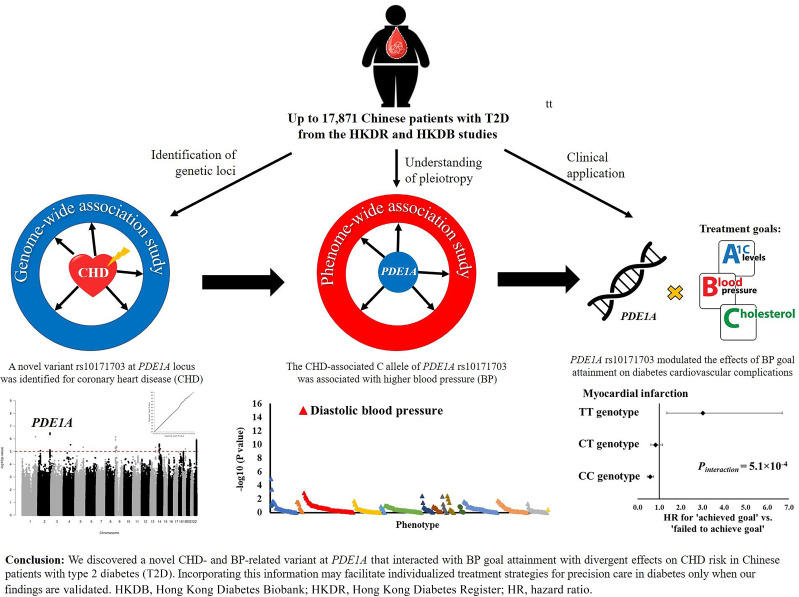
Abstract
OBJECTIVE
In this study we aim to unravel genetic determinants of coronary heart disease (CHD) in type 2 diabetes (T2D) and explore their applications.
RESEARCH DESIGN AND METHODS
We performed a two-stage genome-wide association study for CHD in Chinese patients with T2D (3,596 case and 8,898 control subjects), followed by replications in European patients with T2D (764 case and 4,276 control subjects) and general populations (n = 51,442–547,261). Each identified variant was examined for its association with a wide range of phenotypes and its interactions with glycemic, blood pressure (BP), and lipid controls in incident cardiovascular diseases.
RESULTS
We identified a novel variant (rs10171703) for CHD (odds ratio 1.21 [95% CI 1.13–1.30]; P = 2.4 × 10−8) and BP (β ± SE 0.130 ± 0.017; P = 4.1 × 10−14) at PDE1A in Chinese T2D patients but found only a modest association with CHD in general populations. This variant modulated the effects of BP goal attainment (130/80 mmHg) on CHD (Pinteraction = 0.0155) and myocardial infarction (MI) (Pinteraction = 5.1 × 10−4). Patients with CC genotype of rs10171703 had >40% reduction in either cardiovascular events in response to BP control (2.9 × 10−8 < P < 3.6 × 10−5), those with CT genotype had no difference (0.0726 < P < 0.2614), and those with TT genotype had a threefold increase in MI risk (P = 6.7 × 10−3).
CONCLUSIONS
We discovered a novel CHD- and BP-related variant at PDE1A that interacted with BP goal attainment with divergent effects on CHD risk in Chinese patients with T2D. Incorporating this information may facilitate individualized treatment strategies for precision care in diabetes, only when our findings are validated.
Graphical Abstract
Introduction
Coronary heart disease (CHD) is the leading cause of morbidity and mortality in people with type 2 diabetes (T2D) (1). A recent systematic review indicated that 32.2% of >4 million individuals affected by T2D had cardiovascular disease (CVD), with CHD being the major contributor (2). In a meta-analysis of 858,507 individuals, diabetes conferred a two to three times increased risk of incident CHD (3).
The pathogenesis of CHD is complex and multifactorial. To date, >200 CHD susceptibility loci had been identified in the general population through genome-wide association studies (GWAS) (4–8), although similar genetic associations in patients with T2D remained inconclusive. There is potential genetic heterogeneity in cardiovascular risk between individuals with and without diabetes due to the exacerbating effects of hyperglycemia on CHD such as increased oxidative stress (9,10). Using a candidate gene approach, Doria et al. (10) previously found that the CHD risk associated with the 9p21 locus was increased in patients with T2D, compared with that described in the general population. They also reported a positive interaction between the 9p21 locus and hyperglycemia regarding CHD risk among patients with T2D (10).
