Abstract
Intracellular transport is the basis for the transfer of matter, energy, and information in cells and is critical to many cellular functions. Within the nonequilibrium environment of living cells, the transport behaviours are far from the traditional motion in liquid but are more complex and active. With the advantage of high spatial and temporal resolution, the single-particle tracking (SPT) method is widely utilized and has achieved great advances in revealing intracellular transport dynamics. This review describes intracellular transport from a physical perspective and classifies it into two modes: diffusive motion and directed motion. The biological functions and physical mechanisms for these two transport modes are introduced. Next, we review the principle of SPT and its advances in two aspects of intracellular transport. Finally, we discuss the prospect of near infrared SPT in exploring the in vivo intracellular transport dynamics.
Keywords: Intracellular transport, Single-particle tracking (SPT), Diffusive motion, Directed motion, Near infrared SPT, Dynamics
INTRODUCTION
Intracellular transport dynamics are critical to many cellular functions, such as cell proliferation, motility, and death (Mogre et al. 2020). Almost all biochemical reactions in the cell rely on the intracellular transport of biomolecules. Proteins synthesized in ribosomes usually undergo transportation from the perinuclear regions to the peripheral sites in the cell or are secreted outside the cell membrane (Schwarz and Blower 2016). As the energy unit in cells, ATP is produced in the mitochondria and transported to different sites within the cell for consumption. External signals received by the cell membrane usually need to be transmitted to the nucleus to initiate transcriptional responses. In short, intracellular transport is the basis for the transfer of matter, energy, and information.
The living cell is a nonequilibrium system in which many energy-consuming reactions give rise to complex and active transport behaviours (Gallet et al. 2009; Wilhelm 2008). In addition, macromolecule crowding and spatial heterogeneity have significant impacts on intracellular transport. As such, molecule transport dynamics in living cells is far from traditional motion in liquid. According to the motion type, transport in eukaryotic cells can be divided into random diffusion and directed transport from a physical perspective (Fig. 1).
Figure 1.
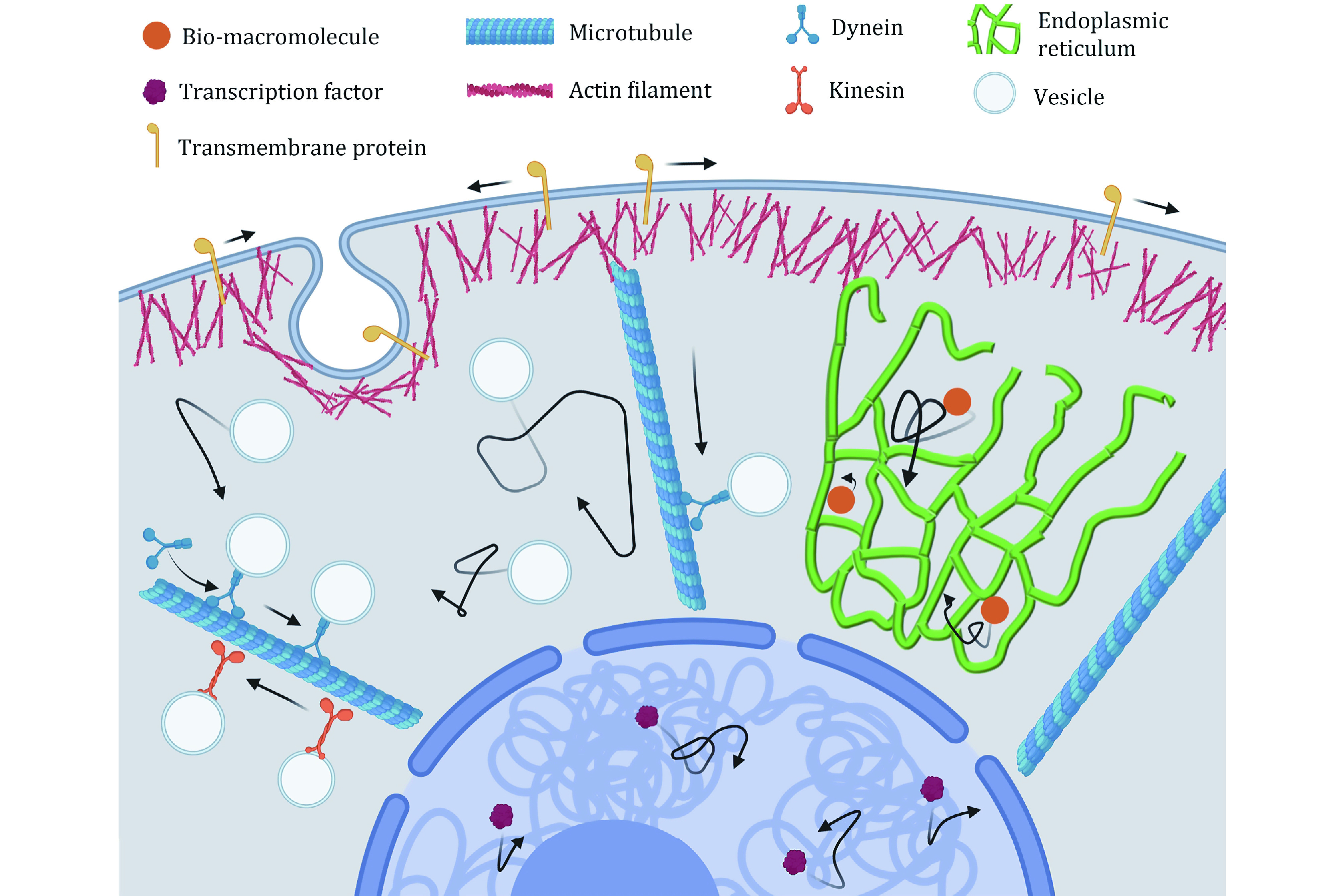
Schematic of intracellular transport. In cytoplasm, the diffusion of vesicles and bio-macromolecules, and the directed motion of vesicles driven by motor proteins along microtubules. In nucleus, the diffusion of the transcription factors
Exploring the dynamics of intracellular transport requires advanced technologies. In recent decades, the development of noninvasive techniques for visualizing dynamic processes in living cells has greatly promoted our understanding of intracellular transport, including fluorescence recovery after photobleaching (FRAP) (Lippincott-Schwartz et al. 2003; Reits and Neefjes 2001) and fluorescence correlation spectroscopy (FCS) (Bulseco and Wolf 2007; Elson 2011; Haustein and Schwille 2007; Kim et al. 2007; Tudor et al. 2007; Vukojevic et al. 2007). However, the above techniques provide ensemble average results, lacking the spatiotemporal dynamics of individual molecules. Single-particle tracking (SPT) enables us to visualize individual molecules in living cells, locate single molecules with nanoscale precision, and measure their individual transport dynamics as a function of time. In terms of the spatiotemporal information from single molecule trajectories, we can further probe the heterogeneous environment within the cell. Many advances have been made in the study of intracellular transport dynamics through SPT (Balint et al. 2013; Cognet et al. 2014; Ge et al. 2021; Hui et al. 2017; Jiang et al. 2020; Kusumi et al. 2014; Li et al. 2015b, 2016b, 2018a; von Diezmann et al. 2017; Xu et al. 2021).
This review is organized as follows. First, we introduce the biological functions of intracellular transport. Next, we describe two types of intracellular transport and their physical mechanisms. Then, we concentrate on the fundamental principle of SPT technology, including imaging and data analysis. After that, we focus on the applications of SPT in intracellular studies. Finally, we discuss future studies of intracellular transport dynamics, especially strategies for developing in vivo SPT methods.
BIOLOGICAL MEANING OF INTRACELLULAR TRANSPORT
Matter translocation
Matter translocation plays an important role in maintaining the physiological functions of cells. The secretions synthesized by cells are transported from intracellular to extracellular space. For example, synthesized proteins are translocated from perinuclear to peripheral sites or eventually secreted outside the cell membrane. In eukaryotes, proteins are first synthesized in ribosomes attached to the rough endoplasmic reticulum (ER) (Schwarz and Blower 2016). Then, after further modifications in the Golgi (Orci et al. 2000), proteins are transported in vesicles to the plasma membrane or secreted into the extracellular matrix. Conversely, extracellular biomolecules that enter the membrane undergo intracellular transport in the cytoplasm. For example, extracellular small molecules such as amino acids cross the cell membrane by diffusion, while macromolecules such as growth factors, membrane proteins, and lipids pass through the cell membrane via endocytosis and are then translocated within the cell (Basturea 2019). Viruses, such as influenza, enter the cell through endocytosis and undergo intracellular transport to their destinations (Lakadamyali et al. 2003).
In cells, diffusion is the major mechanism for matter translocations. However, the diffusion coefficients of large cargos such as the vesicles in oocytes and lysosomes in epithelial cells are only 0.003 and 0.071 μm2/s, respectively (Bandyopadhyay et al. 2014; Drechsler et al. 2017; Koslover et al. 2016). At such slow diffusion rates, it would take several hours for the large cargos to cross the cell. To be more efficient, cells utilize motor-driven active transport, which is essential in eukaryotic cells (De Matteis and Luini 2008). For example, in neurons which can be up over 1-metre long, axon cargos are transported along microtubules by motor proteins, including kinesin and dynein (Mudrakola et al. 2009).
Energy transfer
Energy is stored in the form of ATP or nutrients such as carbohydrates and fats, playing indispensable roles in intracellular metabolism and various life activities. Small molecules such as ATP and glucose are rapidly transported within the cell by diffusion, with diffusion rates of approximately 145 and 200 μm2/s, respectively (Riley et al. 1999; Vendelin et al. 2000). Due to the low efficiency of diffusion in large-scale transport, actively directed transport prevails in large cells. For example, in individual hyphae, nutrients such as N-rich amino acids are translocated by both diffusion and vesicle transport; however, in the mycelial network, vesicle transport dominates (Fricker et al. 2017, 2008).
Signal transduction
When the cell membrane senses and receives signals from the extracellular environment, the signals in the form of active small molecules are usually transmitted to the nucleus, in which certain transcriptional regulation takes place to trigger cellular responses. Intracellular signal transduction involves the diffusion of activated proteins from the cell membrane to cytoplasmic targets or other cellular sites. For example, MEK kinase is activated at the membrane and then diffuses into the cytoplasm to activate downstream kinases (Kholodenko et al. 2000). In addition, vesicle transport driven by motor proteins is an efficient alternative. A typical example is the internalization and endocytic trafficking of nerve growth factors in neuronal signalling pathways (Harrington and Ginty 2013). Besides, the cellular mechanotransduction as a growing area is also related to the intracellular transport (He et al. 2020; Liu et al. 2019; Wang et al. 2020).
