Abstract
Pseudomonas aeruginosa uses quorum sensing (QS) to coordinate the expression of multiple genes necessary for establishing and maintaining infection. It has previously been shown that lasR QS mutations frequently arise in cystic fibrosis (CF) lung infections, however, there has been far less emphasis on determining whether other QS system mutations arise during infection or in other environments. To test this, we utilized 852 publicly available sequenced P. aeruginosa genomes from the Pseudomonas International Consortium Database (IPCD) to study P. aeruginosa QS mutational signatures. To study isolates by source, we focused on a subset of 654 isolates collected from CF, wounds, and non-infection environmental isolates, where we could clearly identify their source. We also worked with a small collection of isolates in vitro to determine the impact of lasR and pqs mutations on isolate phenotypes. We found that lasR mutations are common across all environments and are not specific to infection nor a particular infection type. We also found that the pqs system proteins PqsA, PqsH, PqsL and MexT, a protein of increasing importance to the QS field, are highly variable. Conversely, RsaL, a negative transcriptional regulator of the las system, was found to be highly conserved, suggesting selective pressure to repress las system activity. Overall, our findings suggest that QS mutations in P. aeruginosa are common and not limited to the las system; however, LasR is unique in the frequency of putative loss-of-function mutations.
Keywords: cystic fibrosis, LasR, Pseudomonas aeruginosa, quorum sensing
Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that can be problematic in a number of infection types and environments [1]. One of the major adaptations of P. aeruginosa during chronic infection is the loss of quorum sensing (QS) [2–8]. In P. aeruginosa, QS regulates the expression of hundreds of genes, including those that encode for secreted products and virulence factors [9, 10]. It is regulated via a complex hierarchical network, composed of two N-acyl-homoserine lactone (AHL) circuits known as LasR-LasI and RhlR-RhlI, two orphan regulators termed QscR and VqsR, and a negative transcriptional regulator of the las system, RsaL [9–19]. The Las and Rhl systems are composed of LuxR-LuxI pairs, which are homologous to other Gram-negative bacterial QS systems. The LuxR-type receptors (LasR, RhlR) act as transcriptional regulators, and the LuxI-type proteins (LasI, RhlI) are signal synthases. LasI produces 3-oxo-dodecanoyl-l-homoserine lactone (3OC12-HSL), and RhlI produces N-butanoyl-l-homoserine lactone (C4-HSL). Both signals can function in a combinatorial manner to synergistically regulate genes [20, 21]. Working in conjunction with the two AHL systems, is an alkyl-quinolone (AQ) system, comprising the pqsABCDE operon and pqsH, pqsL and pqsR (mvfR) genes. These genes drive the synthesis and response of 2-heptyl-3-hydroxy-4-quinolone (the Pseudomonas quinolone signal; PQS) [22, 23].
LasR was first identified in 1991 as a regulator of the lasB (elastase) gene [24]. It has since been described as a key QS regulator in the well-studied laboratory strains PAO1 and PA14, where it has been shown to sit at the top of the QS hierarchy, regulating both the rhl and pqs systems [9, 11–15]. lasR mutants have frequently been isolated from cystic fibrosis (CF) lungs and more recently, it has been shown that some CF strains use RhlR to regulate the rhl and pqs systems in the absence of functional LasR [25–30]. The decoupling of the AHL QS hierarchy reportedly requires the inactivation of MexT [25, 27, 31], a regulator of the multi-drug efflux pump operon MexEF-OprN [32, 33]. It has also been shown that mexT mutation in PAO1 can decouple public rhlR-regulated traits from private metabolic lasR-regulated traits [20]. PqsE and RhlR have also been suggested to function as a ligand:receptor pair in some QS ‘re-wired’ strains [34–36].
Previous work has shown that lasR mutations are also found outside the CF lung, but the extent and variation of these mutations is still being elucidated [26, 30]. The degree of mutation in other QS genes in infection and environmental strains of P. aeruginosa remains unknown. In this study, we explored the diversity and frequency of QS mutations across a range of ecologically distinct environments to determine (i) which QS genes are frequently mutated; (ii) mutational signatures, or patterns in other genes associated with lasR mutations; (iii) lasR gene mutation frequency specific to isolate source; and (iv) the phenotypic outcome of QS mutations.
Methods
Querying QS genes from the International Pseudomonas Consortium Database
Using nucleotide sequences from PAO1 (GCF_000006765.1), we queried QS genes using blastn for isolates from the IPCD [37]. We chose this strain because it is a fully sequenced, frequently used lab strain. We first compared strains with nucleotide mutations relative to PAO1 in each of the QS genes of interest, to determine how frequently strains exhibit nucleotide polymorphisms across multiple QS genes. We then translated these sequences into protein sequences calculating putative amino acid sequence similarities using BLOSUM80 [38]. First, we compared genes found in each isolate against our reference strain, PAO1, normalized against the similarity of the reference against itself. We then calculated the mean dissimilarity score of all isolates compared to PAO1. Some isolates were missing genes due to sequencing errors or true truncations, the number of isolates with a given gene present was under 852 for all genes. All analyses, including translation steps were conducted in R version 4.3. All code and files are available on a publicly available database: https://github.com/SPDiggleLab/QSFrequency2022
Creating an IPCD database using blastn
We pulled IPCD data from GenBank from the PRJNA325248 BioProject (https://www.ncbi.nlm.nih.gov/bioproject/325248), and downloaded contigs as a multifasta file. We used the makeblastdb/ command to generate a database of all isolate contigs.
Using blastn to find QS genes for each isolate
Using our generated database, we queried the PAO1 sequence for each gene found on Pseudomonas.com [39], against the database. We generated csv files for each gene, which included the gene sequences for each isolate.
Comparing strains with nucleotide polymorphisms in multiple QS genes
We used the csv files for each gene generated from the blastn analysis to isolate all accession information of strains with <100 % query coverage and <100 % identity to the PAO1 sequence of each QS gene. For each QS gene, we used this list of NCBI accession IDs, corresponding to unique strains, to visualize the number of strains with nucleotide mutations in one or more genes with InteractiVenn [40]. We then calculated the number of strains and the percentage of strains in the IPCD database with at least one mutation in each QS gene. We generated accession lists of mutated strains for each QS gene using a custom R script (v.4.0.2) and generated Fig. 1 with InteractiVenn. All analyses were conducted in R version 4.3.
