Abstract
Objectives
To determine how the intrinsic severity of successively dominant SARS-CoV-2 variants changed over the course of the pandemic.
Methods
A retrospective cohort analysis in the NHS Greater Glasgow and Clyde (NHS GGC) Health Board. All sequenced non-nosocomial adult COVID-19 cases in NHS GGC with relevant SARS-CoV-2 lineages (B.1.177/Alpha, Alpha/Delta, AY.4.2 Delta/non-AY.4.2 Delta, non-AY.4.2 Delta/Omicron, and BA.1 Omicron/BA.2 Omicron) during analysis periods were included. Outcome measures were hospital admission, ICU admission, or death within 28 days of positive COVID-19 test. We report the cumulative odds ratio; the ratio of the odds that an individual experiences a severity event of a given level vs all lower severity levels for the resident and the replacement variant after adjustment.
Results
After adjustment for covariates, the cumulative odds ratio was 1.51 (95% CI: 1.08–2.11) for Alpha versus B.1.177, 2.09 (95% CI: 1.42–3.08) for Delta versus Alpha, 0.99 (95% CI: 0.76–1.27) for AY.4.2 Delta versus non-AY.4.2 Delta, 0.49 (95% CI: 0.22–1.06) for Omicron versus non-AY.4.2 Delta, and 0.86 (95% CI: 0.68–1.09) for BA.2 Omicron versus BA.1 Omicron.
Conclusions
The direction of change in intrinsic severity between successively emerging SARS-CoV-2 variants was inconsistent, reminding us that the intrinsic severity of future SARS-CoV-2 variants remains uncertain.
Keywords: Intrinsic severity, COVID-19, SARS-CoV-2, Alpha variant, Delta variant, Omicron variant
Introduction
Since the SARS-CoV-2 pandemic started in late 2019, a succession of variants have achieved dominance, each replacing the previous dominant variant. From late 2020 these were designated variants of concern (VOCs); variants that exhibit increased transmission rates, antigenic differences, and/or case severity.1 The three VOCs that most impacted both the pandemic and epidemic in Scotland were Alpha (Pango lineage B.1.1.7), which emerged in the UK in September 2020,2 Delta (Pango lineage B.1.617.2), which emerged in India prior to October 20201 and spread globally in May 2021, with> 1000 introductions to the UK,3 and most recently Omicron (Pango lineage B.1.1.529), which emerged in Africa in November 2021,1 and spread very rapidly around the globe. Before the Omicron variant emerged, the Delta sublineage AY.4.2 was on course to replace the other Delta lineages, with growth rate estimates4 implying that it would become dominant in the UK in early 2022. This spread was arrested by the emergence of the more transmissible and immune-evading Omicron variant, which supplanted nearly all non-Omicron diversity5 and has itself diversified into several subvariants.
Understanding any change in disease severity associated with infection by new variants of a virus (especially one that has newly entered the human population) is critical from a clinical, public health, and basic science perspective. For example, knowledge of the severity of a new variant is vitally important in the decision-making process regarding the stringency of control measures and the roll out of vaccination and other treatments. It is expected that upon entry to a new host species from a zoonotic reservoir, the consequences of infection will be unpredictable with the severity of disease caused by the pathogen likely to be far from its evolutionary optima.6 SARS-CoV-2 is on average associated with low virulence in younger age groups whilst severe outcomes manifest in older age groups and those with comorbidities. Evolution of virulence may therefore not be strongly constrained, rather, factors governing transmission and immune evasion are likely to determine the direction of evolution.7
Our aim was to determine how the intrinsic severity of successively dominant SARS-CoV-2 variants has changed over the course of the Coronavirus disease 2019 (COVID-19) pandemic in Scotland, by making use of data from the largest health board in the country, Greater Glasgow and Clyde. To explore this, we used detailed clinical metadata and viral genomic data from National Health Service Greater Glasgow and Clyde (NHS GGC) to analyse relative case severity within 28 days of diagnosis between successive dominant lineages ( Fig. 1); B.1.177 versus Alpha, Alpha versus Delta, non-AY.4.2 versus AY.4.2 Delta, non-AY.4.2 Delta versus Omicron, and BA.1 Omicron versus BA.2 Omicron. This series of comparisons allowed us to assess trends in severity, removing this bias and accounting for other critical variables including detailed comorbidity and changes in the availability of new treatments and vaccination.
Fig. 1.
SARS-CoV-2 sequencing through time in Scotland.
Figure adapted and extended from Pascall et al.26
Patients and methods
This is a cohort study comparing the clinical outcomes of adults infected with a resident dominant SARS-CoV-2 lineage, with those infected with the SARS-CoV-2 lineage that would subsequently displace it.
Genome sequencing
Sequences were generated using the ARTIC Network protocol, originally developed for Oxford Nanopore-based sequencing.8 and derived versions adapted to Illumina and ARTIC-unrelated amplicon-based protocols. The COVID-19 Genomics UK Consortium (COG-UK) pipeline was used for alignment and Pango lineage assignment.9
Data inclusion/exclusion
This work was originally performed in real time for the COVID-19 response, as such, the time windows begin at the start of the first month where we believed we had enough sequences of both the resident and replacement lineages to perform the analysis, and end at the end of last month meeting the same criteria.
