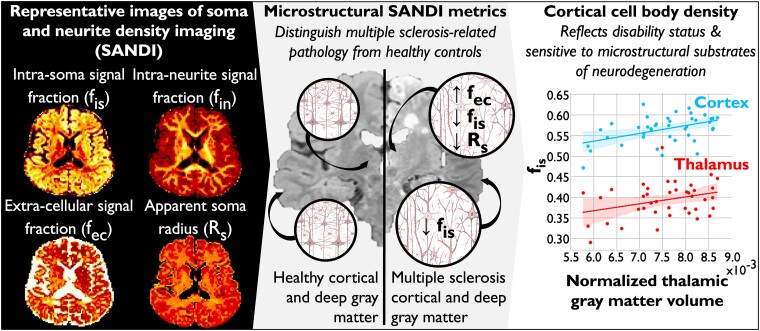
Abstract
Multiple sclerosis features complex pathological changes in grey matter that begin early and eventually lead to diffuse atrophy. Novel approaches to image grey-matter microstructural alterations in vivo are highly sought after and would enable more sensitive monitoring of disease activity and progression. This cross-sectional study aimed to assess the sensitivity of high-gradient diffusion MRI for microstructural tissue damage in cortical and deep grey matter in people with multiple sclerosis and test the hypothesis that reduced cortical cell body density is associated with cortical and deep grey-matter volume loss. Forty-one people with multiple sclerosis (age 24–72, 14 females) and 37 age- and sex-matched healthy controls were scanned on a 3 T Connectom MRI scanner equipped with 300 mT/m gradients using a multi-shell diffusion MRI protocol. The soma and neurite density imaging model was fitted to high-gradient diffusion MRI data to obtain estimates of intra-neurite, intra-cellular and extra-cellular signal fractions and apparent soma radius. Cortical and deep grey-matter microstructural imaging metrics were compared between multiple sclerosis and healthy controls and correlated with grey-matter volume, clinical disability and cognitive outcomes. People with multiple sclerosis showed significant cortical and deep grey-matter volume loss compared with healthy controls. People with multiple sclerosis showed trends towards lower cortical intra-cellular signal fraction and significantly lower intra-cellular and higher extra-cellular signal fractions in deep grey matter, especially the thalamus and caudate, compared with healthy controls. Changes were most pronounced in progressive disease and correlated with the Expanded Disability Status Scale, but not the Symbol Digit Modalities Test. In multiple sclerosis, normalized thalamic volume was associated with thalamic microstructural imaging metrics. Whereas thalamic volume loss did not correlate with cortical volume loss, cortical microstructural imaging metrics were significantly associated with thalamic volume, and not with cortical volume. Compared with the short diffusion time (Δ = 19 ms) achievable on the Connectom scanner, at the longer diffusion time of Δ = 49 ms attainable on clinical scanners, multiple sclerosis-related changes in imaging metrics were generally less apparent with lower effect sizes in cortical and deep grey matter. Soma and neurite density imaging metrics obtained from high-gradient diffusion MRI data provide detailed grey-matter characterization beyond cortical and thalamic volumes and distinguish multiple sclerosis–related microstructural pathology from healthy controls. Cortical cell body density correlates with thalamic volume, appears sensitive to the microstructural substrate of neurodegeneration and reflects disability status in people with multiple sclerosis, becoming more pronounced as disability worsens.
Keywords: multiple sclerosis, grey matter, neurodegeneration, advanced diffusion imaging
Krijnen et al. evaluated a novel in vivo diffusion imaging model for grey-matter microstructural damage in people with multiple sclerosis using high-gradient MRI. Grey-matter cellular changes appear sensitive to detect the microstructural substrate of neurodegeneration and reflect clinical disability status.
Graphical Abstract
Graphical abstract.
Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating and neurodegenerative disease of the CNS. In addition to characteristic white matter (WM) changes, MS features complex pathological changes in grey matter (GM), eventually leading to generalized atrophy. Whole-brain atrophy is a major determinant of clinical and cognitive worsening and is most pronounced in progressive MS (PMS).1-3 Whole-brain atrophy is used as a primary outcome measure in Phase 2 PMS clinical trials and is driven in part by neuroaxonal degeneration in the GM.4-6
While GM volume loss is an important predictor of clinical outcomes in people with MS and explains long-term disability better than WM atrophy,7,8 the exact microstructural underpinnings of atrophy and their temporal association with volume loss are still largely unexplored. Findings from histopathology and longitudinal imaging studies suggest that neurodegeneration begins at disease onset alongside neuroinflammation.9,10 Neuropathology studies have shown that chronic inflammation in the CNS plays a central role in the dysregulation of neuronal and axonal metabolism, resulting in neuroaxonal damage through apoptosis and necrosis, among other mechanisms.11,12 Neuroaxonal degeneration occurs not only at sites of overt demyelination but also throughout the brain parenchyma, including in normal-appearing (NA) tissue without evidence of focal inflammation.13-15
Neuronal and axonal loss is highly heterogeneous across GM but follows a reasonably consistent sequence throughout the disease course.1,2,16 In people with relapsing-remitting MS (RRMS), GM volume loss presents early in the posterior cingulate gyrus as well as in the precuneus and thalamus.16 In particular, volume loss in the thalamus and other deep GM nuclei, e.g. the caudate and putamen, have gained increased attention in recent years due to their relevance to clinical outcomes.17-23 Many studies have shown that the deep GM exhibits a disproportionate degree of MS-specific atrophy over time and is associated with an increased risk of disability progression.3,24 In people with MS, non-relapsing clinical progression may reflect a point beyond which compensatory mechanisms are no longer able to counteract the effects of irreversible neuroaxonal injury.11,25In vivo imaging markers of GM microstructural alterations prior to overt volume loss are needed to provide more sensitive monitoring of disease activity and progression prior to irreversible disability.
The advent of high-performance gradients on human MRI scanners has enabled the translation of promising diffusion-weighted (DW) microstructural imaging methods from animal and ex vivo studies to the investigation of tissue microstructure in the in vivo human brain.26-32 Current DW imaging modelling approaches are limited in their representation of GM microstructure as they cannot distinguish water in the extra-cellular space from water in cell bodies. To overcome this issue, a novel DW compartment-based model for apparent cell body, i.e. soma, and neurite density imaging (SANDI) has been proposed, in which the soma of any brain cell type is explicitly included as a spherical compartment of finite radius.33 The SANDI model was initially demonstrated in high-gradient diffusion MRI data from the MGH-USC Human Connectome Project and has been shown to define new contrasts reflecting brain cyto- and myelo-architecture.33 Another complication in the non-invasive characterization of tissue microstructure by DW-MRI is the concept of water exchange across the neurite membrane. It has been suggested that at reasonably short diffusion times (Δ < 20 ms), water exchange between neurites and soma, but also and more importantly, water exchange between neurites and the extra-cellular compartment can be considered negligible,33-35 supporting the importance of the high-gradient systems for characterizing microstructural changes in cortical and deep GM, although not yet available in clinical settings.
The goal of this study was to assess the sensitivity of SANDI on dedicated high-gradient diffusion MRI data acquired on the 3 T Connectome scanner for microstructural tissue alterations in cortical and deep GM in people with MS compared with healthy controls (HCs), and to evaluate microstructural tissue properties such as cell body and neurite density relative to GM volume, clinical disability scores and MS phenotype. For reference, we also assessed microstructural tissue alterations in NAWM and lesional WM in people with MS. We hypothesized that SANDI metrics, particularly cell body density in GM, would be reduced in cortical and deep GM of people with MS, reflecting cellular and neuronal loss in the MS brain, and that these metrics would be associated with GM volume loss. As SANDI and other DW microstructural models of GM gain traction in MS and other neurological diseases, we also sought to explore the impact of diffusion time on SANDI metrics in MS, taking advantage of the short diffusion times accessible on the Connectome scanner. We postulated that inter-compartmental water exchange would result in a shift of signal fractions from the intra-cellular to extra-cellular space at longer diffusion times, resulting in better sensitivity of SANDI at relatively short diffusion times (Δ = 19 ms) to cell body density compared with longer diffusion times (Δ = 49 ms). These short diffusion times are achievable with the use of cutting-edge hardware currently in development for the clinical setting.36,37
Materials and methods
Participants
This cross-sectional study was approved by the Massachusetts General Hospital institutional review board and is compliant with the Health Insurance Portability and Accountability Act guidelines. All participants provided written informed consent according to the Declaration of Helsinki. Forty-one people with a clinical diagnosis of MS who visited the MS Clinic at the Massachusetts General Hospital between 2015 and 2019 and 37 age- and sex-matched HC were included. Of the 41 people with MS, 32 had RRMS and 9 had PMS (3 primary- and 6 secondary-PMS). Inclusion criteria for people with MS were a diagnosis of clinically definite MS, being relapse free for at least 3 months, and receiving stable disease-modifying treatment or no treatment for at least 6 months. Exclusion criteria for all included participants were presence of other structural brain disease and contraindication to MRI. The current study includes a targeted re-analysis of data that were acquired on the Connectome scanner as part of a larger longitudinal imaging study in MS, and a subset of the data has been previously published with an emphasis on WM analyses.30,38,39
The following patient characteristics were extracted from the electronic medical records of the MS participants: age, sex, disease duration and type of disease-modifying therapy. All MS participants underwent a standard clinical examination within 1 week of the MRI scan. Neurological disability was assessed by a board-certified neurologist who was blinded to the imaging results using the Symbol Digit Modalities Test (SDMT) and Expanded Disability Status Scale (EDSS).
