Abstract
Natural products are a major source of new antibiotics. Here we utilize biosynthetic instructions contained within metagenome-derived congener biosynthetic gene clusters (BGCs) to guide the synthesis of improved antibiotic analogs. Albicidin and cystobactamid are the first members of a new class of broad spectrum ρ-aminobenzoic acid (PABA)-based antibiotics. Our search for PABA specific adenylation domain sequences in soil metagenomes revealed that BGCs in this family are common in nature. Twelve BGCs that are bioinformatically predicted to encode 6 new congeners were recovered from soil metagenomic libraries. Synthesis of these 6 predicted structures led to the identification of potent antibiotics with changes in spectrum of activity and the ability to circumvent resistance conferred by endopeptidase cleavage enzymes.
Keywords: Metagenome, Evolution, Antibiotic, Albicidin, Cystobactamid, Syn-BN
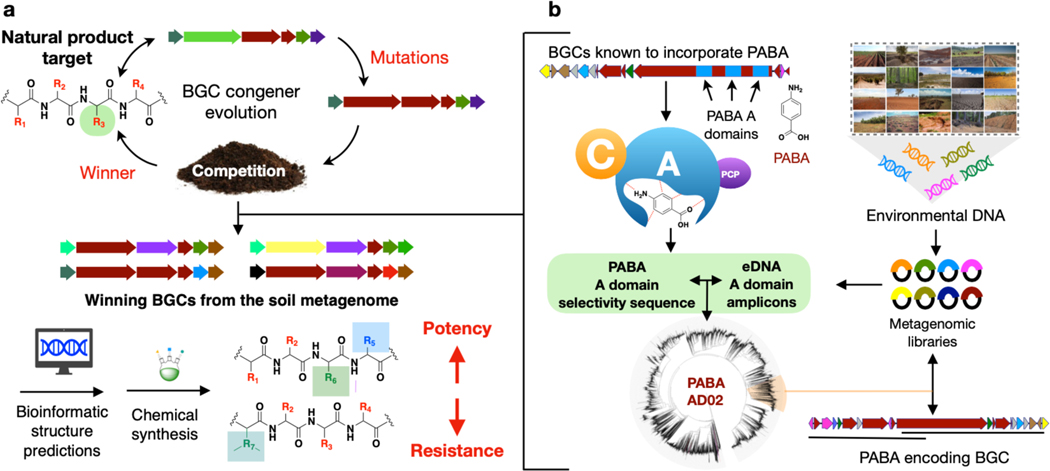
In many instances a characterized natural product represents only one example of a larger family of congener structures. These natural congeners may arise as a result of selective pressures to have different potencies, responses to resistance, and spectra of activity. Soil metagenomes contain large collections of bacteria, including many difficult-to-culture species making them rich sources of congener biosynthetic gene clusters (BGCs). In this study, we combine soil metagenomics with bioinformatic natural product structure prediction methods to intelligently guide the synthesis of albicidin and cystobactamid analogs with improved antibacterial activity and the ability to overcome naturally occurring resistance, two likely outcomes of diverse selective pressures that drive the evolution of natural antibiotic congeners (Figure 1).
Figure 1.
a) Overview of metagenome guided biosynthesis. Natural selection is predicted to result in congeners with varied potency, spectrum of activity, and response to resistance. Bioinformatic analysis of BGCs that encode these congeners can guide the synthesis of improved antibiotics. b) To identify albicidin and cystobactamid congener BGCs, A domain sequences amplified from soil metagenomic libraries were searched for sequences that grouped with domains known to incorporate PABA. These sequences were used to guide the recovery of clones containing albicidin and cystobactamid congener BGCs.
