Abstract
A select group of patients with hepatocellular carcinomas (HCC) benefit from surgical, radiologic, and systemic therapies that include a combination of anti-angiogenic and immune-checkpoint inhibitors. However, because HCC is generally asymptomatic in its early stages, this not only leads to late diagnosis, but also to therapy resistance. The nucleoside analogue 6-thio-dG (THIO) is a first-in-class telomerase-mediated telomere-targeting anticancer agent. In telomerase expressing cancer cells, THIO is converted into the corresponding 5′-triphosphate, which is efficiently incorporated into telomeres by telomerase, activating telomere damage responses and apoptotic pathways. Here, we show how THIO is effective in controlling tumor growth and, when combined with immune checkpoint inhibitors, is even more effective in a T-cell-dependent manner. We also show telomere stress induced by THIO increases both innate sensing and adaptive antitumor immunity in HCC. Importantly, the extracellular high-mobility group box 1 protein acts as a prototypical endogenous DAMP (Damage Associated Molecular Pattern) in eliciting adaptive immunity by THIO. These results provide a strong rationale for combining telomere-targeted therapy with immunotherapy.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignant tumor of the liver and is the third leading cause of cancer-related deaths including a low 5-year survival rate and limited therapeutic options (1, 2). Although early-stage patients with HCC receive therapies with curative intent such as liver resection, radiotherapy, and liver transplantation, patients with advanced-stage liver cancer almost universally develop recurrence or metastases within 5 years even after first- and second-line therapies (3, 4). For many years, the commonly used therapeutic regimens for advanced HCC in clinical practice has been targeted therapies based on tyrosine kinase inhibitors such as the multi-kinase inhibitor sorafenib, anti-angiogenic drugs, radiotherapy, and other chemotherapies. However, these treatments are characterized by low response rates, high recurrence rates, and low overall survival (5).
HCC is considered an immunogenic tumor, containing infiltrating tumor-specific T-cell lymphocytes and other immune cells (6). In recent years, application of immune checkpoint inhibitors (ICI) such as programmed cell death protein 1 (PD1), programmed cell death ligand 1 (PDL1), and cytotoxic T lymphocyte antigen 4 (CTLA-4) mAbs against immune regulatory checkpoints have partially improved treatment of solid tumors including HCC (7, 8). Because pembrolizumab and nivolumab (anti-PD1 humanized antibodies) have shown efficacy in a subset of relapsed and refractory patients with HCC who previously received sorafenib, these two drugs were approved by the FDA as a second-line treatment. However, objective response rates remained relatively low for nivolumab and pembrolizumab (9, 10). Patients who fail to respond to immune monotherapy search for new therapeutic strategies to improve the effect of immunotherapy against HCC. The combination of atezolizumab (PDL1 inhibitor) and bevacizumab (angiogenic inhibitor) has been one of the most successful treatments to date and currently is approved by the FDA as a first-line treatment for patients with unresectable HCC (5, 11–13). However, even with these advances less than 30% of patients have durable remissions and often become resistant to these therapies.
Antigen-presenting cells (APC) such as dendritic cells (DC), macrophages, and Langerhans cells mediate the cellular immune response by processing and presenting antigens for recognition by lymphocytes such as natural killer (NK) cells, B cells, and T cells. DCs are considered an important APC with a vital role in the activation of immune system (14). Adaptive antitumor immunity is highly dependent on innate immunity. Innate immunity, as a first host immune barrier, can sense non-self-material by various pattern recognition receptors such as cytosolic DNA sensors (15, 16). Formation of cytosolic chromatin fragments and micronuclei are the source of cancer cell-derived DNA that is taken up by DCs. This tumor-derived cytosolic DNA activates cyclic GMP-AMP (cGAMP) synthase-stimulator of IFN genes (cGAS–STING) pathway. Cytosolic dsDNA directly binds to cGAS and catalyzes the production of cGAMP, which eventually stimulates the conformation change of the STING molecule. This activates TANK-binding kinase 1 (TBK1), phosphorylates IFN regulatory transcription factor 3 (IRF3), and stimulates type I IFN expression in cancer cells or DCs, initiating innate anticancer immunity (17–21). Type I IFN signaling is essential to the cross-priming of tumor-specific T cells by activating DCs (22, 23).
Small molecules that cause tumor cell death may activate innate sensing pathways or target other immunosuppressive factors in the tumor microenvironment to trigger antitumor immunity. Therefore, combining immunotherapy with small molecules may induce stronger antitumor effects and achieve increased long-term patient survival (24). Recently, we reported that 6-thio-2′deoxyguanosine (6-thio-dG, also known as THIO), a telomerase-mediated telomere targeting agent, is preferentially incorporated into de novo synthesized telomeres by telomerase and causes rapid telomere uncapping, which further induces telomere-associated DNA damage in cancer cells (25). Additionally, 6-thio-dG (THIO) overcomes targeted-therapy and chemotherapy resistance in advanced preclinical lung cancer models (26). Since telomerase is overexpressed in ∼85% of tumors, but remains silent in normal cells, telomerase is a highly attractive, almost universal, target to develop new anticancer drugs (27–29). Modified nucleoside analogues that target cancer telomeres such as 6-thio-dG (THIO) are a new therapeutic approach in preclinical/clinical studies. We have recently shown that damaged DNA fragments induced by 6-thio-dG (THIO) are taken up by DCs and trigger the cGAS-STING pathway and enhance adaptive immune responses in preclinical studies of colon and lung cancer (30). Moreover, sequential administration of 6-thio-dG (THIO) with anti-PDL1 reduces tumor burden compared with THIO monotherapy by enhancing innate sensing and adaptive immunity (30).
In this study, we evaluated THIO to determine its therapeutic effects in vitro and in vivo together with ICIs in preclinical models of liver cancer. Our results provide an experimental rationale for a potentially new HCC treatment approach based on the administration of a telomere targeting agent in sequential combination with ICI. This approach represents a different paradigm for treatment of telomerase-positive tumors, based on the dual mechanism; directly killing tumor cells and activating innate sensing pathways or reversing the immunosuppressive microenvironment to enhance the therapeutic effect of immunotherapies. This strategy could result in broad therapeutic effects of modified THIO analogues in advanced telomerase-expressing tumors.
Materials and Methods
Cell lines
HCC53N murine liver cancer cells (new cell line, FVB background, RRID: IMSR_JAX:001800) were grown in a Medium X (DMEM:199, 4:1; Hyclone) supplemented with 10% cosmic calf serum (Hyclone). The Hep55–1C cell line (RRID: CVCL_5766) was kindly provided by Tim Greten (NIH). Hep55–1C murine liver cancer cells (C57BL/6 background) were grown in DMEM with 4.5 g/L glucose and sodium pyruvate without L-glutamine (Corning, Catalog No. 15–013-CV) supplemented with 2 mmol/L L-glutamine (Corning, Catalog No. 25–005-CI) and 10% FBS. Hep1C7 murine liver cancer cells (C57BL/6 background) were kindly provided by Hao Zhu (UTSW). Hep1C7 cells were grown in DMEM supplemented with 10% FBS. RIL175 cells (RRID: CVCL_B7TK) were grown in DMEM (Corning, Catalog No. 15–013-CV) supplemented with 10% FBS, 1% non-essential amino acids (NEAA; Cyagen, Catalog No. NEAA-10201–50), and 1% sodium pyruvate (Gibco, Catalog No. 11360–070). Huh7 (RRID: CVCL_0336) and Hep3B cells (RRID: CVCL_0326) were grown in DMEM (Gibco, Catalog No. 11965–092) supplemented with 10% FBS (Gibco, Catalog No. 26140–079), HepG2 cells (RRID: CVCL_0027) were grown in EMEM media (ATCC, Catalog No. 30–2003) supplemented with 10% FBS. SNU-387 (RRID: CVCL_0250), SNU-475 (RRID: CVCL_0497), and SNU-449 cells (RRID: CVCL_0454) were grown at RPMI1640 (Gibco, Catalog No. 11875–093) supplemented with 10% FBS. All the cells in this study were grown without antibiotics and incubated in a humidified atmosphere with 5% CO2 at 37°C. THLE2 primary normal liver cells (RRID: CVCL_3803) were grown in bronchial epithelial cell growth basal medium (BEGM) without gentamycin/amphotericin (GA) but included epinephrine (Lonza, Catalog No. CC3170) supplemented with 5 ng/mL EGF, 70 ng/mL phosphoethanolamine, and 10% FBS. THLE2 telomerase silent cells were grown on precoated plates consisting of a mixture of 0.01 mg/mL fibronectin, 0.03 mg/mL bovine collagen type I, and 0.01 mg/mL BSA dissolved in bronchial epithelial basal medium (BEBM).
Cell lines were expanded upon receipt, frozen in aliquots, and only kept in culture during experiments. Hep55–1C, Hep1C7, Huh7, Hep3B, HepG2, THLE2, and HCC53N cell lines were tested negative for Mycoplasma using the e-Myco plus Mycoplasma PCR Detection Kit (LiliF Diagnostics, 25237). No testing was performed for RIL175, SNU-387, SNU-475, and SNU-449 cell lines.
Drug preparation
6-Thio-dG, Metkinen Oy (THIO) was dissolved in DMSO/water (1:1) to prepare 10 mmol/L stock solution, and in DMSO to prepare 100 mmol/L stock solutions. For mouse in vivo studies, THIO was prepared in a 5% DMSO solution in PBS. Libtayo (anti-PD1, cemiplimab-rwlc) and tecentriq (anti-PDL1, atezolizumab) were kindly provided by UT Southwestern Simmons Cancer Center Pharmacy and was diluted in PBS for a final concentration of 200 μg per mouse. Anti-PDL1 (BioCell, Catalog No. BE0101), anti-VEGFR (BioCell, Catalog No. BE0060) and anti-CD8 (BioCell, Catalog No. BE0061) were diluted in PBS for a final concentration of 200 μg per mouse. r84 (anti-VEGF) was kindly provided by Rolf Brekken at UT Southwestern Medical Center. r84 and anti-VEGFR (BioCell, Catalog No. BE0060) both target VEGF:VEGFR2 interaction.