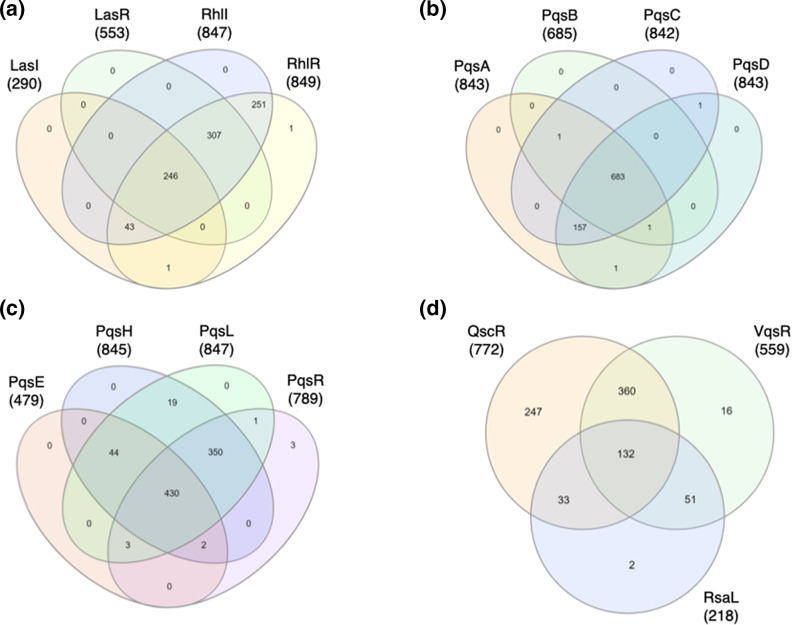
Fig. 1.
Comparison of nucleotide sequence variation among QS genes of P. aeruginosa isolates of the IPCD. There is a high degree of variation in nucleotide sequences in many of the QS genes, obscuring patterns in mutation overlap between genes within an isolate. (a) A Venn diagram of las and rhl system genes are plotted together, showing how many isolates share mutations in multiple genes. In total, 246 isolates are mutated for all four genes, and few isolates have mutations in only a single gene. (b) A Venn diagram for pqsABCD shows a similar pattern and 683 isolates are mutated in all four genes, while few isolates are mutated in only one pqs gene. (c) Few isolates have only one pqs gene mutation, and 430 isolates have mutations in all pqsEHLR genes. (d) A Venn diagram of the negative QS regulators vqsR, rsaL and qscR show that 132 isolates share mutations in all three genes.
Translating nucleotide to amino acid sequence
We translated genes to proteins using a custom R script. We first queried only for sequences starting with a canonical ATG start codon. VqsR is an exception as it begins with the alternative start codon ‘GTG’. We translated the sequences meeting these criteria using the translate function from the BioStrings R package (v.2.58.0) [41].
Calculating dissimilarity scores for isolates’ QS proteins. All sequence analyses were performed in R (v.4.0.2) using the Biostrings package v.2.58.0. We compared isolate protein sequences to PAO1 protein sequences using BLOSUM80, a matrix designed to compare protein sequences within species.
Determining truncation rates for LasR and categorizing isolates by location
We determined the length of the reference LasR protein, from PAO1, compared to each isolate protein. If the isolate protein was less than 100 % of the length of the PAO1 protein, we categorized it as truncated. Sequences were categorized as CF-originated (CF), environmental (ENV) or wound (WND). If the sequence was entered into IPCD as environmental, we adopted that label. Additionally, we included sequences labelled from animal hosts as environmental. For CF, we only included sequences with sources explicitly labelled as CF or cystic fibrosis. For wound, we included sequences labelled as wound, ulcer and burn.
Protein mutation analysis using GAMMA
We used GAMMA [42] to find the mutations in QS genes for all isolates. We created a multifasta of genes: lasR,I, rhlR,I, pqsA,B,C,D,E,H,L,R, qscR, rsaL, vqsR, mucA, rpsL, mexT, psdR to query against the IPCD PRJNA325248 multifasta. We ran commands as described in the GAMMA GitHub [42] https://github.com/rastanton/GAMMA, and generated a GAMMA file with mutations reported for all genes in all IPCD isolates. All other sequences are available on our project Github https://github.com/SPDiggleLab/QSFrequency2022
Phenotypic assays
Isolates sequenced as part of the IPCD were donated by the Brown lab at Georgia Tech. We cultured isolates on LB agar plates, and confirmed their identity by sequencing the lasR gene, using colony OneTaq PCR (New England Biolabs). For protease activity, we inoculated colonies in 5 ml of LB broth, and grew strains in biological triplicates for 16–18 h at 37 °C, 200 r.p.m. We pelleted cells and used the supernatant to run the Pierce Fluorescent Protease Assay Kit (Thermo). Readings were quantified using the TPCK Trypsin standard in triplicate, and normalized by strain optical density (OD at 600nm). All readings were compared to NPAO1. For colony autolysis experiments, we inoculated colonies in 5 ml of LB broth, and grew strains for 16–18 h at 37 °C, 200 r.p.m. We tested colony autolysis by spotting 5 µl of overnight cultures on LB agar plates, and growing colonies for 24 h at 37 °C. We scanned images of colonies on plates and quantified lysis by eye, looking for iridescent sheen or an uneven colony surface caused by plaquing.
Results
We utilized the published sequences of 852 P . aeruginosa isolates from the International Pseudomonas Consortium Database (IPCD); a database representing a range of P. aeruginosa strains from different sources including rivers, human infection and plants [37]. We queried key QS genes from the las, rhl and pqs systems, as well as the QS regulators qscR, rsaL and vqsR against gene sequences from PAO1 (GCF_000006765.1) for all 852 isolates. The PAO1 referenced used for our bioinformatics analysis is the closed genome sequence available for PAO1 on blast and Pseudomonas.com. However for in vitro analysis, we used our lab strain Nottingham PAO1 (NPAO1). For some analyses, we additionally looked at mexT and psdR, two genes that have been associated with lasR mutations in vitro [25, 31, 43, 44]. We used two analysis pipelines to look at the variation in QS genes in P. aeruginosa . One pipeline used NCBI BLASTn [45] and BLOSSUM80 [38], and the second analysis utilized GAMMA [42]. All analyses were conducted in R version 4.3 [46]. We report on protein dissimilarity scores generated by BLOSSUM80, which are a measure of how much the amino acid sequence varies from the wild-type PAO1 sequence, with 0 being identical to wild-type. We additionally report codon similarity scores which were generated with GAMMA, with 1.0 being a protein identical to wild-type. Codon similarity is inversely related to the protein dissimilarity score, and is a measure of how similar amino acid sequence is to the wild-type sequence.