Albicidin and cystobactamid are closely related ρ-aminobenzoic acid (PABA)-containing antibiotics that target DNA gyrase.[1, 2] Their potent Gram-negative activity and unique chemical structures make them appealing leads for the potential development of novel therapeutics.[1, 3] However, the identification of an endopeptidase (AlbD) that confers resistance by antibiotic cleavage suggests that this resistance could eventually threaten their therapeutic utility.[4, 5] Here we searched for PABA specific adenylation domain sequences to identify albicidin and cystobactamid BGCs in soil metagenomes. Although they were originally isolated from traditionally underexplored bacterial taxa (Xanthomonas albilineans and Cystobacter spp, respectively)[2, 6], we found that congener BGCs are common in soil metagenomes and that natural structural variations encoded by these BGCs are different from those explored in previous medicinal chemistry studies (Figure 2a). Synthesis of albicidin and cystobactamid analogs predicted from metagenomic BGCs (i.e., synthetic bioinformatic natural products or syn-BNPs)[7] led to our identification of antibiotics with improved potency as well as the ability to circumvent AlbD cleavage without a loss of antibiosis.
Figure 2.
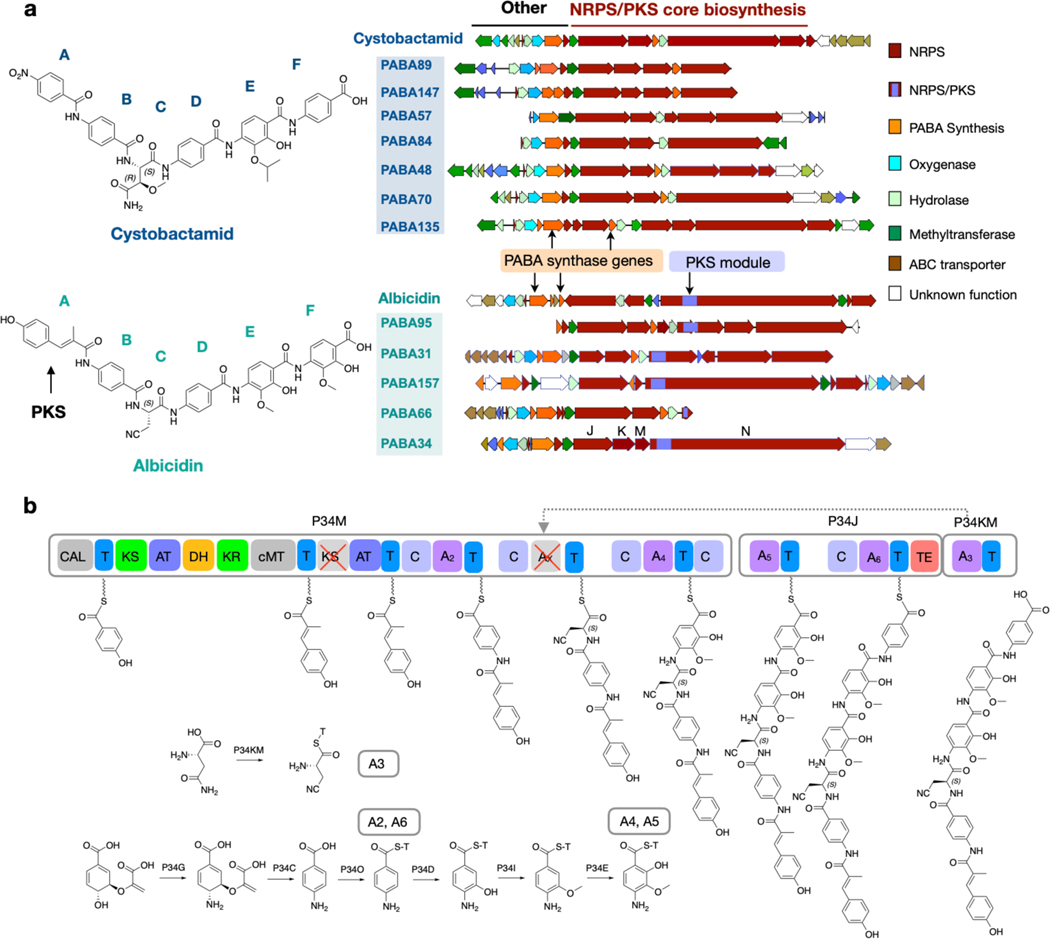
a) Albicidin and cystobactamid congener BGCs isolated from soil metagenomic libraries. b) Proposed biosynthetic pathway for PABA34. Representative analysis used to predict syn-BNP targets.