Image acquisition
All participants underwent brain imaging in a 3 T MRI scanner (Magnetom Connectom; Siemens, Erlangen, Germany) equipped with a maximum gradient strength of 300 mT/m. A custom-built 64-channel-phased array head coil was used for signal reception.40 Diffusion data were acquired using a DW spin-echo single-shot EPI sequence in the sagittal plane [2 × 2 × 2 mm3 voxel size, echo time (TE)/repetition time (TR) = 77/3600 ms, simultaneous multi-slice imaging with slice acceleration factor 2, parallel imaging acceleration factor R = 2, anterior-to-posterior phase encoding, total acquisition time 51 min], following a previously established protocol.30,41 Non-DW images (b = 0) were acquired every 16 images. DW images were acquired at diffusion times of Δ = 19 ms using 8 b-values (b = 50–350–800–1500 s/mm2 in 32 directions, and b = 2400–3450–4750–6000 s/mm2 in 64 directions) and at Δ = 49 ms using 8 b-values (b = 200–950–2300 s/mm2 in 32 directions, and b = 4250–6750–9850–13 500–17 800 s/mm2 in 64 directions). For each diffusion time, a gradient pulse duration of 8 ms was used. Five b = 0 images with a reversed-phase encoding direction were separately acquired to correct for distortions due to susceptibility effects. Anatomical imaging included a T1-weighted multi-echo magnetization prepared rapid acquisition gradient echo (MEMPRAGE) sequence [1 × 1 × 1 mm3 voxel size, TE/TR/inversion time (TI) = 1.15–3.03–4.89–6.75 ms/2530 ms/1100 ms, R = 3, flip angle = 7°, acquisition time 3 min 58 s] and a 3D fluid-attenuated inversion recovery (FLAIR) sequence (0.9 × 0.9 × 0.9 mm3 voxel size, TE/TR/TI = 389 ms/5000 ms/1800 ms, R = 2, acquisition time 5 min 47 s).
Diffusion MRI data processing
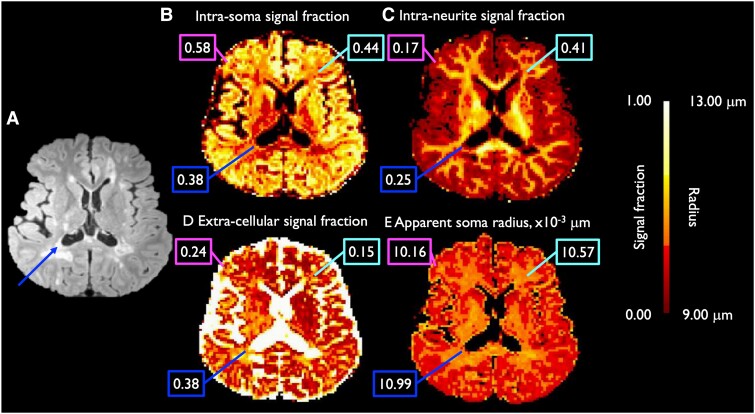
All DW images were pre-processed using an established pipeline.41 In brief, the magnitude DW images were acquired and corrected for gradient non-linearity using in-house software.42 Distortions in the DW images due to susceptibility and eddy current effects were corrected using topup (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/topup) and eddy (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/eddy) in the FMRIB Software Library (FSL version 5.0).43-45 We used the AMICO software46 (https://github.com/daducci/AMICO) to fit the SANDI model to the pre-processed diffusion data, applying a λ2 regularization term of 0.005 and the following default dictionary of atoms: intra-soma diffusivity = 3.0 × 10−3 mm2/s, soma radii = 1.56 × 10−3, 3.44 × 10−3, 4.44 × 10−3, 5.33 × 10−3, 6.00 × 10−3, 6.56 × 10−3, 8.11 × 10−3, 9.56 × 10−3, 1.17 × 10−2 mm, intra-neurite diffusivities = 9.17 × 10−4, 1.69 × 10−3, 3.00 × 10−3 mm2/s, and extra-cellular isotropic mean diffusivities = 3.61 × 10−4, 1.64 × 10−3, 3.00 × 10−3 mm2/s. The regularization factor was empirically chosen based on visual inspection of the fitting results for a range of regularization terms (0.0001 < λ2 < 0.05 in steps of 0.0005) and were in line with values used in previously published papers.34 We initially performed SANDI analysis on DW images acquired at diffusion time of Δ = 19 ms to avoid the potential confound of inter-compartmental exchange at diffusion times of Δ > 20 ms.34,47 Data acquired at diffusion time Δ = 49 ms were also fitted to SANDI and used to assess the impact of diffusion time on SANDI estimates. The model generated maps of the intra-neurite, extra-cellular and intra-soma signal fractions (fin, fec and fis), apparent soma radius (Rs) and intra- and extra-neurite diffusivities (Fig. 1). Based on our hypotheses regarding cell body and neurite density in MS, we chose to include the SANDI metrics of fin, fec, fis and Rs for subsequent analyses. The diffusion tensor model was fitted on the pre-processed diffusion data of b = 800 s/mm2 using the dtifit function in FSL to derive maps of fractional anisotropy, which were used to improve the deep GM segmentation (see below).
Figure 1.
Axial images of SANDI results. Maps include the T2-weighted FLAIR image (A), showing WM lesions (dark blue arrow), of a representative patient with MS, and the corresponding intra-soma (B), intra-neurite (C) and extra-cellular (D) signal fractions and apparent soma radius (E; in μm). Values within a specific voxel consisting cortical GM (magenta; upper left boxes), NAWM (light blue; upper right boxes) and WM lesion (dark blue; lower left boxes) are displayed in boxes. Values displayed in images B, C and D are signal fractions ranging from 0.00 to 1.00, and are therefore unitless.
Registration, segmentation and imaging outcome determination
WM and GM segmentation
WM lesion segmentation of FLAIR hyperintense lesions was performed using a validated FreeSurfer-based automatic segmentation tool.48 A board- and subspecialty-certified neuroradiologist (12 years of experience) manually edited the lesion masks. Lesion volumes were calculated by multiplying lesion area by slice thickness.
Cortical surface and volumetric reconstruction were performed on the T1 MEMPRAGE data using FreeSurfer (version 5.3, http://surfer.nmr.mgh.harvard.edu). An experienced user manually reviewed these reconstructions and filled in WM lesions disrupting the cortical boundary to prevent misclassification of GM.49 After cortical surface reconstruction, we obtained a tissue segmentation map of cortical GM using the 5ttgen hsvs function in MRtrix based on the surface reconstruction of FreeSurfer.50,51 GM and WM volume fractions were computed using the mri_compute_volume_fractions function of FreeSurfer. The cortical GM mask was multiplied by the GM volume fraction to derive a partial volume-weighted cortical GM mask, used in subsequent processing steps to minimize the potential bias from partial volumes.
Fourteen deep GM regions (seven in each hemisphere: accumbens, amygdala, caudate, hippocampus, pallidum, putamen and thalamus) were segmented using the Sequence Adaptive Multimodal SEGmentation52 (SAMSEG) of FreeSurfer, providing an enhanced T1-weighted image as input. The enhanced T1-weighted image was generated by in-house software combining the T1-weighted, FLAIR and fractional anisotropy images. This enhanced T1-weighted image has greater contrast between WM and GM, which can improve segmentation of deep GM regions. A separate FreeSurfer tool was used specifically for segmentation of the thalamus,53 proving the same enhanced T1-weighted image as input in order to better distinguish the lateral border of the thalamus from the surrounding WM. FLAIR images were missing in four HC subjects; hence, in these four subjects, SAMSEG was used providing just the T1-weighted and fractional anisotropy images as inputs. Segmentations were reviewed by an experienced rater for consistency.