PHYSICAL MECHANISMS OF INTRACELLULAR TRANSPORT
To achieve various functional goals of intracellular transport, eukaryotic cells mainly rely on two mechanisms: random diffusion and directed transport. The diffusion is driven by the combination of thermal and intracellular active fluctuations, whereas the directed transport is driven by motor proteins along the cytoskeletons (Fig. 1). Both of them are influenced by the intracellular ATP levels and macromolecule crowding.
Diffusion
The irregular motion of mesoscale particles suspended in a solvent is named Brownian motion and was first found by Robert Brown in 1826, who observed the continuously agitating motion of pollen grains under a light microscope (Brown 1828). Brownian motion is caused by the thermal fluctuations of water molecules. The diffusion coefficient, which represents the range of motion, increases with temperature. The time required for translocation by diffusion is proportional to the square of the distance. Moreover, the diffusion coefficient decreases with particle size or medium viscosity. Although diffusion is nondirectional, it works well for transporting small molecules over short distances (Di Rienzo et al. 2014).
However, intracellular diffusion is different from that observed in dilute solutions (Gregor et al. 2005; Jacobson and Wojcieszyn 1984; Lubyphelps et al. 1987). In some cases, the diffusive motion within the cell appears to be random, although the diffusion rate is much greater than the expected diffusion in solutions (Bursac et al. 2005; Lau et al. 2003). This suggests that other factors contribute to intracellular diffusion in addition to thermal fluctuations. The internal environment of living cells obviously deviates from the equilibrium state, in which many active processes consuming energy exist. Therefore, unlike thermal diffusion in an equilibrium system, the amplitude of diffusive motion in cells is additionally driven by active fluctuations, leading to randomly diffusive motions with increased diffusion rates (Brangwynne et al. 2009; Fakhri et al. 2014; Guo et al. 2014).
Individual molecule trajectories in living cells show nonlinear mean square displacement (MSD) as a function of time, which suggests anomalous diffusion (Saxton and Jacobson 1997; Wieser and Schutz 2008). This anomalous diffusion is attributed to the interactions between molecules and the surrounding intracellular environment, caused by the broad distribution of jump times or jump lengths, or the strong correlations (Bouchaud and Georges 1990). In most cases, the motion of macromolecules and organelles in the cytoplasm is subdiffusion (Hoffman et al. 2006; Li et al. 2016a; Shen et al. 2016; Tolic-Norrelykke et al. 2004; Yamada et al. 2000) due to viscoelastic properties and the crowded environment in the cytoplasm (Hofling and Franosch 2013; Luby-Phelps 2000; Shen et al. 2021; Wang et al. 2013; Weber et al. 2010).
In addition, the distribution of individual step sizes of small molecules in the cytoplasm shows a Laplace distribution, which is different from the desired Gaussian distribution in a uniform medium (Fodor et al. 2015; Gal et al. 2013; He et al. 2016; Lampo et al. 2017). The Laplace distribution is attributed to the wide distribution of diffusivities for individual particles in inhomogeneous environments (Luo and Yi 2018). Such a non-Gaussian distribution indicates the spatial heterogeneity of biomolecule motion in the cytoplasm (Chechkin et al. 2017; Duits et al. 2009). It should be noted that subdiffusion does not necessarily imply a non-Gaussian distribution. For example, fractional Brownian motion is anomalous diffusion but appears as a Gaussian distribution. In contrast, some normal diffusion can exhibit a non-Gaussian distribution (Chechkin et al. 2017; Wang et al. 2012).
Directed motion
In the crowded environment of cells, the diffusion of large molecules and vesicles is physically constrained and is not sufficiently effective for long-distance transport. In this case, directed transport, which relies on motor proteins that hydrolyse ATP and drag the cargos along the cytoskeleton, is more applicable (Brown and Sivak 2020).
Cytoskeletons and molecular motors are essential for directed transport. Cytoskeletons, including microtubules and microfilaments (F-actin), are involved in intracellular transport. Microtubules, the structural backbone of the cytoskeleton, are long hollow tubes with a diameter of 22–25 nm composed of 13 parallel protofilaments. Microtubules are highly dynamic and polarized, alternating between phases of growth and shrinkage (de Forges et al. 2012). Microtubules provide the path for the long-range transport of organelles and membranes. Kinesin and dynein are motor proteins that transport along microtubules (Kikushima et al. 2013; Ross et al. 2008). In general, kinesins move towards the plus ends of microtubules from the perinucleus to the periphery (Duan et al. 2012; Hirokawa et al. 2009), whereas dynein drives retrograde movements towards the minus ends of microtubules from the periphery to the perinucleus (Cianfrocco et al. 2015; Reck-Peterson et al. 2018).
Another important cytoskeleton is the microfilament, which is a solid fibre with a diameter of 4–7 nm distributed beneath cell membranes and in the cytoplasm. Actin filaments are short and polarized. They usually form a randomly oriented network with a mesh size of approximately 50 nm (Barlan et al. 2013). Myosin motors mainly contribute to localized movements of cargo in a short range along actin filaments.
Directed motion has two major advantages. One is to transport intracellular cargo over relatively long distances. For example, in the axons of neurons, the distance can be up to one metre (Hirokawa et al. 2010). Another advantage is that the direction and speed transport dynamics can be well controlled by the cells (Burute and Kapitein 2019). For example, the ratio between kinesin and dynein can affect the direction of cargo transport, which has already been shown by in vitro experiments (Hendricks et al. 2010). A cargo can be driven by multiple motors which cooperate or compete with each other. When the same motors carry cargo, they may share the load and improve performance (Reis et al. 2012). When two different motor proteins attach to a cargo, it may be driven in two directions: if only one motor is active, it determines the cargo's direction of movement; if both motors are active, they will pull in opposite directions, with the stronger determining the direction of transport (Barlan et al. 2013; Gennerich et al. 2007; Hancock 2014; Hendricks et al. 2010; McLaughlin et al. 2016; Muller et al. 2008).
PRINCIPLE OF SINGLE PARTICLE TRACKING
Advanced optical techniques have been applied to explore and understand transport dynamics within the intracellular world. One of the common techniques is fluorescence recovery after photobleaching (FRAP). In FRAP experiments, fluorescent molecules in a small given area are first photobleached by a focused laser beam with high intensity. Then, surrounding unbleached fluorescent molecules diffuse into the photobleached area resulting in fluorescence recovery (Lippincott-Schwartz et al. 2003; Reits and Neefjes 2001). The recovery of the fluorescence intensities is calculated to obtain the molecular diffusivity. Another technique is fluorescence correlation spectroscopy (FCS), which records the fluctuation of fluorescence intensity in a small, illuminated volume. The diffusion coefficient can be determined from the autocorrelation function of the fluorescence intensities (Bulseco and Wolf 2007; Elson 2011; Haustein and Schwille 2007; Kim et al. 2007; Peng et al. 2020; Tudor et al. 2007; Vukojevic et al. 2007).
In contrast to the above ensemble-averaged methods, SPT provides a new perspective of single molecule motion and a better understanding of intracellular transport dynamics. SPT provides spatiotemporal information about the movement of a single biomolecule, which helps to accurately measure different types of biomolecule transport in cells and understand its complexity. In SPT experiments, probes are used to label the molecules of interest, observe their movements under appropriate optical instruments, and analyse their trajectories to explore the intracellular transport dynamics.
Probes
In experiments, the biomolecules first need to be labelled with proper probes, which enables observation and analysis of single-particle motion under microscopy. The probes must have good biocompatibilities with no harmful effects on cell activities. Moreover, the physical properties of the probes are of vital importance, and the probe cannot be so large that it impacts the original motion of biomolecules. To ensure that long trajectories can be followed, the probe should be photostable for long-term imaging.
Researchers have used gold particles (Kusumi et al. 1993), organic dyes (Schmidt et al. 1996), fluorescent proteins (Iino et al. 2001), and quantum dots (QDs) (Dahan et al. 2003) to label particles of interest (Fig. 2A). In 1993, Kusumi et al. labelled receptors on the cell membrane with 40-nm gold particles and observed the diffusive motion of the receptors (Kusumi et al. 1993). However, gold particles cannot be used in multiple-colour imaging.
Figure 2.