Quorum-sensing genes are highly variable at the nucleotide level
We first analysed the diversity of nucleotide sequence for each QS gene for all 852 isolates and looked at which isolates had mutations in multiple QS-related genes. We found a large diversity in sequences and that few isolates had QS genes identical to PAO1 (GCF_000006765.1). The rhl system was especially variable, with 849 and 847 isolates differing from PAO1 for rhlR and rhlI, respectively (Fig. 1a). The las system was more conserved, with 553 and 290 isolates varying from PAO1 for lasR and lasI, respectively (Fig. 1a). Genes involved in 2-alkyl-4-quinolone (AQ) biosynthesis and response showed a large variation in mutations between genes, with pqsE as the least variable (479 mutated isolates), and pqsH and pqsL as the most variable (845, 847, respectively) (Fig. 1c). The orphan QS transcriptional regulators qscR and vqsR were highly variable (772 and 559, respectively). However, the negative las system transcriptional regulator rsaL was relatively conserved (218 mutated isolates) (Fig. 1d).
Negative and positive regulators of the las system are antipodal in sequence variation
We counted the number of unique protein sequences found in our database for each QS gene. We added mucA to our analysis as a reference gene that is frequently mutated in CF lung infections [47, 48], and rpsL, which encodes the 30S ribosomal protein S12; a conserved housekeeping gene [49, 50]. When we queried each QS gene nucleotide sequence against the 852 isolates using blastn, the query returned less than 852 sequences for each gene. This disparity is likely due to gaps in sequences, gene deletions and extensive mutations, preventing blastn from returning a query.
Additionally, for this analysis, we removed all truncated nucleotide sequences from the analysis, to avoid including sequencing near the end of a contig. Fig. 2(a) shows the number of protein sequences analysed for each gene. After MucA, a LasR query returned the fewest number of sequences – 756 out of 852 isolates, suggesting that there are many strains that contain large deletions in LasR, truncations or are lacking the LasR gene entirely. After translating the sequences, we found that LasR had the most unique protein sequences (259) across 852 isolates (Table 1, Fig. 2a) compared to other QS genes and MucA. The next most variable QS proteins, PqsH, PqsA and PqsL contained 189, 170 and 169 unique protein sequences, respectively (Table 1; Fig. 2a). We found that AHL signal synthases were highly conserved; LasI and RhlI had only 61 and 87 unique protein sequences, respectively. RsaL was the most conserved of all studied proteins, with only 18 unique sequences. Compared to LasR, the other key QS proteins were more conserved across isolates. Our protein sequence findings appear to contrast with our nucleotide sequence analysis, but this is likely due to primarily silent mutations in the rhl system. To control for possible bias due to differences in protein length, we normalized gene sizes to PAO1 by dividing the lengths of each gene by the PAO1 reference gene length. We found that the shortest proteins, RspL and RsaL were the most conserved, while the longest genes, PqsH, PqsA and PqsL were all highly variable. The most variable protein, LasR is of intermediate length, compared to RsaL and Pqs proteins (Table 1).
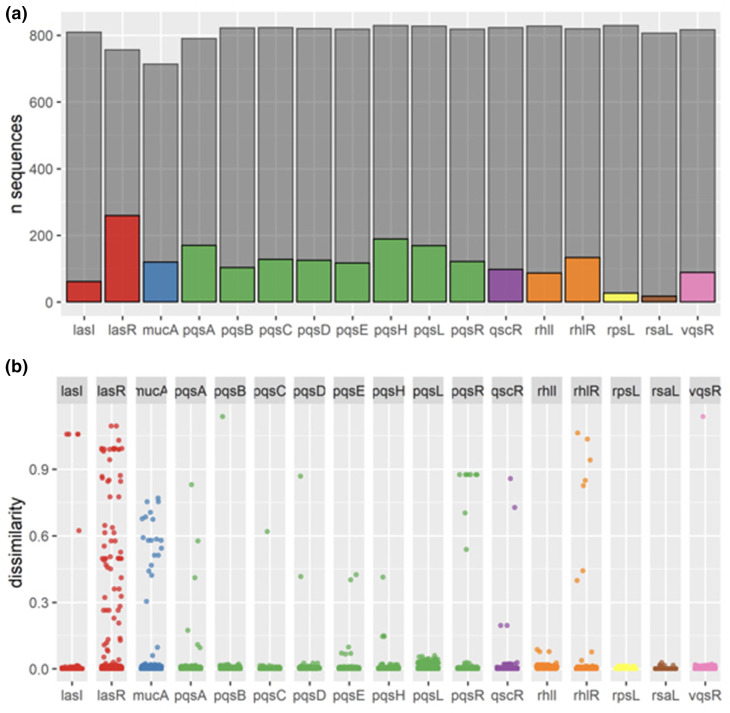
Fig. 2.
Determining variability in QS proteins between P. aeruginosa isolates from the IPCD. (a) We created a database of 852 isolates and used PAO1 to search for the QS proteins of each isolate. Due to the variation in each isolate’s genome and due to gaps in sequencing, each protein queried returned fewer than 852 sequences (shown in grey). We also determined the number of unique sequences for each protein and found that LasR had the highest number of unique sequences (in colour). The next highest were PqsH, PqsA and PqsL. RsaL had the fewest number of unique sequences. (b) Using a custom dissimilarity metric (BLOSUM80), we calculated mean dissimilarity scores. We found that LasR had the highest mean dissimilarity score compared to all QS proteins, and the largest variation.
Table 1.
