Abstract
Acute respiratory distress syndrome (ARDS) is a severe lung condition with high mortality and various causes, including lung infection. No specific treatment is currently available and more research aimed at better understanding the pathophysiology of ARDS is needed. Most lung-on-chip models that aim at mimicking the air-blood barrier are designed with a horizontal barrier through which immune cells can migrate vertically, making it challenging to visualize and investigate their migration. In addition, these models often lack a barrier of natural protein-derived extracellular matrix (ECM) suitable for live cell imaging to investigate ECM-dependent migration of immune cells as seen in ARDS. This study reports a novel inflammation-on-chip model with live cell imaging of immune cell extravasation and migration during lung inflammation. The three-channel perfusable inflammation-on-chip system mimics the lung endothelial barrier, the ECM environment and the (inflamed) lung epithelial barrier. A chemotactic gradient was established across the ECM hydrogel, leading to the migration of immune cells through the endothelial barrier. We found that immune cell extravasation depends on the presence of an endothelial barrier, on the ECM density and stiffness, and on the flow profile. In particular, bidirectional flow, broadly used in association with rocking platforms, was found to significantly delay extravasation of immune cells in contrast to unidirectional flow. Extravasation was increased in the presence of lung epithelial tissue. This model is currently used to study inflammation-induced immune cell migration but can be used to study infection-induced immune cell migration under different conditions, such as ECM composition, density and stiffness, type of infectious agents used, and the presence of organ-specific cell types.
Keywords: Lung infection, Hydrogel, Organ-on-chip, Lung-on-chip, Transmigration, Extravasation, Immune cells, ARDS, Uni- and bidirectional flow, Automated image analysis and segmentation, Lung inflammation
Graphical abstract
1. Introduction
Acute respiratory distress syndrome (ARDS) develops following severe lung injury induced by different agents, such as mechanical ventilation, sepsis, or infection (Ranieri et al., 2012). Clinically, ARDS is defined by its acute onset, hypoxemia, and radiographic opacities (Ferguson et al., 2012; Ranieri et al., 2012). Due to this broad definition, there is significant disease heterogeneity, and diagnosis is based on criteria rather than a single diagnostic test. Similarly, there is no simple ARDS treatment option (Meyer et al., 2021). On the cellular level, acute injury causes an increase in alveolar-capillary barrier permeability, leading to an influx of fluid into the lung and severe lung inflammation (Ware and Matthay, 2000). The recruitment of peripheral immune cells during the inflammatory cascade is a complex, multistep process that is still not completely understood. Through a complex interaction between receptors on immune cells and endothelial cells, immune cells attach to the inflamed endothelium and extravasate into the infected tissue (Abbas et al., 2012; Ley et al., 2007; Maas et al., 2018). Subsequently, immune cells interact with the surrounding extracellular matrix (ECM) in the interstitial space to migrate to the site of infection.
Standard ARDS models are either animal-based (D'Alessio, 2018) or rely on transwell-insert models (Ishii et al., 2021). These models induce direct injury through mechanical ventilation or damage-inducing agents such as LPS, or indirectly through intravenous injection or shock (Russ et al., 2021). Despite advancements, more advanced models resembling human physiology are needed to better understand ARDS in humans, as animal immune systems differ (Masopust et al., 2017; Mestas and Hughes, 2004). Standard in vitro models using human cells have their own limitations, as they tend to be oversimplified and do not capture key aspects of the cellular environment. Organ-on-chip (OOC) models have the ability to imitate specific elements of the organ-specific cellular environments (Bhatia and Ingber, 2014), and recapitulate tissue-level information allowing for comparison with clinical data (Guenat et al., 2020). Lung-on-chip models offer an opportunity to mimic complex multicellular in vitro studies providing phenotypical endpoints (Artzy-Schnirman et al., 2021). Both lung infection (Huh et al., 2010) and lung inflammation (Sengupta et al., 2022) have been modeled in immunocompetent lung-on-chip models. These lung models were used to analyze immune cell attachment and migration in response to LPS-induced inflammation (Sengupta et al., 2022), E. Coli infection (Huh et al., 2010), or to assess SARS-COV2 infection-related damage (Thacker et al., 2021; Zhang et al., 2021) and consist of two compartments, an endothelial and an epithelial channel separated by a porous polydimethylsiloxane (PDMS) membrane in a vertical configuration (Francis et al., 2022; Huh et al., 2010). The visualization of immune cell migration through live imaging in such settings is suboptimal since imaging of an entire z-stack at multiple time-points is required, which is time-consuming and complicates analysis. A more convenient solution is to use an inflammation-on-chip model with two parallel channels separated by a hydrogel barrier in a horizontal orientation enabling convenient visualization of the immune cells migrating through the hydrogel barrier in a single plane (de Haan et al., 2021; Gjorevski et al., 2020; McMinn et al., 2019; Riddle et al., 2022; Wu et al., 2015). In addition, hydrogel barriers used in these models better reflect the ECM than PDMS membranes used in most lung-on-chip systems (Huh et al., 2010; Stucki et al., 2015).
Visualizing immune cell extravasation from a blood vessel during an infection can be done using an inflammation-on-chip model. This involves forming a microvessel alongside (de Haan et al., 2021; Han et al., 2012; Menon et al., 2017; Riddle et al., 2022; Wu et al., 2015) or inside (Hind et al., 2021; McMinn et al., 2019) a hydrogel layer and then inducing an inflammation in a separate compartment. This generates a chemotactic gradient across the hydrogel that attracts immune cells. The extent of immune cell extravasation can be quantified either by imaging (de Haan et al., 2021; Riddle et al., 2022) or by counting the number of cells that have crossed into the hydrogel area (McMinn et al., 2019). To investigate immune cell migration towards tumor tissue, live imaging and automated tracking of immune cells has been carried out (Li et al., 2022). However, this automated analysis of immune cell extravasation and migration has not been carried out in the context of inflammation yet.
A recent study using an inflammation-on-chip model revealed that neutrophil migration is influenced by the composition of the hydrogel, with a higher number of neutrophils crossing into a hydrogel made of a collagen-geltrex mixture compared to collagen I alone (Riddle et al., 2022). This underlines the importance of the ECM composition in inflammatory processes. The combination of multiple cell types (neutrophils-intestinal cells-tissue resident immune cells) was also found to be critical to mimic the complex inflammatory signaling in a gut inflammation-on-chip model (Gjorevski et al., 2020). Another aspect affecting immune cell extravasation during inflammation is whether or not microvessels are perfused. Many studies have used static conditions, based only on the diffusion of the chemoattractant, to create a chemotactic gradient (Han et al., 2012; Hind et al., 2021; McMinn et al., 2019; Wu et al., 2015). More recently, some studies (de Haan et al., 2021; Gjorevski et al., 2020; Riddle et al., 2022) have used rocking platforms that generate bidirectional flow with a hydrostatic pressure difference in the microvessels. Although these systems are popular due to their ease of use – they do not need any pumps nor tubings – the bidirectional flow created is non-physiological. Thacher et al. reported that bidirectional shear stress causes proinflammatory effects on endothelial cells (Thacher et al., 2007). Recent research by Shuler and colleagues (Wang and Shuler, 2018) also revealed that the response of endothelial cells to bidirectional flow differs from that of unidirectional flow, leading to a misalignment of cells with the flow and disruption of VE-cadherin networks. However, to date, no one has demonstrated the effects of bidirectional flow on immune cell extravasation.
This study presents an inflammation-on-chip model composed of endothelial cells, a hydrogel barrier, and lung epithelial cells. Extravasation and migration of immune cells (peripheral blood mononuclear cells (PBMC)) into the hydrogel are investigated for the first time as a function of the following parameters: the flow profile (unidirectional and bidirectional), the density and stiffness of the hydrogel, and the presence or absence of lung epithelial cells. The analysis is based on real-time images, and the barrier functions carried out using permeability assay and immunofluorescence staining. Inflammation is induced with the bacterial chemotactic agent N-Formylmethionyl-leucyl-phenylalanine (fMLP) perfused in the epithelial channel, creating a concentration gradient across the hydrogel barrier. Lung inflammation on chip based on human lung endothelial and epithelial cells is also presented. Finally, a new image analysis tool is presented to rapidly quantify the number and position of migrating immune cells over time. This work highlights the importance of the choice of the perfusion mode, of the hydrogel matrix properties, and of the cell type.
2. Methods
2.1. Design and fabrication of the microfluidic chip
A mold of the chip was designed using Solidworks (Dassault Systems, France). Chips were formed from PDMS 184 (Dow Corning, USA) bonded to standard glass coverslips (VWR, USA). Briefly, a prepolymer was mixed 10:1 with a curing agent, degassed, poured into a mold, and cured at 60 °C overnight. Cured PDMS was lifted from the mold, and reservoirs were punched using biopsy punchers. The PDMS chips were oxygen plasma bonded (Henniker plasma, UK) to glass coverslips. After plasma bonding, chips were stored at 60 °C overnight to increase bonding strength, and, prior to use, chips were sterilized in an ozone/UV chamber (CoolClave, Genlantis, USA) for 15 min.
2.2. Cell culture
Immune cells (PBMCs) were isolated from buffy coat using a ficoll separation gradient, per manufacturer instructions (Lymphoprep, Stemcell Technologies, Canada). After isolation, PBMCs were used fresh or frozen until use. PBMCs were cultured in RPMI media with glutamax (Gibco, USA) supplemented with 10% FBS (Sigma, USA) and 1% Pen/Strep (Sigma, USA). The usage of donor-derived PBMCs was approved by the local blood donation center (SRK Bern).
Human umbilical vein endothelial cells (HUVECs, Gibco, USA) were cultured in endothelial growth media (EGM-2, Lonza, Switzerland) supplemented according to the manufacturer's instructions (EGM-2 BulletKit, Lonza, Switzerland). For passaging, HUVECs were incubated in TrypLE Express (Gibco, USA) for 4 min at 37 °C and centrifuged at 200xg for 5 min. HUVECs were used between passages four and seven.
Human pulmonary microvascular endothelial cells (hPMECs, Promocell, Germany) were cultured in EGM-2MV (Lonza, Switzerland) supplemented according to the manufacturer's instructions (EGM-2MV BulletKit, Lonza, Switzerland). For passaging, hPMECs were incubated in TrypLE Express (Gibco, USA) for 7 min at 37 °C and centrifuged at 200xg for 5 min. hPMECs were used between passages four and seven.
For the culture of 16HBE14o cells (gift from Dr. Gruenert, University of California, San Francisco), flasks were coated with coating solution (0.01% BSA (bovine serum albumin, Sigma, USA) in MEM media supplemented with 30 µg/ml bovine collagen (Advanced Biomatrix, USA) and 10 µg/ml bovine fibronectin (Corning, USA)) for 3 h at 37° The cells were cultured in MEM (Gibco, USA) supplemented with 10% FBS (Sigma, USA), 1% L-Glutamine (Gibco, USA), and 1% Pen/Strep (Gibco, USA). For passaging, the cells were incubated in TrypLE Express (Gibco, USA) for 6 min at 37 °C and centrifuged at 200xg for 3.5 min.
