Abstract
Preliminary work suggested upregulation of inflammatory pathways in patients with common forms of ichthyosis. However, a comprehensive characterization of skin from various ichthyosis subtypes is unavailable, precluding the development of targeted treatments. Thus, we sought to characterize the immune and barrier profiles of common and subtype-specific skin transcriptomes in a large group of patients with ichthyosis. We performed a global RNA-sequencing analysis in 54 patients with ichthyosis (7 with Netherton syndrome, 13 with epidermolytic ichthyosis, 16 with lamellar ichthyosis, and 18 with congenital ichthyosiform erythroderma) and 40 healthy controls. Differentially expressed genes were defined on the basis of fold changes > 2 and false discovery rate < 0.05 criteria. We found robust and significant T helper (Th) 22/Th17 skewing in all subtypes (e.g., IL-17A/C/F, S100A7/8/9/12; P < 0.001) with modest changes in Th2 pathway, primarily in Netherton syndrome, and Th1 skewing in congenital ichthyosiform erythroderma. Across all subtypes (less evident in epidermolytic ichthyosis), lipid metabolism and barrier junction markers were downregulated (e.g., FA2H, CDH10/11/12/2; P < 0.05), whereas epidermal cornification and proliferation measures were upregulated (e.g., SPRR1A/1B/2C/2G, EREG; P < 0.05). Our findings suggest that the common ichthyosis variants share aberrations in Th17/Th22 and barrier function, with minimal Th2 modulation. This may help to elucidate the pathogeneses of these subtypes and inform the development of subtype-specific treatments.
INTRODUCTION
Ichthyoses are a heterogeneous group of more than 50 rare genetic skin disorders (Oji et al., 2010; Sun et al., 2022) that share features of generalized scaling and skin thickening, with varying degrees of erythema and pruritus (Marukian and Choate, 2016). Quality of life can be markedly reduced, especially with moderate-to-severe disease (Dreyfus et al., 2015). Among the most common orphan (i.e., occurring in <1:200,000 individuals) forms of ichthyosis are Netherton syndrome (NS) (resulting from SPINK5 alterations) (Chavanas et al., 2000), epidermolytic ichthyosis (EI) (typically due to alterations in keratin 1 gene K1 or, most often, keratin 10 gene K10) (Takeichi and Akiyama, 2016), and autosomal recessive congenital ichthyoses (Süßmuth et al., 2020). The latter comprises a phenotypic spectrum ranging from harlequin ichthyosis (HI) (most severe and resulting from biallelic variations in ABCA12) to congenital ichthyosiform erythroderma (CIE) (biallelic variants in 11 different genes, most often ALOX12B, ABCA12, and NIPAL4) (Rodríguez-Pazos et al., 2013; Sun et al., 2022; Takeichi and Akiyama, 2016) and lamellar ichthyosis (LI) (due to alterations in TGM1) (Takeichi and Akiyama, 2016).
Phenotypically, NS is characterized by either generalized erythroderma (typical presentation in infants and younger children) or more localized involvement, as well as atopy and sometimes immunodeficiency (Sarri et al., 2017). Patients with EI present with diffuse blistering (epidermolysis) and erythema as infants, evolving over time toward progressive, compensatory hyperkeratosis, often with thick scaling overlying extensor surfaces (Takeichi and Akiyama, 2016). Classified under the umbrella of autosomal recessive congenital ichthyoses, HI and usually CIE and LI present at birth, with marked generalized skin thickening (a cracked armor appearance and deformities with HI and a collodion membrane in CIE and LI). During the first months of life, the characteristic bright red, thickened skin and fine scale of CIE (of variable intensity) and HI (severe) and the large, darker, lamellated scaling of LI become apparent (Takeichi and Akiyama, 2016).
Current treatment for these orphan forms of ichthyosis is not pathogenesis-based (Paller, 2019) and is limited to topical emollients, keratinolytics, and oral retinoids (Hernández-Martin et al., 2013; Mazereeuw-Hautier et al., 2012; Oji and Traupe, 2009). These therapies are associated with safety concerns (Allen et al., 2001; Digiovanna et al., 2013; Halverstam et al., 2007) and are often unsatisfactory. There is a large unmet need for safe and pathogenesis-driven targeted therapies.
Growing evidence suggests that immune aberrations may be involved in ichthyosis pathogenesis, in addition to the barrier defects (e.g., dysfunction of cornified envelope [CE] or lipid metabolism) (Blunder et al., 2017; Elias et al., 2008; Furio et al., 2015; O’Shaughnessy et al., 2010). Increases in various inflammatory markers (e.g. TNF-α, IL-4, IL-13, IL-36, S100A8/9) have been reported in animal models of ichthyosis, in cultured monolayer and three-dimensional keratinocytes (Enjalbert et al., 2020), and in blood/skin samples from patients with ichthyosis (Akagi et al., 2013; Briot et al., 2009; Fontao et al., 2011; Furio et al., 2014; Hosomi et al., 2008; Konishi et al., 2014; Renner et al., 2009; van Gysel et al., 2001). Moreover, our recent RT-qPCR and microarray studies in patients with the major orphan forms of ichthyosis have revealed shared T helper (Th) 17/IL-23 skewing in the skin and/or blood (Malik et al., 2019; Paller et al., 2017), with increases in markers coregulated by IL-17/TNF-α (e.g., IL-17A, IL-17F, IL-36A/B/G) that correlate with levels of disease severity and transepidermal water loss (TEWL) (Malik et al., 2019; Paller et al., 2017). Another study found IL-17/IL-22 activation in peripheral blood from patients with the major orphan ichthyosis subtypes, which also correlated with the severity of clinical measures (Czarnowicki et al., 2018). These observations highlight ichthyosis as a systemic disease (Czarnowicki et al., 2018; Malik et al., 2019; Paller et al., 2017), further suggest a pathogenic role for Th17/IL-17/TNF-α (Malik et al., 2019), and potentially indicate a need for systemic therapy. Indeed, several case reports have shown the potential benefit of ustekinumab (Paller et al., 2018; Poulton et al., 2019; Sun et al., 2021; Volc et al., 2020), secukinumab (Frommherz et al., 2021; Haiges et al., 2019; Hernández-Martín et al., 2019; Luchsinger et al., 2020), infliximab (Fontao et al., 2011), and guselkumab (Steinhoff et al., 2022) in ichthyosis, further supporting the immune involvement in ichthyosis pathogenesis. Previous studies have investigated only a few markers and/or small cohorts, including only a few patients per subtype, owing to the rarity of the condition (Czarnowicki et al., 2018; Malik et al., 2019; Paller et al., 2017), limiting generalizability and hindering therapeutic discovery. Thus, investigations in larger cohorts are needed.
To characterize comprehensively the shared ichthyosis skin transcriptome, we have now performed a global RNA-sequencing (RNA-seq) analysis of skin from the largest cohort to date (n = 54) of patients with the four most prevalent orphan forms of ichthyosis (NS, CIE, LI, and EI) against an analysis of the skin from 40 matched controls. Our data show that these ichthyosis variants share aberrations in Th17/Th22, with Th1 skewing in CIE and minimal Th2 modulation, in addition to global dysregulation of lipid metabolism and epidermal differentiation complex (EDC)/CE-related products.
RESULTS
A total of 54 patients with ichthyosis (age range = 1–70.8 years, mean age = 25.4 years), including those NS (n = 7), LI (n = 16), CIE (n = 18), and EI (n = 13), with known allelic variants (see Supplementary Table S1) were enrolled with 40 healthy control subjects (age range = 1.3–65 years, mean age = 25.6 years) (Table 1). Two of these patients presented with HI but were merged into the CIE group owing to their small sample size. There were no significant differences in age, sex, racial identity, or Ichthyosis Area and Severity Index (IASI) scores among the ichthyosis subtypes and between patients with ichthyosis and controls (Table 1). Principal component analysis revealed strong overlap among the ichthyosis subtypes, which formed a distinct and separate cluster from control samples (Figure 1a).
Table 1.
Patient Demographics
| Patient Metric | Parameter | Control Subjects (n = 40) | Patients with Ichthyosis n = 54) | P-value, Control Subjects Verus Patients with Ichthyosis | Patients with NS (n = 7) | Patients with LI (n = 16) | Patients with CIE (n = 18) | Patients with EI (n = 13) | P-value, Subtypes |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Age (y) | Mean ± SD (range) | 25.6 ± 17 (1.3–65) |
25.4 ± 18 (1–70.8) |
0.954 | 23.5 ± 14 (6.3–43.2) |
30.5 ± 16 (9.2–56.9) |
19.8 ± 16 (1–48.7) | 27.4 ± 22 (1.1–70.8) |
0.355 |
| Sex | F/M | 21/19 | 30/24 | 0.836 | 5/2 | 11/5 | 7/11 | 7/6 | 0.273 |
| Racial identity | AP/As/B/W/other | 0/1/3/33/2 | 1/3/5/39/5 | 0.893 | 0/0/1/5/1 | 0/0/1/13/2 | 1/2/2/11/2 | 0/1/1/10/0 | 0.869 |
| IASI score | Mean ± SD | NA | 24.9 ± 9 (2.4–43.2) |
NA | 26.3 ± 9 (16.2–37.2) |
26.1 ± 7 (12–40.2) |
24.7 ± 10 (10.8–41.4) | 23.1 ± 11 (2.4–43.2) |
0.827 |
Abbreviations: AP, American Indian/Pacific Islander; As, Asian; B, Black; CIE, congenital ichthyosiform erythroderma; EI, epidermolytic ichthyosis; F, female; IASI, Ichthyosis Area Severity Index; LI, lamellar ichthyosis; M, male; NA, not applicable; NS, Netherton syndrome; W, White.
Other denotes other racial identities unknown.