Variation in protein sequence encoded by P. aeruginosa isolates. Using the sequences returned by blastn, we translated genes with full-length nucleotide sequences to determine the number of unique protein sequences encoded by the isolates in the database. We found that LasR had the most unique sequences across queried isolates, and RsaL encoded the fewest unique sequences
|
Gene |
Gene length |
Genes returned |
Unique protein sequences |
Unique sequences/gene length |
|---|---|---|---|---|
|
rsaL |
243 |
806 |
18 |
0.07 |
|
rpsL |
372 |
829 |
27 |
0.07 |
|
lasI |
606 |
809 |
61 |
0.10 |
|
rhlI |
606 |
827 |
87 |
0.14 |
|
vqsR |
807 |
816 |
89 |
0.11 |
|
qscR |
714 |
823 |
98 |
0.14 |
|
pqsB |
852 |
822 |
103 |
0.12 |
|
pqsE |
906 |
818 |
117 |
0.13 |
|
mucA |
585 |
713 |
120 |
0.21 |
|
pqsR |
999 |
818 |
122 |
0.12 |
|
pqsD |
1014 |
820 |
125 |
0.12 |
|
pqsC |
1047 |
823 |
128 |
0.12 |
|
rhlR |
726 |
819 |
133 |
0.18 |
|
pqsL |
1197 |
827 |
169 |
0.14 |
|
pqsA |
1554 |
790 |
170 |
0.11 |
|
pqsH |
1149 |
829 |
189 |
0.16 |
|
lasR |
720 |
756 |
259 |
0.36 |
We next plotted the BLOSUM80 dissimilarity scores for all isolates for each gene and found that LasR had a large number of highly dissimilar sequences (Fig. 2b). The highly dissimilar scores are caused by truncation mutations due to an early stop site leading to a shortened protein. Transcriptional regulators PqsR and RhlR also have a number of truncation mutations, however, there are fewer than LasR. While PqsA, PqsL and PqsH all encode many unique protein sequences, most isolates cluster towards the bottom, indicating that they have dissimilarity scores close to 0, indicating that the proteins have a near identical function.
MexT, PqsL and LasR have the highest mutation rates
We used the protein sequence data from GAMMA to look at the mutation rate of each QS protein (Table 2). For this analysis, we were able to include truncated nucleotide sequences, and only excluded sequences where it was indicated that the gene was located at the contig edge. We included MexT and PsdR proteins in this analysis. For the majority of genes, we compared isolates to the PAO1 (GCF_000006765.1) gene sequence, with the exception of PqsD and RhlI, which we compared to the PAK strain of P. aeruginosa . The MexT sequence for PAO1 has an 8 bp insertion sequence, which was not commonly found in the IPCD database of isolates, as well as many other coding mutations. We instead used the sequence for isolate U0330A as it coded for a protein shared by 128 other isolates, the highest number of strains sharing one MexT sequence. The MexT gene is highly variable, and is mutated in our Nottingham PAO1 lab strain (NPAO1) and the PAO1 reference used (GCF_000006765.1). Among the QS genes, we found that PqsL had the highest mutation rate, followed by LasR. We defined mutation rate in this analysis as the percentage of non-wild-type protein sequences out of queried isolates. RsaL and LasI were highly conserved, with low mutation rates.
Table 2.
Mutation rate and mean codon similarity of QS proteins in P. aeruginosa . We used GAMMA to query QS genes against 852 isolates. We used PAK as a reference for RhlI and PqsD, U0330A as a reference for MexT and PAO1 as a reference for all other genes. GAMMA assigns isolates with a codon similarity measure, which reflects the type of amino acid change or truncation. A low score reflects a likely change in function of the protein, and a score of 1 reflects the isolate is identical to the reference. We found that LasR had the lowest mean codon similarity and RsaL the highest
|
Gene |
Total |
Mutant |
Percent mutant |
Codon similarity |
|---|---|---|---|---|
|
LasR |
826 |
425 |
51.45 % |
0.9343 |
|
MucA |
850 |
257 |
30.24 % |
0.9450 |
|
MexT |
847 |
719 |
84.89 % |
0.9687 |
|
RhlR |
850 |
104 |
12.24 % |
0.9925 |
|
PqsA |
826 |
302 |
36.56 % |
0.9945 |
|
LasI |
832 |
33 |
3.97 % |
0.9949 |
|
PqsE |
844 |
101 |
11.97 % |
0.9951 |
|
PqsD |
847 |
240 |
28.34 % |
0.9953 |
|
PqsL |
850 |
447 |
52.59 % |
0.9953 |
|
VqsR |
849 |
256 |
30.15 % |
0.9955 |
|
QscR |
848 |
101 |
11.91 % |
0.9967 |
|
PqsR |
852 |
200 |
23.47 % |
0.9970 |
|
RhlI |
850 |
198 |
23.29 % |
0.9981 |
|
PqsB |
847 |
260 |
30.70 % |
0.9984 |
|
PqsH |
848 |
336 |
39.62 % |
0.9986 |
|
PsdR |
844 |
38 |
4.50 % |
0.9990 |
|
PqsC |
847 |
164 |
19.36 % |
0.9993 |
|
RpsL |
852 |
21 |
2.46 % |
0.9998 |
|
RsaL |
830 |
6 |
0.72 % |
0.9999 |
Using GAMMA generated protein mutation data, we next calculated the average codon similarity of all isolates to a reference (Table 2). High codon similarity suggests similar protein function, while low codon similarity indicates a change or loss of protein function. For the majority of genes we used PAO1 (GCF_000006765.1) as a reference, however the codon similarity average for several proteins were low. To address this, for RhlI and PqsD we used PAK as the reference strain, and for MexT we used U0330A as these strains were likely to be better models based on higher similarity averages for each protein. Similar to our analysis using BLOSUM80, we found that LasR had the lowest average protein similarity of all QS genes, followed by MexT, which we attributed to the high rate of truncation mutations. All other QS proteins had a codon similarity average of greater than 0.99, indicating that while there is variation in the protein sequence, many of these mutations likely lead to a similarly functioning protein.
LasR protein mutations are common across all environments but are more divergent in human wounds
In agreement with previous studies, LasR was the most variable gene in our protein analysis (Table 1; Fig. 2), and we found that the highly dissimilar LasR sequences were due to protein truncations [2, 30, 51]. To determine if LasR mutations vary by environment, we categorized the strains by source. Using data from the IPCD, we selected a subset of 654 strains labelled as ‘environmental’, ‘cystic fibrosis’ or ‘CF’, and ‘wound’ or ‘ulcer’ or ‘burn’ and reclassified them as environmental (209 strains), CF (396 strains) or wound (wound, ulcer, and burn) (49 strains); 654 total. The remaining 198 strains from the original set of 852 strains were of uncertain origin and therefore excluded from this analysis.
