Abstract
Sexual selection and sexual antagonism are important drivers of eco-evolutionary processes. The evolution of traits shaped by these processes depends on their genetic architecture, which remains poorly studied. Here, implementing a quantitative genetics approach using diallel crosses of the bulb mite, Rhizoglyphus robini, we investigated the genetic variance that underlies a sexually selected weapon that is dimorphic among males and female fecundity. Previous studies indicated that a negative genetic correlation between these two traits likely exists. We found male morph showed considerable additive genetic variance, which is unlikely to be explained solely by mutation-selection balance, indicating the likely presence of large-effect loci. However, a significant magnitude of inbreeding depression also indicates that morph expression is likely to be condition-dependent to some degree and that deleterious recessives can simultaneously contribute to morph expression. Female fecundity also showed a high degree of inbreeding depression, but the variance in female fecundity was mostly explained by epistatic effects, with very little contribution from additive effects. We found no significant genetic correlation, nor any evidence for dominance reversal, between male morph and female fecundity. The complex genetic architecture underlying male morph and female fecundity in this system has important implications for our understanding of the evolutionary interplay between purifying selection and sexually antagonistic selection.
Keywords: quantitative genetics, diallel, dimorphism, genetic architecture, dominance reversal, condition-dependence
Introduction
The nature of genetic variation segregating in natural populations is of considerable interest in evolutionary genetics as standing genetic variance constitutes a major source of a population’s short-term evolutionary potential (Barrett & Schluter, 2008; Barton & Keightley, 2002). Understanding the nature of genetic variation has also been of particular importance in the development of sexual selection theory, as it forms the basis of hypotheses explaining indirect genetic benefits of female mating preferences for costly and exaggerated male signaling structures (Andersson, 1986; Rowe & Houle, 1996), and the phenotypic variation that exists in traits which are used during male contest competition (Berglund et al., 1996). This phenotypic variation may take the shape of discontinuous or discrete expression of the traits that mediate sexual competition and the evolution of alternative reproductive phenotypes adopting different reproductive strategies (Brockmann, 2001; Gross, 1996; Gross & Repka, 1998; Shuster & Wade, 2003; Sinervo & Lively, 1996; Tomkins & Hazel, 2007). Such alternative reproductive phenotypes are found in a wide diversity of taxa (Oliveira et al., 2008) and are hypothesized to exist due to the costliness of these traits where only males of high “quality” (Zahavi, 1975) or “status” (Gross, 1996) can pay the costs of expression.
One of the most likely sources of variation in male genetic quality stems from the continuous influx of deleterious mutations that will segregate in populations at a low frequency under mutation-selection balance (Haldane, 1937; Lande, 1975; Lynch et al., 1999). Such deleterious mutations will be spread across the genome (Andersson, 1986), reducing the amount of resources available for an individual to allocate toward fitness-related traits, commonly referred to as an individual’s “condition” (Rowe & Houle, 1996). Condition-dependence of exaggerated sexually selected trait (SST) expression can become an evolutionary stable strategy (Maynard Smith & Price, 1973), when the survival costs of expressing SSTs in poor-condition individuals exceed their reproductive benefits (Grafen, 1990; Gross & Repka, 1998). The evolution of this condition-dependence thus causes SST expression to be informative of male genetic quality. Furthermore, in at least some taxa it underlies the evolution and expression of alternative reproductive phenotypes, where high-condition males develop into aggressive morphs that engage in contest competition over access to females and express disproportionally large and costly SSTs. In contrast, poor-condition males express disproportionally small or have a complete absence of SSTs and adopt non-aggressive, often “sneaky” mating tactics (Gross, 1996). Implicating loci underlying the genetic variance of condition-dependent SSTs has been challenging due to the nature of this variance, i.e., a large number of loci each with individually small effect sizes (Rowe & Houle, 1996). Despite challenges, a number of recent examples appear consistent with this scenario and include the polygenic determination of antlers in red deer, Cervus elaphus (Peters et al., 2022), mating success in Drosophila melanogaster (Dugand et al., 2019) and the discontinuous expression of a sexually selected weapon in the bulb mite, Rhizoglyphus robini (Parrett et al., 2022).
However, some systems do not conform to this polygenic condition-dependence model, with SST expression determined by relatively few genes or even a single gene (or supergene) of large effect (Hendrickx et al., 2022; Johnston et al., 2013; Küpper et al., 2016; Lamichhaney et al., 2016; Shuster & Wade, 1991; Sinervo & Lively, 1996). The maintenance of variation in such systems with large-effect quantitative trait loci (QTLs) is likely a consequence of balancing selection, for example, negative frequency-dependence (Gross, 1991) or Rock–Paper–Scissor games (Sinervo & Lively, 1996) predicted by evolutionary game theory (Maynard Smith, 1982), or antagonistic pleiotropy and life-history trade-offs (Johnston et al., 2013; Mérot et al., 2020). One other possible widespread form of balancing selection may stem from sexual antagonism, which could have both large-effect QTLs or polygenic underpinnings, and sexually antagonistic polymorphisms maintained due to alternative alleles having opposite fitness consequences in each of the sexes (Connallon & Clark, 2012, 2014). A potentially major source of sexual antagonism may stem from the expression of SSTs, where high-fitness males expressing elaborated SSTs sire low-fitness daughters (Harano et al., 2010; Okada et al., 2021; Plesnar-Bielak et al., 2014). The likelihood of such polymorphisms being maintained by sexually antagonistic selection increases with epistasis between antagonistic loci (Arnqvist et al., 2014), as well as beneficial reversals of dominance between the antagonistic alleles within a given locus (Barson et al., 2015; Connallon & Chenoweth, 2019; Grieshop & Arnqvist, 2018)—both of which reduce the fitness costs of carrying any of the “wrong” alleles for one’s sex. Thus, studying the genetic architecture of SSTs and associated sexually antagonistic effects on female fitness (i.e., partitioning total trait variance into that stemming from additivity, dominance, and epistasis) will help to clarify whether variation in such traits is maintained by balancing selection, mutation-selection balance, or some combination of the two.
The relative contributions of balancing selection and mutation-selection balance to the genetic variance in SST expression have important implications for sexual selection theory and beyond, but remain largely unresolved. For example, rapid adaptation to altered environments may be facilitated if the genetic variation is maintained under balancing selection (Barrett & Schluter, 2008), possibly stemming from sexually antagonistic selection (Connallon & Clark, 2014). Moreover, sexual selection against deleterious mutations can also improve adaptation rates and/or reduce extinction risk (Cally et al., 2019; Fricke & Arnqvist, 2007; Godwin et al., 2020; Jarzebowska & Radwan, 2010; Lorch et al., 2003; Lumley et al., 2015; Martínez-Ruiz & Knell, 2016; Parrett & Knell, 2018; Parrett et al., 2019; Plesnar-Bielak et al., 2012) if the condition-dependent expression of SSTs reveals individuals’ relative share of the population’s mutation load (Grieshop et al., 2021b). On the other hand, by favoring alleles that harm female/population offspring production (Berger et al., 2016; Grieshop et al., 2017; Holland, 2002; Kokko & Brooks, 2003; Rundle et al., 2006), increasing costs associated with SST expression (Bro-Jørgensen, 2014; Doherty et al., 2003; Martins et al., 2018) or by reducing effective population size (Kokko & Brooks, 2003; Parrett et al., 2022) adaptation rates may be hindered and extinction risks increased by sexual selection and sexual conflict.
Here, we implemented a quantitative genetic approach using diallel crosses in the bulb mite, R. robini, in order to partition genetic variance and investigate dominance relationships of a sexually selected weapon, which earlier work implied has sexually antagonistic effects on female fitness (Łukasiewicz et al., 2020; Plesnar-Bielak et al., 2014). Male R. robini are comprised of two morphs distinct in the expression of sexually selected weaponry (Parrett et al., 2022), the aggressive fighters have a thickened third pair of legs and use them while engaging in contest competition, which can be lethal. In contrast, the non-aggressive scramblers have legs with all approximately equal thickness and avoid direct competition (Radwan, 1995; Radwan et al., 2000). Previous work has shown that males in poor phenotypic condition tend to express the scrambler phenotype (Radwan, 1995; Smallegange, 2011), but male morph is nevertheless significantly heritable (Radwan, 1995; Smallegange & Coulson, 2011), with some previous data suggesting the existence of a large-effect QTL (Radwan, 1995). Yet, other evidence suggests that fighters may be associated with a lower load of deleterious mutations (Łukasiewicz et al., 2020; Parrett et al., 2022), such that heritability of morph may result from polygenic condition-dependence. Fighter morphs tend to outcompete scramblers, which incur high mortality in inter-morph competition (Radwan & Klimas, 2001; Radwan et al., 2000), yet the scrambler morph persists. It is plausible that sexual antagonism may also contribute to the maintenance of genetic variance in male morph. Previous work showed that selection for morph results in a correlated response in female fecundity, such that females from fighter-selected treatments have lower fecundity than females from scrambler-selected treatments (Łukasiewicz et al., 2020; Plesnar-Bielak et al., 2014). The relative contribution of each of these candidate mechanisms remains unknown.