All cells were cultured at 37 °C with 5% CO2. As all cells, apart from the PBMCs, are obtained from commercially available sources, no ethical approval was needed.
2.3. Microfluidic culture
Fibrin hydrogels were formed by mixing a solution of 2 U thrombin (Sigma, USA) in EBM-2 (Lonza, Switzerland) with 10 mg/ml, 20 mg/ml or 30 mg/ml fibrinogen (Sigma, USA) at a 1:1 ratio to obtain 5 mg/ml (HydLow), 10 mg/ml (HydMid) and 15 mg/ml (HydHigh) fibrin hydrogels with 1 U thrombin, respectively. Mixing was carried out on ice, and gel solution was added to the chip immediately after mixing. The gels were incubated at 37 °C for 15 min to completely polymerize. After gelation, the cell channels were coated with polydopamine (PDA, Sigma, USA) for 24 h, washed with sterile ddH2O three times, and coated for two hours with a solution containing 50 μg/ml bovine collagen (Advanced Biomatrix, USA) and 100 µg/ml bovine fibronectin (Corning, USA). Chips were washed three times with ddH2O before cell seeding.
1 µl of HUVECs in suspension was seeded on the chip at a concentration of 4 × 107 cells/ml in EGM-2, resulting in a seeding concentration of 4 × 104 cells per chip. The chip was placed in the incubator at a 45° angle for 15 min to allow for endothelial cell attachment at the hydrogel barrier. The chip was then placed on a rocking platform (Mimetas, Netherlands) at a 7° angle with a rocking interval of 8 min for 2.5 h until flushing non-adhered cells. After flushing, endothelial cells in EGM-2 supplemented with 37.5 ng/ml VEGF (Miltenyi Biotech, Germany) were grown to confluency on chip on the rocking platform until experimentation. The bidirectional flow induced by the rocking platform provides the endothelial cells with fresh nutrients from the media reservoirs (Baeyens et al., 2016). Media was exchanged every 24 h.
For coculture experiments, hPMECs and 16HBE epithelial cells were seeded at the same timepoint. First, 1 µl of hPMECs in suspension was seeded at a concentration of 2 × 107 cells/ml, resulting in a seeding concentration of 2 × 104 cells per chip. The seeding procedure was identical to the HUVEC seeding procedure described above. After endothelial cell attachment, 16HBE epithelial cells were seeded in the opposite channel, with 2 × 104 cells per chip. The chip was placed in the incubator for 30 min to allow for epithelial cell attachment and then cell culture media was added to prevent drying out of the chip. After 2.5 h, the channels were flushed and the chip was placed on a rocking platform (Mimetas, Netherlands) at a 7° angle with a rocking interval of 8 min to grow the cells to confluency. Media was exchanged every 24 h.
2.4. Scanning electron microscopy
For scanning electron microscopy (SEM), fibrin hydrogels were produced on gold-coated coverslips. Sample preparation was carried out as previously published (Strässle et al., 2021). Briefly, the hydrogels were hydrated with PBS and subsequently fixed with 2.5% glutaraldehyde (Merck, USA) in 0.1 M cacodylate buffer. After postfixation with 1% OsO4 in 0.1 M cacodylate buffer and dehydration through an ascending ethanol series, samples were dried by evaporation of Hexamethyldisilazane and mounted onto aluminum stubs. Then, a platinum coating by electron beam evaporation was carried out, and samples were analyzed on a Zeiss DSM982 Gemini SEM.
2.5. Rheology
To measure the mechanical properties of the bulk fibrin gel, we performed rheological measurements using a MCR 301 rheometer (Anton Paar, Austria) with a parallel plate geometry of 25 mm diameter (PP25, SN21182) and a gap size of 200 μm. Hydrogels with different concentrations of fibrin (HydLow, HydMid and HydHigh, each of 150 μL volume) were prepared in situ on the rheometer stage, (following the protocol mentioned in Section 2.3) which was maintained at a constant temperature of 37 °C and the excess gel was trimmed before the start of the experiment. To prevent dehydration of the gel during the process of gelation, 200 μL silicone oil with viscosity 9.3 mPa.s (Brookfield viscosity standards, USA) was added to the exposed part of the gel. Oscillatory time-sweep measurements were performed at a strain of 0.5% and a frequency of 0.5 Hz (since the storage and the loss moduli of fibrin are independent of frequency below 1 Hz (Xia et al., 2021)), and the storage (G’) and loss modulus (G’’) were measured up to 15 min after gelation, when a plateau was reached. The G’ and the G’’ are related via Eq. (1), where G* is the complex modulus.
| (1) |
To calculate a Young's modulus, Eq. (2) is used:
| (2) |
To estimate a Young's modulus (Y) from the storage modulus, the following approximations can be used according to literature: 1) At low amplitudes (0.5%) and low frequency (0.5 Hz), the storage modulus (G’) equals the linear shear modulus (G) (Xia et al., 2021) and 2) the Poisson's ratio (ν) for fibrin can be considered to be 0.5 (Becerra et al., 2021). Based on this, Eq. (2) can be simplified to Y = 3*G.
2.6. Flow cytometry
PBMCs were isolated from buffy coat as described above. Freshly isolated PBMCs, frozen and thawed PBMCs, dyed PBMCs, and PBMCs exposed to 100 ng/ml LPS (Sigma, USA) were analyzed using flow cytometry. PBMCs were stained at 4 °C with the following antibodies: anti-CD56 (5.1H11, Biolegend, USA), anti-CD56 (HCD56, Biolegend, USA), anti-CD3 (OKT3, Biolegend, USA), anti-CD16 (3G8, BD Bioscience, USA), anti-CD4 (OKT4, Biolegend, USA), anti-CD8a (SK1, Biolegend, USA), anti-CD14 (M5E2, Biolegend, USA), Live Dead Fixable Blue (Thermofisher, USA). Samples were acquired using a LSRII SORP device (BD Bioscience, USA) and analyzed with Flowjo (version 10.8.1, BD, USA).
2.7. Permeability
To perform permeability testing, chips were taken from the incubator at the desired timepoint, and the media was removed from the media channels. 70 kDa RITC-dextran solution (Sigma, USA, 1 mg/ml in PBS) was added to the endothelial/immune cell channel and allowed to diffuse through the hydrogel. Pictures were taken every 10 s for 5 min on an EVOS M7000 microscope (Thermo Fisher, USA) to obtain a timelapse video of dye diffusion. The apparent permeability coefficient (Papp) was calculated according to Eq. (3) (Ho et al., 2017).
| (3) |
CECM: concentration (measured through dye intensity) of the dye in the ECM
VECM: volume of the ECM compartment
AEC: surface of the endothelial monolayer interfacing with the ECM
CVessel: concentration (measured through dye intensity) of the dye in the vessel (microchannel)
2.8. Inflammation assays
Immediately before the inflammation assays, PBMCs were thawed and stained with PkH67 fluorescent green marker (Sigma, USA) according to the manufacturer's protocol. Two types of inflammation assay were carried out: 1) under unidirectional, constant flow using a peristaltic pump and 2) under bidirectional, non-constant flow using a rocking platform. Pump-controlled flow is unidirectional and a stable, constant flow rate can be maintained, mimicking the unidirectional circulation in the human body. Placement of microfluidic chips on a rocking platform leads to bidirectional, non-constant flow, a flow profile not observed in the human body. In both assays, endothelial and epithelial cells were grown to confluency by culturing the cells on chip on a rocking platform for 48 h until the start of the inflammation assays. During the inflammation assays, stained PBMCs and the fMLP chemoattractant were perfused through the chips, either with a pump or on a rocking platform.
For unidirectional flow experiments with immune cells only, gel-filled chips were connected to a peristaltic pump (Ismatec, Switzerland) and placed under a microscope with an incubation chamber (Nikon Eclipse TI-E, Nikon, Japan) for live imaging. Stained PBMCs (200,000 cells per chip) were perfused through the endothelial/immune cell channel (2 µl/min flow rate) with the peristaltic pump placed after the chip and no recirculation of immune cells, to prevent immune cells stressed by the pressure inside the peristaltic pump from entering the chip. 100 ng/ml fMLP (Sigma, USA) or PBS was perfused through the chemoattractant channel at the same flow rate. The chips were imaged every 5 min for 90 min (PBMC only).
For unidirectional flow experiments in coculture, either hPMECs and 16HBEs (lung model) or HUVECs (endothelial vessel model) were cultured on chip for 48 h until the start of the experiment. The chips were attached to a peristaltic pump (Ismatec, Switzerland) and placed under a microscope with an incubation chamber (Nikon Eclipse TI-E, Nikon, Japan) for live imaging. Stained PBMCs (200,000 cells per chip) were perfused through the endothelial/immune cell channel (2 µl/min flow rate) with the peristaltic pump placed after the chip and no recirculation of immune cells, to prevent immune cells stressed by the pressure inside the peristaltic pump from entering the chip. 100 ng/ml fMLP (Sigma, USA) or PBS was perfused through the epithelial/chemoattractant channel at the same flow rate. The chips were imaged every 5 min for 4 h to obtain timelapse videos.
For bidirectional flow experiments, stained PBMCs (200.000 cells per chip) were seeded into the endothelial/immune cell channel of gel-filled chips with HUVECs. 100 ng/ml fMLP or PBS was added to the chemoattractant channel. The chips were placed on a rocking platform at a 4° angle with rocking intervals of 8 min, to obtain a median flow rate of 2 µl/min (calculations in supplementary Table 1). Chips were imaged after 4, 24 and 48 h of incubation.
2.9. Immunohistochemistry
All steps were carried out at room temperature, except when indicated otherwise. Chips were washed 3 times with PBS, fixed for 12 min with 4% PFA (Invitrogen, USA), and washed again 3 times with PBS. For cell permeabilization, samples were incubated in 0.1% Triton X-100 (Sigma-Aldrich, USA) for 10 min. After washing 3 times with PBS, blocking was done for 1 h with a 2% BSA in PBS solution. Then, the primary antibody (anti PECAM-1, Cell Signaling, USA) diluted 1:100 in blocking solution was added and incubated at 4 °C overnight. After washing 3 times with PBS, a secondary antibody (1:500, anti-mouse 488 or anti-mouse 546, Invitrogen, USA), Hoechst (1:1000, Sigma, USA), and phalloidin 670 (1:150, Acti-Stain, Cytoskeleton, USA) in blocking buffer were added and incubated for 2 h. The chips were washed 4 times with PBS and imaged on a confocal microscope (Zeiss LSM710 and LSM980, Zeiss, Germany).