To transform the partial volume-weighted cortical GM and WM maps, deep GM atlas and WM lesion mask to DWI space, DW images were first non-linearly registered to FreeSurfer space using ANTs registration,54 after which we applied the converted matrix and warp image to all maps. WM lesion segmentations in DW space were subtracted from the WM volume fraction map, yielding a partial volume-weighted NAWM map in DW space. Figure 1A shows the WMSA lesion segmentation in DWI space.
SANDI metric calculations
The cortical GM, NAWM and lesional WM masks and deep GM atlas in DWI space were overlaid on the maps for all SANDI microstructural metrics to calculate mean values of each metric per region of interest. Given the resolution of our DW images (2 mm isotropic), which was comparable with the average cortical thickness, we calculated partial volume-weighted mean values of cortical GM per microstructural metric to account for potential partial volume effects. This accentuated the contribution of voxels containing more cortical GM than voxels containing less GM.
Volume measures
Mean cortical GM volume and intra-cranial volume were calculated based on the T1-weighted images using the mris_anatomical_stats function of FreeSurfer. Deep GM volumes were calculated based on the deep GM segmentation in FreeSurfer space. All volumes are given in millilitre and corrected for intra-cranial volume, yielding normalized volume measures.
Statistical analysis
Statistical analyses were performed with the use of Python v3.7 and IBM SPSS statistics 28.0 (SPSS Inc., Chicago, IL, USA). We used Kolmogorov–Smirnov testing and histogram inspection to test normality of the data. Participant characteristics were expressed as frequency with percentage, mean with standard deviation (SD), or median with inter-quartile range (IQR). Depending on the normality of the data, comparisons between groups were performed using independent samples t-tests or multivariate general linear models. The Mann–Whitney U-test was used as non-parametric alternative, if appropriate. All statistical tests were two-tailed. Effect sizes for SANDI metrics showing a significant difference between MS and HC groups were calculated based on Hedges’ g. P-values were false discovery rate (FDR)-corrected for multiple comparisons (Pcorr) per analysis step with α = 0.05.
Between-group changes in microstructure
Mean values of SANDI metrics were compared between people with MS and HC, separately for cortical and deep GM and NAWM. Deep GM SANDI metrics that showed near-significant differences, defined as unadjusted P < 0.10, were compared for each deep GM region (averaged between the two hemisphere) between people with MS and HC. In WM, near-significant SANDI metrics were compared between NAWM and lesional WM using paired t-tests.
Relation between microstructure and GM volume
In the regions showing significant differences in SANDI metrics between people with MS and HC, we further explored whether the metrics in specific regions of interest were related to volumes of those regions. First, normalized cortical and deep GM volumes were compared between people with MS and HC to determine the presence of atrophy in people with MS. Then, partial correlation coefficients, adjusted for age and sex, were calculated to assess the linear relation between mean SANDI metrics and normalized volumes in MS. Pearson’s or Spearman’s rank correlations were used depending on normality of the data.
Relation between microstructure and clinical disability
In addition, we sought to determine the relation between SANDI metrics in GM and WM, clinical disability and disease subtype. Therefore, SANDI metrics showing significant between-group differences (as described in the second paragraph) were further analysed among MS subtypes: HC versus RRMS, HC versus PMS and RRMS versus PMS. Also, significant correlations between SANDI metrics and GM volumes (as described in the third paragraph) were further assessed within RRMS and PMS separately.
In the total MS cohort, SANDI metrics showing significant between-group differences were correlated with EDSS and SDMT scores in people with MS. Linear correlations between SDMT scores and SANDI metrics were performed using Pearson’s partial correlations, adjusting for age and sex. Due to ordinal data, the rank correlation between EDSS and SANDI metrics was calculated based on Kendall’s Tau correlation. We also correlated SANDI metrics with age using Pearson’s partial correlation adjusting only for sex to assess the relationship of normal aging and potential measures of brain atrophy in MS.
Diffusion time differences
Data acquired at diffusion time Δ = 19 ms, which were used in all main analysis described above, were compared with data acquired at diffusion time Δ = 49 ms. To assess the diffusion time dependence of the SANDI metrics between groups and within the associations with GM volume, additional between-group analyses were performed using the data acquired at diffusion time Δ = 49 ms. Significant correlates of GM volume found at a diffusion time of Δ = 19 ms were assessed at Δ = 49 ms as well.
Results
Demographics
Table 1 shows the characteristics of included participants. Age and sex did not differ significantly between HC and people with MS. Our MS cohort consisted of predominantly people with RRMS [N = 33 (80.5%)] with a mean disease duration of <10 years. People with MS showed significant volume loss of both cortical and deep GM compared with HC [t(76) = 6.54, P = 6.39 × 10−9 and t(76) = 3.81, P = 2.82 × 10−4, respectively]. Of deep GM regions, all except the caudate and pallidum showed MS-related volume loss [F-range(1,76) = 5.51–24.12, P-range = 5.00 × 10−6–0.022], with the largest effect size seen in the thalamus (Hedges’ g = 1.10 compared with g-range = 0.11–0.62 in other deep GM structures). There were no missing data for any of the variables of interest.
Table 1.
Baseline demographics and clinical characteristics of included participants
| Healthy volunteers (N = 37) | People with MS (N = 41) | Group differencesa | |
|---|---|---|---|
| P-value* | |||
| Demographics | |||
| Age, years | 39.3 (14.3) | 45.2 (12.9) | 0.052 |
| Sex, male/female [number (%)] | 16 (43.2) / 21 (56.8) | 11 (26.8) / 30 (73.2) | 0.128 |
| Education, years | 16.2 (2.5) | ||
| Disease characteristics | |||
| Disease duration, years | 9.7 (6.9) | ||
| MS subtype, RR/SP/PP | 33 (80.5)/5 (12.2)/3 (7.3) | ||
| DMT use,b yes [number (%)] | 35 (85.4) | ||
| Clinical characteristics | |||
| EDSS, score | 2.5 [2.0–3.5] | ||
| SDMT, raw score | 51.0 (12.5) | ||
| MR characteristics | |||
| WM lesion load, mL | 4.13 [1.87–10.80] | ||
| Cortical GM volume, mL | 478.9 (57.8) | 421.0 (73.9) | 2.68 × 10−4 |
| Normalized cortical GM volume, mL | 0.30 (0.02) | 0.27 (0.03) | 6.39 × 10−9 |
| Deep GM volume, mL | 44.4 (4.0) | 41.9 (5.0) | 0.016 |
| Normalized deep GM volume, mL | 0.028 (0.002) | 0.027 (0.002) | 2.82 × 10−4 |
Variables are reported as mean (SD) or median (IQR) unless otherwise indicated. DMT, disease-modifying treatment; EDSS, Expanded Disability Status Scale; GM, grey matter; PP, primary progressive; MR, magnetic resonance; MS, multiple sclerosis; RR, relapsing-remitting; SDMT, Symbol Digit Modalities Test; SP, secondary progressive; WM, white matter. aIndependent samples t-test. bSpecific disease-modifying therapies include: nine dimethyl fumarate, eight glatiramer acetate, five fingolimod, five ocrelizumab, three interferon beta (1a/1b), two natalizumab, two rituximab and one teriflunomide. *P-values <0.05 are marked in bold.
The analysis reported in the next three sections focus on the short diffusion time (Δ = 19 ms) data, taking full advantage of the diffusion times accessible using the high-performance gradients on the Connectome scanner.34,47 The last section of the results, referred to as ‘Influence of diffusion time on SANDI metrics’, presents the SANDI analyses using the Δ = 19 ms and Δ = 49 ms data to examine systematically the diffusion time dependence of SANDI metrics in this population and provide results that may be more comparable with what is achievable on current clinical scanners.
Between-group differences in tissue microstructure
Cortical fis was reduced in people with MS compared with HC [F(1,76) = 5.10, P = 0.027, not surviving FDR correction; Table 2]. In deep GM, fis was significantly decreased [F(1,76) = 6.53, Pcorr = 0.039] and fec significantly increased [F(1,76) = 6.43, Pcorr = 0.039] in people with MS compared with HC. Region-specific analysis showed a significantly reduced fis of the caudate and thalamus in people with MS compared with HC [F(1,76) = 13.91, Pcorr = 0.007 and F(1,76) = 7.79, Pcorr = 0.037, respectively]. In addition, fec of the caudate and thalamus were significantly increased, of which only differences in fec of the caudate survived FDR correction [F(1,76) = 12.36, Pcorr = 0.007]. Among all deep GM nuclei, the pallidum was the only deep GM structure to show a significant difference in Rs, which was significantly decreased in people with MS compared with HC [F(1,76) = 10.96, Pcorr = 0.007; Supplementary Table 1]. Based on these results and previous literature suggesting a prominent role of the thalamus and caudate in disease progression and neurodegeneration in MS,24,55 we focused on these two regions in further analyses of SANDI metrics in the deep GM.