For the B.1.177/Alpha analysis, we included all sequenced samples with full data available on all adjustment variables from within the NHS GGC health board between 1st November 2020 and 30th January 2021 (B.1.177: n = 807; Alpha: n = 833). A full demographic breakdown of samples is shown in Table S1. All sequences assigned as B.1.177 and associated sublineages were merged into a single category for the analysis.
For the Alpha/Delta analysis we included all sequenced samples with full metadata between 1st April 2021 and 30th June 2021 (Alpha: n = 2104; Delta: n = 3448). All sequences assigned into B.1.617.2 and associated sublineages were merged into the Delta category for the analysis. A full demographic breakdown of samples is shown in Table S2.
For the Delta/AY.4.2 analysis we included all sequenced samples with full metadata between 1st July 2021 and 31st October 2021 (non-AY.4.2 Delta: n = 8644; AY.4.2 Delta: n = 969). The AY.4.2 category was defined as all sequences assigned as either AY.4.2 or sublineages thereof, all remaining Delta sublineages were combined into the comparison category. A full demographic breakdown of samples is shown in Table S3.
For the non-AY.4.2 Delta/Omicron analysis, we included all sequenced samples with full metadata between 1st December 2021 and 31st December 2021 (non-AY.4.2 Delta: 1164; Omicron: 2694). The Omicron category was defined as all sequences assigned as BA.1 or B.1.1.529, the Delta category was defined as for the Delta/AY.4.2 comparison. Full demographic breakdown of samples is shown in Table S4.
For the BA.1 Omicron/BA.2 Omicron analysis, we included all sequenced samples with full metadata between 1st January 2022 and 31st March 2022 (BA.1 Omicron: 9949; BA.2 Omicron: 12994). The BA.1 Omicron category was defined as all sequences assigned as BA.1 or sublineages thereof, the BA.2 Omicron category was defined as all sequences assigned as BA.2 or sublineages thereof. Full demographic breakdown of samples is shown in Table S5.
Cases with hospital acquired COVID-19, defined as a first positive polymerase chain reaction (PCR) test occurring more than 48 h following admission to hospital, and all cases younger than 18 were excluded.
Data linkage
Cohorts and de-identified linked data for the entire NHS GGC health board (1.2 million people) were prepared by the West of Scotland Safe Haven at NHS GGC. Data used in the analysis included maximum clinical severity at 28 days after the first positive test via a 4-point ordinal scale (1. No hospitalisation; 2. Hospitalisation (excluding elective surgery); 3. Admission to High Dependency Unit (HDU)/Intensive Care Unit (ICU); 4. Death), age at diagnosis, date of positive test, sex, partial postcode, number of vaccine doses, number of relevant comorbidities or risks of ill health (chronic cardiac disease, chronic respiratory disease, chronic renal disease, liver disease, dementia, chronic neurological conditions, connective tissue disease, diabetes, HIV infection, malignant tumours, clinician defined obesity, case shielding, immunosuppressive drugs, chemotherapy) and reinfection. Vaccination, comorbidities, reinfection, age and biological sex at birth were included as they are known to impact clinical outcomes. Note that, in Scotland, vaccination schedules prioritised the elderly and vulnerable. Therefore, throughout, the number of vaccine doses is correlated with age with older individuals having had more doses on average than younger individuals. Date of positive test was included to attempt to adjust for any time varying effects across the study window (e.g., changing clinical practice, hospital load, etc.). Partial postcode was included to attempt to account for spatial clustering of risk mediated though unmeasured variables (e.g., deprivation, ethnicity, etc.).
Severity was scored twice. For severity for “with” analyses, cases were assigned the most severe event that occurred within 28 days of their positive test. For the “of” analyses, events were only counted when they were explicitly linked to COVID-19 infection in the electronic patient records. Different datasets used in the linkage recorded “cause of” event differently. In datasets where ICD-10 codes were used (SMR01, accident and emergency, and deaths), a COVID-19 related ICD-10 code was required (specifically, any code starting U07, U04.9 which corresponds to an incorrect usage of the SARS ICD-10 code, and U10). In datasets where ICD-10 codes were not used (Scottish Intensive Care Society Audit Group data), the string “covid” was searched for case-insensitively in the free text entry. When the number of vaccine doses an individual had been given was calculated, if the last dose had been received less than 14 days before the date of the positive PCR test, it was ignored. Individuals with multiple confirmed episodes of infection (defined as separate positive PCR results more than 90 days apart) were marked as reinfected for any episodes after the first.
Statistical analysis of clinical data
The four-level patient outcome data were analysed using cumulative generalised additive mixed models (GAMMs) with logit links10 fit using Bayesian inference. These GAMMs included lineage, reinfection, patient sex and number of vaccine doses as categorical fixed effects and number of 4C Mortality Score comorbidities identified as a continuous fixed effect, with partial postcode included as a random effect. We included age and date of positive test as non-linear penalised regression splines. The basis dimension of the penalised regression splines was set to the number of unique dates of positive tests minus one and the number of unique ages (rounded to year) minus one respectively, with the intention that regularisation occur through the prior. Given that the pandemic was in its early stages during the first comparison (B.1.177/Alpha) and the vaccination campaign had not yet started, both reinfection and number vaccine doses received were excluded from the first model.