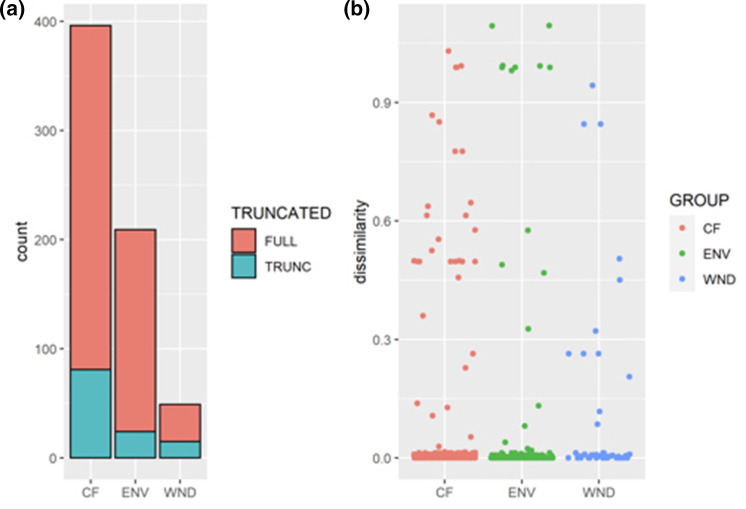
To establish a threshold by which a protein could be deemed functional or not, we looked at truncated LasR proteins within each environment. We compared the amino acid length of LasR in the IPCD strains to the PAO1 LasR protein – which is equal in length to many commonly researched strains including PA14, PAK and the Liverpool epidemic CF strain LESB58. Our assumption was that a truncated protein due to an early stop site, would lead to a nonfunctional protein. We used a stringent 100 % length as a cut-off, and any protein shorter than full-length was considered truncated. Fig. 3(a) shows the proportion of each group that had truncated LasR proteins with CF, environmental and wound isolates having 20, 11 and 30% truncations, respectively. We then plotted the protein sequence dissimilarities categorized by isolate source, and found highly dissimilar isolates, primarily in CF and wounds (Fig. 3b). Overall, we found that lasR mutations are ubiquitous across all environments, but there is a larger percentage of strains with truncated LasR proteins found in infection environments. We additionally calculated the mean and median codon similarity scores for each environment, with one being a protein identical to PAO1. We found that the wound group was the most dissimilar compared to PAO1, with the lowest mean similarity score (0.893), and CF and environmental were similar (CF: 0.953, Env: 0.948). The wound group also had the lowest median similarity score (0.995), followed by CF (0.996) and environmental had the highest median similarity score (1.0).
Fig. 3.
LasR truncations are found in isolates from all sources. To observe the fraction of truncated proteins across all environments, we categorized the isolates into three groups: cystic fibrosis (CF), environmental (ENV) or wound (WND). (a) We show the number of truncated proteins out of the total number of isolates in each group. The protein variation plot (b) depicts the similarity scores for all groups compared, and we see the highly dissimilar truncated proteins consisted primarily of CF and WND isolates.
lasR mutation is not a predictor of exoprotease production or colony autolysis in selected environmental and infection isolates
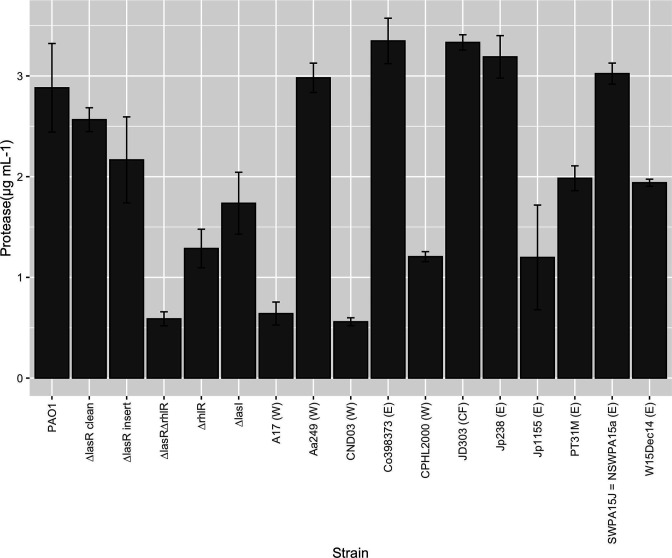
To accompany our genomic analysis, we selected 20 isolates from the IPCD, in order to perform phenotypic assays to assess QS function. We had over 100 of these isolates available from a collection and we chose a selection of isolates to represent wild-type, truncated and mutated LasR proteins from environmental, wound and CF isolates (Table S1, available in the online version of this article). After Sanger sequencing the lasR gene to confirm isolate identification, we found that the eight isolates with ‘truncated’ nucleotide sequences had incorrect sequences (data not shown), likely because the sequence was at the end of a contig rather than a true truncation. We therefore focused on 12 out of 20 isolates with the correct sequence identity. We included our lab strain NPAO1 as well as the clean deletion mutants PAO1∆lasR, PAO1∆rhlR, PAO1∆lasR∆rhlR and PAO1∆lasI. Our PAO1∆lasR strain made high levels of protease that were not significantly lower than NPAO1 (89 % of NPAO1 levels, P=0.338). A lasR gentamicin insertion mutant made 75 % of the protease levels of NPAO1 (P=0.113) (Fig. 4). We found that unlike mutations in LasR, mutations in RhlR had a significant effect on the protease levels produced compared to NPAO1 (PAO1∆rhlR: 45 %, P=0.013; PAO1∆lasR∆rhlR double mutant: 20 %, P=0.011). Compared with both ∆lasR mutants, NPAO1∆lasI made lower amounts of protease (60 %, P=0.025) (Fig. 4). Isolates with mutated LasR proteins that produce significantly less protease than wild-type included A17, CND03, CPHL2000, Jp115 (P values, respectively: 0.01, 0.01, 0.02, 0.01). PT31M, a German environmental isolate contains a truncation mutation in LasR, yet it produced an intermediate amount of protease compared to NPAO1 with an intact LasR (69 %, P=0.062). The Belgian river environmental strain W15Dec14 also produced an intermediate amount of protease (67 %, P=0.065) and is wild-type for LasR, but does contain coding mutations in RhlI. All other isolates with wild-type LasR proteins produced protease levels that were not significantly different from NPAO1 (Fig. 4).
