Abstract
Hydrogen peroxide (H2O2) is a key member of the reactive oxygen species family of transient small molecules that has broad contributions to oxidative stress and redox signaling. The development of selective and sensitive chemical probes can enable the study of H2O2 biology in cell, tissue and animal models. Peroxymycin-1 is a histochemical activity–based sensing probe that responds to H2O2 via chemoselective boronate oxidation to release puromycin, which is then covalently incorporated into nascent proteins by the ribosome and can be detected by antibody staining. Here, we describe an optimized two-step, one-pot protocol for synthesizing Peroxymycin-1 with improved yields over our originally reported procedure. We also present detailed procedures for applying Peroxymycin-1 to a broad range of biological samples spanning cells to animal tissues for profiling H2O2 levels through histochemical detection by using commercially available anti-puromycin antibodies. The preparation of Peroxymycin-1 takes 9 h, the confocal imaging experiments of endogenous H2O2 levels across different cancer cell lines take 1 d, the dot blot analysis of mouse liver tissues takes 1 d and the confocal imaging of mouse liver tissues takes 3–4 d.
Introduction
Reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) are a family of transient and redox-active small molecules that have broad functions in biology as signaling agents and/or markers for oxidative stress and damage1–14. Indeed, H2O2 is a major non-radical ROS that can regulate various downstream targets, including phosphatases, kinases, transcription factors and ion channels, through reversible redox post-translational modifications15–19. Aside from mitochondrial activity, an important source of endogenous production of H2O2 stems from NADPH oxidase proteins, which are stimulated by growth factors, cytokines, hormones and neurotransmitters20–29. NADPH oxidase enzymes play key roles in triggering signaling events that lead to physiological processes such as neural activity and long-term potentiation and depression30–33, stem cell growth and proliferation29,34,35, circadian rhythms36–38 and wound healing39,40.
Deciphering the complex interplay between sources and targets of H2O2 in signaling and stress pathways motivates the development of new methods for probing H2O2 across various biological models. In this context, activity-based sensing with small molecule–based fluorescent and bioluminescent probes provides a powerful method to monitor H2O2 levels in living systems41–44. Our laboratory has advanced this area through the chemoselective H2O2-mediated oxidation of boronate protecting groups41,43,45,46. Many variations, such as multicolor47, ratiometric48,49, two-photon50, organelle-targeting51,52, esterase-sensitive29,53, quinone methide–generating54 and bioluminescent indicators have been developed55,56. These versatile reagents have enabled studies of H2O2 biology, offering advantages that include high selectivity and sensitivity, good spatial and temporal resolution and the ability to track H2O2 in live cell, tissue and animal models29,46,53,57,58. However, cell retention issues minimize the effective use of these reagents in fixed samples.
To address these limitations and expand activity-based sensing of H2O2 to fixed specimens, we reported Peroxymycin-1, a novel histochemical probe for H2O2 detection that is compatible with fixed-cell and mouse tissue samples (Fig. 1)59. Peroxymycin-1 consists of an aryl boronate ester scaffold attached to an alpha-amino group on puromycin, an aminonucleoside antibiotic that can be coupled to nascent proteins synthesized at the ribosome after uncaging the free amine group through H2O2-dependent boronate oxidation (Fig. 2a). In our original report, we demonstrated that Peroxymycin-1 can detect H2O2 with high selectivity and sensitivity across cells and mouse tissues that are compatible with standard fixation protocols. Specifically, Peroxymycin-1 enables profiling of endogenous levels of H2O2 across several types of breast cancer cell lines, showing a correlation between malignancy and higher H2O2 levels. Moreover, application to a diet-induced model of nonalcoholic fatty liver disease shows an increase in H2O2 levels over healthy controls that is consistent with increased expression of cytochrome P450 2E1 and activity of NADPH oxidases in the liver60.
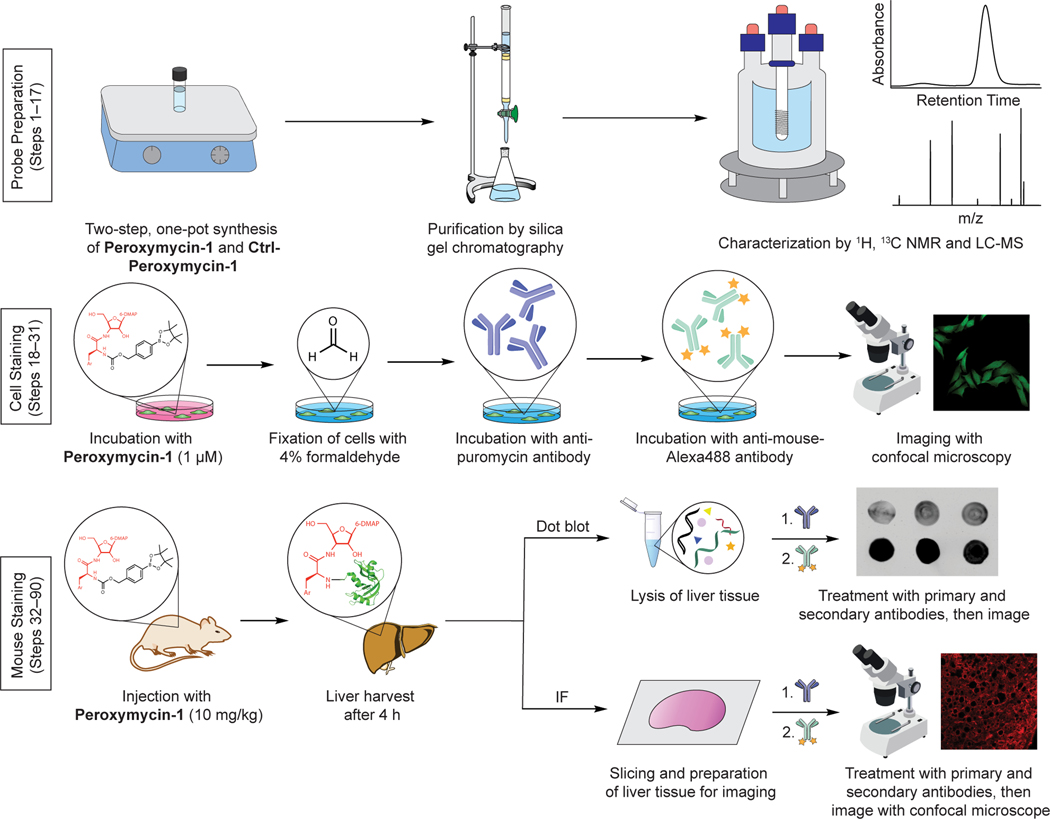
Fig. 1 |. Overview of hydrogen peroxide detection with Peroxymycin-1 in cells and mice.
Ar, 4-methoxyphenyl; IF, immunofluorescence; LC-MS, liquid chromatography-mass spectrometry; 6-DMAP, N,N-dimethyl purin-6-amine. Confocal images reprinted with permission from ref. 59, American Chemical Society.
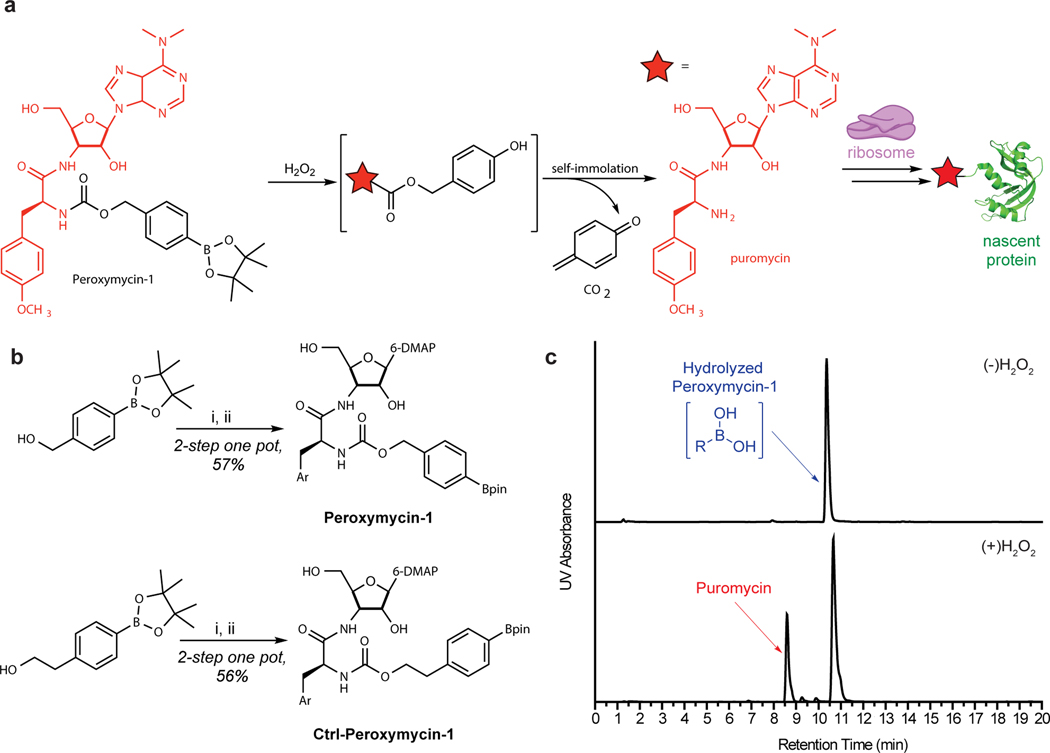
Fig. 2 |. Synthetic and mechanistic scheme of Peroxymycin-1.
a, Mechanism of H2O2 detection by Peroxymycin-1. Oxidative deprotection of the boronate pinacol ester occurs in the presence of H2O2 and releases puromycin, which is incorporated into nascent proteins by the ribosome. Puromycin shown in red. b, Synthetic scheme of Peroxymycin-1 and Ctrl-Peroxymycin-1. Reagents and conditions: (i) 4-nitrophenyl chloroformate, 4-(dimethylamino)pyridine, CH2Cl2, 0–23 °C, 2 h; (ii) puromycin • 2HCl, N,N-diisopropylethylamine, dimethyl formamide, 23 °C, 1 h. c, UV chromatogram (275 nm) of purified Peroxymycin-1 (top) and uncaging in the presence of H2O2 (0.1 mM) to form free puromycin (bottom). Bpin, boronate pinacol ester.
