Abstract
EWS is a member of the FET family of RNA/DNA binding proteins that regulate crucial phases of nucleic acid metabolism. EWS comprises an N-terminal low-complexity domain (LCD) and a C-terminal RNA-binding domain (RBD). The RBD is further divided into three RG-rich regions which flank an RNA-recognition motif (RRM) and a zinc finger (ZnF) domain. Recently, EWS was shown to regulate R-loops in Ewing sarcoma, a pediatric bone and soft-tissue cancer in which a chromosomal translocation fuses the N-terminal LCD of EWS to the C-terminal DNA binding domain of the transcription factor FLI1. Though EWS was shown to directly bind R-loops, the binding mechanism was not elucidated. In the current study, the RBD of EWS was divided into several constructs, which were subsequently assayed for binding to various nucleic acid structures expected to form at R-loops, including RNA stem-loops, DNA G-quadruplexes, and RNA:DNA hybrids. EWS interacted with all three nucleic acid structures with varying affinities and multiple domains contributed to binding each substrate. The RRM and RG2 region appear to bind nucleic acids promiscuously while the ZnF displayed more selectivity for single-stranded structures. With these results, the structural underpinnings of EWS recognition and binding of R-loops and other nucleic acid structures is better understood.
Keywords: EWS, intrinsically disordered protein, R-loop, G-quadruplex, NMR
Introduction
Heterogenous nuclear ribonucleoproteins (hnRNPs) are a diverse class of RNA and DNA binding proteins that play major roles in all aspects of RNA production and processing. The hnRNP family of proteins has gained considerable interest in disease research in recent years owing to the fact that the expression levels of hnRNPs are frequently altered in cancer 1-3 and that mutations in hnRNPs are linked to neurodegenerative diseases 4,5. The hnRNPs share several structural domains such as RNA-recognition motifs (RRM), KH homology domains, Arg-Gly-Gly (RGG) boxes, zinc finger (ZnF) domains as well as intrinsically disordered regions with specific amino acid compositions. However, the arrangement of these domains varies significantly, and considerable functional diversity exists across the hnRNP family. hnRNPs are therefore further classified into 16 subgroups with similar domain arrangements and functions. The hnRNP subgroup P is also referred to as the FET family of proteins after its three members: fused-in-sarcoma, RNA binding protein EWS, and TATA-binding protein associated factor 2N (TAF15).
The exact cellular functions of the FET protein family are not fully understood however they are known to be involved in transcriptional regulation 6,7, RNA processing, transport and alternative splicing 8-10, the DNA damage response and homologous recombination 11,12, regulation of R-loops 13, and telomere length 14-16. These functions have been proposed based primarily on protein interaction studies, which have identified FET protein associations with the transcription initiation complex 7, splicing factors 17-19, other hnRNPs 20 as well as RNA and DNA 14,15,21-23. Discerning the functions of individual members of the FET protein family is difficult in part because all three FET proteins are colocalized intracellularly and are therefore thought to have overlapping functions 20. FET proteins are characterised by an N-terminal low-complexity domain (LCD) enriched with the residues SYGQP, three RG-rich regions, an RRM, a ZnF domain and a nuclear localization sequence (NLS). The RG-rich regions, RRM and ZnF domains are collectively called the RNA-binding domain (RBD) of EWS. While the RRM and ZnF domains are highly conserved, the disordered LCD and RG-rich regions differ in length and sequence composition across the three family members. EWS was discovered as the first FET family member when it was identified as the N-terminal constituent of the EWS-FLI1 fusion oncoprotein that is causative for up to 85% of all Ewing sarcoma (EwS) cases 24. The EWS-FLI1 fusion protein retains the N-terminal LCD of EWS and a C-terminal DNA binding domain from the E Twenty-six (ETS) transcription factor Friend leukemia integration 1 (FLI1). The remaining ~ 15% of EwS cases are caused by related chromosomal translocations in which EWS, FUS or TAF15 are fused with DNA binding domains of transcription factors 25,26, leading to oncogenic transcriptional changes 24,27-29.
Although activation of oncogenic transcriptional pathways is a driver of EwS, in recent years EWS-FLI1 has been shown to support EwS development via a DNA-binding independent mechanism, which has led to the hypothesis that EWS-FLI1 exerts a dominant-negative effect on the normal functions of EWS 13,30-32. In particular, EWS-FLI1 was found to inhibit the ability of EWS to regulate phosphorylation of DNA-directed RNA polymerase II subunit RPB1 (RNA Pol II) by cyclin-dependent kinases (CDK) 7/9, inhibiting breast cancer type 1 susceptibility protein (BRCA1)-mediated DNA repair and causing R-loop accumulation at the transcription bubble 13. Subsequently, EWS was shown to directly interact with R-loops independent of additional binding partners 33. R-loops are three-stranded nucleic acid structures that form under a variety of circumstances including during transcription, telomere lengthening, and at sites of DNA damage. R-loops are stabilized by a variety of factors including C-rich content of the template strand and G-quadruplex (G4) formation on the non-template strand. Consequently, R-loop formation is associated with G4 formation. Further supporting a possible functional role of EWS at R-loops, EWS and FUS have both been demonstrated to directly associate with G4 DNA and RNA structures at telomeres 14,15.
Though the association of FUS and TAF15 with RNA stem-loop structures has been studied at the molecular level, the binding of EWS to stem-loops and other types of nucleic acid structures is relatively unknown. To address this knowledge gap, the nucleic acid binding properties of the RBD of EWS were investigated. Constructs encoding the RRM, RG2, RRM-RG2 and RRM-RG2-ZnF of EWS were recombinantly expressed and purified and assayed for binding to a variety of nucleic acid structures expected to form at R-loops, including RNA stem-loops, DNA G4s and RNA:DNA hybrids. Gel-shift assays revealed that the RRM-only and RG2-only constructs interacted weakly with all nucleic acid structures tested. Binding to RNA-stem-loops was promoted by all three domains (RRM, RG2, and the ZnF). The RRM-only and RG2-only constructs interacted weakly with the DNA G4s, however the RRM-RG2 and RRM-RG2-ZnF constructs bound DNA G4s with low micromolar affinity indicating the weak interactions of the isolated domains act synergistically to bind DNA G4s. Isothermal titration calorimetry (ITC) revealed low micromolar affinity for the binding of EWS RBD to DNA G4. Likewise, weak binding was observed for all constructs to RNA:DNA hybrids, with the RRM-RG2 and RRM-RG2-ZnF similarly having higher affinities. Nuclear magnetic resonance (NMR) spectroscopy demonstrated that the EWS RBD employs a concave surface on the RRM to bind DNA G4s and RNA:DNA hybrids and that residues in RG2 synergize with the RRM to increase affinity for these nucleic acid structures. These results have begun to elucidate the binding preferences of EWS for model R-loops components.