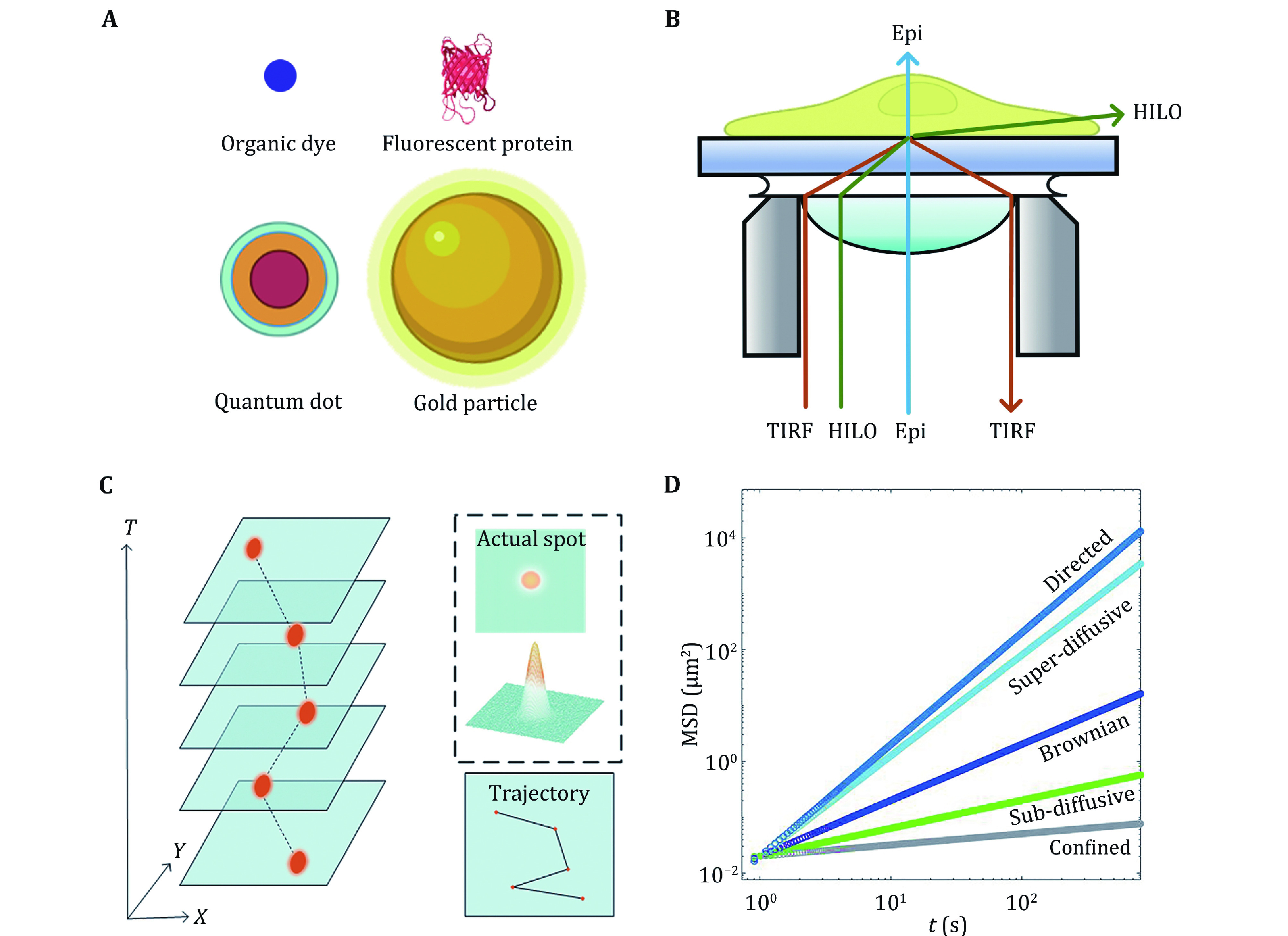
Principle of single particle tracking. A Commonly used probes. B Different illuminations in fluorescence microscopy. C Generation of trajectories. D MSD plots of different motion
In recent years, QDs have been considered ideal probes with excellent optical properties and biocompatibilities for in vivo and in vitro imaging (Dubertret et al. 2002; Larson et al. 2003; Zhou et al. 2015). In 2003, QDs were first used to label glycine receptors on the cell membrane (Dahan et al. 2003). QDs with narrow and symmetric emission spectra have strongly size-dependent emission wavelengths so that they can achieve multicolour SPTs (Zhou et al. 2015). Compared with other probes, QDs are quite photostable for long-term imaging. Although the application scope of QDs is very wide, their fluorescence intermittency (blinking) leads to incomplete molecular motion trajectories (Li et al. 2015c; Nirmal et al. 1996), bringing certain difficulties to data processing.
Optical implementations
Fluorescence microscopy is indispensable to image probes and track their movements. In traditional epifluorescence microscopy (Epi), the laser penetrates the cell vertically. Since all probes across the cell are excited, the overall fluorescence within the cell imposes a strong background on the single probes of interest, making the signal-to-noise ratio relatively low.
To improve the signal-to-noise ratio, total internal reflection fluorescence (TIRF) microscopy was invented to selectively excite fluorescent molecules close to the cover glass (<200 nm) (Axelrod 1981). TIRF is applied to visualize single molecule fluorescence near a surface (Khan et al. 2000; Lu et al. 2018; Sako et al. 2000; Vale et al. 1996; Wu et al. 2020) and especially to observe the diffusive motion of molecules on the cell membrane (Axelrod 2001). It is also used to track secretory granules in secretory processes (Reits and Neefjes 2001; Steyer and Almers 1999; Tsuboi et al. 2001; Zenisek et al. 2000).
Due to the observation depth, TIRF is limited to studying the cell membrane. To observe fluorescent molecules in cells, researchers further invented highly inclined and laminated optical sheet microscopy (HILO) (Tokunaga et al. 2008). The main difference between HILO and TIRF is the incident angle of the excitation light (Fig. 2B). In the HILO microscope, the excitation light no longer undergoes total reflection but penetrates and exits close to the interface, forming a thin layer of excitation light that illuminates the middle layer of the cell (Toomre and Bewersdorf 2010).
Data analysis
Trajectory
In SPT experiments, every probe is observed as a bright submicron spot described by the point spread function (PSF) due to the diffraction limit. We obtain an accurate position of the probe through Gaussian fitting. By linking the same particle’s different positions in consecutive images, particle trajectories are constructed (Fig. 2C). Both time and space information from the trajectories provide an opportunity to understand the characteristics of particle motion and further explore intracellular transport dynamics. Several algorithms are available (Cheezum et al. 2001; Chetverikov and Verestoy 1999; Sbalzarini and Koumoutsakos 2005; Tinevez et al. 2017; Vallotton et al. 2003) to help researchers obtain the trajectory of particles conveniently and efficiently.
Mean square displacement
To analyse the motion, the MSD of the trajectories is generally calculated:
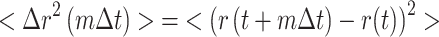 |
where r(t) is the displacement, Δt is the time interval, and < ··· > is the average. Time-averaged MSD is:
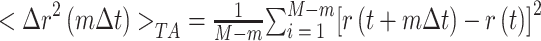 |
where M is the total time length of the trajectory. The ensemble-averaged MSD is:
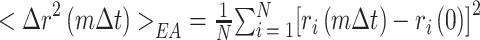 |
where i is the ID of each particle, and N is the total number of all particles. The two MSDs help us understand the motion of target particles, but their results are not always consistent. With increasing Δt, the MSD tends to show an upward trend, which can be described by a remarkable power-law curve MSD ~Δtα. The value of α depends on the motion type of the particle: α = 1 corresponds to Brownian motion, α < 1 refers to subdiffusion, and α > 1 refers to superdiffusion (Fig. 2D).
The diffusion rates are determined from the linear fitting of MSD:
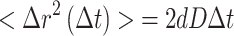 |
where D is the diffusion coefficient of particles, and d represents the dimensionality of space (Dupont et al. 2013; Ning et al. 2019).
Spatial distribution of intracellular diffusion
To describe the spatial heterogeneity of intracellular dynamics, one must focus on the trajectory within a specific spatial range. First, the cell is divided into different grids. Second, the segments of trajectories within a distance threshold to each grid node are chosen. Third, the ensemble-time-averaged MSD is calculated with the trajectory segments, and then the local diffusion rate is calculated by linear fitting of the first three points of the MSD. Fourth, with the diffusion rate at each grid node, the diffusion map of the cell is plotted. The parameters of grid size and distance threshold determine the final resolution of the diffusion map (Fig. 3A).
Figure 3.
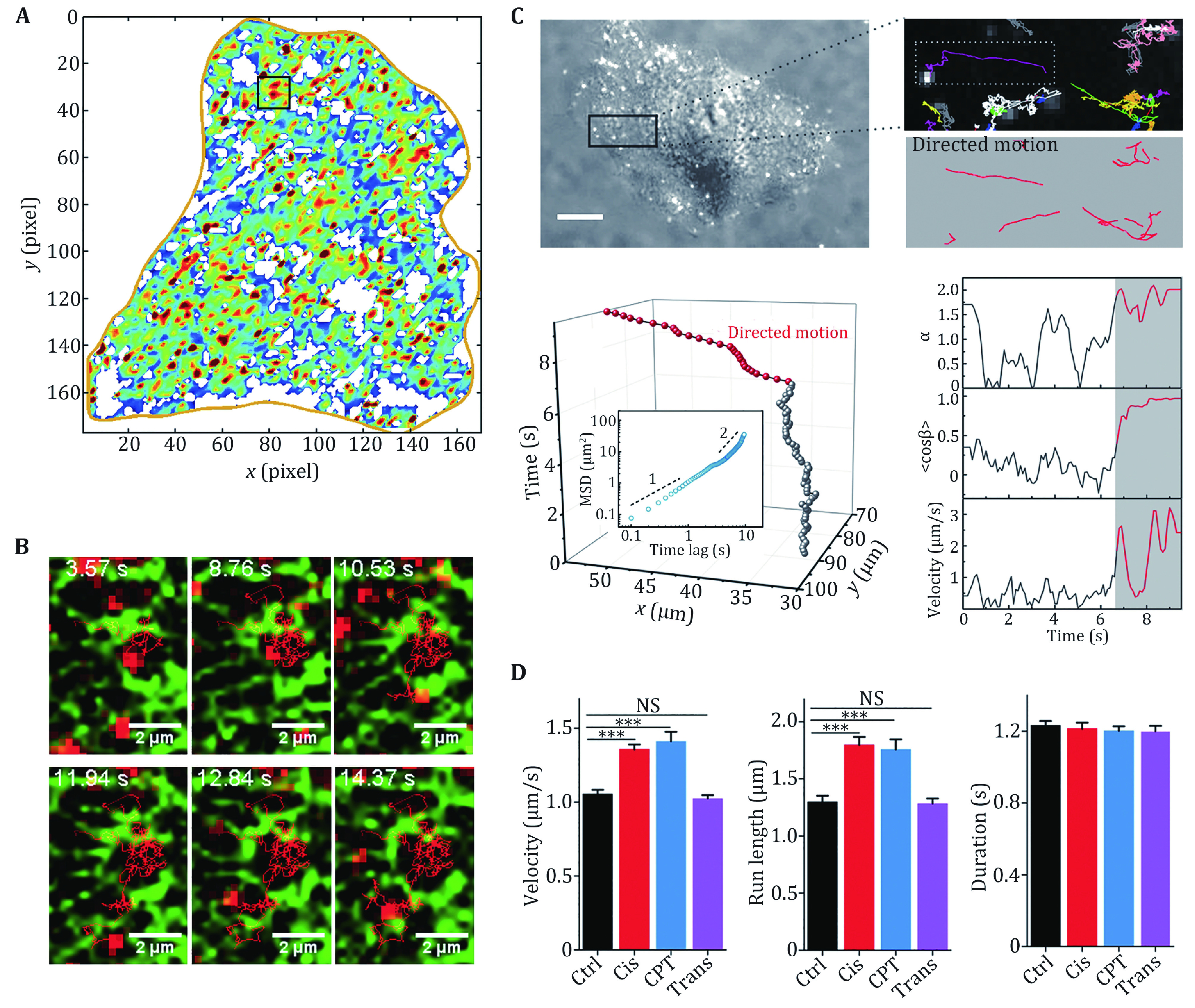
Diffusive motion and direction motion in living cells. A Diffusion map of a A549 human lung cancer cell, in which the local diffusion coefficients from 0 to 0.3 μm2/s are indicated by the colours from blue to red. B Image series of a QD trajectory (red) overlaid with GFP-ER (green). Reprinted with permission from American Chemical Society (Li et al. 2015b). C The trajectories of endocytic vesicles are divided into directed (red) and diffusive motion (grey). D Dynamical parameters of the velocity, run length and duration of the directed motion in early apoptotic cells. Reprinted with permission from National Academy of Sciences (Li et al. 2018a)
Extraction of the directed-motion segments
The typical trajectory of vesicle transport in cells consists of directed and diffusive motions. To extract the segments of directed motion, analysis of both the local MSD and directional persistence is applied to identify the local motion state of single particles in a time window. Compared with the MSD of a whole trajectory, the local MSD uses the particle positions around the time point of interest. Similarly, MSD can be fitted with MSDlocal = AΔtα, where α represents the nonlinear relationship of MSD with time. To further describe the direction information of particle motion, the local directional persistence 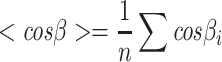 is applied, where βi represents the change in angle between adjacent steps along the trajectory (Li et al.