Cell viability assay for determination of EC50
Murine (Hep1C7, Hep55–1C, HCC53N, RIL175) and human liver cancer cell lines (SNU-449, SNU-475, Hep3B, Huh7, SNU-387, HepG2) were screened with THIO with a dilution series in 9 different points in 96-well plates. Cells were plated 24 hrs prior to the addition of drug, incubated for 4–5 days, and assayed using the CellTiter-Glo Cell Viability Assay according to the manufacturer's instructions (Promega). 3,000 cells for SNU-387, SNU-475, Huh7, Hep3B, Hep1C7, HepG2, SNU-449, THLE2 and 4,000 cells for Hep55–1C, and 5,000 cells for HCC53N were seeded. Dose response curves were generated and EC50s calculated using GraphPad Prism. All samples were analyzed in duplicate or triplicate.
Telomere dysfunction induced foci (TIF)
Hep3B, Huh7 and Hep55–1C cells were seeded onto a slide previously sterilized and placed in a 10 cm Petri dish (Falcon). After 24 hrs, Hep3B, Huh7 and Hep55–1C cells were treated with THIO at EC50 concentrations for each cell line for 48 hrs. Then, slides were rinsed in PBS 1X for 10 minutes on a shaking platform. Slides were fixed in 4% formaldehyde (Thermo Fisher) for 10 minutes on ice and then washed in PBS 1X for 5 minutes, two times. Subsequently, slides were permeabilized with 0.5% Triton X-100 for 10 minutes on ice and then blocked with BSA/PBS 1X for 30 minutes at RT. Anti-mouse primary antibody γH2AX (Millipore) was diluted 1:200 in blocking solution and cells were incubated in a humid chamber at 4°C O/N. Following washes with PBS 1×, cells were incubated with Alexa Fluor 488 conjugated goat anti-mouse, for 45 minutes at RT. After washes with PBS 1×, TIF assay was conducted as previously described (31) with minor modifications. Hep3B, Huh7, and Hep55–1C cells were seeded, treated, and stained with γH2AX. After PBS 1× washes, cells were fixed in 4% formaldehyde in PBS for 20 minutes at RT. The slides were sequentially dehydrated with 70%, 90%, 100% ethanol and subsequently denatured for 3 minutes at 80°C with 20 μL of hybridization mixture contained 70% deionized formamide, 1M Tris pH 7.2, 8.56% buffer MgCl2, 5% maleic blocking reagent, and 25 μg/mL Cy3-conjugated PNA Tel-C (CCCTAA)3 probe (PANAGENE) and incubated overnight at 4°C in a humid chamber. Slides were washed two times for 15 minutes in a wash solution containing 70% formamide, 10 mmol/L Tris pH 7.2, 0.1% BSA, and washed three times for 5 minutes in a solution containing 0.1M Tris pH 7.5, 0.15M NaCl, and 0.08% Tween-20. The slides were dehydrated by ethanol series, air-dried, and counterstained with Vectashield/DAPI (Vector Laboratories). Images were captured at 63× magnification with an Axio Imager Z2 (Carl Zeiss) equipped with an automatic capture system (Metafer; Metasystems) and analyzed with ISIS software (Metasystems).
Droplet digital TRAP assay (telomerase activity)
Quantitation of telomerase enzyme activity was performed as described in ref. 32.
Animal experiments
C57BL/6 female mice (6–8 weeks old) were purchased from Envigo (RRID: IMSR_ENV: HSD-044) or Jackson lab (RRID: IMSR_JAX:000664) and NSG mice were bred at UTSW in a limited-access breeding room staffed by UTSW Animal Resources Center employees. Original breeding pairs for this NSG breeding colony were purchased from Jackson Laboratory (RRID: IMSR_JAX:005557). Batf3−/− mice (RRID: IMSR_JAX:013755) in the C57BL/6 background were kindly provided by Jinming Gao from UT Southwestern Medical Center. Human-myc liver genetically engineered mouse model was provided by Maralice Conacci-Sorrell. All procedures and experiments involving mice were approved by The University of Texas Southwestern Institutional Animal Care and Use Committee and conducted as per institutional guidelines. A total of 3 × 106 Hep3B, 5 × 106 Hep55–1C, and 1 × 106 RIL175 cells were inoculated subcutaneously into the right dorsal flanks of female mice (NSG mice, C57BL/6 mice, C57BL/6 mice, respectively) in 100 μL RPMI:matrigel mixture. Tumor-bearing mice were randomly grouped into treatment groups when tumors grew to around 100 mm3. Tumor volumes were measured by the length (a), width (b), and height (h) and calculated as tumor volume = abh/2.
For Hep3B in vivo experiment, 3 mg/kg THIO was intraperitoneally given 3 consecutive days for over 2 weeks on days 26, 27, 28 and days 33, 34, 35. For anti-CD8 depletion in Hep55–1C in vivo model, anti-CD8 (200 μg) was intraperitoneally given on days 10, 14, 19, and on days 23, 26, and 30 whereas THIO (3 mg/kg) was given on days 11, 12, 13 and 20, 21, 22. For Batf3−/− mice (male and female) Hep55–1C in vivo experiment, THIO (3 mg/kg) was intraperitoneally given 3 consecutive days over 2 weeks on days 10, 11, 12 and days 18, 19, 20.
For sequential in vivo treatment experiments in Hep55–1C, 3 mg/kg THIO alone was intraperitoneally given 3 consecutive days over 2 weeks on days 8, 9, 10 and days 15, 16, 17. Libtayo alone (200 μg) was intraperitoneally given on days 10 and 17. For sequential treatment, 3 mg/kg THIO was given on days 8, 9, 10 and days 15, 16, 17 and 200 μg libtayo was intraperitoneally given on days 12 and 19. Mice weights were measured every 3 to 4 days. Tumor-free mice in THIO and THIO +libtayo groups were rechallenged on the left side of the mice with 107 Hep55–1C on day 114.
For sequential in vivo treatment experiments in Hep55–1C, 3 mg/kg THIO was intraperitoneally given 3 consecutive days over 2 weeks on days 7, 8, 9 and days 20, 21, 22. Anti-PDL1 (200 μg) was intraperitoneally given on days 7, 14, 21, and 28. 200 μg of anti-VEGF was intraperitoneally given on days 9, 12, 16, 19, 23, 26, and 30. 200 μg of anti-CD8 was given on days 6, 10, 13, 17, 20, 24, and 27.
For RIL175 in vivo experiment, 3 mg/kg THIO was intraperitoneally given 3 consecutive days over 2 weeks on days 11, 12, 13 and days 20, 21, 22. Anti-PDL1 (200 μg) was intraperitoneally given on days 11 and 19. For sequential treatments, 3 mg/kg THIO was given on days 11, 12, 13 and days 20, 21, 22 and 200 μg anti-PDL1 or anti-PDL1+anti-VEGF was intraperitoneally given on days 16 and 25. Tumor volumes were measured every 3 to 4 days.
Human-MYC liver cancer mouse model
The MYC-driven tumor model used was a Tet system to conditionally express human MYC in murine hepatocytes (33, 34). This mouse was generated by crossing TRE-MYC, which contains the tetracycline response element adjacent to the human MYC cDNA with the transgenic line LAP-tTA, containing a liver-specific enhancer that drives the expression of the tetracycline-transactivating protein. Male breeders with two copies of LAP-tTA and single copy of TRE-MYC was crossed with WT FVB female mice to generate LAP-tTA/TRE-MYC mice used for the experiments. The breeders were maintained in doxycycline water (1 mg/mL). The females with the litters were transferred to a fresh cage on the day of birth to induce MYC overexpression by removing doxycycline water. Treatment started when the mice were 31 days old and with tumor burden in the liver. 3 mg/kg THIO was intraperitoneally given on days 31, 32, and 33. 200 μg of anti-PDL1 was given on days 35, 42, and 200 μg r84 on days 37, 40, 44, and 47.
IHC
Formalin-fixed, paraffin-embedded tumor tissue sections (5 μmol/L thick) were evaluated using antibodies against CD4 (1:50; Cell Signaling Technology, Catalog No. 25229, RRID: AB_2798898), CD8α (1:200; Cell Signaling Technology, Catalog No. 98941, RRID: AB_2756376), CD11c (1:200; Cell Signaling Technology, Catalog No. 97585, RRID: AB_2800282), F4/80 (1:200; Cell Signaling Technology, Catalog No. 70076, RRID: AB_2799771), granzyme B (1:200; Cell Signaling Technology, Catalog No. 44153, RRID: AB_2857976), HMGB1 (1:200; Cell Signaling Technology, Catalog No. 6893, RRID: AB_10827882), p-STING (1:200; Cell Signaling Technology, Catalog No. 62912, RRID: AB_2799635), or p-TBK1 (1:200; Cell Signaling Technology, Catalog No. 5483, RRID: AB_10693472) according to our previous procedures (35). Briefly, slides were deparaffinized in xylene and rehydrated with decreasing concentrations of ethanol in water. Antigen retrieval was performed by preheating sodium citrate buffer (pH 6.0) in a microwave oven for 2 minutes, then incubating slides at sub-boiling temperature (95°–98°) for 10 minutes and cooling at room temperature for 30 minutes. Endogenous peroxidases were quenched by incubating slides in 3% hydrogen peroxide for 10 minutes. Sections were then washed twice with PBS for 5 minutes each. Endogenous avidin and biotin were blocked using 1× Animal-Free Blocking Solution (Cell Signaling Technology, Catalog No. 15019). Sections were then washed twice with PBS for 5 minutes each. Primary antibodies were applied overnight in a humidified chamber at 4°C. After rinsing slides in PBS, they were incubated in one to three drops of Signal Stain Boost IHC Detection Reagent (Cell Signaling Technology, Catalog No. 8114) for 30 minutes at room temperature. After washing three times with PBS for 5 minutes each, slides were incubated with Signal Stain DAB (Cell Signaling Technology, Catalog No. 8059) for 2 to 5 minutes, depending on the primary antibody. After washing in distilled water, sections were counterstained with hematoxylin (Cell Signaling Technology, Catalog No. 14166), dehydrated by ethanol and xylene, and cover slipped using SignalStain Mounting Medium (Cell Signaling Technology, Catalog No. 14177). Images were collected using an EVOS microscope (Invitrogen).