Materials and Methods
Recombinant protein expression and purification
Gene constructs were optimized for expression in E. coli, synthesized (GenScript) and cloned into pET expression vectors with an N-terminal 8 x His-tag followed by a tobacco etch virus (TEV) protease cleavage site. For the RG2 construct, a maltose-binding protein (MBP) tag was included between the His-tag and the TEV cleavage site. Plasmids were transformed into chemically competent E. coli BL21 Star™ (DE3) (Invitrogen, MA) cells using the heat-shock method and plated on LB agar supplemented with ampicillin (100 μg/mL). To produce proteins with natural abundance isotopes, one colony of the resulting plate was used to inoculate a 100 mL LB starter culture, which was grown at 37 °C overnight with shaking at 225 rpm. The overnight culture (10 – 20 mL) was used to inoculate 1 L of LB, which was then grown in baffled Fernbach flasks at 37 °C with shaking at 225 rpm until OD600 reached ~ 0.6-0.8. At this point, protein expression was induced with 0.5 mM IPTG. For the expression of EWSRRM-RG2-ZnF, ZnCl2 was also added to a final concentration of 0.1 mM at the same time as IPTG was added. Protein expression was then continued for 3 hours at 37 °C (EWSRRM, EWSRG2, and EWSRRM-RG2) or overnight at 22°C (EWSRRM-RG2-ZnF) and cells were then harvested by centrifugation at 4000 x g for 30 minutes at 4 °C. Cell pellets were resuspended in 50 mM Tris pH 8, 1 M NaCl, 20 mM imidazole, 2 mM DTT (only for RRM constructs) with half of one Pierce mini protease inhibitor tablet (Thermo Fisher) and frozen at −20 °C until purification. For EWSRRM-RG2-ZnF, 0.5 mM ZnCl2 was included the resuspension buffer and at all subsequent steps during purification. To produce proteins with isotopic enrichment, one colony of the LB agar plate was used to inoculate a 2 mL LB starter culture that was grown at 37 °C with 225 rpm shaking for ~ 5-6 hours, 1 mL of this culture was used to inoculate a 100 mL M9 starter culture which was grown overnight at 37 °C with shaking at 225 rpm. M9 media was supplemented with 15NH4Cl (1g/L) and 0.02% (w/v) yeast extract for 15N labelling or 15NH4Cl and 13C6 D-glucose (3 g/L) with 0.02% (w/v) Isogro®-13C, 15N (Sigma, MO) for 15N, 13C labelling. The overnight culture (10 – 20 mL) was used to inoculate 1 L M9 culture, and cell growth and protein expression were carried out as described above.
For protein purification, cell resuspensions were first thawed and then sonicated on ice for a total processing time of 3 minutes as 36 cycles of 5 second on pulses followed by 55 second off pulses using a 550 Sonic Dismembrator (Fisher Scientific). The suspension was clarified by centrifugation at 45,000 x g for 30 min at 4 °C. The supernatant was applied to a 5 mL HisTrap HP column (Cytiva, MA) equilibrated with 50 mM Tris pH 8, 1 M NaCl, 20 mM imidazole, 2 mM DTT. The column was washed with 50 mL 20 mM imidazole then with 30 mL of 40 mM imidazole and finally with 30 mL of 50 mM imidazole, all prepared in the above buffer and the target proteins were eluted with 500 mM imidazole prepared in the above buffer. TEV protease was added at ratios between 1:15 and 1:50 (TEV:target protein) and the mixture was dialyzed against 20 mM Tris pH 8, 50 or 100 mM NaCl, 2 mM DTT, 0.5 mM EDTA at room temperature (EWSRRM, EWSRG2, and EWSRRM-RG2) or at 4 °C (EWSRRM-RG2-ZnF) overnight. EDTA was excluded from the dialysis buffer for the EWSRRM-RG2-ZnF and ZnCl2 was included at 0.5 mM. Subsequently, aggregates were removed via centrifugation, NaCl was added to a final concentration of 1 M and imidazole was added to a concentration of 20 mM and the sample was reapplied to the HisTrap column. The flow-through, containing the cleaved target protein was collected, concentrated using Amicon centrifugal concentrators (Millipore, MA) to approximately 2.5 mL and applied to a HiLoad 16/600 Superdex 75 pg column (Cytiva) equilibrated with either 20 mM Tris, pH 7, 50 mM NaCl, 2 mM TCEP, 0.5 mM EDTA, 0.2 mM PMSF (EWSRRM and EWSRRM-RG2), 20 mM potassium phosphate pH 6, 150 mM potassium chloride, 0.2 mM PMSF, 0.5 mM EDTA (EWSRG2), or 20 mM potassium phosphate pH 6, 150 mM potassium chloride, 2 mM TCEP, 0.2 mM PMSF, 0.5 mM ZnCl2 (EWSRRM-RG2-ZnF). Fractions containing the target protein were pooled and concentrated to between 200 μM and 1.2 mM and stored as aliquots at −80°C until use.
Preparation of nucleic acid substrates
Oligonucleotides used in this study are listed in Table 1 and were purchased from IDT. Stem-loop RNA was prepared by resuspending the lyophilized RNA in nuclease-free water to concentrations of 100 μM – 1 mM, heating at 80 °C for 2 minutes then immediately cooling on ice. A DNA G-quadruplex sequence derived from the BLM gene promoter 34, pu20m2, was prepared by resuspending the lyophilized DNA in buffers containing 20 mM potassium phosphate pH 7.5 followed by heating at 95 °C for 5 minutes and then cooling to room temperature. Addition of potassium, which stabilizes G4 structures, before heating and cooling promotes the formation of pu20m2 G4 dimer34. Addition of potassium after cooling promotes G4 formation but minimizes the formation of dimers therefore, KCl was added to a final concentration of 50 mM after cooling to room temperature (Supplementary Fig. 1). Circular dichroism spectroscopy was used to confirm folding of the DNA G4 (Supplementary Fig. 1A). The RNA:DNA hybrids were prepared by first resuspending the component strands in nuclease free water to a concentration of 100 μM – 1 mM. The strands were then mixed together at equimolar concentrations and the mixture was heated to 95 °C for 5 minutes and then cooled to 70 °C over a period of one hour. The mixture was then cooled to room temperature by lowering the temperature 5 °C every 5 minutes. The oligonucleotide substrates were assessed for homogeneity following the annealing/folding protocols using 20% polyacrylamide gels prepared in 0.5 x TBE buffer.
Table 1.
Sequences of nucleic acid oligonucleotide constructs.
Electrophoretic mobility shift assays
For EMSAs performed with the SON-GGU stem-loop and the RNA:DNA hybrid, freshly annealed oligonucleotides (2.5 or 5 μM) were prepared in 20 μL samples containing 8 nM – 80 μM of the appropriate protein construct in 10 mM Tris pH 7.4, 20 mM KCl, 100 mM NaCl, 1 mM MgCl2, 15% glycerol, 1 mM BME. For EMSAs performed with the pu20m2 DNA G4, freshly folded DNA (see above) was diluted to 10 μM in 20 mM Tris pH 7.5, 50 mM KCl. Subsequently, 20 μL samples were prepared containing 8 nM – 80 μM of the appropriate protein construct in 20 mM Tris pH 6, 150 mM KCl, 2 mM TCEP and 15% glycerol and 2.5 μM of folded DNA. The samples were incubated on ice for 1 hour then analyzed using a 6.5% polyacrylamide gel prepared in 1 X TBE (with 0.5 mM ZnCl2 for EWSRRM-RG2-ZnF construct) at 200 V for 60 minutes or 100 V for 2 hours on ice. Gels were stained with 0.4 x SYBR™ Green II RNA stain or SYBR™ Safe DNA gel stain for 15 – 30 minutes in 1 x TBE and imaged using UV transillumination.