If indeed most of the genetic variance underlying morph is due to deleterious mutations, we can expect additive genetic variance to be moderate and comparable to other life-history traits (Mousseau & Roff, 1987). Female fecundity is likely a useful benchmark as it is known to be highly polygenic and therefore a large target for deleterious mutations to act upon (Houle, 1992). Furthermore, if the additive genetic variance is determined by deleterious mutations it should also be accompanied by a comparable portion of dominance variation (Crnokrak & Roff, 1995; Roff & Emerson, 2006), and substantial inbreeding depression (DeRose & Roff, 1999), because deleterious mutations segregating in natural populations are typically recessive (Charlesworth & Willis, 2009). The potential for sexual antagonism to maintain male morph variation would be much enhanced if there were beneficial dominance reversals, that is, if dominant fighter morph (recessive scrambler morph) allele(s) had recessive (dominant) effects on female fecundity. Under such dominance reversal, heterozygous fighter/scrambler genotypes would express the male fighter morph and have high female fecundity, stabilizing the underlying sexually antagonistic polymorphisms (Fry, 2010; Kidwell et al., 1977). Therefore, we investigated dominance relationships for male morph and female fecundity. The relative dominance of each inbred line’s genetic variation over the other lines in the diallel was estimated as the covariance between their mean outcrossed values and the “self-cross” means of the inbred lines they are crossed with (Grieshop & Arnqvist, 2018). If these array covariances are positively correlated between male morph and female fecundity it would indicate that the underlying alleles are either dominant or recessive for both traits, whereas a negative correlation indicates that alleles are dominant for one trait but recessive for the other.
Methods
General husbandry
All mites were reared under standard laboratory conditions. Stock cultures were housed in plastic containers (~7 × 10 cm), large colonies (>10 mites) were reared in small plastic containers (~2.2 cm diameter), and small colonies (10 or less) or individual mites were housed in glass vials (~1 cm diameter). All containers and vials had a base of plaster-of-Paris (~1 cm) which was soaked in water prior to transferring any mites. In order to maintain humidity (> 90%), all mite housing was placed on damp tissue paper and placed within a plastic box containing a ball of soaked tissue paper. Mites were stored in incubators kept at a constant 23°C. Powdered yeast was provided ad libitum for feeding. All housing was checked regularly, to ensure mites had access to yeast and humidity remained high.
Establishing inbred lines
In brief, 41 inbred lines of R. robini, each founded by a single virgin female and male, were established from mites collected from onions in fields close to Mosina, Poland. The morph of each founding male was recorded and in subsequent generations of inbreeding, males of the same morph as the founder male were used to propagate each inbred line (for full details see Łukasiewicz et al., 2020). Inbred lines were developed by full-sib mating for 10 generations, inbreeding was then relaxed due to logistical constraints, and inbred lines were allowed to expand for approximately 3 months (~six generations). Full-sib inbreeding was then resumed for a further four generations, thus giving a total of 14 generations of full-sib inbreeding. Inbred lines were again allowed to expand for approximately 3 months (~six generations) prior to the onset of the current experiment in order to have inbred lines of adequate and stable sizes that could be used for this experiment. During inbreeding protocols, after their initial establishment, each inbred line was maintained with backups. In each generation we reared 20 larvae (or fewer if not available) to adulthood for each inbred line and mated each male of the appropriate morph with a randomly selected virgin female. One to five such pairs were formed, giving up to four backups per inbred line per generation. One of these families was randomly selected to found the next generation, but if this family failed to produce offspring, a random backup was selected to replace it. Before the first expansion (i.e., generation 10) we recorded 233 such cases (counts based on two families, per line, and each generation), of which 199 were due to infertility, and 34 were due to embryonic or larval mortality. Although non-significant (χ2 = 2.89, d.f. = 1, p = 0.089) there was a trend that reproductive failures were observed more often in fighter-founded lines compared to those founded by scramblers, with the average number of observed reproductive failures, per line, in each treatment being 6.42 and 4.40, respectively (Supplementary Figure 1).
Diallel crosses and assays
From those that survived the inbreeding program, we randomly selected 20 inbred lines, 10 founded by a fighter male and 10 by a scrambler male. From each of these inbred lines, we transferred 50 females to a new container for egg-laying for 5 days, after which females were removed from the containers. Three inbred lines, one founded by a fighter and two founded by a scrambler, did not have enough females available, and only 20–30 were therefore placed in egg-laying containers. After a further 6 days, we attempted to isolate approximately 200 larvae and proto-nymphs from each inbred line into individual vials. If on the first day of isolation, we did not achieve adequate numbers, we continued isolating individuals for the next 2 consecutive days. The three inbred lines with low numbers of female parents (from above) were discarded at this stage due to low offspring numbers. In addition, another inbred line (founded by a fighter male) was discarded due to experimenter error. This left us with 16 inbred lines, eight founded by fighter males and eight founded by scrambler males.
From these 16 inbred lines, we created a partial diallel in a “chess-board” design with reciprocal outbred crosses, and all possible inbred crosses. In total, this led to 144 cross combinations: 16 inbred self-crosses (i.e., sires and dams from the same inbred line), and 128 outbred crosses (i.e., sires and dams from different inbred lines) consisting of each inbred line crossed reciprocally with four fighter-founded and four scrambler-founded inbred lines (see Figure 1). For each outbred cross we set up five replicate virgin pairs (P-generation) and for each inbred self-cross we increased this number to 10 replicate virgin pairs (for two self-crosses: IN7 and IN14, it was only possible to establish seven and eight pairs, respectively). The mated pairs were left in vials together and females were allowed to lay eggs for 6 days, after which the adults were removed from the vials. After a further 2 days, each vial was then checked for larvae. We collected F1 larvae for two purposes: (1) to gain virgin females for fecundity assays and (2) to estimate male morph proportion. As it was not logistically feasible to isolate every single larva, and in order to spread our efforts as evenly as possible across the entire diallel, we stopped isolating larvae from an outbred cross when we had isolated our target number of larvae from three out of five replicate pairs (or 6 out of 10 for self-crosses). We aimed to collect 70 larvae per replicate pair, with 10 larvae reared individually to obtain virgin females for fecundity assays, and the remaining reared in groups of 10 individuals per vial (or less if not available) and used to determine morph proportion. These numbers of mites (10 or less) per vial represent low-density rearing conditions. During larvae collection, we prioritized fecundity assays and isolated larvae individually first. Until we reached these targets, we continued isolating larvae throughout the following 10 days from all replicates of a cross combination. In some cases, we were not able to find enough larvae and on occasions, a low level of opportunistic sampling was also performed. Data collected from all replicate pairs were included in our analyses. The number of males used to estimate the morph proportion of each replicate is therefore varied, but any analysis (see below) takes this into account.
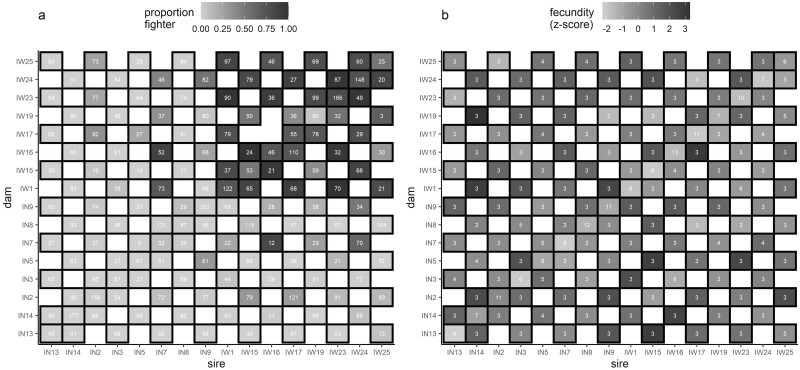
Figure 1.
Heatmaps of (a) morph proportion and (b) female fecundity across the diallel with each sire and dam cross combination. (a) Morph proportion (averaged across replicates), where darker gray indicates a higher proportion of fighter males in that cross combination. Numbers within each square indicate the total number of males from which morph was recorded, with white squares with no numbers indicating no data were collected. (b) Female fecundity (averaged summed Z-scores across replicates) for each cross combination, where darker gray indicates higher summed Z-score (i.e., fecundity controlled for the observer). Numbers within each square indicate the total number of females in which fecundity was recorded, with white squares with no numbers indicating no data were collected. Sire and dam names: IN or IW followed by a number provides inbred lines ID and indicate lines founded by a scrambler or fighter male, respectively (i.e., IN# = scrambler founded inbred line and IW# = fighter founded inbred line).
Morph proportion of F1 adults that eclosed from each group of larvae was determined 8 days after larvae were isolated. F1 adults were sexed and the male morph was recorded before being removed from vials; any nymphs remaining were left in the vials for future morph scoring. Vials were then checked every other day for any remaining mites to mature, with the morph of any males being recorded. Housing mites in groups for morph proportion assays allowed us to sustainably increase sample size compared to isolating individual larvae; the method choice is justified as it was previously shown that colony density does not influence morph determination in this species (Radwan, 1995). Moreover, this better reflects the housing conditions of mites within their pre-experiment conditions compared to individually housed mites. Although in some cases this led to deaths of males, most likely as a consequence of lethal combat, if vials are checked regularly dead mites can be sexed and male morph determined with relative ease: only six dead mites across the entire experiment were unidentifiable. Splitting mites into vials with low-density rearing conditions, rather than rearing all larvae in one pool, allowed us to have similar rearing densities when we did not obtain 60 larvae (the mites were then simply split among fewer groups) or if more offspring were produced and not separated. The numbers of fighters and scramblers were then pooled from these groups of mites for each replicate pair.