HMGB1 release assay
HMGB1 released into cell culture supernatants was evaluated by Western blot analysis or enzyme-linked immuno-absorbent assay kits (Shino-Test Corporation, Catalog No. 326054329) according to the manufacturer's instructions. Serum HMGB1 was evaluated by enzyme-linked immuno-absorbent assay kits from the Shino-Test Corporation.
Western blot analysis
Proteins from cell lysates or supernatants were resolved on 4% to 12% Criterion TX Bis-Tris gels (Bio-Rad, Catalog No. 3450124) and transferred to a nitrocellulose membrane (Bio-Rad, Catalog No. 1620233) using the Trans-Blot Turbo Transfer Pack and System (Bio-Rad). Membranes were blocked with TBST buffer containing 5% skim milk (Cell Signaling Technology, Catalog No. 9999) for 1 hour and incubated overnight at 4°C with various primary antibodies (1:1,000). Following three washes in TBST, membranes were incubated with goat anti-rabbit or anti-mouse IgG HRP secondary antibody (1:1,000; Cell Signaling Technology, Catalog No. 7074, RRID: AB_2099233 or Cell Signaling Technology, Catalog No. 7076, RRID: AB_330924) at room temperature for 1 hour and washed. Chemiluminescence substrate was applied using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Catalog No. 34580) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Catalog No. 34095), and blots were analyzed using the ChemiDoc Touch Imaging System (Bio-Rad) and Image Lab Software (Bio-Rad).
RNA-sequencing (RNA-seq)
RNA-seq was performed by Genewiz and the data processing was done using Trim Galore Trim Galore (RRID: SCR_011847; https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) for quality and adapter trimming. The mouse reference genome sequence and gene annotation data, mm10, were downloaded from Illumina iGenomes (https://support.illumina.com/sequencing/sequencing_software/igenome.html). The quality of RNA-seq libraries was estimated by mapping the reads onto mouse transcript and ribosomal RNA sequences (Ensembl release 89) using Bowtie (v2.3.4.3). STAR (v2.7.2b) and was set to align the reads onto the mouse genome. SAMtools (v1.9; RRID: SCR_002105) was used to sort the alignments, and HTSeq Python package (RRID:SCR_005514) was used to count reads per gene. DESeq2 R Bioconductor package (RRID: SCR_015687) was used to normalize read counts and identify differentially expressed genes. KEGG pathway data were downloaded using KEGG API (https://www.kegg.jp/kegg/rest/keggapi.html) and gene ontology (GO) data were downloaded from NCBI FTP (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2go.gz). The enrichment of DE genes to pathways and GOs were calculated by Fisher exact test in R statistical package. The reads were aligned to mouse RefSeq RNA sequences using the Burrows–Wheeler Aligner (BWA, v0.7.17; RRID: SCR_010910) and the MEM algorithm with the options, -T 19 -h 200 -Y, and primary alignments with proper read pairs were selected by SAMtools with the options, -f 3 -F 2316, to count the reads aligned to ribosomal RNA sequences defined by the sequence descriptions containing the keyword of ribosomal RNA or rRNA. The reads were aligned to human MYC RNA isoform 1 sequence (NM_002467.6) using BWA with MEM algorithm. The alignments were compared with the alignments to mouse genomic and RNA sequences from NCBI Assembly, GRCm38.p6, using a custom Perl script named REMOCON (https://github.com/jiwoongbio/REMOCON) and the reads with better alignments to human MYC were counted.
Digital spatial profiling (DSP) of immuno-oncology-related proteins
Spatial profiling of immuno-oncology-related proteins was performed in the Hep55–1C mouse tumor tissues by using DSP using the GeoMx platform (NanoString; ref. 36) according to the manufacturer's protocol. Mice were treated one cycle with THIO monotherapy (3 mg/kg, days 6, 7, 8), combination of anti-PDL1 (atezolizumab, 200 μg) and anti-VEGF (r84, 200 μg; on day 8) and combination of THIO (days 6, 7, 8), anti-PDL1 (day 10), anti-VEGF (day 10). Tumors were collected 5 days after the last injection for DSP. First, immunofluorescent staining of 5-μm-thick formalin-fixed paraffin-embedded tumor tissues were performed using the following morphology markers for the segment selection: anti-CD3 (Bio-Rad, Catalog No. MCA1477A647, RRID: AB_10841760), anti-CD45 (Novus Biologicals, Catalog No. NBP1–44763AF594), and anti-pan-cytokeratin (Pan-CK; GeoMx Solid Tumor TME Morphology Kit, NanoString). Antibodies for 35 immuno-oncology-related proteins and three housekeeping proteins (S6, Histone H3, GAPDH) conjugated with UV-cleavable capture probes (Supplementary Table S1) were hybridized to the tissues overnight. Three to five representative regions of interest (ROI) were selected in each tissue in the Digital Spatial Profiler software. Within each ROI, areas enriched for cell types of interest were defined as “segments” based on the immunofluorescent staining, and abundance of the immuno-oncology-related proteins was measured in each segment as follows. T-cell-enriched segments were first selected as CD3-positive area (CD3+), from which hybridized capture probes were collected by UV exposure. Subsequently, segments of other leukocytes were selected as CD45-positive segments (CD3−/CD45+) and the capture probes were similarly collected. Finally, Pan-CK-positive or rest of the areas (CD3−/CD45−/Pan-CK±) were profiled as segments of HCC/stromal cells. Raw abundance of each protein was measured by using the nCounter SPRINT Profiler (NanoString) and normalized by scaling with geometric mean of the housekeeping proteins using GeoMx DSP system (NanoString).
Statistical analysis
All the data analyses were performed with GraphPad Prism (RRID: SCR_002798) statistical software. P value determined by two-way ANOVA for tumor growth and shown as mean ± SEM or log-rank test for survival or unpaired two-tailed t tests for other analysis. Welch correction (****, P < 0.0001) was used between control and THIO samples in TIF assay. A value of P < 0.05 was considered statistically significant.
Data availability
The DSP data generated in this study are publicly available in Gene Expression Omnibus (GEO; RRID: SCR_005012) at GSE219108. The raw data are available on request from the corresponding author.
Results
Reduced cell viability and induced telomere dysfunction by THIO in human and mouse liver cancer cells
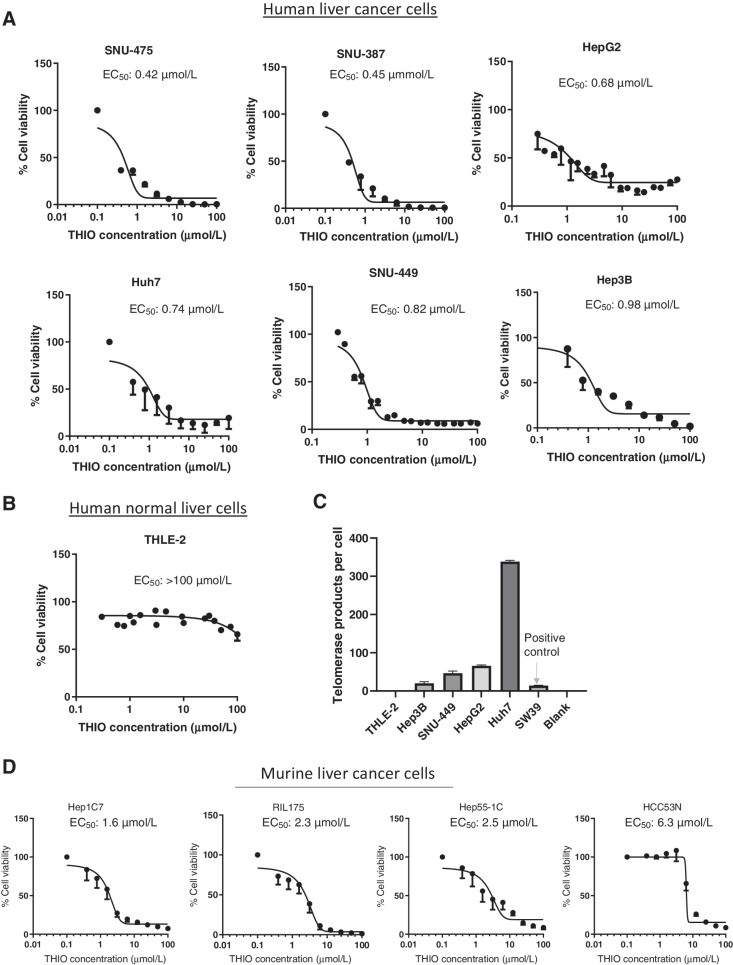
To explore whether THIO is effective in killing liver cancer cells, we first determined the inhibition of cell viability by THIO in a panel of human and mouse liver cancer cells. Although half maximal effective concentration (EC50) was 0.42, 0.45, 0.68, 0.74, 0.82, and 0.98 μmol/L in SNU-475, SNU-387, HepG2, Huh7, SNU-449, and Hep3B in telomerase positive human cancer cell lines, respectively (Fig. 1A), EC50 was over 100 μmol/L in THLE2, a telomerase negative human normal liver epithelial cell line derived from normal liver (Fig. 1B and C). EC50 values were also determined in Hep1C7, RIL175, Hep55–1C, and HCC53N mouse cancer cell lines, and they were 1.6, 2.3, 2.5, and 6.3 μmol/L, respectively (Fig. 1D). To evaluate whether THIO induces damage of telomeres in liver cancer cells, we treated Hep55–1C (mouse cancer cells), Huh7, and Hep3B (human cancer cells) at EC50 concentrations of THIO for each cell line for 48 hours. As expected, THIO significantly induced telomere damage (telomere induced dysfunctional foci, TIF) in all three cell lines tested (Supplementary Fig. S1A).
