision, project administration. K.S. Campbell: Resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, writing–original draft, project administration. P.A. Jones: Conceptualization, resources, supervision, funding acquisition, validation, investigation, visualization, writing–original draft, project administration, writing–review and editing. E.R. Plimack: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, writing–original draft, project administration, writing–review and editing.
References
- 1. von der Maase H, Hansen SW, Roberts JT, Dogliotti L, Oliver T, Moore MJ, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol 2000;18:3068–77. [DOI] [PubMed] [Google Scholar]
- 2. Sternberg CN, de Mulder P, Schornagel JH, Theodore C, Fossa SD, van Oosterom AT, et al. Seven year update of an EORTC phase III trial of high-dose intensity M-VAC chemotherapy and G-CSF versus classic M-VAC in advanced urothelial tract tumors. Eur J Cancer 2006;42:50–4. [DOI] [PubMed] [Google Scholar]
- 3. Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicenter, phase II trial. Lancet 2017;389:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Powles T, Park SH, Voog E, Caserta C, Valderrama BP, Gurney H, et al. Avelumab maintenance therapy for advanced or metastatic urothelial carcinoma. N Engl J Med 2020;383:1218–30. [DOI] [PubMed] [Google Scholar]
- 5. Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol 2015;33:1974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicenter, phase II trial. Lancet 2016;387:1909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zibelman M, Ramamurthy C, Plimack ER. Emerging role of immunotherapy in urothelial carcinoma-advanced disease. Urol Oncol 2016;34:538–47. [DOI] [PubMed] [Google Scholar]
- 8. Zang J, Ye K, Fei Y, Zhang R, Chen H, Zhuang G. Immunotherapy in the treatment of urothelial bladder cancer: insights from single-cell analysis. Front Oncol 2021;11:696716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bellmunt J, Necchi A, Wit RD, Lee J-L, Fong L, Vogelzang NJ, et al. Pembrolizumab (pembro) versus investigator's choice of paclitaxel, docetaxel, or vinflunine in recurrent, advanced urothelial cancer (UC): 5-year follow-up from the phase III KEYNOTE-045 trial. J Clin Oncol 2021;39:4532. [Google Scholar]
- 10. Petrylak DP, Powles T, Bellmunt J, Braiteh F, Loriot Y, Morales-Barrera R, et al. Atezolizumab (MPDL3280A) monotherapy for patients with metastatic urothelial cancer: long-term outcomes from a phase I study. JAMA Oncol 2018;4:537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galsky MD, Saci A, Szabo PM, Han GC, Grossfeld G, Collette S, et al. Nivolumab in patients with advanced platinum-resistant urothelial carcinoma: efficacy, safety, and biomarker analyses with extended follow-up from CheckMate 275. Clin Cancer Res 2020;26:5120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiappinelli KB, Zahnow CA, Ahuja N, Baylin SB. Combining epigenetic and immunotherapy to combat cancer. Cancer Res 2016;76:1683–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet 2016;17:630–41. [DOI] [PubMed] [Google Scholar]
- 15. Topper MJ, Vaz M, Marrone KA, Brahmer JR, Baylin SB. The emerging role of epigenetic therapeutics in immuno-oncology. Nat Rev Clin Oncol 2020;17:75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 2016;354:1160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, et al. The epigenetic landscape of T-cell exhaustion. Science 2016;354:1165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015;162:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 2015;162:961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siebenkas C, Chiappinelli KB, Guzzetta AA, Sharma A, Jeschke J, Vatapalli R, et al. Inhibiting DNA methylation activates cancer testis antigens and expression of the antigen processing and presentation machinery in colon and ovarian cancer cells. PLoS One 2017;12:e0179501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krishnadas DK, Bao L, Bai F, Chencheri SC, Lucas K. Decitabine facilitates immune recognition of sarcoma cells by upregulating CT antigens, MHC molecules, and ICAM-1. Tumour Biol 2014;35:5753–62. [DOI] [PubMed] [Google Scholar]
- 22. Luo N, Nixon MJ, Gonzalez-Ericsson PI, Sanchez V, Opalenik SR, Li H, et al. DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat Commun 2018;9:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nunes SP, Henrique R, Jeronimo C, Paramio JM. DNA methylation as a therapeutic target for bladder cancer. Cells 2020;9:1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2'-deoxycytidine. Cancer Res 2002;62:961–6. [PubMed] [Google Scholar]
- 25. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van der Auwera GA, O'Connor BD, Safari aORMC. Genomics in the cloud: using Docker, GATK, and WDL in Terra. First edition. ed.Sebastopol, CA: O'Reilly Media; 2020. [Google Scholar]