2012a, 2018a). For a perfect unidirectional motion, <cosβ> = 1. The thresholds of α and <cosβ>can be set to judge whether the segments belong to the directional motion. Note that these parameters should be selected according to the experimental conditions. Further control experiments should be performed by eliminating all directed motion with cytoskeleton-disrupting drugs (Fig. 3C).
is applied, where βi represents the change in angle between adjacent steps along the trajectory (Li et al.
2012a, 2018a). For a perfect unidirectional motion, <cosβ> = 1. The thresholds of α and <cosβ>can be set to judge whether the segments belong to the directional motion. Note that these parameters should be selected according to the experimental conditions. Further control experiments should be performed by eliminating all directed motion with cytoskeleton-disrupting drugs (Fig. 3C).
SPT APPLICATIONS IN INTRACELLULAR TRANSPORT
Intracellular transport is the basis of many cellular functions, so it is important to understand the mechanisms of intracellular transport dynamics. In recent decades, due to its high spatial and temporal resolution, SPT has been widely employed to explore intracellular transport and proven to be a powerful and effective tool for measuring intracellular dynamics (Manzo and Garcia-Parajo 2015; Wang et al. 2021). SPT was first applied to study the biomolecule dynamics on the cell membrane, since the TIRF microscopy enables the single-molecule imaging on cell basal membranes with a high signal-to-noise ratio. Moreover, labelling of on-membrane biomolecules by fluorescence probes is relatively easy to achieve. Later on, with the improvements of HILO and 3D imaging techniques and the new generations of fluorescence probes, the SPT studies in the cytoplasm of living cells have been greatly promoted. Recently, the investigations are moving deeper into the nucleus. Despite that the complex intranuclear environment with dense chromatins has brought more difficulties to the labelling and tracking of single molecules, new methods of SPT are being developed. Here, although many advances have been made in SPT of intracellular transport, we can only introduce some representative studies in this review.
Diffusion
Diffusion on the membrane
The membrane of living cells separates the internal and external environment of the cell. Cell membranes not only maintain the intracellular environment of stable cell metabolism but also regulate the exchange of substances between the intracellular and extracellular spaces. The cell membrane is mainly composed of fluid phospholipids and proteins that can move laterally (Jacobson et al. 2019). Moreover, the presence of the cytoskeleton beneath the membrane, lipid rafts, and other factors on the membrane may affect biomolecule motions.
SPT has contributed to important progress in studies of cell membrane dynamics. In 1993, Kusumi et al. used SPT to observe for the first time different movements of receptors on the cell membrane, including stationary mode, simple Brownian diffusion, directed diffusion, and confined diffusion (Kusumi et al. 1993). In 1994, Ghosh et al. observed anomalous diffusion of individual low-density lipoprotein receptors (LDL-Rs) on cell membranes (Ghosh and Webb 1994). Similar phenomena were further observed for other transmembrane receptors, including glycine receptor (GlyR) (Dahan et al. 2003), G-protein-coupled receptor (GPCR) (Calebiro et al. 2013), epidermal growth factor receptor (EGFR) (Chung et al. 2010), and acetylcholine receptors (AChRs) (He et al. 2016), which are attributed to molecular crowding and membrane heterogeneity (Kusumi et al. 2005; Owen et al. 2009; Saxton and Jacobson 1997). Moreover, Lippert et al. found that Wnt3A proteins bind to and diffuse on the plasma membrane of living cells without any receptor binding (Lippert et al. 2017).
The diffusive dynamics of lipids have also been studied by SPT. In 1996, Schmidt et al. applied SPT to analyse the movement of individual lipid molecules in an artificial membrane (Schmidt et al. 1996). It was further discovered by SPT that phospholipids undergo hop diffusion due to the compartmentalization of cell membranes (Fujiwara et al. 2002, 2016; Lagerholm et al. 2017).
It is worth mentioning that in addition to receptors and lipids, the diffusion of viruses and particles on membranes has been investigated by SPT. In 2005, Ewers et al. studied the lateral mobility of murine polyoma virus-like particles (VLPs) on cell membranes and artificial lipid bilayers using SPT (Ewers et al. 2005). Recently, a motion-pattern transition of single nanoparticles on the membrane was revealed (Ge et al. 2021).
Diffusion in the cytoplasm
With the development of SPT technology, it has been widely used to study the diffusion of intracellular molecules. Due to the viscoelastic properties of the cytoplasm and the presence of organelles, diffusion in the cytoplasm is quite complex. In 2013, Tabei et al. observed anomalous diffusion of insulin particles within the cell (Tabei et al. 2013). In 2015, Li et al. introduced a new method based on SPT to rapidly map intracellular diffusion, revealing heterogeneous and compartmentalized diffusion resulting from restriction of the endoplasmic reticulum (ER) (Fig. 3A, 3B) (Li et al. 2015b). In 2018, Zhao et al. further characterized the highly spatiotemporal heterogeneity dynamics of lysosomes in cells (Zhao et al. 2018). With the 3D SPT technique, it was recently found that intracellular diffusion is anisotropic quasi-2D rather than isotropic 3D in adherent cells (Chen 2020; Jiang et al. 2020). Han et al. used the diffusive dynamics of fluorescence beads to explore the intracellular dynamics between benign and malignant breast cancer cells (Han et al. 2020). The combination of SPT and superresolution microscopy enables the study of dynamics in organelles, such as the diffusion properties of proteins in mitochondria (Appelhans et al. 2012).
Diffusion in the nucleus
SPT also has applications in probing diffusive dynamics in the nucleus. In 2005, the Gratton lab tracked interphase chromatin dynamics using a two-photon excitation microscope, showing that chromatin in the nucleus undergoes confined diffusion and diffusional jumps (Levi et al. 2005). Moreover, individual telomeres in the nucleus of eukaryotic cells were found to exhibit anomalous diffusion on a short timescale and normal diffusion on a long timescale (Bronstein et al. 2009). The use of reflected light-sheet microscopy in combination with SPT improves the signal-to-noise ratio and enables the measurement of the diffusive dynamics of individual transcription factors in the nucleus (Gebhardt et al. 2013). Another work studied the diffusive dynamics of transcription factors and found the influence of nuclear architecture on gene regulation (Izeddin et al. 2014). In addition, the diffusion of microinjected viral ribonucleoprotein in the nucleus has been studied (Babcock et al. 2004).
Directed motion
Vesicle trafficking
Endocytic receptor transport is a complex and dynamic process. The transport of endocytic vesicles contains directed motion driven by motor proteins along the cytoskeleton. Endocytic transport after internalization of the QD-ligand-receptor complex in real time has been revealed (Liang et al. 2007; Lidke et al. 2004, 2005; Rajan et al. 2008). Furthermore, the unidirectional and discontinuous transport of nerve growth factors in axons has been shown (Cui et al. 2007). In 2012, a study on EGFR endocytic trafficking found that paclitaxel altered the transport dynamics of endocytic vesicles by interfering with microtubule structures (Li et al. 2012b). In 2018, a study showed that the intracellular transport of endocytic vesicles is accelerated in the early stages of apoptosis due to increased intracellular ATP concentrations. Accelerated transport was demonstrated to be necessary for apoptotic progression (Li et al. 2018a) (Fig. 3C, 3D).
In addition to the translational motion, the rotation of single particles also provides important dynamic information. For example, the rotation of gold nanorods during endocytosis and subsequent intracellular transport was clearly shown (Chen et al. 2017; Xu et al. 2021). Moreover, the rotation of endosomes during neuronal axonal transport was observed by nanorods (Kaplan et al. 2018). In addition, endocytic transport of aptamer-drug conjugates was also characterized by SPT (Lv et al. 2019).
Viral infection
Viral infection is a complex process involving many steps and complex interactions with different subcellular structures (Cheng and Ghany 2020). After entering the cells by endocytosis, the intracellular transport of a single virus to its destinations is critical for viral duplication and other functions. SPT has significantly contributed to the mechanistic understanding of the viral infection process (Brandenburg and Zhuang 2007; Liu et al. 2020b). In 2003, the Zhuang laboratory tracked individual labelled influenza viruses in living cells and determined the internalization and endocytic transport of influenza viruses (Lakadamyali et al. 2003). The intracellular transport of a single virus mainly involves three processes: actin-dependent motion at the periphery of the cell, directed transport by dynein to the perinuclear region, and microtubule-dependent intermittent movement in the perinuclear region. SPT has also been used to elucidate the entry and internalization pathways of other viruses, such as poliovirus (Brandenburg et al. 2007).
Other directed transport
In addition to the directed transport of vesicles and viruses mentioned above, SPT has been used to uncover the directed transport of other intracellular components. In 2006, Courty et al. characterized the in vivo dynamics of individual kinesin motors labelled by QDs (Courty et al. 2006). In 2012, Coppola et al. used 3D-SPT to elucidate the dynamics of cationic liposome-DNA complexes in living cells and found that the complex mainly undergoes directed motion, in which microtubules play important roles (Coppola et al. 2012). In 2017, Katrukha et al. utilized QDs to analyse the role of cytoskeletal modulation in both passive and active intracellular transport (Katrukha et al. 2017). In 2018, a novel type of membraneless organelle named cytoophidium was found to show directed transport in fission yeasts, which is attributed to the myosin V with actin filaments (Li et al. 2018b).