Figure 1.
THIO shows potent activity against telomerase positive human and murine HCC Cells. A and B, Cell viability of human liver cancer and normal epithelial cells treated with the indicated concentrations of THIO for 3 to 5 days. C, Telomerase activity of human liver cancer and normal epithelial cells. D, Cell viability of murine HCC cells treated with the indicated concentrations of THIO for 3 to 5 days. Cell viability was measured using the CellTiter-Glo Assay. Samples were analyzed in duplicate or triplicate, and EC50 values were calculated using Graphpad Prism.
The role of CD8+ T cells and DCs in therapeutic efficacy of THIO
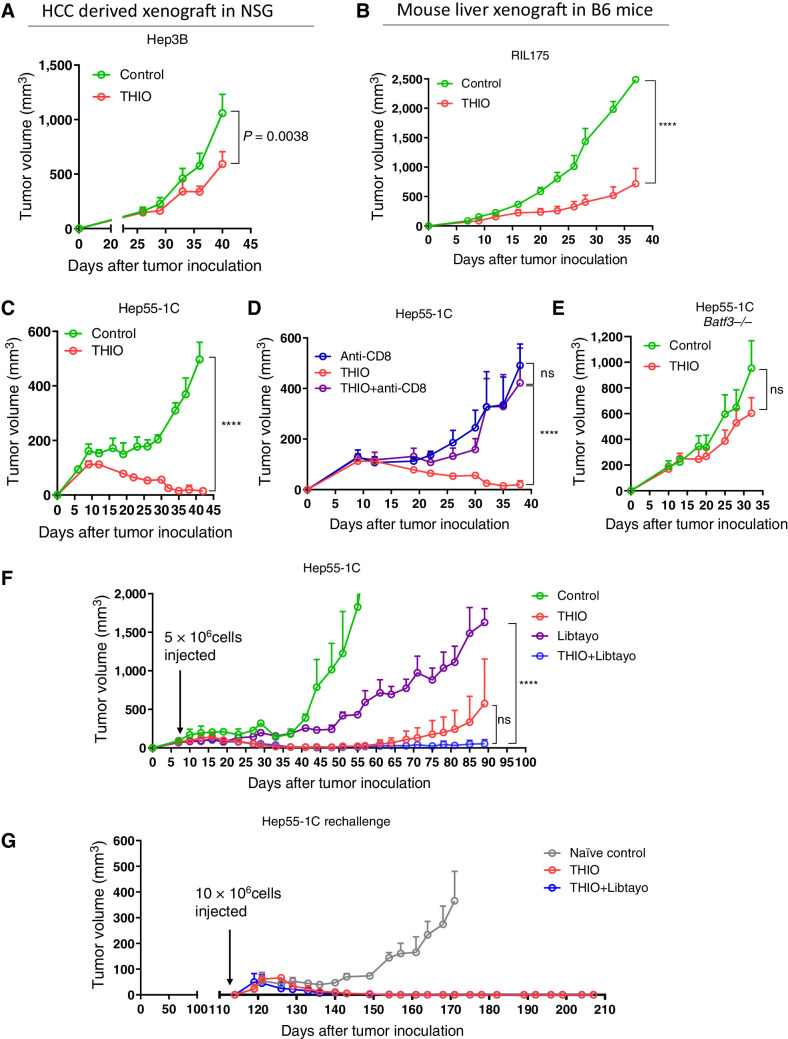
Our previous studies with xenograft models showed tumor growth control with intensive THIO treatment (daily or every other day for over 10 days; refs. 25, 26). However, we recently showed the role of THIO in the adaptive immune system, and how it controls tumor growth by modulating cellular immunity. We used Rag1 knockout mice, which do not generate mature T and B cells. The therapeutic effect of THIO was abolished in the MC38 tumor model in Rag1 knockout mice, indicating adaptive immune cells are largely required for tumor control in vivo. In addition, in the same study, we tested THIO in HCT116 cells derived colon tumor model in immunodeficient mice. In this immunodeficient mouse model, we did not observe any significant differences in THIO treated mice compared with the control group, whereas experiments in humanized mice showed significant difference in tumor growth reduction between the control and THIO-treated groups (30). Here we first evaluated if THIO reduces tumor burden more in syngeneic mouse models compared with immune-deficient HCC mouse models. As an immunocompromised mouse model, we used NSG mice that are immunodeficient, lacking functional/mature T, B, and NK cells, as well as having many defects in innate immunity. As a syngeneic mouse model, we used C57BL/6 mice that are fully immunocompetent. We subcutaneously inoculated Hep3B human liver cancer cells into NSG mice and 26 days after tumor inoculation (when the tumor volume was ∼100 mm3), 3 mg/kg THIO was administered daily intraperitoneally for only 3 consecutive days, and then we analyzed tumor growth for 2 weeks. We subcutaneously inoculated RIL175 and Hep55–1C mouse liver cancer cells into C57BL/6 mice and 8 to 10 days after tumor inoculation (when the tumor volume was ∼100 mm3), 3 mg/kg THIO was administered daily for only 3 consecutive days and tumor volume was monitored for 2 weeks. Tumor growth was significantly reduced in immunocompetent models, whereas tumor growth was only slightly reduced over control levels in the immunocompromised mice (Fig. 2A–C).
Figure 2.
Sequential treatment with THIO and Libtayo Overcomes Checkpoint Inhibitor Resistance and Induces Immune Memory. Antitumor effects of THIO were greater in immunocompetent mice than immunocompromised mice. A total of 3 × 106 Hep3B cells (n = 6; A) or 1 × 106 RIL175 (n = 5; B), or 5 × 106 Hep55–1C (n = 3–5; C--F) were injected subcutaneously into the right dorsal flanks of NSG mice (A) C57BL/6 mice (B–D, F), or C57BL/6 Batf3−/− mice (E). When tumors reached ∼100 mm3, tumor-bearing mice were randomly assigned to control or THIO (3 mg/kg i.p. on 3 consecutive days for 2 weeks) with or without anti-CD8 antibody (200 μg administered 1 day before treatment initiation and then twice/week for 3 weeks; n = 5). F, When tumors reached ∼100 mm3, tumor-bearing mice were randomly assigned to treatment with THIO (3 mg/kg; n = 3), libtayo (cemiplimab, 200 μg; n = 3), or sequential THIO and libtayo (n = 3). THIO was given on days 8, 9, 10 and 15, 16, 17 and libtayo was given on days 12 and 19 after tumor inoculation. G, On day 114, tumor-free mice from THIO (n = 2) and THIO+libtayo (n = 2) groups and control naive mice (n = 3) were re-challenged with 1 × 107 Hep55–1C tumor cells in the opposite (left) flank. Tumor growth was measured every 3 days. Tumor volumes were calculated using the formula abh/2, where a = length, b = width, and h = height of the tumor. P value was determined by two-way ANOVA for tumor growth and shown as mean ± SEM. P = 0.0038, P < 0.0001, and P < 0.05, significant; ns, nonsignificant.
To determine if CD8+ T cells contribute to the THIO-mediated antitumor effect, we depleted CD8+ T cells while giving THIO treatment in wild-type B6 mice. Depletion of CD8+ T cells abolished the therapeutic effect of THIO (Fig. 2D). These indicate that the immune T-cell system contributes to the therapeutic efficacy of THIO. Basic leucine zipper ATF-like transcription factor 3 (BATF3)-dependent DCs are critical for the priming of antigen-specific CD8+ T cells (37). THIO treatment in Batf3−/− mice abolished the effect of THIO, suggesting an important role of BATF3-dependent DCs in the therapeutic effect of THIO in the Hep55–1C syngeneic mouse model (Fig. 2E).
THIO overcomes immunotherapy resistance in advanced tumors
PD1/PDL1 interactions contribute to functional T-cell impairment (38) and immunotherapies against this interaction significantly enhance T-cell responses facilitating antitumor activity (39). Because THIO plays an important role in T-cell activation, we next evaluated the immunomodulatory effects of THIO when sequentially combined with immunotherapy. Thus, we tested the therapeutic efficacy of a mAb targeting PD1 molecule, cemiplimab (libtayo), when sequentially combined with THIO. In such an advanced tumor, there was limited tumor growth inhibition by two treatments with libtayo as a monotherapy. In contrast, THIO alone was able to delay tumor growth resulting in 66.6% of mice becoming completely tumor-free. Sequential treatment with THIO and libtayo completely inhibited tumor growth in all mice (Fig. 2F). Even though statistical differences were not significant between THIO and THIO plus libtayo combination, this trend shows that sequential therapy may improve the efficacy of libtayo treatment. In addition, we did not observe any body weight loss in mice in the sequential treatment group (Supplementary Fig. S2A). We rechallenged tumor-free mice both from THIO alone and sequential treatment groups to check the immune memory response. All mice in both groups spontaneously rejected Hep55–1C tumors and induced durable antitumor immunologic memory in a syngeneic mouse model (Fig. 2G). On the basis of these results, THIO appears to enhance the efficacy of immunotherapy in advanced HCC tumors, and this will potentially benefit PD1/PDL1 blockade-resistant patients with HCC.