The same classes of parameters received the same priors in each model. The intercepts of the models were given t-distribution (location = 0, scale = 2.5, df = 3) priors, fixed effects were given normal (mean = 0, standard deviation = 2.5) priors, random effects and spline standard deviations were given exponential (mean = 2.5) priors. All severity models were fitted using the brms (v. 2.14.4) R package.11 All presented models had no divergent transitions and effective sample sizes of over 200 for all parameters.
We tested the robustness of these estimates to epidemic phase bias. Epidemic phase bias is caused by patient outcomes being correlated with the time from infection to positive test. This correlation makes the estimated odds ratio adjusting for time of positive test a biased estimator of the odds ratio adjusting for time of infection, which is the desired target of estimation.12 When lineages differ in their incidence, this effect can lead to incorrectly concluding that one lineage is associated with more severe disease than the other, when the estimated difference is driven almost entirely by the epidemic phase bias. Sensitivity to this bias was assessed using the method of Seaman et al. 12 We added four days to the population who experienced more extreme outcomes (i.e., hospitalisation, admission to ICU/HCU or death), generating a modified test time for each individual, where if the patient was not hospitalised, it was their original test time and otherwise it was this new test time. We refit the model using the modified times and using individuals whose modified times lie within the inclusion window. The resulting estimate of the cumulate odds ratio was then compared with the cumulate odds ratio estimated from the original model.
Ethical approval
Anonymised clinical data were available using the (NHS GGC SafeHaven platform and included vaccination status (dates and product names for each dose), demographic data (age and sex) comorbidities, hospital records and dates of positive and negative PCR tests, for 1.2 million inhabitants of the (NHS GGC) area over 18 years of age. Data were matched by CHI number and pseudonymised before analysis. Derogated ethical approval for the study was granted by the NHS GGC SafeHaven committee (GSH/21/IM/001).
Results
B.1.177/Alpha
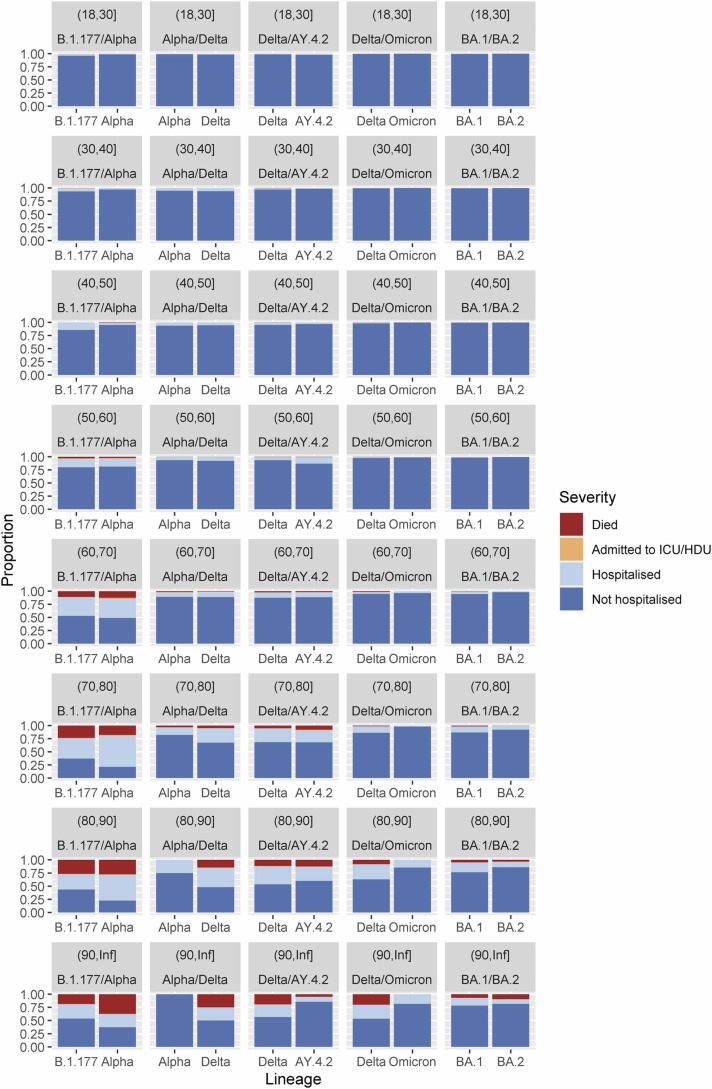
Our first comparison was the severity of the SARS-CoV-2 Alpha VOC versus the previous dominant non-VOC lineage, Pango designation B.1.177 (B.1.177: n = 807; Alpha: n = 833). The replacement of the lineages over time can be seen in Fig. 1. We found that confirmed Alpha cases were associated with more severe infection (“of” - median cumulative odds ratio: 1.60; 95% central interval: 1.10–2.30; probability of association with increased severity:>0.99; “with” – median cumulative odds ratio: 1.51; 95% central interval: 1.08–2.11; probability of association with increased severity: 0.99; breakdown of number events in each group Table S1). The breakdown of “with” severity score by age and lineage can be seen in Fig. 2a. Parameter estimates for all parameters can be found in Table S6 and Fig. S1. We found that, in our epidemic phase bias sensitivity analysis, Alpha remained associated with increased severity, but that the magnitude was reduced, as would be expected, and that in both analyses the probability of the effect being positive was reduced below 95% (“of” - median cumulative odds ratio: 1.31; 95% central interval: 0.91–1.88; probability of association with increased severity: 0.93; “with” – median cumulative odds ratio: 1.24; 95% central interval: 0.88–1.79; probability of association with increased severity: 0.88).