To identify albicidin and cystobactamid congener BGCs, we screened previously archived metagenomic libraries for PABA-specific adenylation (A) domain sequences. In total, these libraries contain ~720 million unique cosmid clones. Their construction and arraying have been described previously.[8] Briefly, environmental DNA (eDNA) extracted directly from diverse soils was cloned into a cosmid vector and then introduced into E. coli using lambda phage.[9] To facilitate the recovery of specific BGCs of interest, each library of >2×107 cosmid clones was arrayed into collections of subpools each containing ~25,000 unique cosmid clones. Library subpools were screened using subpool-specific barcoded A domain degenerate primers. PCR amplicons were then sequenced using Illumina MiSeq technology and the resulting reads from each library subpool were clustered at 95% identity to generate natural product sequence tags (NPSTs) that can be used to guide the discovery of BGCs of interest. Using our environmental surveyor of natural product diversity (eSNaPD) software package,[10] library-derived NPSTs were compared to PABA-specific A domain sequences from the albicidin and cystobactamid BGCs. NPSTs that returned eValues of <10−25 were considered potential PABA-specific A domains and used to construct a PABA A domain phylogenetic tree (Figure 1). NPSTs that grouped most closely with PABA-specific A domains from albicidin and cystobactamid biosynthesis were considered markers for congener BGCs. Sets of overlapping cosmids associated with predicted PABA NPSTs were isolated from the appropriate library subpools and sequenced to reveal 12 complete and partial cystobactamid or albicidin-like BGCs (Figure 2).
The structure of the natural product encoded by each metagenomic BGC was predicted based on the presence or absence of a PKS module, the types of PABA modification enzymes it encodes, and most importantly the predicted substrate specificity of each NRPS A domain. For this analysis, 10 conserved amino acids in the A domain substrate binding pocket were used to determine the substrate specificity (positions 235, 236, 239, 278, 299, 301, 322, 330, 331 and 517).