Abstract
Red alder (Alnus rubra Bong.) is an ecologically significant and important fast-growing commercial tree species native to western coastal and riparian regions of North America, having highly desirable wood, pigment, and medicinal properties. We have sequenced the genome of a rapidly growing clone. The assembly is nearly complete, containing the full complement of expected genes. This supports our objectives of identifying and studying genes and pathways involved in nitrogen-fixing symbiosis and those related to secondary metabolites that underlie red alder's many interesting defense, pigmentation, and wood quality traits. We established that this clone is most likely diploid and identified a set of SNPs that will have utility in future breeding and selection endeavors, as well as in ongoing population studies. We have added a well-characterized genome to others from the order Fagales. In particular, it improves significantly upon the only other published alder genome sequence, that of Alnus glutinosa. Our work initiated a detailed comparative analysis of members of the order Fagales and established some similarities with previous reports in this clade, suggesting a biased retention of certain gene functions in the vestiges of an ancient genome duplication when compared with more recent tandem duplications.
Keywords: Alnus rubra, red alder, genome, actinorhizal plant, nitrogen fixation
Introduction
Red alder (Alnus rubra Bong.) is a tree of pivotal ecological, economic, and cultural importance in the forest ecosystems of western North America. Distributed from Alaska to California, it is found principally on western-facing slopes within a few hundred miles of the coast, with small pockets occurring in Idaho. A pioneer species, red alder establishes rapidly on exposed mineral soil, typically after land disturbances such as logging or flooding. It also grows on so-called marginal lands, which are considered unsuitable for conventional agricultural crops, the sustainable use of which could potentially represent an effective route for expanding the area devoted to growing timber and feedstocks without taking land out of food production. Alders significantly help restore degraded soils, including industrial waste ground. Key to the ability of red alder to improve the soil quality of marginal sites is its symbiotic relationship with the actinobacterium Frankia. Together they form nitrogen-fixing root nodules, which support plant growth in nitrogen-deficient environments and contribute to overall soil fertility (Benson and Silvester 1993; Hart et al. 1997; Deal and Harrington 2006).
From an economic perspective, red alder is mainly used for timber and paper production. In recent years, the annual market value of red alder has exceeded that of Douglas-fir (http://www.westernhardwood.org/Miscellaneous/GIS_hardwood_inventory_6.pdf). Washington State alone has ∼3.7 million acres of annually harvestable/processed hardwoods, with 90% being red alder. In 2002, in Washington State, red alder accounted for >60% of hardwood standing timber available for commercial harvesting (Deal and Harrington 2006). Additional economic potential of red alder comes from its suitability as a biomass feedstock (Gelfand et al. 2013). It grows rapidly, with a wood density of ∼460 kg/m3, as opposed, for example, to 380 kg/m3 maximum in poplar, this being demonstrably economically more valuable (https://www.engineeringtoolbox.com/). It can be coppiced as a short rotation crop, growing into very dense groves (50,000 trees/acre; DeBell 1972), and can produce a high level of biomass at 4–33 dry tons/acre annually (Resch 1988) in different soil types. In particular, as a pioneer species, it often thrives in large numbers on poor land. In this regard, red alder also has a potential role to play in buffering climate change by sequestering carbon: it is estimated that 1 acre of new forest can sequester about 2.5 tons of carbon annually. Indeed, young trees assimilate CO2 at a rate of ∼6 kg/tree/year, this increasing to ∼22 kg/tree/year at about 10 years of age (https://urbanforestrysouth.org/resources/library/citations/method-for-calculating-carbon-sequestration-by-trees-in-urban-and-suburban-settings-1). Growing trees to sequester carbon is viewed as a viable proposition (Cannell 1999).
This study builds on 23 years of clonal selection, initiated by the forestry company Weyerhaeuser and licensed in 2011 to Washington State University (WSU). Historically, clonal red alder variants were carefully selected from the wild, chosen based on their abilities for exceptional growth in adverse conditions, including their ability to thrive under stress, such as high salt, large temperature fluctuations, drought, and differential water use efficiency. The clone chosen for sequencing has an exceptionally high growth rate. The genome sequence reported here will foster an understanding of the basis of this trait, enable the development of molecular markers upon which to build a coherent tree improvement strategy, and stimulate biochemical and other studies that target diverse traits of interest, such as wood chemistry and quality. Moreover, the natural range of red alder is predicted to become hotter and drier, threatening habitat critical to the viability of this important species. The genome reference will also enable the study of genetic variation across the latitudinal range of red alder, allowing the identification of traits that are resilient to these abiotic stresses, and of their associated variants.
Methods
Reference plant material
Alnus rubra clone 639 is a rapidly growing clone developed as part of a clonal selection program carried out by the forestry company Weyerhaeuser. Vegetatively propagated plants were grown in a greenhouse on the WSU campus in Pullman, WA, USA. Requests for clone 639 plants can be made per WSU policy by contacting Prof. Norman G. Lewis, Institute of Biological Chemistry, Plant Sciences Building 101D, WSU, Pullman, WA 99164-7411, USA.
DNA preparation
Washington State University (WSU)
DNA was prepared from newly emerged 1–2″ long apical leaf samples obtained during mid-afternoon hours in May (2017) from 1-year-old greenhouse-grown saplings maintained under environmental conditions of 15 h of 1,000 watt high-pressure sodium lamps, 24°C and 47% relative humidity. The leaves were flash-frozen in liquid nitrogen and stored at −80°C until processed. Frozen leaves were ground to a fine powder in liquid nitrogen using a mortar and pestle. Genomic DNA was isolated from leaves using a DNeasy Plant Mini Kit (Qiagen). Fifty milligrams of the pulverized sample were suspended in Qiagen Buffer AP1 (400 mL) containing β-mercaptoethanol (10 µL). Following incubation with RNase A (400 µg) in a 65°C water bath for 30 min, samples were processed according to the DNeasy Plant Handbook protocol. Purified DNA was eluted from the DNeasy Mini spin columns in Qiagen AE elution buffer (70 μL).