- 28. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. , The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ainscough BJ, Griffith M, Coffman AC, Wagner AH, Kunisaki J, Choudhary MN, et al. DoCM: a database of curated mutations in cancer. Nat Methods 2016;13:806–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 2017;45:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou W, Triche TJ Jr, Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by infinium BeadChips in genomic deletions. Nucleic Acids Res 2018;46:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Triche TJ Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of illumina infinium DNA methylation BeadArrays. Nucleic Acids Res 2013;41:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu T, Liu J, Beck S, Pan S, Capper D, Lechner M, et al. A pan-tissue DNA methylation atlas enables in silico decomposition of human tissue methylomes at cell-type resolution. Nat Methods 2022;19:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in epigenome-wide association Studies. BMC Bioinf 2017;18:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng SC, Breeze CE, Beck S, Dong D, Zhu T, Ma L, et al. EpiDISH web server: epigenetic dissection of intra-sample-heterogeneity with online GUI. Bioinformatics 2019;36:1950–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blume JD, Greevy RA, Welty VF, Smith JR, Dupont WD. An introduction to second-generation P values. Am Stat 2019;73:157–67. [Google Scholar]
- 39. Venables WNR, B. D. Modern Applied Statistics with S. Fourth ed. New York: Springer; 2002. [Google Scholar]
- 40. Koller M. robustlmm: An R package for robust estimation of linear mixed-effects models. J Stat Softw 2016;75:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019;37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 46. Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 2015;31:3593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finotello F, Mayer C, Plattner C, Laschober G, Rieder D, Hackl H, et al. Molecular and pharmacologic modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 2019;11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brooks ME, Kristensen K, van Benthem KJ, Magnusson A, Berg CW, Nielsen A, et al. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. The R Journal 2017;9:378–400. [Google Scholar]
- 49. Adashek JJ, Subbiah IM, Matos I, Garralda E, Menta AK, Ganeshan DM, et al. Hyperprogression and immunotherapy: fact, fiction, or alternative fact? Trends Cancer 2020;6:181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017;32:185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou W, Dinh HQ, Ramjan Z, Weisenberger DJ, Nicolet CM, Shen H, et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet 2018;50:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017;171:540–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fong MHY, Feng M, McConkey DJ, Choi W. Update on bladder cancer molecular subtypes. Transl Androl Urol 2020;9:2881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu S, Yu W, Yang X, Wu C, Cheng F. Traditional classification and novel subtyping systems for bladder cancer. Front Oncol 2020;10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sfakianos JP, Daza J, Hu Y, Anastos H, Bryant G, Bareja R, et al. Epithelial plasticity can generate multi-lineage phenotypes in human and murine bladder cancers. Nat Commun 2020;11:2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 59. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, et al. Macrophage polarization in tumor progression. Semin Cancer Biol 2008;18:349–55. [DOI] [PubMed] [Google Scholar]
- 61. Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci 2019;26:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen RC, et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 2017;171:1284–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lopez-Beltran A, Cimadamore A, Blanca A, Massari F, Vau N, Scarpelli M, et al. Immune checkpoint inhibitors for the treatment of bladder cancer. Cancers 2021;13:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, et al. The epigenetic landscape of T-cell exhaustion. Science 2016;354:1165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T-cell rejuvenation. Cell 2017;170:142–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cibrian D, Sanchez-Madrid F. CD69: from activation marker to metabolic gatekeeper. Eur J Immunol 2017;47:946–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Simon S, Labarriere N. PD-1 expression on tumor-specific T cells: friend or foe for immunotherapy? Oncoimmunology 2017;7:e1364828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol 1996;8:765–72. [DOI] [PubMed] [Google Scholar]
- 69. Goulet CR, Champagne A, Bernard G, Vandal D, Chabaud S, Pouliot F, et al. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of bladder cancer cells through paracrine IL6 signaling. BMC Cancer 2019;19:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Inoue K, Slaton JW, Kim SJ, Perrotte P, Eve BY, Bar-Eli M, et al. Interleukin 8 expression regulates tumorigenicity and metastasis in human bladder cancer. Cancer Res 2000;60:2290–9. [PubMed] [Google Scholar]