Isothermal titration calorimetry
ITC experiments were performed using an Affinity ITC (TA Instruments). Prior to each experiment, EWSRRM, EWSRG2, EWSRRM-RG2 or EWSRRM-RG2-ZnF were dialyzed overnight at room temperature into 20 mM potassium phosphate pH 6, 150 mM potassium chloride, 2 mM TCEP (and with 0.5 mM ZnCl2 for experiments using EWSRRM-RG2-ZnF). After dialysis the protein sample was centrifuged to remove aggregates and the protein concentration was determined by UV spectroscopy. The buffer used for dialysis was filtered and degassed. The protein was then diluted to 50 μM using the filtered dialysis buffer and 350 μL was loaded into the sample cell. For ITC experiments using the DNA G4, pu20m2 DNA was first folded as described above and then dried under vacuum. The DNA was then resuspended directly in the filtered dialysis buffer to between 400 and 500 μM before annealing. The RNA:DNA hybrid was first prepared in nuclease free water, dried under vacuum, and resuspended in the dialysis buffer to between 400 and 500 μM before being subjected to the annealing protocol. Any aggregates were removed via centrifugation and the final concentrations of the nucleic acid substrates were determined by UV spectroscopy. Samples of the nucleic acid substrates (100 μL) were loaded into the titrant syringe. Twenty-five injections of 2 μL (DNA G4) or fifty injections of 1 μL (RNA:DNA hybrid) of the nucleic acid substrates were carried out at 25°C with a stirring rate of 125 rpm, and an injection interval of 200 seconds. Raw heat-profile data underwent baseline correction, integration and was fit to the sum of a blank (constant) model and independent binding model using the NanoAnalyze software.
Nuclear magnetic resonance spectroscopy
NMR experiments were conducted on a Bruker Avance NEO spectrometer (Bruker, MA) operating at a proton Larmor frequency of 700.13 MHz at a temperature of 25 °C. Data were processed using NMRPipe 6 or Topspin 4.1.1 (Bruker) and analyzed with CCPNMR Analysis 3.1 software 7. EWSRRM-RG2-ZnF 1H, 13Ca, 13Cb, 13C’ and 15N backbone resonances were assigned using the same approach as was used for EWSLCD 35 using 1H,15N-HSQC, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO and HCC(CO)NH, recorded on a sample of 400 μM in 20 mM potassium phosphate pH 6, 150 mM potassium chloride, 2 mM TCEP, 0.5 mM ZnCl2. The 1H, 15N-HSQC was recorded using 64* x 1024* complex points in the indirect and direct dimensions, corresponding to acquisition times of 30.1 and 106.5 ms, respectively. The HNCO and HN(CA)CO experiments were recorded using 32* x 32* x 1024* complex points in the indirect (F1, 13C), (F2, 15N) and direct (F3, 1H) dimensions, corresponding to acquisition times of 13.0, 15.0 and 112.6 ms, respectively. The HNCACB and CBCA(CO)NH and HCC(CO)NH experiments were recorded using 100* x 32* x 1024* complex points in the indirect (F1, 13C), (F2, 15N) and direct (F3, 1H) dimensions, corresponding to acquisition times of 8.6, 15.0 and 112.6 ms, respectively. All 3D experiments were recorded using non-uniform sampling (NUS) with a sampling density of 20% for the HNCO and HN(CA)CO experiments and 15% for the HNCACB, CBCA(CO)NH and HCC(CO)NH experiments. Spectra were reconstructed using the SMILE algorithm8 implemented in NMRPipe.
For the titrations of 15N EWSRRM-RG2 and EWSRRM-RG2-ZnF with pu20m2 DNA G4, a 500 μL sample of 75 μM 15N protein was prepared in 20 mM potassium phosphate pH 6, 50 mM potassium chloride, 2 mM TCEP, 0.5 mM ZnCl2, and 0.2 mM PMSF. For EWSRRM-RG2-ZnF 0.5 mM ZnCl2 was used and EDTA was excluded from the buffer. A 1.3 mM stock of pu20m2 DNA G4 folded as described was titrated into the sample to final concentrations of 7.5 and 15 μM, requiring the addition of 5.8 μL of the DNA stock solution. For the titration of 15N EWSRG2 with pu20m2 DNA G4, a 450 μL sample of 50 μM 15N protein was prepared in 20 mM potassium phosphate pH 6, 150 mM potassium chloride, 0.5 mM EDTA, 0.2 mM PMSF. A 460 μM stock of pu20m2 DNA G4 folded as described was titrated into the sample to final concentrations of 5 and 10 μM, requiring the addition of up to 10 μL of DNA. For the titration of 15N EWSRRM-RG2 with pu20m2 DNA G4 a 500 μL sample of 100 μM 15N EWSRRM-RG2 was prepared in 20 mM potassium phosphate pH 6, 50 mM potassium chloride, 2 mM TCEP. A 1 mM stock of pu20m2 DNA G4 was titrated into the sample to final concentrations of 10 μM and 25 μM, requiring the addition of up to 12.5 μL of DNA. For the titration of 15N EWSRRM-RG2 with the RNA:DNA hybrid a 500 μL sample of 50 μM 15N EWSRRM-RG2 was prepared in 20 mM potassium phosphate pH 6, 150 mM potassium chloride, 2 mM TCEP. A 75 μM stock of RNA:DNA hybrid was titrated into the sample to final concentrations of 5 and 10 μM, requiring the addition of 73 μL of RNA:DNA. For the titration of 15N EWSRRM-RG2-ZnF with the RNA:DNA hybrid a 450 μL sample of 75 μM 15N EWSRRM-RG2-ZnF was prepared in 20 mM potassium phosphate pH 6, 150 mM potassium chloride, 2 mM TCEP. A 553 μM stock of RNA:DNA hybrid was titrated into the sample to final concentrations of 7.5 and 15 μM, requiring the addition of 12 μL of RNA:DNA. For all titrations, 1H, 15N-HSQC spectra were recorded at 25°C for each titration point with 64* x 1024* complex data points in the indirect (15N) and direct (1H) dimensions, corresponding to acquisition times of 30.1 and 106.5 ms, respectively.