Figure 1. Overview of ichthyosis subtypes versus controls.
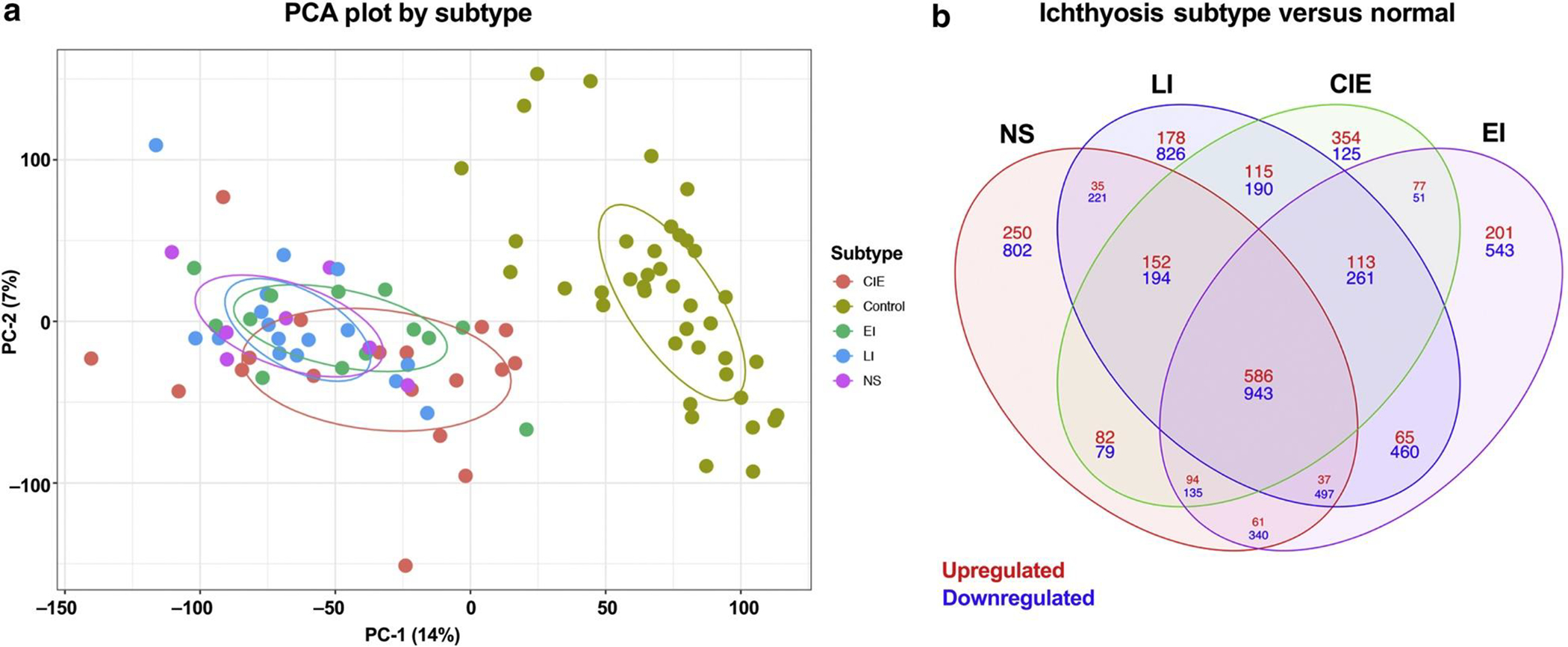
(a) PCA plot showing the ichthyosis subtypes NS, LI, CIE, and EI and controls. (b) Venn diagram showing DEGs comparing ichthyosis subtypes NS, LI, CIE, and EI with normal. Red denotes upregulated, and blue denotes downregulated. CIE, congenital ichthyosiform erythroderma; DEG, differentially expressed gene; EI, epidermolytic ichthyosis; LI, lamellar ichthyosis; NS, Netherton syndrome; PC-1, prinicpal component 1; PC-2, principal component 2; PCA, principal coordinate analysis.
We defined differentially expressed genes (DEGs) with criteria of a false discovery rate (FDR) < 0.05 and a fold change (FCH) > 2, presenting the common and subtype-specific phenotypes (Figure 1b). We found 1,634 DEGs shared across all subtypes (606 upregulated, 1,634 downregulated). We also found unique DEGs within each subtype, with NS showing the highest number of DEGs (239 upregulated, 911 downregulated), followed by LI (190 upregulated, 724 downregulated), EI (212 upregulated, 520 downregulated), and CIE (276 upregulated, 156 downregulated). When compared with previously published data using microarray sequencing in a smaller cohort (Malik et al., 2019), many genes were uniquely detected by RNA-seq within the common phenotype (338 upregulated, 658 downregulated), NS (188 upregulated, 814 downregulated), CIE (249 upregulated, 150 downregulated), EI (195 upregulated, 452 downregulated), and LI (152 upregulated, 588 downregulated) as well as many other unique genes that had not previously reached significance: common phenotype (171 upregulated, 329 downregulated), NS (38 upregulated, 88 downregulated), CIE (23 upregulated, 6 downregulated), EI (8 upregulated, 154 downregulated), and LI (20 upregulated, 80 downregulated) (Supplementary Table S2).
Th17/Th22 is strongly upregulated in all subtypes
We examined the inflammatory profile of the ichthyoses subtypes using previously published immune gene subsets (Dhingra et al., 2014; Guttman-Yassky et al., 2019a; He et al., 2021, 2020a, 2020b; Khattri et al., 2014; Malik et al., 2019, 2017) and expanded on previous work with the detection of, to our knowledge, previously unreported DEGs (Malik et al., 2019). Multiple Th17 and Th22/IL-22–related genes were upregulated across the different subtypes, as depicted in a heatmap (Figure 2). Within the Th17 pathway, elafin/PI3 and CCL20 presented the greatest upregulations in ichthyosis, with an FCH ranging from 12.1 to 223.4, than in the controls across the various ichthyosis subtypes (FDR < 0.001). In addition, LCN2, CXCL1, and IL-17A/F were upregulated in all subtypes (FCH > 2, FDR < 0.05), whereas CAMP was significantly upregulated only in CIE (FCH > 4, FDR < 0.01). Epidermal hyperplasia marker SERPINB4 was strongly upregulated in all subtypes (FCH > 130, FDR < 0.001), particularly in NS and CIE (FCH > 400), and Th22-related IL-22 was significantly enriched in NS and CIE (FCH > 3, FDR < 0.05) (Izuhara et al., 2018). We also found significant global upregulation in many IL-17/TNF-α–mediated markers (Chiricozzi et al., 2011), including KYNU, FOXE1, and S100A7/8/9/12 (FCH > 5, FDR < 0.001), the FCHs for several of which were strikingly higher in NS and CIE than in LI and EI (e.g., PI3, IL-19) or particularly increased in NS, LI, and CIE compared with those in EI (e.g. S100A7/8/9/12, IL-36A, VNN3, SPRR2A/B/D/F) (Figure 2 and Supplementary Figure S1 and Supplementary Material S1). Although the two patients with harlequin ichthyosis were among the highest expressers of several Th17/Th22 genes (e.g., IL36A/G, consistent with a previous report [Enjalbert et al., 2020], PI3, SERPINB4), expression levels for these patients were similar to those of other patients with CIE. In comparison, expression of Th2 markers was much less pronounced. Whereas a few Th2-related genes (CCL18, CCL22, IL4R) (FCH > 2, FDR < 0.05) showed the greatest increases in NS, a subtype linked to atopy (Bitoun et al., 2002; Hannula-Jouppi et al., 2014), the levels of many important genes (IL31, CCL13, CCL17, CCL26, IL13) displayed no statistically significant change relative to those of the controls, across the subtypes, including NS. Of Th1-related genes, the proinflammatory mediators OASL and IL-12RB2 were upregulated across all subtypes (excluding IL-12RB2 in LI) (FCH > 2, FDR < 0.001), whereas other markers (IFN-γ, IL-12B, CXCL9/11) were significantly upregulated primarily in CIE (FCH > 3, FDR < 0.05) (Malik et al., 2019).
Figure 2. Th17/Th22 is strongly upregulated in all subtypes.