Pacific Northwest National Laboratory (PNNL)
DNA was extracted from fully expanded leaves of 1-year-old greenhouse grown seedlings in early summer (2017 May 10). The greenhouse was located at the WSU Tri-Cities campus in Richland, WA. The leaves were quickly cut from the trees, wrapped in damp paper towels, and cooled overnight at 4°C. A Joint Genome Institute plant nuclear DNA protocol was used, consisting of incubation in guanidine-HCl/proteinase K lysis buffer, Qiagen Genomic-tip purification, and isopropanol precipitation (https://jgi.doe.gov/user-programs/pmo-overview/protocols-sample-preparation-information/). Qiagen Genomic-tips (100/G) were used according to the manufacturer's instructions. DNA was suspended in ∼200 µL EtOH solution and stored at −80°C until ready for use.
PacBio sequencing
National Center for Genome Resources (NCGR)
20-kb PacBio libraries were prepared from leaf tissue DNA. The following kits were used according to the manufacturer's instructions: SMRTbell Template Prep Kit 1.0 (catalog number 100-259-100), DNA Sequencing Bundle 4.0 v2 (catalog number 100-676-400), DNA/polymerase-binding kit P6v2 (catalog number 100-372-700), MagBead Kit v2 (catalog number 100-676-500), and DNA Internal Control Complex P6 (catalog number 100-356-500). Libraries were loaded onto SMRT cells and sequenced on a PacBio RSII instrument using P6 polymerase C4 chemistry with 6 h movie times.
Pacific Northwest National Laboratory (PNNL)
Leaf DNA was sheared to 10–20 kb using a Covaris g-Tube, concentrated using AMPure PB magnetic beads, and quality was evaluated using the Qubit dsDNA HS Assay Kit and with the Sage Science Pippin Pulse Electrophoresis system. Size selection was done using the BluePippin size-selection system. These were then used to generate PacBio long-read libraries for sequencing. Primer annealing and polymerase-binding reactions were prepared using the Binding Calculator from PacBio, these being based on available sample volume, concentration, and insert size using default settings. PacBio RSII sequencing was performed with 6 h movies at Yale Center for Genome Analysis.
HDF5, FASTA, and fastq files from both sequencing operations were used for combined analysis at NCGR. The sequence data represent ∼93× genome coverage (presuming an initial genome size of 0.5 Gb, based on other Alnus sp. entries in the Kew Gardens c-values database; Garcia et al. 2014).
Illumina sequencing
Leaf DNA as prepared above (PNNL) was sent to Lucigen Inc. (Middleton, WI, USA), where it was evaluated by agarose gel electrophoresis for RNA contamination and integrity. Having passed both quality checks, a paired-end Illumina TruSeq DNA library was constructed, and the concentration was determined using a Qubit fluorometer (Thermo Fisher). The insert size was estimated by Agilent Bioanalyzer to be 676 bp. The library was sent to the Genomics Service Center at WSU, Spokane, where it was sequenced in paired-end 250 bp configuration on an Illumina HiSeq 2500 instrument. The data were demultiplexed and trimmed by the center before being delivered in FASTQ format.
Genome assembly and annotation
Sequence data were assembled with FALCON (https://github.com/PacificBiosciences/FALCON), while assembly, polishing, and correction were completed using Daligner (https://github.com/cschin/DALIGNER) and Quiver (Chin et al. 2013). Completeness of assembly, compared with other members of the Fagales (Supplementary Table 1), was evaluated using Benchmarking Universal Single-Copy Orthologs (BUSCO) software version 4.01 (Manni et al. 2001) and the eudicots_odb10 lineage data set, containing 31 species and 2,326 BUSCOs. Gene annotation was performed using the MAKER-P pipeline (Cantarel et al. 2007). The first step included generation of a masked Clone 639 genome from repetitive elements and transposable element proteins using Repeatmasker (Tarailo-Graovac and Chen 2009) and Repeatrunner (Smith et al. 2007), respectively. Annotation included ab initio gene predictions using RNA-seq data as species-specific evidence, publicly available ESTs from the order Fagales (including Alnus glutinosa, Betula pendula, and Quercus spp.) as close relatives, and Arabidopsis thaliana as a model species to train the gene prediction software SNAP (Korf 2004) and AUGUSTUS (Stanke et al. 2004). Transfer RNAs were identified and annotated by using tRNAscan-SE (Lowe and Eddy 1997). False positives were discarded by filtering transcripts by their Annotation Edit Distance (AED) and protein homology by running InterProScan (Zdobnov and Apweiler 2001). Sequence repeat annotations were performed by running Repeatmasker, RepeatModeler (Flynn et al. 2020), TransposonsPSI (http://transposonpsi.sourceforge.net/), and LTRharvest (Ellinghaus et al. 2008). Repeats were classified using the MIPS/PGSB Repeat Element Database (Spannagl et al. 2017), as well as TransposonPSI and RepeatModeler.
Flow cytometry
Genome size was estimated by flow cytometry using nuclei from red alder and tomato (Lycopersicon esculentum L.), a standard with a well-known genome size (Doležel et al. 1992). Samples from unexpanded red alder leaves kept at 4°C were chopped with a razor blade together with freshly harvested tomato leaves. Cold chopping buffer (15 mM HEPES, 1 mM EDTA, 80 mM KCl, 20 mM NaCl, 300 mM sucrose, 0.2% Triton X-100, 0.1% DTT, 0.5 mM spermine, and 0.25 mM polyvinylpyrolidone-40; modified from Dart et al. 2004) was added to the leaves prior to chopping, then again before filtering. Once filtered through 2 layers of Miracloth (Millipore Sigma) into a 1.5-mL microtube, the sample was centrifuged at 1,000 × g for 5 min. After removing the supernatant, the pellet was resuspended in 45 μL of 1.68 mM propidium iodide and 0.955 mL of a solution containing 100 mL MgSO4 buffer, 100 mg DTT, and 2.5 mL Triton X100. The MgSO4 buffer consisted of 0.25 g MgSO4·7 H2O, 0.37 g KCl, and 0.12 g of HEPES dissolved in 100 mL H2O with the pH adjusted to 8.0 (Arumuganathan and Earle 1991). The sample was then analyzed using a FACScan Flow Cytometer (BD Biosciences, San Jose, CA, USA). Peak positions corresponding to both sample and standard were recorded and the c-value for the red alder sample was computed.