With the recent advances in CHD genetics, the question of clinical utility is increasingly relevant. Thus far, the majority of studies focused on deriving polygenic risk scores to quantify an individual’s risk for CHD with only modest success in discrimination in both the general population and people with diabetes (11,12). Whether the genetic information has other applications beyond that of risk stratification is also unclear. Clinical trials and epidemiological analysis had confirmed the importance of controlling HbA1c, blood pressure (BP) and LDL cholesterol (LDL-C) to reduce the risk of CVD in diabetes, although their optimal goals remained controversial (13–15). While this multifactorial management is recommended in the American Diabetes Association Standards of Care in Diabetes (the ABC goal), whether this should be modified according to an individual’s biological makeup is unknown.
In this study we therefore had three main goals. First, we aimed to identify novel CHD susceptibility loci in Chinese patients with T2D. We performed a two-stage GWAS in Chinese patients with T2D, followed by replication studies in T2D patients of European descent and in the general population. Second, we examined the pleiotropic effects of top variants on other phenotypes and biological pathways implicated in CHD through phenome-wide association study (PheWAS) and bioinformatic analyses. Finally, we explored the clinical utility of these variants among people with T2D by testing their interaction effects with attainment of treatment goals, if any, on incident cardiovascular events.
Research Design and Methods
Study Design and Participants
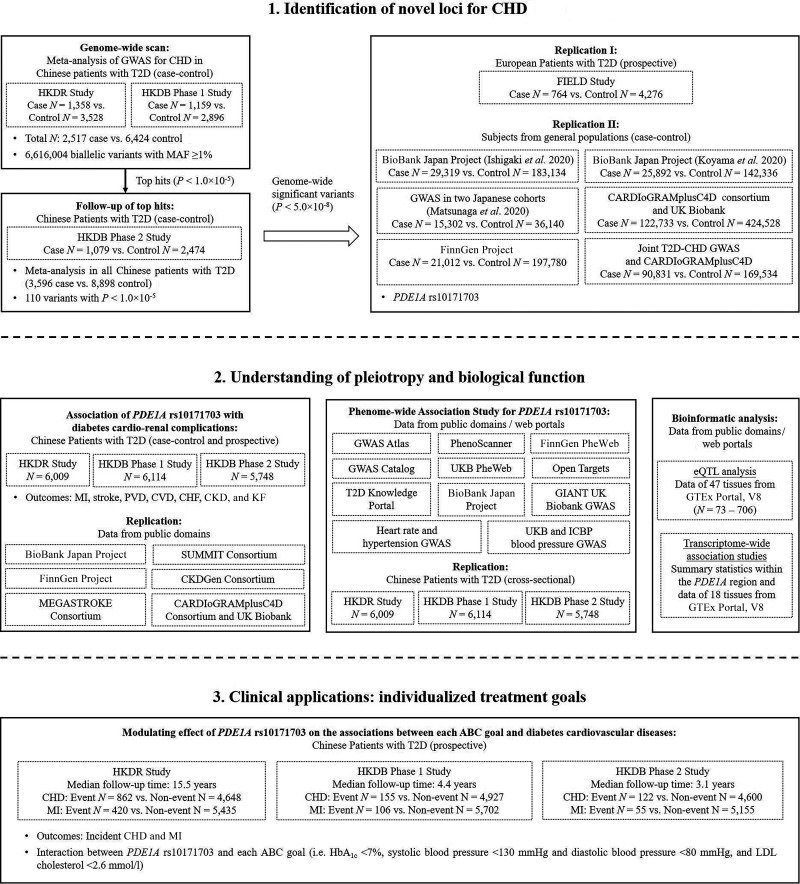
We performed a two-stage analysis to discover novel genetic loci for CHD in patients with T2D. The overall design of the current study is shown in Fig. 1. Details of the design, ascertainment, inclusion criteria, and phenotyping procedures of the study cohorts can be found in Cohort descriptions (Supplementary Material). The clinical characteristics of patients with T2D included in this study are summarized in Supplementary Tables 1 and 2. In the stage of genome-wide scan, we performed a meta-analysis in two independent case-control cohorts of Chinese patients with T2D: 1) 4,886 patients (1,358 case and 3,528 control subjects) from the Hong Kong Diabetes Register (HKDR) (recruitment period 1995–2014) and 2) 4,055 patients (1,159 case and 2,896 control subjects) recruited in 2013–2019 from the Hong Kong Diabetes Biobank (HKDB) phase 1 study (16). The top variants prioritized in the meta-analysis was followed up in 1,079 case and 2,474 control subjects recruited in 2014–2019 from the HKDB phase 2 study (16). All patients in the above three cohorts were southern Han Chinese residing in Hong Kong. The flow diagrams of cohort selection in the HKDR and HKDB studies are shown in Supplementary Fig. 1A–C.