Table 2.
Grey-matter SANDI metrics of included participants
| Δ = 19 ms | Δ = 49 ms | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Healthy volunteers | People with MS | Group differences | Healthy volunteers | People with MS | Group differences | |||||
| Mean (SD) | Mean (SD) | F-testa | P-value* | Effect sizeb | Mean (SD) | Mean (SD) | F-testa | P-value* | Effect sizeb | |
| Cortical grey matter | ||||||||||
| fin | 0.17 (0.02) | 0.17 (0.02) | F(1,76) = 1.50 | 0.224 | 0.28 | 0.11 (0.01) | 0.11 (0.01) | F(1,76) = 1.54 | 0.22 | 0.28 |
| fis | 0.58 (0.03) | 0.56 (0.03) | F(1,76) = 5.10 | 0.027 | 0.51 | 0.32 (0.01) | 0.32 (0.02) | F(1,76) = 1.54 | 0.22 | 0.28 |
| fec | 0.25 (0.02) | 0.26 (0.04) | F(1,76) = 2.85 | 0.095 | 0.38 | 0.57 (0.01) | 0.57 (0.02) | F(1,76) = 0.35 | 0.56 | 0.13 |
| Rs, μm | 10.01 (0.08) | 9.99 (0.12) | F(1,76) = 0.75 | 0.390 | 0.20 | 11.04 (0.12) | 10.92 (0.27) | F(1,76) = 5.97 | 0.017 | 0.55 |
| Deep grey matter | ||||||||||
| fin | 0.29 (0.02) | 0.29 (0.02) | F(1,76) = 0.79 | 0.378 | 0.20 | 0.12 (0.01) | 0.12 (0.01) | F(1,76) = 2.02 | 0.16 | 0.32 |
| fis | 0.52 (0.04) | 0.50 (0.03) | F(1,76) = 6.53 | 0.013 | 0.58 | 0.33 (0.01) | 0.33 (0.02) | F(1,76) = 4.43 | 0.039 | 0.48 |
| fec | 0.20 (0.03) | 0.22 (0.03) | F(1,76) = 6.43 | 0.013 | 0.58 | 0.55 (0.01) | 0.55 (0.02) | F(1,76) = 2.35 | 0.13 | 0.35 |
| Rs, μm | 10.20 (0.10) | 10.14 (0.12) | F(1,76) = 4.60 | 0.035 | 0.49 | 10.11 (0.26) | 9.91 (0.28) | F(1,76) = 10.40 | 0.002 | 0.73 |
| Normal-appearing white matter | ||||||||||
| fin | 0.42 (0.03) | 0.39 (0.04) | F(1,76) = 13.06 | 5.40 × 10−4 | 0.82 | 0.27 (0.02) | 0.25 (0.03) | F(1,76) = 14.62 | 2.68 × 10−4 | 0.87 |
| fis | 0.43 (0.03) | 0.44 (0.03) | F(1,76) = 3.63 | 0.061 | 0.43 | 0.26 (0.01) | 0.27 (0.02) | F(1,76) = 7.11 | 0.009 | 0.60 |
| fec | 0.15 (0.02) | 0.16 (0.02) | F(1,76) = 9.34 | 0.003 | 0.69 | 0.46 (0.02) | 0.48 (0.02) | F(1,76) = 10.13 | 0.002 | 0.72 |
| Rs, μm | 10.23 (0.09) | 10.22 (0.08) | F(1,76) = 0.24 | 0.627 | 0.11 | 10.33 (0.17) | 10.36 (0.15) | F(1,76) = 0.41 | 0.53 | 0.14 |
GM SANDI values in healthy volunteers and people with MS are reported as mean (SD), separately for diffusion time Δ = 19 and 49 ms. GM, grey matter; NAWM, normal-appearing white matter; fin, intra-neurite signal fraction; fis, intra-soma signal fraction; fec, extra-cellular signal fraction; Rs, apparent soma radius. aMultivariate general linear model. bEffect sizes of significant changes between groups are based on Hedges’ g. *P-values <0.05 after FDR correction are marked in bold.
Regarding the WM, people with MS showed significantly decreased fin and increased fec in NAWM compared with HC [F(1,76) = 13.06, Pcorr = 0.004 and F(1,76) = 9.34, Pcorr = 0.014, respectively]. In MS, fis was slightly higher in NAWM compared with HC, but this increase did not reach statistical significance at the short diffusion time. Within people with MS, lesional WM showed lower fin, lower fis and higher fec compared with NAWM [t(40) = 10.28, Pcorr = 8.76 × 10−13, t(40) = 13.94, Pcorr = 9.11 × 10−17, and t(40) = −15.52, Pcorr = 4.74 × 10−18, respectively; Supplementary Table 2].
Relationship between alterations in GM microstructure and volume
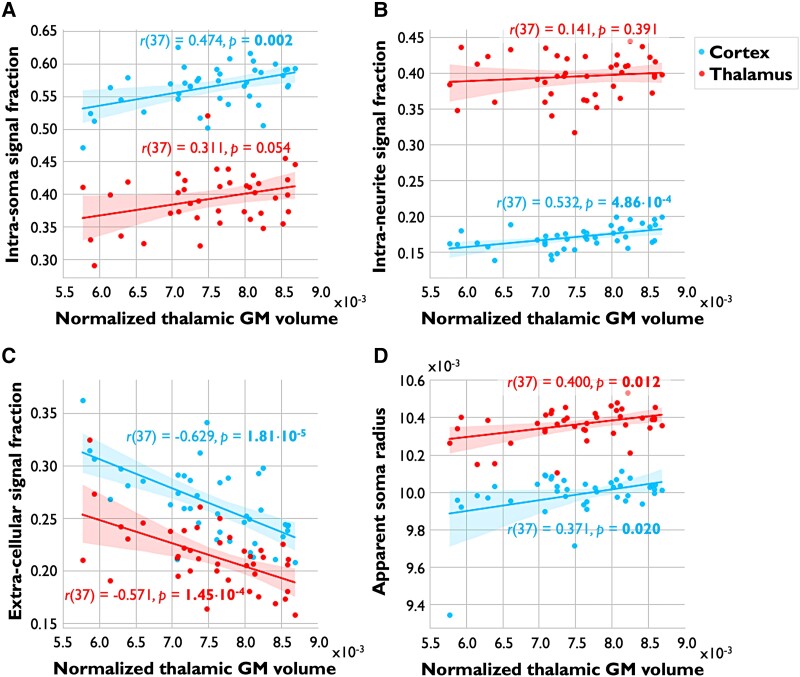
To assess the relationship between MS-related alterations in cell body and neurite signal fractions and GM volume loss, the SANDI metrics that showed significant differences between MS and HC (see Table 2) were correlated with the corresponding normalized GM volume in people with MS. Cortical fis and fec were not associated with normalized cortical volume in people with MS [r(37) = 0.061, P = 0.71, and r(37) = −0.062, P = 0.71, respectively]. In deep GM, people with MS showed a significant association between deep GM fec and deep GM volume [r(37) = 0.481, Pcorr = 0.007]. Focusing on the thalamus, decreased thalamic fis tended to be associated with decreased thalamic volume in people with MS [r(37) = 0.311, P = 0.054]. Normalized thalamic volume was significantly associated with increased thalamic fec [r(37) = −0.571, Pcorr = 0.002] and decreased thalamic Rs [r(37) = 0.400, Pcorr = 0.022; Fig. 2]. In the caudate, we found significant moderate associations between caudate volume and increased fec [r(37) = −0.407, Pcorr = 0.022] and decreased fis and Rs [r(37) = 0.405, Pcorr = 0.022, and r(37) = 0.487, Pcorr = 0.007, respectively; Supplementary Table 3].
Figure 2.
Associations between SANDI metrics and thalamic volume. Imaging metrics in the cortex (blue) and thalamus (red) are shown on the y-axes and normalized thalamic volume on the x-axes in people with MS. Intra-soma (A), intra-neurite (B) and extra-cellular signal fractions (C) and apparent soma radius (D; in μm) are shown in separate plots. Correlation coefficients with corresponding P-values are displayed in representative colours near each plot. P-values <0.05 surviving FDR correction are marked in bold. Values on the y-axis in subplot A, B and C are signal fractions ranging from 0.00 to 1.00, and are therefore unitless. As thalamic volumes were corrected for intra-cranial volume, yielding normalized volume measures, values on the x-axis are unitless as well. GM, grey matter.