Here, we present a detailed protocol for an improved synthesis of Peroxymycin-1. We also detail staining procedures for Peroxymycin-1–treated cell and tissue samples with immunodetection using a commercially available anti-puromycin antibody. The aim of the present protocol is to facilitate the broader use of Peroxymycin-1 by providing a step-by-step procedure with detailed notes for critical steps as well as troubleshooting of potential technical issues. Notably, we have optimized the synthesis of Peroxymycin-1, along with a negative control compound that reacts with H2O2 but does not generate puromycin (Ctrl-Peroxymycin-1), where the two-step reaction sequence described in the original report is now performed in a one-pot reaction procedure with improved yield by omitting aqueous work-up and column chromatography purification steps (Fig. 2). This improved synthetic procedure can yield >60 mg of the final product within 1 d. Peroxymycin-1 synthesized by following the optimized protocol also showed less hydrolysis of the boronate pinacol ester to the boronic acid compared to Peroxymycin-1 synthesized by following the original procedure, confirmed by liquid chromatography-mass spectrometry (LC-MS) analysis (Fig. 2c).
Overview of the procedure
In this protocol, we report an optimized synthesis of Peroxymycin-1 and Ctrl-Peroxymycin-1 and outline detailed procedures for applying the histochemical probe to cell and animal experiments. The H2O2 levels stained by Peroxymycin-1 can be analyzed in cell and animal tissue samples by confocal imaging or dot blot analysis. The first section of this protocol outlines the optimized two-step, one-pot reaction for synthesizing Peroxymycin-1 and Ctrl-Peroxymycin-1 (Steps 1–17). The second section describes the application of Peroxymycin-1 to profile endogenous H2O2 levels in live cells, demonstrating detection of H2O2 by Peroxymycin-1 in live cells and histochemical visualization of the incorporated puromycin with primary and secondary antibodies (Steps 18–31). The use of Peroxymycin-1 in mouse samples is described in the third section, demonstrating the utility of Peroxymycin-1 for profiling H2O2 levels in mouse liver tissue by both dot blot and confocal imaging (Steps 32–90).
Design strategy
The design concept of Peroxymycin-1 relies on functionalizing the primary amine moiety of puromycin with an activity-based sensing group that induces a H2O2-dependent deprotection event that triggers cascade reactions to generate free puromycin for downstream detection. Peroxymycin-1 consists of a puromycin attached to a carbamate-linked aryl boronate pinacol ester (Fig. 2). Oxidative deprotection of the boronate pinacol ester upon treatment with H2O2 leads to linker self-immolation and subsequent release of a 1,4-quinone methide species, CO2 and free puromycin. Puromycin can then incorporate into nascent proteins at the ribosome. The extent of covalent puromycin incorporation thus serves as a proxy for the H2O2 level of the sample and can be visualized by immunofluorescence techniques using a standard anti-puromycin antibody. As a negative control, Ctrl-Peroxymycin-1 bears an extra methylene unit in the self-immolative linker, separating the carbamate-linked puromycin from the aryl boronate pinacol ester. The negative control probe is still vulnerable to oxidative deprotection in the presence of H2O2, but the electron cascade process to release free puromycin cannot occur.
Limitations
Because Peroxymycin-1 requires the activity-based sensing reaction to occur along with puromycin incorporation into nascent proteins, Peroxymycin-1 requires a relatively long probe incubation time for signal processing (up to 4 h) compared to traditional fluorescent probes (5–30 min). As such, Peroxymycin-1 is not designed for real-time detection of dynamic changes of H2O2 in a narrow time window, but rather as a probe for profiling relative H2O2 levels across various samples and/or before and after an external stimulus. Because of the incorporation process of uncaged puromycin at the ribosome, Peroxymycin-1 does not provide localized, subcellular information of H2O2 levels. This attribute is in contrast to genetically encoded fluorescence proteins (e.g., HyPer61 and peroxiredoxin-based probes62) and traditional small-molecule fluorescence probes with an organelle-targeting localization handle (e.g., MitoPY1)51,52,63,64.
Comparison of Peroxymycin-1 to other imaging-based H2O2 probes
In addition to the high selectivity exhibited by Peroxymycin-1 toward H2O2, which is analogous to previously reported boronate-based molecular imaging probes, a key advantage of Peroxymycin-1 is its compatibility with standard immunofluorescence experimental platforms without relying on any genetic manipulation. This feature enables H2O2 detection in live cell and animal samples, including mouse tissue, through signal amplification by a desired secondary antibody–fluorophore conjugate. Unlike traditional fluorescent probes for which dye leakage can decrease the signal-to-noise ratio, puromycin is covalently incorporated into nascent proteins after reaction with H2O2 so that no probe leakage occurs. Comparison of Peroxymycin-1 with other fluorescence-based sensors is summarized in Table 1. For example, Peroxyfluor-2 is a turn-on fluorescent probe for H2O2 with the same boronate trigger as Peroxymycin-1, and its utility has been shown in various live-cell samples47. However, this fluorogenic probe is limited to short-term imaging because of limited probe retention in cells, which also makes it difficult to use in tissue and animal samples. Genetically encoded fluorescence proteins offer a complementary set of tools to monitor H2O2 level reversibly and ratiometrically in biological samples61. Genetic encoding enables expression in specific regions of a cell for organelle-specific H2O2 detection in cells and animal models spanning nematodes, zebrafish and plants. Some limitations of genetically encoded probes are their sensitivity to changes in pH in the physiological range; the need for transfection, which can increase incubation time; and complications related to probe concentration compared to that of small-molecule probes.
Table 1.
Comparison of Peroxymycin-1 with other selected imaging-based H2O2 probes.
| Tool | Application | Pros | Cons | Ref |
|---|---|---|---|---|
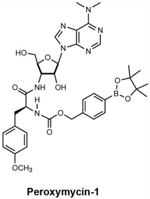
|
Detection of H2O2 in fixed cells and mouse tissue (liver). | Peroxymycin-1 is specific to H2O2, can be used with multiple dyes and in tissue samples, and does not require genetic engineering. | Samples must be 59 fixed to use with Peroxymycin-1, and the probe has relatively long incubation time. Peroxymycin-1 may also have a potential influence on protein synthesis if used in excess doses. | 59 |
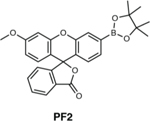
|
Detection of H2O2 in live cells. | PF2 is selective for H2O2 over other ROS species, compatible with live cell imaging, and does not require transfection. | PF2 suffers poor retention inside cells, and unsuitable for tissue and live animal imaging. | 47 |
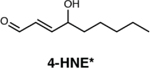
|
Detection of one of the products of ROS in fixed cells and animal tissue (mice, zebrafish, nematodes, etc.). | 4-HNE staining can be used in parallel with other immunostaining techniques and does not require genetic engineering. | 4-HNE staining is not specific to a particular ROS, does not give a direct readout of ROS, is not compatible with live cell imaging. | 65 |
|
Detection of H2O2 in live cells and organisms (mice, nematodes, zebrafish, bacteria, etc.). | This prototypical family of H2O2- specific genetically encoded sensors is suitable for live cells/animals with organelle- or region- specific expression capability. | The protein sensor can be pH sensitive, difficult to control expression, and requires transfection or other genetic methods to introduce. |
61 |
4-HNE is the product formed after reaction with ROS. Other structures (Peroxymycin-1 and PF2) in the table are before the detection of H2O2.
Crystal structure of OxyR (PDB ID: 1I69) is shown.
Finally, immunohistochemical staining of 4-hydroxy-nonenal (4-HNE) is widely used as a marker for oxidative stress and damage in animal tissue samples65. This aldehyde product is produced when any one of a number of ROS, such as O2–, H2O2, hydroxyl radicals and peroxynitrite, acts as a general oxidant. Because of the commercial availability of the anti-4-HNE antibody, this method has seen widespread use. However, the technique is not able to detect ROS directly, much less afford specificity to a particular ROS. 4-HNE staining is also not compatible with live-cell imaging applications.
Applications
Peroxymycin-1 has been successfully used to profile H2O2 levels in human cervical epithelial carcinoma cells (HeLa), human epidermoid carcinoma cells (A-431), human breast carcinoma cells (MDA-MB-231 and MDA-MB-468), non-tumorigenic breast epithelial cells (MCF-10A), invasive breast cancer cells (HS 578T) and normal breast cells (HS 578 Bst)59. Given these results and because puromycin labeling has found widespread use as a staining platform, we anticipate that cell lines and biological specimens compatible with the anti-puromycin antibody should be compatible with Peroxymycin-1 in a similar experimental setup with not only dot blot and confocal imaging analysis as described in this protocol, but also techniques such as western blot66, FACS67 and ELISA68. Blot analyses may be appealing because the technique is inexpensive and does not require use of operationally difficult equipment, such as the confocal microscope. In addition to mouse liver, some examples of cell lines and animal samples that have been studied by using puromycin labeling and detection with the anti-puromycin antibody include neurons69, Drosophila larvae70, and zebrafish71.
Experimental design
Synthesis of Peroxymycin-1 and Ctrl-Peroxymycin-1
We reported the synthesis of Peroxymycin-1 and Ctrl-Peroxymycin-1 in two steps with an aqueous extraction and column chromatography purification process after the first step59. The optimized synthetic procedure reported here takes advantage of a two-step, one-pot reaction setup with only one column chromatography step. Using anhydrous solvent is a key point within this synthesis. Although the reaction is conducted under air, having a large amount of water present in the solution may cause undesired reactivity or decomposition of the starting materials or products. The synthetic procedure is described on the hundred milligrams scale with an overall yield of ~56% for both Peroxymycin-1 and Ctrl-Peroxymycin-1. To conduct the synthesis on a larger scale, users may need to optimize the reaction conditions and purification processes, although in theory, the method outlined in this protocol should be applicable. HPLC may be a viable method for purification, although hydrolysis of the boronate pinacol ester may be observed under aqueous purification conditions.
Applying Peroxymycin-1 and controls for cell experiments
In this protocol, we profiled endogenous H2O2 levels in MDA-MB-231, MDA-MB-468, MCF-10A, HS 578T and HS 578Bst cells, but Peroxymycin-1 has also been successfully used to label A-431 and HeLa cells59. We anticipate that cell lines and conditions compatible with the anti-puromycin antibody will be compatible with Peroxymycin-1 in a similar experimental setup. However, when using different cell lines or conditions than those described in this protocol, exact parameters for the immunostaining processes such as permeabilization, blocking and antibody incubation time/concentration should be optimized experimentally or based on literature precedents.