For female fecundity, vials containing individually housed F1 mites were checked every other day and adults were sexed. Only females < 3 days post-adult-eclosion were used in fecundity assays. From each cross combination, we aimed to assay the fecundity of three F1 females from outbred crosses or six from inbred self-crosses—each from a unique P-generation pair when possible. Isolated F1 females were paired with a random < 3-day-old (post-adult-eclosion) male of either morph from a large outbred stock population, as previous work has shown that male morph does not have direct effects on female fecundity in R. robini (Parrett et al., 2022; Plesnar-Bielak et al., 2014). Females were allowed to oviposit for 10 days, transferring the pair to a new vial on day 5 (replacing any dead males; in both first and second vials n = 19 dead males were observed), and removing the pair on day 10. The intermittent removal of a male during fecundity assays does not lead to detectably lower female fecundity for at least 2 days (Kołodziejczyk & Radwan, 2003), so these rare cases of male death were unlikely to affect our results. The eggs from both vials were counted the same day adults were removed from the vials. On occasions, some eggs had hatched and therefore larvae were also present in the vials, these were included in these counts as eggs. As these were relatively rare and an excess of food remained in tightly plugged vials we do not believe errors were introduced due to some eggs hatching. As female egg-laying rate remains relatively consistent over the first 3 weeks of their lives, after which females have declining fecundity (Tilszer et al., 2006), and because 3 weeks make up a significant proportion of an average female’s lifespan (3–5 weeks depending on the study: Kołodziejczyk & Radwan, 2003; Parrett et al., 2022), such a measure is likely a good estimate of lifetime fitness. In an attempt to exclude the possibility that females did not lay eggs due to a male effect (for example, male sterility or lack of mating), we provided a new male if there were zero eggs in the first vial. If in the second vial also there were zero eggs, we assumed this to be a consequence of the female, but if females produced eggs in the second vial (i.e., with a new male) we assumed the zero count in the first vial to be an effect of the first male. We removed those later situations from our dataset (n = 25), but retained the former as zero eggs laid.
We performed a partial second block specifically targeting cross combinations in which sample sizes were low or completely missing (n = 48). Sample sizes for each cross combination were considered low if we did not have fecundity data for three females from outbred crosses or six females from inbred-self crosses and/or if we had determined the morph from less than 30 males from each outbred cross combination or 60 males from inbred-self crosses. Although an unbalanced sampling effort exists between blocks 1 and 2, repeating a partial block allowed us to have estimates from cross combinations with completely missing data or improve estimates of those with low sample size, and it also allowed us to partition some variance due to environmental factors (i.e., block). It should be noted that our statistical method for variance partitioning is specifically intended to accommodate imbalanced sampling in the diallel (Lenarcic et al., 2012).
Statistical analysis
We performed diallel analysis for sex-specific data separately. In each case, we partitioned the genetic variance of each trait by fitting models using the package litterDiallel (Shorter et al., 2019). This package adapted the BayesDiallel (Lenarcic et al., 2012) Gibbs sampler to also allow generalized linear mixed models (GLMMs) to be fitted using MCMCglmm (Hadfield, 2010). Using this Bayesian modeling approach, we were able to partition the phenotypic variation of male morph and female fecundity (separately) in the F1 offspring of crosses between maternal strain j and paternal strain k into additive effects (), parental-sex effects (), dominance effects (), symmetric and asymmetric epistatic effects (, respectively), and unexplained variance (noise; ), as follows:
Male morph proportion is formulated as a binomial GLMM, with the observed proportion of each male phenotype () modeled as: . Here, is the total number of males and is the proportion of fighters. In MCMCglmm, this is specified as a two-outcome “multinomial2” model with and . The GLMM uses the inverse logit link to the standard BayesDiallel model, while controlling for overall fixed effects of mean () and block, i.e., . Female fecundity is formulated as a Gaussian linear model, with the Z-scores (see below) for each individual modeled as: . Models were iterated 1,000,000 times, with a thinning interval of 1,000 and a burn-in of 50,000. Minimally informative priors (V = 1, nu = 0.002) were used during modeling. The variance components from the diallel random effects are modeled as in Shorter et al. (2019). Block was included as a fixed effect in both models.
Counts of male morph from all vials of a given replicate pair were combined and used to determine male morph proportion. In our experience, humans may systematically differ in their counts of large numbers of small eggs; therefore for female fecundity data, in order to account for any such observer effect (n = 4 observers) we scaled egg counts for each observer individually and took the sum of Z-scores from both egg laying vials. In a number of cases the female died in the first vial (n = 14) and therefore no second vial existed for these females. As we wanted to take this into account in our analysis, we scored the “second vial” as having a count of zero. As a consequence, there was no second observer, so we assigned a Z-score equivalent to the mean Z-score of all observations with counts of zero. In addition, we ran the same fecundity model but took observer effect into account by taking the residuals from a simple generalized model fit to fecundity count data with “observer” as an explanatory variable. A comparison of DIC scores indicates that the model using Z-scores (DIC = 1,898.2) had a substantially better fit compared to modeling residuals (DIC = 3,942.4). We therefore only report the model using Z-scores.
To confirm all chains had good mixing we ran Gelman–Rubin analyses for each model: both had acceptable multivariate psrf (potential scale reduction factor) scores below 1.1 (morph proportion = 1.07, fecundity = 1.06). In addition, we generated null posterior distributions for both models by randomizing the unique cross identifier (within inbred and outbred crosses, separately) and repeating the above analyses, with the assumption that true signals of non-zero variance should be absent/zero in the randomized null distributions.
Diallel crosses also allow for the ordination of strains from those carrying the most dominant alleles to those carrying the most recessive alleles at the loci underlying the trait/s in question (Grieshop & Arnqvist, 2018). The relative amount of dominant/recessive alleles in a given inbred line is estimated by the covariance between its mean outbred values (r; averaged over all outbred combinations of a particular inbred strain) with the mean of the self-cross values of each respective inbred line to which it was crossed (P). This was done separately for all strains, and separately for both male morph (σPM,rM) and female fecundity (σPF,rF). A positive covariance for a given inbred line implies that the alleles it carries are mostly recessive to those carried by the inbred lines it was crossed with (i.e., its outbred values are dependent on the genetic makeup of the inbred lines it is crossed with). Contrastingly, if an inbred line’s outbred values do not covary with the self-cross values of inbred lines it is crossed with (i.e., values of or close to 0), this is an indication that they harbor more dominant alleles than the inbred lines they were crossed with (i.e., their outbred values are independent of the genetic makeup of the inbred lines they are crossed with). This interpretation holds in absence of environmental and epistatic variation (Grieshop & Arnqvist, 2018). To fulfill that assumption, we first fit simple general and generalized linear models to fecundity (Z-score) and morph data, respectively, which modeled environmental effects by fitting block as a fixed effect and modeled epistatic effects by fitting the sire x dam combination as a random effect, using the lme4 package for R (Bates et al., 2015). The residuals from these models were then used to tabulate the mean values for each cross combination, which were used to calculate the array covariances described above (see Supplementary material S1 of Grieshop et al., 2021a for details). Inbred lines are ordinated according to their array covariances—their relative dominance/recessive relationships to one another. The array covariances were also used to test for dominance reversal between male morph and female fecundity: a negative genetic correlation between the traits would indicate that dominant alleles for one trait tend to be recessive for the other, whereas a positive genetic correlation would indicate that alleles tend to be either dominant or recessive for both traits alike—evidence for and against dominance reversal, respectively.
All statistical analysis was performed in R version 3.6 (R Development Core Team, 2020).
Results
In total, we recorded the morph of 8,905 F1 males across all diallel crosses: 1,550 were from inbred crosses, 7,355 were from outbred crosses, 6,572 were scrambler, and 2,333 were fighter males. We also assayed the fecundity of 548 females, 138 from inbred crosses and 410 from outbred crosses. Across both blocks, morph proportion and female fecundity data were collected from 143 and 144 cross combinations, respectively (Figure 1).