Small secreted peptide identification
To identify genes encoding small secreted peptides (SSPs), a bioinformatics pipeline was applied that was recently used for Medicago truncatula (de Bang et al. 2017; Boschiero et al. 2019). First, the SPADA package (Zhou et al. 2013) was used to identify short peptide-coding genes in the red alder Hidden Markov Models (HMMs) from M. truncatula and HMMs from the PlantSSPdb (Ghorbani et al. 2015). New genes identified by SPADA were integrated with general protein gene annotations, redundant genes being removed. Next, the Plant SSP Prediction Tool available at https://mtsspdb.zhaolab.org/database/ was applied to the red alder protein annotations. This applies different approaches to identify SSPs, such as the presence of signal peptide cleavage sites by SignalP server (Petersen et al. 2011), homologies with previously identified known SSPs, protein size, and transmembrane (TM) helix prediction. The combined predictions classify SSPs as “known,” “likely known,” or “putative.” A known SSP has a protein length of ≤200 amino acids, SignalP D-score of >0.25, and homology with previous SSPs, while a putative SSP has a protein length of ≤230 amino acids, SignalP D-score of >0.45, no TM domains, and no significant homologies with known SSPs. A likely known SSP has significant homologies to known SSPs and a small protein length (≤250 amino acids).
K-mer analysis
KAT version 2.4.1 (Mapleson et al. 2017) was applied to 83 Gb of paired-end 250 bp Illumina reads and a FASTA file of the genome assembly. The KAT comp program was run with -m equal to 21, 31, 41, and 51. K-mer spectra were derived from output files using the program kat plot spectra-cn. To prepare input for GenomeScope, k-mers were counted with jellyfish version 2.2.10 using the command jellyfish count -C -m k (21,31,41,51) -s 1000000000 -t 32 and jellyfish histo, respectively. Resulting .histo files were used as input to GenomeScope 2.0 (Ranallo-Benavidez et al. 2020) parameterized as diploid or tetraploid (-p 2 or -p 4), running under R version 3.6.0.
Tandem repeat analysis
To identify tandemly repeated DNA, tandem repeats finder (TRF; Benson 1999) was applied to a collection of PacBio reads in which the polymerase sequenced the template at least 5 times. These reads were generally of high quality. The output of TRF was parsed with a Perl script to extract the repeat features and to write out the repeat monomers in the 50–500 bp range as a FASTA file. Repeats in the most abundant 176–182 bp category were studied further, by comparing them pairwise with one another using BLASTN. Pairs of similar repeats, having an E-value of <1e−10, were grouped together into repeat classes. Arrays of tandem repeats in each PacBio read were curated by hand and split into sets of monomers at a motif common to the borders of all repeats (AGTTTT). Each set of monomers was aligned with Clustal Omega (Sievers and Higgins 2021) and the consensus extracted with the cons program of the EMBOSS package (Rice et al. 2000). Consensus repeat monomers from each PacBio read were aligned with each other, and similarity trees were generated using the software package Seqotron (Fourment and Holmes 2016). The same approach was applied to the A. glutinosa genome assembly (NCBI assembly accession GCA_003254965.1) and B. pendula PacBio reads (SRA accession ERR2003767.1) in order to compare tandem repeats among these related species.
Gene duplication analysis
Protein representations of gene annotations were aligned to one another using BLASTP 2.9.0 with the following parameters: -num_threads 32 -evalue 1e-10 -max_target_seqs 5 -outfmt 6. The output table and the annotations GFF file were used as input to MCScanX (Wang et al. 2012). Output files of MCScanX were parsed in a Perl script to identify start and end coordinates of each duplicate segment and to identify genes therein. Tandemly repeated genes were also evaluated with MCScanX. Protein representations of duplicated genes were aligned to A. thaliana proteins (https://www.arabidopsis.org) using BLASTP. Hits with E-values <1e−10 were analyzed in GOATOOLS (Klopfenstein et al. 2018), to identify gene ontology (GO) terms significantly enriched at P < 0.05 by Fisher's exact test. Further GO-term annotation was done with eggNOG-Mapper (Huerta-Cepas et al. 2017). All red alder annotated proteins and the subsets contained in the self-syntenic and tandemly repeated fractions were submitted to the eggNOG-Mapper server (http://eggnog-mapper.embl.de/). To evaluate the genome nucleotide-level repetitive content, the genome assembly was compared with itself using MUMmer (Delcher et al. 2003). Alignments >30 bp with at least 95% identity were included. Annotated protein-coding genes of 7 other species from the order Fagales were obtained from NCBI and analyzed with red alder protein annotations using OrthoFinder 2.3.3 (Emms and Kelly 2019; Supplementary Table 1). The main use of the OrthoFinder output was to collect genes into paralogous groups for Ks estimation. Nonsynonymous substitution rates (Ks) were computed using a Perl script employing BioPerl modules, in particular Bio::Align::DNAStatistics, which computes Ks using the Nei–Gojobori algorithm (Nei and Kumar 2000).