- 71. Alfaro C, Teijeira A, Onate C, Perez G, Sanmamed MF, Andueza MP, et al. Tumor-produced interleukin-8 attracts human myeloid-derived suppressor cells and elicits extrusion of neutrophil extracellular traps (NETs). Clin Cancer Res 2016;22:3924–36. [DOI] [PubMed] [Google Scholar]
- 72. Tobin RP, Jordan KR, Kapoor P, Spongberg E, Davis D, Vorwald VM, et al. IL6 and IL8 are linked with myeloid-derived suppressor cell accumulation and correlate with poor clinical outcomes in melanoma patients. Front Oncol 2019;9:1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schalper KA, Carleton M, Zhou M, Chen T, Feng Y, Huang SP, et al. Elevated serum interleukin-8 is associated with enhanced intratumor neutrophils and reduced clinical benefit of immune-checkpoint inhibitors. Nat Med 2020;26:688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sanmamed MF, Perez-Gracia JL, Schalper KA, Fusco JP, Gonzalez A, Rodriguez-Ruiz ME, et al. Changes in serum interleukin-8 (IL8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non–small cell lung cancer patients. Ann Oncol 2017;28:1988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Y, Ramachandran V, Sui D, Xu K, Haydu LE, Fang S, et al. Evaluation of plasma IL6 in patients with melanoma as a prognostic and checkpoint immunotherapy predictive biomarker. J Invest Dermatol 2021;142:2046–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Laino AS, Woods D, Vassallo M, Qian X, Tang H, Wind-Rotolo M, et al. Serum interleukin-6 and C-reactive protein are associated with survival in melanoma patients receiving immune checkpoint inhibition. J Immunother Cancer 2020;8:e000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yuen KC, Liu LF, Gupta V, Madireddi S, Keerthivasan S, Li C, et al. High systemic and tumor-associated IL8 correlates with reduced clinical benefit of PD-L1 blockade. Nat Med 2020;26:693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res 2015;21:687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim J, Kwiatkowski D, McConkey DJ, Meeks JJ, Freeman SS, Bellmunt J, et al. The cancer genome atlas expression subtypes stratify response to checkpoint inhibition in advanced urothelial cancer and identify a subset of patients with high survival probability. Eur Urol 2019;75:961–4. [DOI] [PubMed] [Google Scholar]
- 80. Powles T, Kockx M, Rodriguez-Vida A, Duran I, Crabb SJ, Van Der Heijden MS, et al. Clinical efficacy and biomarker analysis of neoadjuvant atezolizumab in operable urothelial carcinoma in the ABACUS trial. Nat Med 2019;25:1706–14. [DOI] [PubMed] [Google Scholar]
- 81. Pauken KE, Shahid O, Lagattuta KA, Mahuron KM, Luber JM, Lowe MM, et al. Single-cell analyses identify circulating antitumor CD8 T cells and markers for their enrichment. J Exp Med 2021;218:e20200920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Prajapati K, Perez C, Rojas LBP, Burke B, Guevara-Patino JA. Functions of NKG2D in CD8(+) T cells: an opportunity for immunotherapy. Cell Mol Immunol 2018;15:470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary TablesSupplementary Table 1: Patient and tumor sample informationSupplementary Table 2: Tumor mutation burdenSupplementary Table 3: Unfiltered whole exome sequencing mutation analysisSupplementary Table 4: Filtered whole exome sequencing mutation analysisSupplementary Table 5: Immunohistochemistry scores of tumorsSupplementary Table 6: FACS panel for PBMC analysisSupplementary Table 7: Geometric mean fluorescence intensity (GMFI) from PBMC FACSSupplementary Table 8: Representativeness of Study Participants
Supplementary Figure 1: Patient recruitment and sample collection overview.
Supplementary Figure 2: Modest treatment-induced DNA methylation changes in UC tumors.
Supplementary Figure 3: Copy number variation detected by SeSAMe from DNA methylation data.
Supplementary Figure 4: Weak correlation of TE expression induction with DNA methylation change and viral mimicry activation.
Supplementary Figure 5: Overall tumor mutation burden and frequency of mutations in cancer-associated genes in UC tumors.
Supplementary Figure 6: Gene Set Enrichment Analysis reveals immune-related pathways discriminate PD and SD tumors.
Supplementary Figure 7: Representative IHC staining of additional PD and SD tumors in pre-treatment and post-treatment timepoint from additional patients.
Supplementary Figure 8: PD-L1 levels in tumor or tumor microenvironment does not associate with tumor progression or control.
Supplementary Figure 9: CD8 infiltration in tumor is associated with survival.
Supplementary Figure 10: Peripheral immune cell dynamics induced by combination therapy.
Supplementary Figure 11: Peripheral immune cells are activated mid-treatment cycle.
Supplementary Figure 12: HLA-DR and NKG2D abundance on peripheral lymphocytes associate with longer progression free survival.
Data Availability Statement
Sequencing data from tumor RNA-seq, WES, and DNA methylome analysis is available on Gene Expression Omnibus repository (GSE222934). H-scores from IHC analysis and data collected from flow cytometry analysis are available in the Supplementary Tables.