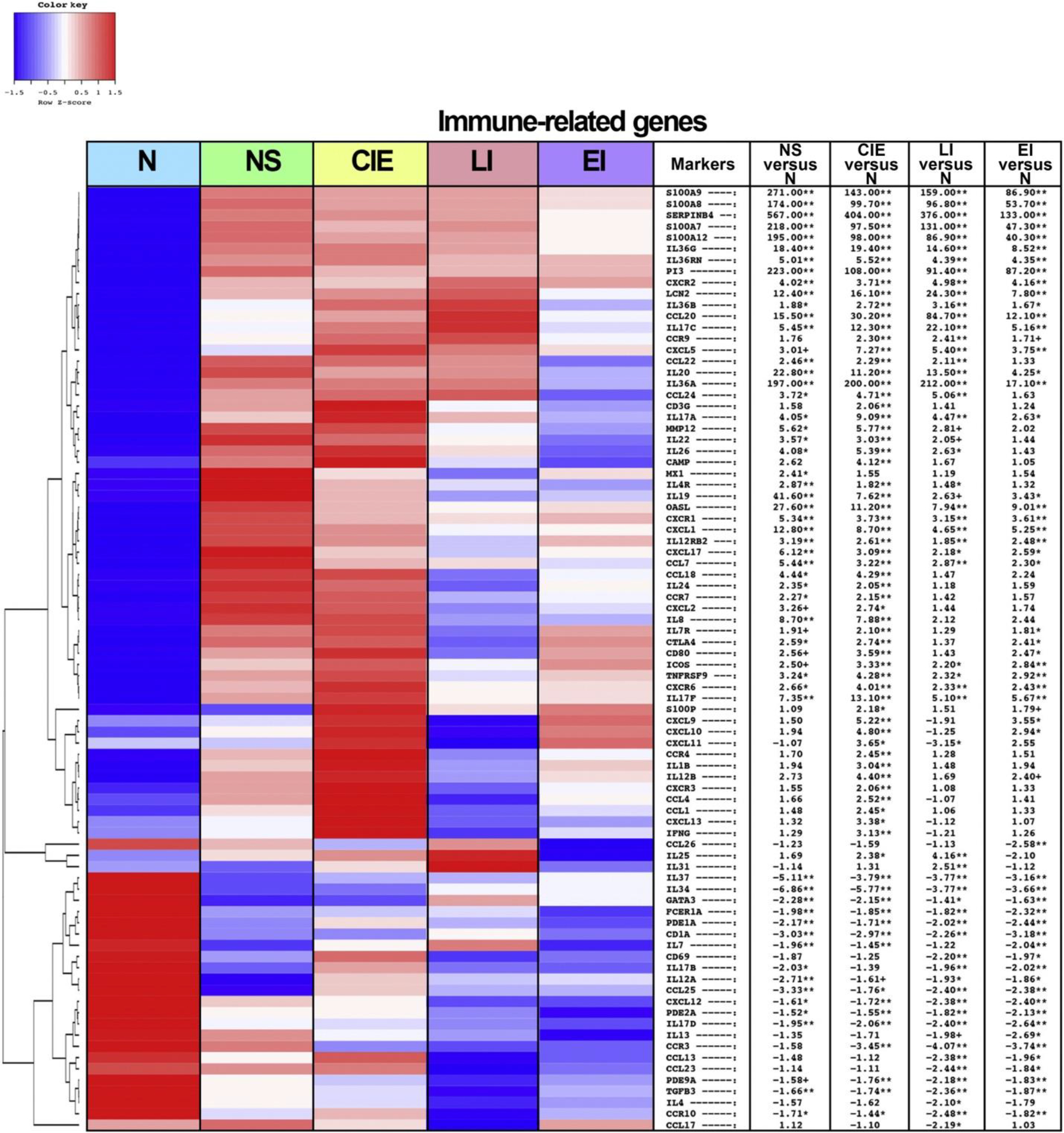
Heatmap representing the differentially expressed immune-related genes from biopsied skin of patients with the various ichthyosis subtypes and healthy controls. Criteria for differential gene expression include FCH > 2 and FDR < 0.05. From left to right, heatmap columns represent N, NS, CIE, LI, and EI. The table shows the FCHs in NS versus N, CIE versus N, LI versus N, and EI versus N. N denotes normal. ∗∗FDR < 0.01, ∗FDR < 0.05, and +FDR < 0.1. CIE, congenital ichthyosiform erythroderma; EI, epidermolytic ichthyosis; FCH, fold change; FDR, false discovery rate; LI, lamellar ichthyosis; NS, Netherton syndrome; Th, T helper.
We also performed a gene set variation analysis (GSVA) utilizing previously published immune-related pathways (Supplementary Figure S2) (Dhingra et al., 2014; Guttman-Yassky et al., 2019a; He et al., 2021, 2020a, 2020b; Malik et al., 2019). GSVA on a Th17-specific gene set, as well as a set of genes synergistically regulated by IL-17 and TNF-α (Chiricozzi et al., 2011), showed significant upregulation in all subtypes (P < 0.001) (Supplementary Figure S2a and b), aligning with a previous study in both ichthyosis and psoriasis (Malik et al., 2019). The Th22 pathway was significantly upregulated in all subtypes (P < 0.001), primarily driven by several highly upregulated markers (SERPINB4, S100A7/8/9/12) (Supplementary Figure S2c). The Th1 pathway was significantly upregulated in all subtypes except in LI, with the highest and most significant upregulation in CIE (Supplementary Figure S2d). Th2 was most robustly and significantly upregulated in NS (Supplementary Figure S2e), followed by CIE. Th2 was not significantly enriched in other subtypes (LI and EI).
Ichthyosis subtypes share global barrier abnormalities, with EI presenting the least disrupted phenotype
Because barrier disruption is a dominant feature of the ichthyoses, we also assessed for barrier-related genes across our cohort, as presented in a heatmap (Figure 3) (Pavel et al., 2020; Renert-Yuval et al., 2021). Global upregulation (FCH > 2, FDR < 0.001) was observed in many genes related to the EDC, including SPRR1A/1B/2C/2G, involucrin gene IVL, PGLYRP3/4, LCE3D, and epiregulin gene EREG (Supplementary Material S1). In addition, genes related to the synthesis/regulation of the CE were upregulated across the ichthyoses (e.g., CNFN, TGM1, CSTA) (FCH > 3, FDR < 0.001) (Supplementary Material S1). Many additional terminal differentiation markers (FLG, PSORS1C2, periplakin gene PPL) were largely preserved, with subtle differences found among the subtypes, such as significant upregulation of FLG in LI and EI (FCH > 2, FDR < 0.01) and significant downregulation of loricrin gene LOR and sciellin gene SCEL in NS (FCH < −2, FDR < 0.05) (Figure 3 and Supplementary Material S1).
Figure 3. Ichthyosis subtypes share global barrier abnormalities, with EI presenting the least disrupted phenotype.
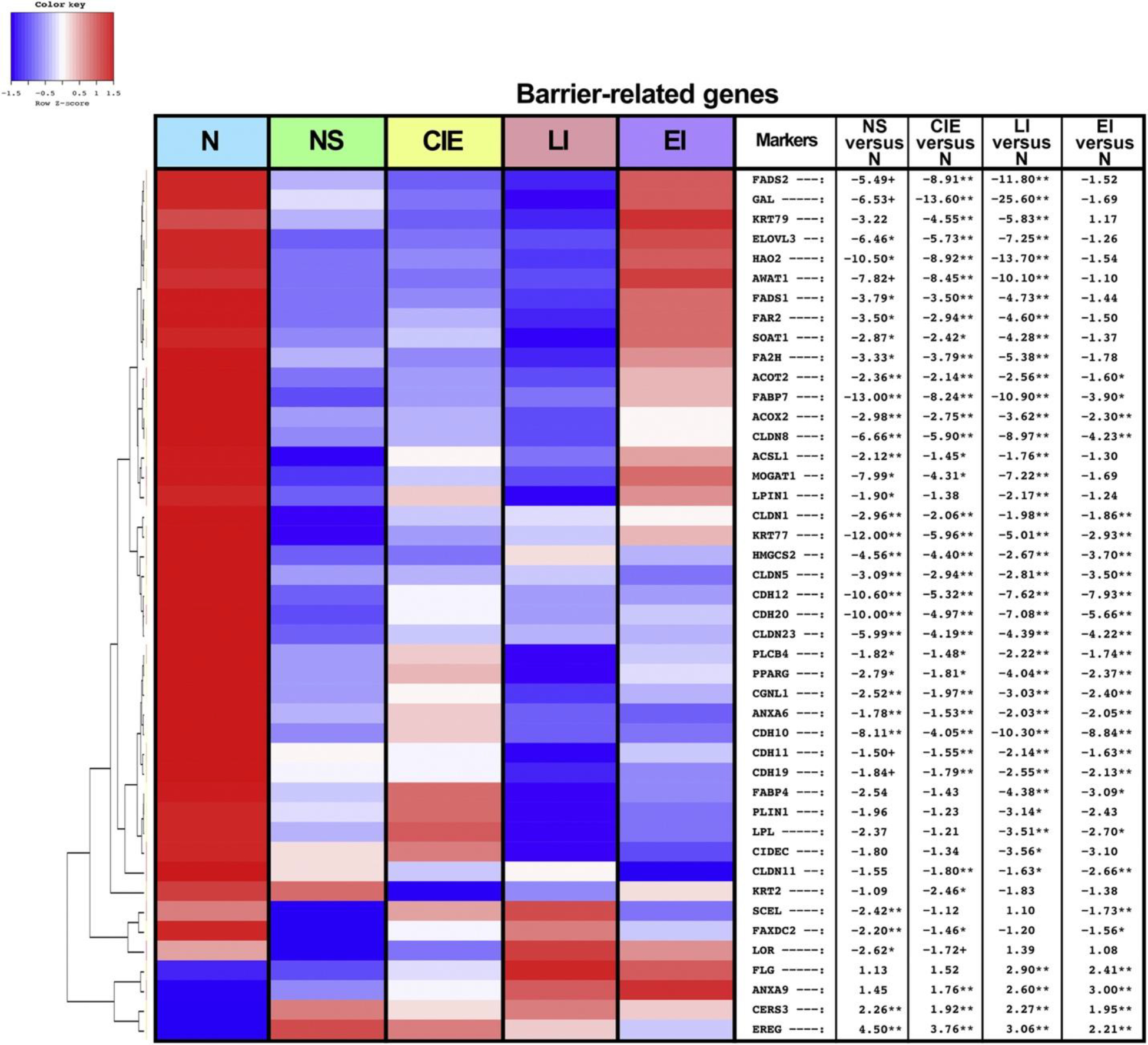
Heatmap representing differentially expressed barrier-related genes from biopsied skin of patients with the various ichthyosis subtypes and healthy controls. Criteria for differential gene expression include FCH > 2 and FDR < 0.05. From left to right, heatmap columns represent N, NS, CIE, LI, and EI. The table shows FCHs in NS versus N, CIE versus N, LI versus N, and EI versus N. N denotes normal. ∗∗FDR < 0.01, ∗FDR < 0.05, and +FDR < 0.1. CIE, congenital ichthyosiform erythroderma; EI, epidermolytic ichthyosis; FCH, fold change; FDR, false discovery rate; LI, lamellar ichthyosis; NS, Netherton syndrome.