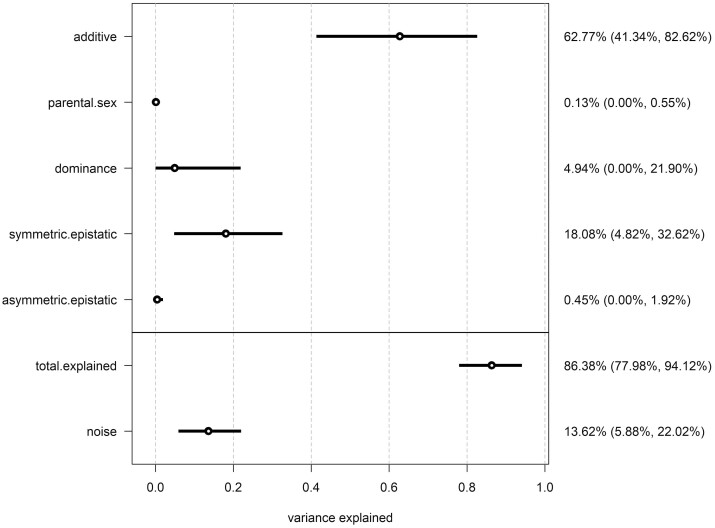
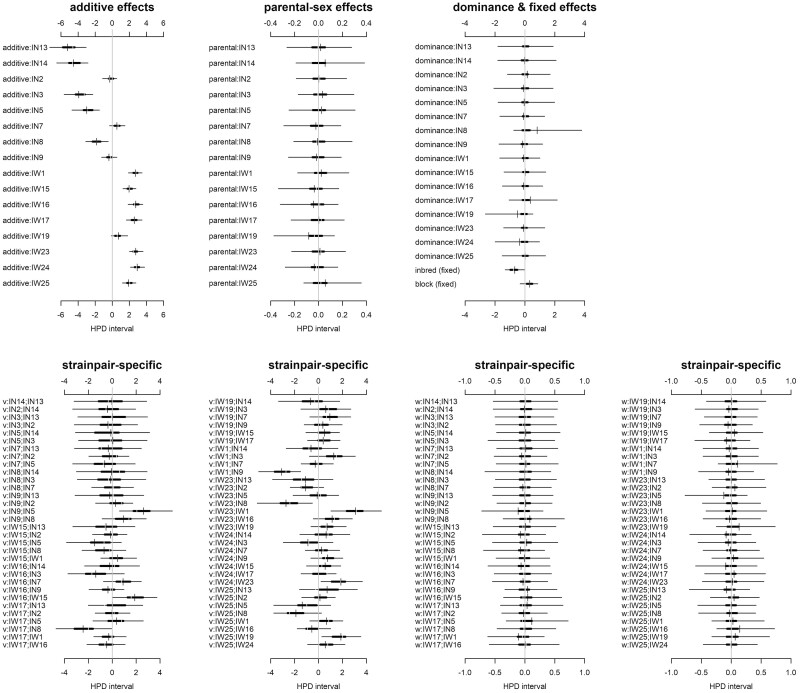
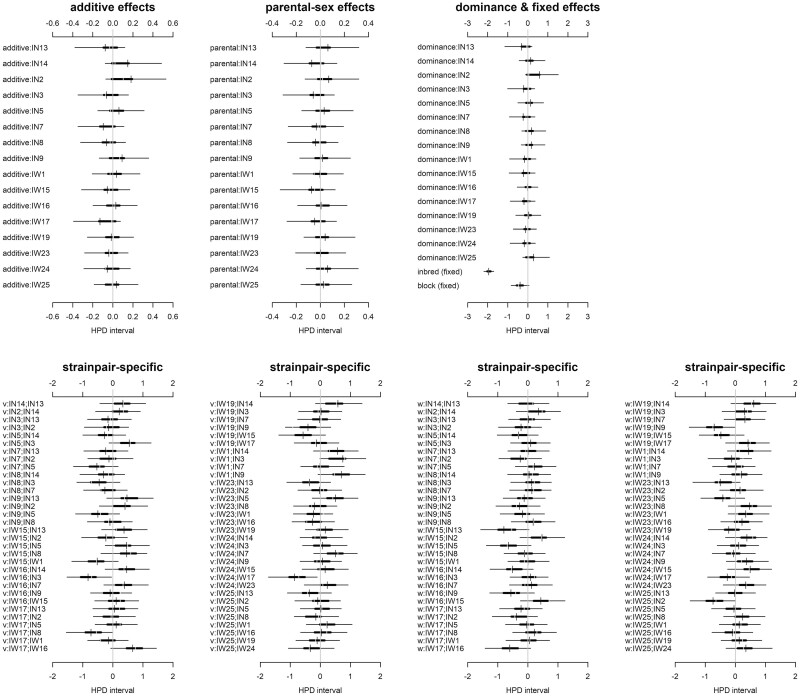
A considerable percentage (86.38%) of the variance in morph proportion was explained by diallel effects, whereas a low percentage (13.62%) of that variance was noise (Figure 2). The majority of variance in morph proportion (62.77%) was explained by additive genetic variance, with symmetric epistatic effects (18.08%) explaining most of the remaining variance. Dominance variance, estimated by strain-specific inbreeding variance, explained only 4.94% of the variance, an order of magnitude lower compared to additive variance. Reciprocal crosses that are autosomally identical but have inherited their cytoplasm and sex-chromosomes from opposite inbred lines revealed almost no influence of parental effects or asymmetric epistasis (0.13% and 0.45% of total variance explained, respectively) on the probability of offspring being a fighter or scrambler. Looking at the highest posterior density (HPD) means and 95% credibility intervals (CIs) of the diallel effects and estimates of inbred line-specific effects provides more detail (Figure 3). The HPD mean for the fixed effect of inbreeding was negative and CIs, close to, but not overlapping zero—indicating inbreeding depression for the expression of the fighter morph. The probability of offspring being either a fighter or scrambler was largely associated with founder morph treatment as seen in their individual additive effects. Seven of the eight fighter-founded inbred lines have positive additive effects (i.e., increased probability of male offspring being fighter) where the HPD interval does not overlap with zero, whereas, five of the eight scrambler-founded inbred lines had negative additive effects (i.e., increased probability of male offspring being a scrambler). A number of specific cross combinations showed symmetric epistatic effects on the probability of male offspring morph beyond their predictive additive effects alone, most notably IN9:IN5, IW1:IN9, IW23:IN8, IW23:IW1, and IW25:IW19 (Figure 3).
Figure 2.
Variance contributions of distinct class effects on morph proportion. Reporting posterior means and 95% highest posterior densities (HPDs) of variance projections.
Figure 3.
Diallel effects on morph proportion of (top row) strain-specific additive, parental sex, and dominance effects, also fixed effects of block and the main effect of inbreeding, and (bottom row) epistatic strainpair-specific effects (labels “v” and “w,” respectively, refer to symmetric and non-symmetric epistatic effects) on morph proportion. Represented for each parameter: thin line: 95% highest posterior density (HPD); thick line: 50% HPD; vertical break: median HPD; dash: mean HPD. The gray vertical line indicates 0, where intervals that exclude 0 have non-negligible effects on the male morph. Positive values indicate an increasing contribution to offspring being fighters and negative values there is decreasing contribution to offspring being a fighter. IN or IW followed by a number provides inbred lines ID and indicates lines founded by a scrambler or fighter male, respectively (i.e., IN# = scrambler founded inbred line and IW# = fighter founded inbred line).
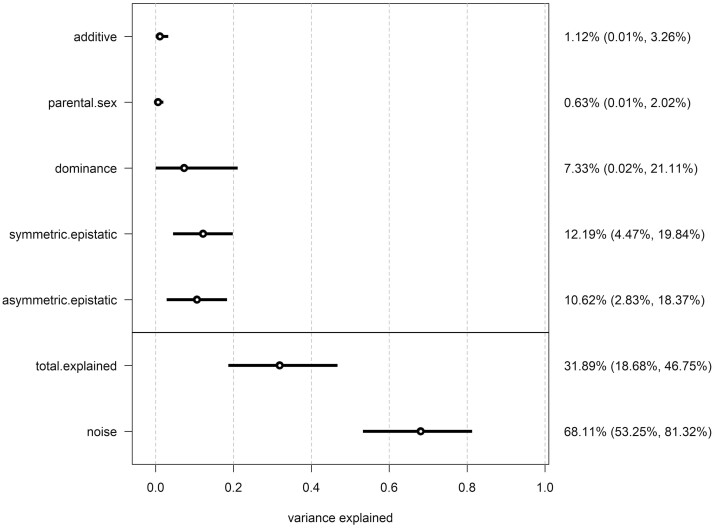
In contrast, considerably more of the variance (68.52%) for fecundity was attributed to noise and a lower percentage (31.48%) was explained by diallel effects (Figure 4). The majority of this variance was explained by epistatic effects (symmetric = 12.37%, asymmetric = 10.36%), with the remaining explained by dominance variance (6.94%) and additive genetic variance (1.13%). As with the male morph data, there was next to no variance explained by parental effects (0.68%). Again, a detailed look at inbred line-specific HPD means and CIs provides further insight (Figure 5). There was a very pronounced fixed effect of inbreeding, with HPD mean being negative with no overlap with zero—indicating substantial inbreeding depression for female fecundity. Additionally, an inspection of how variance in fecundity was explained by symmetric and asymmetric epistatic effects shows three strain-pair specific estimates where the 95% CIs do not overlap zero (v:IW16:IN3, v:IW24:IW17, and w:IW19:IN9) and many others whose HPD means are far from zero but whose 95% CIs nevertheless do overlap zero (Figure 5).
Figure 4.
Variance contributions of distinct class effects on female fecundity (Z-score). Reporting posterior means and 95% HPDs of variance projections.
Figure 5.
Diallel effects on female fecundity (Z-score) of (top row) strain-specific additive, parental sex, and dominance effects, also fixed effects of block and the main effect of inbreeding, and (bottom row) epistatic strainpair-specific effects (labels “v” and “w,” respectively, refer to symmetric and non-symmetric epistatic effects) on female fecundity. Represented for each parameter: thin line: 95% HPD; thick line: 50% HPD; vertical break: median HPD; dash: mean HPD. The gray vertical line indicates 0, where intervals that exclude 0 have non-negligible effects on the male morph. Positive values indicate increasing contribution to female fecundity and negative values decreasing contribution to female fecundity. IN or IW followed by a number provides inbred lines ID and indicates lines founded by a scrambler or fighter male, respectively (i.e., IN# = scrambler founded inbred line and IW# = fighter founded inbred line).
In both cases, randomization of data structure showed that the above patterns are unlikely to be a consequence of random chance (random data structure; morph proportion: total explained variance = 5.52%, noise = 94.48%; female fecundity: total explained variance = 4.75%, noise = 95.25%).