PERSPECTIVES
In this review, we first introduced the biological functions and physical mechanisms of intracellular transport and then briefly reviewed SPT technology and its applications in studying intracellular transport. In the future, more efforts should be made to elucidate the functional roles of intracellular transport dynamics, bridging the gap between physical behaviours and biological functions. As intracellular transport dynamics provide the physical basis for the transfer of matter, energy, and information, which is crucial for cellular activities and functions, cells can regulate their functions by alternating intracellular transport dynamics. For example, intracellular transport dynamics are tightly correlated with apoptotic progression (Li et al. 2018a). Moreover, the functions of diffusion in biochemical reactions and cellular activities remain to be elucidated (Brangwynne et al. 2009). Since SPT has been used to reveal the infection mechanisms of the influenza virus at the single-molecule level, it is expected that SPT will help us to understand COVID-19 infection in the future, which may contribute to designing specific drugs targeting the invasion and intracellular transport of COVID-19 (Ding et al. 2021; Shi 2020).
SPT has promoted the study of intracellular transport dynamics, but there are still some challenges. To date, most SPT studies on intracellular dynamics have been carried out at the cellular level in vitro; however, the environment in Petri dishes is quite different from that in real tissue (Li et al. 2021; Pampaloni et al. 2007), and the characteristics and functions of cells in 3D tissue remain to be investigated in the future (Han et al. 2020; Jiang et al. 2021). Therefore, SPT technology in 3D tissue imaging in vivo is of great significance and has profound prospects. To observe tissues, new 3D imaging techniques are needed. Lattice light-sheet microscopy (LLSM) technology is an effective method for observing deep tissue (Li et al. 2015a; Liu et al. 2018) and provides high spatial-temporal resolution for observing the subcellular dynamics within cells or tissues. In addition to microscope developments, new probes are also needed. Compared with visible light, a fluorescent probe with near-infrared (NIR) emission can achieve deeper penetration and better imaging quality, which is suited for live tissue imaging (Cai et al. 2019; Dai et al. 2021; Li et al. 2019; Smith et al. 2009). Single-walled carbon nanotubes (SWCNTs) have unique intrinsic fluorescence emission in the second NIR window (1000–1700 nm), which makes them candidate fluorescent probes for SPT in deep tissue (Bachilo et al. 2002; Hong et al. 2015; Welsher et al. 2009). In brain tissue, SWCNTs have been tracked to reveal the nanoscale organizational structure of the extracellular space (Godin et al. 2017). In addition, QDs in NIR emission are another promising probe for SPTs in deep tissue (Cassette et al. 2013; Liu et al. 2020a; Zhou et al. 2015). Recently, a new kind of QD emitting at 1600 nm allowed in vivo confocal 3D imaging of tumour vasculatures in mice at a depth of 1.2 mm (Zhang et al. 2018). Although SPT in real tissue is still challenging, it is believed that with the development of optical microscopy and NIR probes, SPT will extend the study of intracellular transport dynamics in vivo, with promising applications in biophysical studies and biomedical diagnosis.
Conflict of interest
Ming-Li Zhang, Hui-Ying Ti, Peng-Ye Wang and Hui Li declare that they have no conflict of interest.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (12074043, 11874415, 12122402), the National Key Research and Development Program (2016YFA0301500), Youth Innovation Promotion Association of CAS (2019006), and the Fundamental Research Funds for the Central Universities (2019NTST26). The schematic figure is created with BioRender.com.
Compliance with Ethical Standards
Human and animal rights and informed consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Contributor Information
Peng-Ye Wang, Email: pywang@iphy.ac.cn.
Hui Li, Email: huili@bnu.edu.cn.
References
- Appelhans T, Richter CP, Wilkens V, Hess ST, Piehler J, Busch KB Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett. 2012;12(2):610–616. doi: 10.1021/nl203343a. [DOI] [PubMed] [Google Scholar]
- Axelrod D Cell-substrate contacts illuminated by total internal-reflection fluorescence. J Cell Biol. 1981;89(1):141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2(11):764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- Babcock HP, Chen C, Zhuang XW Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J. 2004;87(4):2749–2758. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachilo SM, Strano MS, Kittrell C, Hauge RH, Smalley RE, Weisman RB Structure-assigned optical spectra of single-walled carbon nanotubes. Science. 2002;298(5602):2361–2366. doi: 10.1126/science.1078727. [DOI] [PubMed] [Google Scholar]
- Balint S, Vilanova IV, Alvarez AS, Lakadamyali M Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. Proc Natl Acad Scie USA. 2013;110(9):3375–3380. doi: 10.1073/pnas.1219206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay D, Cyphersmith A, Zapata JA, Kim YJ, Payne CK Lysosome Transport as a function of lysosome diameter. PLoS One. 2014;9(1):e86847. doi: 10.1371/journal.pone.0086847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K, Rossow MJ, Gelfand VI The journey of the organelle: teamwork and regulation in intracellular transport. Curr Opin Cell Biol. 2013;25(4):483–488. doi: 10.1016/j.ceb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basturea GN Endocytosis. Mater Methods. 2019;9:2752. doi: 10.13070/mm.en.9.2752. [DOI] [Google Scholar]
- Bouchaud JP, Georges A Anomalous diffusion in disordered media: statistical mechanisms, models and physical applications. Phys Rep. 1990;195(4-5):127–293. doi: 10.1016/0370-1573(90)90099-N. [DOI] [Google Scholar]
- Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang XW, Hogle JM Imaging poliovirus entry in live cells. PLoS Biol. 2007;5(7):1543–1555. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg B, Zhuang XW Virus trafficking - learning from single-virus tracking. Nat Rev Microbiol. 2007;5(3):197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA Intracellular transport by active diffusion. Trends Cell Biol. 2009;19(9):423–427. doi: 10.1016/j.tcb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Bronstein I, Israel Y, Kepten E, Mai S, Shav-Tal Y, Barkai E, Garini Y Transient anomalous diffusion of telomeres in the nucleus of mammalian cells. Phys Rev Lett. 2009;103:018102. doi: 10.1103/PhysRevLett.103.018102. [DOI] [PubMed] [Google Scholar]
- Brown AI, Sivak DA Theory of nonequilibrium free energy transduction by molecular machines. Chem Rev. 2020;120(1):434–459. doi: 10.1021/acs.chemrev.9b00254. [DOI] [PubMed] [Google Scholar]
- Brown R Mikroskopische Beobachtungen über die im Pollen der Pflanzen enthaltenen Partikeln, und über das allgemeine Vorkommen activer Molecüle in organischen und unorganischen Körpern. Annalen der Physik. 1828;90(10):294–313. doi: 10.1002/andp.18280901016. [DOI] [Google Scholar]
- Bulseco DA, Wolf DE Fluorescence correlation spectroscopy: molecular complexing in solution and in living cells. Methods Cell Biol. 2007;81:525–559. doi: 10.1016/S0091-679X(06)81025-3. [DOI] [PubMed] [Google Scholar]
- Bursac P, Lenormand G, Fabry B, Oliver M, Weitz DA, Viasnoff V, Butler JP, Fredberg JJ Cytoskeletal remodelling and slow dynamics in the living cell. Nat Mater. 2005;4(7):557–561. doi: 10.1038/nmat1404. [DOI] [PubMed] [Google Scholar]
- Burute M, Kapitein LC (2019) Cellular logistics: unraveling the interplay between microtubule organization and intracellular transport. Ann Rev Cell Develop Biol, 35: 29-54
- Cai Y, Wei Z, Song C, Tang C, Han W, Dong X Optical nano-agents in the second near-infrared window for biomedical applications. Chem Soc Rev. 2019;48(1):22–37. doi: 10.1039/C8CS00494C. [DOI] [PubMed] [Google Scholar]
- Calebiro D, Rieken F, Wagner J, Sungkaworn T, Zabel U, Borzi A, Cocucci E, Zurn A, Lohse MJ Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc Natl Acad Sci USA. 2013;110(2):743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassette E, Helle M, Bezdetnaya L, Marchal F, Dubertret B, Pons T Design of new quantum dot materials for deep tissue infrared imaging. Adv Drug Deliv Rev. 2013;65(5):719–731. doi: 10.1016/j.addr.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Chechkin AV, Seno F, Metzler R, Sokolov IM Brownian yet non-Gaussian diffusion: from superstatistics to subordination of diffusing diffusivities. Phys Rev X. 2017;7:021002. doi: 10.1103/PhysRevX.7.021002. [DOI] [Google Scholar]
- Cheezum MK, Walker WF, Guilford WH Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys J. 2001;81(4):2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KC, Gu Y, Sun W, Dong B, Wang GF, Fan XX, Xia T, Fang N Characteristic rotational behaviors of rod-shaped cargo revealed by automated five-dimensional single particle tracking. Nat Commun. 2017;8(1):887. doi: 10.1038/s41467-017-01001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS Strong anisotropy of 3D diffusion in living cells. Chin Phys Lett. 2020;37(8):080103. doi: 10.1088/0256-307X/37/8/080103. [DOI] [Google Scholar]
- Cheng X, Ghany MG Key milestones in HCV discovery and therapeutics. The Innovation. 2020;1(3):100067. doi: 10.1016/j.xinn.2020.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverikov D, Verestoy J Feature point tracking for incomplete trajectories. Computing. 1999;62(4):321–338. doi: 10.1007/s006070050027. [DOI] [Google Scholar]
- Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464(7289):783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- Cianfrocco MA, DeSantis ME, Leschziner AE, Reck-Peterson SL Mechanism and regulation of cytoplasmic dynein. Ann Rev Cell Develop Biol. 2015;31:83–108. doi: 10.1146/annurev-cellbio-100814-125438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognet L, Leduc C, Lounis B Advances in live-cell single-particle tracking and dynamic super-resolution imaging. Curr Opin Chem Biol. 2014;20:78–85. doi: 10.1016/j.cbpa.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Coppola S, Estrada LC, Digman MA, Pozzi D, Cardarelli F, Gratton E, Caracciolo G Intracellular trafficking of cationic liposome-DNA complexes in living cells. Soft Matter. 2012;8(30):7919–7927. doi: 10.1039/c2sm25532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M Tracking individual kinesin motors in living cells using single quantum-dot imaging. Nano Lett. 2006;6(7):1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- Cui BX, Wu CB, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S One at a time, live tracking of NGF axonal transport using quantum dots. Proc Natl Acad Sci USA. 2007;104(34):13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302(5644):442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- Dai H, Shen Q, Shao J, Wang W, Gao F, Dong X Small molecular NIR-II fluorophores for cancer phototheranostics. Innovation. 2021;2(1):100082. doi: 10.1016/j.xinn.2021.100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Forges H, Bouissou A, Perez F Interplay between microtubule dynamics and intracellular organization. Int J Biochem Cell Biol. 2012;44(2):266–274. doi: 10.1016/j.biocel.2011.11.009. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Luini A Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9(4):273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- Di Rienzo C, Piazza V, Gratton E, Beltram F, Cardarelli F Probing short-range protein Brownian motion in the cytoplasm of living cells. Nat Commun. 2014;5:5891. doi: 10.1038/ncomms6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding HM, Yin YW, Ni SD, Sheng YJ, Ma YQ Accurate evaluation on the interactions of SARS-CoV-2 with its receptor ACE2 and antibodies CR3022/CB6. Chin Physi Lett. 2021;38(1):018701. doi: 10.1088/0256-307X/38/1/018701. [DOI] [Google Scholar]
- Drechsler M, Giavazzi F, Cerbino R, Palacios IM Active diffusion and advection in Drosophila oocytes result from the interplay of actin and microtubules. Nat Commun. 2017;8(1):1520. doi: 10.1038/s41467-017-01414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan ZW, Xie P, Li W, Wang PY Are coiled-coils of dimeric kinesins unwound during their walking on microtubule? PLoS One. 2012;7(4):e36071. doi: 10.1371/journal.pone.0036071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298(5599):1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- Duits MHG, Li YX, Vanapalli SA, Mugele F Mapping of spatiotemporal heterogeneous particle dynamics in living cells. Phys Rev E. 2009;79(5 Pt 1):051910. doi: 10.1103/PhysRevE.79.051910. [DOI] [PubMed] [Google Scholar]
- Dupont A, Gorelashvili M, Schuller V, Wehnekamp F, Arcizet D, Katayama Y, Lamb DC, Heinrich D Three-dimensional single-particle tracking in live cells: news from the third dimension. New J Phys. 2013;15(7):075008. doi: 10.1088/1367-2630/15/7/075008. [DOI] [Google Scholar]
- Elson EL Fluorescence correlation spectroscopy: past, present, future. Biophys J. 2011;101(12):2855–2870. doi: 10.1016/j.bpj.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P, Helenius A Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc Natl Acad Sci USA. 2005;102(42):15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhri N, Wessel AD, Willms C, Pasquali M, Klopfenstein DR, MacKintosh FC, Schmidt CF High-resolution mapping of intracellular fluctuations using carbon nanotubes. Science. 2014;344(6187):1031–1035. doi: 10.1126/science.1250170. [DOI] [PubMed] [Google Scholar]
- Fodor E, Guo M, Gov NS, Visco P, Weitz DA, van Wijland F Activity-driven fluctuations in living cells. EPL. 2015;110:48005. doi: 10.1209/0295-5075/110/48005. [DOI] [Google Scholar]
- Fricker MD, Heaton LLM, Jones NS, Boddy L The mycelium as a network. Microbiol Spectr. 2017;5(3) doi: 10.1128/microbiolspec.FUNK-0033-2017. [DOI] [PubMed] [Google Scholar]
- Fricker MD, Lee JA, Bebber DP, Tlalka M, Hynes J, Darrah PR, Watkinson SC, Boddy L Imaging complex nutrient dynamics in mycelial networks. J Microsc. 2008;231(2):317–331. doi: 10.1111/j.1365-2818.2008.02043.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol. 2002;157(6):1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara TK, Iwasawa K, Kalay Z, Tsunoyama TA, Watanabe Y, Umemura YM, Murakoshi H, Suzuki KGN, Nemoto YL, Morone N, Kusumi A Confined diffusion of transmembrane proteins and lipids induced by the same actin meshwork lining the plasma membrane. Mol Biol Cell. 2016;27(7):1101–1119. doi: 10.1091/mbc.E15-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal N, Lechtman-Goldstein D, Weihs D Particle tracking in living cells: a review of the mean square displacement method and beyond. Rheol Acta. 2013;52(5):425–443. doi: 10.1007/s00397-013-0694-6. [DOI] [Google Scholar]
- Gallet F, Arcizet D, Bohec P, Richert A Power spectrum of out-of-equilibrium forces in living cells: amplitude and frequency dependence. Soft Matter. 2009;5(15):2947–2953. doi: 10.1039/b901311c. [DOI] [Google Scholar]
- Ge F, Xue J, Du Y, He Y Unmodified single nanoparticles undergo a motion-pattern transition on the plasma membrane before cellular uptake. Nano Today. 2021;39:101158. doi: 10.1016/j.nantod.2021.101158. [DOI] [Google Scholar]
- Gebhardt JCM, Suter DM, Roy R, Zhao ZQW, Chapman AR, Basu S, Maniatis T, Xie XS Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10(5):421–216. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A, Carter AP, Reck-Peterson SL, Vale RD Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131(5):952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh RN, Webb WW Automated detection and tracking of individual and clustered cell-surface low-density-lipoprotein receptor molecules. Biophys J. 1994;66(5):1301–1318. doi: 10.1016/S0006-3495(94)80939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin AG, Varela JA, Gao Z, Danne N, Dupuis JP, Lounis B, Groc L, Cognet L Single-nanotube tracking reveals the nanoscale organization of the extracellular space in the live brain. Nat Nanotechnol. 2017;12(3):238–243. doi: 10.1038/nnano.2016.248. [DOI] [PubMed] [Google Scholar]
- Gregor T, Bialek W, van Steveninck RRR, Tank DW, Wieschaus EF Diffusion and scaling during early embryonic pattern formation. Proc Natl Acad Sci USA. 2005;102(51):18403–18407. doi: 10.1073/pnas.0509483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, Lippincott-Schwartz J, Mackintosh FC, Weitz DA Probing the Stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell. 2014;158(4):822–832. doi: 10.1016/j.cell.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YL, Pegoraro AF, Li H, Li KF, Yuan Y, Xu GQ, Gu ZC, Sun JW, Hao YK, Gupta SK, Li YW, Tang WH, Kang H, Teng LH, Fredberg JJ, Guo M Cell swelling, softening and invasion in a three-dimensional breast cancer model. Nat Phys. 2020;16(1):101–108. doi: 10.1038/s41567-019-0680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock WO Bidirectional cargo transport: moving beyond tug of war. Nat Rev Mol Cell Biol. 2014;15(9):615–628. doi: 10.1038/nrm3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington AW, Ginty DD Long-distance retrograde neurotrophic factor signalling in neurons. Nat Rev Neurosci. 2013;14(3):177–187. doi: 10.1038/nrn3253. [DOI] [PubMed] [Google Scholar]
- Haustein E, Schwille P Fluorescence correlation spectroscopy: novel variations of an established technique. Ann Rev Biophys Biomol Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- He W, Song H, Su Y, Geng L, Ackerson BJ, Peng HB, Tong P Dynamic heterogeneity and non-Gaussian statistics for acetylcholine receptors on live cell membrane. Nat Commun. 2016;7:11701. doi: 10.1038/ncomms11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Gu Z, Wang L, Qu Z, Xu F Coarse-grained molecular dynamics simulation of dendrimer transmembrane transport with temperature-dependent membrane phase states. Int J Heat Mass Transf. 2020;155:119797. doi: 10.1016/j.ijheatmasstransfer.2020.119797. [DOI] [Google Scholar]
- Hendricks AG, Perlson E, Ross JL, Schroeder HW, Tokito M, Holzbaur ELF Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr Biol. 2010;20(8):697–702. doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68(4):610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10(10):682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Hoffman BD, Massiera G, Van Citters KM, Crocker JC The consensus mechanics of cultured mammalian cells. Proc Natl Acad Sci USA. 2006;103(27):10259–10264. doi: 10.1073/pnas.0510348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofling F, Franosch T Anomalous transport in the crowded world of biological cells. Rep Prog Phys. 2013;76(4):046602. doi: 10.1088/0034-4885/76/4/046602. [DOI] [PubMed] [Google Scholar]
- Hong G, Diao S, Antaris AL, Dai H Carbon Nanomaterials for biological imaging and nanomedicinal therapy. Chem Rev. 2015;115(19):10816–10906. doi: 10.1021/acs.chemrev.5b00008. [DOI] [PubMed] [Google Scholar]
- Hui YY, Hsiao WWW, Haziza S, Simonneau M, Treussart F, Chang HC Single particle tracking of fluorescent nanodiamonds in cells and organisms. Curr Opin Solid State Mater Sci. 2017;21(1):35–42. doi: 10.1016/j.cossms.2016.04.002. [DOI] [Google Scholar]
- Iino R, Koyama I, Kusumi A Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophys J. 2001;80(6):2667–2677. doi: 10.1016/S0006-3495(01)76236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, Recamier V, Bosanac L, Cisse II, Boudarene L, Dugast-Darzacq C, Proux F, Benichou O, Voituriez R, Bensaude O, Dahan M, Darzacq X Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife. 2014;3:e02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Liu P, Lagerholm BC The lateral organization and mobility of plasma membrane components. Cell. 2019;177(4):806–819. doi: 10.1016/j.cell.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Wojcieszyn J The translational mobility of substances within the cytoplasmic matrix. Proc Natl Acad Sci USA. 1984;81(21):6747–6751. doi: 10.1073/pnas.81.21.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Li B, Dou SX, Wang PY, Li H Quasi-Two-dimensional diffusion in adherent cells revealed by three-dimensional single quantum dot tracking. Chin Phys Lett. 2020;37(7):078701. doi: 10.1088/0256-307X/37/7/078701. [DOI] [Google Scholar]