The thalamus is known to be a major relay nucleus with extensive cortical anatomic connections. Axonal transection within WM lesions causes disconnections in these tracts projecting into and out of the cortex as well as the thalamus, likely contributing to the reduction in thalamic volume seen on histopathology.19 Thalamic volume loss may therefore reflect the net accumulation of MS-related neurodegenerative damage throughout the entire cortex. To assess the relationship between cortical and thalamic neurodegeneration in people with MS using the novel microstructural metrics presented here, normalized thalamic GM volumes were correlated with cortical SANDI metrics. MS-related cortical atrophy was not associated with normalized thalamic volume [r(37) = 0.264, P = 0.105]. However, thalamic volume was significantly associated with cortical fis, fin and Rs [r-range(37) = 0.371–0.532, Pcorr-range = 0.002–0.050] and inversely associated with cortical fec [r(37) = −0.629, Pcorr = 1.80 × 10−4; Fig. 2].
Relationship between GM microstructure and clinical disability
Trends in SANDI metrics in cortical and deep GM were analysed by disease subtype, i.e. in people with RRMS and PMS. Compared with HC, people with RRMS as well as people with PMS showed similar patterns in SANDI metrics in both GM and WM, with larger effect sizes in PMS (Supplementary Table 4). SANDI metrics in cortex, deep GM and WM as well as normalized GM volumes did not differ between MS subgroups. Both people with RRMS and PMS showed similar associations between normalized GM volumes and SANDI metrics as in the total MS cohort, but with stronger correlations seen in PMS (Supplementary Table 3).
In people with MS, SANDI metrics showing significant differences between MS and HC were correlated with clinical characteristics and disability measures. Older age was significantly associated with decreased fis in cortical GM and NAWM [r(38) = −0.418, Pcorr = 0.042, and r(38) = −0.446, Pcorr = 0.042, respectively]. Increased EDSS as indicator of disease severity was associated with decreased cortical fis and increased fec [r(39) = −0.319, Pcorr = 0.042 and r(39) = 0.327, Pcorr = 0.042, respectively]. SDMT did not show significant associations with SANDI metrics in either GM or WM (Supplementary Table 5).
Influence of diffusion time on SANDI metrics
The main contributors to the total signal fraction in the cortex and deep GM shifted from predominantly fis at Δ = 19 ms (∼0.53 at Δ = 19 ms and ∼0.33 at Δ = 49 ms) to fec at Δ = 49 ms (∼0.24 at Δ = 19 ms and ∼0.56 at Δ = 49 ms) and in NAWM from mainly fin and fis at Δ = 19 ms (∼0.41 at Δ = 19 ms and ∼0.26 at Δ = 49 ms) to fec at Δ = 49 ms (0.16 at Δ = 19 ms and 0.48 at Δ = 49 ms; Table 2). At Δ = 49 ms, between-group alterations in cortical and deep GM SANDI metrics were less apparent with lower effect sizes, except for Rs. Rs showed significant reduction in people with MS in both the cortex and deep GM at Δ = 49 ms [F(1,76) = 5.97, Pcorr = 0.041 and F(1,76) = 10.40, Pcorr = 0.008, respectively] when compared with Δ = 19 ms.
In NAWM, signal fractions at Δ = 49 ms showed MS-related alterations to a similar extent as at Δ = 19 ms, i.e. fin being increased in MS compared with HC [F(1,76) = 13.06, Pcorr = 0.006 at Δ = 19 ms and F(1,76) = 14.62, Pcorr = 0.003 at Δ = 49 ms] and fec being decreased [F(1,76) = 9.34, Pcorr = 0.018 at Δ = 19 ms and F(1,76) = 10.13, Pcorr = 0.008 at Δ = 49 ms; Table 2]. Regarding correlates of GM volume, thalamic atrophy was associated with cortical microstructure at Δ = 49 ms to the same extent as at Δ = 19 ms (Supplementary Table 6).
Discussion
In this work, we evaluated the sensitivity of SANDI, a novel compartment model for GM, for microstructural tissue alterations in cortical and deep GM in people with MS compared with HC and evaluated these changes relative to GM atrophy and MS phenotype. We fitted the SANDI model to high-gradient diffusion MRI data acquired with gradient strengths up to 300 mT/m, which enabled measurements at a relatively short diffusion time to avoid the confounds of inter-compartmental water exchange, as advised in the original implementation of SANDI.33 Our cohort of people with MS showed trends towards decreased fis in both the cortex and deep GM, specifically in the caudate and thalamus, compared with HC. Lower cortical fis and fin were significantly associated with thalamic volume loss in MS.
SANDI has been proposed as a promising biophysical model that captures microstructural features in GM,33,47,56 discriminating not only axonal and dendritic processes as captured by other diffusion models but also cell bodies.33 The application of SANDI to high-gradient diffusion MRI data in MS enabled us to probe alterations in the density of cell bodies and cellular processes corresponding to heterogeneous and complex pathological processes in GM and WM. At a short diffusion time, our cortical SANDI estimates in healthy cortex are in good agreement with previous SANDI work in healthy mouse and human brains.27,33,34,56 Jelescu et al.34 showed cortical fis tends to decrease with increasing diffusion time, which is also reflected in our data, showing a lower cortical fis at Δ = 49 ms. The same applies to the slight increase in cortical Rs estimates with longer diffusion time, consistent with the analyses on diffusion time dependence and exchange by Jelescu et al.34 Our healthy WM estimates at Δ = 49 ms tended to be lower and more comparable with previous high-gradient SANDI work in mouse cortex than our estimates at Δ = 19 ms.27 A recent study by Margoni et al.56 evaluated the SANDI model in people with MS using diffusion data acquired at a longer diffusion time (Δ = 46.9 ms) on a clinical MRI scanner with maximum gradient strength of 80 mT/m. Their reported estimates in WM and GM are in line with our estimates at Δ = 49 ms.56
Cortical atrophy accompanying neuronal and axonal degeneration and demyelination represents one of the most reliable predictors of clinical disability progression in MS.16,57,58 Our findings indicate that microstructural DW imaging using high-performance gradients captures alterations in GM composition at the mesoscopic scale. This may offer a novel predictive biomarker of disability progression beyond cerebral volume loss, which would be expected to postdate neuronal loss and may limit its sensitivity. It highlights the importance of developing and validating imaging markers that are temporally sensitive to the earliest effects of neurodegeneration and response to neuroprotective therapies. In our cohort, people with MS exhibited significant volume loss of GM structures with the exception of the caudate and pallidum, with the largest effect sizes observed in the cortex (g = 1.47) and thalamus (g = 1.10). People with MS also showed lower GM fis compared with HC, whereas the GM fin did not differ, indicating a potential limitation of current DW microstructural models that only account for neurites, e.g. NODDI. Microstructural alterations in the cortex were overall less evident than in deep GM structures compared with HC, i.e. lower effect sizes of changes in SANDI metrics in the cortex compared with deep GM and fewer changes surviving FDR correction. The cortex is known to possess a diversity of cell sizes, density and distribution and is characterized by spatial variations in MS-related atrophy across cortical areas.16,59,60 Histologically, demyelinated cortex shows a reduced cell density of up to 20% compared with NA cortex and seems more extensive in progressive disease with long disease duration.61 Hence, fis would be expected to be more clearly decreased in people with PMS, as suggested by the results found by Margoni et al.56 They showed a significant decrease in both cortical fis and fin in PMS compared with HC and RRMS,56 suggesting that these cortical SANDI metrics are sensitive to cortical pathology in more advanced stages of the disease, highlighting the promise and potential of this method in detecting neuronal and axonal loss in progressive disease. The heterogeneity of cellular composition and pathologic involvement across the cortex may have diminished our sensitivity to MS-related alterations in the cortex within our RRMS-predominant cohort using SANDI; nevertheless, we were able to detect trends of increased cortical fis in MS compared with HC and associations between cortical fis and deep GM volume loss in MS that point to the promise of microstructural DW imaging in identifying GM microstructural alterations early in the disease course. Larger longitudinal studies using high-resolution and high b-value imaging are needed to better characterize the alterations in cortical microstructure throughout the disease course.