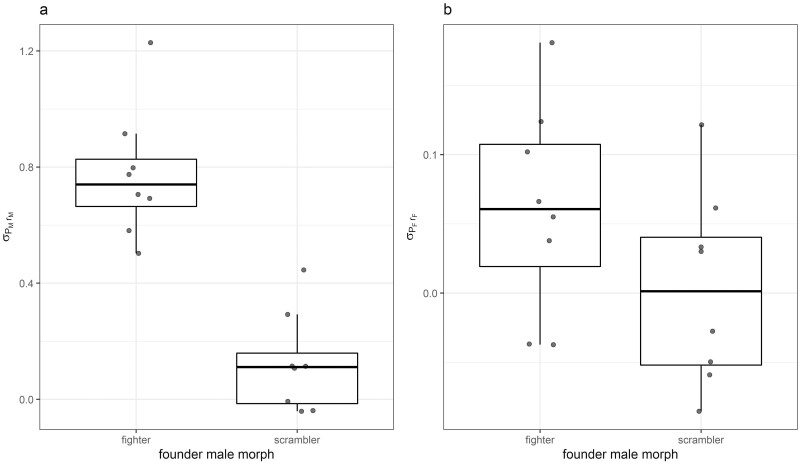
The cross-trait/cross-sex additive genetic correlation between female fecundity and morph proportion was negative but non-significant (r = −0.18, p = .493; Supplementary Figure 2). For morph proportion, dominance ordination revealed that fighter-founded inbred lines had significantly higher mean array covariances (σPM,rM) than scrambler-founded inbred lines (t = 6.56, df = 14, p < .001; Figure 6a), indicating that the fighter morph is recessive to the scrambler morph (Figure 1a). In contrast, no dominance effects were observed for female fecundity with covariance (σPF,rF) not differing between founder morph treatments (t = 1.60, df = 14, p = .131; Figure 6b). Finally, there was no significant genetic correlation between the array covariances for male morph (σPM,rM) and female fecundity (σPF,rF) among inbred lines (Pearson’s r = 0.23, p = .393, Spearman’s r = 0.23, p = .391, Supplementary Figure 3).
Figure 6.
Estimates of the relative amount of recessive allelic variation for (a) morph (σPM,rM) and (b) fecundity (σPF,rF) between founder morph treatments. Boxes are composed of the median and hinge values (25th and 75th percentiles), with whiskers ± interquartile range * 1.5. Individual points denote each inbred line’s mean array covariance for male morph and female fecundity (a: σPM,rM, b: σPF,rF).
Discussion
Here, using diallel crosses of R. robini inbred lines, we investigated the genetic variance components underlying both female fecundity and the expression of a sexually selected weapon that is dimorphic among males. Furthermore, we explored the additive genetic correlation between the two traits, as well as the genetic correlation for the dominance ordinations between the two traits. Our results provide evidence for the inheritance of the weapon being largely explained by additive genetic variation, with contributions from symmetric epistatic effects. We detected a significant inbreeding effect on morph expression, indicating that expression of the fighter phenotype is sensitive to the quantity of exposed deleterious recessives. We also found that fighter-morph alleles tend to be recessive to the scrambler-morph alleles. Female fecundity, by contrast, only had a very small percentage of variance explained by additive genetic effects, with much of the variance explained by symmetric and asymmetric epistatic effects. Fecundity also showed very high inbreeding depression, indicating that fecundity is determined by the quantity of exposed deleterious mutations. Aside from this (likely) polygenic deleterious recessive basis to both traits, as well as some degree of epistasis underlying both traits, there were no direct associations between male morph and female fecundity, including no additive genetic correlation and no evidence of dominance reversal.
The considerable additive genetic variation for morph expression detected here supports earlier work that morph is heritable in R. robini (Radwan, 1995). Building on this work and, by using inbred lines, knowing the genotype of both males and females in the current study, we provide evidence that both sexes appear to contribute equally to the probability of offspring being a fighter or scrambler. This suggests that the additive genetic basis to male morph in this species is predominantly due to autosomal genetic variation. This contrasts a previous study of a con-generic, R. echinopus, which found evidence for paternal morph effects, but overall weaker evidence for additive genetic effects. It was argued that the former is likely linked to variation on the Y-chromosome or an indirect genetic effect (Buzatto et al., 2012). While seemingly at odds, the lack of parental effect in the current study can be explained by differences in sex determination between these two species. R. echinopus has been reported to have XY (Grondziel, 1975), whereas R. robini has XO (Parrett et al., 2022) sex determination, the latter obviously eliminating the scope for any Y-linked paternal effects. Furthermore, the two species show clear differences in their morph determination mode. In R. echinopus, as well as another acarid, Sancassania berlesei (Michalczyk et al., 2018; Radwan, 1993), fighter morph is suppressed by pheromones emanating from dense colonies (Radwan, 2001), whereas no such type of polyphenism is observed in R. robini (Radwan, 1995). Clearly, there is much variation within acarid mites in genetic and environmental contributions to male morph determination, even within the genus Rhizoglyphus. The significant contribution of additive variance found here for R. robini is also in line with some earlier work on this species showing that the proportion of male morph in a population responds to directional selection (Parrett et al., 2022; Plesnar-Bielak et al., 2014; Radwan, 2003a; Smallegange & Coulson, 2011). Our estimate for the additive genetic variance of male morph is over an order of magnitude greater than that for female fecundity—a trait that is often considered an exemplification of a polygenic fitness-related trait in other taxa, with additive variance maintained predominantly via mutation-selection balance (Houle, 1992, also see below). The very high proportion of additive variance for male morph, much-exceeding dominance variance, suggests that mutation-selection balance is unlikely to be the main contributing mechanism. Accordingly, some estimates of heritability obtained in earlier work were close to or exceeded unity (Radwan, 1995), which could be due to segregation of a large effect QTL, although segregation patterns excluded simple Mendelian segregation.
Our estimates of dominance relationships are also inconsistent with morph heritability being mostly due to mutation-selection balance maintaining genetic variance in condition, in turn determining morph expression (Radwan, 1995; Smallegange, 2011). This is because the majority of segregating deleterious mutations, which reduce condition, are expected to be recessive (Charlesworth & Willis, 2009). Thus, the mutation-selection balance hypothesis would not only predict a relatively high proportion of dominance variance that we did not observe but would also predict the scrambler-associated alleles to be recessive—whereas the reverse was observed. Outbred values for fighter-founded inbred lines had considerably higher estimates of covariance with the self-cross value of inbred lines they were outcrossed to, compared to scrambler-founded inbred lines, suggesting that alleles underlying the fighter-morph are recessive. This dominance relationship is consistent with the response to divergent artificial selection of male morph, which led to earlier near-fixation of the fighter-morphs compared to scrambler-morphs (Parrett et al., 2022). This does not exclude the possibility that exposed deleterious alleles could increase the likelihood of expressing the scrambler phenotype, and indeed, we observed a negative overall general inbreeding effect for fighter expression, suggesting morph expression is in some part sensitive to exposed mutational load. Concordantly, Parrett et al., 2022 observed that populations selected for scramblers males have accumulated a large load of putatively recessive mutations spread across the genome. Yet, Parrett et al., 2022 also found a few genomic regions that contained a particularly high density of single nucleotide polymorphisms that differentiated between fighter and scrambler selected populations, and these regions map to the same linkage group (Chmielewski et al., unpublished data), suggesting the existence of a supergene or inversion associated with male morph determination. Thus, it appears that male morph is determined by two overlaying mechanisms: (i) the existence of one or more scrambler-dominant QTL(s) directly influencing the probability (or individuals’ “liability”) of morph expression, possibly residing within a supergene or inversion polymorphism (Parrett et al., 2022), and (ii) male condition and condition-dependent weapon expression, where fighter phenotype expression is inversely associated with polygenic deleterious mutation load (Łukasiewicz et al., 2020; Parrett et al., 2022; present study). Consistent with the latter, manipulating phenotypic condition via food availability and temperature has been shown to influence weapon expression (Plesnar-Bielak et al., 2018; Radwan, 1995; Smallegange, 2011).
In contrast to the estimate for the male morph, we found very small additive genetic variance in female fecundity. Earlier work based on daughter-on-mother regression estimated heritability at 27%, but also showed that it is subject to significant inbreeding depression which is not easily purged and leads to inbred line extinction (Radwan, 2003b). This was interpreted as evidence that the heritability of fecundity is due to a large number of small-effect loci, where it would be hard to purge the deleterious mutation load on fecundity. Here, we also found a large and significant negative general effect of inbreeding, but our estimate of additive genetic variance was considerably lower. It should be noted, however, that additive genetic variance inferred from daughter-on-mother regression ignores epistatic effects (Roff, 1997), which we found to have a pronounced effect on fecundity variance (22.73% in total). Thus, our data are consistent with female fecundity being affected by deleterious recessives (with an only minor contribution to additive effects) that interact epistatically. Our data do not provide further insight into the basis of this epistatic variance in female fecundity, but we note that significant inbreeding depression as well as large contributions of dominance and epistasis are common features of genetic variance in fitness-related traits (DeRose & Roff, 1999; Roff & Emerson, 2006). Our results highlight that epistasis may considerably inflate the heritability of life-history traits when they are estimated using methods that ignore it.
Finally, we found little to no support for sexual antagonism contributing to the maintenance of alternative male morphs. We did not find a significant negative additive genetic correlation between male morph and female fecundity, though the estimate was negative (r = −0.18), as predicted under sexual antagonism. One possibility that could explain why we have not detected that correlation here despite it being evident in the same inbred lines after four generations of inbreeding (Łukasiewicz et al., 2020) could be that the 10 additional generations of inbreeding (see Methods) may have caused the loss of fighter-founded inbred lines with particularly strong fighter-benefit female-detrimental genetic variation (analogous to Grieshop et al., 2017). That is, if sexually antagonistic alleles that decrease female fecundity interacted with recessive deleterious mutations upon inbreeding to cause female infertility and subsequent loss of inbred lines (see Methods), this may have disproportionately purged fighter-benefit/female-detriment sexually antagonistic alleles from our panel of inbred lines, thus affecting our estimates of sexually antagonistic genetic variance. Indeed, in another system (Callosobruchus maculatus), inbred lineage extinction associated with male-benefit/female-detriment sexually antagonistic allelic variation (Grieshop et al., 2017) was likewise accompanied by a reduced additive genetic signal of sexual antagonism in the diallel cross among the extant inbred lines (Grieshop & Arnqvist, 2018) relative to the stronger additive genetic signal of sexual antagonism seen in that population prior to the inbreeding regime (Berger et al., 2014).
