Abstract
Context
Aberrant biosynthesis and secretion of the insulin precursor proinsulin occurs in both type I and type II diabetes. Inflammatory cytokines are implicated in pancreatic islet stress and dysfunction in both forms of diabetes, but the mechanisms remain unclear.
Objective
We sought to determine the effect of the diabetes-associated cytokines on proinsulin folding, trafficking, secretion, and β-cell function.
Methods
Human islets were treated with interleukin-1β and interferon-γ for 48 hours, followed by analysis of interleukin-6, nitrite, proinsulin and insulin release, RNA sequencing, and unbiased profiling of the proinsulin interactome by affinity purification-mass spectrometry.
Results
Cytokine treatment induced secretion of interleukin-6, nitrites, and insulin, as well as aberrant release of proinsulin. RNA sequencing showed that cytokines upregulated genes involved in endoplasmic reticulum stress, and, consistent with this, affinity purification-mass spectrometry revealed cytokine induced proinsulin binding to multiple endoplasmic reticulum chaperones and oxidoreductases. Moreover, increased binding to the chaperone immunoglobulin binding protein was required to maintain proper proinsulin folding in the inflammatory environment. Cytokines also regulated novel interactions between proinsulin and type 1 and type 2 diabetes genome-wide association studies candidate proteins not previously known to interact with proinsulin (eg, Ataxin-2). Finally, cytokines induced proinsulin interactions with a cluster of microtubule motor proteins and chemical destabilization of microtubules with Nocodazole exacerbated cytokine induced proinsulin secretion.
Conclusion
Together, the data shed new light on mechanisms by which diabetes-associated cytokines dysregulate β-cell function. For the first time, we show that even short-term exposure to an inflammatory environment reshapes proinsulin interactions with critical chaperones and regulators of the secretory pathway.
Keywords: type 1 diabetes, type 2 diabetes, human islets, cytokines, proinsulin, protein interactome
All forms of diabetes ultimately result from β-cell failure. Type 1 diabetes (T1D) is a chronic inflammatory disease with autoimmune β-cell destruction, and it is now accepted that type II diabetes (T2D) is also characterized by inflammation with deleterious effects on the β-cell (1–6). Given that more than 400 million people worldwide suffer from diabetes, it is critical to fully understand the physiological consequences of inflammation on the β-cell.
Insulin is initially synthesized as preproinsulin that enters the endoplasmic reticulum (ER) where cleavage of its signal peptide converts it to proinsulin. The ER guides oxidative folding of 3 critical disulfide bonds in proinsulin that are facilitated by chaperones and oxidoreductases [eg, immunoglobulin binding protein (BiP) and protein disulfide isomerase]. We and others identified several ER chaperones and oxidoreductases required for proper proinsulin folding (7–10). Only properly folded proinsulin transits through the ER-Golgi intermediate compartment to the Golgi apparatus from which immature secretory granules are formed. Inside the immature granules, processing enzymes cleave the C-peptide from proinsulin to generate mature insulin that is secreted in response to glucose. Under healthy, β-cell conditions proinsulin is efficiently converted to insulin and little proinsulin is released into the serum.
Three highly conserved pathways regulate ER homeostasis through the unfolded protein response (UPR): protein kinase RNA-like ER kinase (PERK), inositol-requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6). Activated PERK reduces ER burden by transiently attenuating general messenger RNA (mRNA) translation, except for proteins that resolve ER stress. Loss of PERK in vivo causes β-cell failure and permanent neonatal diabetes, and we have shown that even short-term PERK inhibition in isolated human islets induces proinsulin misfolding (10). IRE1 activation leads to splicing of the transcription factor X-box-binding protein (XBP1) that induces genes encoding ER protein translocation, ER chaperones, and ER effectors (11). ATF6 processing and activation leads to its nuclear translocation to induce expression of genes encoding functions complementary, as well as overlapping, with spliced XBP1. Under homeostatic conditions, ER stress signaling through the 3 UPR sensors is repressed by their interaction with BiP. However, when BiP binds misfolded proteins, it is released from PERK, IRE1, and ATF6, thereby activating the UPR signaling (12). Thus increased protein binding to BiP is considered a gold standard as an indicator of increased unfolded/misfolded protein. Misfolded proteins that do not bind BiP do not activate the UPR (13, 14). Discriminating which additional factors promote productive proinsulin folding from defective proinsulin forms that activate the UPR is critical, because chronic UPR activation is linked to β-cell demise.
Β-cells produce up to 6000 molecules of proinsulin per second (15), making them susceptible to ER stress and misfolding under conditions of increased demand for insulin, as occurs upon depletion of β-cell mass or in response to insulin resistance. There is evidence of ER stress in islets from patients with T1D and T2D (16), and ER stress precedes T1D in the nonobese diabetic mouse islets (17–19). We showed that high glucose alone (as occurs in both forms of diabetes) is sufficient to induce high molecular weight aggregates of misfolded proinsulin in normal human islets (10). We also observed proinsulin misfolding in islets from patients with T2D (10) and in islets from mice fed a high fat diet (20). Strikingly, proinsulin misfolding preceded hyperglycemia in the db/db model of T2D (20). Together, the data suggest a connection among inflammation, ER stress, and proinsulin misfolding.
A second common feature of islet pathology in T1D and T2D is secretion of unprocessed proinsulin (21). In fact, detectable serum proinsulin persists in a majority of individuals with longstanding T1D, even those who no longer have detectable serum C-peptide (22). Moreover, proinsulin in the bloodstream is a predictor of T2D, obesity, and cardiovascular disease, and the ratio of secreted proinsulin to insulin is a measure of β-cell dysfunction associated with T2D (23). Notably however, circulating proinsulin also provides clear evidence of β-cell survival in patients with T1D and T2D. Therefore, deciphering new strategies to restore optimal performance in these β-cells is critically important.
To gain insight into the effects of an inflammatory environment on β-cell function, transcriptomic, proteomic, and epigenetic analyses of human islets treated with diabetes-associated cytokines interleukin-1β (IL-1β) and interferon-γ (IFN-γ) have cataloged genes/proteins regulated by cytokines (24, 25). While these data illuminate the composition of islets, they do not identify the proteins that interact with proinsulin to coordinate its folding, trafficking, and processing. We recently used proinsulin affinity purification-mass spectrometry (AP-MS) to comprehensively define the proinsulin interactome in normal human islets (10). Here human islets treated with IL-1β and IFN-γ were similarly subjected to proinsulin AP-MS for comparative analysis with the reference interactome.