[11] We compared the A domain substrate binding pockets found in each metagenomic NRPS with characterized A domain substrate binding pockets (Tables S9–S16). Signature sequences for PABA as well as modified PABAs [4-amino-2-hyroxy-3-isopropoxybenzoic acid (AHIBA) or 4-amino-2-hydroxy-3-methoxylbenzoic acid (AHMBA)] were derived from A domains found in the albicidin and cystobactamid BGCs (Tables S9–S10). An analysis of the tailoring genes that encode the differential functionalization of PABA allowed us to distinguish between the presence of AHIBA and AHMBA in the predicted product of each BGC.
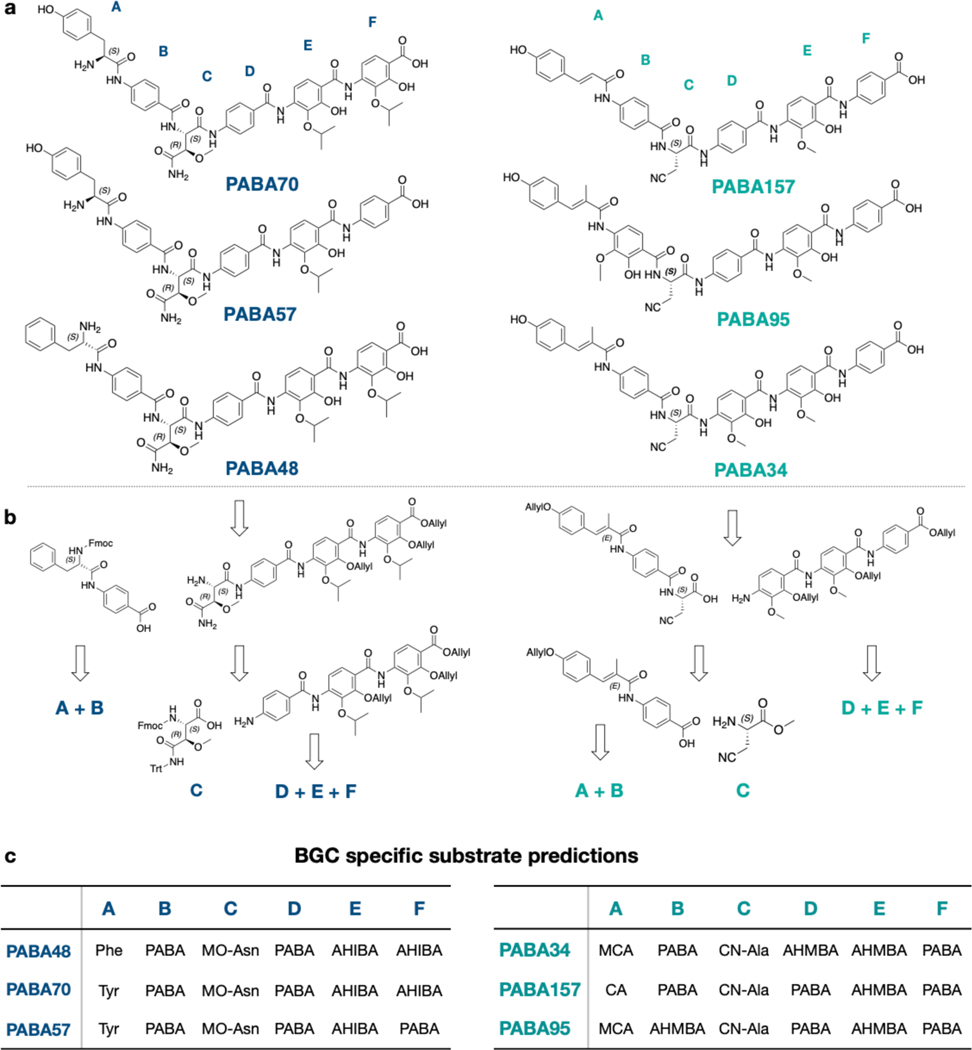
Among the seven cystobactamid-like BGCs we identified, we could predict three different structural analogs, which are embodied by BGCs PABA48, PABA70 and PABA57. Each unique predicted cystobactamid congener BGC encodes 6 NRPS modules. A domain substrate specificity analysis of the 6 A domains in each BGC together with an analysis of the tailoring genes in these BGCs indicates that the central four residues (positions B, C, D, E) are conserved across this family of congeners (Figures 2a and 3a): PABA, β-methoxyasparagine (MO-Asn), PABA, and AHIBA. In place of the PABA seen at the N-terminus of known cystobactamids, our analysis predicted that congener BGCs incorporate either tyrosine or phenylalanine at this position. The C-terminal residue is predicted to be an AHIBA in two cystobactamid congeners (PABA48 and PABA70) and PABA in the third (PABA57). As would be expected for a gene cluster that encodes a metabolite containing AHIBA, cystobactamid congener BGCs are predicted to encode a B12-dependent radical SAM enzyme (CysS homolog) that introduces the t-butyl functionality onto AHIBA.[12]
Figure 3.
Bioinformatically predicted structures and representative retrosynthetic analyses. a) Bioinformatically predicted congener structures. b) Representative synthetic analysis. c) BGC specific substrate predictions.