Expanding on previous work (Malik et al., 2019), we were able to detect additional lipid metabolism–related markers that were downregulated across all subtypes except EI (e.g., ELOVL3, FA2H, FAR2) (FCH < −3, FDR < 0.05). Downregulated barrier genes found in EI were largely composed of cadherins and claudins (CDH10/12/19/20, CLDN5/8/11/23), most of which were also downregulated in other subtypes (FCH > 2, FDR < 0.05). Overall, LI skin displayed the greatest dysregulation of lipid metabolism–related genes, with the largest decreases in galanin gene GAL, HAO2, and FADS2 (FCH < −11, FDR < 0.001). On the contrary, EI presented modest modulation of lipid-related markers, with many genes not reaching significance (e.g., FADS2, galanin gene GAL, AWAT1). Among the barrier-related genes shared across two or more ichthyoses subtypes, FABP7, keratin 77 gene K77, CDH12/20, CLDN23, and MOGAT1 were most downregulated in NS (FCH < −5, FDR < 0.05), whereas FADS2, galanin gene GAL, and AWAT1 were most downregulated in CIE and LI (FCH < −8, FDR < 0.01).
RT-qPCR analysis supports RNA-seq data
To further validate our results, we measured mRNA levels of 45 markers representing Th-pathway and barrier-related genes, some of which may be poorly detected by RNA-seq owing to relatively low expression, using RT-qPCR (Supplementary Figure S3) (Dhingra et al., 2014; Guttman-Yassky et al., 2019a; He et al., 2021, 2020a, 2020b; Khattri et al., 2014; Malik et al., 2019, 2017; Pavel et al., 2020; Renert-Yuval et al., 2021). Paralleling our RNA-seq findings, several Th17 markers (e.g., IL-36A/G, KYNU, VNN1) (P < 0.05) were significantly upregulated in all subtypes, along with other Th17/Th22-related genes, including IL17A/F, IL19 (significant in all except in LI with RNA-seq), S100A7/9/12, and IL22 (P < 0.01) (Supplementary Figures S3 and S4). Furthermore, as in RNA-seq, Th1-associated markers, including IFN-γ and IL-12B/IL-12/23p40 were upregulated only in CIE (P < 0.01), and CXCL10 was significantly upregulated in CIE and EI (P < 0.05). Overall, Th2 marker expression in all subtypes was similar to that in the controls, with few genes showing significant downregulation in one of the various subtypes (e.g., CCL17 in LI, CCL26 in EI, CCL18 in CIE) (P < 0.05). The terminal differentiation marker EREG was upregulated in NS, CIE, and EI, and FLG was upregulated in LI and EI (P < 0.01).
Immune and barrier markers correlate with disease severity in ichthyosis
We also performed Spearman correlations between mRNA markers and clinical severity scores—total IASI with its subscores (IASI-erythema [IASI-E] and IASI-scaling) and epidermal barrier disruption, measured by TEWL—(Figure 4, Supplementary Table S3, and Supplementary Materials S2). Significant positive correlations were noted between immune markers and total IASI (e.g., FOXE1), IASI-scaling (e.g., SOX7, implicated in tumorigenesis) (Stovall et al., 2014), and IASI-E (e.g., PI3, SERPINB4, SAA4, and psoriasis-associated SLPI) (Skrzeczynska-Moncznik et al., 2020) (all r > 0.34, P < 0.05). Significant negative correlations were observed between total IASI and barrier-related CETP (r = 0.49, P < 0.001), between IASI-scaling and CETP and CDH19 (r = −0.4 and −0.38, respectively, P < 0.01), as well as between IASI-E and IL-34 (a negative regulator of inflammation) (Guttman-Yassky et al., 2019b) and FABP7 (r < −0.34, P < 0.05). TEWL (excluded five patients for whom TEWL was not recorded) was most highly and positively correlated with tumor-promoting FURIN (Fu et al., 2012) as well as keratin-related (EPN3), gap junction–related (GJB2/6), and terminal differentiation–related (CALML5) markers (r > 0.43, P < 0.01) and most negatively correlated with CLDN1, lipid metabolism–related marker ACER2, and keratin-related KRTDAP (r < −0.34, P < 0.05).
Figure 4. Th17/Th22 enrichment in skin correlates with clinical indices and with CD4+/CD8+ IL-22+ T-cell populations in the blood.
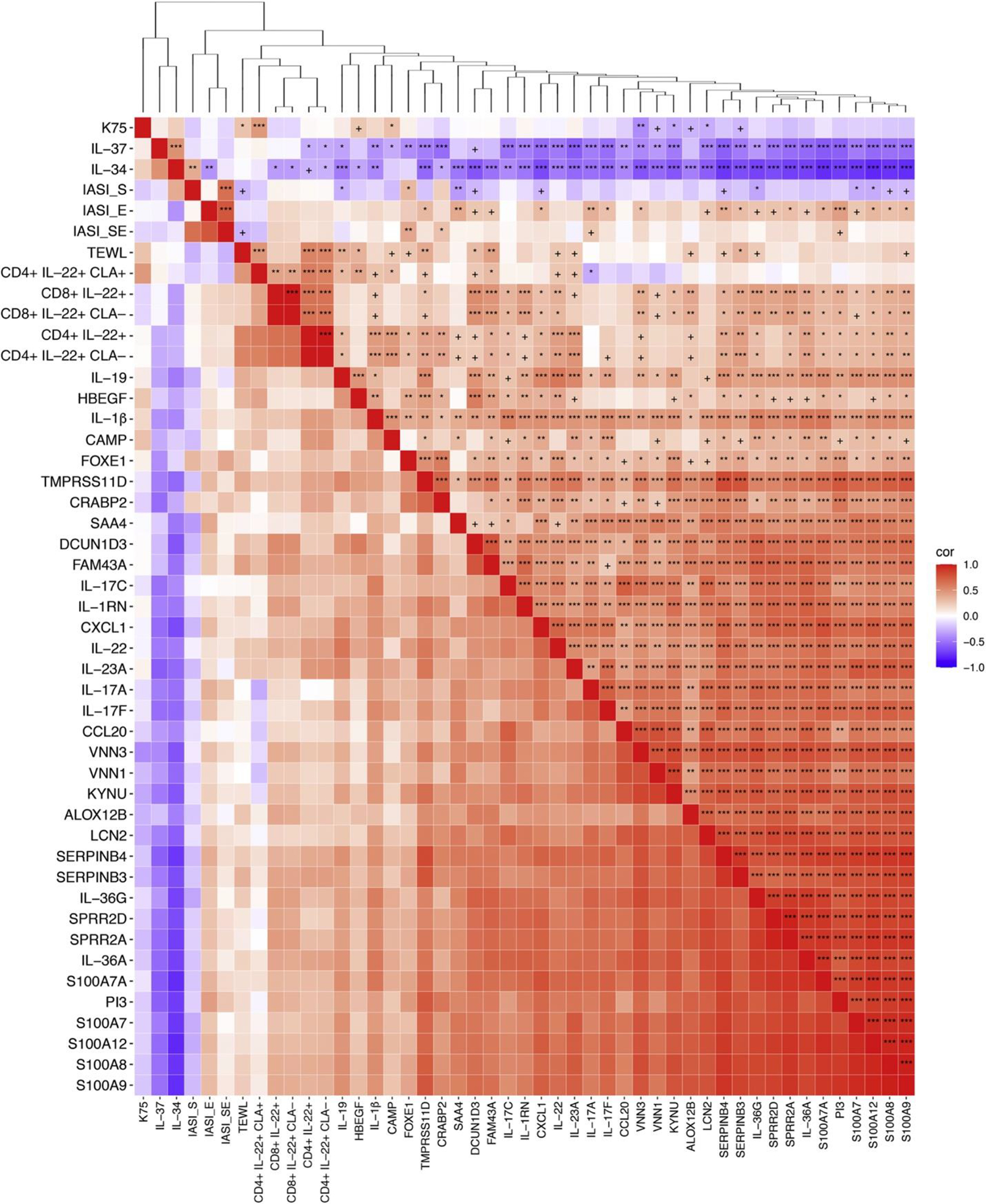
Spearman correlation heat map of Th17-, Th22-, and IASI-S/IASI-E/IASI-SE in the skin and CD4+/CD8+ IL-22+ in the blood of patients with ichthyosis. The dendrogram represents the k-means clustering of marker/variables. +P < 0.1, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Cor, correlation; IASI-E, Ichthyosis Area Severity Index-Erythema; IASI-S, Ichthyosis Area Severity Index-Scaling; IASI-SE, Total Ichthyosis Area Severity Index; K, keratin; Th, T helper.
Th17/Th22 enrichment in skin correlates with CD4+/CD8+ IL-22+ T-cell populations in blood
We additionally sought to integrate a systemic metric of inflammation into our analysis by performing Spearman correlations between mRNA markers and CD4+ and CD8+ T-cell populations in the blood using previously published flow cytometry data from 32 of our 54 patients with ichthyosis (n = 5 for NS, n = 10 for CIE, n = 12 for LI, and n = 5 for EI) (Czarnowicki et al., 2018). We observed strong, positive correlations (r > 0.35, P < 0.05) between several Th17/Th22 and synergistically upregulated IL-17/TNF-α markers (e.g., S100A8/9, IL-22, SERPINB4, IL-23A, DCUN1D3) and IL-22+ T-cell populations (CD4+ and CD8+), notably those that did not express cutaneous lymphocyte-associated antigen (CLA), a skin-homing marker (Akdis et al., 1997) (see Supplementary Table S4, Figure 4, and Supplementary Materials S3). These patterns held at the pathway level (r > 0.35, P < 0.05), where CD8+ IL-22+ T-cell counts correlated positively with GSVA scores of previously published Th22/IL-22, Th17-specific, and synergistically upregulated IL17/TNFα gene sets (Supplementary Table S4) (Chiricozzi et al., 2011; Dhingra et al., 2014; Guttman-Yassky et al., 2019a; He et al., 2021, 2020a, 2020b; Malik et al., 2019).