The AP-MS data revealed that cytokine treatment modified proinsulin binding to multiple ER chaperones and oxidoreductases while increasing a propensity to proinsulin misfolding. Novel proinsulin interactions with proteins previously identified as diabetes genome-wide association studies (GWAS) candidates were also regulated by cytokines. Finally, cytokines enhanced proinsulin interaction with a protein trafficking network including microtubule-regulatory proteins. We show that microtubules limit cytokine-induced proinsulin release, which is enhanced upon destabilization of microtubules by Nocodazole. Together the data reveal that even short-term exposure to inflammatory cytokines has profound effects on proinsulin biosynthesis.
Materials and Methods
Human Islets
Islet preparations averaging 85% to 95% purity by dithizone and 95% viability by ethidium bromide/fluorescein diacetate staining were procured from Prodo labs. The donors had no history of diabetes and had normal hemoglobin A1c. The islets were cultured for 48 hours in Prodo media of Dulbecco’s modified Eagle medium (105 mg/dL) with or without cytokines IL-1β (50 U/mL) and IFN-γ (1000 U/mL). Nocodazole 5 µg/mL was added to some cultures for 48 hours. PERK inhibitor GSK157 (2 µM) was added to some cultures for 18 to 24 hours. Subtilase cytotoxin (SubAB) and mutant SubAB (2.5 µg/mL) were added to some cultures for 6 hours.
RNA Sequencing
RNA was prepared from cytokine untreated and treated islets using Rneasy kit (Qiagen) and total RNA was ribosomal depleted to remove ribosomal RNA. The complementary DNA was ligated with index adapters for each sample followed by purification and then enriched with polymerase chain reaction (PCR) to create the final library. The quality and quantity of the libraries were detected by Agilent Bioanalyzer and Kapa Biosystems quantitative PCR (qPCR). Multiplexed libraries were pooled and single-end 50 bp sequencing was performed on 1 flow-cell of an Illumina Next Gen Sequencing platform. For analysis, reads were mapped to human genome (version Homo sapiens.GRCh37.75), and 87% overlap with exons was obtained [Supplementary Table 1 (26)]. Canonical pathways were mapped using ingenuity pathway analysis. Reverse Transcription (RT)-qPCR validation primers [Supplementary Table 2 (26)].
Proinsulin AP-MS
Methods are described in detail in (10). Briefly, human islets were lysed in 50 mM tris(hydroxymethyl)-aminomethane (Tris) pH 7.4, 150 mM sodium chloride, and 1% TX-100 with protease inhibitor cocktail (Thermo Fisher Scientific). Lysates were precleared with protein G agarose beads and immunoprecipitated with beads cross-linked to mouse immunoglobulin (IgG) or proinsulin antibody (20G11) overnight at 4°C. A fraction of the beads were removed for protein elution to confirm successful immunoprecipitation (IP) by Western blot and silver staining. The majority of the beads were subjected to denaturation, reduction, and trypsin digestion followed by 2-dimensional liquid chromatography-tandem mass spectrometry analysis. The mass spectrometer was operated in positive data dependent acquisition mode with up to 5 second-stage mass spectrometry spectra triggered per duty cycle. Tandem mass spectrometry (MS/MS) spectra were searched against the Homo sapiens UniProt protein sequence database (January 2015 version) using MaxQuant (version 1.5.0.25). Label-free intensity was normalized by Loess method using Normalyzer. MSstats was used to calculate a confidence (P-value) and fold change of proinsulin IP-to-control IgG IP intensity ratio for each protein in each donor preparation separately [Supplementary Table 3 (26)].
Data Filtering
Datasets were filtered based on 3 criteria: fold change ≥ 2, P ≤ 0.05, and delta MS/MS counts ≥ 10 in either untreated or cytokine-treated conditions. We then performed enriched ontology analyses on the interactors that were common to both groups or enriched in “treated” or “untreated” conditions to identify proteins belonging to “protein targeting” and “processing” functional groups.
Western Blotting
Isolated islets were lysed in radioimmunoprecipitation assay buffer (10 mM Tris pH 7.4, 150 mM sodium chloride, 0.1% sodium dodecyl sulfate, 1% NP-40, 2 mM EDTA) with protease and phosphatase inhibitors (Thermo Fisher Scientific) on ice for 30 minutes, and lysates were collected after centrifugation at 4°C for 15 minutes at 12000 g. Samples were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (4-12% Bis-Tris gel; Bio-Rad Laboratories, Inc.). Primary antibodies and dilutions are shown in Supplementary Table 4 (26). For secondary antibodies, goat anti-mouse and goat anti-rabbit were used in 1:5000 (Li-Cor, IRDye®-800CW RRID:AB_2687553 or IRDye®-680RD RRID:AB_10974977). After wash, membranes were imaged on Licor Odyssey CLX with ImageStudio software, and, for quantitation, the band intensities were analyzed by ImageJ.
Immunohistochemistry
Human islets incubated for 48 hours ± cytokines were harvested and fixed in 4% paraformaldehyde. Paraffin embedding, sectioning, and slide preparations were done in the Sanford Burnham Prebys Histopathology Core Facility. Sections were stained with primary antibodies (1:200) specific for proinsulin (20G11, a gift of Dr. William Balch), kinesin family member 2A (Abcam, ab197988 RRID: AB_2921234), and 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific). For secondary antibodies, Alexa Fluor 594 goat α-rabbit IgG (Thermo Fisher Scientific, RRID: AB_2313921) and Alexa Fluor 488 goat α-mouse IgG were used (Invitrogen, RRID: AB_2534069). Microscope images were taken at 200×.
Griess Assay
Nitrite release into the media was tested using the Griess reagent (Sigma) used per manufacturer’s recommendation.
Enzyme-linked Immunoassays
Interleukin 6 (IL-6; Abcam), proinsulin, and insulin (Mercodia) were run according to manufacturer’s instructions.
Results
Cytokines Promote Inflammatory Signaling and Increase Both Insulin and Proinsulin Release From Human Islets
To investigate the effects of inflammation on β-cell function, human islets (preparations from 10 individuals) were treated with the diabetes-associated cytokines IL-1β (50 U/ml) and IFN-γ (1000 U/ml) for 48 hours. These conditions were previously shown to emulate many aspects of diabetes-associated inflammation without inducing significant cell death, and, consistent with this, cytokine-treated islets appeared of normal size and morphology (Fig. 1A) (24, 27, 28).