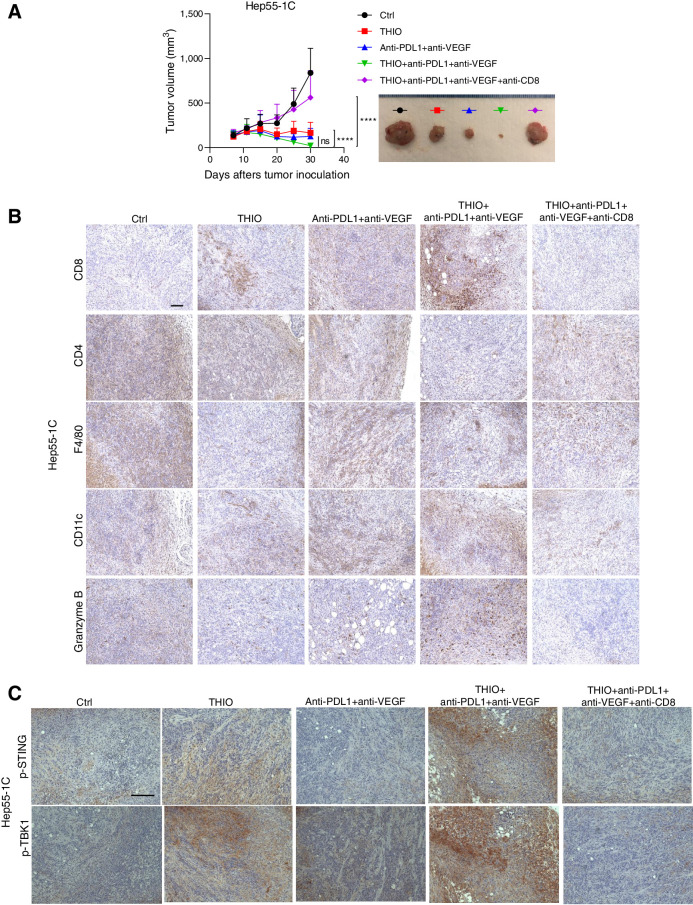
Atezolizumab and bevacizumab is approved by the FDA for patients with HCC. We next determined if THIO improves the efficacy of anti-PDL1+anti-VEGF in vivo. We used the Hep55–1C liver syngeneic mouse model and compared anti-PDL1+anti-VEGF with THIO+anti-PDL1+anti-VEGF. Although the difference between groups was not statistically significant, THIO trended to improve the efficacy of anti-PDL1+anti-VEGF (Fig. 3A). Moreover, this combination therapy significantly increased the expression of CD8 and granzyme B, showing the dependency of antitumor effect of THIO and anti-PDL1/anti-VEGF to CD8+ T cells (Fig. 3B; Supplementary Fig. S3A).
Figure 3.
THIO reduces tumor burden in vivo through activation of a CD8+ antitumor immune response. A, A total of 5 × 106 Hep55–1C cells were injected subcutaneously into the right dorsal flanks of C57BL/6 mice (n = 10/group). When tumors reached ∼100 mm3, tumor-bearing mice were randomly assigned to control, THIO or anti-PDL1 plus anti-VEGF or THIO plus anti-PDL1 plus anti-VEGF, or THIO plus anti-PDL1 plus anti-VEGF plus anti-CD8. 3 mg/kg THIO was intraperitoneally given 3 consecutive days for 2 weeks on days 7, 8, 9, and 20, 21, and 22; 200 μg anti-PDL1 (BioCell, Catalog No. BE0101) was intraperitoneally given on days 7, 14, 21, and 28; 200 μg anti-VEGF (BioCell, Catalog No. BE0060) was intraperitoneally given on days 9, 12, 16, 19, 23, 26, and 30; 200 μg anti-CD8 was given on days 6, 10, 13, 17, 20, 24, and 27. IHC staining of CD8, CD4, F4/80, CD11c, Granzyme B (B) and p-STING and p-TBK1 (C). Scale bars of all panels in these B and C are the same, 100 μm. Tissues are from mice treated with THIO or anti-PDL1 plus anti-VEGF or THIO plus anti-PDL1 plus anti-VEGF or THIO plus anti-PDL1 plus anti-VEGF plus anti-CD8. P value determined by two-way ANOVA for tumor growth and shown as mean ± SEM. P < 0.0001, significant.
We previously showed that tumor-derived DNA damage triggers the cytosolic DNA sensing cGAS/STING pathway and activates the IFN-I pathway (30), suggesting an essential role of the STING pathway in THIO triggered innate sensing. In previous studies, we also showed the essential role of host STING signaling in THIO-treated innate sensing (30). Here, we further evaluated the role of STING activation in a HCC model. We observed an increase of STING and TBK1 phosphorylation in THIO treated Hep55–1C tumors and even more pronounced phosphorylation observed in THIO+anti-PDL1+anti-VEGF tumors. Surprisingly, these effects were completely diminished when we depleted CD8+ T cells while giving THIO or THIO plus anti-PDL1 plus anti-VEGF treatment (Fig. 3C; Supplementary Fig. S3B), indicating an essential role of CD8+ T cells in mediating HCC tumor control.
To gain insight into immune landscape underlying the therapeutic response, DSP of 35 immuno-oncology-related proteins was performed in the implanted Hep55–1C tissues from the mice treated with THIO alone, anti-PDL1/anti-VEGF, or THIO in combination with the anti-PDL1/anti-VEGF therapy (Supplementary Fig. S4A). Compared with the vehicle control group, immune checkpoint proteins, PD1/PDL1 and CTLA4-related proteins were induced in the treatment arms. THIO monotherapy increased CTLA-4 in T cells, potentially limiting T-cell activation. However, combination of THIO with anti-PDL1/anti-VEGF did not increase CTLA-4 and can be one of the reasons of increased antitumor immunity. Interestingly, another set of immune checkpoint proteins, that is, T-cell immunoglobulin mucin-3 (Tim-3), lymphocyte activating 3 (LAG3), and glucocorticoid-induced tumor necrosis factor receptor-related protein (GITR), were distinctly induced when THIO was combined with anti-PDL1/anti-VEGF therapy (Supplementary Figs. S4B and S5A). Of note, abundance of the indicators of T-cell-mediated cytotoxicity, that is, granzyme B (GZMB), CD40L, and inducible T-cell costimulator (ICOS, a.k.a. CD278), was the highest in the THIO plus anti-PDL1/anti-VEGF group, providing an additional rationale of the triple combination therapy. Markers of immune-suppressive regulatory T cells [forkhead box P3 (FOXP3)] and myeloid-derived-suppressor cell (MDSC; CD11b and Ly6G/C) were reduced in THIO monotherapy and combination therapies. These observations collectively support enhanced T-cell cytotoxicity and mitigated immune-suppressive tissue microenvironment with the combination therapies with THIO. The distinct pattern of induced immune checkpoint proteins may inform further improvement of therapeutic efficacy in future studies. However, it is important to note that analysis of large datasets with screening technologies produce noise and require independent validation using additional technologies.
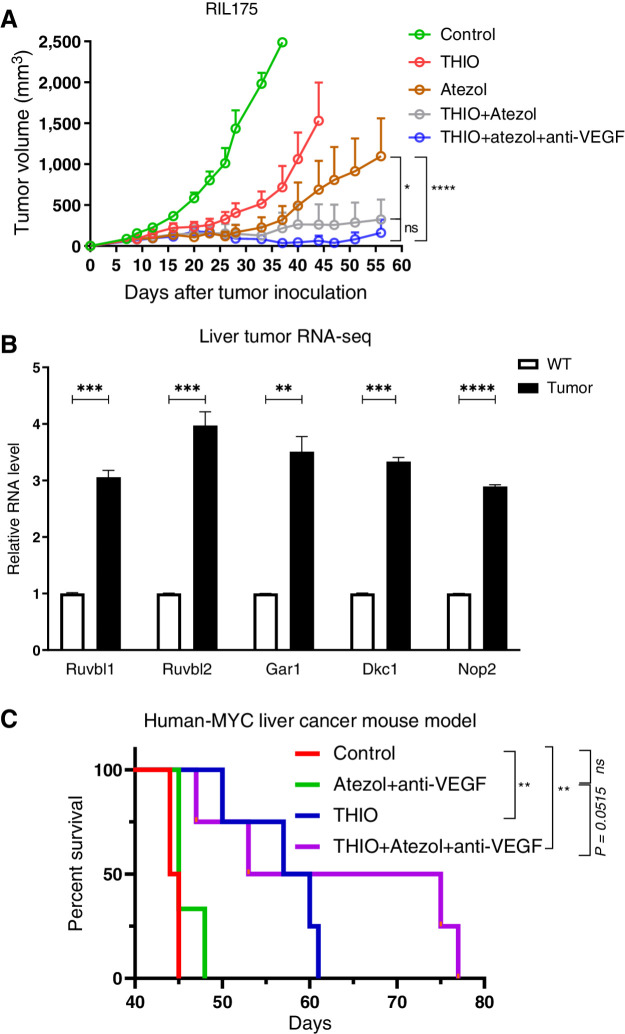
We next tested another liver syngeneic mouse model RIL175 to determine the therapeutic response of sequential therapy. We showed reduced tumor burden, higher tumor-free mice (80%) following THIO plus atezolizumab plus anti-VEGF and/or THIO plus atezolizumab (60% tumor-free mice) compared with atezolizumab (40% tumor-free mice) and THIO (20% tumor-free mice) groups in the RIL175 syngeneic mouse model at the end of day 56. Even though the number of tumor-free mice was higher in THIO plus atezolizumab plus anti-VEGF, the statistical difference between THIO plus atezolizumab plus anti-VEGF and THIO plus atezolizumab was not significant, showing the efficacy of both treatment options in the RIL175 liver model (Fig. 4A). We continued to monitor tumor-free mice in both treatment groups and found one tumor in THIO plus atezolizumab plus anti-VEGF group started to come back at day 77. However, 60% of mice were still tumor-free in both THIO plus atezolizumab and THIO plus atezolizumab plus anti-VEGF groups.