Figure 1.
Study design and workflow. Step 1: to identify novel loci for increased risk of diabetes cardiovascular complications, we conducted a meta-analysis of two GWAS for CHD in Chinese patients with T2D. We followed up the top signals in an additional cohort of Chinese patients with T2D and further performed replication in multiple populations. Finally, the PDE1A locus was identified. Step 2: to understand the pleiotropy and the underlying biological function of the PDE1A locus, we examined this locus in associations with 1) seven types of diabetes cardio-renal complications using two different study designs (i.e., case-control and prospective designs) and 2) a wide spectrum of phenotypes using the publicly available data. We also performed bioinformatic analyses for the PDE1A locus, including the eQTL analysis, and the transcriptome-wide association study. Step 3: to explore the potential clinical utility of personal genetic information, we investigated the effect of each ABC goal on diabetes cardiovascular risk according to different genotypes of PDE1A rs10171703. CHF, congestive heart failure; GIANT, Genetic Investigation of ANthropometric Traits; MAF, minor allele frequency; UKB, UK Biobank.
In the first-stage in silico replication, we included a prospective cohort of European patients with T2D from the Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study (764 case and 4,276 control subjects) (17). The second-stage replication included case-control samples from several published CHD GWAS in general populations. These included the BioBank Japan (BBJ) project reported by Ishigaki et al. (6) (29,319 case and 183,134 control subjects) and Koyama et al. (7) (25,892 case and 142,336 control subjects), another Japanese GWAS reported by Matsunaga et al. (8) (15,302 case and 36,140 control subjects), the meta-analysis of Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) plus The Coronary Artery Disease (C4D) Genetics (CARDIoGRAMplusC4D) and UK Biobank data (122,733 case and 424,528 control subjects) (5), the FinnGen project (21,012 case and 197,780 control subjects) (18), and the joint T2D-CHD GWAS (90,831 case and 169,534 control subjects) (4).
Since kidney function is an important mediator for CVD (19), we analyzed the associations of variants with cardiovascular-renal complications (case-control and prospective setting) and quantitative cardiometabolic traits (cross-sectional data at baseline) in up to 17,871 Chinese patients with T2D from the HKDR and HKDB studies. We compared our results with genetic variants associated with cardiovascular and renal diseases in general populations using publicly available data contributed by the BBJ project (6,20), the FinnGen project (18), the MEGASTROKE consortium (21), the CARDIoGRAMplusC4D consortium and UK Biobank GWAS (22), the SUrrogate markers for Micro- and Macrovascular hard endpoints for Innovative diabetes Tools (SUMMIT) consortium (23,24), and the Chronic Kidney Disease Genetics (CKDGen) Consortium (25). All participants gave written informed consent for DNA collection and data analysis for research purposes at the time of assessment. Each study was approved from the appropriate institutional review boards of the respective institutions.