Pathway enrichment analysis highlights cellular proliferation signaling
For a broader, unbiased perspective on ichthyosis-related changes, all DEGs in ichthyosis versus controls were also analyzed using function-based pathway databases (canonical/Kyoto Encyclopedia of Genes and Genomes/Reactome/BioCarta pathways) (Supplementary Table S5) (Fang et al., 2016). We found that across all ichthyosis subtypes, the top 20 enriched pathways (FDR < 0.05) were related to cell cycle and replication (e.g., aurora B signaling, FOXM1 transcription factor network, DNA replication), along with pathways related to extracellular matrix–associated genes (Supplementary Figure S5a). We also assessed the cellular structures in which ichthyosis-related differentially expressed pathways are localized to function. An analysis of gene ontology by cellular component terms identified the CE as the primary cellular location for ichthyosis-related differentially expressed pathways, followed by structures specific to cell replication and division (e.g., kinetochore, spindle microtubule, midbody) (Supplementary Figure S5b). We also conducted a GSVA of previously published barrier gene sets (Guttman-Yassky et al., 2019a; Malik et al., 2019; Pavel et al., 2020; Renert-Yuval et al., 2021; Sanyal et al., 2019), which further validated that the EDC/CE geneset was significantly enriched across all ichthyosis subtypes against normal skin (P < 0.01), with comparable levels of enrichment across the ichthyoses (Supplementary Figure S2f).
DISCUSSION
We present, to our knowledge, a previously unreported comprehensive RNA-seq study in the skin from the largest cohort of patients with ichthyosis to date, with the greatest representation of each of the four most common orphan ichthyosis variants (NS, EI, LI, and CIE) alongside healthy control subjects. Our study also integrates flow cytometry data from the blood compartment into an RNA-seq analysis of the skin in patients with ichthyosis. This study largely expands on our previous work with fewer patients (Bitoun et al., 2002; Chamcheu et al., 2011; Hannula-Jouppi et al., 2014; Marukian and Choate, 2016; Oji and Traupe, 2006), showing that the ichthyoses have a Th17 component similar to that of psoriasis (Malik et al., 2019; Paller et al., 2017) and not the Th2-predominant component of atopic dermatitis, another inflammatory skin disorder with increased TEWL, barrier dysfunction, and abnormal epidermal lipid metabolism (Proksch et al., 2009; Renert-Yuval et al., 2021).
Our RNA-seq analysis uniquely captured key Th17/Th22 markers that did not reach significance in previous microarray studies (Malik et al., 2019). These include IL-23R (upregulated in CIE, EI, and LI), IL-17A/C (upregulated in all subtypes), and IL-22 (upregulated in NS and CIE). Further expanding our previous work with microarrays (Malik et al., 2019), we were able to measure the expression of more genes coding for epithelial junction proteins (cadherins, claudins), which revealed significant downregulation in nearly all genes in most subtypes. These changes were accompanied by significant upregulation of EDC markers and genes contributing to CE formation in our ichthyosis cohort compared to controls, a pattern that was less pronounced in our previous study with microarrays. Notably, our analysis found significant global upregulation in small proline-rich protein genes SPRR2A/B/D/E/F, which are expressed during keratinocyte terminal differentiation to produce CE precursors (Fischer et al., 1998, 1996), and that SPRR2A and SPPR2F also positively and significantly correlated with IASI-E.
In line with past work with microarrays and PCR (Malik et al., 2019; Paller et al., 2017), the ichthyosis subtypes exhibited strong global upregulation of IL-17/IL-23–regulated genes relative to controls, including many markers that are synergistically and additively upregulated with TNF-α (IL-17A/C, IL-19, IL-23R, PI3, CCL20) (Chiricozzi et al., 2011) and are classically enriched in psoriasis (e.g. IL-36A/G, KYNU, VNN1, VNN3). We also observed remarkable upregulation in markers upregulated by both IL-22 and IL-17, notably the antimicrobial S100s (S100A7/8/9/12) and the epidermal hyperplasia marker SERPINB4, which have also been linked to psoriasis (Nograles et al., 2008; Suárez-Fariñas et al., 2010; Takahashi and Yamasaki, 2020). Also consistent with previous studies were the modest upregulation in NS of some Th2 markers, but not IL-13 or CCL17/TARC, and the variable and often insignificant modulation in the other subtypes, differentiating the ichthyosis molecular profile from that of atopic dermatitis (Ewald et al., 2015; Guttman-Yassky et al., 2019a, 2019b) and potentially reflecting reciprocal counter-regulation with Th17 (Lynch et al., 2016). We were able to validate many of our immune markers with RT-qPCR analysis. Several IL-17/IL-23–related markers also significantly and positively correlated with IASI-E (e.g. SERPINB4, PI3, IL-17A/F, S100A8/9/12) and TEWL (e.g. FAM43A, SERPINB3, IL-19), suggesting that IL-17 and IL-22 pathway upregulation may be implicated in the cutaneous inflammation and barrier dysfunction observed in ichthyosis, similar to psoriasis (Carrier et al., 2011; Enjalbert et al., 2020; Madonna et al., 2019; Nograles et al., 2008; Pfaff et al., 2017; Suárez-Fariñas et al., 2010).
In addition, we showed downregulation of lipid homeostasis genes in all subtypes, particularly in LI, with more preserved expression in EI, which is consistent with our previous study and the common observation that the ichthyoses are characterized by barrier dysregulation (Elias et al., 2008; Honda et al., 2018; Malik et al., 2019; Süßmuth et al., 2020; Yamamoto et al., 2020). Disruption of lipid metabolism and lipid barrier formation, with concomitant inflammation, has been hypothesized to trigger epidermal hyperplasia and hyperkeratosis in ichthyoses, resulting in a compensatory increase in EDC and CE expression (Elias et al., 2008; Vahlquist and Törmä, 2020; Zaenglein et al., 2021), as suggested by our report.
Using validated function-based pathway databases, we found that all subtypes exhibited enrichment of genes related to cellular proliferation and extracellular matrix synthesis as well as increased production of CE and cellular replication components. These results corroborate the upregulation of markers in EDC/CE seen in our GSVA analysis and also lend support to the role of epidermal hyperplasia in the disease process.
Because other inflammatory skin diseases (e.g., atopic dermatitis, psoriasis) have been shown to exhibit systemic inflammation with potential cardiovascular morbidities (Brunner et al., 2017; He et al., 2020a; Hu and Lan, 2017; Puig, 2017), we additionally extended our analysis to the blood compartment in our ichthyosis cohort, who have already shown increased serum IL-17A and greater populations of CD4+ and CD8+ T cells (CLA+ and CLA−) expressing IL-22 and of CD4+ CLA+ T cells expressing IL-17 than of the controls (Czarnowicki et al., 2018; Malik et al., 2019). We found that the expression of IL-22 and IL-17/TNF-α–driven markers in the skin correlated strongly and positively with counts of CD4+ and CD8+ T cells expressing IL-22+, notably in CLA− populations. This pattern was also found between IL-22+ T-cell (CD4+/CD8+) counts and the expression of terminal differentiation markers SPRR2A/D, LCE3D, and the innate inflammation marker IL-1B. These results suggest a potential association between circulating IL-22+ T-cell numbers and IL-17/IL-22–driven inflammation and barrier dysregulation in the skin, especially because both of these parameters also positively correlated with IASI-E and/or TEWL (Czarnowicki et al., 2018). Although the data on nondermatological morbidities of ichthyoses remain limited, systemic targeting of IL-17/IL-22–driven inflammation may provide benefit in disease management for all subtypes in this study and requires further investigation. Indeed, preliminary case reports have already shown initial promise in results with some immune-targeting treatments, including ustekinumab, a mAb against IL-12 and IL-23; secukinumab, a mAb against IL-17A; guselkumab, specifically targeting IL-23; and infliximab, a chimeric mAb against TNF-α, displaying potential in treating NS (Fontao et al., 2011; Luchsinger et al., 2020; Volc et al., 2020), CIE (Poulton et al., 2019; Steinhoff et al., 2022), and other rare ichthyotic disorders (Frommherz et al., 2021; Haiges et al., 2019; Hernández-Martín et al., 2019; Paller et al., 2018; Sun et al., 2021). Agents targeting other immune components may also provide promising routes for further studies, such as nitric oxide synthase 2 inhibitor 1400W or the Jak inhibitor tofacitinib, which were found to improve various metrics of barrier function in models of HI, the most severe subtype within the autosomal recessive congenital ichthyoses spectrum (Enjalbert et al., 2020).
This study has a few limitations. First, although we were able to recruit the largest cohort of patients within each ichthyosis subtype to date, sample sizes remained relatively small owing to the rarity of the diseases. We also recognize a predominance of patients identifying as white in both our ichthyosis and control groups, potentially limiting the applicability of these results to patients of other races. Finally, this is an observational study, and clinical trials targeting specific biomarkers are needed to further validate the therapeutic potential of our findings.