2.10. Automated analysis of immune cell migration
Migration of immune cells inside the hydrogel was analyzed with an automated analysis pipeline. The migration distance was measured as the vertical distance between the immune cell and the endothelial barrier. The endothelial barrier was detected as a bright line in the phase contrast image and modeled as a line. Immune cells were detected in the phase-contrast image using multi-template matching (in order to detect in- and out-of-focus cells) and false-positives filtered out based on the fluorescence intensity measured inside the cells. Using the locations of the endothelial barrier and of the immune cells, the migrated distance was measured for each time point and summarized in histograms. The Python analysis pipeline can be found at https://github.com/guiwitz/mfdiffusion and uses numerous open-source packages including jupyter (Kluyver et al., 2016), numpy (Harris et al., 2020), pandas (Pandas Development Team, 2020), scikit-image (Van Der Walt et al., 2014), scikit-learn (Pedregosa et al., 2012), matplotlib (Hunter, 2007) and seaborn (Waskom et al., 2020).
2.11. Statistics
Statistical analysis was performed using Graphpad Prism 9 software (Graphpad, USA). Sample size, test type, and data representation is indicated in each graph.
3. Results
3.1. Chip design
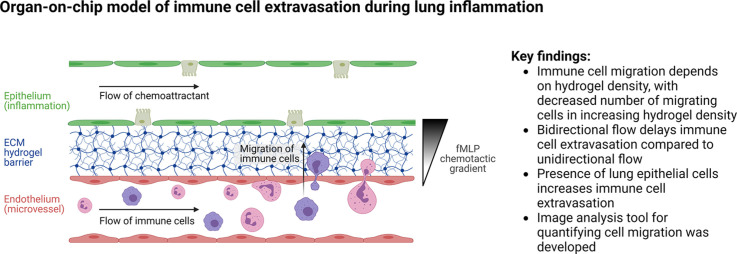
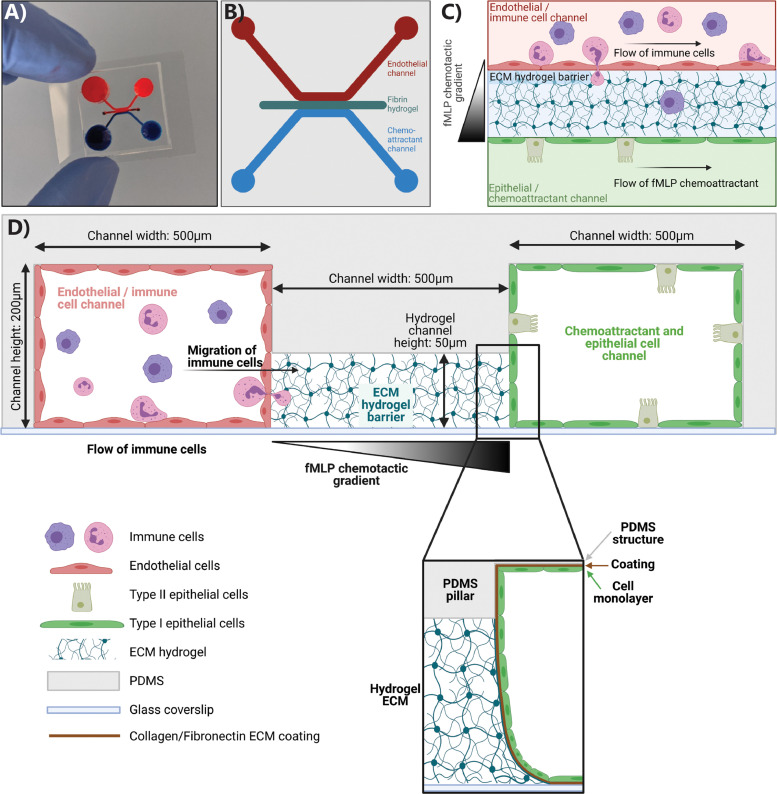
The inflammation-on-chip model is a three-channel PDMS-based microfluidic model (Fig. 1 ). Fig. 1A shows a picture of a chip filled with dye; Fig. 1B-D show a schematic overview of the chip from various angles. The hydrogel is contained in the 50 µm high central channel by capillary barriers formed on either sides of the channel at the interface with the higher adjacent channels (200 µm high, Fig. 1D). This abrupt geometrical change leads to a sharp increase of the contact angle, which stops the hydrogel flow at the channel edge during filling and thus contains the hydrogel in the central channel. The hydrogel channel is flanked by two equally sized, perfusable side channels, allowing for bidirectional fluid flow when placed on a rocking platform or unidirectional flow using a peristaltic pump. The blood vessel space is modeled in the endothelial/immune cell channel, with an endothelial lining and immune cell perfusion (Fig. 1D). The opposite channel represents the inflamed area, with perfusion of a chemoattractant and culture of lung epithelial cells (named chemoattractant channel from here). A stable chemotactic gradient across the hydrogel is established by constant pump-based perfusion of immune cells and chemoattractant at an equal flow rate.
Fig. 1.
Inflammation-on-chip design. The inflammation-on-chip model consists of three PDMS-molded channels. (A) Picture of a chip with dye inside the channels. (B) Schematic design of the whole chip, top view. (C) Schematic of the three fluidic channels from a top view. (D) Schematic of the chip from a side view. The 50 µm central hydrogel channel is flanked by two side channels: the endothelial/immune cell channel and the epithelial/chemoattractant channel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Hydrogel barrier formation
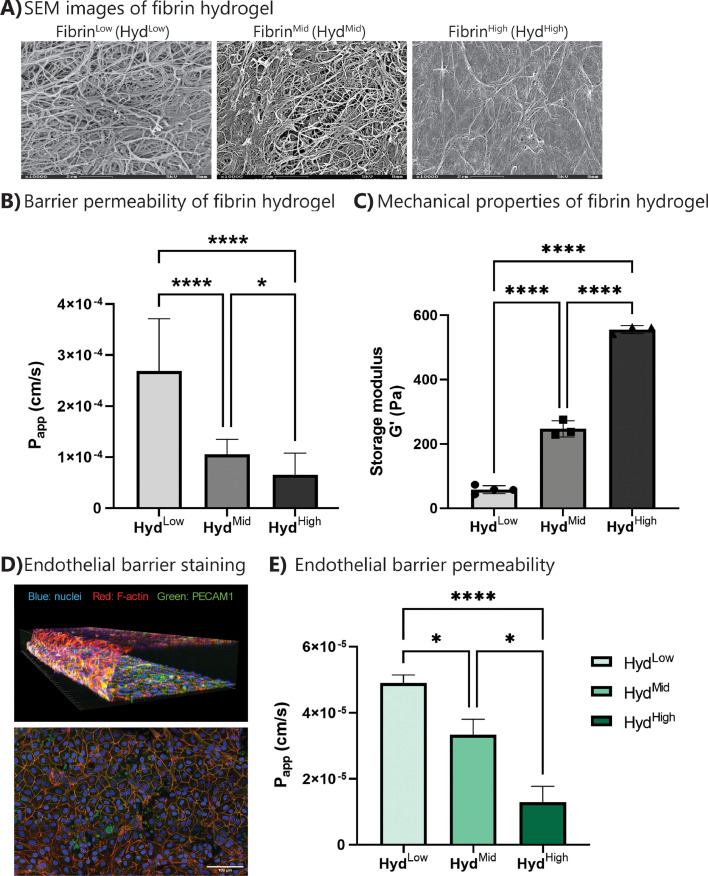
Three different fibrin hydrogels (HydLow, HydMid and HydHigh) were used. SEM imaging and a permeability assay with 70 kDa RITC-dextran showed a significant difference in hydrogel morphology and apparent permeability between these three hydrogels (Fig. 2 A and 2B). In addition, rheological testing revealed an increase in storage and loss modulus with increasing fibrin protein content inside the hydrogel (Fig. 2C and supplementary figure 1). Immune cells were seeded on HydLow and HydHigh hydrogels and exposed to standard culture media (RPMI) or culture media supplemented with 100 ng/ml of the chemotactic peptide N-formyl-methionyl-leucyl-phenylalanine (fMLP). SEM imaging showed that fMLP exposure, but not hydrogel type, altered immune cell morphology (supplementary figure 2). Overall, fibrin hydrogels could be added to the chip allowing the study of immune cell migration through different hydrogels.
Fig. 2.
Barrier formation in inflammation-on-chip. Hydrogels (HydLow, HydMid and HydHigh) were processed for SEM and permeability measurements. HUVECs were cultured on chip in the endothelial/immune cell channel and processed for immunohistochemistry and permeability measurements. (A) SEM images of hydrogels display different morphologies. Scale bar: 2 µm. N = 2 gels per condition (B) A RITC-dextran-based apparent permeability assay shows decreased Papp in HydHigh compared to HydMid and HydLow hydrogels. N = 15 chips per condition. Graphs show mean with SD. (C) Rheological testing shows an increase in storage modulus with increasing fibrin protein content in the hydrogel. N = 3–4 hydrogels per condition. Graph shows mean with SD and individual data points. (D) Immunohistochemistry shows a confluent endothelial layer (right) in a 3D structure alongside the hydrogel and lining the channel (left). Green: PECAM-1, red: actin, blue: nuclei. Scale bar: 100 µm. (E) With HUVECs present, Papp depends on hydrogel type. N = 8–12 chips per condition, at least 3 independent experiments. Graph shows mean with SEM. Statistics: ANOVA with Tukey's multiple comparison test. *0.05 < p < 0.01. **** p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Endothelial barrier formation
HUVECs were seeded in the endothelial/immune cell channel and cultured on a rocking platform to form a continuous, tight endothelial barrier alongside the hydrogel. Staining with PECAM-1 and actin displayed a continuous barrier alongside the hydrogel, which was visualized in 3D (Fig. 2C and supplementary video 1). Furthermore, adding HUVECs into the channel decreased barrier permeability (supplementary figure 3) in all hydrogel conditions to values in the range of 10−5 cm/s, but also with HUVECs, barrier permeability was hydrogel-dependent, with HUVECs alongside HydHigh hydrogels showing the lowest permeability. In addition, HUVECs barrier permeability was stable over the entire culture period (supplementary figure 3C).