Depending on the cell line and conditions, the generation of free puromycin within a cell sample may affect its protein synthesis rate, and Peroxymycin-1 should be normalized by a control puromycin signal to account for possible differences in the protein synthesis rate between samples. A control sample incubated with puromycin should be set up alongside the Peroxymycin-1 and Ctrl-Peroxymycin-1 samples, and the fluorescence intensity of the control should be used to normalize the Peroxymycin-1 and Ctrl-Peroxymycin-1 signals (experiment described in Anticipated results, and normalization method described in Supplementary information, Data analysis). When using Peroxymycin-1 for the first time with cell lines not reported within this protocol, the puromycin control experiment should always be carried out to ensure that the possible differences in protein synthesis rate between samples does not cause bias in results.
Materials
Biological materials
Cells: MDA-MB-231, MDA-MB-468, MCF-10A, HS 578T and HS 578Bst cells (UC Berkeley Tissue Culture Facility, RRIDs CVCL_0062, CVCL_0419, CVCL_0598, CVCL_0332 and CVCL_0807, respectively) !CAUTION Cells should be regularly checked to ensure that they are authentic and are not infected with mycoplasma ▲ CRITICAL The cells used in this protocol were cultured in the UC Berkeley Tissue Culture Facility, but cells from other companies are a suitable alternative. We recommend cells from American Type Culture Collection (e.g., HeLa cells, cat. no. ATCC CCL-2).
Mice: 8-week-old male C57BL/6 mice (Jackson Laboratory) !CAUTION All animal studies were approved by and performed according to the guidelines of the Animal Care and Use Committee of the University of California, Berkeley. Any experiments involving live mice must conform to relevant institutional and national regulations.
Reagents
Synthesis of Peroxymycin-1 and Ctrl-Peroxymycin-1
!CAUTION All reagents in this section are harmful. Handle with gloves and avoid contact with eyes and skin. Volatile materials should be used inside a fume hood.
4-(4,4,5,5-Tetramethyl-1,3,2-dioxaborolan-2-yl)benzene ethanol (Combi Blocks, cat. no. 651030–55-6) !CAUTION 4-(4,4,5,5-Tetramethyl-1,3,2-dioxaborolan-2-yl)benzene ethanol is harmful.
4-(4,4,5,5-Tetramethyl-1,3,2-dioxaborolan-2-yl)benzyl alcohol (TCI, cat. no. T3496)
4-Dimethylaminopyridine (DMAP; Oakwood Chemical, cat. no. 001704) !CAUTION DMAP is harmful.
4-Nitrophenyl chloroformate, 97% (Gas Chromatography) (AK Scientific, cat. no. A614) !CAUTION 4-Nitrophenyl chloroformate is corrosive.
Methanol for chromatography (Fisher Scientific, cat. no. A452–4) !CAUTION Methanol is harmful and volatile.
Methylene chloride for chromatography (CH2Cl2; anhydrous; Fisher Scientific, cat. no. D143–4) !CAUTION Methylene chloride is harmful and volatile.
N,N-Diisopropylethylamine (DIPEA; Sigma Aldrich, cat. no. 387649) !CAUTION DIPEA is corrosive and volatile.
N,N-Dimethylformamide (DMF; anhydrous; Fisher Scientific, cat. no. D143–4) !CAUTION DMF is harmful.
Puromycin dihydrogen chloride (Adipogen, cat. no. AG-CN2–0078) !CAUTION Puromycin dihydrogen chloride is harmful.
Cell staining
Donkey anti-mouse antibody–Alexa Fluor 488 conjugate (Invitrogen, cat. no. A21202; RRID: AB_141607)
DMEM (Gibco, cat. no. 10566–016)
FBS (VWR, cat. no. 97068–085)
Hoechst 33342 (Invitrogen, cat. no. H21492)
Mouse anti-puromycin antibody (Kerafast, cat. no. EQ0001; RRID: AB_2620162)
PBS (Corning, cat. no. 21–040-CM)
Mouse breeding
High-fat diet (HFD; 60% calories from fat; Research Diets, cat. no. D12492)
Regular chow (18% calories from protein; Teklad, cat. no. 2018)
Mouse dot blot experiments
Bicinchoninic acid (BCA) protein assay reagent A (Thermo Scientific, cat. no. 23223)
BCA protein assay reagent B (Thermo Scientific, cat. no. 23224); URL for BCA protein assay protocol: https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0011430_Pierce_BCA_Protein_Asy_UG.pdf&title=VXNlciBHdWlkZTogUGllcmNlIEJDQSBQcm90ZWluIEFzc2F5IEtpdA==
Donkey anti-mouse antibody–Alexa Fluor 647 conjugate (Invitrogen, cat. no. A-31571; RRID: AB_162542)
Mouse anti-lamin B antibody (Abcam, cat. no. ab16048; RRID: AB_10107828)
Nonfat dry milk (Bio-Rad, cat. no. 1706404)
RIPA buffer (1×) (Boston BioProducts, cat. no. BP-116X) !CAUTION RIPA buffer is harmful. Handle with gloves to avoid contact with eyes.
Tris buffered saline with Tween 20 (20×) (TBST; Santa Cruz Biotechnology, cat. no. sc-362311)
Mouse tissue immunofluorescence
16% formaldehyde aqueous solution (Electron Microscopy Sciences, cat. no.15710-S) !CAUTION Formaldehyde is harmful. Handle with gloves to avoid contact with eyes and skin.
Goat anti-mouse IgG Fab fragment (Jackson ImmunoResearch, cat. no. 115–007-003; RRID: AB_2338476)
BSA (Fisher Scientific, cat. no. BP1600)
Donkey anti-mouse Alexa Fluor 647 (LifeTech, cat. no. A31571; RRID: AB_162542)
Normal donkey serum (Jackson Immunoresearch, cat. no. 017–000-121; RRID: AB_2337258)
Sucrose (Fisher Scientific, cat. no. 57–50-1)
Tissue-freezing medium (TFM; General Data, cat. no. TFM-5)
Triton X-100 (Fisher Scientific, cat. no. BP151–100) !CAUTION Triton X-100 is harmful. Handle with gloves to avoid contact with eyes and skin.
Equipment
Synthesis of Peroxymycin-1 and Ctrl-Peroxymycin-1
4-ml Wheaton clear glass vials with caps (VWR, cat. no. 66011–548)
Analytical balance (Denver Instrument Co., A-200DS)
Analytical thin-layer chromatography plates (250-μm thickness) (Silicycle, cat. no. NC0898660)
Biotage automated column (Isolera One Accelerated Chromatographic Isolation)
Biotage SNAP KP-Sil cartridge, silica, 10 g
Clamps
Crystallization dish
Disposable glass Pasteur pipettes (VWR, cat. no.14673–043)
Disposable scintillation vials, 20 ml (Wheaton, cat. no. 66011–548)
Dry solvent purification system
Gravity convection oven
LC-MS (Agilent 1220 LC System, Advion expression compact mass spectrometer)
Magnetic stir plate
Magnetic stir bars (<1 cm)
Micropipette, 20–200 μl (Sartorius mechanical pipette)
Needles (Air-Tite veterinary needles 22 gauge (G) × 4 inch thin, cat. no. 89219–270)
Nitrile gloves (VWR, cat. no. 32934–080)
Nitrogen gas (N2)
Pie block (Chemglass, cat. no. CG-1991-P-13)
Pipette bulbs
Rotary evaporator
Rubber septa
Sartorius Biohit Optifit tips (Fisher Scientific, cat. no. 14559469)
Single-neck, round-bottomed flasks (100–500 ml)
Spatula
Syringes (HSW Norm-Ject 1-ml Luer-slip syringes, cat. no. 53548–001)
VWR disposable culture tubes, 16 × 100 mm (VWR, cat. no. 47729–572)
Whatman Mini-UniPrep transparent syringeless filters (Fisher Scientific, cat. no. 09–923-102)
Zorbax SB-C18 column (Agilent, 5.0-μm pore size, 4.6 mm × 250 mm)
XBridge C18 column (Waters, 3.5-μm pore size, 4.6 × 100 mm)
Cell staining
Eight-well borosilicate chambered cover glass slide (Lab Tek, cat. no. 155409) or 96-well plate (sterile, clear, flat-bottom polystyrene Tissue Culture (TC)-treated microplate; Corning, cat. no. 3598)
CO2 incubator (Thermo Scientific, 3586 Napco series 8000 WJ)
Confocal microscope (Zeiss LSM 710 or Perkin Elmer Opera Phenix high-content screening system)
Mouse dot blot experiments
Homogenizer (Fisher Scientific, Bel-Art F650000000)
Imaging system (Licor Odyssey)
Nitrocellulose membrane (Bio-Rad, cat. no. 162–0146)
Orbital shaker (Benchmark Scientific Orbi-blotter low-speed shaker)
Plastic container for dot blot (dimensions: 3.5 × 5 × 3 cm)
Mouse tissue immunofluorescence
Confocal microscope (Zeiss LSM 710)
Cover glass (Fisher Scientific, cat. no. 12–544-18)
Cryostat microtome (Thermo Scientific Microm HM550)
Dry ice
Glass staining jar
Hydrophobic pen (Vector, cat. no. H-4000)
Intermediate cryomold (Sakura Finetek, cat. no. 25608–924)
Kimwipes (VWR, cat. no. 89218–057)
Microscope slide (Fisher Scientific, cat. no. 12–550-343)
Nail polish (Fisher Scientific, cat. no. 50–949-071)
Superfrost microscope slides (Fisher Scientific, cat. no. 12–550-15)
Surgical scissors
Tweezers
Reagent setup
Peroxymycin-1, Ctrl-Peroxymycin-1, puromycin stock solution
Make a 1 mM stock (1,000×) of Peroxymycin-1 (molecular weight: 731.5 g/mol), Ctrl-Peroxymycin-1 (molecular weight: 745.4 g/mol) or puromycin dihydrochloride (molecular weight: 544.43) in dimethyl sulfoxide (DMSO) in an Eppendorf tube. No degradation was observed when this solution was stored at −20 °C for 1 year.