Definition for CHD Outcome
In the HKDR and HKDB studies, the CHD end point was defined according to the ICD-9. Ascertainment of CHD was based on a composite of acute myocardial infarction (MI), nonfatal ischemic heart disease, or angina pectoris. For case-control analysis, patients with either prevalent or incident CHD were classified as case subjects. Patients were classified as control subjects if they had duration of diabetes ≥10 years and were free from CVD. CVD was defined as the occurrence of CHD, stroke (defined as the occurrence of ischemic stroke except transient ischemic attack, hemorrhagic stroke, or acute but ill-defined cerebrovascular disease), or peripheral vascular disease (PVD) (defined as the occurrence of amputation, gangrene, or peripheral revascularization). For prospective analysis, we excluded prevalent cases and examined only incident events. Follow-up time was calculated as the period from enrollment to the first occurrence of end point, the date of death, or 31 December 2019—whichever came first. In the FIELD study, CHD was defined as a composite of nonfatal MI, coronary death, and revascularization.
Statistical Analysis
Within-cohort, logistic regression and Cox proportional hazards regression were used to examine the associations between genetic variants (additive model) and diabetes cardio-renal complications, with different covariate adjustments in the case-control and prospective analysis, respectively. Meta-analysis was performed with use of the inverse variance–weighted method under a fixed-effects model. Cochran Q test was used to assess the heterogeneity of effect between studies. We accounted for population stratification with adjustment for principal components (PCs) and genomic control in GWAS.
The gene-(ABC achievement) interaction effects on new-onset diabetes cardiovascular end points were tested withy Cox regression in the combined cohort of Chinese patients with T2D (HKDR study, HKDB phase 1 and 2 studies). For each significant interaction effect, a subgroup analysis was performed with Cox regression to examine the association between the goal attainment and end point, with stratification by different genotypes.
Detailed methods and materials for 1) genotyping, quality controls, and imputation; 2) defining outcome variables; 3) PheWAS and bioinformatic analyses; 4) construction of polygenic risk scores; and 5) statistical analysis can be found in Supplementary Methods (Supplementary Material).