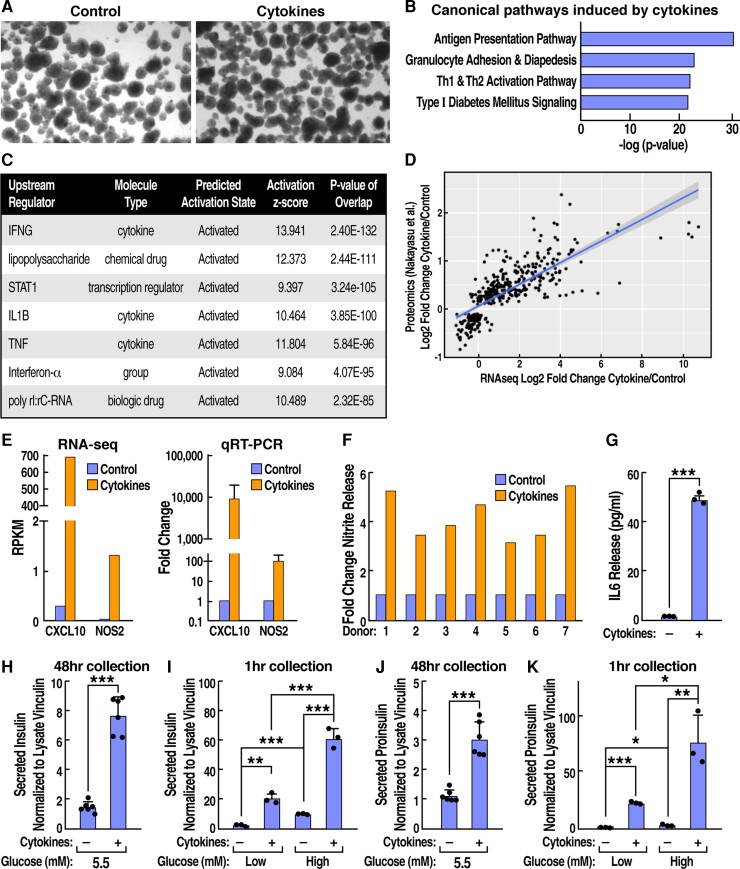
Figure 1.
Cytokine-treated human islets exhibit upregulated chemokine expression with release of interleukin 6 and nitric oxide, altered glucose-stimulated insulin secretion, and aberrant proinsulin release. Cytokine-treated and untreated islets were assayed for (A) morphology by light microscopy; (B) canonical pathways induced by cytokines in RNA sequencing (RNA-seq) data revealed by ingenuity pathway analysis; (C) upstream pathways predicted by ingenuity pathway analysis to produce the observed cytokine-regulated expression profile in RNA-seq data; (D) comparison of cytokine vs untreated human islets in our RNA-seq profile vs published whole proteome data (25); (E) RNA-seq and real-time quantitative reverse transcription-polymerase chain reaction analysis of C-X-C motif chemokine ligand 10 and nitric oxide synthase 2 (inducible nitric oxide synthase 2) expression; (F) nitrite release by Griess assay; ELISA data for secreted human (G) IL6, (H and I) insulin, and (J and K) proinsulin. Data represent the mean of 3 to 6 independent biological replicates analyzed by unpaired t-test. Error bars are ± SD; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
To measure inflammatory signaling induced by cytokines, we used a combination of RNA sequencing (RNA-seq), RT-qPCR, Western blots, the Griess assay for nitrite release, and enzyme-linked immunosorbent assay (ELISAs). Analysis of RNA-seq [Supplementary Table 1 (26)] by ingenuity pathway analysis revealed that “antigen presentation” and “type I diabetes signaling” were 2 of the top 4 pathways induced by the cytokines versus untreated islets (Fig. 1B). Among the “antigen presentation” genes upregulated by cytokines, proteasome subunit beta type-9 is a proteosomal protease that produces peptides that the cytokine-induced transporter 1 and transporter 2 (TAP2) transfer into the ER. Among genes in the “type I diabetes signaling” category, cytokine-induced myeloid differentiation primary response 88 recruits (also induced) tumor necrosis factor receptor-associated factor 6, and together they activate nuclear factor kappa B. Strikingly, analysis of upstream signaling effectors predicted to generate the observed RNA-seq profiles correctly identified IL-1β, and IFN-γ (Fig. 1C). We next compared our RNA-seq data to a recent proteomic study of human islets treated with the same cytokine cocktail, identifying a strong correlation between the RNA and protein profiles (Pearson correlation: 0.77, P-value < 2.2 × 10−16) (Fig. 1D) (25). RNA-seq and RT-qPCR revealed that cytokines induced inflammatory effectors (eg, the chemokine C-X-C motif chemokine ligand 10 and inducible nitric oxide synthase (from the NOS2 gene) (Fig. 1D), suggesting that cytokines induced the release of inflammatory factors from islets. Indeed, the Griess assay revealed that cytokines triggered nitrite release from 7 individual donor islet preparations (P = 7 × 10−4) (Fig. 1F) and induced a 32-fold increase in secreted IL-6, as measured by an ELISA. Shown are results representative of 3 preparations, tested in triplicate, P <0.001 (Fig. 1G).
We next examined the effect of cytokines on insulin and proinsulin release from human islets. We collected media from samples of 200 islets that were incubated in 5.5 mM glucose ± IL-1β and IFN-γ treatment for 48 hours. To measure glucose-stimulated insulin secretion (GSIS), the same islet samples were then washed (removing cytokines), followed by sequential 1-hour incubations in low (2.2 mM), and then high (22 mM), glucose. Insulin ELISA of the 48-hour media samples revealed that cytokines induced a marked increase in insulin release (control = 3.4 nM, + cytokines = 17.7 nM, P < 0.001) (Fig. 1H). The cytokine treatment also increased insulin release at both low and high glucose as well as GSIS (the ratio of high/low glucose values) (Fig. 1I). Under normal conditions very little proinsulin is released from β-cells and instead is processed to insulin. To determine whether the cytokine-treated islets exhibited aberrant proinsulin release associated with β-cell dysfunction in diabetes, aliquots of the same media samples collected previously were also assayed for proinsulin by ELISA. In islets (5.5 mM glucose) treated with cytokines for 48 hours, a nearly 3-fold increase in proinsulin release was observed by ELISA (baseline without cytokines= 2.1 nM) (Fig. 1J), as well as by Western blot [Supplementary Figure 1 (26)]. Proinsulin secretion was also increased by cytokines in both the low-glucose (control = 4 pM, + cytokines = 8.5 pM) and high-glucose 1-hour incubations (control =70 pM, + cytokines = 233 pM). All ELISA values in figures were normalized to vinculin from corresponding islet lysates (Fig. 1H-K), and data are representative of experiments with 10 donor islet preparations. To ensure that insulin and proinsulin in the media emanated from intact islets, media was analyzed by Western blot for 2 of the 10 most highly induced cytoplasmic proteins by cytokines, guanylate binding protein 5, and indoleamine 2,3-dioxygenase 1. The 2 proteins were detected only in cell lysates and not in media samples, arguing against cell lysis as a mechanism for insulin or proinsulin release with cytokines [Supplementary Table 2 (26)].