Key differences between cystobactamid and albicidin-like BGCs include the presence of a specific polyketide synthase module in albicidin BGCs and genes for the biosynthesis of 4-amino-2-hydroxy-3-isopropoxybenzoic acid (AHIBA) biosynthesis in cystobactamid BGCs (Figure 3). Among the five hybrid PKS/NRPS BGCs identified, we could predict 3 distinct albicidin congeners. BGCs PABA34, PABA157 and PABA95 are representative of these three new structures. Three residues are positionally conserved across these congeners. This includes a central β-L-cyanoalanine (CN-Ala), a 4-amino-2-hydroxy-3-methoxybenzoic acid (AHMBA) at the E position, and a PABA at the F position. Positions B and D contain different arrangements of PABA or AHMBA. The N-terminal methyl-coumaric acid (MCA) seen in albicidin is introduced by the PKS module that contains a ketosynthase (KS), dehydratase (DH), ketoreductase (KR), and methyltransferase (MT) domain. The corresponding PKS module in the PABA34 and PABA95 BGCs contain the same collection of domains (Figures S4, S6); however, in BGC PABA157 this module is missing the MT domain, suggesting the biosynthesis of the coumaric acid (CA) in place of MCA (Figure S5). This led to our prediction that the products of PABA34 and PABA95 contain MCA at position A, while PABA157 incorporates a CA as the N-terminal building block. Interestingly, no residue remains constant across these six predicted natural congeners. Although a growing number of analogs have been produced in synthetic efforts to improve the potency and resistance profile of this class of antibiotics, none of them match the structures of these naturally selected congeners encoded by these metagenomic BGCs.[13–15] Based on our bioinformatic predictions, the total synthesis of each predicted congener was undertaken. These structures almost exclusively arise from the coupling of a shared set of substituted and unsubstituted PABA monomers. A convergent synthesis was therefore envisioned that would enable facile access to all six structures from a minimum number of monomer building blocks. In keeping with previous synthetic routes,[16] each congener was disconnected retrosynthetically into three fragments: the N-terminal dipeptide, the central β-methoxyasparagine or β-cyanoalanine unit, and the C-terminal tripeptide. Figure 3b shows a representative retrosynthetic analysis of both a predicted albicidin and cystobactamid congener. Utilization of this general strategy allowed us to easily incorporate different predicted PABA building blocks at positions B, D, E and F as well as incorporate diverse building blocks at the N-terminal position. Integration of the central β-L-cyanoalanine or β-methoxyasparagine units occurred strategically in each synthesis to minimize opportunities for racemization. The final deprotected products, which we have called synthetic bioinformatic natural products (syn-BNPs) were HPLC purified, and their structures were confirmed by HRMS as well as 1H and 13C NMR spectroscopy.
All syn-BNPs, as well as the parent antibiotic albicidin were assayed for antimicrobial activity against the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), which are a collection of bacteria most commonly associated with antibacterial resistant nosocomial infections. Compared to albicidin, syn-BNPs showed improved potency against a number of pathogens (Table 1, S1, S18) and differences in spectrum of activity, as might be expected for congeners that have evolved under different natural selective pressures. PABA34 showed the broadest potent Gram-negative activity with an MIC of ≤2 μg/mL against A. baumannii, P. aeruginosa, E. cloacea and E. coli (Table 1). PABA48 was the most potent broad-spectrum antibiotic we identified. It had the same MIC as albicidin against A. baumannii, but was 2, 16, and >64 fold more potent than albicidin against S. aureus, E. faecium and K. pneumoniae, respectively. Interestingly, unlike albicidin, all three cystobactamid analogs (PABA48, 70 and 57) showed potent activity against K. pneumoniae with MICs ranging from 0.125 to 1 μg/mL. Based on a time-dependent killing analysis against E. coli, all 6 syn-BNPs caused rapid cell death (Figure S9).
Table 1.
Syn-BNP MICs against ESKAPE pathogens and E. coli.
| Gram-Positive | Gram-negative | ||||||
|---|---|---|---|---|---|---|---|
| ESKAPE & E. coli | E. faecium com15 | S. aureus SH1000 | K. pneumoniae 10031 | A. baumannii 17978 | P. aeruginosa PA01 | E. cloacae 13047 | E. coli 25922 |
|
| |||||||
| PABA Compounds | MIC (μg/mL) | ||||||
|
| |||||||
| Albicidin | 2 | 0.5 | >8 | 0.125 | 1 | 8 | 0.015 |
| PABA48 | 0.125 | 0.25 | 0.125 | 0.125 | >8 | >8 | 0.125 |
| PABA70 | 1 | 1 | 1 | 4 | >8 | >8 | 2 |
| PABA57 | >8 | >8 | 1 | >8 | >8 | >8 | 2 |
| PABA34 | >8 | >8 | >8 | 2 | 2 | 2 | 0.0075 |
| PABA157 | >8 | 0.5 | >8 | 0.125 | 1 | 8 | 0.03 |
| PABA95 | 8 | 4 | >8 | 0.5 | 8 | 8 | 0.25 |
The most variable position among BGC predicted analogs was the A position, which involved four different building blocks (Figure 3). PABA48, 70 and 57 are the first examples of a proteinogenic amino acid appearing at this position. This places a positively charged amine at the N-terminus of the antibiotic. In contrast, all previously characterized congeners contain a neutral PABA or MCA at this position. The difference in potency between PABA48 and PABA70, which differ only by the proteinogenic amino acid at their N-termini, suggests this position is particularly important for potency. Traditional SAR studies have found that a small H-bond acceptor at the para position of the phenyl ring at this position is important for antibacterial activity.[17, 18] As this is not found in the most potent broad spectrum analog PABA48, it appears that nature has found an orthogonal strategy for increasing potency in this family of antibiotics.