All Spectra were processed in Topspin 4.1.1 apodized with a sine bell function, zero filled to twice the number of acquired points and analyzed using CCPNMR Analysis 3.1 software. Chemical shift perturbations (CSPs) were calculated by weighting the 1H and 15N chemical shifts with respect to their gyromagnetic ratio using the following equation:
Results and Discussion
EWS RRM, RG2 and ZnF contribute to binding the SON RNA stem-loop
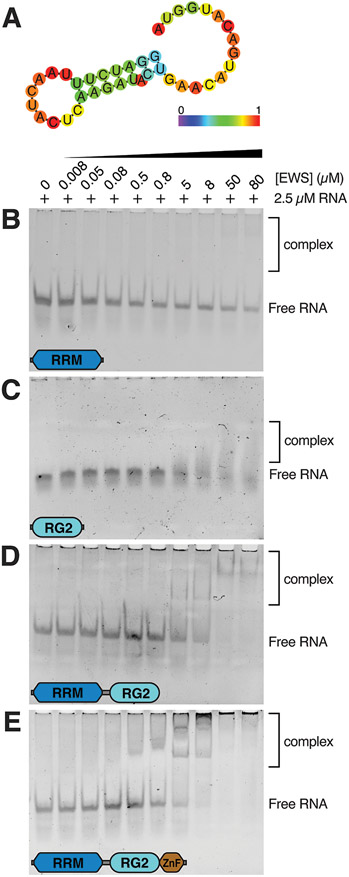
The FET family of proteins are known to associate with RNA in vivo 36 and the interaction of FUS and TAF15 with RNA stem-loops has already been characterized at the molecular level using NMR 22,23. These studies demonstrated that the RRM domain of FUS and TAF15 bind to loop regions of RNA stem-loops from the gene SON in a manner dependent on the conformation of the loop but without sequence specificity 22,23. For FUS, the RG2 region also contributed to binding the duplex part of the stem-loop, while the ZnF domain interacted with ssRNA at the 3’ end of the oligonucleotide. This was not observed for TAF15; however, this is likely because TAF15 was assayed for binding to a stem-loop structure without the additional ssRNA sequence at the 3’ end. Sequence homology implies that EWS is likely to interact with RNA stem-loops in a similar manner. In this work, constructs encoding EWSRRM, EWSRG2, EWSRRM-RG2 and EWSRRM-RG2-ZnF were assayed for their ability to bind an RNA stem-loop structure with a ssRNA sequence at the 3’ end from the gene SON (SON-GGU, Fig. 1A). Gel-shift assays demonstrated that EWSRRM and EWSRG2 interacted weakly with the SON-GGU stem-loop as evidenced by free RNA being observed at all protein concentrations tested. For EWSRRM, only a very faint band corresponding to protein-RNA complexes being formed was observed at the highest protein concentrations (Fig. 1B). For EWSRG2, diffuse bands with small changes in electrophoretic mobility were also observed only at the highest protein concentrations tested (Fig. 1C). Both the The EWSRRM-RG2 and EWSRRM-RG2-ZnF constructs interacted with higher affinity with the RNA stem-loop as evidenced by a complete loss of the band corresponding to free RNA at the highest protein concentrations tested and the appearance of a higher molecular weight complexes being formed (Figs. 1D,E). The EWSRRM-RG2-ZnF construct appeared to interact with a slightly higher affinity than the EWSRRM-RG construct because bands corresponding to protein-RNA complexes were observed earlier in the titration than for EWSRRM-RG (Figs. 1D,E). Therefore, this experiment indicates that all three domains appear to contribute to RNA stem-loop binding in agreement with the expected mode of interaction of EWS RBD with RNA stem-loops in which the RRM likely engages the looped-out region of the stem-loop, the RG2 region might interact with the minor groove of the duplex region of the stem-loop and the ZnF might engage the ssRNA at the 3’ end 22. Importantly, the isolated RRM and RG2 regions do not display strong binding to this RNA stem-loop suggesting that the higher affinity interaction observed for the EWSRRM-RG2 and EWSRRM-RG2-ZnF constructs is the result of increased avidity due to multiple low-affinity binding sites.
Figure 1. EWS RBD binds to the SON-GGU RNA stem-loop.
A) Predicted secondary structure of the SON-GGU oligonucleotide using the RNAfold web server 46. Nucleotides are color-coded according to their likelihood of forming base pairs. EMSA of 2.5 μM SON-GGU RNA stem-loop titrated with 8 nM – 80 μM of (B)EWSRRM, (C) EWSRG2, (D) EWSRRM-RG2 or (E) EWSRRM-RG2-ZnF.
EWS RBD binds to DNA G-quadruplexes with low micromolar affinity via RRM and RG2
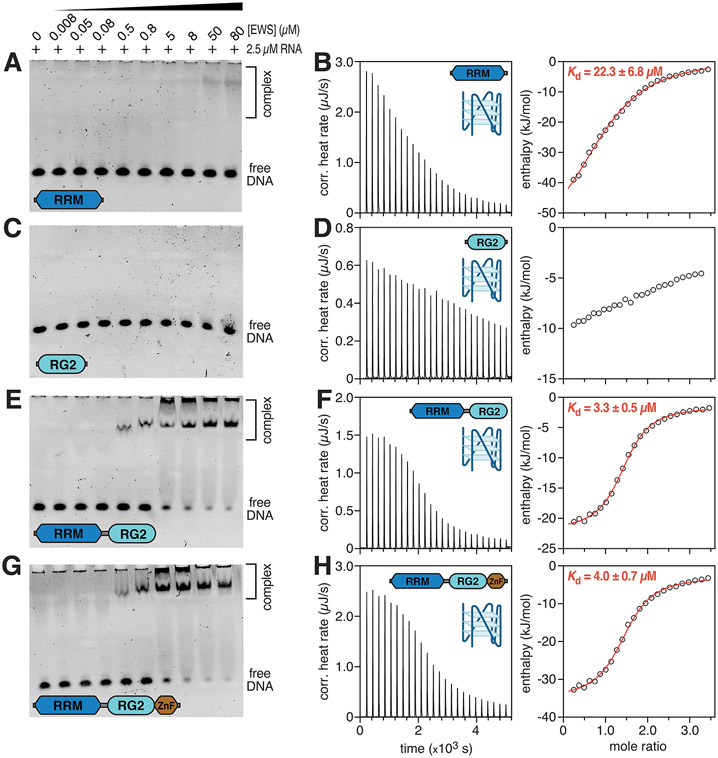
EWS and FUS are also known to bind G4 DNA and RNA sequences found at telomeres 14,15. Interestingly, these studies proposed that G4 binding was encoded by just the RG3 region based on gel shift assays conducted with various constructs of EWS and FUS 15,37. However, some binding by constructs comprising RG1, RRM, RG2, and ZnF was also observed yet the relative affinities of the various constructs for DNA G4s was not assessed in these studies because only a single protein concentration was used in the gel shift assays. G4 formation is associated with R-loops because the non-template strand is often enriched with guanine nucleotides that promote G4 folding. To determine whether the structured domains of EWS (RRM and ZnF), as well as RG2 contribute to binding DNA G4s, the four EWS RBD constructs were tested for their ability to bind to a model parallel intramolecular DNA G4 34. The EWSRRM was found to interact weakly with the G4, with free DNA being observed across the entire titration series, however a band corresponding to a protein-DNA complex was observed at the two highest protein concentrations (Fig. 2A). ITC further confirmed that the EWSRRM associates weakly with the DNA G4, yielding an apparent dissociation constant of 22.3 ± 6.8 μM (Fig. 2B, Table 2). EMSAs and ITC indicated that EWSRG2 displayed little to no binding to the DNA G4 (Fig. 2C,D). The ITC data could not be accurately fit to derive an apparent dissociation constant for this construct due to extremely weak binding. The EWSRRM-RG2 and EWSRRM-RG2-ZnF constructs appeared to share a similar affinity for the DNA G4 (Figs. 2E-H), with almost complete loss of the band corresponding to free DNA being observed at the two highest protein concentrations tested in the titration series (Fig. 2E,G). ITC confirmed that the affinity of EWSRRM-RG for the DNA G4 was essentially identical to that of EWSRRM-RG2-ZnF with apparent dissociation constants of 3.3 ± 0.5 μM and 4.0 ± 0.7 μM, respectively (Fig. 2F,H and Table 2). Interestingly, these apparent dissociation constants are almost identical to those reported for FUSRRM-RG for the SON RNA stem-loop structure, which was identified as the top RNA hit for FET proteins by photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP)36, supporting the hypothesis that DNA G4 binding by FET proteins likely occurs in vivo. For EWSRRM, an n value of 1.12 + 0.06 was obtained indicating close to a 1:1 complex being formed. For EWSRRM-RG and EWSRRM-RG2-ZnF, n values closer to 1.4 were obtained indicating that the observed stoichiometry is higher than 1:1 (Table 2). Close inspection of the native-PAGE gels indicates that these EWS constructs seem to form at least two discrete species with the DNA G4 (evidenced by a band that barely enters the gel just below the wells and another band corresponding to protein-DNA complexes approximately one fifth of the way down the gels). This observation likely accounts for the greater than 1:1 stoichiometry observed in the ITC experiments.