In conclusion, we found significant Th17/Th22-driven inflammation, particularly with genes regulated by IL-17/TNF-α and IL-22, that correlated with circulating IL-22+ T-cell populations in all ichthyosis subtypes compared with that in the controls. We identified significant downregulation in lipid metabolism and tight junction genes, accompanied by significant upregulation of EDC, CE, and epidermal hyperproliferation–related markers. Our findings expand on the similarities in inflammatory and barrier dysfunction drawn between the pathogenesis of ichthyosis and psoriasis, suggesting a path forward in specific treatments for ichthyosis with psoriasis as a roadmap, namely the systemic antagonism of IL-17, IL-23, IL-36, and/or IL-22. Identifying additional serum markers of inflammation with potential roles in cutaneous pathology could broaden our arsenal of systemic treatments, affording more customizability to individual patients. Further studies on these fronts would help to address a desperately unmet need for effective therapy and a better understanding of the pathogenesis of these rare diseases.
MATERIALS AND METHODS
Patient characteristics and samples
This study was reviewed and approved by local Institutional Review Boards at Icahn School of Medicine at Mount Sinai (New York, NY) and Northwestern University (Chicago, IL). All subjects (if aged ≥12 years) in addition to their parents (if aged <18 years) provided informed written consent. Demographics, medical history, skin examination, clinical severity scores (IASI also called total IASI) (Paller et al., 2017), pruritus scores (5-D itch scale) (Elman et al., 2010), and TEWL were obtained, as reported (Paller et al., 2017). The IASI score consists of erythema (IASI-E) and scaling (IASI-scaling) subscores and quantifies score intensity within body regions (Paller et al., 2017). The 4.5-mm biopsy specimens were obtained from the upper arm.
RNA extraction and gene expression analyses
RNA was extracted from skin biopsies using Qiagen miRNeasy Mini kits (Qiagen, Hilden, Germany), as previously described (Sanyal et al., 2019; Suárez-Fariñas et al., 2015). The libraries were generated with TruSeq Stranded messenger RNA Library Prep kits (Illumina, San Diego, CA), as reported (Pavel et al., 2020). Next-generation sequencing was performed on Illumina Hiseq 4000 (Illumina, San Diego, CA) with single-ended 100 red cycles. RNA-seq sample quality was assessed using FastQC (Cambridge, United Kingdom) and MultiQC (Ewels et al., 2016). Samples were aligned to the human reference genome on the basis of the STAR RNA-seq aligner (Dobin et al., 2013), and sequencing reads were assigned to genomic features by featureCounts (Liao et al., 2014). The data were subsequently log2 transformed with voom transform (Law et al., 2014).
For RT-qPCR, 500 pg of RNA per sample was subjected to reverse transcription, preamplification, and RT-qPCR, as previously described (Sanyal et al., 2019). TaqMan Low-Density Array cards (Thermo Fisher Scientific, Waltham, MA) were used for RT-qPCR, as previously reported (Noda et al., 2015; Sanyal et al., 2019). Ct values were normalized to the housekeeping gene RPLP0. The primers used are listed in Supplementary Table S6.
Statistical analysis
Statistical analysis was performed with R software (www.R-project.org) and packages available through Bioconductor (www.bioconductor.org). We estimated FCHs between comparisons of interest and conducted hypothesis testing using contrasts under the general framework for linear models in R software limma package, adjusting for age. P-values from the moderated (paired) t-test were adjusted for multiple hypotheses using the Benjamini–Hochberg procedure. Genes with FCH > 2 and a FDR < 0.05 were considered differentially expressed. Hierarchical clustering of samples/conditions used a McQuitty agglomeration algorithm.
For RT-qPCR data, the RT-qPCR values were normalized to RPLP0 by negatively transforming Ct values to –dCt. Undetected –dCt values were estimated for each gene as 20% of the minimum across all samples. Flow cytometry data were acquired from a previously published dataset (Czarnowicki et al., 2018).
GSVA was performed using unsupervised sample-wise enrichment in the R software GSVA package, and Spearman correlations were performed with the R stats package, with clustering executed through a k-means algorithm (Hänzelmann et al., 2013). Gene set overexpression analysis was performed with XGR software using canonical/Kyoto Encyclopedia of Genes and Genomes/Reactome/BioCarta pathways and gene ontology by cellular components (Fang et al., 2016).
Supplementary Material
Acknowledgments
We are grateful to the Foundation for Ichthyosis and Related Skin Types for allowing this research to be performed in part at its Family Conferences in 2014 and 2016. We acknowledge the core resources provided by the Northwestern University Skin Disease Research Center (National Institute of Arthritis and Musculoskeletal and Skin Diseases P30AR057216 and P30AR075049). This research received funding from AnaptysBio.
Abbreviations:
- CE
cornified envelope
- CIE
congenital ichthyosiform erythroderma
- CLA
cutaneous lymphocyte-associated antigen
- DEG
differentially expressed gene
- EDC
epidermal differentiation complex
- EI
epidermolytic ichthyosis
- FCH
fold change
- FDR
false discovery rate
- GSVA
gene set variation analysis
- HI
harlequin ichthyosis
- IASI
Ichthyosis Area and Severity Index
- IASI-E
Ichthyosis Area and Severity Index-erythema
- LI
lamellar ichthyosis
- NS
Netherton syndrome
- RNA-seq
RNA-sequencing
- TEWL
transepidermal water loss
- Th
T helper
Footnotes
Conflict of Interest
JK declares receiving consulting/honoraria from AbbVie, Aclaris, Allergan, Almirall, Amgen, Arena, Aristea, Asana, Aurigene, Biogen Idec, Boehringer-Ingelheim, Bristol-Myers Squibb, Escalier, Galapagos, Janssen, Lilly, MoonLake Immunotherapeutics, Nimbus, Novartis, Pfizer, Sanofi, Sienna Biopharmaceuticals, Sun Pharma, Target-Derm, UCB, Valeant, and Ventyx and grant support (to The Rockefeller University) from AbbVie, Akros, Allergan, Amgen, Avillion, Biogen, Botanix, Boehringer-Ingelheim, Bristol-Myers Squibb, Exicure, Innovaderm, Incyte, Janssen, Kyowa Kirin, Lilly, Nimbus Lackshmi, Novan, Novartis, PAREXEL, Pfizer, Regeneron, UCB, and Vitae Pharmaceuticals. EGY is an employee of Icahn School of Medicine at Mount Sinai and has received research funds (grants paid to the institution) from AbbVie, Almirall, Amgen, AnaptysBio, Asana Biosciences, AstraZeneca, Boehringer-Ingelheim, Celgene, Dermavant, DS Biopharma, Eli Lilly, Galderma, Glenmark/Ichnos Sciences, Innovaderm, Janssen, Kiniksa, Kyowa Kirin, Leo Pharma, Medimmune/Astra Zeneca, Novan, Novartis, Pfizer, Ralexar, Regeneron, Sienna Biopharma, UCB, Union Therapeutics/Antibiotx, Vitae, and Dermira. EGY is also a consultant for Sanofi Aventis, Regeneron, Stiefel/GlaxoSmithKline, MedImmune, Celgene, Anacor, AnaptysBio, Dermira, Galderma, Glenmark, Novartis, Pfizer, Vitae, Leo Pharma, AbbVie, Eli Lilly, Kyowa, Mitsubishi Tanabe, Asana Biosciences, Promius, Aditum Bio, Almirall, Alpine, Amgen, Arena, AstraZeneca, Bluefin Biomedicine, Boehringer-Ingelheim, Boston Pharmaceuticals, Botanix, Bristol-Meyers Squibb, Cara Therapeutics, Clinical Outcome Solutions, DBV, Dermavant, Douglas Pharmaceutical, DS Biopharma, EMD Serono, Evelo Bioscience, Evidera, FIDE, Haus Bioceuticals, Ichnos Sciences, Incyte, Larrk Bio, Medicxi, Medscape, Neuralstem, Noble Insights, Novan, Okava Pharmaceuticals, Pandion Therapeutics, Principia Biopharma, RAPT Therapeutics, Realm, SATO Pharmaceutical, Sienna Biopharma, Seanegy Dermatology, Seelos Therapeutics, Serpin Pharma, Siolta Therapeutics, Sonoma Biotherapeutics, Sun Pharma, Target PharmaSolutions and Union Therapeutics, Vanda Pharmaceuticals, Ventyx Biosciences, Vimalan, Concert, and Sienna Biopharma. ASP has received research funds (paid to institution) from AbbVie, AnaptysBio, Eli Lilly, Incyte, Janssen, KrystalBio, Regeneron, and UCB; is a consultant for AbbVie, Abeona, Alcimed, Almirall, Amagma, AnaptysBio, Arena, Azitra, BiomX, Boehringer-Ingelheim, Castle Biosciences, Catawba, Dermira, Eli Lilly, Exicure, Forte, Kamari, Leo, Lifemax, NAOS, Novartis, Pfizer, Phoenix, Pierre Fabre, Regeneron, Sanofi/Genzyme, Seanergy, Trifecta, and UCB; and serves on a data safety monitoring board for AbbVie, Bausch, Galderma, and Novan. ABP is an employee of the University of Mississippi and has a research contract with Icahn School of Medicine at Mount Sinai.
Supplementary Material
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2022.03.022.
Data availability statement
The data discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through Gene Expression Omnibus Series accession number GSE192832 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192832).