Fig. 4.
Exoprotease secretion by environmental and clinical isolates. We measured the supernatant exoprotease levels produced by stationary phase isolates, compared to PAO1, PAO1∆lasR and PAO1∆lasR∆rhlR. We normalized the readings by optical density (OD 600nm) and compared all isolates and clean mutants to PAO1. * denotes a strain is significantly different from NPAO1, P<0.05. Strain origin abbreviated are listed after strain names as follows: environmental (E), wound, burn or ulcer (W), cystic fibrosis (CF).
Lysis in QS mutants has long been observed in the lab, specifically in lasR, lasI and/or pqs mutants [51–54]. The lysis phenotype is characterized by an iridescent sheen on the colony surface, and an uneven lysed texture; some pqs mutants alternatively show plaquing similar to phage activity. The iridescent sheen typically seen in lasR mutants has previously been attributed to HHQ accumulation [51]. We used QS deletion mutant strains to determine how QS mediates colony autolysis. PAO1∆lasR and PAO1∆pqsL showed similar lysis phenotypes and a metallic sheen, while PAO1∆pqsH caused colony plaquing (Fig. 5). PAO1∆pqsA∆lasR, PAO1∆pqsA∆pqsH and PAO1∆pqsA∆pqsL mutations led to a loss of colony autolysis (Fig. 5). We found that all LasR wild-type strains we tested from the IPCD showing lysis or plaquing, contained multiple mutations in the pqs quinolone system, as follows: SWPA15J=NSWPA15 a (PqsC,D,E), 5BR2 (PqsD,L), Co398373 (PqsD,L), Jp238 (PqsB,D,L), W15D14 (PqsE,D,L), Aa249 (PqsB,D,L), JD303 (PqsA,B,D). Strains with wild-type LasR showing lysis were from mixed environmental, wound and CF sources. There were two lasR mutant isolates that showed no lysis – A17 and CPHL2000, both sourced from human wounds (Fig. 5). Isolates A17 and CPHL2000 have one and two SNPs in LasR, respectively, both creating a change in amino acid sequence (A17: L110P, CPHL2000: E196G). As expected, the three isolates with truncated LasR (CND03, Jp1155, and PT31M) demonstrated lysis.
Fig. 5.
Colony autolysis phenotypes of isolates compared to lab strain QS mutants. We spotted overnight cultures on LBA plates to observe the colony autolysis phenotypes exhibited by clean QS deletion mutants alongside environmental and clinical isolates. Colony autolysis and colony plaquing were quantified by eye, by looking for the characteristic iridescent sheen or uneven colony texture normally associated with phage activity. Strain SWPA15J=NSWPA15 a is labelled as SWPA15J. Strain origin abbreviated are listed after strain names as follows: environmental (E),wound, burn or ulcer (W),cystic fibrosis (CF).
Discussion
In P. aeruginosa , lasR QS mutants are frequently isolated from human chronic infection and environmental sources, but less is known about other QS genes [2, 51, 55]. Using a publicly available database of 852 fully sequenced isolates from CF, wounds and non-human associated (environmental) strains [37], we determined the mutation frequency and variation of lasR and other QS genes in P. aeruginosa . To determine how QS genotype impacts phenotypic function, we used 12 strains that were sequenced as part of the IPCD study [37]. We found that (i) by multiple metrics, LasR is the most variable QS protein; (ii) LasR mutations are found in isolates across all environments, suggesting that any environment can drive the evolution of these mutations; (iii) the negative las system regulator RsaL is well-conserved; (iv) signal synthases LasI and RhlI are conserved compared to their transcriptional regulator pairs; (v) coding mutations in pqs system genes are common and may impact colony autolysis, however they are less divergent than LasR mutations; (vi) MexT is highly variable and mutations are common.
Our work supports the long-held belief that LasR is a commonly mutated QS protein as we showed that LasR has the highest number of unique protein sequences compared to other QS genes. A high number of isolates carried mutations in LasR compared to the PAO1 reference protein sequence. The IPCD strains also had the lowest average codon similarity for the LasR gene when compared to other QS regulon genes, which we attributed to nonsense mutations leading to protein truncation, and other large changes in amino acid sequence. Our results also highlight that LasR mutations are found in environmental isolates and are not specific to human infections. Truncation mutations are more commonly found in human wound infections, suggesting that there might be some selective pressure to evolve loss of function mutations. Mean and median similarity scores suggest there is not a large difference between CF and environmental isolates, and instead we found that wound isolates are enriched for loss of function mutations. The wound group was the smallest group of isolates we queried thus a larger exploration of wound strains would provide a deeper understanding of this phenomenon.
Interestingly, the most highly conserved QS protein was RsaL, which negatively regulates the las QS system [16, 56, 57]. RsaL and LasR are antipodal in variation across isolates and perform opposite jobs in regulating the las system. There may be a benefit to tightly conserving the ‘off’ switch of the las system while frequently mutating and losing the function of the ‘on’ switch, suggesting that a key element of the QS regulatory cascade is down-regulating las-controlled QS in P. aeruginosa . Given this complexity, it remains a challenge to understand why the las system evolved and what fitness benefits it provides P. aeruginosa in different environments. There may be evolutionary benefits for both the maintenance and loss of LasR so that both lasR positive and negative strains can stably coexist in heterogenous populations and contribute to an overall community function. In support of this idea, it has been shown that (i) lasR- strains overproduce Rhl-associated factors and cross-feed wild-type cells in low iron environments, which will likely impact infection dynamics of mixed populations [58]; (ii) mixed lasR +/- populations display decreased virulence in mouse models of infection [59]; and (iii) mixed populations exhibit enhanced tolerance to beta-lactam antibiotics [60]. These data suggest there are likely considerable fitness advantages to cells growing in heterogeneous QS populations, perhaps as a bet-hedging mechanism for future disturbance events.