- Jiang SJ, Li H, Zeng Q, Xiao ZH, Zhang XH, Xu MM, He Y, Wei Y, Deng XL The dynamic counterbalance of RAC1-YAP/OB-Cadherin coordinates tissue spreading with stem cell fate patterning. Adv Sci. 2021;8(10):2004000. doi: 10.1002/advs.202004000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan L, Ierokomos A, Chowdary P, Bryant Z, Cui BX Rotation of endosomes demonstrates coordination of molecular motors during axonal transport. Sci Adv. 2018;4(3):e1602170. doi: 10.1126/sciadv.1602170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrukha EA, Mikhaylova M, van Brakel HX, van Bergen En Henegouwen PM, Akhmanova A, Hoogenraad CC, Kapitein LC Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots. Nat Commun. 2017;8:14772. doi: 10.1038/ncomms14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Pierce D, Vale RD Interactions of the chemotaxis signal protein CheY with bacterial flagellar motors visualized by evanescent wave microscopy. Curr Biol. 2000;10(15):927–930. doi: 10.1016/S0960-9822(00)00629-1. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Brown GC, Hoek JB Diffusion control of protein phosphorylation in signal transduction pathways. Biochem J. 2000;350:901–907. doi: 10.1042/bj3500901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikushima K, Kita S, Higuchi H A non-invasive imaging for the in vivo tracking of high-speed vesicle transport in mouse neutrophils. Sci Rep. 2013;3:1913. doi: 10.1038/srep01913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Heinze KG, Schwille P Fluorescence correlation spectroscopy in living cells. Nat Methods. 2007;4(11):963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- Koslover EF, Chan CK, Theriot JA Disentangling Random motion and flow in a complex medium. Biophys J. 2016;110(3):700–709. doi: 10.1016/j.bpj.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Ann Rev Biophys Biomol Struct. 2005;34:351–354. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Sako Y, Yamamoto M Confined lateral diffusion of membrane-receptors as studied by single-particle tracking (nanovid microscopy) −− Effects of calcium-induced differentiation in cultured epithelial-cells. Biophys J. 1993;65(5):2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Tsunoyama TA, Hirosawa KM, Kasai RS, Fujiwara TK Tracking single molecules at work in living cells. Nat Chem Biol. 2014;10(7):524–532. doi: 10.1038/nchembio.1558. [DOI] [PubMed] [Google Scholar]
- Lagerholm BC, Andrade DM, Clausen MP, Eggeling C Convergence of lateral dynamic measurements in the plasma membrane of live cells from single particle tracking and STED-FCS. J Phys D Appl Phys. 2017;50(6):063001. doi: 10.1088/1361-6463/aa519e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, Zhuang XW Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA. 2003;100(16):9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampo TJ, Stylianidou S, Backlund MP, Wiggins PA, Spakowitz AJ Cytoplasmic RNA-protein particles exhibit non-Gaussian subdiffusive behavior. Biophys J. 2017;112(3):532–542. doi: 10.1016/j.bpj.2016.11.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, Webb WW Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science. 2003;300(5624):1434–1436. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- Lau AWC, Hoffman BD, Davies A, Crocker JC, Lubensky TC Microrheology, stress fluctuations, and active behavior of living cells. Phys Rev Lett. 2003;91(19):198101. doi: 10.1103/PhysRevLett.91.198101. [DOI] [PubMed] [Google Scholar]
- Levi V, Ruan QQ, Plutz M, Belmont AS, Gratton E Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys J. 2005;89(6):4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dou SX, Yuan JW, Liu YR, Li W, Ye FF, Wang PY, Li H Intracellular transport is accelerated in early apoptotic cells. Proc Natl Acad Sci USA. 2018a;115(48):12118–12123. doi: 10.1073/pnas.1810017115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Shao L, Chen BC, Zhang X, Zhang MS, Moses B, Milkie DE, Beach JR, Hammer JA, Pasham M, Kirchhausen T, Baird MA, Davidson MW, Xu PY, Betzig E Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015a;349(6251):aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DF, He SQ, Wu YF, Liu JQ, Liu Q, Chang BS, Zhang Q, Xiang ZH, Yuan Y, Jian C, Yu AX, Cheng Z Excretable lanthanide nanoparticle for biomedical imaging and surgical navigation in the second near-infrared window. Adv Sci. 2019;6(23):1902042. doi: 10.1002/advs.201902042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Dou SX, Liu YR, Li W, Xie P, Wang WC, Wang PY Mapping intracellular diffusion distribution using single quantum dot tracking: compartmentalized diffusion defined by endoplasmic reticulum. J Am Chem Soc. 2015b;137(1):436–444. doi: 10.1021/ja511273c. [DOI] [PubMed] [Google Scholar]
- Li H, Duan ZW, Dou SX, Wang PY The study of EGFR transport in single cell using an automatic method of trajectory identification. Acta Phys Sin. 2012a;61(6):068701. doi: 10.7498/aps.61.068701. [DOI] [Google Scholar]
- Li H, Duan ZW, Xie P, Liu YR, Wang WC, Dou SX, Wang PY Effects of paclitaxel on EGFR endocytic trafficking revealed using quantum dot tracking in single cells. PLoS One. 2012b;7(9):e45465. doi: 10.1371/journal.pone.0045465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Huang Y, Wang PY, Ye F, Liu JL Data on dynamic study of cytoophidia in Saccharomyces cerevisiae. Data Brief. 2016a;8:40–44. doi: 10.1016/j.dib.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ye FF, Ren JY, Wang PY, Du LL, Liu JL Active transport of cytoophidia in Schizosaccharomyces pombe. FASEB J. 2018b;32(11):5891–5898. doi: 10.1096/fj.201800045RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zheng Y, Han YL, Cai SQ, Guo M Nonlinear elasticity of biological basement membrane revealed by rapid inflation and deflation. Proc Natl Acad Sci USA. 2021;118(11):e2022422118. doi: 10.1073/pnas.2022422118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu RW, Ma L, Fan SN, Li H, Hu SX, Li M (2015c) Redox-mediated reversible modulation of the photoluminescence of single quantum dots. Chin Phys B 24(7): 078202. https://robots.iopscience.iop.org/article/10.1088/1674-1056/24/7/078202/meta
- Li YM, Shang L, Nienhaus GU Super-resolution imaging-based single particle tracking reveals dynamics of nanoparticle internalization by live cells. Nanoscale. 2016b;8(14):7423–7429. doi: 10.1039/C6NR01495J. [DOI] [PubMed] [Google Scholar]
- Liang ZY, Xu N, Guan YH, Xu M, He QH, Han QD, Zhang YY, Zhao XS The transport of alpha(1A)-adrenergic receptor with 33-nm step size in live cells. Biochem Biophys Res Commun. 2007;353(2):231–237. doi: 10.1016/j.bbrc.2006.11.116. [DOI] [PubMed] [Google Scholar]
- Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol. 2005;170(4):619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22(2):198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- Lippert A, Janeczek AA, Furstenberg A, Ponjavic A, Moerner WE, Nusse R, Helms JA, Evans ND, Lee SF Single-Molecule Imaging of Wnt3A Protein Diffusion on Living Cell Membranes. Biophys J. 2017;113(12):2762–2767. doi: 10.1016/j.bpj.2017.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH (2003) Photobleaching and photoactivation: following protein dynamics in living cells. Nat Cell Biol Suppl:
- Liu HJ, Li CW, Qian Y, Hu L, Fang J, Tong W, Nie RR, Chen QW, Wang H Magnetic-induced graphene quantum dots for imaging-guided photothermal therapy in the second near-infrared window. Biomaterials. 2020a;232:119700. doi: 10.1016/j.biomaterials.2019.119700. [DOI] [PubMed] [Google Scholar]
- Liu S, Yang H, Lu TJ, Genin GM, Xu F Electrostatic switching of nuclear basket conformations provides a potential mechanism for nuclear mechanotransduction. J Mech Phys Solids. 2019;133:103705. doi: 10.1016/j.jmps.2019.103705. [DOI] [Google Scholar]
- Liu SL, Wang ZG, Xie HY, Liu AA, Lamb DC, Pang DW Single-Virus tracking: from imaging methodologies to virological applications. Chem Rev. 2020b;120(3):1936–1979. doi: 10.1021/acs.chemrev.9b00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, Kohrman AQ, Medwig TN, Dambournet D, Forster R, Cunniff B, Ruan Y, Yashiro H, Scholpp S, Meyerowitz EM, Hockemeyer D, Drubin DG, Martin BL, Matus DQ, Koyama M, Megason SG, Kirchhausen T, Betzig E Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science. 2018;360(6386):eaaq1392. doi: 10.1126/science.aaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XM, Li H, You J, Li W, Wang PY, Li M, Dou SX, Xi XG Folding dynamics of parallel and antiparallel G-triplexes under the influence of proximal DNA. J Phys Chem B. 2018;122(41):9499–9506. doi: 10.1021/acs.jpcb.8b08110. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K Cytoarchitecture and physical properties of cytoplasm: volume, viscosity. diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Lubyphelps K, Castle PE, Taylor DL, Lanni F Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3t3 cells. Proc Natl Acad Sci USA. 1987;84(14):4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Yi M Non-Gaussian diffusion in static disordered media. Phys Rev E. 2018;97(4-1):042122. doi: 10.1103/PhysRevE.97.042122. [DOI] [PubMed] [Google Scholar]
- Lv C, Yang C, Ding D, Sun Y, Wang RW, Han D, Tan WH Endocytic pathways and intracellular transport of aptamer-drug conjugates in live cells monitored by single-particle tracking. Analyt Chem. 2019;91(21):13818–13823. doi: 10.1021/acs.analchem.9b03281. [DOI] [PubMed] [Google Scholar]
- Manzo C, Garcia-Parajo MF A review of progress in single particle tracking: from methods to biophysical insights. Rep Prog Phys. 2015;78(12):124601. doi: 10.1088/0034-4885/78/12/124601. [DOI] [PubMed] [Google Scholar]
- McLaughlin RT, Diehl MR, Kolomeisky AB Collective dynamics of processive cytoskeletal motors. Soft Matter. 2016;12(1):14–21. doi: 10.1039/C5SM01609F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogre SS, Brown AI, Koslover EF Getting around the cell: physical transport in the intracellular world. Phys Biol. 2020;17(6):061003. doi: 10.1088/1478-3975/aba5e5. [DOI] [PubMed] [Google Scholar]