Peroxymycin-1 (or Ctrl-Peroxymycin-1 or puromycin) incubation medium
Dilute the 1,000× DMSO stock solution of Peroxymycin-1, Ctrl-Peroxymycin-1 or puromycin in DMEM (no FBS) to obtain a concentration of 1 μM for a total volume of 1 ml. The solution should be freshly prepared before each experiment.
PBS with 4% (wt/vol) formaldehyde
Make a 4% (wt/vol) formaldehyde solution by mixing 3 ml of PBS and 1 ml of 16% (wt/vol) formaldehyde solution. The solution should be freshly prepared before each experiment.
PBS with 0.1% (vol/vol) Triton X-100
Mix 0.1 ml of Triton X-100 and 99.9 ml of PBS. Shake the solution on a rotator for ≥20 min at room temperature (RT; 19–22 °C). The solution can be stored at RT for >1 week unless obvious precipitation is observed. However, we recommend freshly preparing the solution for each experiment.
Permeabilization buffer
Add 0.1 ml of Triton X-100 to 99.9 ml of PBS. Mix the solution in a rotator for ≥20 min at RT. The solution can be stored at RT for >1 week unless obvious precipitation is observed. However, we recommend freshly preparing the solution for each experiment.
Anti-puromycin antibody incubation solution for cell experiments
Make an anti-puromycin antibody solution (0.3 μl, 1:500 (vol/vol) dilution) in FBS (15 μl, 10% (vol/vol)) and PBS with 0.1% (vol/vol) Triton X-100 (134 μl). The solution should be freshly prepared before each experiment.
Anti-mouse antibody incubation solution for cell experiments
Make an anti-mouse antibody–Alexa Fluor 488 conjugate incubation solution (1.5 μl, 1:100 (vol/vol) dilution) in FBS (15 μl, 10% (vol/vol)), 1 μM Hoechst 33342 and PBS with 0.1% (vol/vol) Triton X-100 (134 μl). The solution should be freshly prepared before each experiment.
Mice preparation
Feed 8-week-old male C57BL/6 mice (Jackson Laboratory) regular chow or an HFD for 20 weeks. Measure body weight and food intake weekly. !CAUTION All the experiments using mice should be performed in agreement with institutional and governmental guidelines. The procedures in this protocol were approved by and performed according to the guidelines of the Animal Care and Use Committee of the University of California, Berkeley. Any experiments involving live mice must conform to relevant institutional and national regulations.
Peroxymycin-1 stock solution for injection into mice
For dosing mice at 10 mg/kg, prepare a stock solution at 2 mg/ml in PBS/PET (PET: 60% (vol/vol) poly(ethylene glycol) 400, 30% (vol/vol) ethanol and 10% (vol/vol) Tween-80) solution mixture (1:1 (vol/vol)). It is important to first dissolve the Peroxymycin-1 solid in PET completely (sonication and vortex can be used) and then add an equal volume of PBS. The solution should be transparent and must be freshly prepared before each experiment.
TBST solution
Dilute 50 ml of the 20× TBST solution in 950 ml of MilliQ water. The solution can be stored at RT for >1 year unless obvious precipitation is observed.
Anti-puromycin antibody incubation solution for dot blot
Dilute 2 μl of the anti-puromycin antibody in 2 ml of the tissue permeabilization buffer (1:1,000 (vol/vol) dilution). The antibody solution can be reused for future experiments (usually a total of two to three blot experiments). After the experiment, transfer the antibody solution to an Eppendorf tube and add sodium azide solution (0.02–0.05% (wt/vol) final). Store at 4 °C until obvious precipitation is observed.
Anti-lamin B1 antibody incubation solution for dot blot
Dilute 2 μl of the anti-lamin B1 antibody in 2 ml of the tissue permeabilization buffer (1:1,000 (vol/vol) dilution). The antibody solution can be reused for future experiments (usually a total of two to three blot experiments). After the experiment, transfer the antibody solution to an Eppendorf tube and add sodium azide solution (0.02–0.05% (wt/vol) final). Store at 4 °C until obvious precipitation is observed.
Anti-mouse antibody incubation solution for dot blot
Dilute 1.0 μl of the anti-mouse antibody–Alexa Fluor 647 conjugate in 10 ml of the tissue permeabilization buffer (1:10,000 (vol/vol) dilution). The antibody solution can be reused for future experiments (usually a total of two to three blot experiments). After the experiment, transfer the antibody solution to an Eppendorf tube and add sodium azide solution (0.02–0.05% (wt/vol) final). Store at 4 °C until obvious precipitation is observed.
30% (wt/vol) sucrose solution
Dissolve 300 mg of sucrose in 1 ml of MilliQ water. The solution should be freshly prepared before each experiment.
Tissue permeabilization buffer
Dissolve 50 mg of BSA in 1 ml of PBS solution containing 0.5% (vol/vol) Triton X-100. The solution should ideally be freshly prepared before each experiment but can also be stored for 1 month at 4 °C.
Mouse tissue blocking buffer
Dilute 20 μl of the anti-mouse IgG Fab fragment solution in 200 μl of the tissue permeabilization buffer (1:10 (vol/vol) dilution). The solution should be freshly prepared before each experiment.
Tissue blocking buffer
Dilute 200 μl of the donkey serum in 2,000 μl of the tissue permeabilization buffer (1:10 (vol/vol) dilution). The solution should be freshly prepared before each experiment.
Anti-puromycin antibody incubation solution for tissue imaging
Dilute 2 μl of the anti-puromycin antibody in 200 μl of the tissue permeabilization buffer (1:100 (vol/vol) dilution). The solution should be freshly prepared before each experiment.
Anti-mouse antibody incubation solution for tissue imaging
Dilute 0.8 μl of the anti-mouse antibody–Alexa Fluor 647 conjugate in 200 μl of the tissue permeabilization buffer (1:250 (vol/vol) dilution). The solution should be freshly prepared before each experiment.
Procedure
Synthesis of Peroxymycin-1 and Ctrl-Peroxymycin-1 ● Timing 7–9 h
-
1
Transfer 4-nitrophenyl chloroformate (33.7 mg, 0.167 mmol, 1 equiv.) and DMAP (20.4 mg, 0.167 mmol, 1 equiv.) into a 4-ml glass vial by using a spatula. Charge the vial with a small, oven-dried stir bar. (4-Nitrophenyl chloroformate should be stored in a refrigerator and warmed to RT before weighing.)
-
2
Transfer 4-(hydroxymethyl) phenylboronic acid pinacol ester (39.1 mg, 0.167 mmol, 1 equiv.) for Peroxymycin-1 or 4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl) benzene ethanol (41.4 mg, 0.167 mmol, 1 equiv.) for Ctrl-Peroxymycin-1 into a different 4-ml vial by using a spatula.
-
3
Add anhydrous CH2Cl2 (0.54 ml, obtained from a dry solvent purification system) to both vials by using a syringe and needle. White precipitate will form in the solution of 4-nitrophenyl chloroformate and DMAP.
▲ CRITICAL STEP Anhydrous solvent is used, but the reaction is carried out in air, because the reaction is not extremely sensitive to air and moisture.
-
4
Cap both vials and place in a pie block. Cool to 0 °C for 5 min in an ice-water bath while stirring.
▲ CRITICAL STEP Both solutions should be completely cooled to 0 °C before they are mixed with each other.
-
5
Transfer the 4-nitrophenyl chloroformate and DMAP mixture quickly to the stirring 4-(hydroxymethyl) phenylboronic acid pinacol ester solution on the ice-water bath by using a Pasteur pipette. The white precipitate should solubilize when the two solutions are combined.
-
6
While stirring, warm the reaction to RT over 2 h.
-
7
Evaporate volatiles by using a gentle flow of N2 gas at RT (<30 min).
-
8
Re-dissolve the off-white chunky powder in anhydrous DMF (0.54 ml, obtained from a dry solvent purification system).
-
9
Weigh puromycin dihydrogen chloride (100 mg, 0.184 mmol, 1.1 equiv.) into a separate 4-ml vial.
-
10
Dissolve puromycin dihydrochloride in anhydrous DMF (0.54 ml, obtained from a dry solvent purification system).
-
11
Add DIPEA (0.19 ml, 1.1 mmol, 6.5 equiv.) to the puromycin solution by using a micropipette.
-
12
Add the puromycin solution by using a Pasteur pipette into a stirred solution of the carbonate intermediate from Step 8 at RT.
-
13
Stir the reaction solution at RT for 1 h.
? TROUBLESHOOTING
-
14
Confirm completion of the reaction by LC-MS analysis. A major peak at ~11 min in the UV chromatogram at 275.0 nm (when running a gradient of 5–95% (vol/vol) acetonitrile in water with constant 0.05% (vol/vol) formic acid over 20 min at 1 ml/min by using the XBridge C18 column denoted in Equipment) corresponds to hydrolyzed Peroxymycin-1 (650 mass-to-charge ratio (m/z)) (Fig. 2c).
-
15
Remove volatiles under a gentle flow of N2 gas (~2 h) or reduced pressure to obtain a clear yellow solid.
■ PAUSE POINT No noticeable decomposition was observed after storing the crude product in the dark at −20 °C for 3 d. However, it is recommended that the product be purified as soon as possible to minimize decomposition.
-
16
Purify the product by silica gel column chromatography (column diameter: 2 cm, silica gel height: 8–10 cm) by using CH2Cl2 as an eluent by increasing methanol concentration (0–5% (vol/vol)). Load crude material onto the column by dissolving in regular CH2Cl2 (0.2–0.4 ml). A Biotage flash purification system was used to run an automated silica chromatography column, but the purification process and solvent gradient yielded the same result when performed on a manual silica column. The retention factor on silica thin-layer chromatography plates was 0.3 in 95:5 CH2Cl2/methanol.
-
17
Confirm the identity and purity of the product by NMR spectroscopy (1H NMR and 13C NMR) and LC-MS. This procedure yielded 70 mg of Peroxymycin-1 and 70 mg of Ctrl-Peroxymycin-1 as off-white solids (overall yields of 57% and 56% for Peroxymycin-1 and Ctrl-Peroxymycin-1, respectively)59.
■ PAUSE POINT The solid form of the product can be stored in the dark at −20 °C for several years. Stock solution of product in DMSO (10 mM) can also be stored at −20 °C (>1-year shelf life).