SNP discovery
Eighty-three giga base pairs of paired-end 250 bp genomic DNA Illumina reads were aligned to the haploid genome assembly with HISAT2 (Kim et al. 2019) with parameters: --no-spliced-alignment--threads 32 --no-unal. Duplicate BAM file reads were marked using sambamba (https://lomereiter.github.io/sambamba/docs/sambamba-markdup.html) and variant calling was done with FreeBayes (Garrison and Marth 2012) using 2 parameterizations: -P 0.001 -p 2 -i -X -u -C 10 -m 30 -q 20 -! 30, and -P 0.001 -p 2 -i -X -u -C 5 -m 30 -q 20 -! 10. The resulting SNP calls were used to evaluate the quality of the genome assembly by running the BCFtools (Danecek et al. 2021) command: bcftools stats --samples on the VCF files. The PSC section of each statistics report was consulted to determine the number of homozygous reference and homozygous nonreference SNP calls, which should both be low if the assembly quality is high.
Results and discussion
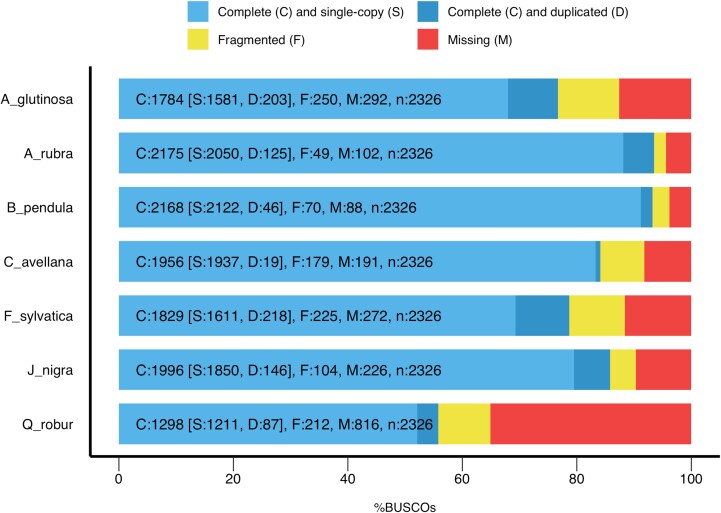
Genome assembly statistics are in Table 1. The primary assembly was treated as a typical haploid, as commonly used for annotation and genome size estimation. The associated contigs are probably sequences allelic to primary contigs, useful for variant discovery and evaluating allelic expression differences. Overall genome GC composition was estimated to be 37%. Genome assembly completeness was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO v.4.01; Manni et al. 2021), a quantitative assessment of genome assembly and annotation completeness based on evolutionarily informed expectations of gene content across lineages. BUSCO analysis using the eudicot lineage revealed that 2,050 (88.13%) of 2,326 total single-copy ortholog genes were present in the A. rubra genome assembly and an additional 125 (5.37%) genes were duplicated, resulting in a completeness estimate of 93.5%. This genome completeness is comparable or superior to that of several other genomes in the Fagales available at the time this research was conducted (Fig. 1), including additional members of the Betulaceae, such as silver birch (B. pendula), black alder (A. glutinosa), and European hazelnut (Corylus avellana L.); the Fagaceae including European beech (Fagus sylvatica) and English oak (Quercus robur); and the Juglandaceae, such as black walnut (Juglans nigra) with completeness estimates of 93.2, 76.7, 84.1, 78.7, 55.8, and 85.8%, respectively (Fig. 1). In support of the BUSCO analysis of conserved single copy genes, we evaluated the representation of an assembled transcriptome in the genome assembly. About 92.4% of the assembled transcripts were found in the genome assembly (not shown).
Table 1.
Red alder genome assembly statistics, including the complete assembly (primary and associated contigs), and primary contigs only.
| Primary contigs | Primary and associated contigs | |
|---|---|---|
| Number of contigs | 1,363 | 2,717 |
| Contig N50 | 1.74 Mb | 1.53 Mb |
| Longest contig | 8.25 Mb | 8.25 Mb |
| Mean contig length | 0.356 Mb | 0.204 Mb |
| Assembly length | 485.60 Mb | 552.90 Mb |
| GC content | 36.83% | 36.70% |
Associated contigs are from the regions of the assembly graph where there was sufficient variation to infer the haplotypes of primary contigs.
Fig. 1.
Assembly completeness assessment with BUSCO for Alnus rubra vs select members of the order Fagales.
Genome annotation
A total of 52,758 protein coding genes were predicted after filtering using an AED of <1, and comparison with known protein domains. Because of our interest in nitrogen-fixing symbiosis, further annotation focused specifically on SSPs, a signaling molecule class that participates in a vast range of plant growth and development processes, including root development and nodulation (Djordjevic et al. 2015; Kereszt et al. 2018). A total of 2,494 of the genes predicted by the MAKER pipeline were identified as SSPs. Additionally, 1,043 new SSP genes were predicted, bringing the total number of genes to 53,801, within the predicted ranges of other Fagales members (Supplementary Table 1). Supplementary Table 3 shows all of the predicted SSPs, along with their annotation details and whether they are classified as already known SSP (311), likely SSP (219), or putative SSP (1,968).
Repetitive DNA content
Annotation of transposable elements used RepeatMasker (Tarailo-Graovac and Chen 2009), TransposonsPSI (http://transposonpsi.sourceforge.net/), and LTRharvest (Ellinghaus et al. 2008), with the MIPS/PGSB Repeat Element Database applied for their classification (Spannagl et al. 2017). The estimated genome total repeat content was 130.5 Mb, averaged from different approaches employed, representing 23–31% (lower and upper size estimates) of the genome. Gypsy and Copia LTR retrotransposons were the most abundant repeats, occupying 3.5 and 4% of the genome (Supplementary Table 2). Equivalent proportions in silver birch, the closest relative with data available, were 8.5 and 2.3%, respectively (Salojärvi et al. 2017).