Defining the Proinsulin Interactome in Cytokine-Treated Islets
We recently reported on the proinsulin interactome in normal human islets that was highly conserved across a diverse group of 6 donors that included both sexes and spanned 3 ethnicities (10). To provide mechanistic insight into the effects of proinflammatory cytokines on proinsulin biosynthetic pathways, we treated equal portions of the same 6 islet preparations with IL-1β and IFN-γ for proinsulin AP-MS. The islets were lysed and immunoprecipitated with beads conjugated with a mouse mAb specific for human proinsulin or with control mouse IgG-conjugated beads. A small amount of each sample was used to confirm that total proinsulin recovery was similar in cytokine-treated and untreated samples [Supplementary Table 2A (26)]. The bulk of each sample was then subjected to on-bead denaturation, reduction, trypsin digestion, and liquid chromatography-tandem mass spectrometry analyses.
The liquid chromatography-tandem mass spectrometry runs were conducted concurrently with the corresponding untreated islet samples as previously described (10). MS/MS spectra were searched against the human Uni-Prot database and analyzed with MaxQuant. Three of the islet samples were also assayed in duplicate, to ensure technical reproducibility [Supplementary Figure 2B (26)]. Similar numbers of proteins were identified in all MS runs [Supplementary Figure 2C (26)]. As in our previous study, Loess-R was used for normalization and MSstats was used to calculate a statistical confidence score (P-value) for each protein based on intensity and reproducibility of detection across samples. Comparison of MS/MS counts for proinsulin (bait) across the 6 islet samples confirmed the remarkable consistency in proinsulin recovery with and without cytokines.
To identify the most highly conserved cytokine-induced alterations in the proinsulin biosynthetic pathway, data were analyzed using the 4 criteria we described for normal islets (10): (i) total intensity in proinsulin IP vs IgG IP ≥ 2, (ii) P ≤ 0.05, (iii) MS/MS ≥ 10, and (iv) gene expression in single-cell RNA-seq of human β-cells >1 per million reads (29). This stringent analysis produced a dataset of 648 identified proteins, 140 enriched in cytokine treatment, 108 enriched in untreated islets, and 400 shared between conditions (10) [Supplementary Table 2 (26)]. For visualization of the data, each protein was assigned to a compartment or function based on Genecards, Uniprot, or literature (Fig. 2). For simplicity, proteins assigned a primarily ribosomal, nuclear, mitochondrial cellular function are not included in Figure 2. A selection of identified proinsulin interactors were validated by proinsulin affinity purification-Western blot [Supplementary Figure 3 (26)].
Figure 2.
Defining the proinsulin interaction network regulated by cytokines. Affinity purification-mass spectrometry-identified proinsulin interactions in cytokine-treated islets were compared with recently published proinsulin interactome in untreated islets (10). Icon size reflects significance of interaction with proinsulin vs control immunoglobulin; increasing icon size = increasing significance (smallest icons P = 0.05). Deepening red reflects proinsulin interaction increased by cytokines and deepening blue reflects proinsulin interactions decreased by cytokines; palest for each color = 2-fold difference. Protein categories were defined by the Human Protein Atlas. For simplicity, proinsulin interacting proteins primarily in nucleus, mitochondria, ribosome, or undetermined categories are not visualized in this figure but are included in uploaded dataset.
As reported for the proinsulin interactome in untreated human islets (10), MS results for cytokine-treated islets displayed striking sensitivity and specificity. The sensitivity is exemplified by identification of the processing enzyme proprotein convertase subtilisin/kexin type 1 (PCSK1), despite the fact that it interacts only transiently with proinsulin. In contrast, specificity was demonstrated by a lack of nonspecific proinsulin interaction with amylin, which is expressed at high levels and cosecreted with insulin in β-cells, and proinsulin did not interact with chemokines that were among the proteins most highly induced by cytokines at the expression level (25).
Cytokines Alter Proinsulin Folding and Interactions With ER Protein Folding Machinery
Unbiased cluster analysis of all proinsulin interactions in Cytoscape identified “protein folding in ER” as a pathway significantly altered by cytokines. The proinsulin-interacting proteins in this group are shown with their interactions with each other in Figure 3A. Among ER folding factors, the chaperone BiP was the most significant proinsulin interactor (proinsulin IP/IgG IP) under cytokine conditions (P = 0) (Fig. 2) and also exhibited the most significant P-value for the increase in binding in cytokine treated relative to control islets (P = <0.001), validated in Supplementary Figure 3 (26). We previously showed that BiP plays a critical role in proinsulin folding in human and murine islets (8, 10, 20, 30), suggesting that increased BiP might be required to maintain proinsulin folding in an inflammatory environment. To test this, islets ± cytokines were treated with the Shiga-toxigenic Escherichia coli virulence factor SubAB that cleaves and inactivates BiP. Indeed, cytokine treatment exacerbated SubAB-induced proinsulin misfolding, identified by accumulation of high molecular weight multimers on a nonreducing gel (Fig. 3B, lanes 4 vs 8). As a control, islets were treated with enzymatically inactive mutant toxin SubAA272B, which did not induce misfolding (31). We previously reported PERK inhibition, which stimulates proinsulin synthesis (32), also triggers proinsulin misfolding in human islets (10). Therefore, we examined the effect of cytokines on islets treated with the PERK inhibitor glycogen synthase kinase 157 (GSK157). Islets incubated with GSK157 in the presence of cytokines exhibited increased proinsulin misfolding compared to GSK157 treatment alone (Fig. 3B, lanes 2 vs 6). Together, the data suggest that islets in an inflammatory environment are in a precarious position with regards to proinsulin folding. This may result from a combination of both gained and lost interactions; for example, in addition to BiP, cytokines increased proinsulin binding to the oxidoreductase protein disulfide isomerase (Figs. 2 and 3A) that we recently showed is required for efficient proinsulin maturation and β-cell health in response to stress in vivo (diet-induced obesity) (8). In contrast however, peroxiredoxin-4, which we showed was required for protein folding in oxidant stressed β-cells (as with cytokine treatment), exhibited lower proinsulin binding in cytokine-treated islets (Fig. 2). Taken together, the data are consistent with a response to cytokines that is both adaptive and maladaptive.
Figure 3.