? TROUBLESHOOTING
Live-cell H2O2 detection with Peroxymycin-1 and cell fixation ● Timing 5–6 h
-
18
Culture adherent cells in a 96-well plate (70–80% confluency). Remove cell culture medium from each well in a laminar hood or biosafety cabinet. Wash cells two times with 100 μl of DMEM (no FBS).
▲ CRITICAL STEP Steps 18 and 19 should be performed by using sterile reagents and equipment.
-
19
Add Peroxymycin-1 or puromycin incubation medium (a total volume of 100 μl in each well) and then place the plate in a 5% CO2 humidified incubator at 37 °C for 4 h.
▲ CRITICAL STEP Additional control experiments with puromycin are highly recommended because the incorporation rate of free puromycin to nascent proteins (i.e., protein synthesis rate) may differ from one sample to another. The normalization process discussed in Supplementary information, Data analysis should be performed accordingly.
-
20
Remove the medium and rinse cells with 150 μl of PBS per well. Aspirate the PBS subsequently. PBS can be replaced by Hank’s buffered saline solution with calcium and magnesium. This wash process can be performed without the sterile setup if cells of interest can tolerate short-term, non-sterile conditions.
-
21
Rinse one additional time with 150 μl of PBS.
-
22
To fix cells, add PBS with 4% (wt/vol) formaldehyde solution (150 μl per well) and place the plate on a bench at RT for 10 min. Remove solution from the wells.
-
23
Rinse three times with 150 μl of PBS.
-
24
Add 150 μl of PBS.
■ PAUSEPOINT The fixed cells can be stored at 4 °C for several weeks if a sufficient amount of PBS is maintained in each well to cover the cells.
Immunodetection of the incorporated puromycin ● Timing 3–6 h
-
25
Remove the PBS. To permeabilize cells, add PBS with 0.1% (vol/vol) Triton X-100 solution (150 μl) and place cells on a bench at RT for 5 min.
▲ CRITICAL STEP The permeabilization conditions and the following antibody incubation should be optimized for different cell lines, and an additional blocking step after the permeabilization step may be required for certain cell lines.
-
26
Remove the PBS with 0.1% (vol/vol) Triton X-100. Add anti-puromycin antibody incubation solution for cell experiments (150 μl) and place the plate in a 37 °C incubator for 30 min.
-
27
Aspirate the solution and add 150 μl of PBS in each well. Repeat this process three times in total.
-
28
Add secondary anti-mouse antibody incubation solution in each well for cell experiments (150 μl) and place the plate in a 37 °C incubator for 30 min.
-
29
Aspirate the solution and add 150 μl of PBS in each well. Repeat this process three times in total.
-
30
Add in PBS and image cells by using an inverted confocal microscope according to the instrument’s protocol. Alexa Fluor 488 was excited at 488 nm, and emission was collected at 500–550 nm. Hoechst 33342 was excited at 375 nm, and emission was collected at 435–480 nm.
? TROUBLESHOOTING
-
31
Analyze the images by using ImageJ or Harmony high-content imaging and analysis software by measuring fluorescence intensity, following Supplementary information, Data analysis.
H2O2 detection in mice with Peroxymycin-1 and harvesting of liver ● Timing 6–7 h
-
32
Inject 10 mg/kg Peroxymycin-1 stock solution in PBS/PET intraperitoneally into regular- or highfat-diet mice.
-
33
Place mice in cages for 4 h.
-
34
Fill the chamber with CO2 until mice are completely euthanized.
-
35
Cut the mouse belly and harvest liver tissues.
■ PAUSE POINT Harvested liver tissues can be stored at −80 °C, although mounting the tissues in TFM is necessary for the imaging.
Dot blot–based visualization of H2O2 level in Peroxymycin-1–treated mouse liver ● Timing 20 h
Tissue lysis
-
36
Cool down RIPA buffer (1 ml) with ice and add the chilled solution to ~10 mg of the harvested liver sample from Step 35.
-
37
Homogenize the liver sample on ice by using a homogenizer.
-
38
Separate the supernatant (the lysate solution) and insoluble materials by centrifugation (16,000g, 15 min, 4 °C).
-
39
Transfer the supernatant to another centrifuge tube.
▲ CRITICAL STEP Steps 37 and 38 may be repeated multiple times because insoluble materials tend to heterogeneously spread in the lysate solution and may be difficult to completely separate in a single centrifugation process.
? TROUBLESHOOTING
-
40
Quantify the total protein amount (mg/ml) in the lysate by BCA assay, following the manufacturer’s protocol (URL available in Materials).
-
41
Dilute the lysate solution to 1 mg/ml with RIPA buffer.
Dot blot preparation and blocking
-
42
Spot 2 μl of the lysate solution onto a nitrocellulose membrane, pre-cut to 2 × 4 cm for 15 spots.
-
43
Place the membrane on a bench at RT for 20 min, until the spotted droplet dries out.
-
44
Transfer the membrane into a western blot incubation container.
-
45
Add milk TBST solution (1 ml, 5% (wt/vol)) to the container.
-
46
Place the container on an orbital shaker at RT for 30 min.
-
47
Remove the solution from the container.
-
48
Add TBST solution (1 ml) and place the container on the orbital shaker at RT for 3 min.
-
49
Repeat Steps 47 and 48 three times and remove TBST solution from the container.
Antibody staining of dot blot
-
50
Add anti-puromycin or anti-lamin B1 antibody incubation solution for dot blot to the container with the membrane.
-
51
Place the container on the orbital shaker at 4 °C overnight.
-
52
Repeat Steps 47 and 48 five times (for a total of five washes) and remove TBST solution from the container.
-
53
Add anti-mouse antibody incubation solution for dot blot.
-
54
Place the container at RT for 1 h.
-
55
Repeat Steps 47 and 48 five times (for a total of five washes) and remove TBST solution from the container.
-
56
Image the membrane with a blot-imaging system.
Confocal microscopy visualization of H2O2 level in Peroxymycin-1–treated mouse liver ● Timing 3–4 d
Tissue fixation, preservation and cryosectioning
-
57
Add PBS with 4% (wt/vol) formaldehyde solution to the liver tissue sample (Step 36) in a 15-ml Falcon tube.
-
58
Place the tube in a 4 °C refrigerator overnight.
-
59
Remove the PBS with 4% (wt/vol) formaldehyde solution and add 30% (wt/vol) sucrose solution (10 ml).
-
60
Place the tube in a 4 °C refrigerator overnight.
-
61
Trim liver tissue with surgical scissors to isolate the desired region of interest.
-
62
Transfer the liver tissue to the cryomold by using tweezers.
-
63
Fill the cryomold with TFM so that the entire liver tissue is fully covered.
-
64
Place the cryomold on a block of dry ice to freeze the TFM and embedded liver tissue.
■ PAUSE POINT The cryomold was stored at −80 °C for 1 d before moving to the next step. Although longer storage may be possible, we generally recommend moving on to the next step as soon as possible.
-
65
Set the cryostat to −20 °C and thaw the TFM/liver tissue block in the cryostat chamber.
-
66
Section the liver tissue to 30 μm on a slide by using a cryostat. Collect the tissue slice directly onto a superfrost slide.
-
67
Allow all slides to dry at RT for ≥30 min.
■ PAUSE POINT The slides were stored at −80 °C for 1 d before moving to the next step. Although longer storage may be possible, we generally recommend moving on to the next step as soon as possible. After removing from −80 °C, rehydrate slides by adding enough PBS to fully cover the tissue on the slide.
-
68
Blot the area around the tissue with a Kimwipe to remove excess moisture.
-
69
With a hydrophobic pen, delineate a boundary around the tissue sample, leaving a small space around the liver tissue. Dry the boundary at RT for 5 min.
▲ CRITICAL STEP Even when the boundary is being dried, the tissue sample should not be allowed to completely dry out, and a minimal amount of buffer should be applied directly onto the tissue sample with a pipette.
-
70
Place the slide in a staining jar containing PBS and incubate at RT for 3 min.
-
71
Transfer the slide into another staining jar containing fresh PBS and incubate at RT for 3 min.
-
72
Repeat Step 71 two times.
Tissue permeabilization and blocking
-
73
Remove the slide from the jar and add tissue permeabilization buffer (200 μl) directly on the slide. Place a coverslip on the slide to prevent the sample from drying out. Place the slide on a bench at RT for 20 min.
-
74
Remove the tissue permeabilization buffer.
-
75
Add mouse tissue blocking buffer (200 μl), place a coverslip on the slide to prevent the sample from drying out, and place the slide on a rocker at RT for 2 h.
▲ CRITICAL STEP The incubation with the mouse tissue blocking buffer is critical when the sample of interest is derived from mouse or when anti-mouse secondary antibody–fluorophore conjugate will be used. If a sample is from different species or the anti-puromycin–fluorophore conjugate is used, Step 75 should be omitted.
-
76
Remove the mouse tissue blocking buffer.
-
77
Place the slide in a staining jar containing enough tissue permeabilization buffer to completely cover the sample and incubate at RT for 3 min; then, remove the slide from the jar.
-
78
Repeat Step 77 twice.
-
79
Add tissue blocking buffer (200 μl) and place the slide on the bench at RT for 2 h.
-
80
Remove the tissue blocking buffer.
Antibody staining of the tissue sample
-
81
Add anti-puromycin antibody incubation solution for tissue imaging (200 μl) and place the slide covered with a coverslip on a bench at RT for 1 h.
-
82
Remove the anti-puromycin antibody incubation solution.
-
83
Repeat Step 77 twice.
-
84
Add anti-mouse antibody incubation solution for tissue imaging (200 μl) and place the slide covered with a coverslip on a bench at RT for 1 h.
-
85
Remove the anti-mouse antibody incubation solution.
-
86
Repeat Step 77 twice. Additional staining processes such as DAPI staining may be performed during or after Step 84. A staining/wash protocol for the staining of interest should be followed.
-
87
Add mounting medium (25 μl).
-
88
Place a coverslip onto the slide. Remove excess mounting medium coming out of the coverslip by using a Kimwipe.
-
89
Seal the coverslip with nail polish and dry at RT for ≥5 min.
-
90
Image the sample by using a confocal microscope. Alexa Fluor 647 was excited with a 633-nm HeNe laser, and emission was collected on a META detector between 638 and 759 nm.