Fig. 2.
Comparison of case severity by age between pairs of co-circulating SARS-CoV-2 lineages Clinical severity was measured on a four-level ordinal scale based on observed outcomes within 28 days of a positive test: no hospitalisation, hospitalisation, admission to Intensive Care Unit/High Dependency Unit (ICU/HDU), death. The first column compares B.1.177 and Alpha between 1st November 2020 and 30th January 2021. The second column compares Alpha and Delta between 1st April 2021 and 30th June 2021. The third column compares Delta lineages against AY.4.2 between 1st July 2021 and 30th September 2021. The fourth column compares non-AY.4.2 Delta and Omicron between 1st December 2021 and 31st December 2021. The final column compares BA.1 Omicron and BA.2 Omicron between 1st January 2022 and 31st March 2022.
Alpha/Delta
Our second comparison was the severity of the SARS-CoV-2 Delta VOC versus the previous dominant Alpha VOC lineage (Alpha: n = 2104; Delta: n = 3448). We estimate a substantial increase in case severity associated with Delta infections relative to Alpha (“of” - median cumulative odds ratio: 2.19; 95% central interval: 1.48–3.32; probability of association with increased severity:>0.99; “with” - median cumulative odds ratio: 2.09; 95% central interval: 1.42–3.08; probability of association with increased severity:>0.99; breakdown of number events in each group Table S2). The breakdown of “with” severity score by age and lineage can be seen in Fig. 2b. Parameter estimates for all parameters can be found in Table S7 and Fig. S1. This effect is large enough that in epidemic phase bias sensitivity bias model, we still estimate a positive effect with a probability of positivity of over 0.95 (“of” - median cumulative odds ratio: 1.45; 95% central interval: 0.98–2.16; probability of association with increased severity: 0.96; “with” - median cumulative odds ratio: 1.40; 95% central interval: 0.97–2.02; probability of association with increased severity: 0.97).
Non-AY.4.2 Delta/AY.4.2 Delta
Our third comparison was the severity of the SARS-CoV-2 Delta sublineage AY.4.2 versus the other Delta sublineages it was replacing (non-AY.4.2 Delta: n = 8644; AY.4.2 Delta: n = 969). We estimate that the AY.4.2 lineage infections are associated with similar case severity to that seen in other Delta sublineage infections (“of” - median cumulative odds ratio: 1.05; 95% central interval: 0.79–1.39; probability of association with increased severity: 0.64; “with” - median cumulative odds ratio: 0.99; 95% central interval: 0.76–1.27; probability of association with increased severity: 0.46; breakdown of number events in each group Table S3). The breakdown of “with” severity score by age and lineage can be seen in Fig. 2c. Parameter estimates for all parameters can be found Table S8 and Fig. S1. In this case, there is no noticeable effect of epidemic phase bias, likely as the growth rate difference between the two variants was small (“of” - median cumulative odds ratio: 1.02; 95% central interval: 0.76–1.35; probability of association with increased severity: 0.57; “with” - median cumulative odds ratio: 0.98; 95% central interval: 0.75–1.27; probability of association with increased severity: 0.44).
Non-AY.4.2 Delta/Omicron
Our fourth comparison was the severity of the SARS-CoV-2 Omicron VOC versus the non-AY.4.2 Delta sublineages (non-AY.4.2 Delta: 1164; Omicron: 2694). We find that Omicron (BA.1 sublineage) infection is associated with substantially less severe disease (“of” - median cumulative odds ratio: 0.15; 95% central interval: 0.01–1.48; probability of association with increased severity: 0.06; “with” - median cumulative odds ratio: 0.49; 95% central interval: 0.22–1.06; probability of association with increased severity: 0.04; breakdown of number events in each group Table S4). The breakdown of “with” severity score by age and lineage can be seen in Fig. 2d. Parameter estimates for all parameters can be found in Table S9 and Fig. S1. Both the “with” and “of” analyses show a strong impact of epidemic phase bias. Omicron was the faster growing lineage, so the estimates of Omicron are driven more negative (“of” - median cumulative odds ratio: 0.07; 95% central interval:<0.01–0.68; probability of association with increased severity: 0.01; “with” - median cumulative odds ratio: 0.19; 95% central interval: 0.08–0.42; probability of association with increased severity:<0.01).