Another possibility is that the 16 inbred lines used here simply did not capture a significant proportion of segregating morph-specific sexually antagonistic variation present within an outbred population. While we are limited in the degree to which we can extrapolate beyond these 16 inbred lines, we note that the number of inbred lines used in the current study is equal to or greater than many comparable experiments using diallel crosses (e.g., Buzatto et al., 2012; Grieshop & Arnqvist, 2018; Grieshop et al., 2021b; Lüpold et al., 2016; Maurizio et al., 2018; Shorter et al., 2019), and such limitation did not prevent Grieshop and Arnqvist (2018) from detecting significant sexual antagonism. In the case of our experiment, if sexual antagonism was associated with genetic variants underlying male dimorphism as previously hypothesized (Łukasiewicz et al., 2020; Plesnar-Bielak et al., 2014), these variants should be present in our sample representing equal numbers of inbred lines derived from fighter and scrambler males. We, therefore, consider the loss of sexually antagonistic variation during inbreeding as a more likely explanation of why we have not observed similar morph-specific sexual antagonism reported by earlier studies.
While we might have underestimated the additive signal of sexual antagonism, we still did not find any evidence for dominance reversal either between male morph and female fecundity, which would help maintain polymorphisms underlying the two traits under sexually antagonistic balancing selection (see Reid, 2022 for a common mechanism by which dominance reversal could ensue for both major-effect QTLs and polygenic underpinnings). A presumably polygenic signal of dominance reversal was previously reported for male and female fitness in seed beetles using a similar quantitative genetic approach as that used here (Grieshop & Arnqvist, 2018), but other methods have revealed dominance reversals for cases of major-effect QTLs and supergenes (Barson et al., 2015; Pearse et al., 2019). In R. robini however, dominance reversals between the male morph and female fecundity do not seem to be contributing to the stable maintenance of polymorphisms underlying genetic variance in the male morph.
Overall, our study shows high additive genetic variance for the dimorphic expression of a weapon, which when taken with dominance and inbreeding results strongly suggest two overlaying mechanisms for morph determination exist. We propose that one (or more) large effect scrambler-dominant QTL(s) directly influences male morph expression which is simultaneously affected by polygenic condition. Contrastingly, we did not detect much additive genetic variance for female fecundity, although considerable epistatic effects were found, highlighting models not accounting for epistasis may inflate estimates of heritability. Our study revealed that the genetic architecture of the male morph is very distinct from that underlying female fecundity, with beneficial dominance reversal unlikely to be contributing to maintaining polymorphisms for the male morph. The maintenance of male polyphenism in R. robini remains to be fully resolved, with complex genetic and environmental effects yet to be fully teased apart.
Supplementary Material
Acknowledgments
We thank Mateusz Konczal for the useful discussion. We are grateful for laboratory assistance from Małgorzata Niśkiewicz, Karolina Przesmycka, Issy Bolitho, and Martyna Wilczek. Work from this project was funded by the National Science Centre (2017/27/B/NZ8/00077 to J.R. and 2020/39/D/NZ8/00069 to J.M.P.), Swedish Research Council (2018-06775 to K.G.), and National Institutes of Health (F32-AG064883 to P.L.M.).
Contributor Information
Jonathan M Parrett, Evolutionary Biology Group, Faculty of Biology, Adam Mickiewicz University, Poznań, Poland.
Aleksandra Łukasiewicz, Evolutionary Biology Group, Faculty of Biology, Adam Mickiewicz University, Poznań, Poland.
Sebastian Chmielewski, Evolutionary Biology Group, Faculty of Biology, Adam Mickiewicz University, Poznań, Poland.
Agnieszka Szubert-Kruszyńska, Evolutionary Biology Group, Faculty of Biology, Adam Mickiewicz University, Poznań, Poland.
Paul L Maurizio, Department of Medicine, Section of Genetic Medicine, University of Chicago, Chicago, Illinois, United States.
Karl Grieshop, Department of Ecology and Evolutionary Biology, University of Toronto, Toronto, Canada; Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Stockholm, Sweden.
Jacek Radwan, Evolutionary Biology Group, Faculty of Biology, Adam Mickiewicz University, Poznań, Poland.
Data availability
Data used in this manuscript have been deposited on Dryad doi:10.5061/dryad.18931zd2b
Author contributions
J.R. and K.G. conceived the idea. J.R., J.M.P., A.Ł., and S.C. designed the experiment. J.M.P., A.Ł., S.C., A.S.-K., and J.R. performed data collection. J.M.P., P.L.M., and K.G. carried out the analysis. J.M.P. and J.R. wrote the manuscript with input from P.L.M. and K.G.
Conflict of interest: The authors have no conflict of interest to declare.
References
- Andersson, M. (1986). Evolution of condition-dependent sex ornaments and mating preferences: Sexual selection based on viability differences. Evolution, 40(4), 804–816. 10.1111/j.1558-5646.1986.tb00540.x [DOI] [PubMed] [Google Scholar]
- Arnqvist, G., Vellnow, N., & Rowe, L. (2014). The effect of epistasis on sexually antagonistic genetic variation. Proceedings of the Royal Society B: Biological Science, 281, 20140489. 10.1098/rspb.2014.0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, R. D. H., & Schluter, D. (2008). Adaptation from standing genetic variation. Trends in Ecology and Evolution, 23, 38–44. 10.1016/j.tree.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Barson, N. J., Aykanat, T., Hindar, K., Baranski, M., Bolstad, G. H., Fiske, P., Jacq, C., Jensen, A. J., Johnston, S. E., Karlsson, S., Kent, M., Moen, T., Niemelä, E., Nome, T., Næsje, T. F., Orell, P., Romakkaniemi, A., Sægrov, H., Urdal, K., … Primmer, C. R. (2015). Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature, 528(7582), 405–408. 10.1038/nature16062 [DOI] [PubMed] [Google Scholar]
- Barton, N. H., & Keightley, P. D. (2002). Understanding quantitative genetic variation. Nature Reviews Genetics, 3(1), 11–21. 10.1038/nrg700 [DOI] [PubMed] [Google Scholar]
- Bates, D., Mächler, M., Bolker, B. M., & Walker, S. C. (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67, 1–51. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Berger, D., Grieshop, K., Lind, M. I., Goenaga, J., Maklakov, A. A., & Arnqvist, G. (2014). Intralocus sexual conflict and environmental stress. Evolution, 68(8), 2184–2196. 10.1111/evo.12439 [DOI] [PubMed] [Google Scholar]
- Berger, D., Martinossi-Allibert, I., Grieshop, K., Lind, M. I., Maklakov, A. A., & Arnqvist, G. (2016). Intralocus sexual conflict and the tragedy of the commons in seed beetles. American Naturalist, 188(4), E98–E112. 10.1086/687963 [DOI] [PubMed] [Google Scholar]
- Berglund, A., Bisazza, A., & Pilastro, A. (1996). Armaments and ornaments: An evolutionary explanation of traits of dual utility. Biological Journal of the Linnean Society, 58(4), 385–399. 10.1111/j.1095-8312.1996.tb01442.x [DOI] [Google Scholar]
- Brockmann, H. J. (2001). The evolution of alternative strategies and tactics. Advances in the Study of Behavior, 30, 1–51. 10.1016/S0065-3454(01)80004-8 [DOI] [Google Scholar]
- Bro-Jørgensen, J. (2014). Will their armaments be their downfall? Large horn size increases extinction risk in bovids. Animal Conservation, 17, 80–87. 10.1111/acv.12062 [DOI] [Google Scholar]
- Buzatto, B. A., Simmons, L. W., & Tomkins, J. L. (2012). Paternal effects on the expression of a male polyphenism. Evolution, 66(10), 3167–3178. 10.1111/j.1558-5646.2012.01662.x [DOI] [PubMed] [Google Scholar]
- Cally, J. G., Stuart-Fox, D., & Holman, L. (2019). Meta-analytic evidence that sexual selection improves population fitness. Nature Communications, 10, 2017. 10.1038/s41467-019-10074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., & Willis, J. H. (2009). The genetics of inbreeding depression. Nature Reviews Genetics, 10(11), 783–796. 10.1038/nrg2664 [DOI] [PubMed] [Google Scholar]
- Connallon, T., & Chenoweth, S. F. (2019). Dominance reversals and the maintenance of genetic variation for fitness. PLoS Biology, 17, e3000118. 10.1371/journal.pbio.3000118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon, T., & Clark, A. G. (2012). A general population genetic framework for antagonistic selection that accounts for demography and recurrent mutation. Genetics, 190(4), 1477–1489. 10.1534/genetics.111.137117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon, T., & Clark, A. G. (2014). Balancing selection in species with separate sexes: Insights from fisher’s geometric model. Genetics, 197(3), 991–1006. 10.1534/genetics.114.165605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnokrak, P., & Roff, D. A. (1995). Dominance variance: Associations with selection and fitness. Heredity, 75(5), 530–540. 10.1038/hdy.1995.169 [DOI] [Google Scholar]