Troubleshooting
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 13 | Low reaction yields | Hydrolysis of 4-nitrophenyl chloroformate | In-situ formation of the 4-nitrophenyl carbonate intermediate is key for coupling of the aryl-boronate trigger to puromycin. A deep yellow color change upon addition of the puromycin dihydrochloride solution to the generated 4-nitrophenyl carbonate solution indicates the liberation of nitrophenol upon coupling. Absence of this color change indicates that coupling of the aryl-boronate trigger to puromycin has not occurred and can be fixed by using anhydrous solvents. |
| 17 | Multiple peaks were observed in LC analysis after purification | Formation of hydrolyzed boronate (i.e. Pinacol ester was hydrolyzed) | Hydrolyzed Peroxymycin-1 should have 651 m/z. If the hydrolyzed Peroxymycin-1 is the only additional species found, then the mixture is sufficiently pure for use in cell or mouse staining, as the hydrolyzed compound is also active in staining experiments, and is formed when aqueous solution is added to Peroxymycin-1 in the biological experiments and LC-MS characterization experiments. However, if significant concentrations of other compounds such as DMAP (123 m/z), p-nitrophenol (140 m/z), or puromycin (472 m/z), or other unknown comopounds are present then the mixture should be repurified by column chromatography. The extra impurities may be due to the decomposition of 4-nitrophenyl chloroformate, although this was not observed by the authors. Using anhydrous solvent is recommended to decrease the probability of side reactions occurring. |
| 30 | Strong background signal was observed for mouse-derived cell lines | Anti-mouse secondary antibody binds to mouse-specific epitopes. | Block the sample with anti-mouse Fab fragment at step 25. Alternatively, use anti-puromycin antibody– fluorophore conjugates, so that use of the secondary antibody can be omitted. |
| 30 | Variation in fluorescence intensity between replicate samples observed. | Puromycin incorporation rate may differ between samples. | Perform the cell staining experiment outlined in the Anticipated results section and normalize the Peroxymycin-1 signal by control puromycin signal (see Data Analysis section for details). |
| 39 | Tissue lysate solution is still opaque. | The centrifugation process is insufficient. | Repeat the centrifugation multiple times until the solution becomes clearer. |
Timing
With commercially available precursors in hand, the synthesis and purification of Peroxymycin-1 is expected to take 7–9 h. The H2O2 detection or visualization is expected to take 7 h for cell samples, 25 h for mouse tissue samples in the dot blot platform and >4 d for mouse tissue samples in the confocal imaging platform.
Steps 1–17, synthesis of Peroxymycin-1 and Ctrl-Peroxymycin-1: 7–9 h including purification
Steps 18–24, live-cell endogenous H2O2 detection with Peroxymycin-1 and cell fixation: 5–6 h
Steps 25–31, immunodetection of incorporated puromycin: 3–6 h
Steps 32–35, H2O2 detection in mice with Peroxymycin-1 and harvesting of liver: 6–7 h
Steps 36–56, dot blot–based visualization of H2O2 level in Peroxymycin-1–treated mouse liver: 20 h Steps 57–90, visualization of H2O2 level in Peroxymycin-1–treated mouse liver by confocal microscopy: 3–4 d
Anticipated results
Reactivity and selectivity of Peroxymycin-1 toward H2O2 in a test tube
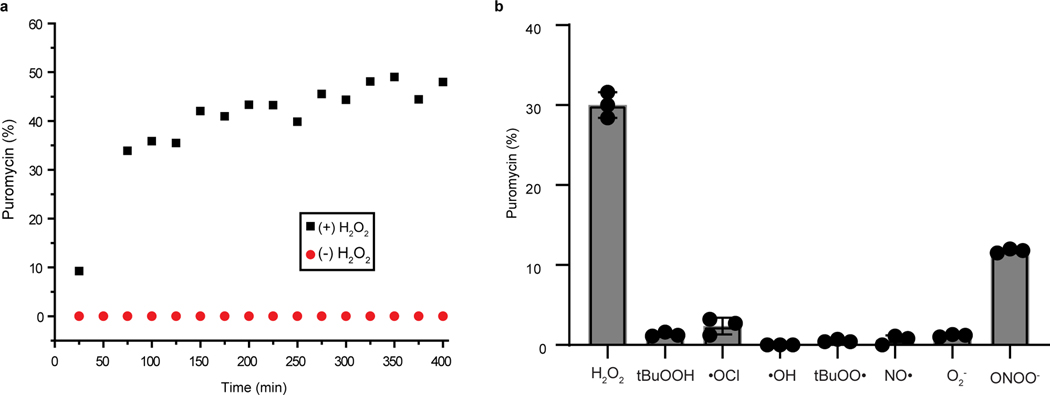
The reaction between Peroxymycin-1 (0.3 mM) and H2O2 (0.1 mM) can be carried out in PBS (20 mM, pH 7.4), methanol and DMSO solution mixture (64% PBS, 33% methanol and 3% DMSO (vol/ vol/vol) and analyzed by LC-MS at different time intervals (Fig. 3). For reactivity experiments, LC-MS analysis was performed on an XBridge C18 column by using a gradient elution from 5% (vol/vol) to 95% (vol/vol) acetonitrile in water (constant 0.05% (vol/vol) formic acid) over 20 min at 1 ml/min (see Materials for column specifications) (Figs. 2c and 3a). After 25 min of reaction, the ratio of Peroxymycin-1 to puromycin was 91% to 9%. After 6 h of reaction, the ratio of Peroxymycin-1 to puromycin was 56% to 44% (Fig. 3a). The peak corresponding to the hydrolyzed free boronic acid Peroxymycin-1 product was observed at 11.0 min (650 m/z), and puromycin was observed at 8.9 min (472 m/z) (Fig. 2c). For selectivity experiments, LC-MS analysis was performed on an SB-C18 column by using a gradient elution from 5% (vol/vol) to 95% (vol/vol) methanol in water (constant 0.05% (vol/vol) formic acid) over 12 min and an isocratic elution with 95% (vol/vol) methanol from 12 to 15 min at 0.5 ml/min (see Materials for column specifications) (Fig. 3b). For ROS selectivity studies, the peak corresponding to the hydrolyzed free boronic acid Peroxymycin-1 product was observed at 14.2 min, and puromycin was observed at 11.5 min.
Fig. 3 |. In vitro analysis of Peroxymycin-1 reactivity and selectivity.
a, Reaction kinetics of Peroxymycin-1 (0.3 mM) treated with H2O2 (0.1 mM) in PBS analyzed by LC-MS. The percentage of puromycin liberated by the reaction was obtained from its UV absorbance and plotted over 6 h. b, Selectivity of Peroxymycin-1 toward various reactive oxygen and nitrogen species. Selectivity of Peroxymycin-1 (0.3 mM) for H2O2 (0.1 mM) over other reactive oxygen and nitrogen species was analyzed by LC-MS after treatment with species at 0.1 mM (except ONOO– at 0.05 mM) for 1 h at room temperature. Error bars denote s.d. (n = 5). tBuOOH, tert-butyl hydroperoxide.
Analytical data
Peroxymycin-1
Peroxymycin-1 was obtained in 57% yield (70 mg) as an off-white solid.
1H NMR (500 MHz, CDCl3): δ 8.09 (s, 1H), 8.06 (s, 1H), 7.75 (d, J = 7.6 Hz, 2H), 7.24 (d, J = 7.9 Hz, 2H), 7.10 (d, J = 8.0 Hz, 2H), 6.84 (s, 1H), 6.81 (d, J = 8.2 Hz, 2H), 5.77 (d, J = 7.5 Hz, 1H), 5.64 (s, 1H), 5.08 (d, J = 12.6 Hz, 1H), 4.99 (d, J = 12.8 Hz, 1H), 4.65 (m, 1H), 4.49 (m, 1H), 4.41 (m, 1H), 3.98 (m, 1H), 3.88 (d, J = 12.4 Hz, 1H), 3.74 (s, 3H), 3.65 (d, J = 12.2 Hz, 1H), 3.60–3.26 (br s, 6H), 3.01 (m, 1H), 2.96 (m, 1H), 1.32 (s, 12H).
13C NMR (126 MHz, CDCl3): δ 172.3, 158.7, 156.1, 154.4, 150.0, 146.8, 139.1, 137.8, 134.9, 130.2, 128.1, 126.9, 120.8, 114.1, 91.0, 84.7, 83.9, 73.3, 66.9, 61.9, 56.5, 55.3, 50.8, 38.1, 24.8.
Ctrl-Peroxymycin-1
Ctrl-Peroxymycin-1 was obtained in 56% yield (70 mg) as an off-white solid.
1H NMR (500 MHz, CDCl3): δ 8.11 (s, 1H), 8.06 (s, 1H), 7.71 (d, J = 8.0 Hz, 2H), 7.16 (d, J = 7.6 Hz, 2H), 7.10 (d, J = 8.1 Hz, 2H), 6.83 (d, J = 8.6 Hz, 2H), 6.76 (s, 1H), 5.64 (s, 1H), 5.57 (d, J = 8.0 Hz, 1H), 4.66 (s, 1H), 4.44 (m, 1H), 4.40 (m, 1H), 4.22 (m, 2H), 4.01 (m, 1H), 3.89 (d, J = 12.7 Hz, 1H), 3.74 (s, 3H), 3.67 (d, J = 12.8 Hz, 1H), 3.58–3.32 (br s, 6H), 3.01 (m, 1H), 2.92 (m, 1H), 2.88 (m, 2H), 1.31 (s, 12H).
13C NMR (126 MHz, CDCl3): δ 172.4, 158.8, 156.3, 154.7, 150.1, 141.2, 137.9, 135.1, 130.4, 128.4, 128.4, 120.9, 114.3, 91.2, 84.9, 83.9, 73.4, 65.8, 62.1, 56.5, 55.4, 53.6, 50.9, 38.2, 35.7, 24.9.