3.4. Immune cell characterization
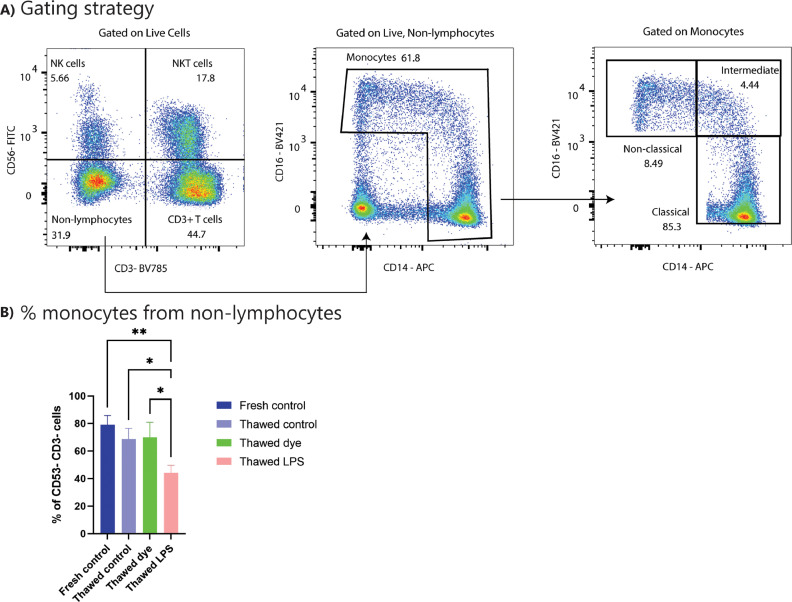
Since this study focused on the migration of peripheral blood immune cells, peripheral blood mononuclear cells (PBMCs) were used as an immune cell population. Gating strategies to distinguish major immune lineages (monocyte subsets and lymphocytes) are illustrated in Fig. 3 A. There was no observable shift in major PBMC populations between fresh and frozen PBMCs (Fig. 3B, supplementary figure 4). In addition, staining of PBMCs using the PkH67 labeling dye did not change PBMC composition (Fig. 3B, supplementary figure 4). However, activating PBMCs with LPS led to a reduction in monocyte frequencies (Fig. 3B). The concentration of fMLP chemoattractant used in the ensuing migration assays did not alter the composition of the different PBMC populations (supplementary figure 4). In general, the percentage of classical monocytes was elevated compared to literature (Lee et al., 2017), with 85% classical monocytes reported in literature compared to 95% classical monocytes in our patient samples. However, the fraction of classical monocytes was similar between patients, indicating a consistent shift of populations compared to literature that does not affect the migration assay. Taken together, these data show that freeze/thaw and/or PkH67 labeling does not affect PBMC composition, thereby pinpointing the suitability of the experimental setting used in our study to mimic in vivo physiological conditions.
Fig. 3.
Characterization of PBMC subpopulations. PBMCs were isolated from buffy coat and either used fresh or frozen for later use. Different treatments of PBMCs were compared. All cells were stained and analyzed by flow cytometry. (A) Gating strategy used in the flow cytometry analysis to distinguish the indicated immune populations among PBMCs. (B) Fraction (%) of monocytes present in the non-lymphocyte population following different treatments. N = 3 individual donors. Graphs show mean with SD. Statistics: ANOVA with Tukey's multiple comparison test. *0.05 < p < 0.01. **0.01 < p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Endothelial barrier is necessary for immune cell extravasation
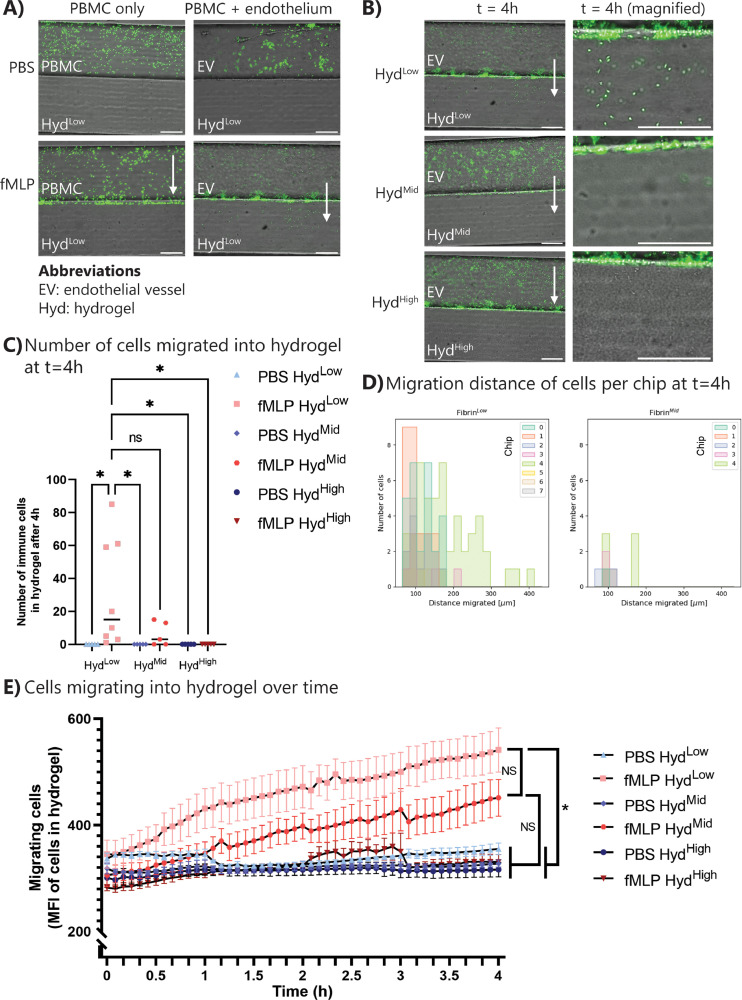
Next we tested how immune cells react to exposure to a chemotactic gradient without the presence of other cell types or with an endothelial barrier present. To this end, immune cells were first perfused through the uncoated endothelial/immune cell channel, with simultaneous perfusion of PBS or the chemoattractant fMLP through the chemoattractant channel. In a static system, the chemoattractant will diffuse through the chip over time, and the gradient is lost. Thus, to create a stable chemoattractant gradient, fMLP was perfused through the system simultaneously with the immune cells using a peristaltic pump. Live imaging for 90 min showed non-specific attachment of immune cells to the chip when exposed to PBS (Fig. 4 A (top left) and supplementary videos 2 and 4). However, when exposed to fMLP, immune cells migrated towards, but not into, the hydrogel barrier (Fig. 4A, supplementary figure 5A and supplementary videos 3 and 5). The attachment of dyed immune cells to the hydrogel was assessed by measuring the mean fluorescence intensity at the hydrogel barrier (supplementary figure 5B/C). Analysis revealed an increase in attachment over time (supplementary figure 5D), with both HydLow and HydHigh hydrogels. In contrast, when an endothelial barrier was present, immune cells extravasated from the endothelial vessel into the hydrogel space (Fig. 4A and supplementary figure 6). Thus, without an endothelial vessel, immune cells were able to sense the chemotactic gradient and migrated towards the gradient, but not into the hydrogel, but presence of an endothelium was necessary to induce transmigration into the hydrogel environment.
Fig. 4.
Immune cells extravasate during inflammation in a hydrogel-dependent manner. PBMCs were thawed and dyed with green fluorescent dye. PBMCs were perfused through the endothelial/immune cell channel, while PBS or fMLP was perfused through the opposite channel. Migration was followed for up to 4 h. (A) Representative images of the inflammation assay without (left) and with (right) endothelial cells, without (top) and with (bottom) chemoattractant fMLP. Scale bar: 200 µm. White arrow: direction of migration. (B) Representative images of PBMC attachment and transmigration across the endothelial barrier at t = 4 h. Scale bar: 200 µm. White arrow: direction of migration. (C) Quantification of the number of immune cells extravasated into the hydrogel at t = 4 h. Graph shows individual data points with median. (D) Migration distance of immune cells inside the hydrogel at t = 4 h. Data represented per chip sample. (E) Quantification of mean fluorescent intensity in the hydrogel over time shows extravasation only with HydLow and HydMid with fMLP. N = 4–8 chips per condition, at least three independent experiments. Graph shows mean with SEM. Statistics: ANOVA with Tukey's multiple comparison test. *0.05 < p < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Immune cells transmigrate into the hydrogel dependent on hydrogel type
For the inflammation assay, immune cells were perfused for 4 h through the endothelial vessel, either in healthy (PBS) or inflamed (fMLP) conditions. When exposed to PBS, a fraction of the immune cells interacted with the endothelial cells through temporary attachment but did not show any specific (trans)migration (supplementary figure 6A and supplementary videos 6, 8 and 10). On the other hand, chemoattractant exposure induced immune cell migration towards the chemoattractant channel (Fig. 4B and supplementary video 7 and 9). Transmigration of immune cells into the hydrogel depended on hydrogel type, with immune cells transmigrating into HydLow and HydMid but not into HydHigh (Fig. 4B and supplementary videos 7, 9 and 11). Quantifying the number of immune cells inside the hydrogel at t = 4 h showed variability in the number of immune cells transmigrating into the fMLP-exposed chips with HydLow and HydMid (Fig. 4C). This is likely due to the inherent variation of PBMCs as a population leading to a difference in the amount of migrating cells added to a chip. Although not significant due to the variability in number of extravasated immune cells, a trend can be observed with a lower number of immune cells extravasating into the HydMid condition compared to HydLow, corresponding with the lower permeability of HydMid hydrogels. The mean fluorescence intensity of the PBMCs inside the hydrogel was measured using timelapse imaging to assess immune cell extravasation (Fig. 4E). An increase in fluorescence intensity, indicating localization of immune cells in the hydrogel, over time was observed in the HydLow and HydMid with fMLP conditions (Fig. 4E). Through automated detection of immune cells, the distance of immune cells from the endothelial barrier after 4 h of fMLP exposure was assessed. Fig. 4D shows the migration distance of immune cells per chip sample. It is visible that there is variability in the number of immune cells extravasated per chip and their migration distance, but a general trend of more and further migration in HydLow compared to HydMid ECM can be observed. Automated detection of migration distance was also carried out over time (supplementary figure 6B). Here, all immune cells and their migration distance at each imaged timepoint are depicted, but from one chip sample only. This shows that very detailed migration data can be extracted from this migration assay. In summary, hydrogel-dependent transmigration experiments indicated that immune cell transmigration is dependent not solely on chemoattractant concentration and immune cell-endothelial cell interactions but also on the ECM environment behind the endothelial barrier.
To investigate the effect of endothelial barrier tightness on immune cell migration, endothelial cells cultured for 24 and ???48 h were exposed to PBMCs and fMLP, and migration was quantified. After 24 h of culture, the endothelial barrier was not yet confluent, which it was after 48 h of culture. Thus, more immune cells extravasated after 24 h than 48 h in the fMLP HydLow condition (supplementary figure 7A). However, no immune cells extravasated into the HydHigh hydrogel (supplementary figure 7A). These results were verified by measuring the mean fluorescence intensity inside the hydrogel over time and automated analysis of immune cell migration distance (supplementary figure 7B-D). Overall, these results showed that even though the endothelial barrier was not confluent at the earlier timepoint, the hydrogel-dependent extravasation of immune cells still persisted.
3.7. The endothelial barrier is affected by immune cell media
To minimize immune cell activation, PBMCs were perfused in RPMI media, the standard culture media for PBMCs.(Hønge et al., 2017) The RPMI media is different from the EGM-2 media HUVECs are normally cultured in. Thus, HUVECs were grown to confluency in EGM-2, and RPMI media with or without PBMCs was added to the endothelial/immune cell channel to assess the effect of RPMI on HUVEC barrier formation. PBS or fMLP was then added to the chemoattractant channel, and chips were incubated on a rocking platform for 4 h. PBMC-free RPMI media significantly changed HUVEC morphology (supplementary figure 8A) compared to the endothelial growth media EGM-2. Furthermore, RPMI decreased the levels of actin and PECAM-1, a cell barrier protein in HUVECs (supplementary figure 8B/C). However, when HUVECs were treated with PBMCs in RPMI media, there was no apparent effect on actin and PECAM-1 expression, most likely due to staining of the PBMCs masking the endothelial staining (supplementary figure 8D/E). The RPMI-induced decrease in HUVEC barrier protein expression caused an increase in apparent permeability (supplementary figure 8F), which was further enhanced when combining RPMI with PBMCs and fMLP (supplementary figure 8 G). Thus, endothelial cells form a tight barrier on the chip which is altered by exposure to RPMI media. The decreased barrier tightness can be further enhanced by adding PBMC perfusion and fMLP exposure.