BA.1 Omicron/BA.2 Omicron
Our final comparison was the severity of the BA.1 Omicron sublineage versus the BA.2 Omicron sublineage (BA.1 Omicron: 9949; BA.2 Omicron: 12994). We find that BA.2 Omicron infection is associated with less severe disease than BA.1 Omicron (“of” - median cumulative odds ratio: 0.83; 95% central interval: 0.63–1.12; probability of association with increased severity: 0.11; “with” - median cumulative odds ratio: 0.86; 95% central interval: 0.68–1.09; probability of association with increased severity: 0.11; breakdown of number events in each group Table S5). The breakdown of “with” severity score by age and lineage can be seen in Fig. 2e. Parameter estimates for all parameters can be found in Table S10 and Fig. S1. This association is uncertain, but both the “with” and “of” analyses show a strong impact of epidemic phase bias. As BA.2 Omicron was the faster growing lineage, so the estimates of BA.2 Omicron are driven more negative, and our certainty of an association with a reduction of severity becomes higher (“of” - median cumulative odds ratio: 0.51; 95% central interval: 0.38–0.69; probability of association with increased severity:<0.01; “with” - median cumulative odds ratio: 0.57; 95% central interval: 0.49–0.72; probability of association with increased severity:<0.01).
Discussion
The principal findings of this study of the relative severity of COVID-19 cases caused by successive SARS-CoV-2 variant waves in Scotland was that Alpha was associated with more severe disease than B.1.177, Delta was associated with more severe disease than Alpha, non-AY.4.2 Delta and AY.4.2 Delta were associated with similar disease severity, Omicron was associated with much less severe disease than non-AY.4.2 Delta, and that BA.2 Omicron was associated with less severe disease than BA.1 Omicron. These conclusions were after accounting for comorbidities, date of positive test (to adjust for time varying effects across the study window), previous infection and vaccine availability, and robust to epidemic phase bias and the possibility of coincidental SARS-CoV-2 infection at admission to hospital. The successive replacements that we studied were not consistent in the direction of change in case severity.
Our study design has several strengths. It is the first study to our knowledge to analyse the sequential replacement of variants throughout the pandemic with respect to the progression of severity attributable to virus evolution, and to use a consistent analytical approach across sequential SARS-CoV-2 lineages. Lauring et al.13 is, in spirit, similar to our work, but their primary focus is not on the trajectory of severity, and they do not consider multiple time matched comparisons.
Secondly, we test the robustness of the severity analysis to epidemic phase bias and the impact of differences in the definition of severity outcomes (“of” analysis versus “with” analysis) across sequential variants, showing that these different definitions lead to qualitatively similar results.
Also, our approach takes a broad view of the definition of disease severity, by including community- and hospital-based cases and by considering a wider variety of clinical outcomes than those considered in most previous severity analyses. This is both a strength and limitation of our study as it provides a more accurate assessment of disease severity in the adult population than studies that have concentrated solely on hospitalised patients, or on the risk of hospitalisation, however due to our sample size the numbers of deaths and admissions to ICU in our cohort were low.
Our study does, however, have some limitations. Firstly, we only include cases with sequenced genomes, and thus our sample is biased towards cases with lower cycle threshold (Ct), because these cases are more likely to have been sequenced. This is likely to be particularly important when the Ct distribution of infections differs between variants, such as the higher Ct values seen with Omicron infection compared to Delta infection.14, 15 The direction of the sampling bias on our results depends on which of the variants is associated with the lower Ct infections. Requiring sequences for our samples also limits us to the set of individuals who have been tested by PCR, likely to represent hospitalised patients more than those in the community and assumes that this these proportions are stable across our sub-periods, an assumption which is untestable from the data we have available. We did not have access to biomarker data, and as such could not adjust for correlates of immunity (e.g., T cell counts, antibody titre) directly. Additionally, our sample size was not large enough to adjust for all the factors we would have liked to (e.g., fitting a dose by vaccine brand interaction, given that different brands are known to provide differential protection against different variants,7, 16, 17 or accounting for the differential protection induced by infection prior to vaccination).
There have been a series of other studies investigating each of the comparisons in our study individually. For Alpha versus previous variants, most previous studies in most regions have also estimated an increase in severity over extant diversity. A wide variety of end points have been used, 28-day mortality, hospitalisation, and an ordinal scale based around supplemental oxygen.18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 Our estimates are consistent with the majority of these studies.
When considering the Delta variant, two UK community analyses found that Delta (or an S-gene proxy) infections were associated with a higher risk of admission to hospital than with Alpha.31, 32 Comparable results were observed in Danish, US, and Canadian populations with a study in a Norwegian population being the exception.33, 34, 35, 36 The US and Canadian studies also found that Delta was associated with increased risk of ICU admission and death.34, 35 Most of these studies are therefore consistent with our results.
Our estimate that AY.4.2 is associated with approximately the sample severity as other Delta sublineages is inconsistent with an analysis of the English population which found that confirmed AY.4.2 cases are associated with lower hospitalisation risk than cases associated with non-AY.4.2 Delta.37 This inconsistency between our study and others may be explained by differences in the adjustment variables used, or because the larger sample size in Nyberg et al. 37 allowed precise isolation of a small negative effect, with their effect estimate falling within our credible interval.