- Mudrakola HV, Zhang K, Cui BX Optically resolving individual microtubules in live axons. Structure. 2009;17(11):1433–1441. doi: 10.1016/j.str.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MJI, Klumpp S, Lipowsky R Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc Natl Acad Sci USA. 2008;105(12):4609–4614. doi: 10.1073/pnas.0706825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning L, Liu P, Zong Y, Liu R, Yang M, Chen K Universal Scaling law for colloidal diffusion in complex media. Phys Rev Lett. 2019;122(17):178002. doi: 10.1103/PhysRevLett.122.178002. [DOI] [PubMed] [Google Scholar]
- Nirmal M, Dabbousi BO, Bawendi MG, Macklin JJ, Trautman JK, Harris TD, Brus LE Fluorescence intermittency in single cadmium selenide nanocrystals. Nature. 1996;383(6603):802–804. doi: 10.1038/383802a0. [DOI] [Google Scholar]
- Orci L, Ravazzola M, Volchuk A, Engel T, Gmachl M, Amherdt M, Perrelet A, Sollner TH, Rothman JE Anterograde flow of cargo across the Golgi stack potentially mediated via bidirectional "percolating" COPI vesicles. Proc Natl Acad Sci USA. 2000;97(19):10400–10405. doi: 10.1073/pnas.190292497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DM, Williamson D, Rentero C, Gaus K Quantitative microscopy: protein dynamics and membrane organisation. Traffic. 2009;10(8):962–971. doi: 10.1111/j.1600-0854.2009.00908.x. [DOI] [PubMed] [Google Scholar]
- Pampaloni F, Reynaud EG, Stelzer EHK The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8(10):839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Peng SJ, Li WP, Yao YR, Xing WJ, Li PL, Chen CL Phase separation at the nanoscale quantified by dcFCCS. Proc Natl Acad Sci USA. 2020;117(44):27124–27131. doi: 10.1073/pnas.2008447117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan SS, Liu HY, Vu TQ Ligand-bound quantum dot probes for studying the molecular scale dynamics of receptor endocytic trafficking in live cells. ACS Nano. 2008;2(6):1153–1166. doi: 10.1021/nn700399e. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, Carter AP The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol. 2018;19(6):382–398. doi: 10.1038/s41580-018-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis GF, Yang G, Szpankowski L, Weaver C, Shah SB, Robinson JT, Hays TS, Danuser G, Goldstein LSB Molecular motor function in axonal transport in vivo probed by genetic and computational analysis in Drosophila. Mol Biol Cell. 2012;23(9):1700–1714. doi: 10.1091/mbc.e11-11-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits EAJ, Neefjes JJ From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol. 2001;3(6):E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- Riley MR, Muzzio FJ, Reyes SC Experimental and modeling studies of diffusion in immobilized cell systems −− A review of recent literature and patents. Appl Biochem Biotechnol. 1999;80(2):151–188. doi: 10.1385/ABAB:80:2:151. [DOI] [PubMed] [Google Scholar]
- Ross JL, Ali MY, Warshaw DM Cargo transport: molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20(1):41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Minoguchi S, Yanagida T Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2(3):168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K Single-particle tracking: applications to membrane dynamics. Ann Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151(2):182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Schutz GJ, Baumgartner W, Gruber HJ, Schindler H Imaging of single molecule diffusion. Proc Natl Acad Sci USA. 1996;93(7):2926–2929. doi: 10.1073/pnas.93.7.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Blower MD The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci. 2016;73(1):79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QJ, Kassim H, Huang Y, Li H, Zhang J, Li G, Wang PY, Yan J, Ye FF, Liu JL Filamentation of metabolic enzymes in Saccharomyces cerevisiae. J Genet Genomics. 2016;43(6):393–404. doi: 10.1016/j.jgg.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YA, Wu HY, Lu PJ, Wang DZ, Shayegan M, Li H, Shi WC, Wang ZZ, Cai LH, Xia J, Zhang M, Ding RH, Herrmann H, Goldman R, MacKintosh FC, Moncho-Jorda A, Weitz DA Effects of Vimentin intermediate filaments on the structure and dynamics of in vitro multicomponent interpenetrating cytoskeletal networks. Phys Rev Lett. 2021;127:108101. doi: 10.1103/PhysRevLett.127.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y New Virus, New Challenge. Innovation. 2020;1(1):100005. doi: 10.1016/j.xinn.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Mancini MC, Nie S Bioimaging: second window for in vivo imaging. Nat Nanotechnol. 2009;4(11):710–711. doi: 10.1038/nnano.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer JA, Almers W Tracking single secretory granules in live chromaffin cells by evanescent-field fluorescence microscopy. Biophys J. 1999;76(4):2262–2271. doi: 10.1016/S0006-3495(99)77382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabei SMA, Burov S, Kim HY, Kuznetsov A, Huynh T, Jureller J, Philipson LH, Dinner AR, Scherer NF Intracellular transport of insulin granules is a subordinated random walk. Proc Natl Acad Sci USA. 2013;110(13):4911–4916. doi: 10.1073/pnas.1221962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5(5):455–455. doi: 10.1038/nmeth0508-455. [DOI] [PubMed] [Google Scholar]
- Tolic-Norrelykke IM, Munteanu EL, Thon G, Oddershede L, Berg-Sorensen K (2004) Anomalous diffusion in living yeast cells. Phys Rev Lett93(7): 078102. https://doi.org/10.1103/PhysRevLett.93.078102
- Toomre D, Bewersdorf J A New Wave of Cellular Imaging. Ann Rev Cell Develop Biol. 2010;26:285–314. doi: 10.1146/annurev-cellbio-100109-104048. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Kikuta T, Warashina A, Terakawa S Protein kinase C-dependent supply of secretory granules to the plasma membrane. Biochem Biophys Res Commun. 2001;282(2):621–628. doi: 10.1006/bbrc.2001.4603. [DOI] [PubMed] [Google Scholar]
- Tudor C, Feige J, Gelman L, Wahli W, Desvergne B, Engelborghs Y (2007) Dynamics and interactions of nuclear receptors in living cells measured by fluorescence correlation spectroscopy. Biophys J : 325a-325a
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380(6573):451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P, Ponti A, Waterman-Storer CM, Salmon ED, Danuser G Recovery, visualization, and analysis of actin and tubulin polymer flow in live cells: a fluorescent speckle microscopy study. Biophys J. 2003;85(2):1289–1306. doi: 10.1016/S0006-3495(03)74564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendelin M, Kongas O, Saks V Regulation of mitochondrial respiration in heart cells analyzed by reaction-diffusion model of energy transfer. Am J Physiol. 2000;278(4):C747–764. doi: 10.1152/ajpcell.2000.278.4.C747. [DOI] [PubMed] [Google Scholar]
- von Diezmann A, Shechtman Y, Moerner WE Three-Dimensional localization of single molecules for super resolution imaging and single-particle tracking. Chem Rev. 2017;117(11):7244–7275. doi: 10.1021/acs.chemrev.6b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V, Ming Y, D'Addario C, Johansson B, Rigler R, Terenius L Fluorescence correlation spectroscopy (FCS) study of mu-opioid receptor dynamics in live cells. FEBS J. 2007;274:107–107. [Google Scholar]
- Wang B, Kuo J, Bae SC, Granick S When Brownian diffusion is not Gaussian. Nat Mater. 2012;11(6):481–485. doi: 10.1038/nmat3308. [DOI] [PubMed] [Google Scholar]
- Wang B, Kuo J, Granick S Bursts of active transport in living cells. Phys Rev Lett. 2013;111(20):208102. doi: 10.1103/PhysRevLett.111.208102. [DOI] [PubMed] [Google Scholar]
- Wang M, Yang Y, Han L, Xu F, Li F Cell mechanical microenvironment for cell volume regulation. J Cell Physiol. 2020;235(5):4070–4081. doi: 10.1002/jcp.29341. [DOI] [PubMed] [Google Scholar]
- Wang ZA, Wang XJ, Zhang Y, Xu WL, Han XJ Principles and applications of single particle tracking in cell research. Small. 2021;17(11):e2005133. doi: 10.1002/smll.202005133. [DOI] [PubMed] [Google Scholar]
- Weber SC, Spakowitz AJ, Theriot JA Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104(23):238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsher K, Liu Z, Sherlock SP, Robinson JT, Chen Z, Daranciang D, Dai HJ A route to brightly fluorescent carbon nanotubes for near-infrared imaging in mice. Nat Nanotechnol. 2009;4(11):773–780. doi: 10.1038/nnano.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser S, Schutz GJ Tracking single molecules in the live cell plasma membrane-Do's and Don't's. Methods. 2008;46(2):131–140. doi: 10.1016/j.ymeth.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Wilhelm C Out-of-equilibrium microrheology inside living cells. Phys Rev Letters. 2008;101(2):028101. doi: 10.1103/PhysRevLett.101.028101. [DOI] [PubMed] [Google Scholar]
- Wu WQ, Zhang ML, Song CP A comprehensive evaluation of a typical plant telomeric G-quadruplex (G4) DNA reveals the dynamics of G4 formation, rearrangement, and unfolding. J Biol Chem. 2020;295(16):5461–5469. doi: 10.1074/jbc.RA119.012383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Du Y, Ge F, He Y, Yeung ES (2021) Unveiling cellular internalization dynamics of single gold nanorods by tracking their orientational and translational motions. CCS Chem. https://doi.org/ 10.31635/ccschem.021.202000729:995-1004
- Yamada S, Wirtz D, Kuo SC Mechanics of living cells measured by laser tracking microrheology. Biophys J. 2000;78(4):1736–1747. doi: 10.1016/S0006-3495(00)76725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenisek DP, Steyer JA, Almers W Imaging exocytosis of single synaptic vesicles with evanescent field microscopy. Biophys J. 2000;78(1):260a–260a. [Google Scholar]
- Zhang M, Yue J, Cui R, Ma Z, Wan H, Wang F, Zhu S, Zhou Y, Kuang Y, Zhong Y, Pang DW, Dai H Bright quantum dots emitting at approximately 1, 600 nm in the NIR-IIb window for deep tissue fluorescence imaging. Proc Natl Acad Sci USA. 2018;115(26):6590–6595. doi: 10.1073/pnas.1806153115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HS, Zhou QM, Xia MC, Feng JX, Chen Y, Zhang SC, Zhang XR Characterize collective lysosome heterogeneous dynamics in live cell with a space- and time-resolved method. Analyt Chem. 2018;90(15):9138–9147. doi: 10.1021/acs.analchem.8b01563. [DOI] [PubMed] [Google Scholar]
- Zhou J, Yang Y, Zhang CY Toward biocompatible semiconductor quantum dots: from biosynthesis and bioconjugation to biomedical application. Chem Rev. 2015;115(21):11669–11717. doi: 10.1021/acs.chemrev.5b00049. [DOI] [PubMed] [Google Scholar]