The signal synthases of the two AHL QS systems in P. aeruginosa are conserved compared to their transcriptional regulator pairs. It has been hypothesized that there could be evolutionary mechanisms to conserve signal synthases while mutating transcriptional regulators [61]. We saw a significant difference in protease production by the clean deletion of the regulator mutant LasR and the signal synthase mutant LasI, where deletion of LasI resulted in a larger decrease in protease production. The reason for this could be further explored and may be impacted by the MexT mutation in NPAO1, common to other PAO1 lab strains, or the fact that the 3′ end of RsaL is also deleted in a clean LasR deletion mutation [62]. There could also be an evolutionary benefit to maintaining some protease function in the absence of fully functioning QS, supporting the maintenance of signal synthases while mutating positive transcriptional regulators.
Recent studies on QS in P. aeruginosa has revealed that the complex and intertwined las, rhl and pqs systems can be re-wired if lasR becomes mutated [25, 27, 31, 34–36]. It is not always clear whether these strains are entirely QS-null or if they have re-wired their QS systems to circumvent the loss of lasR. In our in vitro work, we looked at lysis and protease secretion as indicators of QS function, which have previously been used to screen for LasR function [51, 63]. We found an environmental isolate, PT31M, with a truncated lasR that produces high levels of protease. PT31M may be an example of a re-wired isolate showing QS independent of LasR, suggesting this adaptation is not specific to CF lung infections. Colony lysis has been used as a predictor for las system function, however this study has shown little correlation between lysis and lasR genotype, potentially due to pqs mutation mediated lysis seen in NPAO1∆pqsH and NPAO1∆pqsL.
Overall, our work shows that LasR is uniquely mutated compared to other QS genes; future work should more strongly focus on the ecology of mixed QS-phenotypes to better understand QS-involvement in infection and other environments. With ongoing work identifying QS-inhibitors targeting the las QS system, the frequency of lasR mutated strains found in our study suggests that this particular pursuit might be improved by targeting a less variable QS gene, such as rhl system genes or RsaL.
Supplementary Data
Funding information
We wish to thank The Cystic Fibrosis Foundation (DIGGLE20G0) and The National Institute for Health (R01AI153116) to SPD for funding.
Acknowledgement
We also thank members of the Diggle Lab and Jon Gerhart for helpful discussion. We thank the Brown Lab at Georgia Tech for donating strains from the IPCD collection.
Author contribution
S.P.D. and K.O. designed the study. K.O., C.Y.Z. and M.M. performed the in silico analysis of the data. K.O. performed the in vitro analysis. All authors contributed to the writing of the manuscript.
Conflicts of interest
The authors declare no competing interests.
Footnotes
A supplementary table is available with the online version of this article.
Abbreviations: AHL, N-acyl homoserine lactone; AQ, alkyl quinolone; CF, cystic fibrosis; C4-HSL, N-butanoyl-L-homoserine lactone; ENV, environmental; IPCD, International Pseudomonas Consortium Database; 3OC12-HSL, 3-oxo-dodecanoyl-L-homoserine lactone; PQS, Pseudomonas quinolone signal; QS, quorum sensing; WND, wound.
References
- 1.Diggle SP, Whiteley M. Microbe Profile: Pseudomonas aeruginosa: opportunistic pathogen and lab rat. Microbiology. 2021;167 doi: 10.1099/mic.0.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaFayette SL, Houle D, Beaudoin T, Wojewodka G, Radzioch D, et al. Cystic fibrosis–adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci Adv. 2015;1:1–15. doi: 10.1126/sciadv.1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen SK, Rau MH, Johansen HK, Ciofu O, Jelsbak L, et al. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J. 2012;6:31–45. doi: 10.1038/ismej.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology. 2010;156:1108–1119. doi: 10.1099/mic.0.033993-0. [DOI] [PubMed] [Google Scholar]
- 6.Williams P, Cámara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Galán-Vásquez E, Luna B, Martínez-Antonio A. The regulatory network of Pseudomonas aeruginosa . Microb Inform Exp. 2011;1:3. doi: 10.1186/2042-5783-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa . Protein Cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azimi S, Klementiev AD, Whiteley M, Diggle SP. Bacterial quorum sensing during infection. Annu Rev Microbiol. 2020;74:201–219. doi: 10.1146/annurev-micro-032020-093845. [DOI] [PubMed] [Google Scholar]
- 11.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding F, Oinuma K-I, Smalley NE, Schaefer AL, Hamwy O, et al. The Pseudomonas aeruginosa orphan quorum sensing signal receptor QscR regulates global quorum sensing gene expression by activating a single linked operon. mBio. 2018;9:e01274-18. doi: 10.1128/mBio.01274-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa . Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 16.Rampioni G, Bertani I, Zennaro E, Polticelli F, Venturi V, et al. The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter. J Bacteriol. 2006;188:815–819. doi: 10.1128/JB.188.2.815-819.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rampioni G, Schuster M, Greenberg EP, Zennaro E, Leoni L. Contribution of the RsaL global regulator to Pseudomonas aeruginosa virulence and biofilm formation. FEMS Microbiol Lett. 2009;301:210–217. doi: 10.1111/j.1574-6968.2009.01817.x. [DOI] [PubMed] [Google Scholar]
- 18.Liang H, Deng X, Ji Q, Sun F, Shen T, et al. The Pseudomonas aeruginosa global regulator VqsR directly inhibits QscR to control quorum-sensing and virulence gene expression. J Bacteriol. 2012;194:3098–3108. doi: 10.1128/JB.06679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juhas M, Wiehlmann L, Huber B, Jordan D, Lauber J, et al. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa . Microbiology. 2004;150:831–841. doi: 10.1099/mic.0.26906-0. [DOI] [PubMed] [Google Scholar]
- 20.Gurney J, Azimi S, Brown SP, Diggle SP. Combinatorial quorum sensing in Pseudomonas aeruginosa allows for novel cheating strategies. Microbiology. 2020;166:777–784. doi: 10.1099/mic.0.000941. [DOI] [PubMed] [Google Scholar]