Our results are consistent with studies from England, Scotland, Denmark, Norway, South Africa, Canada, and the US suggesting that Omicron infections are less severe than infections with Delta.13, 38, 39, 40, 41, 42, 43, 44 This reduction in case severity resulted in increased numbers of patients being admitted to hospital with a coincidental positive SARS-CoV-2 test rather than due to COVID-19, but this did not seem to overly impact our estimate of the severity of the variant relative to Delta. Contrary to some reporting,45, 46 we find that BA.2 is infection is likely associated with a small further decrease in severity relative to BA.1 infection, potentially because we had access to genomically confirmed infections and did not have to use proxies of lineage.
It is important to appreciate measures of disease severity are highly context dependent. The clinical situation of the pandemic has shifted dramatically in Scotland during the study period, from a time with very little prior immunity to one with widespread vaccine and prior infection mediated immunity. Treatment availability with steroids, antivirals and antithrombotic agents has had a huge impact on reducing length of hospital stay and mortality.47, 48, 49 Testing patterns have also changed dramatically across the study, with periods of higher and lower rates of testing, as well as changes in the proportion of positives being sequenced. All these factors may impact the relative severity of variants, as well as the meanings of estimates of the association between lineage and severity, as the populations being compared are not the same between comparisons. For this reason, our results cannot be used to compare the intrinsic case severity of variants that did not co-circulate.
Our results demonstrate that successive variants of SARS-CoV-2 are associated with inconsistent directions of change in intrinsic disease severity after other factors are accounted for in the analysis, including comorbidities, vaccination, previous infection, and date of positive test. In keeping with this finding, we have recently demonstrated fundamental changes in the life cycle of the Omicron variants BA.1 and BA.2 that may provide a biological explanation for the substantial drop in severity associated with this variant7; Omicron is associated with less cell-to-cell fusion and tropism for nasal epithelial cells rather than cells present in the lungs as well as an endosomal rather than a direct cell entry pathway.
Given that the progression of SARS-CoV-2 virulence has not been consistent over time and that the entry pathway of SARS-CoV-2 may be changed by single amino acid changes within the S2 domain of the spike protein,50 historical trends in severity should not be used to predict the severity of future variants. This is especially true as there is no guarantee a newly emerging lineage will descend from the current Omicron diversity, and thus may have very different biological characteristics. However, once a variant has emerged, the likelihood of immune evasion and the method of cell entry may be estimated from the genome sequence. The relative reduction in severity seen with the Omicron variant should not make us complacent to the potential risks of future SARS-CoV-2 variants. Any increase to intrinsic severity in a variant with similar transmissibility to Omicron could be devastating to health systems and communities.
Results from this study were made available in a timely manner to national government agencies, and informed decisions on public health restrictions, vaccination strategies, and healthcare service planning. This study provides an important baseline for future research monitoring the relative severity of new variants and highlights the importance of ongoing genomic “early-warning” surveillance to detect new variants of concern promptly as they emerge.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Thomas C Williams is Principal Investigator for the BronchStart project, which is funded by the Respiratory Syncytial Virus Consortium in Europe (RESCEU), with data collection supported by the National Institute for Health Research.
Acknowledgements
The authors would like to acknowledge the work of the West of Scotland Safe Haven team in supporting extractions and linkage to de-identified NHS patient datasets. COG-UK is supported by funding from the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute.
This work was supported by UKRI through the JUNIPER consortium (grant number MR/V038613/1, https://maths.org/juniper/); the Medical Research Council (MRC) (grant numbers MC UU 12014/12, MC UU 00002/11, https://www.ukri.org/councils/mrc/); the G2P-UK National Virology Consortium (grant number MR/W005611/1, https://www.ukri.org). The funders had no input on the study design and analysis.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jinf.2023.05.019.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.World Health Organization [Internet]. Tracking SARS-CoV-2 variants. [cited 2022 March 23] Available from: 〈https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/〉.
- 2.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations; 2020 [cited 2022 March 23]. Available from: 〈https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563〉.
- 3.McCrone J.T., Hill V., Bajaj S., Pena R.E., Lambert B.C., Inward R., et al. Context-specific emergence and growth of the SARS-CoV-2 Delta variant. Nature. 2022;610:154–160. doi: 10.1038/s41586-022-05200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.UK Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England: technical briefing 29; 2021 [cited 2022 March 23]. Available from: 〈https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1036501/Technical_Briefing_29_published_26_November_2021.pdf〉.
- 5.GISAID [Internet]. Tracking of variants. [cited 2022 March 23]. Available from: 〈https://www.gisaid.org/hcov19-variants/〉.
- 6.Visher E., Evensen C., Guth S., Lai E., Norfolk M., Rozins C., et al. The three Ts of virulence evolution during zoonotic emergence. Proc R Soc B. 2021;288:20210900. doi: 10.1098/rspb.2021.0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willett B.J., Grove J., MacLean O.A., Wilkie C., De Lorenzo G., Furnon W., et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quick J. nCoV-2019 sequencing protocol v3 (LoCost) V.3. protocols.io; 2020 [cited 2022 March 23]. Available from: 〈https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bp2l6n26rgqe/v3〉.