Cytokines alter human proinsulin interactions with endoplasmic reticulum (ER) factors and induce propensity to proinsulin misfolding. (A) Cluster analysis using MCODE algorithm in Cytoscape identifies “protein folding in ER” as altered by cytokines. (B) Human islets treated ± cytokines, ± protein kinase RNA-like ER kinase inhibitor, and ± subtilase cytotoxin (that cleaves immunoglobulin binding protein) analyzed by Western blot for proinsulin on nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blots for proinsulin, immunoglobulin binding protein, and glyceraldehyde 3-phosphate dehydrogenase shown were analyzed by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (C) Real-time quantitative reverse transcription-polymerase chain reaction of unfolded protein response genes in cytokine-treated vs untreated human islets. Error bars are ± standard error of the mean; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
We next examined the expression of ER stress response genes by RT-qPCR. Cytokines modestly increased the levels of the UPR genes C/EBP homologous protein, ATF6, and IRE1 and spliced XBP1, among others (Fig. 3C). ER stress appeared to be submaximal as we did not observe phosphorylation of the translation initiation factor eukaryotic translation initiation factor 2A (eIF2α) at Ser51, an event that leads to translation attenuation upon PERK activation. MIN6 cells treated with thapsigargin, a noncompetitive inhibitor of the sarco/ER calcium ion ATPase, served as a positive control for induction of eIF2α phosphorylation [Supplementary Figure 4 (26)].
Diabetes GWAS Candidate Proteins Interact With Proinsulin and With Each Other
We observed that cytokines regulated proinsulin interactions with several GWAS candidates associated with T1D and T2D (Fig. 4). Among T1D candidates, cytokines increased proinsulin interactions with TAP2 and serine/threonine-protein kinase D2 (PRKD2), while interactions with cordon-bleu protein (COBL) and tumor suppressor candidate 3 (TUSC3) were decreased. Among T2D candidates, cytokine exposure induced proinsulin interactions with stromal cell-derived factor 2-like 1 (SDF2L1), SCY1-like pseudokinase 1 (SYCL1), insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), and ubiquitin protein ligase EC3 (UBE3C) and reduced interactions with spectrin repeat containing nuclear envelope protein 1 (SYNE1) (33, 34). Two GWAS candidates implicated in both T1D and T2D are Ataxin-2, which exhibited increased proinsulin interactions in cytokine conditions, and pleckstrin homology domain containing A1 (PLEKHA1), which exhibited reduced proinsulin interactions by cytokines (type2diabetesgenetics.org) (35, 36). To investigate whether proinsulin binding GWAS proteins formed a network, we used unsupervised analysis of published protein:protein interactions curated in BioGrid (BioGrid 4.4) for each candidate. Strikingly, the GWAS factors formed a tight matrix of interactions with each other and with a small number of additional proinsulin interactors we identified by AP-MS (Fig. 4). Taken together, the proinsulin AP-MS data suggest β-cell roles for diabetes GWAS candidates for which little has been known.
Figure 4.
Cytokines regulate proinsulin interactions with diabetes genome-wide association studies candidate proteins. Of the connections shown here, STX4, STXBP1, and STXBPL5 are the only proteins that were not identified as proinsulin interactors.
The proinsulin interactome data also provided clues to islet autoimmunity that contributes to T1D pathogenesis. Proinsulin itself is an autoantigen in T1D (37), and we found that cytokines induced proinsulin interactions with the additional autoantigens islet cell autoantigen 69 kDa (ICA69) and chromogranin A (38).
Cytokines Induce Proinsulin Interactions With ER-Golgi Transport and Microtubule Associated Proteins
Ingenuity Pathway analysis revealed that the proinsulin interaction network includes a cluster of Golgi-ER trafficking proteins regulated by cytokines. Among these, cytokines significantly induced proinsulin interactions with several kinesins, a family of microtubule motor proteins [ie, KIF1A, KIF21A, and, most significantly, KIF2A (fragment-crystallizable cytokine phosphatidylinositol/IgG = 4.2, P = 1.1 × 10−6)] (Fig. 5A). KIF2A binding to proinsulin was verified by proinsulin AP-Western blot (Fig. 5B). Increased KIF2A localization with proinsulin in cytokine-treated human islets was also validated by immunohistochemistry (Fig. 5C). The immunohistochemistry showed that KIF2A protein expression in β-cells was heterogeneous, and analysis of previously published single-cell RNA-seq data also showed KIF2A heterogeneity at the level of transcription (29) [Supplementary Figure 5 (26)]. KIF2A has plus-end microtubule depolymerizing activity. Because microtubule depolarization was shown to increase insulin release during GSIS (39, 40), we investigated whether microtubule dynamics play a role in cytokine-induced proinsulin release. Samples of 200 human islets incubated in 5.5 mM glucose ± cytokines for 48 hours were treated ± the microtubule destabilizing drug nocodazole for the final 24 hours. At the 48-hour point, media was collected (48-hour samples) and islets were washed for 1 hour in 2.2 mM glucose prior to a 1-hour incubation in 2.2 mM (low glucose), followed by 1-hour incubation in 22 mM (high glucose) media, with a sample collected from the latter at 10 minutes to gauge first-phase release (high glucose, 10 minutes) and the remainder collected at 1 hour (high glucose, 1 hour). At all glucose concentrations, nocodazole alone stimulated insulin and proinsulin release, as did cytokines alone (cytokines also shown in Fig. 1). Moreover, nocodazole acted in concert with cytokines to induce insulin and proinsulin secretion (Fig. 5D). Values in the figure are shown as fold change over controls. For reference, actual control values for insulin and proinsulin, respectively, were 188 pM versus 29 pM in low glucose/1 hour, 729 pM vs 42 pM for high glucose/1 hour and 186 pM vs 36 pM for high glucose/first 10 minutes. All experiments were repeated 3 times.
Figure 5.
Cytokines induce proinsulin interactions with microtubule-associated trafficking proteins, and microtubule destabilization promotes proinsulin secretion. (A) Cytokine-induced interactome changes in proinsulin Golgi- endoplasmic reticulum transport network identified by clustering analysis using MCODE algorithm in Cytoscape. (B) Validation of proinsulin binding to KIF2A in human islets by proinsulin immunoprecipitation and Western blot. (C) Immunohistochemistry of paraffin embedded human islets ± cytokine treatment, immunostained for KIF2A and proinsulin. (D) Media was harvested from human islets and assayed for proinsulin and insulin by enzyme-linked immunosorbent assay following 48-hour incubation ±cytokines in 5.5 mM glucose, then incubations for 1 hour in 2.2 mM glucose, 10 minutes in 22 mM glucose, and 1 hour in 22 mM glucose. At the conclusion of all incubations, islet lysates were prepared for Western blotting of vinculin that was used for normalization. Data represent the mean of 3 independent biological replicates analyzed by unpaired t-test. Error bars are ± standard error of the mean; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Discussion
Recently we identified the proinsulin biosynthesis network in normal human islets (10). The data set the stage for comparative analyses with interactomes from islets under perturbed conditions. Here we sought to understand how inflammatory cytokines associated with T1D and T2D affect the network of human proinsulin protein interactions along the secretory pathway. One of the cytokines, IL-1β, long studied in T1D, is also a documented islet perturbagen in T2D. In fact, elevated levels of IL-1β are predictive of T2D, and IL-1 antagonism improves glycemia and β-cell function in patients with T2D (1, 2). Nevertheless, there are limits to our understanding of inflammation in islets.