REFERENCES
- Akagi A, Kitoh A, Moniaga CS, Fujimoto A, Fujikawa H, Shimomura Y, et al. Case of Netherton syndrome with an elevated serum thymus and activation-regulated chemokine level. J Dermatol 2013;40:752e3. [DOI] [PubMed] [Google Scholar]
- Akdis M, Akdis CA, Weigl L, Disch R, Blaser K. Skin-homing, CLA+ memory T cells are activated in atopic dermatitis and regulate IgE by an IL-13-dominated cytokine pattern: IgG4 counter-regulation by CLA-memory T cells. J Immunol 1997;159:4611e9. [PubMed] [Google Scholar]
- Allen A, Siegfried E, Silverman R, Williams ML, Elias PM, Szabo SK, et al. Significant absorption of topical tacrolimus in 3 patients with Netherton syndrome. Arch Dermatol 2001;137:747e50. [PubMed] [Google Scholar]
- Bitoun E, Chavanas S, Irvine AD, Lonie L, Bodemer C, Paradisi M, et al. Netherton syndrome: disease expression and spectrum of SPINK5 mutations in 21 families. J Invest Dermatol 2002;118:352e61. [DOI] [PubMed] [Google Scholar]
- Blunder S, Rühl R, Moosbrugger-Martinz V, Krimmel C, Geisler A, Zhu H, et al. Alterations in epidermal eicosanoid metabolism contribute to inflammation and impaired late differentiation in FLG-mutated atopic dermatitis. J Invest Dermatol 2017;137:706e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briot A, Deraison C, Lacroix M, Bonnart C, Robin A, Besson C, et al. Kallikrein 5 induces atopic dermatitiselike lesions through PAR2-mediated thymic stromal lymphopoietin expression in Netherton syndrome. J Exp Med 2009;206:1135e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner PM, Suárez-Fariñas M, He H, Malik K, Wen HC, Gonzalez J, et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci Rep 2017;7:8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier Y, Ma HL, Ramon HE, Napierata L, Small C, O’Toole M, et al. Interregulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: implications in psoriasis pathogenesis. J Invest Dermatol 2011;131: 2428e37. [DOI] [PubMed] [Google Scholar]
- Chamcheu JC, Siddiqui IA, Syed DN, Adhami VM, Liovic M, Mukhtar H. Keratin gene mutations in disorders of human skin and its appendages. Arch Biochem Biophys 2011;508:123e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet 2000;25:141e2. [DOI] [PubMed] [Google Scholar]
- Chiricozzi A, Guttman-Yassky E, Suárez-Fariñas M, Nograles KE, Tian S, Cardinale I, et al. Integrative responses to IL-17 and TNF-a in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol 2011;131:677e87. [DOI] [PubMed] [Google Scholar]
- Czarnowicki T, He H, Leonard A, Malik K, Magidi S, Rangel S, et al. The major orphan forms of ichthyosis are characterized by systemic T-cell activation and Th-17/Tc-17/Th-22/Tc-22 polarization in blood. J Invest Dermatol 2018;138:2157e67. [DOI] [PubMed] [Google Scholar]
- Dhingra N, Shemer A, Correa da Rosa J, Rozenblit M, Fuentes-Duculan J, Gittler JK, et al. Molecular profiling of contact dermatitis skin identifies allergen-dependent differences in immune response. J Allergy Clin Immunol 2014;134:362e72. [DOI] [PubMed] [Google Scholar]
- Digiovanna JJ, Mauro T, Milstone LM, Schmuth M, Toro JR. Systemic retinoids in the management of ichthyoses and related skin types. Dermatol Ther 2013;26:26e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus I, Pauwels C, Bourrat E, Bursztejn AC, Maruani A, Chiaverini C, et al. Burden of inherited ichthyosis: a French national survey. Acta Derm Venereol 2015;95:326e8. [DOI] [PubMed] [Google Scholar]
- Elias PM, Williams ML, Holleran WM, Jiang YJ, Schmuth M. Pathogenesis of permeability barrier abnormalities in the ichthyoses: inherited disorders of lipid metabolism. J Lipid Res 2008;49:697e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elman S, Hynan LS, Gabriel V, Mayo MJ. The 5-D itch scale: a new measure of pruritus. Br J Dermatol 2010;162:587e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert F, Dewan P, Caley MP, Jones EM, Morse MA, Kelsell DP, et al. 3D model of harlequin ichthyosis reveals inflammatory therapeutic targets. J Clin Invest 2020;130:4798e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald DA, Malajian D, Krueger JG, Workman CT, Wang T, Tian S, et al. Meta-analysis derived atopic dermatitis (MADAD) transcriptome defines a robust AD signature highlighting the involvement of atherosclerosis and lipid metabolism pathways. BMC Med Genomics 2015;8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32(19):3047e8. 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Knezevic B, Burnham KL, Knight JC. XGR software for enhanced interpretation of genomic summary data, illustrated by application to immunological traits. Genome Med 2016;8:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer DF, Gibbs S, van De Putte P, Backendorf C. Interdependent transcription control elements regulate the expression of the SPRR2A gene during keratinocyte terminal differentiation. Mol Cell Biol 1996;16: 5365e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer DF, van Drunen CM, Winkler GS, van de Putte P, Backendorf C. Involvement of a nuclear matrix association region in the regulation of the SPRR2A keratinocyte terminal differentiation marker. Nucleic Acids Res 1998;26:5288e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontao L, Laffitte E, Briot A, Kaya G, Roux-Lombard P, Fraitag S, et al. Infliximab infusions for Netherton syndrome: sustained clinical improvement correlates with a reduction of thymic stromal lymphopoietin levels in the skin. J Invest Dermatol 2011;131:1947e50. [DOI] [PubMed] [Google Scholar]
- Frommherz LH, Schempp CM, Has C. Secukinumab for the treatment of SAM syndrome associated with desmoglein-1 deficiency. Br J Dermatol 2021;184:770e2. [DOI] [PubMed] [Google Scholar]
- Fu J, Bassi DE, Zhang J, Li T, Nicolas E, Klein-Szanto AJ. Transgenic overexpression of the proprotein convertase furin enhances skin tumor growth. Neoplasia 2012;14:271e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furio L, de Veer S, Jaillet M, Briot A, Robin A, Deraison C, et al. Transgenic kallikrein 5 mice reproduce major cutaneous and systemic hallmarks of Netherton syndrome. J Exp Med 2014;211:499e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furio L, Pampalakis G, Michael IP, Nagy A, Sotiropoulou G, Hovnanian A. KLK5 inactivation reverses cutaneous hallmarks of Netherton syndrome. PLOS Genet 2015;11:e1005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman-Yassky E, Bissonnette R, Ungar B, Suárez-Farinãs M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol 2019a;143:155e72. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Diaz A, Pavel AB, Fernandes M, Lefferdink R, Erickson T, et al. Use of tape strips to detect immune and barrier abnormalities in the skin of children with early-onset atopic dermatitis. JAMA Dermatol 2019b;155:1358e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiges D, Fischer J, Hörer S, Has C, Schempp CM. Biologic therapy targeting IL-17 ameliorates a case of congenital ichthyosiform cornification disorder. J Dtsch Dermatol Ges 2019;17:70e2. [DOI] [PubMed] [Google Scholar]
- Halverstam CP, Vachharajani A, Mallory SB. Cushing syndrome from percutaneous absorption of 1% hydrocortisone ointment in Netherton syndrome. Pediatr Dermatol 2007;24:42e5. [DOI] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Laasanen SL, Heikkilä H, Tuomiranta M, Tuomi ML, Hilvo S, et al. IgE allergen component-based profiling and atopic manifestations in patients with Netherton syndrome. J Allergy Clin Immunol 2014;134:985e8. [DOI] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Bissonnette R, Wu J, Diaz A, Saint-Cyr Proulx E, Maari C, et al. Tape strips detect distinct immune and barrier profiles in atopic dermatitis and psoriasis. J Allergy Clin Immunol 2021;147:199e212. [DOI] [PubMed] [Google Scholar]
- He H, Li R, Choi S, Zhou L, Pavel A, Estrada YD, et al. Increased cardiovascular and atherosclerosis markers in blood of older patients with atopic dermatitis. Ann Allergy Asthma Immunol 2020a;124:70e8. [DOI] [PubMed] [Google Scholar]
- He H, Olesen CM, Pavel AB, Clausen ML, Wu J, Estrada Y, et al. Tape-strip proteomic profiling of atopic dermatitis on dupilumab identifies minimally invasive biomarkers. Front Immunol 2020b;11:1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Martin A, Aranegui B, Martin-Santiago A, Garcia-Doval I. A systematic review of clinical trials of treatments for the congenital ichthyoses, excluding ichthyosis vulgaris. J Am Acad Dermatol 2013;69: 544e9.e8. [DOI] [PubMed] [Google Scholar]
- Hernández-Martín A, Kennedy-Batalla R, Cañedo E, Bernaldo-de-Quirós E, Carazo-Gallego B, Vera A, et al. Imbalance in T-helper 17 cells and targeted therapy in an infant with SAM-like syndrome. N Engl J Med 2019;381:2176e8. [DOI] [PubMed] [Google Scholar]
- Honda Y, Kitamura T, Naganuma T, Abe T, Ohno Y, Sassa T, et al. Decreased skin barrier lipid acylceramide and differentiation-dependent gene expression in ichthyosis gene Nipal4-knockout mice. J Invest Dermatol 2018;138:741e9. [DOI] [PubMed] [Google Scholar]
- Hosomi N, Fukai K, Nakanishi T, Funaki S, Ishii M. Caspase-1 activity of stratum corneum and serum interleukin-18 level are increased in patients with Netherton syndrome. Br J Dermatol 2008;159:744e6. [DOI] [PubMed] [Google Scholar]