- 9.Nicholls S.M., Poplawski R., Bull M.J., Underwood A., Chapman M., Abu-Dahab K., et al. CLIMB-COVID: continuous integration supporting decentralised sequencing for SARS-CoV-2 genomic surveillance. BMC Genom. 2021;22:196. doi: 10.1186/s13059-021-02395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bürkner P.-C., Vuorre M. Ordinal regression models in psychology: a tutorial. Adv Methods Pract Psychol Sci. 2019;2(1):77–101. [Google Scholar]
- 11.Bürkner P.-C. brms: an R package for Bayesian multilevel models using stan. J Stat Softw. 2017;80(1):1–28. [Google Scholar]
- 12.Seaman S.R., Nyberg T., Overton C.E., Pascall D.J., Presanis A.M., De Angelis D. Adjusting for time of infection or positive test when estimating the risk of a post-infection outcome in an epidemic. Stat Methods Med Res. 2022;31(10):1942–1958. doi: 10.1177/09622802221107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauring A.S., Tenforde M.W., Chappell J.D., Gaglani M., Ginde A.A., McNeal T. Clinical severity of, and effectiveness of mRNA vaccines against, covid-19 from omicron, delta, and alpha SARS-CoV-2 variants in the United States: prospective observational study. BMJ. 2022;376 doi: 10.1136/bmj-2021-069761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tassetto M., Garcia-Knight M., Anglin K., Lu S., Zhang A., Romero M., et al. Detection of higher cycle threshold values in culturable SARS-CoV-2 Omicron BA.1 sublineage compared with pre-omicron variant specimens — San Francisco Bay Area, California, July 2021—March 2022. MMWR Morb Mortal Wkly Rep. 2022;71:1151–1154. doi: 10.15585/mmwr.mm7136a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sentis C., Billaud G., Bal A., Frobert E., Bouscambert M., Destras G., et al. SARS-CoV-2 omicron variant, lineage BA.1, is associated with lower viral load in nasopharyngeal samples compared to delta variant. Viruses. 2022;14(5):919. doi: 10.3390/v14050919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;206:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UK Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England: technical briefing 34; 2022 [cited 2022 March 23]. Available from: 〈https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050236/technical-briefing-34-14-january-2022.pdf〉.
- 18.Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group, Edmunds W.J., Jewell N.P., Diaz-Ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Challen R., Brooks-Pollock E., Read J., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grint D.J., Wing K., Williamson E., McDonald H.I., Evans D., Evans S.J.W., et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Eur Surveill. 2021;26(11):2100256. doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyberg T., Twohig K.A., Harris R.J., Seaman S.R., Flannagan J., Allen H., et al. Risk of hospital admission for patients with SARS-CoV-2 variant B.1.1.7: cohort analysis. BMJ. 2021;373:1412. doi: 10.1136/bmj.n1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dabrera G., Allen H., Zaidi A., Twohig K., Thelwall S., Marchant E., et al. Assessment of mortality and hospital admissions associated with confirmed infection with SARS-CoV-2 variant of concern VOC-202012/01 (B.1.1.7) a matched cohort and time-to-event analysis. SSRN. 2021 doi: 10.2807/1560-7917.ES.2022.27.20.2100377. 〈https://ssrn.com/abstract=3802578〉 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grint D.J., Wing K., Houlihan C., Gibbs H.P., Evans S.J.W., Williamson E., et al. Severity of severe acute respiratory system coronavirus 2 (SARS-CoV-2) alpha variant (B.1.1.7) in England. Clin Infect Dis. 2021;75(1):e1120–e1127. doi: 10.1093/cid/ciab754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snell L.B., Wang W., Alcolea-Medina A., Charalampous T., Batra R., de Jongh L., et al. Descriptive comparison of admission characteristics between pandemic waves and multivariable analysis of the association of the Alpha variant (B.1.1.7 lineage) of SARS-CoV-2 with disease severity in inner London. BMJ Open. 2022;12 doi: 10.1136/bmjopen-2021-055474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021;21:1246–1256. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong S.W.X., Chiew C.J., Ang L.W., Mak T.-M., Cui L., Toh M.P.H.S., et al. Clinical and virological features of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617.2 (Delta) Clin Infect Dis. 2022;75(1):e1128–e1136. doi: 10.1093/cid/ciab721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stirrup O., Boshier F., Venturini C., Guerra-Assunção A., Alcolea-Medina A., Beckett A., et al. SARS-CoV-2 lineage B.1.1.7 is associated with greater disease severity among hospitalised women but not men: multicentre cohort study. BMJ Open Resp Res. 2021;8 doi: 10.1136/bmjresp-2021-001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascall D.J., Vink E., Blacow R., Bulteel N., Campbell A., Campbell R., et al. The SARS-CoV-2 Alpha variant was associated with increased clinical severity of COVID-19 in Scotland: A genomics-based retrospective cohort analysis. PLOS ONE. 2023;18(14) doi: 10.1101/2021.08.17.21260128e0284187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veneti L., Seppälä E., Larsdatter Storm M., Valcarcel Salamanca B., Alnes Buanes E., Aasand N., et al. Increased risk of hospitalisation and intensive care admission associated with reported cases of SARS-CoV-2 variants B.1.1.7 and B.1.351 in Norway, December 2020 -May 2021. PLOS ONE. 2021;16(10) doi: 10.1371/journal.pone.0258513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funk T., Pharris A., Spiteri G., Bundle N., Melidou A., Carr M., et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Eur Surveill. 