Another class of repetitive DNA consists of tandemly repeated units. These can be associated with important functional elements of the chromosome, such as centromeres (Melters et al. 2013). To identify tandemly repeated DNA, we selected a subset of 15,433 high-accuracy PacBio reads having at least 5 circular consensus reads. Tandemly repeated DNA arrays were identified by applying TRF (Benson 1999). The most common repeat unit identified was ∼180 bp long, typical of centromeric DNA (Melters et al. 2013), found in 43 PacBio reads. Mapping this repeat back to the haploid assembly showed that it tends to be found in contiguous arrays; DNA segments with at least 95% repetitive content averaged 8,795 bp. These arrays were found concentrated in smaller contigs with a median size of 9,614 bp. The longest array observed was 59 kb (Supplementary Table 4). A comparative approach was applied to the genomes of A. rubra, B. pendula, and A. glutinosa. Red alder and B. pendula tandem repeats revealed no similarities. The major class of red alder tandem repeats was, however, closely related to a small family of repeats of similar length in A. glutinosa. The relationships among the repeat consensus units are shown in a neighbor-joining tree (Supplementary Fig. 1). The consensus sequences of these repeats, which are viable candidates for centromeric DNA, are in Supplementary Table 5.
Ploidy and genome size
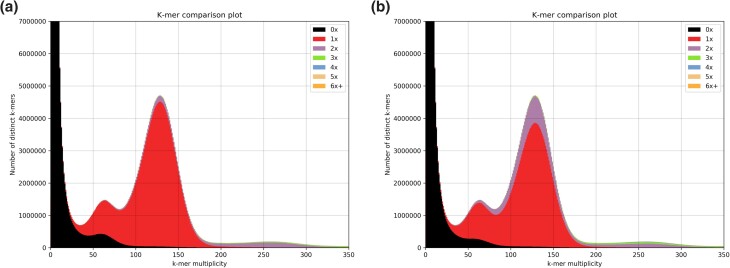
Loveless (2021) used molecular genetic methods, in some cases confirmed by microscopy, to show that red alder diploids and tetraploids were present in 9 out of 10 sampling locations in Washington, Oregon, and Idaho. We explored the ploidy of Clone 639 using k-mer analysis. K-mer spectra (k = 21) from analysis with the k-mer analysis toolkit (Mapleson et al. 2017) are shown in Fig. 2. The associated contigs contributed most of the duplicated k-mers in the genome, i.e. those mapping twice to the reference genome (purple shading), further evidence that associated contigs represent allelic segments of primary contigs. The k-mer multiplicities of 65 and 130 (Fig. 2) represent heterozygous and homozygous portions of the genome assembly. This analysis strongly suggests that our red alder clone is diploid, although autotetraploidy cannot be ruled out. Analysis of the primary assembly with k = 31, 41, and 51 did not differ appreciably from k = 21 (Supplementary Fig. 2). In support of diploidy, red alder k-mers were also analyzed using GenomeScope 2.0 (Ranallo-Benavidez et al. 2020), parameterized as diploid or tetraploid (Supplementary Fig. 3). The data fitted the GenomeScope diploid models closely at all values of k, whereas the data deviated from the tetraploid models.
Fig. 2.
K-mer analysis toolkit (KAT) analysis of 21-mers. a) Primary (haploid) genome assembly. b) Primary contigs plus associated contigs.
Five sources of data were used to estimate the genome size to be in the range 415–563 Mb. (1) The primary contig assembly has a total length of ∼486 Mb. (2) Our flow cytometry analysis indicated 2C = 1.14 pg/nucleus which provides a genome size estimate of 563 Mb. Three samples yielded the same flow cytometry estimate. (3) GenomeScope estimated the genome size at 414 Mb (k = 21) to 423 Mb (k = 51). (4) Extrapolating read coverage of single copy BUSCO genes to the haploid assembly gives a genome size estimate of 452 Mb. (5) The findGSE algorithm (Sun et al. 2018) provided estimates of 494 (k = 21) to 557 (k = 51) Mb. These estimates span a wide range of values, but are in the range reported for other alders (Kew Gardens c-values database, cited by Garcia et al. 2014).
SNP discovery
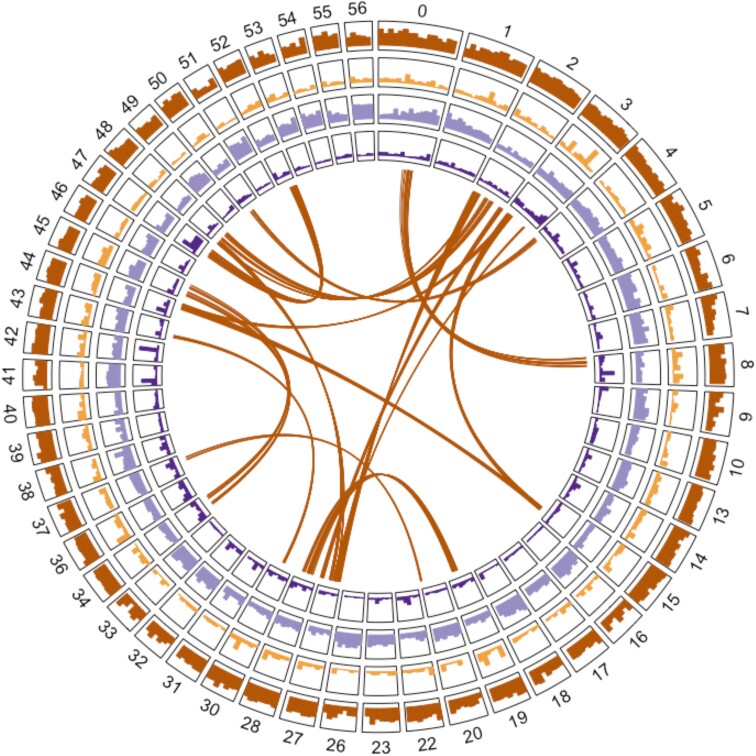
Genome heterozygosity was evaluated by aligning Illumina genomic DNA sequence reads from the same clone to the haploid assembly. Paired-end 250 bp Illumina reads (83 Gb total) were aligned to the haploid genome assembly using HISAT2 (Kim et al. 2019), with variant calling utilizing FreeBayes (Garrison and Marth 2012). Parameters using a minimum sequencing depth of 30, a minimum of 10 reads for the minor allele, a minimum mapping quality of 20, and a minimum sequence quality of 20, gave 257,020 SNPs. Relaxing the read number having a minor allele to 5 gave 281,163 SNPs. Using the more conservative criteria and the largest genome size estimate (563 Mb), this gave a SNP approximately every 2,600 bp. With more relaxed SNP calling criteria and the smallest genome size estimate (415 Mb), the inter-SNP distance was 1,570 bp. These SNPs are by definition heterozygous and do not address population-level variation. Nonetheless, they will be useful in future selection, breeding, and population studies. The distribution of SNPs in the 50 largest contigs is illustrated in Fig. 3. The BAM file statistics are also supportive of a high-quality assembly; of 172,567,868 read pairs, only 1,355,595 (0.79%) had partners mapping to different contigs with high-quality alignments (mapQ > 5). Because we aligned short reads from the same plant to the haploid assembly, all SNPs detected are expected to be heterozygous. Accordingly, 2 important metrics are the numbers of homozygous reference and homozygous “alt” SNP calls. If the assembly quality is high, both of these should be very low. In the case of the more stringent SNP calling parameters, these numbers were 0 and 607 (of 257,020), respectively. In the case of the more relaxed parameters, they were 0 and 1,304 (of 281,163), respectively. These statistics support the conclusion that the assembly is accurate.