- DeRose, M. A., & Roff, D. A. (1999). A comparison of inbreeding depression in life-history and morphological traits in animals. Evolution, 53(4), 1288–1292. 10.1111/j.1558-5646.1999.tb04541.x [DOI] [PubMed] [Google Scholar]
- Doherty, P. F., Sorci, G., Royle, J. A., Hines, J. E., Nichols, J. D., & Boulinier, T. (2003). Sexual selection affects local extinction and turnover in bird communities. Proceedings of the National Academy of Sciences, 100, 5858–5862. 10.1111/1365-2656.12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugand, R. J., Tomkins, J. L., & Kennington, W. J. (2019). Molecular evidence supports a genic capture resolution of the lek paradox. Nature Communications, 10(1), 1359. 10.1038/s41467-019-09371-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke, C., & Arnqvist, G. (2007). Rapid adaptation to a novel host in a seed beetle (Callosobruchus maculatus): The role of sexual selection. Evolution, 61(2), 440–454. 10.1111/j.1558-5646.2007.00038.x [DOI] [PubMed] [Google Scholar]
- Fry, J. D. (2010). The genomic location of sexually antagonistic variation: Some cautionary comments. Evolution, 64(5), 1510–1516. 10.1111/j.1558-5646.2009.00898.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin, J. L., Lumley, A. J., Michalczyk, L., Martin, O. Y., & Gage, M. J. G. (2020). Mating patterns influence vulnerability to the extinction vortex. Global Change Biology, 26(8), 4226–4239. 10.1111/gcb.15186 [DOI] [PubMed] [Google Scholar]
- Grafen, A. (1990). Biological signals as handicaps. Journal of Theoretical Biology, 144(4), 517–546. 10.1016/s0022-5193(05)80088-8 [DOI] [PubMed] [Google Scholar]
- Grieshop, K., & Arnqvist, G. (2018). Sex-specific dominance reversal of genetic variation for fitness. PLoS Biology, 16, e20068101–e20068121. 10.1371/journal.pbio.2006810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshop, K., Berger, D., & Arnqvist, G. (2017). Male-benefit sexually antagonistic genotypes show elevated vulnerability to inbreeding. BMC Evolutionary Biology, 17, 1–10. 10.1186/s12862-017-0981-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshop, K., Ho, E. K., & Kasimatis, K. (2021a). Dominance reversals, antagonistic pleiotropy, and the maintenance of genetic variation. arXiv, arXiv:2109.01571, preprint: not peer reviewed. [Google Scholar]
- Grieshop, K., Maurizio, P. L., Arnqvist, G., & Berger, D. (2021b). Selection in males purges the mutation load on female fitness. Evolution Letters, 5(4), 328–343. 10.1002/evl3.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondziel, E. (1975). Sex chromosomes in Rhizoglyphus echinopus (F, & R.) (Acarina, Acaridae). Folia Biologica, 23(1), 69–72. [PubMed] [Google Scholar]
- Gross, M. R. (1991). Evolution of alternative reproductive strategies: Frequency-dependent sexual selection in male bluegill sunfish. Philosophical Transactions of the Royal Society London B, 332, 59–66. [Google Scholar]
- Gross, M. R. (1996). Alternative reproductive strategies and tactics: Diversity within sexes. Trends in Ecology and Evolution, 11(2), 92–98. 10.1016/0169-5347(96)81050-0 [DOI] [PubMed] [Google Scholar]
- Gross, M. R., & Repka, J. (1998). Stability with inheritance in the conditional strategy. Journal of Theoretical Biology, 192(4), 445–453. 10.1006/jtbi.1998.0665 [DOI] [PubMed] [Google Scholar]
- Hadfield, J. D. (2010). MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R Package. Journal of Statistical Software, 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S. (1937). The effect of variation of fitness. American Naturalist, 71(735), 337–349. 10.1086/280722 [DOI] [Google Scholar]
- Harano, T., Okada, K., Nakayama, S., Miyatake, T., & Hosken, D. J. (2010). Intralocus sexual conflict unresolved by sex-limited trait expression. Current Biology, 20(22), 2036–2039. 10.1016/j.cub.2010.10.023 [DOI] [PubMed] [Google Scholar]
- Hendrickx, F., De Corte, Z., Sonet, G., Van Belleghem, S. M., Köstlbacher, S., & Vangestel, C. (2022). A masculinizing supergene underlies an exaggerated male reproductive morph in a spider. Nature Ecology and Evolution, 6(2), 195–206. 10.1038/s41559-021-01626-6 [DOI] [PubMed] [Google Scholar]
- Holland, B. (2002). Sexual selection fails to promote adaptation to a new environment. Evolution, 56(4), 721–730. 10.1554/0014-3820(2002)056[0721:ssftpa]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- Houle, D. (1992). Comparing evolvability and variability. Genetics, 130(1), 195–204. 10.1093/genetics/130.1.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzebowska, M., & Radwan, J. (2010). Sexual selection counteracts extinction of small populations of the bulb mites. Evolution, 64(5), 1283–1289. 10.1111/j.1558-5646.2009.00905.x [DOI] [PubMed] [Google Scholar]
- Johnston, S. E., Gratten, J., Berenos, C., Pilkington, J. G., Clutton-Brock, T. H., Pemberton, J. M., & Slate, J. (2013). Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature, 502(7469), 93–95. 10.1038/nature12489 [DOI] [PubMed] [Google Scholar]
- Kidwell, J. F., Clegg, M. T., Stewart, F. M., & Prout, T. (1977). Regions of stable equilibria for models of differential selection in the two sexes under random mating. Genetics, 85(1), 171–183. 10.1093/genetics/85.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokko, H., & Brooks, R. (2003). Sexy to die for? Sexual selection and the risk of extinction. Annales Zoologici Fennici, 40, 207–219. [Google Scholar]
- Kołodziejczyk, M., & Radwan, J. (2003). The effect of mating frequency on female lifetime fecundity in the bulb mite, Rhizoglyphus robini (Acari: Acaridae). Behavior, Ecology and Sociobiology, 53(2), 110–115. 10.1007/s00265-002-0557-0 [DOI] [Google Scholar]
- Küpper, C., Stocks, M., Risse, J. E., Dos Remedios, N., Farrell, L. L., McRae, S. B., Morgan, T. C., Karlionova, N., Pinchuk, P., Verkuil, Y. I., Kitaysky, A. S., Wingfield, J. C., Piersma, T., Zeng, K., Slate, J., Blaxter, M., Lank, D. B., & Burke, T. (2016). A supergene determines highly divergent male reproductive morphs in the ruff. Nature Genetics, 48(1), 79–83. 10.1038/ng.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney, S., Fan, G., Widemo, F., Gunnarsson, U., Thalmann, D. S., Hoeppner, M. P., Kerje, S., Gustafson, U., Shi, C., Zhang, H., Chen, W., Liang, X., Huang, L., Wang, J., Liang, E., Wu, Q., Lee, S. M. Y., Xu, X., Höglund, J., … Andersson, L. (2016). Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nature Genetics, 48, 84–88. 10.1038/ng.3430 [DOI] [PubMed] [Google Scholar]
- Lande, R. (1975). The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genetical Research, 26(3), 221–235. 10.1017/s0016672300016037 [DOI] [PubMed] [Google Scholar]
- Lenarcic, A. B., Svenson, K. L., Churchill, G. A., & Valdar, W. (2012). General Bayesian approach to analyzing diallel crosses of inbred strains. Genetics, 190(2), 413–435. 10.1534/genetics.111.132563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch, P. D., Proulx, S., Rowe, L., & Day, T. (2003). Condition dependent sexual selection can accelerate adaptation. Evolutionary Ecology Research, 5, 867–881. [Google Scholar]
- Łukasiewicz, A., Niśkiewicz, M., & Radwan, J. (2020). Sexually selected male weapon is associated with lower inbreeding load but higher sex load in the bulb mite. Evolution, 74(8), 1851–1855. 10.1111/evo.14033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley, A. J., Michalczyk, L., Kitson, J. J. N., Spurgin, L. G., Morrison, C. A., Godwin, J. L., Dickinson, M. E., Martin, O. Y., Emerson, B. C., Chapman, T., & Gage, M. J. G. (2015). Sexual selection protects against extinction. Nature, 522(7557), 470–473. 10.1038/nature14419 [DOI] [PubMed] [Google Scholar]
- Lüpold, S., Manier, M. K., Puniamoorthy, N., Schoff, C., Starmer, W. T., Luepold, S. H. B., Belote, J. M., & Pitnick, S. (2016). How sexual selection can drive the evolution of costly sperm ornamentation. Nature, 533(7604), 535–538. 10.1038/nature18005 [DOI] [PubMed] [Google Scholar]
- Lynch, M., Blanchard, J., Houle, D., Kibota, T., Schultz, S., Vassilieva, L., & Willis, J. (1999). Perspective: Spontaneous deleterious mutation. Evolution, 53(3), 645–663. 10.1111/j.1558-5646.1999.tb05361.x [DOI] [PubMed] [Google Scholar]
- Martínez-Ruiz, C., & Knell, R. J. (2016). Sexual selection can both increase and decrease extinction probability: Reconciling demographic and evolutionary factors. Journal of Animal Ecology, 86(1), 117–127. 10.1111/1365-2656.12601 [DOI] [PubMed] [Google Scholar]
- Martins, M. J. F., Puckett, T. M., Lockwood, R., Swaddle, J. P., & Hunt, G. (2018). High male sexual investment as a driver of extinction in fossil ostracods. Nature, 556(7701), 366–369. 10.1038/s41586-018-0020-7 [DOI] [PubMed] [Google Scholar]