Figure 4.
Sequential treatment with THIO or anti-PDL1 monotherapy, anti-PDL1+anti-VEGF, or THIO+anti-PDL1+anti-VEGF improve therapeutic responses and overall survival. A, A total of 1 × 106 RIL175 were injected subcutaneously into the right dorsal flanks of C57BL/6 mice. When tumors reached ∼100 mm3, tumor-bearing mice were randomly assigned to control, THIO or anti-PDL1 (Atezol) monotherapy, THIO plus anti-PDL1, or THIO plus anti-PDL1 plus anti-VEGF (r84; n = 5/group). 3 mg/kg THIO was intraperitoneally given 3 consecutive days over 2 weeks on days 11, 12, 13 and 20, 21, 22; 200 μg anti-PDL1 and 200 μg anti-VEGF was intraperitoneally given 3 to 4 days after last injection of THIO. Tumor growth was measured every 3 days. Tumor volumes were calculated using the formula abh/2, where a is the length, b is the width, and h is the height of the tumor. The final tumor measurement from the mice that were euthanized due to tumor size are included in the graph as other tumors were continued to be measured in the same group until day 56. P value determined by two-way ANOVA for tumor growth and shown as mean ± SEM (*, P = 0.0414; ****, P < 0.0001, significant; ns, nonsignificant). B, Relative mRNA expression of indicated genes plotted from RNA-seq in control (n = 2) and MYC-expressing livers (n = 3). P values were determined by multiple unpaired t tests. P values for Ruvbl1, Ruvbl2, Gar1, Dkc1, Nop2 between WT and tumor are ***P = 0.000192, ***P = 0.000481, **P = 0.001096, ***P = 0.000029, and ***P = 0.000004, respectively. C, Treatment started when the mice were 31 days old. 3 mg/kg THIO was intraperitoneally given on days 31, 32, and 33. 200 μg of anti-PDL1 was given on days 35, 42 and 200 μg r84 on days 37, 40, 44, 47; P value determined by log-rank test for survival analysis. **P = 0.0084 between control (n = 4) vs. THIO+atezol+anti-VEGF (n = 4), P = 0.0515 (ns) between atezol+anti-VEGF (n = 3) vs. THIO plus atezol plus anti-VEGF (n = 4), **P = 0.0084 between control (n = 4) vs. THIO (n = 4), P > 0.9999 (ns) between control vs. atezol+anti-VEGF.
MYC-driven tumors display an upregulation in the mRNA levels of genes that encode for telomerase accessory proteins (Ruvbl1, Ruvbl2, Gar1, Dkc1, Nop2; Fig. 4B), suggesting that MYC-driven liver tumors have an upregulation in telomerase activity. Given that increased in MYC levels is correlated with poor outcome of patients with HCC. Strategies to use telomerase-mediated telomere targeted therapy alone (e.g., THIO) or in combination with other drugs may enhance treatment of these patients. To test this hypothesis, we used a human-MYC liver cancer mouse model and tested THIO alone and together with atezolizumab plus anti-VEGF. We showed an increased overall survival following THIO plus atezolizumab plus anti-VEGF treatment compared with THIO monotherapy and/or atezolizumab plus anti-VEGF groups in the human-MYC liver cancer mouse model (Fig. 4C).
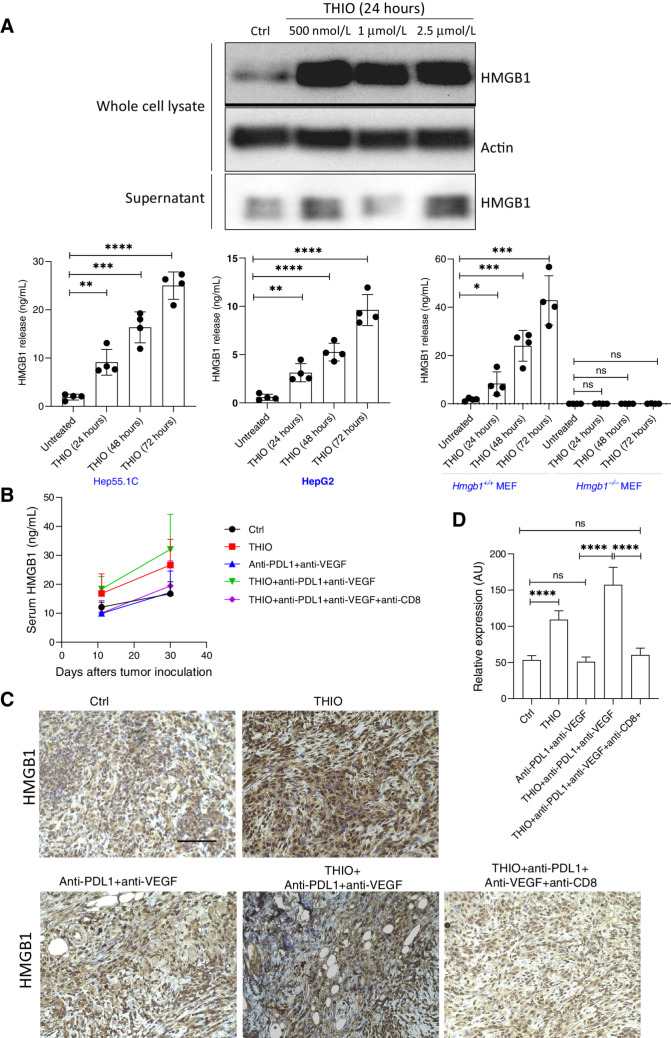
Induced HMGB1 release in THIO-treated tumors
High-mobility group box 1 protein (HMGB1) release from dead or dying cells is characterized with immunogenic cell death induced by anticancer therapies (40). HMGB1 is the most abundant nonhistone nuclear protein and extracellular HMGB1 acts as a prototypical endogenous damage associated molecular pattern (DAMP) in eliciting adaptive immunity (41). Once HMGB1 is secreted, it engages the Toll-like receptor (TLR4) on DCs to accelerate the processing of phagocytic cargo in the DC and facilitate antigen presentation by DCs to T cells (42). Here, we investigated the role of HMGB1 in THIO-treated tumors due to the role of THIO in antitumor immune responses. We found that THIO induced the release of HMGB1 in the Hep55–1 murine and HepG2 human liver cancer cells in a time-dependent manner (Fig. 5A). HMGB1 release was not observed in HMGB1−/− MEFs in vitro (Fig. 5A). The loss of HMGB1 inhibited THIO-induced HMGB1 release, suggesting a potential role of HMGB1 in THIO-induced cell death. HMGB1 released by cancer cells is known to be processed by APCs, such as DCs and macrophages, and they produce interleukins such as IL12 and IL15 to promote the cytotoxic effect of NK cells, cytotoxic T lymphocytes, and induce IFNγ release (43). Our in vitro results indicated that HMGB1 released from Hep55–1C and HepG2 dying cells may mediate immunogenic cell death during THIO treatment. Therefore, we next determined if THIO also induces HMGB1 release in vivo by comparing serum HMGB1 release and HMGB1 expression in control, THIO, anti-PDL1 plus anti-VEGF and THIO plus anti-PDL1 plus anti-VEGF cohorts. We observed that THIO and THIO plus anti-PDL1 plus anti-VEGF treatments induced HMGB1 release (Fig. 5B) and HMGB1 protein expression (Fig. 5C and D) compared with control and anti-PDL1 plus anti-VEGF treatments. However, the statistical differences between groups in Fig. 5B are not significant, but there is a trend that THIO and THIO plus anti-PDL1 plus anti-VEGF treatments induce HMGB1 release. Interestingly, when we depleted CD8+ T cells, the HMGB1 level decreased to untreated control or anti-PDL1 plus anti-VEGF serum/expression baseline levels. Because interactions between HMGB1 released by dying tumor cells and TLR4 in DCs are important for the cross-presentation of tumor antigens to T cells and success of anticancer therapies, these results suggest that HMGB1 can be one of the important DAMP in THIO-mediated anticancer immune responses.
Figure 5.