- 21.Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, et al. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci. 2014;111:4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubern JF, Diggle SP. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol Biosyst. 2008;4:882–888. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 23.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, et al. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostylev M, Kim DY, Smalley NE, Salukhe I, Greenberg EP, et al. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci. 2019;116:7027–7032. doi: 10.1073/pnas.1819796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feltner JB, Wolter DJ, Pope CE, Groleau M, Smalley NE, et al. LasR variant cystic fibrosis isolates reveal an adaptable quorum. mBio. 2016;7:e01513–16. doi: 10.1128/mBio.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz RL, Asfahl KL, Van den Bossche S, Coenye T, Crabbé A, et al. RhlR-regulated acyl-homoserine lactone quorum sensing in a cystic fibrosis isolate of Pseudomonas aeruginosa . mBio. 2020;11:e00532-20. doi: 10.1128/mBio.00532-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomason MK, Voichek M, Dar D, Addis V, Fitzgerald D, et al. A rhlI 5’ UTR-derived sRNA regulates RhlR-dependent quorum sensing in Pseudomonas aeruginosa . mBio. 2019;10:e02253-19. doi: 10.1128/mBio.02253-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R, Déziel E, Groleau MC, Schaefer AL, Greenberg EP. Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc Natl Acad Sci. 2019;116:7021–7026. doi: 10.1073/pnas.1819801116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groleau M-C, Taillefer H, Vincent AT, Constant P, Déziel E. Pseudomonas aeruginosa isolates defective in function of the LasR quorum sensing regulator are frequent in diverse environmental niches. Environ Microbiol. 2021;00:1–14. doi: 10.1101/2021.03.25.437011. [DOI] [PubMed] [Google Scholar]
- 31.Oshri RD, Zrihen KS, Shner I, Omer Bendori S, Eldar A. Selection for increased quorum-sensing cooperation in Pseudomonas aeruginosa through the shut-down of a drug resistance pump. ISME J. 2018;12:2458–2469. doi: 10.1038/s41396-018-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schweizer HP. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet Mol Res. 2003;2:48–62. [PubMed] [Google Scholar]
- 33.Köhler T, Epp SF, Curty LK, Pechère JC. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa . J Bacteriol. 1999;181:6300–6305. doi: 10.1128/JB.181.20.6300-6305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee S, Moustafa DA, Stergioula V, Smith CD, Goldberg JB, et al. The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa . Proc Natl Acad Sci. 2018;115:E9411–E9418. doi: 10.1073/pnas.1814023115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrow JM, 3rd, Sund ZM, Ellison ML, Wade DS, Coleman JP, et al. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-Reyes S, Cocotl-Yañez M, Soto-Aceves MP, González-Valdez A, Servín-González L, et al. PqsR-independent quorum-sensing response of Pseudomonas aeruginosa ATCC 9027 outlier-strain reveals new insights on the PqsE effect on RhlR activity. Mol Microbiol. 2021;116:1113–1123. doi: 10.1111/mmi.14797. [DOI] [PubMed] [Google Scholar]
- 37.Freschi L, Vincent AT, Jeukens J, Emond-Rheault J-G, Kukavica-Ibrulj I, et al. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol. 2019;11:109–120. doi: 10.1093/gbe/evy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–53. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:1–7. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pagès H, Aboyoun P, Gentleman RDS. Biostrings: efficient manipulation of biological strings n.d.
- 42.Stanton RA, Vlachos N, Halpin AL, Birol I. GAMMA: a tool for the rapid identification, classification and annotation of translated gene matches from sequencing data. Bioinformatics. 2022;38:546–548. doi: 10.1093/bioinformatics/btab607. [DOI] [PubMed] [Google Scholar]
- 43.Asfahl KL, Walsh J, Gilbert K, Schuster M. Non-social adaptation defers a tragedy of the commons in Pseudomonas aeruginosa quorum sensing. ISME J. 2015;9:1734–1746. doi: 10.1038/ismej.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smalley NE, Schaefer AL, Asfahl KL, Perez C, Greenberg EP, et al. Evolution of the quorum sensing regulon in cooperating populations of Pseudomonas aeruginosa. mBio. 2022;13:1–19. doi: 10.1128/mbio.00161-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Team Rs RStudio: integrated development environment for R n.d.
- 47.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, et al. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deretic V, Martin DW, Schurr MJ, Mudd MH, Hibler NS, et al. Conversion to mucoidy in Pseudomonas aeruginosa . Biotechnology. 1993;11:1133–1136. doi: 10.1038/nbt1093-1133. [DOI] [PubMed] [Google Scholar]
- 49.Weir TL, Stull VJ, Badri D, Trunck LA, Schweizer HP, et al. Global gene expression profiles suggest an important role for nutrient acquisition in early pathogenesis in a plant model of Pseudomonas aeruginosa infection. Appl Environ Microbiol. 2008;74:5784–5791. doi: 10.1128/AEM.00860-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dumas J-L, van Delden C, Perron K, Köhler T. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett. 2006;254:217–225. doi: 10.1111/j.1574-6968.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 51.D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hazan R, Que YA, Maura D, Strobel B, Majcherczyk PA, et al. Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr Biol. 2016;26:195–206. doi: 10.1016/j.cub.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Groleau M-C, Taillefer H, Vincent AT, Constant P, Déziel E. Pseudomonas aeruginosa isolates defective in function of the LasR quorum sensing regulator are frequent in diverse environmental niches. Microbiology. doi: 10.1101/2021.03.25.437011. [DOI] [PubMed]
- 56.de Kievit T, Seed PC, Nezezon J, Passador L, Iglewski BH. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa . J Bacteriol. 1999;181:2175–2184. doi: 10.1128/JB.181.7.2175-2184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, et al. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa . Mol Microbiol. 2007;66:1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- 58.Mould DL, Botelho NJ, Hogan DA. Intraspecies signaling between common variants of Pseudomonas aeruginosa increases production of quorum-sensing-controlled virulence factors. mBio. 2020;11:1–16. doi: 10.1128/mBio.01865-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, et al. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 60.Azimi S, Roberts AEL, Peng S, Weitz JS, McNally A, et al. Allelic polymorphism shapes community function in evolving Pseudomonas aeruginosa populations. ISME J. 2020;14:1929–1942. doi: 10.1038/s41396-020-0652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eldar A. Social conflict drives the evolutionary divergence of quorum sensing. Proc Natl Acad Sci. 2011;108:13635–13640. doi: 10.1073/pnas.1102923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.LoVullo ED, Schweizer HP. Pseudomonas aeruginosa mexT is an indicator of PAO1 strain integrity. J Med Microbiol. 2020;69:139–145. doi: 10.1099/jmm.0.001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.