Detection of endogenous H2O2 in live cells with Peroxymycin-1 and immunostaining
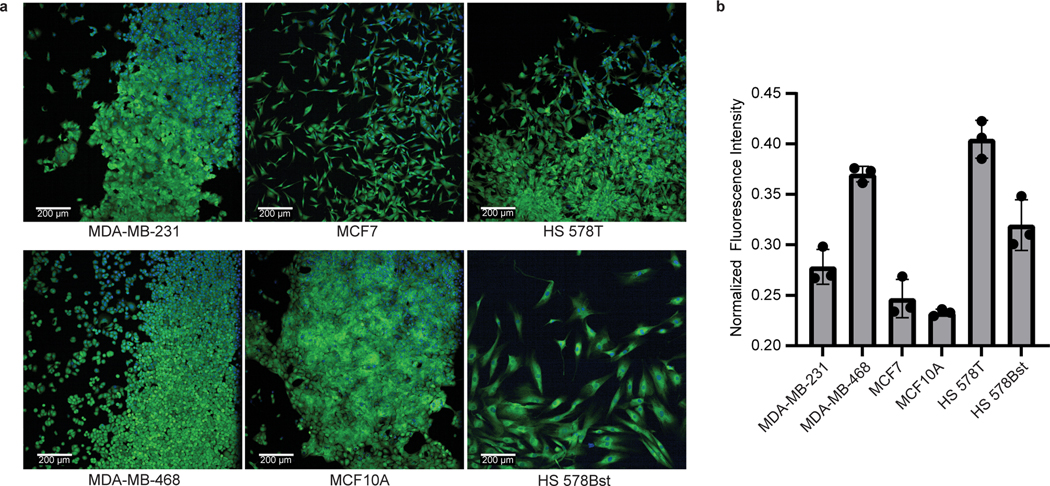
Prepare a 96-well plate with six wells of each of the following cell types: MDA-MB-231, MDA-MB-468, MCF7, MCF10A, HS 578T and HS 578Bst. Follow Steps 18–31 of the above protocol. At Step 19, treat three wells of each cell type with puromycin incubation medium and three wells with Peroxymycin-1 incubation medium. When imaging the cells, stronger fluorescence should be observed for puromycin-treated cells, and the mean fluorescence intensity measurements of the puromycin-treated cells should be used to normalize the fluorescence signal of Peroxymycin-1–treated samples (Fig. 4).
Fig. 4 |. Confocal microscopy images of endogenous H2O2 detection by using Peroxymycin-1.
a, Confocal images of Peroxymycin-1–treated human cell lines (1 μM). Hoechst: nuclear staining (blue channel). Anti-puromycin: staining with mouse anti-puromycin antibody, visualized by donkey anti-mouse antibody–Alexa Fluor 488 conjugate (green channel). Scale bar: 200 μm for all images. b, Quantification of fluorescence intensity of the images of Peroxymycin-1–treated cell lines. Values are plotted as Peroxymycin-1 fluorescence intensity ± s.d. (n = 3 technical replicates, with 2 fields examined per replicate), normalized to puromycin-treated control cells (for details of the normalization process, see Supplementary information, data analysis).
Detection of H2O2 in mouse tissue with Peroxymycin-1 and immunostaining
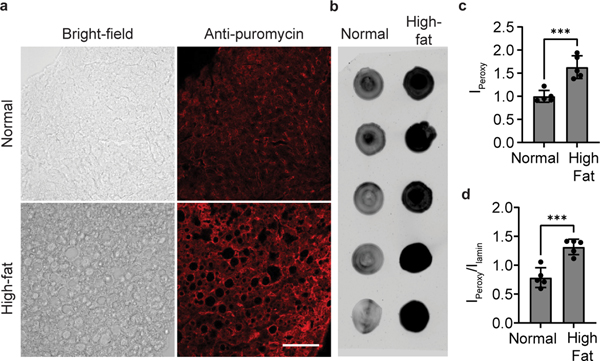
Feed 8-week-old male C57BL/6 mice with normal chow (NC) or anHFD for 20 weeks. Following Steps 32–35, inject the mice with Peroxymycin-1 and harvest the liver tissue. For analysis by dot blot, follow Steps 36–56. A higher fluorescence ratio should be observed for anti-puromycin to anti-lamin B1 staining in the HFD mice than in the NC control mice (Fig. 5b,d). For immunofluorescence imaging using confocal microscopy, complete Steps 57–90. Again, a higher fluorescence signal should be observed for HFD mice than for NC control mice (Fig. 5a,c).
Fig. 5 |. Profiling H2O2 levels with Peroxymycin-1 in liver tissue of mice fed a normal diet and a high-fat diet.
10 mg/kg of Peroxymycin-1 were injected intraperitoneally into mice, and the liver was harvested after 4 h. IPeroxy was divided by Ilamin for normalization of the total protein amount in the lysate sample. a, Confocal microscopy images of liver tissues of Peroxymycin-1–treated mice, stained with mouse anti-puromycin and anti-mouse–Alexa Fluor 647. Scale bar: 100 μm. b, Dot blots of liver tissue lysates probed by using mouse anti-puromycin antibody and then anti-mouse antibody–Alexa Fluor 488. c and d, Quantification of intensities of a and b, respectively. Error bars denote s.d. (n = 5). ***, P < 0.001. Institutional approval for the experiments shown in this figure was obtained from the University of California, Berkeley.High fat, mice raised on high-fat chow; Ilamin, fluorescence intensity of puromycin–treated sample stained with mouse anti-lamin antibody and anti-mouse antibody–Alexa Fluor 647 conjugates; IPeroxy, fluorescence intensity of puromycin-treated sample stained with mouse anti-puromycin antibody and anti-mouse antibody–Alexa Fluor 647 conjugates; Normal, mice raised on normal chow. Confocal and dot blot images reprinted with permission from ref. 59, American Chemical Society.
Supplementary Material
Acknowledgements
We thank the NIH (R01 GM 79465, R01 GM 139245 and R01 ES 28096 to C.J.C.) for research support. K.H. thanks the College of Chemistry for a summer undergraduate research fellowship. J.O. thanks the Japan Society for the Promotion of Science for a postdoctoral fellowship. C.Y.-S.C. thanks the Croucher Foundation for a postdoctoral fellowship. M.S.M. thanks the UC President’s Postdoctoral Fellowship Program, Chinook-Berkeley Postdoctoral Fellowship Program and an NIH MOSAIC K99/R00 (1K99GM143573-01) award for funding. C.J.C. is a CIFAR Fellow. We thank Alison Killilea and Carissa Tasto (UC Berkeley Tissue Culture Facility) for expert technical assistance.
Footnotes
Competing interests
A patent application has been filed for the Peroxymycin-1 probe. The patent application number is PCT/US2019/023242.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596-022-00694-7.
Peer review information Nature Protocols thanks Deju Ye and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Data are available through figshare: Fig. 2c, https://doi.org/10.6084/m9.figshare.16639975; Fig. 3a, https://doi.org/10.6084/m9.figshare.16639570; Fig. 4, https://doi.org/10.6084/m9.figshare.16640434; Fig. 5, https://doi.org/10.6084/m9.figshare.16639537; Supplementary Fig. 1, https://doi.org/10.6084/m9.figshare.18480879; Supplementary Fig. 2, https://doi.org/10.6084/m9.figshare.18480885; Supplementary Fig. 3, https://doi.org/10.6084/m9.figshare.18480888; Supplementary Fig. 4, https://doi.org/10.6084/m9.figshare.18480891; Supplementary Fig. 5, https://doi.org/10.6084/m9.figshare.18480894; Supplementary Fig. 6, https://doi.org/10.6084/m9.figshare.18480897; Supplementary Fig. 7, https://doi.org/10.6084/m9.figshare.18480900; Supplementary Fig. 8, https://doi.org/10.6084/m9.figshare.18480903. Source data are provided with this paper.