3.8. Immune cells sense flow profile and react differently on chip
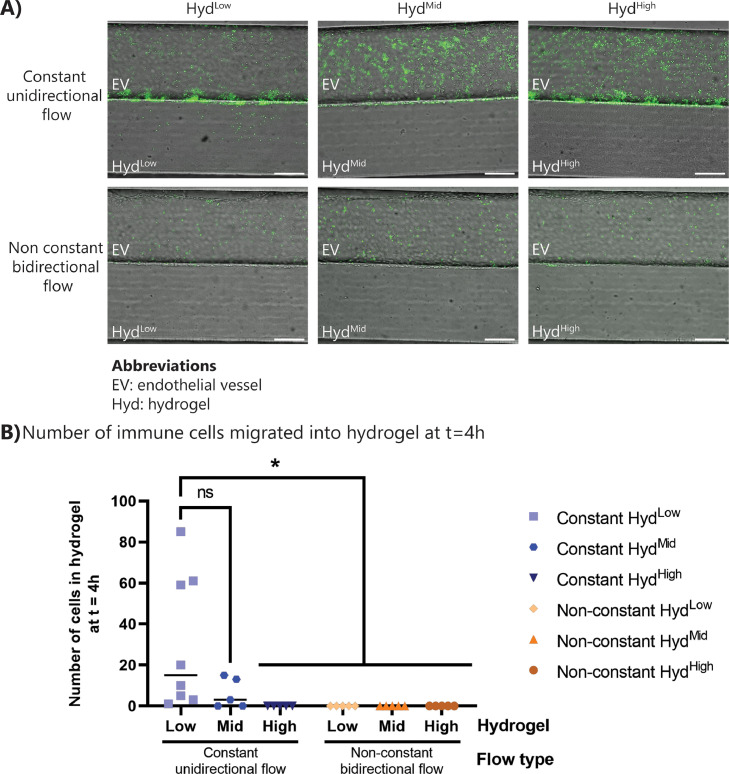
A main advantage of the inflammation-on-chip design is the possibility to flexibly adapt the flow profile inside the microfluidic channels. To perform the inflammation assays depicted in Fig. 4, unidirectional, pump-based flow was applied. For establishing the endothelial barrier, the cells were cultured under bidirectional, non-constant flow on a rocking platform. Though unidirectional flow is more similar to the in vivo situation than a non-constant, bidirectional flow, the ease of use of placing a collection of chips on a rocking platform has led to widespread usage of bidirectional flow as a method to apply shear stress in microphysiological systems, and to investigate immune cell migration. Thus, the inflammation assay was carried out under unidirectional and bidirectional flow. The number of PBMCs, exposure of fMLP and median flow rate was kept the same between the two conditions, to assess the effect of flow type only. After 4 h of exposure to the fMLP chemoattractant under bidirectional flow, immune cell attachment to the endothelium was observed, but no immune cells had transmigrated into the hydrogel (Fig. 5 ). In contrast, after 4 h of exposure to the fMLP chemoattractant under unidirectional flow, the acute inflammation led to acute extravasation and migration of immune cells (Figs. 4 and 5). An inflammatory stimulus was still detected by the immune cells under bidirectional, non-constant flow, as shown by the immune cell migration observed after 24 and 48 h in the fMLP-stimulated inflamed condition (supplementary figure 9). The flow type did not have a significant effect on endothelial barrier proteins (supplementary figure 10), indicating that the delayed onset of extravasation is an effect of the bidirectional non-constant flow profile on the immune cells. This result highlights the importance of physiological flow conditions for investigating immune cell migration during inflammation, specifically with regards to flow direction and constancy of flow profiles.
Fig. 5.
Bidirectional flow showed reduced migration of immune cells. The inflammation assay was carried out under unidirectional constant flow with a pump and under bidirectional non-constant flow on a rocking platform for 4 h. (A) Representative images of fMLP-exposed chips under unidirectional and bidirectional flow after 4 h. (B) Quantification of the number of transmigrated immune cells in the inflammation assay under bidirectional and unidirectional flow. N = 4–8 chips per condition, at least three independent experiments. Statistics: ANOVA with Tukey's multiple comparison test. *0.05 < p < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.9. Immune cells transmigrate during inflammation in a lung-on-chip
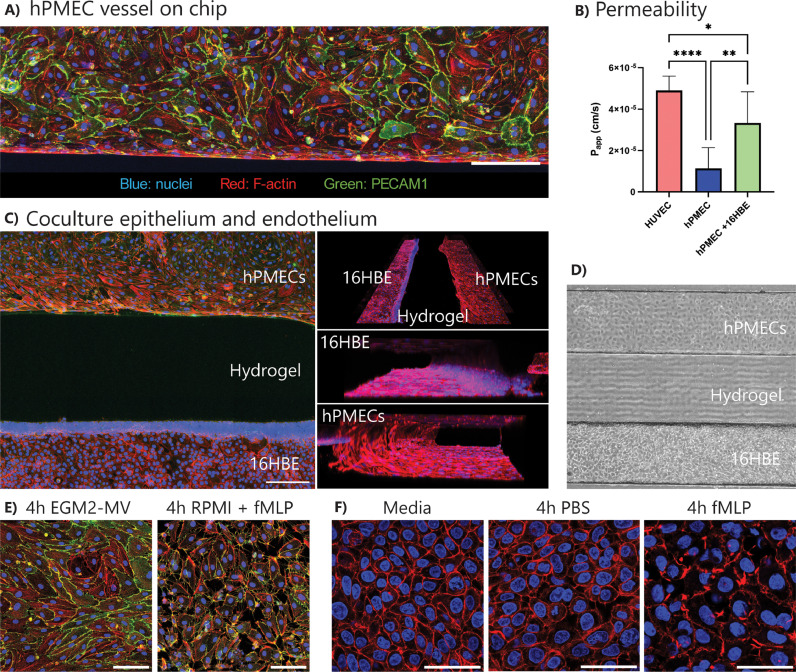
We have shown that the inflammation-on-chip can be used to model immune cell transmigration from a blood vessel toward an ECM environment. The inflammation-on-chip was further developed to investigate lung inflammation-on-chip. hPMECs were seeded in the endothelial/immune cell channel and co-cultured with 16HBE lung epithelial cells in the chemoattractant channel, separated by a HydLow hydrogel. Staining and permeability assay showed a tight endothelial barrier in the endothelial/immune cell channel (Fig. 6 A/B and supplementary video 12). Apparent permeability of the endothelial vessel was lower than HUVEC apparent permeability and did not alter significantly over culture time (Fig. 6B and supplementary figure 11A), but increased when adding 16HBE epithelial cells in the opposite channel (Fig. 6B). As supplementation of 37.5 ng/ml VEGF improved HUVEC growth and barrier formation (based on previous experiments, data not shown), hPMEC media was also supplemented with VEGF. Here, adding 37.5 ng/ml VEGF induced the differentiation of hPMECs towards a mesenchymal-like phenotype (supplementary figure 11B). Thus, for further experiments, hPMECs were cultured without adding VEGF.
Fig. 6.
Lung-on-chip to model inflammation. hPMECs and 16HBEs were seeded on the chip and grown to confluency. (A) Representative image of a pulmonary microvessel-on-chip stained for F-actin and PECAM1. Scale bar: 200 µm. (B) Papp of a pulmonary microvessel compared to gel only and a HUVEC vessel. N = 6–12 chips per condition from at least 4 individual experiments. C + D) Representative images of co-culture of pulmonary endothelial and lung epithelial cells. Immunohistochemistry (C; red = actin, green = PECAM-1, blue = nuclei) and brightfield image (D). Scale bar: 200 µm. (E) Representative images of confluent pulmonary endothelium exposed to normal culture media (left) or RPMI immune cell culture media (right). Red = actin, blue = nuclei. Scale bar: 100 µm. N = 3 chips per condition. (F) Representative images of confluent epithelial cells exposed to normal culture media (left), PBS (center), or fMLP (right). Red = actin, blue = nuclei. Scale bar: 50 µm. N = 3 chips per condition. Graphs show mean with SD. Statistics: ANOVA with Tukey's multiple comparison test. ****p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A confluent epithelial layer was cultured in the chemoattractant channel, which could be visualized in 3D (Fig. 6C/D and supplementary video 12). Where the endothelial cells attached to all four walls of the channel, the epithelial cells mainly attached to the coated glass bottom and the hydrogel, but not to the coated PDMS, as seen using F-actin staining (Fig. 6C). Similar to the HUVEC cells, 4-hour direct RPMI exposure to hPMECs in combination with fMLP in the opposite channel induced a breakdown of barrier proteins (Fig. 6E). Additionally, confluent epithelial barrier actin structures were disrupted when exposed to fMLP for 4-hours but not to PBS (Fig. 6F).