- Hu SC-S, Lan C-CE. Psoriasis and cardiovascular comorbidities: focusing on severe vascular events, cardiovascular risk factors and implications for treatment. Int J Mol Sci 2017;18:2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuhara K, Yamaguchi Y, Ohta S, Nunomura S, Nanri Y, Azuma Y, et al. Squamous cell carcinoma antigen 2 (SCCA2, SERPINB4): an emerging biomarker for skin inflammatory diseases. Int J Mol Sci 2018;19:1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri S, Shemer A, Rozenblit M, Dhingra N, Czarnowicki T, Finney R, et al. Cyclosporine in patients with atopic dermatitis modulates activated inflammatory pathways and reverses epidermal pathology [published correction appears in J Allergy Clin immunol 2014;134:764] J Allergy Clin Immunol 2014;133:1626e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Tsuda T, Sakaguchi Y, Imai Y, Ito T, Hirota S, et al. Upregulation of interleukin-33 in the epidermis of two Japanese patients with etherton syndrome. J Dermatol 2014;41:258e61. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smith GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923e30. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Luchsinger I, Knöpfel N, Theiler M, Bonnet des Claustres M, Barbieux C, Schwieger-Briel A, et al. Secukinumab therapy for Netherton syndrome. JAMA Dermatol 2020;156:907e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Ferreira MA, Phipps S. Th2/Th17 reciprocal regulation: twists and turns in the complexity of asthma phenotypes. Ann Transl Med 2016;4:S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madonna S, Girolomoni G, Dinarello CA, Albanesi C. The significance of IL-36 hyperactivation and IL-36R targeting in psoriasis. Int J Mol Sci 2019;20:3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik K, He H, Huynh TN, Tran G, Mueller K, Doytcheva K, et al. Ichthyosis molecular fingerprinting shows profound TH17 skewing and a unique barrier genomic signature. J Allergy Clin Immunol 2019;143:604e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik K, Ungar B, Garcet S, Dutt R, Dickstein D, Zheng X, et al. Dust mite induces multiple polar T cell axes in human skin. Clin Exp Allergy 2017;47: 1648e60. [DOI] [PubMed] [Google Scholar]
- Marukian NV, Choate KA. Recent advances in understanding ichthyosis pathogenesis. F1000Res 2016;5. F1000 Faculty Rev-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazereeuw-Hautier J, Dreyfus I, Barbarot S, Serrentino L, Bourdon-Lanoy E, Ezzedine K, et al. Factors influencing quality of life in patients with inherited ichthyosis: a qualitative study in adults using focus groups. Br J Dermatol 2012;166:646e8. [DOI] [PubMed] [Google Scholar]
- Noda S, Suárez-Fariñas M, Ungar B, Kim SJ, de Guzman Strong C, Xu H, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol 2015;136:1254e64. [DOI] [PubMed] [Google Scholar]
- Nograles KE, Zaba LC, Guttman-Yassky E, Fuentes-Duculan J, Suárez-Fariñas M, Cardinale I, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol 2008;159:1092e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oji V, Tadini G, Akiyama M, Blanchet Bardon C, Bodemer C, Bourrat E, et al. Revised nomenclature and classification of inherited ichthyoses: results of the first ichthyosis Consensus Conference in Sorèze 2009. J Am Acad Dermatol 2010;63:607e41. [DOI] [PubMed] [Google Scholar]
- Oji V, Traupe H. Ichthyoses: differential diagnosis and molecular genetics. Eur J Dermatol 2006;16:349e59. [PubMed] [Google Scholar]
- Oji V, Traupe H. Ichthyosis: clinical manifestations and practical treatment options. Am J Clin Dermatol 2009;10:351e64. [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy RF, Choudhary I, Harper JI. Interleukin-1 alpha blockade prevents hyperkeratosis in an in vitro model of lamellar ichthyosis. Hum Mol Genet 2010;19:2594e605. [DOI] [PubMed] [Google Scholar]
- Paller AS. Profiling immune expression to consider repurposing therapeutics for the ichthyoses. J Invest Dermatol 2019;139:535e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paller AS, Czarnowicki T, Renert-Yuval Y, Holland K, Huynh T, Sadlier M, et al. The spectrum of manifestations in desmoplakin gene (DSP) spectrin repeat 6 domain mutations: immunophenotyping and response to ustekinumab. J Am Acad Dermatol 2018;78:498e505.e2. [DOI] [PubMed] [Google Scholar]
- Paller AS, Renert-Yuval Y, Suprun M, Esaki H, Oliva M, Huynh TN, et al. An IL-17-dominant immune profile is shared across the major orphan forms of ichthyosis. J Allergy Clin Immunol 2017;139:152e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavel AB, Zhou L, Diaz A, Ungar B, Dan J, He H, et al. The proteomic skin profile of moderate-to-severe atopic dermatitis patients shows an inflammatory signature. J Am Acad Dermatol 2020;82:690e9. [DOI] [PubMed] [Google Scholar]
- Pfaff CM, Marquardt Y, Fietkau K, Baron JM, Lü scher B. The psoriasis-associated IL-17A induces and cooperates with IL-36 cytokines to control keratinocyte differentiation and function. Sci Rep 2017;7:15631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton C, Gration D, Murray K, Baynam G, Halbert A. Autosomal recessive congenital ichthyosis due to homozygous variants in NIPAL4 with a dramatic response to ustekinumab. Pediatr Dermatol 2019;36:1002e3. [DOI] [PubMed] [Google Scholar]
- Proksch E, Fölster-Holst R, Bräutigam M, Sepehrmanesh M, Pfeiffer S, Jensen JM. Role of the epidermal barrier in atopic dermatitis. J Dtsch Dermatol Ges 2009;7:899e910. [DOI] [PubMed] [Google Scholar]
- Puig L Cardiometabolic comorbidities in psoriasis and psoriatic arthritis. Int J Mol Sci 2017;19:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renert-Yuval Y, Del Duca E, Pavel AB, Fang M, Lefferdink R, Wu J, et al. The molecular features of normal and atopic dermatitis skin in infants, children, adolescents, and adults. J Allergy Clin Immunol 2021;148:148e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner ED, Hartl D, Rylaarsdam S, Young ML, Monaco-Shawver L, Kleiner G, et al. Comèl-Netherton syndrome defined as primary immunodeficiency [published correction appears in J Allergy Clin Immunol 2009;124:1318] J Allergy Clin Immunol 2009;124:536e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Pazos L, Ginarte M, Vega A, Toribio J. Autosomal recessive congenital ichthyosis. Actas Dermosifiliogr 2013;104:270e84. [DOI] [PubMed] [Google Scholar]
- Sanyal RD, Pavel AB, Glickman J, Chan TC, Zheng X, Zhang N, et al. Atopic dermatitis in African American patients is TH2/TH22-skewed with TH1/ TH17 attenuation. Ann Allergy Asthma Immunol 2019;122:99e110.e6. [DOI] [PubMed] [Google Scholar]
- Sarri CA, Roussaki-Schulze A, Vasilopoulos Y, Zafiriou E, Patsatsi A, Stamatis C, et al. Netherton syndrome: a genotype-phenotype review. Mol Diagn Ther 2017;21:137e52. [DOI] [PubMed] [Google Scholar]
- Skrzeczynska-Moncznik J, Zabieglo K, Osiecka O, Morytko A, Brzoza P, Drozdz L, et al. Differences in staining for neutrophil elastase and its controlling inhibitor SLPI reveal heterogeneity among neutrophils in psoriasis. J Invest Dermatol 2020;140:1371e8.e3. [DOI] [PubMed] [Google Scholar]
- Steinhoff M, Al-Marri F, Al Chalabi R, Gieler U, Buddenkotte J. Recalcitrant erythrodermic ichthyosis with atopic dermatitis successfully treated with Dupilumab in combination with Guselkumab. Skin Health Dis 2022;2: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovall DB, Cao P, Sui G. SOX7: from a developmental regulator to an emerging tumor suppressor. Histol Histopathol 2014;29:439e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Lowes MA, Zaba LC, Krueger JG. Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA). PLoS One 2010;5:e10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Ungar B, Correa da Rosa J, Ewald DA, Rozenblit M, Gonzalez J, et al. RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. J Allergy Clin Immunol 2015;135:1218e27. [DOI] [PubMed] [Google Scholar]
- Sun Q, Burgren NM, Cheraghlou S, Paller AS, Larralde M, Bercovitch L, et al. The genomic and phenotypic landscape of ichthyosis: an analysis of 1000 kindreds. JAMA Dermatol 2022;158:16e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Wine Lee L, Hall EK, Choate KA, Elder RW. Hair and skin predict cardiomyopathies: Carvajal and erythrokeratodermia cardiomyopathy syndromes. Pediatr Dermatol 2021;38:31e8. [DOI] [PubMed] [Google Scholar]
- Süßmuth K, Traupe H, Metze D, Oji V . Ichthyoses in everyday practice: management of a rare group of diseases. J Dtsch Dermatol Ges 2020;18:225e43. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Yamasaki K. Psoriasis and antimicrobial peptides. Int J Mol Sci 2020;21:6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi T, Akiyama M. Inherited ichthyosis: non-syndromic forms. J Dermatol 2016;43:242e51. [DOI] [PubMed] [Google Scholar]
- Vahlquist A, Törmä H. Ichthyosis: A road model for skin research. Acta Derm Venereol 2020;100. adv00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gysel D, Koning H, Baert MR, Savelkoul HF, Neijens HJ, Oranje AP. Clinico-immunological heterogeneity in Comel-Netherton syndrome. Dermatology 2001;202:99e107. [DOI] [PubMed] [Google Scholar]
- Volc S, Maier L, Gritsch A, Aichelburg MC, Volc-Platzer B. Successful treatment of Netherton syndrome with ustekinumab in a 15-year-old girl. Br J Dermatol 2020;183:165e7. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Hattori M, Chamulitrat W, Ohno Y, Kihara A. Skin permeability barrier formation by the ichthyosis-causative gene FATP4 through formation of the barrier lipid u-O-acylceramide. Proc Natl Acad Sci USA 2020;117:2914e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaenglein AL, Levy ML, Stefanko NS, Benjamin LT, Bruckner AL, Choate K, et al. Consensus recommendations for the use of retinoids in ichthyosis and other disorders of cornification in children and adolescents. Pediatr Dermatol 2021;38:164e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through Gene Expression Omnibus Series accession number GSE192832 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192832).