In deep GM, decreases in fis were driven by reduced fis in the thalamus and caudate—regions that are known for a disproportionate degree of MS-specific atrophy over time.24 Interestingly, our population of predominantly RRMS did not show significant volume loss of the caudate compared with HC, whereas microstructural changes in cell body density were clearly detectable. Based on histology, deep GM demyelination is especially prominent in the caudate and medial and anterior thalamic nuclei, and these regions show significantly reduced neuronal density in non-demyelinated, i.e. NA GM, and to a greater extent in demyelinated GM.62,63 These findings support the observed reductions in fis, particularly in the caudate and thalamus. Studying these microstructural changes in the light of neurodegeneration, thalamic volume loss only trended towards association with decreased thalamic fis in people with MS, whereas it was significantly positively correlated with thalamic Rs. Recent histopathological observations by Mahajan et al.64 in a predominantly PMS cohort found that the loss of neurons in the thalamus did not occur in proportion to loss of thalamic volume. Comparing neuronal density in the thalamic nuclei with the least and most volume, Mahajan et al.64 found an overall reduction in neuronal density of 17.6% in the nuclei with the least volume, suggesting neurons were lost in excess to volume loss. This could explain the absence of a clear association between thalamic neuronal density, i.e. fis, and thalamic volume in our study. Interestingly, Mahajan et al.64 also found that excessive neuronal loss seemed to be driven by a decrease in density of smaller neurons (200–400 μm2) instead of larger neurons (>400 μm2), suggesting a preferential loss of smaller inter-neurons relative to the larger relay neurons. Theoretically, this would indicate a relative increase in the radius of the cell bodies in a voxel within the thalamus with declining thalamic volume, yielding a negative correlation between Rs and thalamic volume. Conversely, Vercellino et al.62 studied the inflammatory and neurodegenerative changes in deep GM in a population consisting of ∼50% RRMS patients and found reductions in neuronal size in both the caudate and thalamus that were associated with longer disease duration in people with MS. Given the high degree of MS-specific atrophy over time in the caudate and thalamus,24 these findings would suggest a lower neuronal radius, Rs, with declining deep GM volumes, in line with our observations of a positive correlation between thalamic and caudate volume and their respective Rs. Previous high-gradient diffusion MRI measurements using the Connectome scanner fitted to a multi-compartment model of axonal diameter revealed a clinically relevant increase in apparent axon diameter in the corpus callosum of people with MS,65 suggesting that information on axonal and cellular size and density may provide added information beyond what can be gleaned from conventional diffusion tensor imaging. Nevertheless, for a detailed comparison of Rs and cellular size, further experiments comparing MRI and histology are needed.
The observed relationship between thalamic microstructure and volume loss suggests thalamic SANDI metrics, specifically Rs, might be relevant surrogate measures of irreversible thalamic tissue loss. Interestingly, thalamic volume loss in people with MS was not only related to tissue microstructure in the thalamus itself but also in the cortex. Thus, cortical microstructural changes, as reflected here by decreased fis, might occur in proportion to thalamic volume loss, which has been shown to be a marker of neurodegeneration in the early stages of MS.16 Current results would therefore support the theory that thalamic volume loss reflects the net accumulation of MS-related neurodegenerative damage throughout the brain, making it a sensitive and appealing biomarker in people with MS. The cross-sectional study design limits us in drawing bold conclusions about causality and the sequential order of events, which warrants longitudinal studies. The absence of a correlation between cortical SANDI metrics and cortical volume could also be due to the contribution of other cortical pathology associated with heterogeneous cell populations, e.g. within demyelinated cortex, as described above, thereby confounding the expected correlation between cortical fis and volume loss. Also, in contrast to deep GM, we studied the cortex as a whole, instead of focusing on specific parts of the cortex, e.g. the precuneus and cingulate gyrus, which are known to show early atrophy.16 Future work that evaluates the cortex on a regional basis may strengthen the association between cortical volume loss and SANDI metrics.
In this study, assessment of the WM demonstrated that people with MS showed significantly decreased fin and increased fec in NAWM when compared with HC especially in progressive phenotypes; these findings are in line with those reported by other recent studies.56 Histopathological studies support these results and have shown diffuse axonal damage in the NAWM of people with MS.7,66 We did not observe significant MS-related changes in NAWM fis estimates, as seen in GM, consistent with prior studies.56 The absence of a significant change in fis in NAWM might be explained by the co-existence of an inflammatory process including microglial activation and neurodegenerative processes including loss of oligodendrocytes,66,67 counterbalancing each other’s effect on fis. In MS, lesional WM showed significantly lower fin and fis compared with NAWM, which is consistent with previous literature.56,66 After the acute phase of inflammation dissipates, inactive WM lesions are characterized by severe demyelination and axonal loss, and there is near-complete absence of macrophages and microglia.66 This corresponds with current findings of a lower cell body density, i.e. lower fis, and fewer axons and dendrites, i.e. lower fin. Further, histopathological evidence suggests that diffuse axonal damage and chronic active WM lesions are more profound in people with PMS than in acute or relapsing disease.7,67,68 In this study, the degree of microstructural loss (i.e. decrease in NAWM fin and lesional fin and fis) between PMS and RRMS was not significantly different, which may be due to the small sample size of our cohort. Nevertheless, current findings support the hypothesis that a similar neuropathological process occurs in RRMS and PMS as a continuum in the disease course.56,69
Finally, we assessed the relevance of SANDI metrics in relation to clinical disability. We did not find significant differences in SANDI metrics nor GM volumes between people with RRMS and PMS, likely due the small size of our PMS cohort. However, we found significant associations between EDSS as an indicator of disease severity and SANDI metrics in cortical GM, even in this relatively mildly affected MS cohort (median EDSS of 2.5). Our findings are in good agreement with the findings of Margoni et al.,56 whose MS cohort consisted of more severely disabled people with MS with a median EDSS of 6.5. Our results support the hypothesis that loss of microstructural integrity occurs early in the disease, tracks closely with disability status and becomes more pronounced as disability worsens.
Our study is limited by its cross-sectional study design and small sample size. Furthermore, in this study, we did not evaluate cortical lesions due to the limited set of contrasts and spatial resolution available at 3 T sensitive to cortical demyelination. A regional evaluation to cortical pathology in relationship to cortical SANDI metrics is an important follow-up study and may increase the effect size of alterations compared with healthy individuals. Another limitation raises from the potential bias in the SANDI results due to partial volume effects, in particular with the cerebrospinal fluid. Although we tried to limit this bias by generating partial volume-weighted microstructural maps, partial volume effects might not be fully eliminated, especially given the resolution of the DW images. Future work might harness free-water elimination methods to mitigate this bias. In our study, we analysed the magnitude DW images, which were available in all participants. Fitting the SANDI model to magnitude DW images may incur a bias in the estimation of intra-neurite signal fraction when compared with real-valued DW images due to the Rician noise distribution of magnitude data.27 Given that our main findings centred on the relationship between thalamic volume and cortical intra-cellular signal fraction, which is less affected by the Rician noise bias, we decided to make use of the data at our disposal and analyse the magnitude data in this study. Future work will focus on systematic examination of the differences in parameter estimation between magnitude and real-valued DW images in the subset of people of MS who had real-valued data available. The SANDI model has not yet been fully histopathologically validated in MS, having only been studied in a combined pathological MRI study evaluating the spinal cord of two people with MS and two non-neurological participants at 9.4 T.70 Both fin and fis showed significant correlations with the intensities of myelin and astrocyte staining, respectively. Further histopathological validation of the SANDI model in GM and WM in MS is needed to gain a better understanding of contributors to the derived metrics at different disease stages and within different tissues.
Assessment of neurodegeneration involving GM is clinically relevant and important for advancement of clinical trials. GM atrophy is widely used in clinical trials as an outcome measure for treatment response. However, the absolute rates for volume loss in the MS brain are small,71 requiring large sample sizes to demonstrate and complicated by the intra-subject variability in brain volume measures.72 This has hindered the uniform application of GM atrophy as an imaging biomarker in clinical and research settings. The increasing availability of high-performance gradients on clinical 3 T MRI scanners41,73,74 underscores the potential of advanced diffusion MRI models like SANDI for uncovering the microstructural substrate of neurodegeneration in MS and other neurological disorders, with the expectation that this would occur prior to GM volume loss. Further investigations of GM pathology using high-performance gradients are timely and necessary to assess the potential of DW microstructural metrics as highly sensitive measures of neuroprotection in clinical trials.
Conclusion
Our study demonstrates that SANDI metrics obtained using high-gradient diffusion MRI are sensitive to cellular changes in the cortex and deep GM in the MS brain compared with healthy tissue, providing a detailed GM characterization that track with cortical and thalamic volumes. Application of in vivo biomarkers to detect early cellular loss could provide an advantage over assessment of volumetric changes in development of neuroprotective therapy in MS.