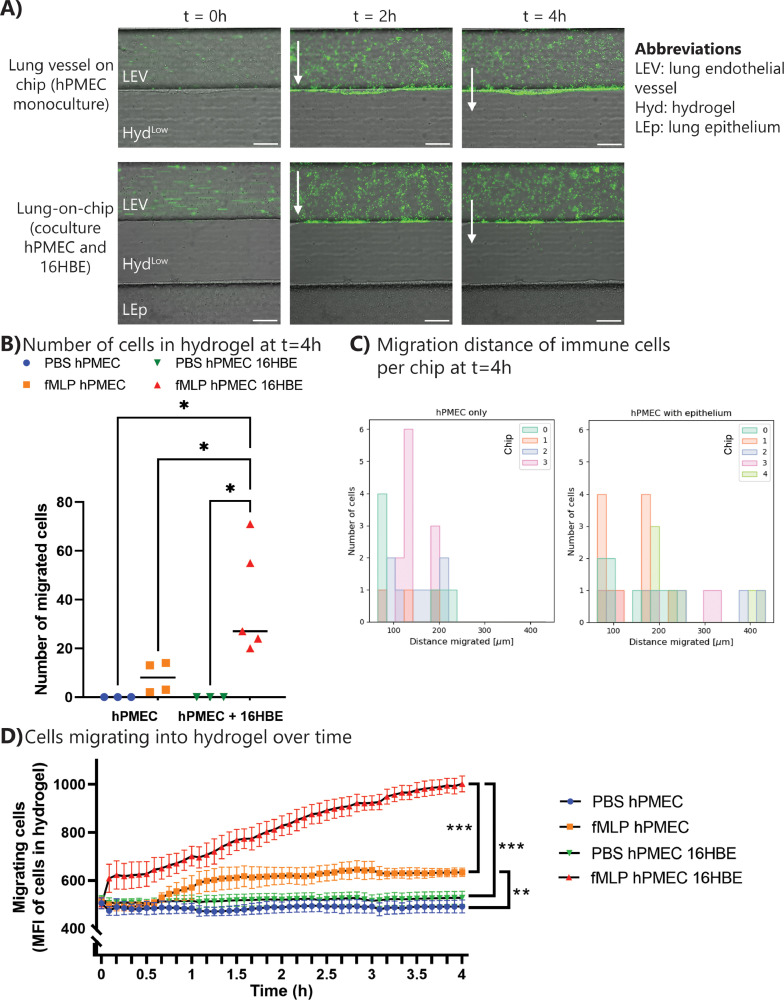
To investigate lung inflammation on chip, immune cells were perfused through the lung endothelial vessel and the epithelium was exposed to PBS/fMLP (supplementary figure 12A). Corresponding with the lower barrier permeability of pulmonary endothelial cells compared to HUVECs, immune cell extravasation upon fMLP exposure was minimal in chips with pulmonary endothelial cells only (Fig. 7 A/B, supplementary video 14). On the other hand, when adding lung epithelial cells to model lung inflammation on chip, a significant increase in immune cell extravasation could be observed (Fig. 7A/B), also over time (Fig. 7D and suppelementary video 16). With an epithelium present, immune cells migrated further than with an endothelium only (Fig. 7C and supplementary figure 12C), and a low number of immune cells migrated through the complete hydrogel to interact with the epithelial barrier (supplementary figure 12B). Overall, the inflammation-on-chip model can be utilized to investigate immune cell migration during acute lung inflammation.
Fig. 7.
Immune cells extravasate during inflammation in the lung-on-chip. PBS or fMLP was perfused through the chemoattractant channel, while PBMCs were perfused through the endothelial/immune cell channel. Migration was observed upon fMLP exposure. (A) Representative images of immune cells (green) extravasating during inflammation in the lung-on-chip model. Scale bar: 200 µm. (B) Number of cells transmigrated into hydrogel at t = 4 h. Graph shows individual data points with median. (C) Migration distance of immune cells inside the hydrogel at t = 4 h. Data represented per chip sample. (D) Transmigration of immune cells inside hydrogel over time quantified with mean fluorescent intensity. Graph shows mean with SEM. N = 3–5 chips per condition. Statistics: ANOVA with Tukey's multiple comparison test. *0.05 < p < 0.01. **0.01 < p < 0.001. ***0.001 < p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
4.1. Unidirectional vs bidirectional flow affects immune cell extravasation
This paper proposes a new inflammation-on-chip model to investigate immune cell extravasation during inflammation in real-time. We showed that immune cell extravasation strongly depends on the flow profile exerted on the endothelial cells. Our research indicates that a bidirectional flow delays extravasation of immune cells, occurring 24 h after the start of the assay rather than 2–3 h under a unidirectional flow. In vivo, immune cells extravasate within minutes to hours after detection of pathogen signals, due to upregulation of E-selection and P-selection on the endothelium (Strell and Entschladen, 2008). This result of delayed immune cell extravasation has important implications for the organs-on-chip community, who, in their efforts to create user-friendly systems, have widely adopted the use of rocking platforms that do not require tubing or pumping systems. This discovery supports the previous findings of Shuler and colleagues, who demonstrated that bidirectional flow in contrast to unidirectional flow can lead to significant changes in the endothelium, including the formation of poorly defined VE-cadherin networks and limited F-actin realignment (Wang and Shuler, 2018). To overcome this, they utilized tilting platforms and innovative microfluidic chips to establish unidirectional flow systems (Wang and Shuler, 2018). Others have used bidirectional flow to investigate the extravasation of T cells, only taking place after 24 h (de Haan et al., 2021).
The cause of slow activation and delayed extravasation of immune cells under non-constant, bidirectional flow, which is non-physiological in nature, is unknown and requires more research. The impact of bidirectional flow on endothelial cells, the relationship between immune and endothelial cells, and the immune cells themselves (as they are known to be affected by shear stress (Mitchell et al., 2014; Mondadori et al., 2020)) could all cause the delayed extravasation. In our experiments, we exposed endothelial and epithelial cells to a shear stress of 0.6 dyne/cm2 by using bidirectional flow during initial culture to refresh the media and provide sufficient nutrients to the cells. The tilting angle and duration were calculated to completely renew the media before changing the tilting direction. During inflammation experiments, PBMCs and fMLP were perfused at a flow rate of 2 µl/min, corresponding to a shear stress of 0.1 dyne/cm2, with a constant unidirectional flow, or with a non-constant bidirectional flow with a median flow rate of 2 µl/min. Most immune cells extravasate in postcapillary venules (Ley et al., 2007) where shear stress can reach 1–6 dyne/cm2, which is higher than the shear stress experienced in the microfluidic chip (Roux et al., 2020). However, the low shear stress in the chip allows immune cells more time to adhere to the endothelium and transmigrate.
4.2. ECM-dependent immune cell migration
Another important finding of this research is that the density and stiffness of the hydrogel barrier impact the immune cell migration. More immune cells migrated into HydLow than into HydMid, but did not migrate into the HydHigh upon fMLP stimulation. This impaired migration could be explained by differences in fiber density or hydrogel stiffness. Previous research has shown that fibrin hydrogel stiffness depends on thrombin and fibrinogen concentration in the hydrogel (Duong et al., 2009; Kim et al., 2020). In this study, fibrin hydrogel mechanical properties were measured using parallel plate rheology, and found that the storage and loss modulus values were consistent with previous research (Ryan et al., 1999). Although different mechanical testing methods can complicate comparisons between studies (Polio et al., 2018), the calculation of Young's Modulus from the storage modulus (explained in Section 2.5) allowed or an estimation of the stiffness of the HydLow, HydMid and HydHigh hydrogels at approximately 174 Pa, 740 Pa and 1666 Pa, respectively. These values fall within the range as Young's modulus values of lung ECM measured by atomic force microscopy (Booth et al., 2012). It has been shown that immune cells normally migrate further on stiffer substrates, indicating that the lack of migration observed in our experiments is likely not due to a change in stiffness (Hsieh et al., 2019; Jannat et al., 2010; Oakes et al., 2009). In contrast, collagen matrix density affects neutrophil transmigration and gene expression related to immune regulation and chemoattraction (Han et al., 2012; Larsen et al., 2020). Neutrophil extravasation decreases with increasing concentrations of collagen in a microfluidic system (Han et al., 2012). In another study, it was shown that when macrophages were cultured inside a higher density 3D collagen matrix, the transcriptional profile of these macrophages changed towards a more anti-inflammatory profile (Larsen et al., 2020), promoted Tregs and myeloid-derived suppressor cells attraction, and were less efficient at attracting CD8+ T cells. A similar mechanism could occur in this situation, where increased fiber density decreases pro-inflammatory gene expression and impairs extravasation. Recently, another model was developed to investigate neutrophil transmigration into collagen and geltrex hydrogels (Riddle et al., 2022). The authors showed that hydrogel protein concentration and protein composition affect immune cell extravasation. Unfortunately, they worked with bidirectional flow using a rocking platform and thus could not do live imaging. In order to fully understand the lack of immune cell extravasation into the high-density hydrogel in our model, it would be of interest to test their different hydrogel types to investigate immune cell migration in real time with unidirectional flow.
4.3. Lung inflammation-on-chip
Pulmonary microvascular endothelial cells and lung epithelial cells were seeded on the chip and immune cells were perfused through to create a lung inflammation-on-chip model. The cultured endothelial cells reached confluence and exhibited typical endothelial markers, along with a permeability of 10−5 cm/s, which is consistent with previous findings reported by both our group (Zeinali et al., 2021) and others (de Haan et al., 2021). The experiments showed that epithelial cells react to fMLP stimulation by increasing immune cell migration. This is most likely due to the secretion of cytokines and chemokines by the epithelial cells, which attracts more immune cells (Stadnyk, 1994). Our lung-on-chip model was cultured in a submerged condition. To create a more physiological environment, epithelial cells should be cultured at an air-liquid interface (ALI). The alveolar basement membrane thickness ranges between 0.1–2 µm in vivo (Jain et al., 2021; Weibel, 2015). Most lung-on-chips include a thin culture membrane representing this basement membrane, although usually still slightly thicker than the natural basement membrane (5–10 µm, (Huh et al., 2010; Stucki et al., 2018; Zamprogno et al., 2021)), with the exception of a novel 1 µm thickness basement membrane model (Doryab et al., 2022). In this model, the space between the epithelial and endothelial barrier is a 500 µm wide fibrin hydrogel, which is not representative of the thin alveolar basement membrane. The chip was designed in this manner on purpose to better visualize immune cell migration through the ECM area and create a stable chemotactic gradient, which would not be possible in the standard lung-on-chip configuration. In order to maintain a stable chemotactic gradient over time, a low shear stress was applied to the pulmonary epithelium. While this level of shear stress is not considered physiological, it was necessary to establish the chemotactic gradient, and did not affect the epithelium negatively. Moreover, by separating the endothelium and epithelium in this chip, the secretory effect of epithelial cells on immune cell migration can be investigated without direct contact in the initial phase, and eventually, a few immune cells even reached the epithelial barrier. However, as a next step, it would be interesting to adapt the basement membrane compartment to mimic the thin basement membrane more accurately and potentially add air into the epithelial channel (Cao et al., 2022). Additionally, physiological stretch could be added to simulate the continuous mechanical stress that pulmonary cells are exposed to.
4.4. Automated quantification of immune cell migration in the hydrogel
An important development presented in this study is the automated quantification of the number of migrating immune cells inside the hydrogel over time. It is essential to be able to compare migrating cells between different experiments. The migration of immune cells depends on many factors, in particular their sensitivity to subtle changes in the ECM microenvironment (Ford et al., 2019; Pérez-Rodríguez et al., 2022). Thus, experimental results involving immune cells often show high variability within a condition, as seen in this and earlier studies (de Haan et al., 2021; Pérez-Rodríguez et al., 2022). It is thus important to be able to quantify their migration. Our goal here was to be able to quickly grasp the distribution of migration distances in the ECM at a given timepoint and not to perform single-cell tracking. This reduces the constraint on segmentation accuracy and allowed us to develop a simple and robust analysis workflow based on classical image processing algorithms such as template matching. Through this unique combination, the complexity (annotation, training etc.) of deep learning methods can be avoided. The software is open-source and can be used and adapted for further research.