Data and Resource Availability
The data sets generated during or analyzed in the current study are available from the corresponding author on reasonable request.
Results
Two-Stage GWAS Analysis in Chinese Patients With T2D
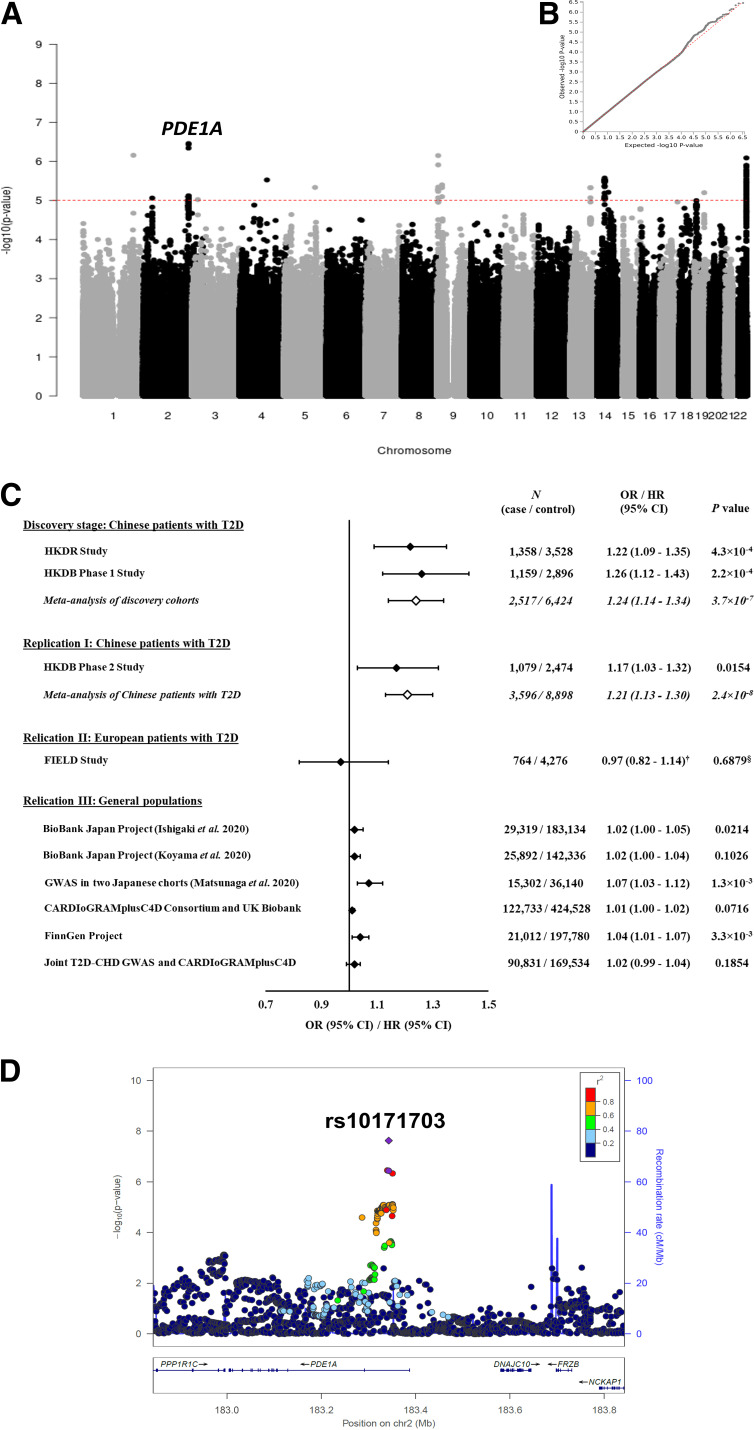
First, we performed a meta-analysis of two GWAS for CHD in 8,941 Chinese patients with T2D (2,517 case and 6,424 control subjects). Figure 2A and B shows the Manhattan plots and the corresponding quantile-quantile plots for the results of the genome-wide scan. Among 6,616,004 biallelic and autosomal single nucleotide polymorphisms (SNPs) available in the HKDR and HKDB phase 1 cohorts, 110 SNPs from 13 novel and distinct genomic regions (within ± 250 kb of the sentinel SNP) were prioritized based on the meta-analysis P value <1.0 × 10−5 (Supplementary Table 3). Correction of genomic control did not considerably change the results (Supplementary Table 3). We then followed up these 110 top SNPs in the HKDB phase 2 cohort consisting of 1,079 case and 2,474 control subjects. A novel signal located at the PDE1A gene showed consistent associations with CHD (1.9 × 10−3 < P < 0.0181 for 20 SNPs within the PDE1A loci selected for replication), with comparable effect estimates across the three cohorts (Supplementary Table 3). The meta-analysis of all three cohorts in Chinese patients with T2D (3,596 case and 8,898 control subjects) demonstrated a genome-wide significant association and a 1.21-fold (95% CI 1.13–1.30; P = 2.4 × 10−8) increased risk of CHD per copy of C allele of the sentinel SNP rs10171703 (Fig. 2C and D and Supplementary Table 4). This association was independent of other baseline covariates including duration of diabetes, smoking status, and obesity traits (Supplementary Table 5). After further adjustment for BP, HbA1c, and concomitant use of medications, the odds ratio (OR) was attenuated to 1.14 (95% CI 1.05–1.25; P = 2.9 × 10−3). Adjustment for lipid levels or renal function did not further change the OR. There was no residual association after conditioning on rs10171703 at PDE1A locus, suggesting that this association was driven by one or more strongly linked SNPs (data not shown).
Figure 2.