Fig. 3.
Annotation summary of the fifty largest contigs in the red alder genome. In order from the outside: all protein coding genes, SSP genes, LTR transposons, SNPs, and duplicated segments displayed as ribbons.
Gene duplications
Genome complexity was analyzed using MCScanX (Wang et al. 2012), which classifies genes into paralogous groups, and identifies tandem gene duplications and duplications present in self-syntenic (collinear) genome segments. This analysis identified 211 duplicated segments averaging 561 kb in length, the largest being 5 Mb and the smallest 5.6 kb. Together, these account for 238 Mb or more than half of the genome (Supplementary Table 6). Duplicated genes per self-syntenic segment ranged from 4 to 41, averaging 9. In total, 1,931 duplicated gene pairs residing in self-syntenic segments were identified. MCScanX also identified 1,220 tandemly repeated gene pairs. To explore potential relationships between duplicated regions and possible evolutionary events, further analysis utilized the dissect_multiple_alignment program of the MCScanX package to estimate gene numbers present in blocks of differing depths. The great majority were present in blocks of depth 1, 2, and 3 (40,205, 10,767, and 3,033 genes, respectively). Duplicated segments in the largest 50 contigs (shown as ribbons in the center of Fig. 3) most likely represent vestiges of the γ whole-genome duplication (WGD) in the eudicot lineage (Vekemans et al. 2012), now mostly represented by segment pairs. Very few genes are present in blocks of greater depth. The 76 genes in blocks of depth 4 probably represent tandem duplications within collinear segments. Investigation of the 15 genes in blocks of depth 8 assisted with quality control of the genome assembly, since all were found to be present on contigs containing plastid DNA. Genome self-comparison at the nucleotide level using MUMmer (Delcher et al. 2003) compared 30-mers with a threshold of 95% for identity. About 127 Mb (23–30% of the red alder genome, depending on the size estimate) is composed of repeated DNA segments. This is similar to the fraction of genes (26%) present in the combined collinear and tandemly repeated classes.
Prior work (Salojärvi et al. 2017) established that in B. pendula, syntenically and tandemly duplicated genes were enriched for different GO terms, implying selective retention of transcription factors in syntenic segments, and expansion by tandem duplication of genes involved in secondary metabolism and host defense, among others. That work (Klopfenstein et al. 2018) identified GO terms significantly enriched at P < 0.05 by Fisher's exact test following correction for multiple testing. We applied the same tool to Arabidopsis annotations of our syntenic and tandemly repeated genes (GOATOOLS version 1.0.3). The results, shown in Supplementary Tables 7 and 8, reflect these findings: enriched genes in self-syntenic segments emphasized responses to chemical entities, including hormones, protein kinase activity, and, in particular, transcription factors. Tandemly repeated genes were enriched for GO terms including environmental responses, i.e. wounding, oxidoreductase activity, responses to external stimuli (including other organisms), and secondary metabolism.
Assuming non-neutrality of nonsynonymous nucleotide substitutions, determining the Ks rate can assist with determining the relative ages of gene duplications (Blanc and Wolfe 2004; Maere et al. 2005; Tang et al. 2008). We determined the distribution of pairwise gene Ks values for all paralogous gene pairs and for gene pairs in the collinear and tandemly duplicated sets (Supplementary Fig. 4). Genes remaining from the γ-duplication are most likely to be those in the secondary peak at Ks of ∼0.8–1.0. The Ks spectrum of genes present in the self-syntenic regions was consistent with some recent tandem duplication of genes themselves remaining in multiple copies from the γ duplication. Ks for gene families in other members of the order Fagales was also computed (Supplementary Fig. 5), based on ortho groups as above, with the gene annotation sources for each species indicated in Supplementary Table 1. Gene duplication histories, as deduced from Ks distributions, were similar for A. rubra, A. glutinosa, F. sylvatica, Q. robur, and Casuarina glauca. By comparison, B. pendula and C. avellana retained greater numbers of gene duplications from the γ-WGD, compared with recent tandem duplications. Juglans nigra, a sister to the branch containing Betula, Alnus, Corylus, and Casuarina (Li et al. 2004) contained evidence of a much more recent WGD, as indicated by the large number of gene families with Ks of ∼0.3. This duplication is absent from other Fagales members we analyzed. In support of this observation, the recent report of the Juglans regia genome (Marrano et al. 2020) illustrated extensive collinearity of whole chromosomes. Cumulatively, the evidence suggests remnants of the γ duplication in red alder and ongoing tandem gene duplications with deletion of duplicates over time are not under selective constraints (Blanc and Wolfe 2004).