2021;26(16):2100348. doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheikh A., McMenamin J., Taylor B., Robertson C., Public Health Scotland, EAVE II Collaborators SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397(10293):2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Twohig K.A., Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S., et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22(1):35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bager P., Wohlfahrt J., Rasmussen M., Albertsen M., Krause T.G. Hospitalisation associated with SARS-CoV-2 delta variant in Denmark. Lancet Infect Dis. 2021;21(10):1351. doi: 10.1016/S1473-3099(21)00580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bast E., Tang F., Dahn F., Palacio A. Increased risk of hospitalisation and death with the delta variant in the USA. Lancet Infect Dis. 2021;21(12):1629–1630. doi: 10.1016/S1473-3099(21)00685-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisman D.N., Tuite A.R. Evaluation of the relative virulence of novel SARS-CoV-2 variants: a retrospective cohort study in Ontario, Canada. CMAJ. 2021;193(42):E1619–E1625. doi: 10.1503/cmaj.211248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veneti L., Salamanca B.V., Seppälä E., Starrfelt J., Storm M.L., Bragstad K., et al. No difference in risk of hospitalization between reported cases of the SARS-CoV-2 Delta variant and Alpha variant in Norway. Int J Infect Dis. 2022;115:178–184. doi: 10.1016/j.ijid.2021.12.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyberg T., Harman K., Zaidi A., Seaman S.R., Andrews N., Nash S.G., et al. Hospitalization and mortality risk for COVID-19 cases with SARS-CoV-2 AY.4.2 (VUI-21OCT-01) compared to non-AY.4.2 delta variant sublineages. J Infect Dis. 2022:Jiac063. doi: 10.1093/infdis/jiac063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyberg T., Ferguson N.M., Nash S.G., Webster H.H., Flaxman S., Andrews N., et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399(10332):1303–1312. doi: 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulloa A.C., Buchan S.A., Daneman N., Brown K.A. Estimates of SARS-CoV-2 Omicron variant severity in Ontario, Canada. JAMA. 2022 doi: 10.1001/jama.2022.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheikh A., Kerr S., Woolhouse M., McMenamin J., Robertson C. Severity of omicron variant of concern and effectiveness of vaccine boosters against symptomatic disease in Scotland (EAVE II): a national cohort study with nested test-negative design. Lancet Infect Dis. 2022;22:959–966. doi: 10.1016/S1473-3099(22)00141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bager P., Wohlfahrt J., Bhatt S., Stegger M., Legarth R., Moller C.H., et al. Risk of hospitalisation associated with infection with SARS-CoV-2 omicron variant versus delta variant in Denmark: an observational cohort study. Lancet Infect Dis. 2022;22(7):967–976. doi: 10.1016/S1473-3099(22)00154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veneti L., Boas H., Brathen Kristoffersen A., Stalcrantz J., Bragstad K., Hungnes O., et al. Reduced risk of hospitalisation among reported COVID-19 cases infected with the SARS-CoV-2 Omicron BA.1 variant compared with the Delta variant, Norway, December 2021 to January 2022. Eur Surveill. 2022;27(4) doi: 10.2807/1560-7917.ES.2022.27.4.2200077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward I.L., Bermingham C., Ayoubkhani D., Gethings O.J., Pouwels K.B., Yates T., et al. Risk of covid-19 related deaths for SARS-CoV-2 omicron (B.1.1.529) compared with delta (B.1.617.2): retrospective cohort study. BMJ. 2022;378 doi: 10.1136/bmj-2022-070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolter N., Jassat W., Walaza S., Welch R., Moultrie H., Groome M., et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399(10323):437–446. doi: 10.1016/S0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolter N., Jassat W., The DATCOV-Gen author group, von Gottberg A., Cohen C. Clinical severity of omicron lineage BA.2 infection compared with BA.1 infection in South Africa. Lancet. 2022;400:93–96. doi: 10.1016/S0140-6736(22)00981-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewnard J.A., Hong V.X., Patel M.M., Kahn R., Lipsitch M., Tartof S.Y. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat Med. 2022;28(9):1933–1943. doi: 10.1038/s41591-022-01887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021;384(3):238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sholzberg M., Tang G.H., Rahhal H., Al Hamzah M., Kreuziger L.B., Áinle F.N., et al. Effectiveness of therapeutic heparin versus prophylactic heparin on death, mechanical ventilation, or intensive care unit admission in moderately ill patients with covid-19 admitted to hospital: RAPID randomised clinical trial. BMJ. 2021;375:n2400. doi: 10.1136/bmj.n2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peacock T.P., Brown J.C., Zhou J., Thakur N., Sukhova K., Newman J., et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv. 2022 doi: 10.1101/2021.12.31.474653. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material