Supplementary Material
Abbreviations
- DW =
diffusion-weighted
- EDSS =
Expanded Disability Status Scale
- f ec =
extra-cellular signal fractions
- f in =
intra-neurite signal fractions
- f is =
intra-soma signal fractions
- FDR =
false discovery rate
- FLAIR =
fluid-attenuated inversion recovery
- GM =
grey matter
- HC =
healthy control
- MEMPRAGE =
multi-echo magnetization prepared rapid acquisition gradient echo
- MS =
multiple sclerosis
- NA =
normal-appearing
- PMS =
progressive multiple sclerosis
- R s =
apparent soma radius
- RRMS =
relapsing-remitting multiple sclerosis
- SAMSEG =
Sequence Adaptive Multimodal SEGmentation
- SANDI =
soma and neurite density imaging
- SDMT =
Symbol Digit Modalities Test
- TE =
echo time
- TI =
inversion time
- TR =
repetition time
- WM =
white matter
Contributor Information
Eva A Krijnen, Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114, USA; MS Center Amsterdam, Anatomy and Neurosciences, Amsterdam Neuroscience, Amsterdam UMC location VUmc, 1081 HV Amsterdam, The Netherlands.
Andrew W Russo, Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114, USA.
Elsa Salim Karam, Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114, USA.
Hansol Lee, Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA 02129, USA.
Florence L Chiang, Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA 02129, USA.
Menno M Schoonheim, MS Center Amsterdam, Anatomy and Neurosciences, Amsterdam Neuroscience, Amsterdam UMC location VUmc, 1081 HV Amsterdam, The Netherlands.
Susie Y Huang, Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA 02129, USA.
Eric C Klawiter, Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114, USA.
Supplementary material
Supplementary material is available at Brain Communications online.
Funding
This work was supported by The McCourt Foundation and National Institutes of Health under grant numbers P41-EB015896, P41-EB030006, U01-EB026996, R01-NS118187 and K23-NS096056.
Competing interests
E.A.K., A.W.R., E.S.K., H.L. and F.L.C. report no conflicts of interest. M.M.S. serves on the editorial board of Neurology and Frontiers in Neurology, receives research support from the Dutch MS Research Foundation, Eurostars-EUREKA, ARSEP, Amsterdam Neuroscience, MAGNIMS and ZonMW and has served as a consultant for or received research support from Atara Biotherapeutics, Biogen, Celgene/Bristol Meyers Squibb, Genzyme, MedDay and Merck. S.Y.H. has received consulting fees and research grants from Siemens Healthineers. E.C.K. has received consulting fees from Banner Life Sciences, Galen/Atlantica, Genentech, Greenwich Biosciences and OM1, and research funds from Abbvie, Biogen and Genentech.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- 1. Rahmanzadeh R, Lu PJ, Barakovic M, et al. Myelin and axon pathology in multiple sclerosis assessed by myelin water and multi-shell diffusion imaging. Brain. 2021;144(6):1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Solana E, Martinez-Heras E, Montal V, et al. Regional grey matter microstructural changes and volume loss according to disease duration in multiple sclerosis patients. Sci Rep. 2021;11(1):16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eshaghi A, Prados F, Brownlee WJ, et al. Deep gray matter volume loss drives disability worsening in multiple sclerosis. Ann Neurol. 2018;83(2):210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Angelis F, Connick P, Parker RA, et al. Amiloride, fluoxetine or riluzole to reduce brain volume loss in secondary progressive multiple sclerosis: The MS-SMART four-arm RCT. Efficacy Mech Eval. 2020;7(3). [PubMed] [Google Scholar]
- 5. Chataway J, Schuerer N, Alsanousi A, et al. Effect of high-dose simvastatin on brain atrophy and disability in secondary progressive multiple sclerosis (MS-STAT): A randomised, placebo-controlled, phase 2 trial. Lancet. 2014;383(9936):2213–2221. [DOI] [PubMed] [Google Scholar]
- 6. Geurts JJ, Calabrese M, Fisher E, Rudick RA. Measurement and clinical effect of grey matter pathology in multiple sclerosis. Lancet Neurol. 2012;11(12):1082–1092. [DOI] [PubMed] [Google Scholar]
- 7. Filippi M, Brück W, Chard D, et al. Association between pathological and MRI findings in multiple sclerosis. Lancet Neurol. 2019;18(2):198–210. [DOI] [PubMed] [Google Scholar]
- 8. Fisher E, Lee JC, Nakamura K, Rudick RA. Gray matter atrophy in multiple sclerosis: A longitudinal study. Ann Neurol. 2008;64(3):255–265. [DOI] [PubMed] [Google Scholar]
- 9. Lucchinetti CF, Popescu BF, Bunyan RF, et al. Inflammatory cortical demyelination in early multiple sclerosis. N Engl J Med. 2011;365(23):2188–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Louapre C, Govindarajan ST, Giannì C, et al. Beyond focal cortical lesions in MS: An in vivo quantitative and spatial imaging study at 7 T. Neurology. 2015;85(19):1702–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friese MA, Schattling B, Fugger L. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat Rev Neurol. 2014;10(4):225–238. [DOI] [PubMed] [Google Scholar]
- 12. Dutta R, Trapp BD. Pathogenesis of axonal and neuronal damage in multiple sclerosis. Neurology. 2007;68(22 Suppl 3):S22–S31; discussion S43-S54. [DOI] [PubMed] [Google Scholar]
- 13. Kutzelnigg A, Lucchinetti CF, Stadelmann C, et al. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain. 2005;128(Pt 11):2705–2712. [DOI] [PubMed] [Google Scholar]
- 14. Bitsch A, Schuchardt J, Bunkowski S, Kuhlmann T, Brück W. Acute axonal injury in multiple sclerosis. Correlation with demyelination and inflammation. Brain. 2000;123(Pt 6):1174–1183. [DOI] [PubMed] [Google Scholar]
- 15. Frischer JM, Bramow S, Dal-Bianco A, et al. The relation between inflammation and neurodegeneration in multiple sclerosis brains. Brain. 2009;132(Pt 5):1175–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eshaghi A, Marinescu RV, Young AL, et al. Progression of regional grey matter atrophy in multiple sclerosis. Brain. 2018;141(6):1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ontaneda D, Raza PC, Mahajan KR, et al. Deep grey matter injury in multiple sclerosis: A NAIMS consensus statement. Brain. 2021;144(7):1974–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bakshi R, Czarnecki D, Shaikh ZA, et al. Brain MRI lesions and atrophy are related to depression in multiple sclerosis. Neuroreport. 2000;11(6):1153–1158. [DOI] [PubMed] [Google Scholar]
- 19. Cifelli A, Arridge M, Jezzard P, Esiri MM, Palace J, Matthews PM. Thalamic neurodegeneration in multiple sclerosis. Ann Neurol. 2002;52(5):650–653. [DOI] [PubMed] [Google Scholar]
- 20. Houtchens MK, Benedict RH, Killiany R, et al. Thalamic atrophy and cognition in multiple sclerosis. Neurology. 2007;69(12):1213–1223. [DOI] [PubMed] [Google Scholar]
- 21. Azevedo CJ, Cen SY, Khadka S, et al. Thalamic atrophy in multiple sclerosis: A magnetic resonance imaging marker of neurodegeneration throughout disease. Ann Neurol. 2018;83(2):223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zivadinov R, Havrdová E, Bergsland N, et al. Thalamic atrophy is associated with development of clinically definite multiple sclerosis. Radiology. 2013;268(3):831–841. [DOI] [PubMed] [Google Scholar]
- 23. Shiee N, Bazin PL, Zackowski KM, et al. Revisiting brain atrophy and its relationship to disability in multiple sclerosis. PLoS One. 2012;7(5):e37049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Azevedo CJ, Cen SY, Jaberzadeh A, Zheng L, Hauser SL, Pelletier D. Contribution of normal aging to brain atrophy in MS. Neurol Neuroimmunol Neuroinflamm. 2019;6(6):e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andravizou A, Dardiotis E, Artemiadis A, et al. Brain atrophy in multiple sclerosis: Mechanisms, clinical relevance and treatment options. Auto Immun Highlights. 2019;10(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ong HH, Wright AC, Wehrli SL, et al. Indirect measurement of regional axon diameter in excised mouse spinal cord with q-space imaging: Simulation and experimental studies. Neuroimage. 2008;40(4):1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ianuş A, Carvalho J, Fernandes FF, et al. Soma and Neurite Density MRI (SANDI) of the in-vivo mouse brain and comparison with the Allen Brain Atlas. Neuroimage. 2022;254:119135. [DOI] [PubMed] [Google Scholar]