References
- 1.Baynes JW Role of oxidative stress in development of complications in diabetes. Diabetes 40, 405–412 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Multhaup G. et al. Reactive oxygen species and Alzheimer’s disease. Biochem. Pharmacol 54, 533–539 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Stone JR & Yang S. Hydrogen peroxide: a signaling messenger. Antioxid. Redox Signal 8, 243–270 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Rhee SG H2O2, a necessary evil for cell signaling. Science 312, 1882–1883 (2006). [DOI] [PubMed] [Google Scholar]
- 5.D’Autréaux B. & Toledano MB ROS as signaling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol 8, 813–824 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Finkel T, Serrano M. & Blasco MA The common biology of cancer and ageing. Nature 448, 767–774 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Winterbourn CC Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol 4, 278–286 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Dickinson BC & Chang CJ Chemistry and biology of reactive oxygen species in signaling or stressresponses. Nat. Chem. Biol 7, 504–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy MP et al. Unraveling the biological roles of reactive oxygen species. Cell Metab. 13, 361–366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schieber M. & Chandel NS ROS function in redox signaling and oxidative stress. Curr. Biol 24, R453–R462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichmann D, Voth W. & Jakob U. Maintaining a healthy proteome during oxidative stress. Mol. Cell 69, 203–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sies H. & Jones DP Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol 21, 363–383 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Inoguchi T. et al. High glucose level and free fatty acid stimulate reactive oxygen species production throughprotein kinase C–dependent activation of NAD(P)H oxidase in cultured vascular cells. Diabetes 49, 1939–1945 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Park L. et al. Nox2-derived radicals contribute to neurovascular and behavioral dysfunction in mice overexpressing the amyloid precursor protein. Proc. Natl Acad. Sci. USA 105, 1347–1352 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt KN, Amstad P, Cerutti P. & Baeuerle PA The roles of hydrogen peroxide and superoxide asmessengers in the activation of transcription factor NF-κB. Chem. Biol 2, 13–22 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Guyton KZ, Liu Y, Gorospe M, Xu Q. & Holbrook NJ Activation of mitogen-activated protein kinaseby H2O2: role in cell survival following oxidant injury. J. Biol. Chem 271, 4138–4142 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Lee S-R, Kwon K-S, Kim S-R & Rhee SG Reversible inactivation of protein-tyrosine phosphatase 1B inA431 cells stimulated with epidermal growth factor. J. Biol. Chem 273, 15366–15372 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Salmeen A. et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423, 769–773 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Avshalumov MV & Rice ME Activation of ATP-sensitive K+ (KATP) channels by H2O2 underlies glutamate-dependent inhibition of striatal dopamine release. Proc. Natl Acad. Sci. USA 100, 11729–11734 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambeth JD NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol 4, 181–189 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Dinauer MC, Orkin SH, Brown R, Jesaitis AJ & Parkos CA The glycoprotein encoded by theX-linked chronic granulomatous disease locus is a component of the neutrophil cytochrome b complex. Nature 327, 717–720 (1987). [DOI] [PubMed] [Google Scholar]
- 22.Volpp B, Nauseef W. & Clark R. Two cytosolic neutrophil oxidase components absent in autosomalchronic granulomatous disease. Science 242, 1295–1297 (1988). [DOI] [PubMed] [Google Scholar]
- 23.Clark RA et al. Genetic variants of chronic granulomatous disease: prevalence of deficiencies of two cytosolic components of the NADPH oxidase system. N. Engl. J. Med 321, 647–652 (1989). [DOI] [PubMed] [Google Scholar]
- 24.Ohba M, Shibanuma M, Kuroki T. & Nose K. Production of hydrogen peroxide by transforming growthfactor-beta 1 and its involvement in induction of egr-1 in mouse osteoblastic cells. J. Cell Biol 126, 1079–1088 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundaresan M, Yu Z-X, Ferrans VJ, Irani K. & Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 270, 296–299 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Kimura T, Okajima F, Sho K, Kobayashi I. & Kondo Y. Thyrotropin-induced hydrogen peroxideproduction in FRTL-5 thyroid cells is mediated not by adenosine 3′, 5′-monophosphate, but by Ca2+ signaling followed by phospholipase-A2 activation and potentiated by an adenosine derivative. Endocrinology 136, 116–123 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Bae YS et al. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGFreceptor-mediated tyrosine phosphorylation. J. Biol. Chem 272, 217–221 (1997). [PubMed] [Google Scholar]
- 28.Mukhin YV et al. 5-Hydroxytryptamine1A receptor/Giβγ stimulates mitogen-activated protein kinase via NAD(P)H oxidase and reactive oxygen species upstream of src in chinese hamster ovary fibroblasts. Biochem. J 347, 61–67 (2000). [PMC free article] [PubMed] [Google Scholar]
- 29.Dickinson BC, Peltier J, Stone D, Schaffer DV & Chang CJ Nox2 redox signaling maintains essentialcell populations in the brain. Nat. Chem. Biol 7, 106–112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamsler A. & Segal M. Hydrogen peroxide modulation of synaptic plasticity. J. Neurosci 23, 269–276 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tejada-Simon MV et al. Synaptic localization of a functional NADPH oxidase in the mouse hippocampus.Mol. Cell. Neurosci 29, 97–106 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennan AM et al. NADPH oxidase is the primary source of superoxide induced by NMDA receptoractivation. Nat. Neurosci 12, 857–863 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Pasquale R, Beckhauser TF, Hernandes MS & Giorgetti Britto LR LTP and LTD in the visualcortex require the activation of NOX2. J. Neurosci 34, 12778–12787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Belle JE et al. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewaland neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell 8, 59–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C, Luo J, He L, Montell C. & Perrimon N. Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca2+ signaling in the Drosophila midgut. eLife 6, e22441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Neill JS & Reddy AB Circadian clocks in human red blood cells. Nature 469, 498–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wible RS et al. NRF2 regulates core and stabilizing circadian clock loops, coupling redox and timekeepingin Mus musculus. eLife 7, e31656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pei J-F et al. Diurnal oscillations of endogenous H2O2 sustained by p66Shc regulate circadian clocks. Nat. Cell Biol 21, 1553–1564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niethammer P, Grabher C, Look AT & Mitchison TJ A tissue-scale gradient of hydrogen peroxidemediates rapid wound detection in zebrafish. Nature 459, 996–999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hervera A. et al. Reactive oxygen species regulate axonal regeneration through the release of exosomalNADPH oxidase 2 complexes into injured axons. Nat. Cell Biol 20, 307–319 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Lippert AR, Van de Bittner GC & Chang CJ Boronate oxidation as a bioorthogonal reaction approachfor studying the chemistry of hydrogen peroxide in living systems. Acc. Chem. Res 44, 793–804 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan J, Dodani SC & Chang CJ Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat. Chem 4, 973–984 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brewer TF, Garcia FJ, Onak CS, Carroll KS & Chang CJ Chemical approaches to discovery andstudy of sources and targets of hydrogen peroxide redox signaling through NADPH oxidase proteins. Annu. Rev. Biochem 84, 765–790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruemmer KJ, Crossley SWM & Chang CJ Activity-based sensing: a synthetic methods approach forselective molecular imaging and beyond. Angew. Chem. Int. Ed 59, 13734–13762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang MCY, Pralle A, Isacoff EY & Chang CJ A selective, cell-permeable optical probe for hydrogenperoxide in living cells. J. Am. Chem. Soc 126, 15392–15393 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller EW, Tulyathan O, Isacoff EY & Chang CJ Molecular imaging of hydrogen peroxide producedfor cell signaling. Nat. Chem. Biol 3, 263–267 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Dickinson BC, Huynh C. & Chang CJ A palette of fluorescent probes with varying emission colors for imaging hydrogen peroxide signaling in living cells. J. Am. Chem. Soc 132, 5906–5915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srikun D, Miller EW, Domaille DW & Chang CJ An ICT-based approach to ratiometric fluorescence imaging of hydrogen peroxide produced in living cells. J. Am. Chem. Soc 130, 4596–4597 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Albers AE, Okreglak VS & Chang CJ A FRET-based approach to ratiometric fluorescence detection of hydrogen peroxide. J. Am. Chem. Soc 128, 9640–9641 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Chung C, Srikun D, Lim CS, Chang CJ & Cho BR A two-photon fluorescent probe for ratiometric imaging of hydrogen peroxide in live tissue. Chem. Commun 47, 9618–9620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dickinson BC & Chang CJ A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J. Am. Chem. Soc 130, 9638–9639 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dickinson BC, Tang Y, Chang Z. & Chang CJ A nuclear-localized fluorescent hydrogen peroxide probe for monitoring sirtuin-mediated oxidative stress responses in vivo. Chem. Biol 18, 943–948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller EW, Dickinson BC & Chang CJ Aquaporin-3 mediates hydrogen peroxide uptake to regulatedownstream intracellular signaling. Proc. Natl Acad. Sci. USA 107, 15681–15686 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwashita H, Castillo E, Messina MS, Swanson RA & Chang CJ A tandem activity-based sensing andlabeling strategy enables imaging of transcellular hydrogen peroxide signaling. Proc. Natl Acad. Sci. USA 118, e2018513118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van de Bittner GC, Dubikovskaya EA, Bertozzi CR & Chang CJ In vivo imaging of hydrogenperoxide production in a murine tumor model with a chemoselective bioluminescent reporter. Proc. Natl Acad. Sci. USA 107, 21316–21321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van de Bittner GC, Bertozzi CR & Chang CJ Strategy for dual-analyte luciferin imaging: in vivobioluminescence detection of hydrogen peroxide and caspase activity in a murine model of acute Inflammation. J. Am. Chem. Soc 135, 1783–1795 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin L. et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulateredox homeostasis and tumor growth. Cancer Cell 27, 257–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoenfeld JD et al. O2•− and H2O2–mediated disruption of Fe metabolism causes the differential susceptibility of NSCLC and GBM cancer cells to pharmacological ascorbate. Cancer Cell 31, 487–500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung CY-S, Timblin GA, Saijo K. & Chang CJ Versatile histochemical approach to detection ofhydrogen peroxide in cells and tissues based on puromycin staining. J. Am. Chem. Soc 140, 6109–6121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhibi M. et al. The intake of high fat diet with different trans fatty acid levels differentially induces oxidative stress and non alcoholic fatty liver disease (NAFLD) in rats. Nutr. Metab 8, 65–77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bilan Dmitry S. & Belousov Vsevolod V. In vivo imaging of hydrogen peroxide with HyPer probes.Antioxid. Redox Signal 29, 569–584 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Morgan B. et al. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat. Chem. Biol 12, 437–443 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Srikun D, Albers AE, Nam CI, Iavarone AT & Chang CJ Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-Tag protein labeling. J. Am. Chem. Soc 132, 4455–4465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickinson BC, Lin VS & Chang CJ Preparation and use of MitoPY1 for imaging hydrogen peroxide inmitochondria of live cells. Nat. Protoc 8, 1249–1259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szweda PA, Tsai L. & Szweda LI Immunochemical detection of a fluorophore derived from the lipid peroxidation product 4-hydroxy-2-nonenal and lysine. In Oxidants and Antioxidants: Ultrastructure and Molecular Biology Protocols (ed. Armstrong D) Vol. 196 277–290 (Humana Press, 2002). [DOI] [PubMed] [Google Scholar]
- 66.Spangler B. et al. A reactivity-based probe of the intracellular labile ferrous iron pool. Nat. Chem. Biol 12, 680–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidt EK, Clavarino G, Ceppi M. & Pierre P. SUnSET, a nonradioactive method to monitor proteinsynthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Su K-H et al. HSF1 critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promotegrowth. Nat. Cell Biol 18, 527–539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.tom Dieck S. et al. Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deliu LP, Ghosh A. & Grewal SS Investigation of protein synthesis in Drosophila larvae using puromycin labelling. Biol. Open 6, 1229–1234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bielczyk-Maczyńska E. et al. The ribosome biogenesis protein Nol9 is essential for definitive hematopoiesis and pancreas morphogenesis in zebrafish. PLoS Genet. 11, e1005677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available through figshare: Fig. 2c, https://doi.org/10.6084/m9.figshare.16639975; Fig. 3a, https://doi.org/10.6084/m9.figshare.16639570; Fig. 4, https://doi.org/10.6084/m9.figshare.16640434; Fig. 5, https://doi.org/10.6084/m9.figshare.16639537; Supplementary Fig. 1, https://doi.org/10.6084/m9.figshare.18480879; Supplementary Fig. 2, https://doi.org/10.6084/m9.figshare.18480885; Supplementary Fig. 3, https://doi.org/10.6084/m9.figshare.18480888; Supplementary Fig. 4, https://doi.org/10.6084/m9.figshare.18480891; Supplementary Fig. 5, https://doi.org/10.6084/m9.figshare.18480894; Supplementary Fig. 6, https://doi.org/10.6084/m9.figshare.18480897; Supplementary Fig. 7, https://doi.org/10.6084/m9.figshare.18480900; Supplementary Fig. 8, https://doi.org/10.6084/m9.figshare.18480903. Source data are provided with this paper.