DNA gyrase resistance:
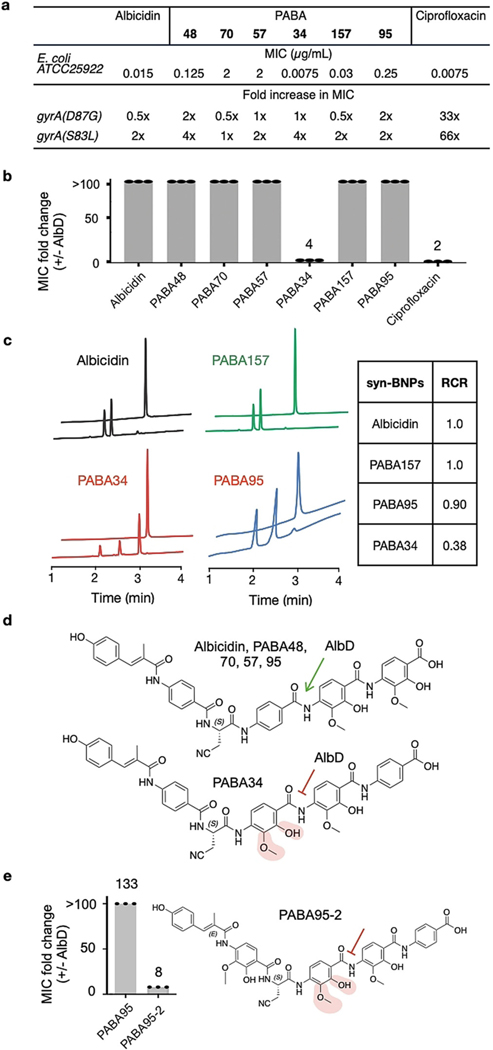
Consistent with cystobactamid and albicidin, all syn-BNPs inhibited DNA gyrase in vitro (Figure S8). As mutations in DNA gyrase commonly confer resistance to DNA gyrase inhibitors, we tested each syn-BNP against E. coli containing two different GyrA mutations (S83L, D87G) that are commonly seen in fluoroquinolone (i.e., ciprofloxacin) resistant clinical isolates.[19] Neither mutation conferred cross resistance to any syn-BNP (Figure 4a). While other mutations would likely arise with clinical introduction of this class of antibiotics, it is appealing that our current analogs retain activity against existing problematic DNA gyrase variants.
Figure 4.
Syn-BNPs have different resistance profiles. a) Activity of syn-BNPs against ciprofloxacin-resistant E. coli. b) MIC fold difference between E. coli that either expresses or does not express AlbD.c) HPLC analysis of PABA34 and albicidin digested by AlbD. d) Schematic representation of AHMBA protection from AlbD cleavage. e) APABA95 analogue containing AHMBA is protected from AlbD cleavage.