Figure 2. EWS RBD binds to the pu20m2 DNA G4.
EMSAs of 2.5 μM pu20m2 DNA G4 titrated with 8 nM – 80 μM of (A) EWSRRM, (C) EWSRG2, (E) EWSRRM-RG2 and (G) EWSRRM-RG2-ZnF. Raw ITC heat profile and extracted enthalpy from a titration of pu20m2 DNA G4 with 50 μM of (B) EWSRRM, (D) EWSRG2, (F) EWSRRM-RG2 and (H) EWSRRM-RG2-ZnF. Fits to the raw enthalpy data obtained using the NanoAnalyze software are shown by the red curve in (B), (F) and (H).
Table 2.
Thermodynamic parameters for the binding of EWS RBD constructs to a DNA G-quadruplex.
| EWSRRM | EWSRRM-RG | EWSRRM-RG2-ZnF | |
|---|---|---|---|
| (μM) | 22.3 ± 6.8 | 3.28 ± 0.5 | 3.96 ± 0.7 |
| n | 1.12 + 0.06 | 1.4 ± 0.02 | 1.43 ± 0.03 |
| (kJ/mol) | −63.32 ± 9.7 | −20.53 ± 0.59 | −32.34 ± 1.24 |
| 4.48 | 3.04e+005 | 2.53e+005 | |
| (kJ/mol) | 36.77 | −10.77 | 1.5 |
| ΔG (kJ/mol) | −26.55 | −31.30 | −30.84 |
| ΔS (J/mol·K) | −123.30 | 36.14 | −5.03 |
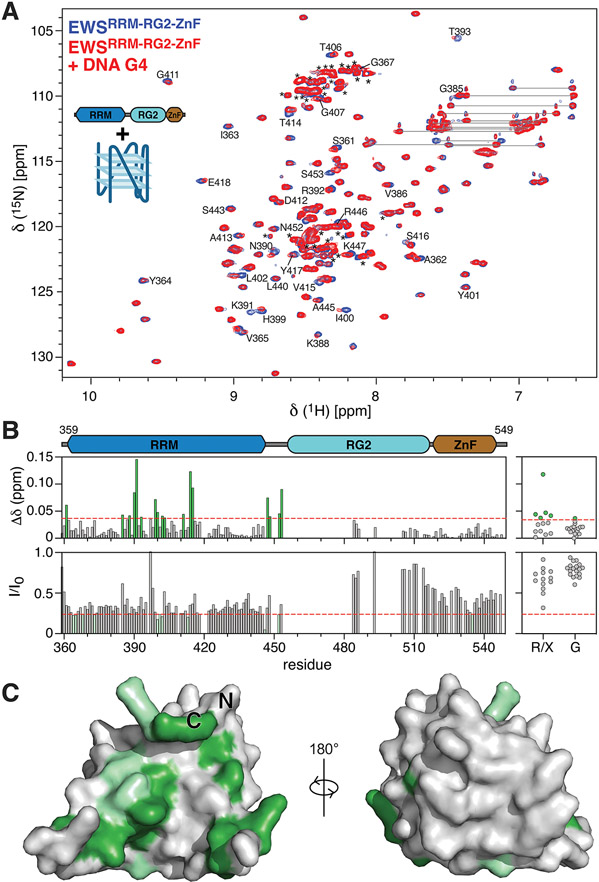
NMR spectroscopy was also employed to determine which regions of EWS RBD bind the DNA G4. The 1H-15N HSQC spectrum of EWSRRM-RG2-ZnF was assigned to 75% of all non-proline residues (Supplemental Fig. 2). However, most of the unassigned residues cluster to the RG2 region, which is characterized by a highly repetitive sequence enriched with Arg, Gly and Pro residues with limited signal dispersion, precluding unambiguous assignment by traditional approaches. Consequently, the RRM and ZnF regions are assigned at 98% and 91% of all non-proline residues, respectively, while RG2 is unambiguously assigned at only 29%. Nevertheless, unassigned peaks in the glycine region of the spectra (106-111 ppm) and in the arginine region of the spectra (118-124 ppm) were picked and their chemical shifts and signal intensities were tracked along with all assigned residues upon titration of 15N EWSRRM-RG2-ZnF with the DNA G4 to assess whether the unassigned peaks arising from RG2 contribute to DNA G4 binding. Titration of EWSRRM-RG2-ZnF with DNA G4 resulted in significant spectral changes manifesting as both CSPs as well as signal broadening (Fig. 3A,B), both of which indicate an interaction with the DNA G4. The signal broadening was not due to aggregation as the sample remained completely clear throughout the titration and is instead attributed to intermediate exchange between the free and DNA-bound form of EWSRRM-RG2-ZnF. Consequently, dissociation constants were not able to be derived from NMR titrations, but the use of sub-stoichiometric concentrations of DNA enabled site-specific mapping of the residues involved in the DNA G4 interaction. Residues assigned to the RRM and some unassigned residues arising from RG2 both displayed larger chemical shifts than those observed for the ZnF domain (Fig. 3B), supporting the findings from both the gel shift assays and the ITC that the RRM and RG2 both contribute to binding DNA G4s but that the ZnF does not appear to contribute to this interaction. Further supporting this conclusion, titration of EWSRRM-RG2 with the DNA G4 revealed an identical pattern of CSPs, with large shifts identified both on the concave surface of the RRM as well as for unassigned peaks arising from RG2 (Supplementary Fig. 3). Additionally, titration of 15N EWSRG2 with the pu20m2 DNA G4 revealed only extremely small CSPs, consistent with the results of the EMSAs, which indicated that the isolated RG2 binds extremely weakly to the DNA G4 (Supplementary Fig. 4). Residues in the EWS RRM with the largest chemical shifts as well as the most significant signal broadening (Fig. 3B) were mapped to the AlphaFold 38 structural model (Fig. 3C), and were clearly localized to the concave surface of the RRM formed by the four-stranded β-sheet as well as loops 3, 4 and 8. Therefore the putative binding site for the DNA G4 was essentially identical to the RNA stem-loop binding site identified in FUS and TAF15 22,23.
Figure 3. EWS RBD binds to the pu20m2 DNA G4 via the RRM and RG2 regions.
A) Overlay of 1H15N-HSQC spectra of 75 μM 15N EWSRRM-RG2-ZnF alone (blue) or in the presence of 15 μM DNA G4 (red). Selected peaks are assigned using the one-letter amino acid code, resonance pairs corresponding to asparagine and glutamine sidechains are indicated by lines, unassigned resonances arising mostly from RG2 are indicated by asterisks. B) CSPs (top panels) and the ratio of signal intensity (I/I0, bottom panels) were calculated for all assigned resonances (left panels) and for unassigned glycine resonances (G) or unassigned resonances of unknown type (R/X, right-panels) upon titration of the DNA G4 with EWSRRM-RG2-ZnF. Red dashed lines indicate a CSP of 0.036 or I/I0 of 0.24 and were used as the threshold for identifying peaks with the most significant changes. C) Assigned resonances within the RRM with the most significant CSPs (green) and signal broadening (light green) were mapped to the AlphaFold 38 structural model of EWSRRM.