- 28. Barazany D, Basser PJ, Assaf Y. In vivo measurement of axon diameter distribution in the corpus callosum of rat brain. Brain. 2009;132(Pt 5):1210–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McNab JA, Edlow BL, Witzel T, et al. The Human Connectome Project and beyond: Initial applications of 300 mT/m gradients. Neuroimage. 2013;80:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang SY, Tian Q, Fan Q, et al. High-gradient diffusion MRI reveals distinct estimates of axon diameter index within different white matter tracts in the in vivo human brain. Brain Struct Funct. 2020;225(4):1277–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang SY, Tobyne SM, Nummenmaa A, et al. Characterization of axonal disease in patients with multiple sclerosis using high-gradient-diffusion MR imaging. Radiology. 2016;280(1):244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fan Q, Nummenmaa A, Witzel T, et al. Axon diameter index estimation independent of fiber orientation distribution using high-gradient diffusion MRI. Neuroimage. 2020;222:117197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palombo M, Ianus A, Guerreri M, et al. SANDI: A compartment-based model for non-invasive apparent soma and neurite imaging by diffusion MRI. Neuroimage. 2020;215:116835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jelescu IO, de Skowronski A, Geffroy F, Palombo M, Novikov DS. Neurite Exchange Imaging (NEXI): A minimal model of diffusion in gray matter with inter-compartment water exchange. Neuroimage. 2022;256:119277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olesen JL, Østergaard L, Shemesh N, Jespersen SN. Diffusion time dependence, power-law scaling, and exchange in gray matter. Neuroimage. 2022;251:118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan Q, Eichner C, Afzali M, et al. Mapping the human connectome using diffusion MRI at 300 mT/m gradient strength: Methodological advances and scientific impact. Neuroimage. 2022;254:118958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vachha B, Huang SY. MRI with ultrahigh field strength and high-performance gradients: Challenges and opportunities for clinical neuroimaging at 7 T and beyond. Eur Radiol Exp. 2021;5(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ngamsombat C, Tian Q, Fan Q, et al. Axonal damage in the optic radiation assessed by white matter tract integrity metrics is associated with retinal thinning in multiple sclerosis. Neuroimage Clin. 2020;27:102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu F, Fan Q, Tian Q, et al. Imaging G-ratio in multiple sclerosis using high-gradient diffusion MRI and macromolecular tissue volume. AJNR Am J Neuroradiol. 2019;40(11):1871–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keil B, Blau JN, Biber S, et al. A 64-channel 3 T array coil for accelerated brain MRI. Magn Reson Med. 2013;70(1):248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tian Q, Fan Q, Witzel T, et al. Comprehensive diffusion MRI dataset for in vivo human brain microstructure mapping using 300 mT/m gradients. Sci Data. 2022;9(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fan Q, Witzel T, Nummenmaa A, et al. MGH-USC Human Connectome Project datasets with ultra-high b-value diffusion MRI. Neuroimage. 2016;124(Pt B):1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: Application to diffusion tensor imaging. Neuroimage. 2003;20(2):870–888. [DOI] [PubMed] [Google Scholar]
- 44. Andersson JLR, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. [DOI] [PubMed] [Google Scholar]
- 46. Daducci A, Canales-Rodríguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran JP. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage. 2015;105:32–44. [DOI] [PubMed] [Google Scholar]
- 47. Ianus A, Alexander DC, Zhang H, Palombo M. Mapping complex cell morphology in the grey matter with double diffusion encoding MR: A simulation study. Neuroimage. 2021;241:118424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lindemer ER, Salat DH, Smith EE, et al. White matter signal abnormality quality differentiates mild cognitive impairment that converts to Alzheimer’s disease from nonconverters. Neurobiol Aging. 2015;36(9):2447–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Govindarajan KA, Datta S, Hasan KM, et al. Effect of in-painting on cortical thickness measurements in multiple sclerosis: A large cohort study. Hum Brain Mapp. 2015;36(10):3749–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith RE, Tournier JD, Calamante F, Connelly A. Anatomically-constrained tractography: Improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage. 2012;62(3):1924–1938. [DOI] [PubMed] [Google Scholar]
- 51. Smith R, Skoch A, Bajada CJ, Caspers S, Connelly A. Hybrid surface-volume segmentation for improved anatomically-constrained tractography. In: Proceedings of the Organisation for Human Brain Mapping. Virtual, June 23-July 3, 2020. Abstract Nr 1280.
- 52. Puonti O, Iglesias JE, Van Leemput K. Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling. Neuroimage. 2016;143:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Iglesias JE, Insausti R, Lerma-Usabiaga G, et al. A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology. Neuroimage. 2018;183:314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nourbakhsh B, Azevedo C, Maghzi AH, Spain R, Pelletier D, Waubant E. Subcortical grey matter volumes predict subsequent walking function in early multiple sclerosis. J Neurol Sci. 2016;366:229–233. [DOI] [PubMed] [Google Scholar]
- 56. Margoni M, Pagani E, Preziosa P, et al. In vivo quantification of brain soma and neurite density abnormalities in multiple sclerosis. J Neurol. 2023; 270(1):433–445. [DOI] [PubMed] [Google Scholar]
- 57. Popescu V, Klaver R, Voorn P, et al. What drives MRI-measured cortical atrophy in multiple sclerosis? Mult Scler. 2015;21(10):1280–1290. [DOI] [PubMed] [Google Scholar]
- 58. Eijlers AJC, van Geest Q, Dekker I, et al. Predicting cognitive decline in multiple sclerosis: A 5-year follow-up study. Brain. 2018;141(9):2605–2618. [DOI] [PubMed] [Google Scholar]
- 59. Jeon T, Mishra V, Uh J, et al. Regional changes of cortical mean diffusivities with aging after correction of partial volume effects. Neuroimage. 2012;62(3):1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fukutomi H, Glasser MF, Zhang H, et al. Neurite imaging reveals microstructural variations in human cerebral cortical gray matter. Neuroimage. 2018;182:488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vercellino M, Plano F, Votta B, Mutani R, Giordana MT, Cavalla P. Grey matter pathology in multiple sclerosis. J Neuropathol Exp Neurol. 2005;64(12):1101–1107. [DOI] [PubMed] [Google Scholar]
- 62. Vercellino M, Masera S, Lorenzatti M, et al. Demyelination, inflammation, and neurodegeneration in multiple sclerosis deep gray matter. J Neuropathol Exp Neurol. 2009;68(5):489–502. [DOI] [PubMed] [Google Scholar]
- 63. Haider L, Simeonidou C, Steinberger G, et al. Multiple sclerosis deep grey matter: The relation between demyelination, neurodegeneration, inflammation and iron. J Neurol Neurosurg Psychiatry. 2014;85(12):1386–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mahajan KR, Nakamura K, Cohen JA, Trapp BD, Ontaneda D. Intrinsic and extrinsic mechanisms of thalamic pathology in multiple sclerosis. Ann Neurol. 2020;88(1):81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang SY, Fan Q, Machado N, et al. Corpus callosum axon diameter relates to cognitive impairment in multiple sclerosis. Ann Clin Transl Neurol. 2019;6(5):882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kuhlmann T, Ludwin S, Prat A, Antel J, Brück W, Lassmann H. An updated histological classification system for multiple sclerosis lesions. Acta Neuropathol. 2017;133(1):13–24. [DOI] [PubMed] [Google Scholar]
- 67. Kutzelnigg A, Lassmann H. Cortical demyelination in multiple sclerosis: A substrate for cognitive deficits? J Neurol Sci. 2006;245(1-2):123–126. [DOI] [PubMed] [Google Scholar]
- 68. Luchetti S, Fransen NL, van Eden CG, Ramaglia V, Mason M, Huitinga I. Progressive multiple sclerosis patients show substantial lesion activity that correlates with clinical disease severity and sex: A retrospective autopsy cohort analysis. Acta Neuropathol. 2018;135(4):511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Correale J, Gaitán MI, Ysrraelit MC, Fiol MP. Progressive multiple sclerosis: From pathogenic mechanisms to treatment. Brain. 2017;140(3):527–546. [DOI] [PubMed] [Google Scholar]
- 70. Palombo M, Grussu F, Schneider T, et al. New potential MRI markers of glial scarring and tissue damage in multiple sclerosis spinal cord pathology using diffusion MRI. In: Proceedings of the International Society for Magnetic Resonance in Medicine. Virtual, August 8-14, 2020. Abstract Nr 1184.
- 71. Popescu V, Agosta F, Hulst HE, et al. Brain atrophy and lesion load predict long term disability in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2013;84(10):1082–1091. [DOI] [PubMed] [Google Scholar]
- 72. Rocca MA, Battaglini M, Benedict RH, et al. Brain MRI atrophy quantification in MS: From methods to clinical application. Neurology. 2017;88(4):403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Foo TKF, Tan ET, Vermilyea ME, et al. Highly efficient head-only magnetic field insert gradient coil for achieving simultaneous high gradient amplitude and slew rate at 3.0 T (MAGNUS) for brain microstructure imaging. Magn Reson Med. 2020;83(6):2356–2369. [DOI] [PubMed] [Google Scholar]
- 74. Huang S, Rosen B, Paech D. Life at the edge—Exploring the limits of our “fields”. Editorial comment RSNA edition. MAGNETOM Flash. 2022;82:4-10.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.