THIO induces HMGB1 release in vivo and in vitro. A, Western blot analysis of HMGB1 in THIO-treated Hep55–1C samples (500 nmol/L, 1 μmol/L, and 2.5 μmol/L at 24 hours). Whole cell lysate and supernatant were collected following THIO treatment at indicated concentrations and time. Actin was used a loading control. HMGB1 release in THIO-treated samples at 24, 48 and 72 hours with ELISA Kit in Hep55–1C, HepG2, Hmgb1+/+MEF, and Hmgb1−/−MEF. P values were determined by unpaired t tests. P values between untreated vs. THIO (24, 48, and 72 hours) in Hep55–1C are **, P = 0.0019; ***, P = 0.0001; ****, P < 0.0001; respectively. P values between untreated vs. THIO (24, 48, and 72 hours) in HepG2 are **, P = 0.0020; ****, P < 0.0001; ****, P < 0.0001; respectively. P values between untreated vs. THIO (24, 48, and 72 hours) in Hmgb1+/+ MEF are *, P = 0.0355; ***, P = 0.0004; ***, P < 0.0002; respectively. P values between untreated vs. THIO (24, 48, and 72 hours) in Hmgb1−/− MEF are not significant (ns). B, HMGB1 release in THIO or anti-PDL1 (BioCell, Catalog No. BE0101) plus anti-VEGF (BioCell, Catalog No. BE0060) or THIO plus anti-PDL1 plus anti-VEGF or THIO plus anti-PDL1 plus anti-VEGF plus anti-CD8 treated samples with ELISA Kit (n = 4/group). The statistical differences between groups in this figure are not significant. C, IHC staining of HMGB1 in tumor tissues that were treated with THIO, anti-PDL1 plus anti-VEGF, THIO plus anti-PDL1 plus anti-VEGF or THIO plus anti-PDL1 plus anti-VEGF plus anti-CD8. Scale bar of all panels in this figure is the same, 50 μm. D, The quantification of IHC data on HMBG1 expression level is shown. P values were determined by unpaired t tests. P values between control vs. THIO, control vs. anti-PDL1+anti-VEGF, anti-PDL1+anti-VEGF vs. THIO+anti-PDL1+anti-VEGF, control vs. THIO+anti-PDL1+anti-VEGF+anti-CD8+, THIO+anti-PDL1+anti-VEGF vs. THIO+anti-PDL1+anti-VEGF+anti-CD8+ are ****P < 0.0001, P = 0.5410 (ns), ****P < 0.0001, P = 0.2086 (ns), and P = 0.5410 (ns), respectively.
Discussion
Telomeres are (TTAGGG)n DNA repeats located at the ends of chromosomes where with shelterin proteins they protect chromosome ends from being recognized as DNA damage structures (44). The telomere-synthesizing enzyme telomerase is silenced in most human somatic cells, ultimately resulting in replicative senescence, whereas most (>85%) human tumors exhibit telomerase activation, allowing continued cell proliferation. Therefore, telomerase is a highly attractive target for anticancer therapy (reviewed in ref. 45). We recently showed that a modified nucleoside analogue 6-thio-2′deoxyguanosine (6-thio-dG, THIO) is a telomerase-mediated telomere targeting small molecule. Thus, the 5′-triphosphate of THIO is efficiently recognized by telomerase and incorporated into de novo synthesized telomeres inducing telomere damage, resulting in cell death for almost all cancer cells, but not normal telomerase silent cells (25). THIO also induces immunogenic cell death in colon and lung cancer mouse models in vivo (30). Here, we investigated the therapeutic effect of THIO in HCC. HCC is the most common primary liver cancer type in the United States. Although the combination of atezolizumab (PDL1 inhibitor) and bevacizumab (angiogenesis inhibitor) has become a successful treatment for advanced HCC, disease recurrence and metastasis remain the foremost concern for almost all patients with liver cancer (46).
Tyrosine kinase inhibitors are used for treatment of patients with advanced HCC. Sorafenib and Lenvatinib are approved as first-line and regorafenib and cabozantinib are used as second-line therapies. THIO is currently in phase II clinical trials in where it is used in sequential combination with an immune checkpoint inhibitor. One of our goals is to make immunologically “cold” tumors into “hot” tumors, by changing the tumor microenvironment and activating innate and adaptive patient immune systems with THIO. Therefore, in clinical trials we lead with THIO treatment, and then follow with immune checkpoint inhibitor(s). We do not believe that tyrosine kinase inhibitors will add to the immune activation functions of THIO. However, we previously showed THIO can overcome EGFR tyrosine kinase inhibitor resistance in NSCLC (26).
The concept of immunogenic cell death in cancer that is triggered by cytotoxic agents has become the interest of researchers due to antitumor specific T-cell immune responses in preclinical studies (30, 47, 48). Some anticancer treatments, such as ionizing radiation, chemotherapy, and telomere targeted therapy are far more efficient when applied to immunocompetent mice compared with immunodeficient mice developing tumors from the same cellular origin. This concept prompted the search for the molecular links between tumor cell death, stimulation of pathogen recognition receptors, innate, and adaptive immunity (30, 49). In this study, we showed antitumor immune effects of THIO in HCC using immunocompetent mice. DCs and CD8+ T cells have an important role in THIO-associated activity. We also showed the effect of sequential treatment of THIO and/or anti-PD1, anti-PDL1, anti-VEGF in different mouse models and importantly that this effect was abolished when CD8+ T cells were depleted. Moreover, tumor-free mice from these treatment groups with THIO developed antitumor immune memory, indicating acquired cellular immunity against rechallenging liver tumor cell injections.
The cGAS–STING pathway is an important DNA-sensing machinery in innate immunity. cGAS (cyclic GMP–AMP synthase) is an innate cytosolic DNA sensor and activation of cGAS stimulates STING (stimulator of interferon genes) protein to trigger IFN signaling. STING recruits TANK binding kinase-1 (TBK1) and IFN regulatory factor 3 (IRF3). IRF3 translocates to the nucleus and exerts its transcriptional function in immune-stimulated genes (ISG) and type I IFN (50). We showed the phosphorylation of STING and TBK1 in Hep55–1C murine liver tumors following THIO and THIO plus anti-PDL1 plus anti-VEGF. This activation was lost following CD8+ T-cell depletion, showing the role of the cGAS–STING pathway and the interaction of innate sensing and adaptive immunity in THIO treatment.
Preclinical studies showed that dying tumor cells can release a series of danger signals, namely DAMPs. These signals contribute to the recruitment and activation of antigen presenting cells leading to propagation of antitumoral immunity to enhance therapeutic efficacy (40). The release of HMGB1 is a well-characterized endogenous mediator of anticancer immune responses. Such an immune response involves a complex hierarchy of immune effectors, including monocyte-derived DCs producing IL1β, γ/δ T cells producing IL17 and conventional CD8+ α/β T cells producing IFNγ (42, 51–53). HMGB1 is a highly conserved chromosome-binding protein that acts as a DNA chaperone to maintain genome stability. In addition to its nuclear role, HMGB1 protein can also be actively secreted by inflammatory cells or passively released by dead or dying cells to mediate immune responses (54–56). Previous studies also suggest that HMGB1 can amplify cGAS/STING activation during DNA stress (57). Telomere function is tightly linked to chromatin structure and many of the chromatin structural changes are mediated by HMG proteins, including HMGB1, HMGB2, HMGB3, and HMGB4. Previous studies showed reduced telomerase activity and telomere dysfunction in HMGB1 knockout mouse embryonic fibroblasts (58). The absence of HMGB1 expression by dying tumor cells exposed to anthracyclines or oxaliplatin compromises DC-dependent T-cell priming by tumor-associated antigens (49). Because we showed DC-dependent T-cell priming by THIO, we tested for the presence of HMGB1 in dying tumor cells exposed to THIO. We showed HMGB1 presence in THIO treated samples, suggesting that HMGB1 may play a role in DC-dependent T-cell priming.
There are other factors which play a role in the anticancer effects of THIO. One of them is the conversion of THIO into corresponding 5′-triphosphate, as well as biochemical stability of this triphosphate in cells. 6-Thio-2′-deoxyguanosine 5′-triphosphate (6-thio-dGTP) is the actual telomerase substrate, which is incorporated into de novo synthesized DNA. As a result of this process, guanine bases are being substituted with 6-thioguanine bases, and this substitution leads to DNA damage and cell death. In addition, the induction of both general DNA damage and telomere damage may potentiate the overall effect of THIO in telomerase positive cancer cells and is another reason why we do not see a direct correlation between EC50 values and telomerase activity in HCC cells.
Overall, our results reveal the anti-tumor immune response role of THIO as a telomerase-dependent telomere targeting therapeutic in HCC. THIO induces telomere damage, activates the cGAS–STING pathway, enhances the cross-priming capacity of DCs and tumor-specific T-cell activation. Moreover, we showed a potential role of HMGB1 in THIO induced T-cell activation. In addition, our study showed efficacy of sequential administration of THIO and anti-PD1, anti-PDL1, and anti-VEGF in advanced tumors, thus providing a strong scientific rationale to translate combination therapy into clinical trials in HCC. Importantly, THIO has received FDA Orphan Drug Designation for HCC and small cell lung cancer (SCLC), indicating the importance and clinical relevance of our current studies. The sequential treatment of THIO and immune checkpoint inhibitors are also planned to be evaluated in HCC, SCLC, colorectal cancer, and other types of solid tumors.
Supplementary Material
Supplementary Data
Supplementary Table1
Supplementary Figure Legends
Acknowledgments
J. Shay holds the Southland Financial Corporation Distinguished Chair in Geriatrics Research. This work was in part supported by P50CA070907, P30CA142543, and MAIA Biotechnology for J. Shay. This work was also supported by the Simmons Comprehensive Cancer Center support grant 2P30CA142543. Research by D. Tang. and R. Kang was supported by grants from the NIH (R01CA160417, R01CA229275, and R01CA211070). M. Conacci-Sorrell was supported by Cancer Prevention and Research Institute of Texas (CPRIT; RP220046), American Cancer Society 724003, Welch foundation I-2058–20210327, NCI R01CA245548, NIGMS GM145744–01, and the Circle of Friend's award. A. Flusche was supported by the PB2PHD Program, UT Southwestern Medical Center Graduate School of Biomedical Sciences.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 691
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
I. Mender reports a patent for 6-thio-dG licensed to MAIA Biotechnology. S. Barron reports grants from NCI during the conduct of the study. Y. Hoshida reports grants from NIH, European Commission, and Cancer Prevention & Research Institute of Texas; other support from Alentis Therapeutics and Espervita Therapeutics; personal fees from Helio Genomics and Roche Diagnostics outside the submitted work. S. Gryaznov reports a patent for US 10463685 issued and licensed to UTSW. J.W. Shay reports grants and personal fees from Maia Biotechnology during the conduct of the study; also has a patent for 6-thio-dG licensed to Maia Biotechnology; and Jerry Shay is a founding scientist and SAB member of Maia Biotechnology. No disclosures were reported by the other authors.