AlbD encoded resistance:
One key naturally occurring resistance mechanism for this class of antibiotics is the cleavage of the amide bond between the D and E monomers by AlbD-like endopeptidases.[4, 20] As circumventing common antibiotic resistance mechanisms is likely to play a key role in the natural selection of antibiotic congeners, we reasoned that some of the congeners might have evolved to evade AlbD-mediated cleavage. To determine the susceptibility of our BGC inspired analogs to AlbD encoded resistance, the MIC of each syn-BNP against E. coli engineered to express AlbD was compared to that of E. coli containing an empty expression vector. All syn-BNPs with the exception of PABA34, showed at least a 100-fold increase in MIC against the AlbD expressing strain (Figure 4b, Table S2), suggesting that PABA34 was resistant to proteolytic cleavage by AlbD. To test this hypothesis in vitro, AlbD was overexpressed in E. coli BL21 and purified as a 6-His protein. Recombinant AlbD was then used to digest each albicidin-like syn-BNP. The amount of AlbD-mediated cleavage after 20 minutes, beyond which we saw little additional cleavage, was determined for each syn-BNP by HPLC-UV (Figure 4c). The extent to which AlbD cleaved each substrate was compared to the extent it cleaved albicidin to give a relative cleavage ratio (RCR) for all three albicidin-like syn-BNPs. Both PABA95 and PABA157 had RCRs close to 1. PABA34 on the other hand, had an RCR of 0.38 indicating, as anticipated from the in vivo resistance experiment, that PABA34 was less susceptible than albicidin to AlbD cleavage. PABA34 is unique in that it contains the modified PABA, AHMBA, at the D position directly adjacent to the endopeptidase cleavage site, suggesting that this modification prevents hydrolysis by AlbD, thereby circumventing AlbD resistance (Figure 4d). To test this hypothesis, we synthesized a version of PABA95 (PABA95–2) where the PABA at the D position was replaced with AHMBA. As seen with PABA34, this structure was not susceptible to AlbD encoded resistance (Figure 4e). Against E. coli, the MIC for PABA95 increased by 133-fold upon expression of the albD gene. Against E. coli, the MIC for PABA95 increased by 133-fold upon expression of the albD gene. In the case of PABA95–2, the MIC only increased by 8-fold (i.e., 0.06 μg/mL to 0.5 μg/ ml) (Table S2).
In summary, bioinformatic analysis of PABA encoding BGCs cloned from soil metagenomes guided our synthesis of albicidin and cystobactamid analogs that demonstrated increased potency and different spectra of activity as well as the ability to circumvent resistance conferred by AlbD endopeptidase degradation. Traditional synthetic efforts to address AlbD resistance have focused primarily on replacing the amide bond at the AlbD cleavage site with bioisosteres.[13, 15, 18] Although this provided resistance to AlbD cleavage, these structures often showed reduced antibiosis against a number of Gram-negative pathogens.[13, 18] In contrast, PABA34 was not only resistant to AlbD cleavage but also retained a high affinity for DNA gyrase (Figure S8) and showed potent activity against most Gram-negative ESKAPE pathogens. Future synthetic efforts to further optimize resistance to AlbD cleavage while maximizing activity against Gram-negative pathogens will likely benefit from mimicking the natural incorporation of a highly substituted PABA monomer (like AHMBA) at the D position. Our results highlight the potential utility of metagenome derived BGCs to guide the synthesis of improved natural product variants, especially variants that overcome clinically relevant resistance while maintaining potent antibiosis.
Supplementary Material
Acknowledgements:
This work was supported by the National Institutes of Health (grant 1U19AI142731 and 5R35GM122559)
References:
- [1].Cociancich S, Pesic A, Petras D, Uhlmann S, Kretz J, Schubert V, Vieweg L, Duplan S, Marguerettaz M, Noell J, Pieretti I, Hugelland M, Kemper S, Mainz A, Rott P, Royer M, Sussmuth RD, Nat Chem Biol 2015, 11, 195–197. [DOI] [PubMed] [Google Scholar]