4.5. Outlook: ARDS-on-chip
The development of this inflammation-on-chip model represents a crucial first step towards replicating the pathology of ARDS on chip. This model successfully incorporates two key features of ARDS, including epithelial damage (as demonstrated in Fig. 6F) and an infiltration of peripheral immune cells (Fig. 7as previously reported by Ware et al. (Ware and Matthay, 2000). To further enhance this model, the introduction of a pathogen could be considered to simulate lung infection on chip. For bringing the inflammation-on-chip model towards a model of ARDS, the epithelium should be cultured in air-liquid interface (ALI) to not only observe epithelial damage and an influx of peripheral immune cells, but also the formation of an edema. Having a model that mimics these critical features of ARDS is a crucial step towards gaining a deeper understanding of the cellular processes involved in the disease. In order to effectively treat ARDS, it is necessary to address these pathological changes. Our model could be used for testing of new ARDS drugs by measuring the development of these features over time. For example, immune cell influx is necessary to combat pathogens but can also be detrimental and contribute to further epithelial damage. Specifically, neutrophils act as the first line of defense and become functionally distinct upon pulmonary infection, with increased chemotaxis and metabolic activity (Meyer et al., 2021). In the current system, neutrophils are not present in the PBMC population, but they can be added. This chip could be used to research severe immune cell-induced damage observed in ARDS and investigate novel drugs that can enhance necessary immune function but prevent severe pulmonary damage.
4.6. Conclusion
Overall, this research shows that immune cell extravasation is dependent upon a multitude of factors, not only the interaction between immune cells and endothelial cells but also the flow profile and the structure of the underlying ECM. With the unique combination of tissue-specific cells, live monitoring of immune cell migration and automated analysis of cell migration, in the future, this model could be utilized to assess immune cell migration in lung infection or for conditions such as ARDS with the potential to test drugs to inhibit the massive influx of immune cells and the correlated tissue damage.
Author contributions
LO, SZ, AS and OG conceived and designed the study. LO and JY performed the experiments. JY and PK carried out the FACS experiments and analysis. YC carried out rheology measurements and analysis. DF aided in data analysis. GW developed analysis software. LO analyzed and interpreted the data and wrote the initial draft of the manuscript. AS, SZ and OG interpreted the results and thoroughly reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This project has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No 812954 and from the Swiss National Science Foundation, project No 185365.
CRediT authorship contribution statement
Lisette van Os: Conceptualization, Methodology, Formal analysis, Investigation, Validation, Writing – original draft, Visualization. Jeremy Yeoh: Investigation, Resources. Guillaume Witz: Formal analysis, Software, Data curation. Dario Ferrari: Data curation. Philippe Krebs: Supervision, Resources. Yashoda Chandorkar: Investigation, Resources. Soheila Zeinali: Conceptualization, Writing – review & editing. Arunima Sengupta: Conceptualization, Writing – review & editing. Olivier T. Guenat: Conceptualization, Supervision, Funding acquisition, Resources, Writing – review & editing.
Acknowledgements
The authors thank Prof. emeritus Dr. Michael Stoffel and Helga Mogel for their contribution to the SEM imaging of hydrogels. In Fig. 1, drawings were created with BioRender.com.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ejps.2023.106485.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Abbas A.K., Lichtman A.H., Pillai S. Cellular and Molecular Immunology. 7th ed. Elsevier; 2012. Cellular and molecular immunology. [Google Scholar]
- Artzy-Schnirman A., Arber Raviv S., Doppelt Flikshtain O., Shklover J., Korin N., Gross A., Mizrahi B., Schroeder A., Sznitman J. Advanced human-relevant in vitro pulmonary platforms for respiratory therapeutics. Adv. Drug Deliv. Rev. 2021;176 doi: 10.1016/J.ADDR.2021.113901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens N., Bandyopadhyay C., Coon B.G., Yun S., Schwartz M.A. Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Invest. 2016;126:821. doi: 10.1172/JCI83083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra N.Y., Restrepo L.M., Galeano Y., Tobón A.C., Turizo L.F., Mesa M. Improving fibrin hydrogels’ mechanical properties, through addition of silica or chitosan-silica materials, for potential application as wound dressings. Int. J. Biomater. 2021;2021 doi: 10.1155/2021/9933331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Booth A.J., Hadley R., Cornett A.M., Dreffs A.A., Matthes S.A., Tsui J.L., Weiss K., Horowitz J.C., Fiore V.F., Barker T.H., Moore B.B., Martinez F.J., Niklason L.E., White E.S. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am. J. Respir. Crit. Care Med. 2012;186:866–876. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T., Shao C., Yu X., Xie R., Yang C., Sun Y., Yang S., He W., Xu Y., Fan Q., Ye F. Biomimetic Alveolus-on-a-Chip for SARS-CoV-2 infection recapitulation. Research. 2022;2022:1–12. doi: 10.34133/2022/9819154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio F.R. Mouse models of acute lung injury and ARDS. Methods Mol. Biol. 2018;1809:341–350. doi: 10.1007/978-1-4939-8570-8_22/FIGURES/3. [DOI] [PubMed] [Google Scholar]
- de Haan L., Suijker J., van Roey R., Berges N., Petrova E., Queiroz K., Strijker W., Olivier T., Poeschke O., Garg S., van den Broek L.J. A microfluidic 3D endothelium-on-a-chip model to study transendothelial migration of T cells in health and disease. Int. J. Mol. Sci. 2021;22:8234. doi: 10.3390/IJMS22158234. 2021Page 8234 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doryab Ali, Taskin Mehmet Berat, Stahlhut Philipp, Groll Jürgen, Schmid Otmar, Doryab A., Schmid O., Taskin M.B., Stahlhut P., Groll J. Real-time measurement of cell mechanics as a clinically relevant readout of an in vitro lung fibrosis model established on a bioinspired basement membrane. Adv. Mater. 2022;34 doi: 10.1002/ADMA.202205083. [DOI] [PubMed] [Google Scholar]
- Duong H., Wu B., Tawil B. Modulation of 3D fibrin matrix stiffness by intrinsic fibrinogen–thrombin compositions and by extrinsic cellular activity. Tissue Eng. Part A. 2009;15:1865. doi: 10.1089/TEN.TEA.2008.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N.D., Fan E., Camporota L., Antonelli M., Anzueto A., Beale R., Brochard L., Brower R., Esteban A., Gattinoni L., Rhodes A., Slutsky A.S., Vincent J.L., Rubenfeld G.D., Taylor Thompson B., Marco Ranieri V. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive Care Med. 2012;38:1573–1582. doi: 10.1007/s00134-012-2682-1. [DOI] [PubMed] [Google Scholar]
- Ford A.J., Orbach S.M., Rajagopalan P. Fibroblasts stimulate macrophage migration in interconnected extracellular matrices through tunnel formation and fiber alignment. Biomaterials. 2019;209:88–102. doi: 10.1016/j.biomaterials.2019.03.044. [DOI] [PubMed] [Google Scholar]
- Francis I., Shrestha J., Paudel K.R., Hansbro P.M., Warkiani M.E., Saha S.C. Recent advances in lung-on-a-chip models. Drug Discov. Today. 2022;27:2593–2602. doi: 10.1016/J.DRUDIS.2022.06.004. [DOI] [PubMed] [Google Scholar]
- Gjorevski N., Avignon B., Gérard R., Cabon L., Roth A.B., Bscheider M., Moisan A. Neutrophilic infiltration in organ-on-a-chip model of tissue inflammation. Lab. Chip. 2020;20:3365–3374. doi: 10.1039/D0LC00417K. [DOI] [PubMed] [Google Scholar]
- Guenat O.T., Geiser T., Berthiaume F. Clinically relevant tissue scale responses as new readouts from organs-on-a-chip for precision medicine. Annu. Rev. Anal. Chem. (Palo Alto. Calif.) 2020;13:111–133. doi: 10.1146/ANNUREV-ANCHEM-061318-114919. [DOI] [PubMed] [Google Scholar]
- Han S., Yan J.J., Shin Y., Jeon J.J., Won J., Jeong H.E., Kamm R.D., Kim Y.J., Chung S. A versatile assay for monitoring in vivo-like transendothelial migration of neutrophils. Lab. Chip. 2012;12:3861–3865. doi: 10.1039/c2lc40445a. [DOI] [PubMed] [Google Scholar]
- Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., Kern R., Picus M., Hoyer S., van Kerkwijk M.H., Brett M., Haldane A., del Río J.F., Wiebe M., Peterson P., Gérard-Marchant P., Sheppard K., Reddy T., Weckesser W., Abbasi H., Gohlke C., Oliphant T.E. Array programming with NumPy. Nature. 2020:357–362. doi: 10.1038/s41586-020-2649-2. 2020 5857825 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind L.E., Giese M.A., Schoen T.J., Beebe D.J., Keller N., Huttenlocher A. Immune cell paracrine signaling drives the neutrophil response to a. fumigatus in an infection-on-a-chip model. Cell. Mol. Bioeng. 2021;14:133–145. doi: 10.1007/s12195-020-00655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y.T., Adriani G., Beyer S., Nhan P.T., Kamm R.D., Kah J.C.Y. A facile method to probe the vascular permeability of nanoparticles in nanomedicine applications. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-00750-3. 2017 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hønge B.L., Petersen M.S., Olesen R., Møller B.K., Erikstrup C. Optimizing recovery of frozen human peripheral blood mononuclear cells for flow cytometry. PLoS ONE. 2017;12 doi: 10.1371/JOURNAL.PONE.0187440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J.Y., Keating M.T., Smith T.D., Meli V.S., Botvinick E.L., Liu W.F. Matrix crosslinking enhances macrophage adhesion, migration, and inflammatory activation. APL Bioeng. 2019;3 doi: 10.1063/1.5067301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Yuan Hsin H., Ingber D.E. Reconstituting organ-level lung functions on a chip. Science. 2010;328(80–):1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- Ishii M., Tsuchiya T., Doi R., Morofuji Y., Fujimoto T., Muto H., Suematsu T., Mori R., Matsumoto K., Miyazaki T., Tomoshige K., Watanabe H., Iwatake M., Nagayasu T. Increased in vitro intercellular barrier function of lung epithelial cells using adipose-derived mesenchymal stem/stromal cells. Pharmaceutics. 2021;13 doi: 10.3390/PHARMACEUTICS13081264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P., Nishiguchi A., Linz G., Wessling M., Ludwig A., Rossaint R., Möller M., Singh S. Reconstruction of ultra-thin alveolar-capillary basement membrane mimics. Adv. Biol. 2021;5 doi: 10.1002/adbi.202000427. [DOI] [PubMed] [Google Scholar]