Authors' Contributions
I. Mender: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Siteni: Investigation, visualization, methodology. S. Barron: Investigation. A.M. Flusche: Investigation. N. Kubota: Investigation, visualization, methodology. C. Yu: Investigation. C. Cornelius: Investigation. E. Tedone: Investigation. M. Maziveyi: Investigation, writing–review and editing. A. Grichuk: Investigation. N. Venkateswaran: Investigation. M. Conacci-Sorrell: Writing–review and editing. Y. Hoshida: Data curation, methodology, writing–review and editing. R. Kang: Investigation. D. Tang: Data curation, visualization, methodology, writing–review and editing. S. Gryaznov: Conceptualization, funding acquisition, visualization, writing–review and editing. J.W. Shay: Conceptualization, resources, supervision, funding acquisition, investigation, visualization, writing–review and editing.
References
- 1. Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941–53. [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 3. Yegin EG, Oymaci E, Karatay E, Coker A. Progress in surgical and nonsurgical approaches for hepatocellular carcinoma treatment. Hepatobiliary Pancreat Dis Int 2016;15:234–56. [DOI] [PubMed] [Google Scholar]
- 4. Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2020;17:139–52. [DOI] [PubMed] [Google Scholar]
- 5. Xing R, Gao J, Cui Q, Wang Q. Strategies to improve the antitumor effect of immunotherapy for hepatocellular carcinoma. Front Immunol 2021;12:783236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lawal G, Xiao Y, Rahnemai-Azar AA, Tsilimigras DI, Kuang M, Bakopoulos A, et al. The immunology of hepatocellular carcinoma. Vaccines (Basel) 2021;9:1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greten TF, Lai CW, Li G, Staveley-O'Carroll KF. Targeted and immune-based therapies for hepatocellular carcinoma. Gastroenterology 2019;156:510–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Finn RS, Ryoo B-Y, Merle P, Kudo M, Bouattour M, Lim H-Y, et al. Results of KEYNOTE-240: phase 3 study of pembrolizumab (Pembro) vs best supportive care (BSC) for second line therapy in advanced hepatocellular carcinoma (HCC). J Clin Oncol 2019;37(15_suppl):4004. [Google Scholar]
- 10. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet North Am Ed 2017;389:2492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galle PR, Finn RS, Qin S, Ikeda M, Zhu AX, Kim TY, et al. Patient-reported outcomes with atezolizumab plus bevacizumab versus sorafenib in patients with unresectable hepatocellular carcinoma (IMbrave150): an open-label, randomised, phase 3 trial. Lancet Oncol 2021;22:991–1001. [DOI] [PubMed] [Google Scholar]
- 12. Rizzo A, Ricci AD, Brandi G. Atezolizumab in advanced hepatocellular carcinoma: good things come to those who wait. Immunotherapy 2021;13:637–44. [DOI] [PubMed] [Google Scholar]
- 13. Rizzo A, Ricci AD, Brandi G. Immune-based combinations for advanced hepatocellular carcinoma: shaping the direction of first-line therapy. Future Oncol 2021;17:755–7. [DOI] [PubMed] [Google Scholar]
- 14. Mellman I. Dendritic cells: master regulators of the immune response. Cancer Immunol Res 2013;1:145–9. [DOI] [PubMed] [Google Scholar]
- 15. Sayour EJ, Mitchell DA. Manipulation of innate and adaptive immunity through cancer vaccines. J Immunol Res 2017;2017:3145742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature 2016;535:65–74. [DOI] [PubMed] [Google Scholar]
- 17. Khoo LT, Chen LY. Role of the cGAS-STING pathway in cancer development and oncotherapeutic approaches. EMBO Rep 2018;19:e46935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 2016;17:1142–9. [DOI] [PubMed] [Google Scholar]
- 19. Margolis SR, Wilson SC, Vance RE. Evolutionary origins of cGAS-STING signaling. Trends Immunol 2017;38:733–43. [DOI] [PubMed] [Google Scholar]
- 20. Ma Z, Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 2016;19:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015;347:aaa2630. [DOI] [PubMed] [Google Scholar]
- 22. Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends Immunol 2013;34:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li A, Yi M, Qin S, Song Y, Chu Q, Wu K. Activating cGAS-STING pathway for the optimal effect of cancer immunotherapy. J Hematol Oncol 2019;12:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han C, Zhang A, Liu Z, Moore C, Fu YX. Small molecular drugs reshape tumor microenvironment to synergize with immunotherapy. Oncogene 2021;40:885–98. [DOI] [PubMed] [Google Scholar]
- 25. Mender I, Gryaznov S, Dikmen ZG, Wright WE, Shay JW. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2'-deoxyguanosine. Cancer Discov 2015;5:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mender I, LaRanger R, Luitel K, Peyton M, Girard L, Lai TP, et al. Telomerase-mediated strategy for overcoming non-small cell lung cancer targeted therapy and chemotherapy resistance. Neoplasia 2018;20:826–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mender I, Gryaznov S, Shay JW. A novel telomerase substrate precursor rapidly induces telomere dysfunction in telomerase positive cancer cells but not telomerase silent normal cells. Oncoscience 2015;2:693–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shay JW, Wright WE. Telomerase activity in human cancer. Curr Opin Oncol 1996;8:66–71. [DOI] [PubMed] [Google Scholar]
- 29. Shay JW, Wright WE. Telomeres and telomerase: implications for cancer and aging. Radiat Res 2001; 155(1 Pt 2):188–93. [DOI] [PubMed] [Google Scholar]
- 30. Mender I, Zhang A, Ren Z, Han C, Deng Y, Siteni S, et al. Telomere stress potentiates STING-dependent anti-tumor immunity. Cancer Cell 2020; 38:400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mender I, Shay JW. Telomere dysfunction induced foci (TIF) analysis. Bio Protoc 2015;5:e1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ludlow AT, Robin JD, Sayed M, Litterst CM, Shelton DN, Shay JW, et al. Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic Acids Res 2014;42:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci U S A 1996;93:10933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 1999;4:199–207. [DOI] [PubMed] [Google Scholar]
- 35. Liu K, Huang J, Liu J, Li C, Kroemer G, Tang D, et al. HSP90 mediates IFNgamma-induced adaptive resistance to anti-PD-1 immunotherapy. Cancer Res 2022;82:2003–18. [DOI] [PubMed] [Google Scholar]
- 36. Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol 2020;38:586–99. [DOI] [PubMed] [Google Scholar]
- 37. Ataide MA, Komander K, Knopper K, Peters AE, Wu H, Eickhoff S, et al. BATF3 programs CD8(+) T cell memory. Nat Immunol 2020;21:1397–407. [DOI] [PubMed] [Google Scholar]
- 38. Shi L, Chen S, Yang L, Li Y. The role of PD-1 and PD-L1 in T-cell immune suppression in patients with hematological malignancies. J Hematol Oncol 2013;6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J, Chen Z, Li Y, Zhao W, Wu J, Zhang Z. PD-1/PD-L1 checkpoint inhibitors in tumor immunotherapy. Front Pharmacol 2021;12:731798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Galluzzi L, Vitale I, Warren S, Adjemian S, Agostinis P, Martinez AB, et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J Immunother Cancer 2020;8:e000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L, et al. HMGB1 in health and disease. Mol Aspects Med 2014;40:1–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med 2007;13:1050–9. [DOI] [PubMed] [Google Scholar]
- 43. Briukhovetska D, Dorr J, Endres S, Libby P, Dinarello CA, Kobold S. Interleukins in cancer: from biology to therapy. Nat Rev Cancer 2021;21:481–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005;19:2100–10. [DOI] [PubMed] [Google Scholar]
- 45. Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov 2016;6:584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. [DOI] [PubMed] [Google Scholar]
- 47. Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013;31:51–72. [DOI] [PubMed] [Google Scholar]
- 48. Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer 2012;12:860–75. [DOI] [PubMed] [Google Scholar]
- 49. Yamazaki T, Hannani D, Poirier-Colame V, Ladoire S, Locher C, Sistigu A, et al. Defective immunogenic cell death of HMGB1-deficient tumors: compensatory therapy with TLR4 agonists. Cell Death Differ 2014;21:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov 2020;10:26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1β-dependent adaptive immunity against tumors. Nat Med 2009;15:1170–8. [DOI] [PubMed] [Google Scholar]
- 52. Ma Y, Aymeric L, Locher C, Mattarollo SR, Delahaye NF, Pereira P, et al. Contribution of IL-17-producing gamma delta T cells to the efficacy of anticancer chemotherapy. J Exp Med 2011;208:491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ma Y, Adjemian S, Mattarollo SR, Yamazaki T, Aymeric L, Yang H, et al. Anticancer chemotherapy-induced intratumoral recruitment and differentiation of antigen-presenting cells. Immunity 2013;38:729–41. [DOI] [PubMed] [Google Scholar]
- 54. Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 2010;1799:101–13. [DOI] [PubMed] [Google Scholar]
- 55. Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 2000;405:354–60. [DOI] [PubMed] [Google Scholar]
- 56. Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev 2003;13:170–8. [DOI] [PubMed] [Google Scholar]
- 57. Andreeva L, Hiller B, Kostrewa D, Lassig C, de Oliveira Mann CC, Jan Drexler D, et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 2017;549:394–8. [DOI] [PubMed] [Google Scholar]
- 58. Polanska E, Dobsakova Z, Dvorackova M, Fajkus J, Stros M. HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma 2012;121:419–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Table1
Supplementary Figure Legends
Data Availability Statement
The DSP data generated in this study are publicly available in Gene Expression Omnibus (GEO; RRID: SCR_005012) at GSE219108. The raw data are available on request from the corresponding author.