We found that cytokines altered proinsulin interactions with numerous ER resident proteins in what appears to be a mix of adaptive and maladaptive changes. For example, significantly increased proinsulin binding to the major ER chaperone BiP was observed in cytokine-treated islets. Inactivation of BiP with the toxin SubAB provoked more proinsulin misfolding in cytokine conditions than in normal human islets, suggesting that increased BiP binding is required to maintain proinsulin folding in cytokine-treated cells. In contrast, cytokine treatment lowered proinsulin interactions with peroxiredoxin 4 (PRDX4), a peroxiredoxin that we recently showed promotes proper proinsulin folding, particularly in stress conditions. PRDX4, when overexpressed, was also shown to protect β-cells against streptozotocin-induced injury insulitis (41). In summary, proinsulin folding appears to be maintained in cytokine conditions through recruitment of additional chaperones and oxidoreductases, yet reduced interactions with PRDX4 and cochaperones (eg, ERDJ3 and ERDJ5) may explain the increased propensity to misfolding.
We noted modest ER stress in cytokine-treated islets. The relationship between ER stress and protein misfolding may result in a vicious cycle leading to β-cell failure. It is well known that ER stress (eg, induced by thapsigargin or inhibition of the UPR sensor PERK) results in proinsulin misfolding. Interestingly, however, a lesson from patients with mutant INS-gene-induced diabetes of youth is that proinsulin misfolding can also be the primary event that drives ER stress. Mutant INS-gene-induced diabetes of youth is characterized by a misfolding-inducing mutation in only 1 allele encoding insulin. The mutant proinsulin is unable to fold and entraps the nonmutant proinsulin in high molecular weight complexes that cannot leave the ER, and ER stress ensues. Thus the data suggest that, regardless of the initiating event, ER stress and proinsulin misfolding generate a propagative cycle leading to β-cell failure.
ER and Golgi localized proteins were among GWAS candidates identified as proinsulin binding. In the ER, the T2D candidate SDF2L1 is involved in endoplasmic reticulum-associated degradation (42). The T1D candidate TUSC3 is an ER glycosyltransferase (43, 44), and the T1D candidate TAP2 plays a role in antigen presentation (Figs. 2 and 4). Proinsulin binding GWAS factors localized to the Golgi included SCYL1 (45) and the T1D candidate PRKD2, a trans-Golgi network protein required for the release of chromogranin-A-containing secretory granules (46). Moreover, BioGrid analysis of the GWAS candidates’ protein:protein interactions yielded a tight network of GWAS factors binding to each other and to a small number of additional proinsulin interactors. The data therefore suggest that the GWAS candidates sit at a nexus of a highly conserved matrix of proteins involved in proinsulin biosynthesis. To the best of our knowledge, other than TAP2 and SDF2L1, these GWAS candidates have not previously been reported to interact with proinsulin.
Consistent with other reports, cytokine treatment induced proinsulin release into the media (47), mimicking the pathological proinsulin release observed in sera from patients with both T1D and T2D. The proinsulin release occurred in the context of increased proinsulin binding to the microtubule-associated kinesin motor proteins KIF1A and KIF21A, KIF2A. KIF2A, a microtubule depolymerase, was also identified in a proinsulin interactome study we conducted in MIN6 cells, which employed a different antibody to immunoprecipitate proinsulin (48). The association of insulin granules with microtubules has been known for some time (49), and Golgi-derived microtubules are critical for replenishment of the insulin granule pool and proper insulin secretion (39, 40). Given the increased binding of proinsulin with KIF proteins, we tested whether microtubules played an essential role in cytokine-induced proinsulin. Indeed, the microtubule destabilizer nocodazole promoted proinsulin secretion and cooperated with cytokines to further induce proinsulin secretion. To the best of our knowledge, this is the first demonstration that cytokines and microtubule destabilization work together to promote proinsulin, as well as insulin, release.
One suggested mechanism for proinsulin secretion in T1D is that immature granules are released in which there is reduced expression of PCSK1 and carboxypeptidase E that process proinsulin maturation to insulin (22). Consistent with this model, we find that cytokines induced proinsulin secretion and increased the interaction between proinsulin and the autoantigen ICA1 (ICA69) that resides on the membranes of immature but not mature secretory granules (50). In contrast to a model in which processing enzymes are limiting, however, cytokines did not alter proinsulin interaction with PCSK1. Thus there may be additional avenues for proinsulin escape.
The proinsulin AP-MS data suggested an alternate microtubule dependent pathway leading to proinsulin secretion through assembly and trafficking of stress granules (51). We found that cytokines strongly promoted proinsulin interactions with 3 highly conserved stress granule assembly factors: the diabetes GWAS factors Ataxin-2 and IGF2BP2, as well as the quintessential stress granule marker stress granule assembly factor 1 (G3BP1) (33, 34, 52). Stress granules are non-membranous cytoplasmic condensates formed via interactions among intrinsically disordered regions of proteins (53). They contain translationally halted transcripts as well as protein cargo that form spontaneously by liquid-liquid phase separation as part of the integrated stress response. How might proinsulin protein incorporation into stress granules result in its secretion? Notably: (i) stress granule protein components have been identified in extracellular vesicles (eg, exosomes) from multiple cell types lines (54); (ii) human islets secrete exosomes containing both proinsulin and insulin protein (55); and (iii) IL-1β and IFN-γ treatment of human islets altered the composition of RNAs in exosomes, suggesting that changes in the protein composition may occur as well (56). Thus the interactome data suggest that stress granules may also play a role in aberrant proinsulin trafficking/release.
The cytokine-dependent proinsulin interactome data also provides clues related to T1D autoimmunity. In cytokine conditions, proinsulin interacted with TAP2 and calnexin, which together form a major histocompatibility complex intermediate complex during antigen presentation (57). Further, autoantigens ICA69 and chromogranin A bound proinsulin in cytokine conditions (38). Chromogranin A is a particularly intriguing proinsulin partner given that highly immunogenic peptides comprised of small regions of proinsulin and chromogranin A fused together were recently identified in T1D (58). It was suggested that molecular crowding in granules may contribute to the peptide-fusion phenomenon, and to the best of our knowledge the AP-MS data presented here provide the first demonstration that proinsulin and chromogranin A physically interact in human β-cells.
Both T1D and T2D are characterized by circulating levels of inflammatory cytokines. Our data reveal that even short-term exposure to inflammatory conditions reshapes the proinsulin interactome in ways that affect proinsulin folding, trafficking, and secretion.