- Jannat, R., Dembo, M., Hammer, D., 2010. Neutrophil adhesion and chemotaxis depend on substrate mechanics 22. 10.1088/0953-8984/22/19/194117.Neutrophil. [DOI] [PMC free article] [PubMed]
- Kim J.S., Kim T.H., Kang D.L., Baek S.Y., Lee Y., Koh Y.G., Kim Y.Il. Chondrogenic differentiation of human ASCs by stiffness control in 3D fibrin hydrogel. Biochem. Biophys. Res. Commun. 2020;522:213–219. doi: 10.1016/j.bbrc.2019.11.049. [DOI] [PubMed] [Google Scholar]
- Kluyver T., Ragan-Kelley B., Pérez F., Granger B., Bussonnier M., Frederic J., Kelley K., Hamrick J., Grout J., Corlay S., Ivanov P., Avila D., Abdalla S., Willing C. Positioning and Power in Acadimic Publishing: Player Agents and Agendas. 2016. Jupyter Notebooks – a publishing format for reproducible computational workflows; pp. 87–90. Proc. 20th Int. Conf. Electron. Publ. ELPUB 2016. [DOI] [Google Scholar]
- Larsen A.M.H., Kuczek D.E., Kalvisa A., Siersbæk M.S., Thorseth M.-L., Johansen A.Z., Carretta M., Grøntved L., Vang O., Madsen D.H. Collagen density modulates the immunosuppressive functions of macrophages. J. Immunol. 2020;205:1461–1472. doi: 10.4049/jimmunol.1900789. [DOI] [PubMed] [Google Scholar]
- Lee J., Tam H., Adler L., Ilstad-minnihan A., Macaubas C., Mellins E.D. The MHC class II antigen presentation pathway in human monocytes differs by subset and is regulated by cytokines. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0183594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K., Laudanna C., Cybulsky M.I., Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- Li J., Chen J., Bai H., Wang H., Hao S., Ding Y., Peng B., Zhang J., Li L., Huang W. An overview of organs-on-chips based on deep learning. Research. 2022;2022:1–20. doi: 10.34133/2022/9869518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S.L., Soehnlein O., Viola J.R. Organ-specific mechanisms of transendothelial neutrophil migration in the lung, liver, kidney, and aorta. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.02739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Sivula C.P., Jameson S.C. Of mice, dirty mice, and men: using mice to understand human immunology. J. Immunol. 2017;199:383–388. doi: 10.4049/jimmunol.1700453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn P.H., Hind L.E., Huttenlocher A., Beebe D.J. Neutrophil trafficking on-a-chip: an: in vitro, organotypic model for investigating neutrophil priming, extravasation, and migration with spatiotemporal control. Lab. Chip. 2019;19:3697–3705. doi: 10.1039/c9lc00562e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N.V., Tay H.M., Wee S.N., Li K.H.H., Hou H.W. Micro-engineered perfusable 3D vasculatures for cardiovascular diseases. Lab. Chip. 2017;17:2960–2968. doi: 10.1039/c7lc00607a. [DOI] [PubMed] [Google Scholar]
- Mestas J., Hughes C.C.W. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Meyer N.J., Gattinoni L., Calfee C.S. Acute respiratory distress syndrome. Lancet. 2021;398:622. doi: 10.1016/S0140-6736(21)00439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M.J., Lin K.S., King M.R. Fluid shear stress increases neutrophil activation via platelet-activating factor. Biophys. J. 2014;106:2243. doi: 10.1016/J.BPJ.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondadori C., Crippa M., Moretti M., Candrian C., Lopa S., Arrigoni C. Advanced microfluidic models of cancer and immune cell extravasation: a systematic review of the literature. Front. Bioeng. Biotechnol. 2020;8:907. doi: 10.3389/FBIOE.2020.00907/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes P.W., Patel D.C., Morin N.A., Zitterbart D.P., Fabry B., Reichner J.S., Tang J.X. Neutrophil morphology and migration are affected by substrate elasticity. Blood. 2009;114:1387–1395. doi: 10.1182/blood-2008-11-191445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandas Development Team, T., 2020. pandas-dev/pandas: Pandas 1.1.3. 10.5281/ZENODO.7549438. [DOI]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay É. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2012;12:2825–2830. doi: 10.48550/arxiv.1201.0490. [DOI] [Google Scholar]
- Pérez-Rodríguez S., Borau C., García-Aznar J.M., Gonzalo-Asensio J. A microfluidic-based analysis of 3D macrophage migration after stimulation by Mycobacterium, Salmonella and Escherichia. BMC Microbiol. 2022;22:1–14. doi: 10.1186/S12866-022-02623-W/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polio S.R., Kundu A.N., Dougan C.E., Birch N.P., Ezra Aurian-Blajeni D., Schiffman J.D., Crosby A.J., Peyton S.R. Cross-platform mechanical characterization of lung tissue. PLoS ONE. 2018;13 doi: 10.1371/JOURNAL.PONE.0204765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri V.M., Rubenfeld G.D., Thompson B.T., Ferguson N.D., Caldwell E., Fan E., Camporota L., Slutsky A.S. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307:2526–2533. doi: 10.1001/JAMA.2012.5669. [DOI] [PubMed] [Google Scholar]
- Riddle R.B., Jennbacken K., Hansson K.M., Harper M.T. Endothelial inflammation and neutrophil transmigration are modulated by extracellular matrix composition in an inflammation-on-a-chip model. Sci. Rep. 2022;12:1–14. doi: 10.1038/s41598-022-10849-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux E., Bougaran P., Dufourcq P., Couffinhal T. Fluid shear stress sensing by the endothelial layer. Front. Physiol. 2020;11:1–17. doi: 10.3389/fphys.2020.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ M., Boerger E., von Platen P., Francis R.C.E., Taher M., Boemke W., Lachmann B., Leonhardt S., Pickerodt P.A. Surfactant depletion combined with injurious ventilation results in a reproducible model of the acute respiratory distress syndrome (ARDS) JoVE (J. Vis. Exp. 2021;2021:e62327. doi: 10.3791/62327. [DOI] [PubMed] [Google Scholar]
- Ryan E.A., Mockros L.F., Weisel J.W., Lorand L. Structural origins of fibrin clot rheology. Biophys. J. 1999;77:2813. doi: 10.1016/S0006-3495(99)77113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A., Roldan N., Kiener M., Froment L., Raggi G., Imler T., de Maddalena L., Rapet A., May T., Carius P., Schneider-Daum N., Lehr C.-M., Kruithof-de Julio M., Geiser T., Marti T.M., Stucki J.D., Hobi N., Guenat O.T. A new immortalized human alveolar epithelial cell model to study lung injury and toxicity on a breathing lung-on-chip system. Front. Toxicol. 2022;4:1–18. doi: 10.3389/ftox.2022.840606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadnyk A.W. Cytokine production by epithelial cells1. FASEB J. 1994;8:1041–1047. doi: 10.1096/FASEBJ.8.13.7926369. [DOI] [PubMed] [Google Scholar]
- Strässle M., Laloli L., Gultom M., V'kovski P., Stoffel M.H., Crespo Pomar S., Chanfon Bätzner A., Ebert N., Labroussaa F., Dijkman R., Jores J., Thiel V. Establishment of caprine airway epithelial cells grown in an air-liquid interface system to study caprine respiratory viruses and bacteria. Vet. Microbiol. 2021;257 doi: 10.1016/J.VETMIC.2021.109067. [DOI] [PubMed] [Google Scholar]
- Strell C., Entschladen F. Extravasation of leukocytes in comparison to tumor cells. Cell Commun. Signal. 2008;6:1–13. doi: 10.1186/1478-811X-6-10/FIGURES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki A.O., Stucki J.D., Hall S.R.R., Felder M., Mermoud Y., Schmid R.A., Geiser T., Guenat O.T. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab. Chip. 2015;15:1302–1310. doi: 10.1039/c4lc01252f. [DOI] [PubMed] [Google Scholar]
- Stucki J.D., Hobi N., Galimov A., Stucki A.O., Schneider-Daum N., Lehr C.M., Huwer H., Frick M., Funke-Chambour M., Geiser T., Guenat O.T. Medium throughput breathing human primary cell alveolus-on-chip model. Sci. Rep. 2018;8:1–13. doi: 10.1038/s41598-018-32523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacher T., Gambillara V., Da Silva R., Montorzi G., Stergiopulos N., Silacci P. Oscillatory shear stress and reduced compliance impair vascular functions. Clin. Hemorrheol. Microcirc. 2007;37:121–130. [PubMed] [Google Scholar]
- Thacker V.V., Sharma K., Dhar N., Mancini G., Sordet-Dessimoz J., McKinney J.D. Rapid endotheliitis and vascular damage characterize SARS-CoV-2 infection in a human lung-on-chip model. EMBO Rep. 2021;22 doi: 10.15252/EMBR.202152744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T. Scikit-image: image processing in Python. PeerJ. 2014;2014 doi: 10.7717/PEERJ.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.I., Shuler M.L. UniChip enables long-term recirculating unidirectional perfusion with gravity-driven flow for microphysiological systems. Lab. Chip. 2018;18:2563–2574. doi: 10.1039/C8LC00394G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware L.B., Matthay M.A. The acute respiratory distress syndrome. N. Engl. J. Med. 2000;342:1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- Waskom, M., Botvinnik, O., Ostblom, J., Gelbart, M., Lukauskas, S., Hobson, P., Gemperline, D.C., Augspurger, T., Halchenko, Y., Cole, J.B., Warmenhoven, J., Ruiter, J., de, Pye C., Hoyer, S., Vanderplas, J., Villalba, S., Kunter, G., Quintero, E., Bachant, P., Martin, M., Meyer, K., Swain, C., Miles, A., Brunner, T., O'Kane, D., Yarkoni, T., Williams, M.L., Evans, C., Fitzgerald, C., Brian, 2020. mwaskom/seaborn: v0.10.1 (April 2020). 10.5281/ZENODO.3767070. [DOI]
- Weibel E.R. On the tricks alveolar epithelial cells play to make a good lung. Am. J. Respir. Crit. Care Med. 2015;191:504–513. doi: 10.1164/rccm.201409-1663OE. [DOI] [PubMed] [Google Scholar]
- Wu X., Newbold M.A., Haynes C.L. Recapitulation of in vivo-like neutrophil transendothelial migration using a microfluidic platform†. Analyst. 2015;140:5055–5064. doi: 10.1039/C5AN00967G. [DOI] [PubMed] [Google Scholar]
- Xia J., Cai L.H., Wu H., MacKintosh F.C., Weitz D.A. Anomalous mechanics of Zn2+-modified fibrin networks. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/PNAS.2020541118/SUPPL_FILE/PNAS.2020541118.SM04.AVI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamprogno P., Wüthrich S., Achenbach S., Thoma G., Stucki J.D., Hobi N., Schneider-Daum N., Lehr C.M., Huwer H., Geiser T., Schmid R.A., Guenat O.T. Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane. Commun. Biol. 2021;4:1–10. doi: 10.1038/s42003-021-01695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeinali S., Thompson E.K., Gerhardt H., Geiser T., Guenat O.T. Remodeling of an in vitro microvessel exposed to cyclic mechanical stretch. APL Bioeng. 2021;5(2):026102. doi: 10.1063/5.0010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wang P., Luo R., Wang Y., Li Z., Guo Y., Yao Y., Li M., Tao T., Chen W., Han J., Liu H., Cui K., Zhang X., Zheng Y., Qin J. Biomimetic human disease model of SARS-CoV-2-induced lung injury and immune responses on organ chip system. Adv. Sci. 2021;8:1–14. doi: 10.1002/advs.202002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.