- [2].Baumann S, Herrmann J, Raju R, Steinmetz H, Mohr KI, Huttel S, Harmrolfs K, Stadler M, Muller R, Angew Chem Int Ed Engl 2014, 53, 14605–14609. [DOI] [PubMed] [Google Scholar]
- [3].Hashimi SM, Wall MK, Smith AB, Maxwell A, Birch RG, Antimicrob Agents Chemother 2007, 51, 181187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vieweg L, Kretz J, Pesic A, Kerwat D, Gratz S, Royer M, Cociancich S, Mainz A, Sussmuth RD, J Am Chem Soc 2015, 137, 7608–7611. [DOI] [PubMed] [Google Scholar]
- [5].Planke T, Cirnski K, Herrmann J, Muller R, Kirschning A, Chemistry 2020, 26, 4289–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].BIRCH R, PATIL S, Phytopathology 1983, 73, 1368–1374. [Google Scholar]
- [7].Chu J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, Gordon EA, Reddy BV, Charlop-Powers Z, Zebroski HA, Gallardo-Macias R, Jaskowski M, Satish S, Park S, Perlin DS, Freundlich JS, Brady SF, Nat Chem Biol 2016, 12, 1004–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brady SF, Nat Protoc 2007, 2, 1297–1305; [DOI] [PubMed] [Google Scholar]; Chang FY, Ternei MA, Calle PY, Brady SF, J Am Chem Soc 2015, 137, 6044–6052; [DOI] [PMC free article] [PubMed] [Google Scholar]; Kang HS, Brady SF, Angewandte Chemie (International ed 2013, 52, 11063–11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Owen JG, Reddy BV, Ternei MA, Charlop-Powers Z, Calle PY, Kim JH, Brady SF, Proc Natl Acad Sci U S A 2013, 110, 11797–11802; [DOI] [PMC free article] [PubMed] [Google Scholar]; Owen JG, Charlop-Powers Z, Smith AG, Ternei MA, Calle PY, Reddy BV, Montiel D, Brady SF, Proc Natl Acad Sci U S A 2015, 112, 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reddy BV, Milshteyn A, Charlop-Powers Z, Brady SF, Chem Biol 2014, 21, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stachelhaus T, Mootz HD, Marahiel MA, Chem Biol 1999, 6, 493–505. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y, Schnell B, Muller R, Begley TP, Methods Enzymol 2018, 606, 199–216; [DOI] [PubMed] [Google Scholar]; Wang Y, Begley TP, J Am Chem Soc 2020, 142, 9944–9954. [DOI] [PubMed] [Google Scholar]
- [13].Behroz I, Durkin P, Gratz S, Seidel M, Rostock L, Spinczyk M, Weston JB, Sussmuth RD, Chem Eur J 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kerwat D, Gratz S, Kretz J, Seidel M, Kunert M, Weston JB, Sussmuth RD, ChemMedChem 2016, 11, 1899–1903. [DOI] [PubMed] [Google Scholar]
- [15].Planke T, Cirnski K, Herrmann J, Muller R, Kirschning A, Chem Eur J 2020, 26, 4289–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Elgaher WAM, Hamed MM, Baumann S, Herrmann J, Siebenburger L, Krull J, Cirnski K, Kirschning A, Bronstrup M, Muller R, Hartmann RW, Chem Eur J 2020, 26, 7219–7225; [DOI] [PMC free article] [PubMed] [Google Scholar]; Huttel S, Testolin G, Herrmann J, Planke T, Gille F, Moreno M, Stadler M, Bronstrup M, Kirschning A, Muller R, Angew Chem Int Ed Engl 2017, 56, 12760–12764; [DOI] [PubMed] [Google Scholar]; Moeller M, Norris MD, Planke T, Cirnski K, Herrmann J, Muller R, Kirschning A, Org Lett 2019, 21, 8369–8372; [DOI] [PubMed] [Google Scholar]; Planke T, Moreno M, Huttel S, Fohrer J, Gille F, Norris MD, Siebke M, Wang L, Muller R, Kirschning A, Org Lett 2019, 21, 1359–1363. [DOI] [PubMed] [Google Scholar]
- [17].Testolin G, Cirnski K, Rox K, Prochnow H, Fetz V, Grandclaudon C, Mollner T, Baiyoumy A, Ritter A, Leitner C, Chemical Science 2020, 11, 1316–1334; [DOI] [PMC free article] [PubMed] [Google Scholar]; Planke T, Cirnski K, Herrmann J, Müller R, Kirschning A, Chemistry 2020, 26, 4289–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Testolin G, Cirnski K, Rox K, Prochnow H, Fetz V, Grandclaudon C, Mollner T, Baiyoumy A, Ritter A, Leitner C, Chem Sci 2020, 11, 1316–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bansal S, Tandon V, Int J Antimicrob Agents 2011, 37, 253–255. [DOI] [PubMed] [Google Scholar]
- [20].Walker MJ, Birch RG, Pemberton JM, Mol Microbiol 1988, 2, 443–454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.