Conclusion
We report the annotated genome of a rapidly growing clone of A. rubra, a tree of significant ecological, cultural and economic importance. Although fragmented, the assembly is as or more complete than other sequenced genomes in the order Fagales available at the time this research was conducted, and the annotated genes, repeats, and variants provide the necessary resources for understanding red alder's many interesting traits, as well as for future breeding and selection endeavors, and population studies. This study initiated comparative genomics analysis of the order Fagales; the addition of the red alder genome to this collection facilitates such work, which is ongoing.
Supplementary Material
Acknowledgments
The authors would like to thank Kristin Engbrecht and Richard White for logistical and sample preparation support and valuable discussions, as well as Barri Herman and Julie Thayer for the maintenance of red alder clones at the WSU Puyallup and WSU greenhouses, respectively.
Contributor Information
Kim K Hixson, Institute of Biological Chemistry, Washington State University (WSU), Pullman, WA 99164, USA; Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory (PNNL), Richland, WA 99352, USA.
Diego A Fajardo, National Center for Genome Resources (NCGR), Santa Fe, NM 87505, USA.
Nicholas P Devitt, National Center for Genome Resources (NCGR), Santa Fe, NM 87505, USA.
Johnny A Sena, National Center for Genome Resources (NCGR), Santa Fe, NM 87505, USA.
Michael A Costa, Institute of Biological Chemistry, Washington State University (WSU), Pullman, WA 99164, USA.
Qingyan Meng, Institute of Biological Chemistry, Washington State University (WSU), Pullman, WA 99164, USA.
Clarissa Boschiero, Noble Research Institute, Ardmore, OK 73401, USA.
Patrick Xuechun Zhao, Noble Research Institute, Ardmore, OK 73401, USA.
Eric J Baack, Biology Department, Luther College, Decorah, IA 52101, USA.
Vanessa L Paurus, Biological Science Division, Pacific Northwest National Laboratory (PNNL), Richland, WA 99352, USA.
Laurence B Davin, Institute of Biological Chemistry, Washington State University (WSU), Pullman, WA 99164, USA.
Norman G Lewis, Institute of Biological Chemistry, Washington State University (WSU), Pullman, WA 99164, USA.
Callum J Bell, National Center for Genome Resources (NCGR), Santa Fe, NM 87505, USA.
Data availability
The PacBio and Illumina genomic sequence reads are deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA689849. The genome assembly has been deposited at GenBank under the accession JAJPGS000000000. Supplementary files that fully describe the data reported herein have been uploaded to Figshare and can be accessed at https://doi.org/10.6084/m9.figshare.17532155. These include the assembly, annotated genes, proteins and repeats, and VCF files containing SNPS.
Supplemental material available at G3 online.
Funding
This work was supported by the National Science Foundation (award number 1547842) and partly by the United States Department of Agriculture (grant 2011-68005-30416). A portion of this research was supported by the Intramural Program at EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Biological and Environmental Research program and operated under Contract No. DE-AC05-76RL01830. A portion of this research was also conducted under the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory, a multiprogram national laboratory operated by Battelle for the US Department of Energy.
Author contributions
C.J.B., K.K.H., L.B.D., and N.G.L. conceived and designed the experiments. C.J.B., D.A.F., and J.A.S. performed the annotation of the red alder genome. C.B. and P.X.Z. performed the annotation of the SSPs. E.B. performed the flow cytometry and analysis. M.A.C., and V.L.P. performed DNA extractions. C.J.B., D.A.F., J.A.S., K.K.H., L.B.D., M.A.C., N.G.L., N.P.D., and Q.M. analyzed the data. C.J.B., K.K.H., L.B.D., and N.G.L. wrote the paper.
Literature cited
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9(3):208–218. doi: 10.1007/BF02672069. [DOI] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DR, Silvester WB. Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev. 1993;57(2):293–319. doi: 10.1128/mr.57.2.293-319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16(7):1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschiero C, Lundquist PK, Roy S, Dai X, Zhao PX, Scheible W-R. Identification and functional investigation of genome-encoded, small, secreted peptides in plants. Curr Protoc Plant Biol. 2019;4(3):e20098. doi: 10.1002/cppb.20098. [DOI] [PubMed] [Google Scholar]
- Cannell MGR. Growing trees to sequester carbon in the UK: answers to some common questions. Forestry. 1999;72(3):237–247. doi: 10.1093/forestry/72.3.237. [DOI] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10(2):2078. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart S, Kron P, Mable BK. Characterizing polyploidy in Arabidopsis lyrata using chromosome counts and flow cytometry. Can J Bot. 2004;82(2):185–197. doi: 10.1139/b03-134. [DOI] [Google Scholar]
- Deal RL, Harrington CA. Red Alder: A State of Knowledge. Portland (OR): US Department of Agriculture, Forest Service, Pacific Northwest Research Station Report No.: PNW-GTR-669; 2006. [Google Scholar]
- de Bang TC, Lundquist PK, Dai X, Boschiero C, Zhuang Z, Pant P, Torres-Jerez I, Roy S, Nogales J, Veerappan V, et al. Genome-wide identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 2017;175(4):1669–1689. doi: 10.1104/pp.17.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBell DS. Potential productivity of dense, young thickets of red alder. Res Note PNW-RN-2 Portland US Dep Agric For Serv Pac Northwest For Range Exp Stn 6 P. 2; 1972. [accessed 2022 Dec 5].https://www.fs.usda.gov/research/treesearch/40615.
- Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003;00(1):10:3:1. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Mohd-Radzman NA, Imin N. Small-peptide signals that control root nodule number, development, and symbiosis. J Exp Bot. 2015;66(17):5171–5181. doi: 10.1093/jxb/erv357. [DOI] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol Plant. 1992;85(4):625–631. doi: 10.1111/j.1399-3054.1992.tb04764.x. [DOI] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9(1):18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020;117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourment M, Holmes EC. Seqotron: a user-friendly sequence editor for Mac OS X. BMC Res Notes. 2016;9(1):106. doi: 10.1186/s13104-016-1927-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S, Leitch IJ, Anadon-Rosell A, Canela MÁ, Gálvez F, Garnatje T, Gras A, Hidalgo O, Johnston E, de Xaxars G M, et al. Recent updates and developments to plant genome size databases. Nucleic Acids Res. 2014;42(D1):D1159–D1166. doi: 10.1093/nar/gkt1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G.. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907. doi: 10.48550/arXiv.1207.3907, 20 July 2012, preprint: not peer reviewed. [DOI]
- Gelfand I, Sahajpal R, Zhang X, Izaurralde RC, Gross KL, Robertson GP. Sustainable bioenergy production from marginal lands in the US Midwest. Nature. 2013;493(7433):514–517. doi: 10.1038/nature11811. [DOI] [PubMed] [Google Scholar]
- Ghorbani S, Lin Y-C, Parizot B, Fernandez A, Njo MF, de Peer YV, Beeckman T, Hilson P. Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. J Exp Bot. 2015;66(17):5257–5269. doi: 10.1093/jxb/erv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SC, Binkley D, Perry DA. Influence of red alder on soil nitrogen transformations in two conifer forests of contrasting productivity. Soil Biol Biochem. 1997;29(7):1111–1123. doi: 10.1016/S0038-0717(97)00004-7. [DOI] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Mergaert P, Montiel J, Endre G, Kondorosi É. Impact of plant peptides on symbiotic nodule development and functioning. Front Plant Sci. 2018;9:1026. doi: 10.3389/fpls.2018.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DV, Zhang L, Pedersen BS, Ramírez F, Vesztrocy AW, Naldi A, Mungall CJ, Yunes JM, Botvinnik O, Weigel M, et al. GOATOOLS: a python library for gene ontology analyses. Sci Rep. 2018;8(1):10872. doi: 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5(1):59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R-Q, Chen Z-D, Lu A-M, Soltis DE, Soltis PS, Manos PS. Phylogenetic relationships in Fagales based on DNA sequences from three genomes. Int J Plant Sci. 2004;165(2):311–324. doi: 10.1086/381920. [DOI] [Google Scholar]
- Loveless JB. 2021. Next generation sequencing identifies population structure and signatures of local adaptation in red alder (Alnus rubra Bong.). PhD Thesis Portland State University.
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 2005;102(15):5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Zdobnov EM. BUSCO: assessing genomic data quality and beyond. Curr Protoc. 2021;1(12):e323. doi: 10.1002/cpz1.323. [DOI] [PubMed] [Google Scholar]
- Mapleson D, Garcia Accinelli G, Kettleborough G, Wright J, Clavijo BJ. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33(4):574–576. doi: 10.1093/bioinformatics/btw663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrano A, Britton M, Zaini PA, Zimin AV, Workman RE, Puiu D, Bianco L, Di Pierro EA, Allen BJ, Chakraborty S, et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. GigaScience. 2020;9(5):giaa050. doi: 10.1093/gigascience/giaa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14(1):R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. Illustrated Edition. Oxford (NY): Oxford University Press; 2000. [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11(1):1432. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch H. Red alder: opportunities for better utilization of a resource; 1988. College of Forestry, Oregon State University. https://ir.library.oregonstate.edu/concern/technical_reports/tm70mw13b
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Salojärvi J, Smolander O-P, Nieminen K, Rajaraman S, Safronov O, Safdari P, Lamminmäki A, Immanen J, Lan T, Tanskanen J, et al. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat Genet. 2017;49(6):904–912. doi: 10.1038/ng.3862. [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. The Clustal Omega multiple alignment package. Methods Mol Biol. 2021;2231:3–16. doi: 10.1007/978-1-0716-1036-7_1. [DOI] [PubMed] [Google Scholar]
- Smith CD, Edgar RC, Yandell MD, Smith DR, Celniker SE, Myers EW, Karpen GH. Improved repeat identification and masking in Dipterans. Gene. 2007;389(1):1–9. doi: 10.1016/j.gene.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannagl M, Nussbaumer T, Bader K, Gundlach H, Mayer KFX. MPGSB/MIPS PlantsDB Database framework for the integration and analysis of plant genome data. Methods Mol Biol. 2017;1533:33–44. doi: 10.1007/978-1-4939-6658-5_2. [DOI] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32(Web Server):W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ding J, Piednoël M, Schneeberger K. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics. 2018;34(4):550–557. doi: 10.1093/bioinformatics/btx637. [DOI] [PubMed] [Google Scholar]
- Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18(12):1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009;25(1):4.10.1–4.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Vekemans D, Proost S, Vanneste K, Coenen H, Viaene T, Ruelens P, Maere S, Van de Peer Y, Geuten K. Gamma paleohexaploidy in the stem lineage of core eudicots: significance for MADS-box gene and species diversification. Mol Biol Evol. 2012;29(12):3793–3806. doi: 10.1093/molbev/mss183. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-H, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zhou P, Silverstein KAT, Gao L, Walton JD, Nallu S, Guhlin J, Young ND. Detecting small plant peptides using SPADA (small peptide alignment discovery application). BMC Bioinformatics. 2013;14(1):335. doi: 10.1186/1471-2105-14-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PacBio and Illumina genomic sequence reads are deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA689849. The genome assembly has been deposited at GenBank under the accession JAJPGS000000000. Supplementary files that fully describe the data reported herein have been uploaded to Figshare and can be accessed at https://doi.org/10.6084/m9.figshare.17532155. These include the assembly, annotated genes, proteins and repeats, and VCF files containing SNPS.
Supplemental material available at G3 online.