Acknowledgments
We thank Alexandre Rosa Campos in the Sanford Burnham Prebys proteomics core for helpful discussion and William Balch at Scripps Research Institute for the 20G11 antibody.
Abbreviations
- AP-MS
affinity purification-mass spectrometry
- ATF6
activating transcription factor 6
- BiP
immunoglobulin binding protein
- GSIS
glucose-stimulated insulin secretion
- GWAS
genome-wide association studies
- ELISA
enzyme-linked immunosorbent assay
- ER
endoplasmic reticulum
- IFN-γ
interferon-γ
- IgG
immunoglobulin
- IL-1β
interleukin-1β
- IL-6
interleukin 6
- IP
immunoprecipitation
- IRE1
inositol-requiring enzyme 1
- mRNA
messenger RNA
- MS/MS
tandem mass spectrometry
- PCSK1
proprotein convertase subtilisin/kexin type 1
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- PCR
polymerase chain reaction
- PRDX4
peroxiredoxin 4
- qPCR
real-time polymerase chain reaction
- RNA-seq
RNA sequencing
- RT
reverse transcription
- SDF2L1
stromal cell-derived factor 2-like 1
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SubAB
subtilase cytotoxin
- TAP2
transporter 2
- Tris
tris(hydroxymethyl)-aminomethane
- UPR
unfolded protein response
- XBP1
X-box-binding protein
Contributor Information
Duc T Tran, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA; Plexium, San Diego, CA, USA.
Anita Pottekat, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA; Illumina, San Diego, CA, USA.
Kouta Lee, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA.
Megha Raghunathan, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA.
Salvatore Loguercio, Scripps Research Institute, La Jolla, CA, USA.
Saiful A Mir, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA; University of Calcutta, West Bengal, India.
Adrienne W Paton, The University of Adelaide, Adelaide, Australia.
James C Paton, The University of Adelaide, Adelaide, Australia.
Peter Arvan, University of Michigan Medical School, Ann Arbor, MI, USA.
Randal J Kaufman, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA.
Pamela Itkin-Ansari, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA.
Funding
This work was supported by National Institutes of Health grant National Institutes of Health R24 DK110973-01 (P.I.-A., P.A., and R.J.K.) and Juvenile Diabetes Research Foundation 2-SRA-2015-47-M-R (P.I.-A., and R.J.K.). The work was also supported by Helmsley Charitable Trust/University of Miami Award Number 2018-T1D060 (P.I.-A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Helmsley Charitable Trust or the University of Miami.
Disclosures
The authors have no conflicts of interest relevant to this article to disclose.
Data Availability
The data sets generated and analyzed in the current study are available in the ProteomeXchange repository (data set identifier PXD014476; (http://proteomecentral.proteomexchange.org/cgi/GetDataset) and at NDEx (https://public.ndexbio.org/#/network/0c0a451c-88b1-11ea-aaef-0ac135e8bacf).
References
- 1. Dinarello CA, Donath MY, Mandrup-Poulsen T. Role of IL-1beta in type 2 diabetes. Curr Opin Endocrinol Diabetes Obes. 2010;17(4):314‐321. [DOI] [PubMed] [Google Scholar]
- 2. Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11(2):98‐107. [DOI] [PubMed] [Google Scholar]
- 3. Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol. 2009;5(4):219‐226. [DOI] [PubMed] [Google Scholar]
- 4. Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29(3):317‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32(4):457‐467. [DOI] [PubMed] [Google Scholar]
- 6. Ziegler AG, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity. 2010;32(4):468‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grunnet LG, Mandrup-Poulsen T. Cytokines and type 1 diabetes: a numbers game. Diabetes. 2011;60(3):697‐699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jang I, Pottekat A, Poothong J, et al. PDIA1/P4HB is required for efficient proinsulin maturation and ss cell health in response to diet induced obesity. Elife. 2019;8:e44528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ladiges WC, Knoblaugh SE, Morton JF, et al. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54(4):1074‐1081. [DOI] [PubMed] [Google Scholar]
- 10. Tran DT, Pottekat A, Mir SA, et al. Unbiased profiling of the human proinsulin biosynthetic interaction network reveals a role for peroxiredoxin 4 in proinsulin folding. Diabetes. 2020;69(8):1723‐1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hassler JR, Scheuner DL, Wang S, et al. The IRE1alpha/XBP1s pathway is essential for the glucose response and protection of beta cells. PLoS Biol. 2015;13(10):e1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326‐332. [DOI] [PubMed] [Google Scholar]
- 13. Dorner AJ, Wasley LC, Kaufman RJ. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992;11(4):1563‐1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng DT, Watowich SS, Lamb RA. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol Biol Cell. 1992;3(2):143‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schuit FC, Kiekens R, Pipeleers DG. Measuring the balance between insulin synthesis and insulin release. Biochem Biophys Res Commun. 1991;178(3):1182‐1187. [DOI] [PubMed] [Google Scholar]
- 16. Marchetti P, Bugliani M, Lupi R, et al. The endoplasmic reticulum in pancreatic beta cells of type 2 diabetes patients. Diabetologia. 2007;50(12):2486‐2494. [DOI] [PubMed] [Google Scholar]
- 17. Marhfour I, Lopez XM, Lefkaditis D, et al. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia. 2012;55(9):2417‐2420. [DOI] [PubMed] [Google Scholar]
- 18. Sahin GS, Lee H, Engin F. An accomplice more than a mere victim: the impact of beta-cell ER stress on type 1 diabetes pathogenesis. Mol Metab. 2021;54:101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tersey SA, Nishiki Y, Templin AT, et al. Islet beta-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes. 2012;61(4):818‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arunagiri A, Haataja L, Pottekat A, et al. Proinsulin misfolding is an early event in the progression to type 2 diabetes. Elife. 2019;8:e44532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Freese J, Al-Rawi R, Choat H, et al. Proinsulin to C-peptide ratio in the first year after diagnosis of type 1 diabetes. J Clin Endocrinol Metab. 2021;106(11):e4318‐e4326. [DOI] [PubMed] [Google Scholar]
- 22. Sims EK, Syed F, Nyalwidhe J, et al. Abnormalities in proinsulin processing in islets from individuals with longstanding T1D. Transl Res. 2019;213:90‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roder ME, Porte D Jr, Schwartz RS, Kahn SE. Disproportionately elevated proinsulin levels reflect the degree of impaired B cell secretory capacity in patients with noninsulin-dependent diabetes mellitus. J Clin Endocrinol Metab. 1998;83(2):604‐608. [DOI] [PubMed] [Google Scholar]