- Maurizio, P. L., Ferris, M. T., Keele, G. R., Miller, D. R., Shaw, G. D., Whitmore, A. C., West, A., Morrison, C. R., Noll, K. E., Plante, K. S., Cockrell, A. S., Threadgill, D. W., de Villena, F. P. M., Baric, R. S., Heise, M. T., & Valdar, W. (2018). Bayesian diallel analysis reveals MX1-dependent and MX1-independent effects on response to influenza A virus in mice. G3: Genes, Genomes, Genetics, 8, 427–445. 10.1534/g3.117.300438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J. (1982). Evolution and the theory of games. Cambridge University Press. [Google Scholar]
- Maynard Smith, J., & Price, G. R. (1973). The logic of animal conflict. Nature, 246, 15–18. 10.1038/246015A0 [DOI] [Google Scholar]
- Mérot, C., Llaurens, V., Normandeau, E., Bernatchez, L., & Wellenreuther, M. (2020). Balancing selection via life-history trade-offs maintains an inversion polymorphism in a seaweed fly. Nature Communications, 11, 670. 10.1038/s41467-020-14479-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalczyk, L., Dudziak, M., Radwan, J., & Tomkins, J. L. (2018). Fitness consequences of threshold trait expression subject to environmental cues. Proceedings of the Royal Society B: Biological Sciences, 285, 20180783. 10.1098/rspb.2018.0783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau, T. A., & Roff, D. A. (1987). Natural selection and the heritability of fitness components. Heredity, 59(2), 181–197. 10.1038/hdy.1987.113 [DOI] [PubMed] [Google Scholar]
- Okada, K., Katsuki, M., Sharma, M. D., Kiyose, K., Seko, T., Okada, Y., Wilson, A. J., & Hosken, D. J. (2021). Natural selection increases female fitness by reversing the exaggeration of a male sexually selected trait. Nature Communications, 12, 3420. 10.1038/s41467-021-23804-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, R., Taborsky, M., & Brockmann, H. J. (2008). Alternative reproductive tactics: An integrative approach. Cambridge University Press. [Google Scholar]
- Parrett, J. M., Chmielewski, S., Aydogdu, E., Łukasiewicz, A., Rombauts, S., Szubert-Kruszyńska, A., Babik, W., Konczal, M., & Radwan, J. (2022). Genomic evidence that a sexually selected trait captures genome-wide variation and facilitates the purging of genetic load. Nature Ecology and Evolution, 6(9), 1330–1342. 10.1038/s41559-022-01816-w [DOI] [PubMed] [Google Scholar]
- Parrett, J. M., & Knell, R. J. (2018). The effect of sexual selection on adaptation and extinction under increasing temperatures. Proceedings of the Royal Society B: Biological Sciences, 285, 20180303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrett, J. M., Mann, D. J., Chung, A. Y. C., Slade, E. M., & Knell, R. J. (2019). Sexual selection predicts the persistence of populations within altered environments. Ecology Letters, 22(10), 1629–1637. 10.1111/ele.13358 [DOI] [PubMed] [Google Scholar]
- Pearse, D. E., Barson, N. J., Nome, T., Gao, G., Campbell, M. A., Abadía-Cardoso, A., Anderson, E. C., Rundio, D. E., Williams, T. H., Naish, K. A., Moen, T., Liu, S., Kent, M., Moser, M., Minkley, D. R., Rondeau, E. B., Brieuc, M. S. O., Sandve, S. R., Miller, M. R., … Lien, S. (2019). Sex-dependent dominance maintains migration supergene in rainbow trout. Nature Ecology and Evolution, 3(12), 1731–1742. 10.1038/s41559-019-1044-6 [DOI] [PubMed] [Google Scholar]
- Peters, L., Huisman, J., Kruuk, L. E. B., Pemberton, J. M., & Johnston, S. E. (2022). Genomic analysis reveals a polygenic architecture of antler morphology in wild red deer (Cervus elaphus). Molecular Ecology, 31(4), 1281–1298. 10.1111/mec.16314 [DOI] [PubMed] [Google Scholar]
- Plesnar-Bielak, A., Skrzynecka, A. M., Miler, K., & Radwan, J. (2014). Selection for alternative male reproductive tactics alters intralocus sexual conflict. Evolution, 68, 2137–2144. 10.1111/evo.12409 [DOI] [PubMed] [Google Scholar]
- Plesnar-Bielak, A., Skrzynecka, A. M., Prokop, Z. M., & Radwan, J. (2012). Mating system affects population performance and extinction risk under environmental challenge. Proceedings of the Royal Society B: Biological Sciences, 279, 4661–4667. 10.1098/rspb.2012.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesnar-Bielak, A., Skwierzyńska, A. M., Hlebowicz, K., & Radwan, J. (2018). Relative costs and benefits of alternative reproductive phenotypes at different temperatures—Genotype-by-environment interactions in a sexually selected trait. BMC Evolutionary Biology, 18, 1–10. 10.1186/s12862-018-1226-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Radwan, J. (1993). The adaptive significance of male polymorphism in the acarid mite Caloglyphus berlesei. Behavior, Ecology and Sociobiology, 33(3), 201–208. 10.1007/bf00216601 [DOI] [Google Scholar]
- Radwan, J. (1995). Male morph determination in two species of acarid mites. Heredity, 74(6), 669–673. 10.1038/hdy.1995.91 [DOI] [Google Scholar]
- Radwan, J. (2001). Male morph determination in Rhizoglyphus echinopus (Acaridae). Experimental and Applied Acarology, 25(2), 143–149. 10.1023/a:1010688516704 [DOI] [PubMed] [Google Scholar]
- Radwan, J. (2003a). Heritability of male morph in the bulb mite, Rhizoglyphus robini (Astigmata, Acaridae). Experimental and Applied Acarology, 29(1–2), 109–114. 10.1023/a:1024260719013 [DOI] [PubMed] [Google Scholar]
- Radwan, J. (2003b). Inbreeding depression in fecundity and inbred line extinction in the bulb mite, Rhizoglyphus robini. Heredity, 90(5), 371–376. 10.1038/sj.hdy.6800254 [DOI] [PubMed] [Google Scholar]
- Radwan, J., Czyz, M., Konior, M., & Kołodziejczyk, M. (2000). Aggressiveness in two male morphs of the bulb mite Rhizoglyphus robini. Ethology, 106(1), 53–62. 10.1046/j.1439-0310.2000.00498.x [DOI] [Google Scholar]
- Radwan, J., & Klimas, M. (2001). Male dimorphism in the bulb mite, Rhizoglyphus robini: Fighters survive better. Ethology Ecology and Evolution, 13(1), 69–79. 10.1080/08927014.2001.9522788 [DOI] [Google Scholar]
- Reid, J. M. (2022). Intrinsic emergence and modulation of sex-specific dominance reversals in threshold traits. Evolution, 76(9), 1924–1941. 10.1111/evo.14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff, D. A. (1997). Evolutionary quantitative genetics. Chapman and Hall. [Google Scholar]
- Roff, D. A., & Emerson, K. (2006). Epistasis and dominance: Evidence for differential effects in life-history versus morphological traits. Evolution, 60(10), 1981–1990. [PubMed] [Google Scholar]
- Rowe, L., & Houle, D. (1996). The Lek paradox and the capture of genetic variance by condition dependent traits. Proceedings of the Royal Society B: Biological Sciences, 263, 1415–1421. [Google Scholar]
- Rundle, H. D., Chenoweth, S. F., & Blows, M. W. (2006). The roles of natural and sexual selection during adaptation to a novel environment. Evolution, 60(11), 2218–2225. [PubMed] [Google Scholar]
- Shorter, J. R., Maurizio, P. L., Bell, T. A., Shaw, G. D., Miller, D. R., Gooch, T. J., Spence, J. S., McMillan, L., Valdar, W., & de Villena, F. P. M. (2019). A diallel of the mouse collaborative cross founders reveals strong strain-specific maternal effects on litter size. G3: Genes, Genomes, Genetics, 9, 1613–1622. 10.1534/g3.118.200847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster, S. M., & Wade, M. J. (1991). Equal mating success among male reproductive strategies in a marine isopod. Nature, 350(6319), 608–610. 10.1038/350608a0 [DOI] [Google Scholar]
- Shuster, S. M., & Wade, M. J. (2003). Mating systems and strategies. Princeton University Press. [Google Scholar]
- Sinervo, B., & Lively, C. M. (1996). The rock-paper-scissors game and the evolution of alternative male strategies. Nature, 380(6571), 240–243. 10.1038/380240a0 [DOI] [Google Scholar]
- Smallegange, I. M. (2011). Complex environmental effects on the expression of alternative reproductive phenotypes in the bulb mite. Evolutionary Ecology, 25, 857–873. 10.1007/s10682-010-9446-6 [DOI] [Google Scholar]
- Smallegange, I. M., & Coulson, T. (2011). The stochastic demography of two coexisting male morphs. Ecology, 92(3), 755–764. 10.1890/09-2069.1 [DOI] [PubMed] [Google Scholar]
- Tilszer, M., Antoszczyk, K., Sałek, N., Zajac, E., & Radwan, J. (2006). Evolution under relaxed sexual conflict in the bulb mite Rhizoglyphus robini. Evolution, 60(9), 1868–1873. 10.1554/06-060.1 [DOI] [PubMed] [Google Scholar]
- Tomkins, J. L., & Hazel, W. (2007). The status of the conditional evolutionarily stable strategy. Trends in Ecology and Evolution, 22(10), 522–528. 10.1016/j.tree.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Zahavi, A. (1975). Mate selection—A selection for a handicap. Journal of Theoretical Biology, 53(1), 205–214. 10.1016/0022-5193(75)90111-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this manuscript have been deposited on Dryad doi:10.5061/dryad.18931zd2b