Collectively, the data demonstrates that the RRM is capable of binding to DNA G4s, and that RG2 increases the affinity of the RRM for the G4 perhaps due to increased avidity, while the ZnF domain does not appear to contribute to the binding to DNA G4s. It is unsurprising that RG2 supports the interaction with the DNA G4 because RG-rich sequences have been demonstrated to bind G4 structures 39-41. Based on the studies of the FUS RRM interactions with an RNA stem-loop, it is possible that EWSRRM may engage the DNA G4 via one or more of the loops that form between the guanine nucleotides 22. However, FUSRRM interacts with the loop of the RNA stem-loop via a concave surface that accommodates four nucleotide bases in a tight turn conformation using both hydrophobic and hydrogen bonding interactions 22. In the model DNA G4 structure tested here, the looped-out regions are expected to consist of just a single base 34, therefore the RRM may instead bind the DNA G4 via the short stretches of ssDNA at its 5’ and 3’ ends according to the canonical mode of interaction of RRMs with ssRNA 42. Varying the lengths of the loops between the guanine nucleotides in the model DNA G4 will help to determine whether the RRM interacts with the G4 via the loops or via ssDNA structures. Interestingly, the RG3 region of EWS was found to associate with higher affinity with DNA G4s with at least three nucleotides separating the guanine repeats compared to G4s with just a single intervening nucleotide 43. Future studies will expand the series of protein constructs tested to elucidate the roles of RG1/3 in promoting interactions with DNA G4s. Additionally, DNA G4s with varying loop-lengths/conformations should be tested for binding by all of the EWS RBD constructs to assess the role of the loops in recruiting EWS.
EWS RBD binds to RNA:DNA hybrids via RRM and RG2
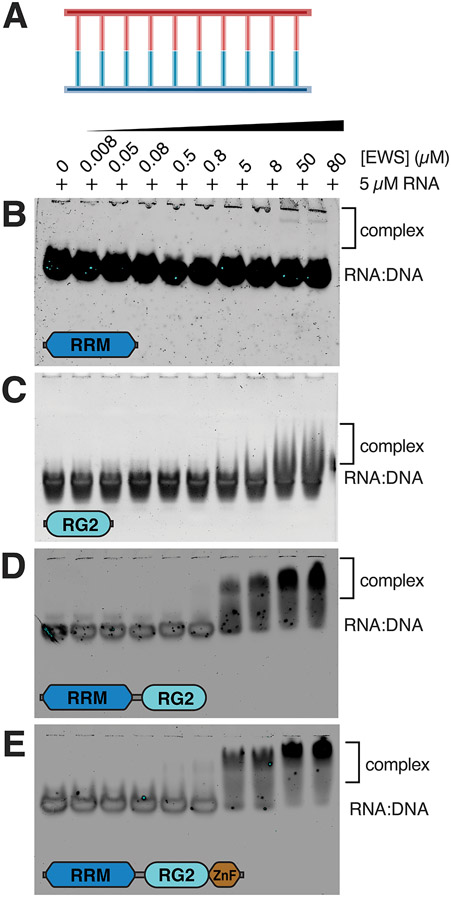
In addition to RNA stem-loops and DNA G4s, RNA:DNA hybrids are the defining features of R-loops (Fig. 4A). FUS has previously been characterized as displaying weaker affinity for double-stranded nucleic acid sequences than for ssRNA or ssDNA 21 and although very little is known about double-stranded nucleic acid binding by EWS, Takahama et al demonstrated that EWS preferentially interacts with G4 DNA at telomeres rather than duplex DNA 15. Gel shift assays revealed that the EWSRRM construct does not interact with the RNA:DNA hybrid or that the interaction is extremely weak, as only an extremely faint band corresponding to bound RNA:DNA hybrid is seen at the highest two protein concentrations (Fig. 4B). Slightly stronger binding was observed for EWSRG2 with the RNA:DNA hybrid as some diffuse bands corresponding to protein-RNA:DNA complexes were observed at the two highest protein concentrations (Fig. 4C). The EWSRRMand EWSRRM-RG2-ZnF constructs both bound to the RNA:DNA hybrid with higher affinity as evidenced by the appearance of higher molecular weight smears corresponding to protein-bound RNA:DNA at the four highest protein concentrations in the titration (Fig. 4D,E). The ZnF domain did not appear to contribute binding affinity of EWS for the hybrid, suggesting that the tighter binding observed by these constructs relative to EWSRRM and EWSRG is due to synergy between the multiple weak binding sites on both the RRM and RG2 domains. This hypothesis is consistent with several crystal structures and solution NMR structures that demonstrate that RG-rich regions bind to double-stranded duplex regions of nucleic acids 22,40,44.
Figure 4. EWS RBD binds to the RNA:DNA hybrid.
A) Schematic for the RNA:DNA hybrid. EMSA of 5 μM RNA:DNA hybrid titrated with 8 nM – 80 μM of (B) EWSRRM, (C) EWSRG2, (D) EWSRRM-RG2 or (E) EWSRRM-RG2-ZnF.
ITC experiments were conducted to measure dissociation constants for the binding of EWSRRM-RG2 and EWSRRM-RG2-ZnF to RNA:DNA hybrids, however the very small heat changes upon injection indicated that the binding is either considerably weaker than the binding to DNA G4s or that binding occurs non-specifically at multiple sites (Supplementary Fig. 5). The smeared appearance of the shifted bands in the gel-shift assays (Fig. 4D,E) might also indicate weak binding at multiple sites or a faster rate of complex dissociation (and a lower affinity) in the case of the RNA:DNA hybrid when compared with the DNA G4, however further ITC experiments carried out at different temperatures or with different buffer conditions may resolve these issues and enable dissociation constants to be measured.