- 24. Eizirik DL, Sammeth M, Bouckenooghe T, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8(3):e1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakayasu ES, Syed F, Tersey SA, et al. Comprehensive proteomics analysis of stressed human islets identifies GDF15 as a target for type 1 diabetes intervention. Cell Metab. 2020;31(2):363‐374.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tran D, Pottekat A, Lee K, et al. Supplementary data for: Inflammatory cytokines rewire the proinsulin interaction in human islets. Uploaded August 11, 2022. Figshare. 10.6084/m9.figshare.20361855 [DOI] [PMC free article] [PubMed]
- 27. Lopes M, Kutlu B, Miani M, et al. Temporal profiling of cytokine-induced genes in pancreatic beta-cells by meta-analysis and network inference. Genomics. 2014;103(4):264‐275. [DOI] [PubMed] [Google Scholar]
- 28. Padgett LE, Broniowska KA, Hansen PA, Corbett JA, Tse HM. The role of reactive oxygen species and proinflammatory cytokines in type 1 diabetes pathogenesis. Ann N Y Acad Sci. 2013;1281(1):16‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang YJ, Schug J, Won KJ, et al. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65(10):3028‐3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scheuner D, Vander Mierde D, Song B, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11(7):757‐764. [DOI] [PubMed] [Google Scholar]
- 31. Paton AW, Beddoe T, Thorpe CM, et al. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature. 2006;443(7111):548‐552. [DOI] [PubMed] [Google Scholar]
- 32. Harding HP, Zeng H, Zhang Y, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153‐1163. [DOI] [PubMed] [Google Scholar]
- 33. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536(7614):41‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma RB, O’Donnell AC, Stamateris RE, et al. Insulin demand regulates beta cell number via the unfolded protein response. J Clin Invest. 2015;125(10):3831‐3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xue A, Wu Y, Zhu Z, et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9(1):2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Musthaffa Y, Hamilton-Williams EE, Nel HJ, et al. Proinsulin-specific T-cell responses correlate with estimated c-peptide and predict partial remission duration in type 1 diabetes. Clin Transl Immunology. 2021;10(7):e1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pietropaolo M, Castano L, Babu S, et al. Islet cell autoantigen 69 kD (ICA69). Molecular cloning and characterization of a novel diabetes-associated autoantigen. J Clin Invest. 1993;92(1):359‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trogden KP, Zhu X, Lee JS, Wright CVE, Gu G, Kaverina I. Regulation of glucose-dependent Golgi-derived microtubules by cAMP/EPAC2 promotes secretory vesicle biogenesis in pancreatic beta cells. Curr Biol. 2019;29(14):2339‐2350.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu X, Hu R, Brissova M, et al. Microtubules negatively regulate insulin secretion in pancreatic beta cells. Dev Cell. 2015;34(6):656‐668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ding Y, Yamada S, Wang KY, et al. Overexpression of peroxiredoxin 4 protects against high-dose streptozotocin-induced diabetes by suppressing oxidative stress and cytokines in transgenic mice. Antioxid Redox Signal. 2010;13(10):1477‐1490. [DOI] [PubMed] [Google Scholar]
- 42. Hayes MG, Pluzhnikov A, Miyake K, et al. Identification of type 2 diabetes genes in Mexican Americans through genome-wide association studies. Diabetes. 2007;56(12):3033‐3044. [DOI] [PubMed] [Google Scholar]
- 43. Huttlin EL, Bruckner RJ, Navarrete-Perea J, et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell. 2021;184(11):3022‐3040.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeon YJ, Kim T, Park D, et al. miRNA-mediated TUSC3 deficiency enhances UPR and ERAD to promote metastatic potential of NSCLC. Nat Commun. 2018;9(1):5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bone RN, Oyebamiji O, Talware S, et al. A computational approach for defining a signature of beta-cell Golgi stress in diabetes. Diabetes. 2020;69(11):2364‐2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bergholdt R, Brorsson C, Palleja A, et al. Identification of novel type 1 diabetes candidate genes by integrating genome-wide association data, protein-protein interactions, and human pancreatic islet gene expression. Diabetes. 2012;61(4):954‐962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hostens K, Pavlovic D, Zambre Y, et al. Exposure of human islets to cytokines can result in disproportionately elevated proinsulin release. J Clin Invest. 1999;104(1):67‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pottekat A, Becker S, Spencer KR, et al. Insulin biosynthetic interaction network component, TMEM24, facilitates insulin reserve pool release. Cell Rep. 2013;4(5):921‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Farshori PQ, Goode D. Effects of the microtubule depolymerizing and stabilizing agents nocodazole and taxol on glucose-induced insulin secretion from hamster islet tumor (HIT) cells. J Submicrosc Cytol Pathol. 1994;26(2):137‐146. [PubMed] [Google Scholar]
- 50. Cao M, Mao Z, Kam C, et al. PICK1 and ICA69 control insulin granule trafficking and their deficiencies lead to impaired glucose tolerance. PLoS Biol. 2013;11(4):e1001541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chernov KG, Barbet A, Hamon L, Ovchinnikov LP, Curmi PA, Pastre D. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J Biol Chem. 2009;284(52):36569‐36580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Markmiller S, Soltanieh S, Server KL, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172(3):590‐604.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Franchini DM, Lanvin O, Tosolini M, et al. Microtubule-driven stress granule dynamics regulate inhibitory immune checkpoint expression in T cells. Cell Rep. 2019;26(1):94‐107.e7. [DOI] [PubMed] [Google Scholar]
- 54. Jimenez L, Yu H, McKenzie AJ, et al. Quantitative proteomic analysis of small and large extracellular vesicles (EVs) reveals enrichment of adhesion proteins in small EVs. J Proteome Res. 2019;18(3):947‐959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cianciaruso C, Phelps EA, Pasquier M, et al. Primary human and rat beta-cells release the intracellular autoantigens GAD65, IA-2, and proinsulin in exosomes together with cytokine-induced enhancers of immunity. Diabetes. 2017;66(2):460‐473. [DOI] [PubMed] [Google Scholar]
- 56. Krishnan P, Syed F, Jiyun Kang N, Mirmira RG, Evans-Molina C. Profiling of RNAs from human islet-derived exosomes in a model of type 1 diabetes. Int J Mol Sci. 2019;20(23):5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Diedrich G, Bangia N, Pan M, Cresswell P. A role for calnexin in the assembly of the MHC class I loading complex in the endoplasmic reticulum. J Immunol. 2001;166(3):1703‐1709. [DOI] [PubMed] [Google Scholar]
- 58. Delong T, Wiles TA, Baker RL, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351(6274):711‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and analyzed in the current study are available in the ProteomeXchange repository (data set identifier PXD014476; (http://proteomecentral.proteomexchange.org/cgi/GetDataset) and at NDEx (https://public.ndexbio.org/#/network/0c0a451c-88b1-11ea-aaef-0ac135e8bacf).