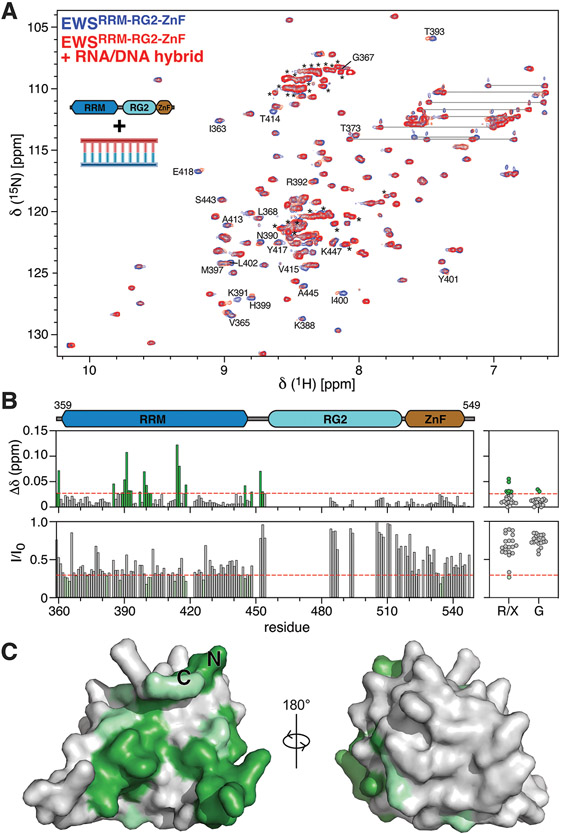
NMR spectroscopy was also employed to confirm the hypothesis that the RRM and RG2 regions synergistically contribute to binding the RNA:DNA hybrid. EWSRRM-RG2-ZnF was titrated with the RNA:DNA hybrid and as for the DNA G4, significant spectral changes were observed including both CSPs as well as signal broadening (Fig. 5A,B). Due to signal broadening, dissociation constants were again not able to be derived via NMR, however site-specific CSPs were mapped at sub-stoichiometric concentrations of the RNA:DNA hybrid (Fig. 5B). Large chemical shifts as well as residues with the most significant signal broadening (Fig. 5B) were mapped to the concave surface of the RRM (Fig. 5C) and were consistent with those identified as the binding site for the DNA G4 (Fig. 3B). Additional large shifts arose from unassigned peaks from the RG2 region (Fig. 5B). Titration of EWSRRM-RG with the RNA:DNA hybrid revealed an identical pattern of shifts and signal broadening in both the RRM and RG2 domains (Supplementary Fig. 6). This indicates that although the isolated RRM and isolated RG2 constructs have a very weak affinities for the RNA:DNA hybrid (Fig. 4B,C), the two domains appear to act synergistically to bind the hybrid with a higher affinity. No significant shifts were observed on the ZnF domain (Fig. 5B), supporting the gel-shift assays which indicated that the ZnF domain does not increase the affinity of EWS RBD for the RNA:DNA hybrid (compare Figs. 4D and C). Based on existing structures of homologous ZnF domains from FUS and RAN-binding protein 2 (RanBP2) bound to ssRNA, it is likely that the ZnF domain binds single-stranded nucleic acid substrates via a trinucleotide GGU motif 22,45, in contrast to the RRM and RG2 domains which appear to be able to interact with nucleic acid substrates adopting a variety of conformations with little sequence specificity. Consequently, it will be interesting to test the binding of EWSRRM-RG2-ZnF to RNA:DNA hybrids with single stranded nucleic acid overhangs as well as DNA G4s with longer single stranded DNA sequences at both the 5’ and 3’ ends. The ZnF may engage these single-stranded sequences and confer specificity to the interactions of EWS with DNA G4s and RNA:DNA hybrids.
Figure 5. EWS RBD binds to the RNA:DNA hybrid via the RRM and RG2 regions.
A) Overlay of 1H15N-HSQC spectra of 75 μM 15N EWSRRM-RG2-ZnF alone (blue) or in the presence of 15 μM RNA:DNA hybrid (red). Peaks are assigned using the one-letter amino acid code, resonance pairs corresponding to asparagine and glutamine sidechains are indicated by lines, unassigned resonances arising mostly from RG2 are indicated by asterisks. B) CSPs (top panels) and the ratio of signal intensity (I/I0, bottom panels) were calculated for all assigned resonances (left panels) and for unassigned glycine resonances (G) or unassigned resonances of unknown type (R/X, right-panels) upon titration of the RNA:DNA hybrid with EWSRRM-RG2-ZnF. Red dashed lines indicate a CSP of 0.028 or I/I0 of 0.29 and were used as the threshold for identifying peaks with the most significant changes. D) Assigned resonances within the RRM with the most significant CSPs (green) and signal broadening (light green) were mapped to the AlphaFold 38 structural model of EWSRRM.
Conclusions
This study has begun to uncover the nucleic acid binding preferences of the RBD of EWS. Consistent with previous studies of FUS and TAF15, EWS RBD was found to bind RNA stem-loops and this binding was promoted by the RRM, RG2 and ZnF regions. Prompted by recent studies that have demonstrated a role of EWS in regulating R-loop dynamics, the ability of the RBD of EWS to bind to nucleic acid structures that are expected to be formed at R-loops was investigated. Expanding on previous work that demonstrated that the RG3 region of EWS can bind DNA and RNA G4s at telomeres, this study demonstrated that various domains of the EWS RBD can also bind DNA G4s. NMR indicated that this binding appeared to be conferred by both the RRM and RG2 region, however gel-shift assays and ITC revealed that the isolated RRM and RG2 domains interact only weakly with the DNA G4 and therefore that both the RRM and RG2 are required for the interaction with the DNA G4. ITC confirmed that the ZnF domain did not contribute to the binding affinity for DNA G4s. Lastly, the RBD of EWS was also shown to interact with RNA:DNA hybrids, this interaction appeared to be dependent on both the RRM and RG2 because gel shift assays did not demonstrate binding to the RNA:DNA hybrid by EWSRRM and only weak binding was observed for EWSRG2. However, large CSPs were observed for both the RRM and RG2 region upon titration of EWSRRM-RG2-ZnF with an RNA:DNA hybrid, supporting the hypothesis that both the RRM and RG2 may be required for binding this nucleic acid conformation. The ZnF did not appear to contribute to the binding affinity for the RNA:DNA hybrid. Therefore, it appears that both the RRM and RG2 region of EWS can bind nucleic acid structures in a promiscuous manner. The RRM appears to bind G4s with a higher affinity than RG2 and conversely, RG2 appears to bind RNA:DNA hybrids with a slightly higher affinity than the RRM. The ZnF appeared to only contribute to binding the RNA stem-loop and likely does so via an interaction with ssRNA at the 3’ end as was shown for FUS and TAF15. Therefore, the ZnF probably binds single-stranded nucleic acid conformations with less promiscuity than the RRM or RG2 and may be important for conferring specificity to the interactions of EWS with RNA and DNA in vivo. Further studies will expand both the series of protein constructs tested and also the conformations and sequences of the nucleic acid constructs tested to further elucidate the mechanism by which EWS engages with R-loops.
Supplementary Material
Acknowledgements
This study was funded in part by NIGMS R01GM140127 (to DSL), GCCRI Startup Funds (DSL) and The Welch Foundation BN-0032 (to REB and the UIW Department of Chemistry and Biochemistry). DSL is the Shohet Family Fund for Ewing Sarcoma Research St. Baldrick’s Scholar and acknowledges the support of the St. Baldrick’s Foundation (634706). EES was supported by a CPRIT Research Training Award (RP170345). UIW provided research support for RB, MRW, SA, and RDH (from Department of Chemistry and Biochemistry) and CNT (from School of Medicine) The authors would like to thank Drs. Antione Baudin and Karen Lewis for advice and technical assistance with the ITC measurements and EMSAs respectively. Figures were in part created with BioRender.com. This work is based upon research conducted in the Structural Biology Core Facilities, a part of the Institutional Research Cores at the University of Texas Health Science Center at San Antonio supported by the Office of the Vice President for Research and the Mays Cancer Center Drug Discovery and Structural Biology Shared Resource (NIH P30 CA05417X4).
Abbreviations:
- hnRNP
heterogeneous nuclear ribonucleoprotein
- RRM
RNA-recognition motif
- RGG
Arg-Gly-Gly
- ZnF
zinc finger
- FUS
fused in sarcoma
- EWS
RNA-binding protein, EWS
- TAF15
TATA-binding protein associated factor 2N
- LCD
low-complexity domain
- RG-rich
Arg-Gly rich
- NLS
nuclear localization sequence
- RBD
RNA-binding domain
- EwS
Ewing sarcoma
- ETS
E-twenty-six transformation-specific
- FLI1
Friend leukemia integration 1
- RNA Pol II
DNA-directed RNA polymerase II subunit RPB1
- CDK
cyclin-dependent kinase
- BRCA1
breast cancer type 1 susceptibility protein
- G4
G-quadruplex
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
- TEV
Tobacco Etch Virus
- MBP
Maltose binding protein
- EDTA
ethylenediaminetetraacetic acid
- PMSF
phenylmethylsulfonyl fluoride
- TBE
Tris-borate-EDTA
- EMSA
electrophoretic mobility shift assay
- NUS
non-uniform sampling
- PAR-CLIP
photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation
- CSP
chemical shift perturbation
- RanBP2
ran-binding protein 2
Footnotes
Conflict of Interest statement
The authors declare no competing interests.
Data availability statement
NMRPipe processing scripts are available upon reasonable request, expression plasmids encoding EWSRRM, EWSRG2, EWSRRM-RG2, and EWSRRM-RG2-ZnF were deposited with Addgene (188045, 199439, 188046, and 195868) respectively. The backbone resonance assignments for the EWSRRM-RG2-ZnF were deposited in the BMRB (51741).
References
- 1.David CJ, Chen M, Assanah M, Canoll P & Manley JL HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clower CV et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A 107, 1894–1899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golan-Gerstl R et al. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res 71, 4464–4472 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Hutten S & Dormann D hnRNPA2/B1 Function in Neurodegeneration: It’s a Gain, Not a Loss. Neuron 92, 672–674 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kim HJ et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotti A, Lutz Y, Heard DJ, Chambon P & Tora L hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J 15, 5022–5031 (1996). [PMC free article] [PubMed] [Google Scholar]
- 7.Bertolotti A et al. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol Cell Biol 18, 1489–1497 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paronetto MP Ewing sarcoma protein: a key player in human cancer. Int J Cell Biol 2013, 642853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan AY & Manley JL The TET family of proteins: functions and roles in disease. J Mol Cell Biol 1, 82–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paronetto MP, Miñana B & Valcárcel J The Ewing Sarcoma Protein Regulates DNA Damage-Induced Alternative Splicing. Molecular Cell 43, 353–368 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Baechtold H et al. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem 274, 34337–34342 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Li H et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest 117, 1314–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorthi A et al. EWS–FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 555, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahama K et al. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem Biol 20, 341–350 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Takahama K, Kino K, Arai S, Kurokawa R & Oyoshi T Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J 278, 988–998 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Takahashi A et al. EWS/ETS Fusions Activate Telomerase in Ewing’s Tumors. Cancer Research 63, 8338–8344 (2003). [PubMed] [Google Scholar]
- 17.Yang L, Embree LJ, Tsai S & Hickstein DD Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem 273, 27761–27764 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Chansky HA & Hickstein DD EWS·Fli-1 Fusion Protein Interacts with Hyperphosphorylated RNA Polymerase II and Interferes with Serine-Arginine Protein-mediated RNA Splicing *. Journal of Biological Chemistry 275, 37612–37618 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Chansky HA, Hu M, Hickstein DD & Yang L Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res 61, 3586–3590 (2001). [PubMed] [Google Scholar]
- 20.Pahlich S, Quero L, Roschitzki B, Leemann-Zakaryan RP & Gehring H Analysis of Ewing sarcoma (EWS)-binding proteins: interaction with hnRNP M, U, and RNA-helicases p68/72 within protein-RNA complexes. J Proteome Res 8, 4455–4465 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Schwartz JC & Cech TR Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res 43, 7535–7543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loughlin FE et al. The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity. Molecular Cell 73, 490–504.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Kashyap M, Ganguly AK & Bhavesh NS Structural delineation of stem-loop RNA binding by human TAF15 protein. Sci Rep 5, 17298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delattre O et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359, 162–165 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Picard C et al. Identification of a novel translocation producing an in-frame fusion of TAF15 and ETV4 in a case of extraosseous Ewing sarcoma revealed in the prenatal period. Virchows Arch 481, 665–669 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Shing DC et al. FUS/ERG gene fusions in Ewing’s tumors. Cancer Res 63, 4568–4576 (2003). [PubMed] [Google Scholar]
- 27.Guillon N et al. The Oncogenic EWS-FLI1 Protein Binds In Vivo GGAA Microsatellite Sequences with Potential Transcriptional Activation Function. PLOS ONE 4, e4932 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gangwal K et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. PNAS 105, 10149–10154 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangwal K, Close D, Enriquez CA, Hill CP & Lessnick SL Emergent Properties of EWS/FLI Regulation via GGAA Microsatellites in Ewing’s Sarcoma. Genes Cancer 1, 177–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaishankar S, Zhang J, Roussel MF & Baker SJ Transforming activity of EWS/FLI is not strictly dependent upon DNA-binding activity. Oncogene 18, 5592–5597 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Embree LJ, Azuma M & Hickstein DD Ewing sarcoma fusion protein EWSR1/FLI1 interacts with EWSR1 leading to mitotic defects in zebrafish embryos and human cell lines. Cancer Res 69, 4363–4371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boone MA et al. The FLI portion of EWS/FLI contributes a transcriptional regulatory function that is distinct and separable from its DNA-binding function in Ewing sarcoma. Oncogene 40, 4759–4769 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan H et al. Cohesin SA1 and SA2 are RNA binding proteins that localize to RNA containing regions on DNA. Nucleic Acids Res 48, 5639–5655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K-B et al. Oxidative Damage Induces a Vacancy G-Quadruplex That Binds Guanine Metabolites: Solution Structure of a cGMP Fill-in Vacancy G-Quadruplex in the Oxidized BLM Gene Promoter. J. Am. Chem. Soc 144, 6361–6372 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson CN, Xu X, Holloway SP & Libich DS The 1H, 15N and 13C resonance assignments of the low-complexity domain from the oncogenic fusion protein EWS-FLI1. Biomol NMR Assign 16, 67–73 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoell JI et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol 18, 1428–1431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagi R, Miyazaki T & Oyoshi T G-quadruplex binding ability of TLS/FUS depends on the β-spiral structure of the RGG domain. Nucleic Acids Res 46, 5894–5901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuzawa T & Oyoshi T Roles of the RGG Domain and RNA Recognition Motif of Nucleolin in G-Quadruplex Stabilization. ACS Omega 5, 5202–5208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasilyev N et al. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc Natl Acad Sci U S A 112, E5391–5400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozdilek BA et al. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res 45, 7984–7996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maris C, Dominguez C & Allain FH-T The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272, 2118–2131 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Takahama K, Sugimoto C, Arai S, Kurokawa R & Oyoshi T Loop Lengths of G-Quadruplex Structures Affect the G-Quadruplex DNA Binding Selectivity of the RGG Motif in Ewing’s Sarcoma. Biochemistry 50, 5369–5378 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Phan AT et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol 18, 796–804 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loughlin FE et al. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5′ splice site-like sequences. Proceedings of the National Academy of Sciences 106, 5581–5586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenz R et al. ViennaRNA Package 2.0. Algorithms Mol Biol 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMRPipe processing scripts are available upon reasonable request, expression plasmids encoding EWSRRM, EWSRG2, EWSRRM-RG2, and EWSRRM-RG2-ZnF were deposited with Addgene (188045, 199439, 188046, and 195868) respectively. The backbone resonance assignments for the EWSRRM-RG2-ZnF were deposited in the BMRB (51741).