Results for two-stage genome-wide association and replication studies. A: Manhattan plot for the meta-analysis of GWAS from the HKDR study and the HKDB phase 1 study. The y-axis represents the −log10 P value, and the x-axis represents the 6,616,004 analyzed biallelic SNPs. The dashed red horizontal line corresponds to the threshold of significance (P < 1 × 10−5). There are 110 points with P < 1 × 10−5, and the label localizes the novel susceptibility locus to CHD discovered in the current study. B: Quantile-quantile (Q-Q) plot for the meta-analysis of GWAS from the HKDR study and the HKDB phase 1 study. The dotted line corresponds to the null hypothesis. C: Forest plot for the association between PDE1A rs10171703 and CHD for all studied populations. ORs and 95% CIs were reported according to the C allele of rs10171703 (i.e., the CHD-associated risk allele). †HR and 95% CIs were reported according to the C allele of rs10171703. P value was obtained from logistic regression model. §P value was obtained from Cox regression model. D: Regional plot of the PDE1A locus. The purple circle and diamond represent the SNP rs10171703 identified from the meta-analysis of GWAS in the HKDR study and the HKDB phase 1 study (genome-wide scan), and the meta-analysis in the HKDR study and the HKDB phase 1 and 2 studies (all cohorts of Chinese patients with T2D), respectively. Other SNPs are colored according to level of LD, which is measured by r2, with the sentinel SNP. The recombination rates estimated from the 1000 Genomes project Asian data are shown. The genes in the interval are indicated in the bottom panel. chr, chromosome.
Replication Studies in Other Populations
Next, we sought further support for our finding, in silico replication of the PDE1A signal for CHD, in 1) European patients of T2D from the FIELD study (764 case and 4,276 control subjects) and 2) multiple studies of general populations (n = 51,442–547,261). The PDE1A rs10171703 variant was not replicated in the FIELD study (P = 0.6879). It gave P value ≤0.05, with association smaller than but direction of association concordant with that in Chinese patients with T2D in general populations of Japanese (OR 1.02 [95% CI 1.00–1.05] in the study by Ishigaki et al. [6] and 1.07 [1.03–1.12] in the study by Matsunaga et al. [8]) and Europeans (1.04 [1.01–1.07] in FinnGen project) (Fig. 2C and Supplementary Table 4). Although no significant association was seen in other samples of general population including the study by Koyama et al. (7), meta-analysis of CARDIoGRAMplusC4D consortium and UK Biobank, and joint T2D-CHD GWAS, all were in the same direction for rs10171703 (Fig. 2C and Supplementary Table 4). The overall meta-analysis of all study cohorts was not conducted due to the differences in study designs and study populations, as well as the overlapping samples between these cohorts.
Association for Diabetes Cardiovascular-Renal Complications
We examined associations for cardiovascular-renal complications in two sets of analyses using case-control and prospective design in Chinese patients with T2D (Supplementary Table 6). In the case-control analysis, in addition to CHD, the C allele of rs10171703 was associated with all cardiovascular-renal outcomes including MI, stroke, PVD, CVD, congestive heart failure, chronic kidney disease (CKD), and kidney failure (KF), with ORs ranging from 1.12 to 1.22 (2.0 × 10−8 < P < 0.0182). Only associations for MI, stroke, CVD, CKD, and KF remained significant after Bonferroni correction for testing seven outcomes or after exclusion of the CHD cases. In the prospective analysis, each copy of the rs10171703 C allele raised the hazards for CHD, stroke, CVD, CKD, and KF by 9–14% (2.7 × 10−3 < P < 0.0343) (Supplementary Table 6). Associations for MI, PVD, and congestive heart failure were not significant.
To see whether these associations were found in the general population, we used the publicly available data and observed consistent evidence for the association of rs10171703 with MI in the BBJ project, the FinnGen project, and the meta-analysis of UK Biobank and CARDIoGRAMplusC4D GWAS (OR 1.02–1.04; 4.7 × 10−3 < P < 0.0127), as well as its association with stroke in the MEGASTROKE consortium (1.03 [95% CI 1.01–1.04]; P = 6.5 × 10−3) (Supplementary Table 7). However, the associations for renal complications were not replicated.
PheWAS and Bioinformatic Analysis
We conducted PheWAS to explore the pathways underlying the association of PDE1A locus with CVD. European data available from the T2D Knowledge Portal showed that the PDE1A rs10171703 had an established adverse association with BP. The C allele that increases CHD risk was associated with elevated diastolic BP (DBP) (β ± SE 0.130 ± 0.017; P = 4.1 × 10−14) and systolic BP (SBP) (0.098 ± 0.030; P = 1.1 × 10−3) in the UK Biobank and International Consortium of Blood Pressure (ICBP) BP GWAS (29), as well as a higher risk for hypertension in the heart rate and hypertension GWAS (37) (OR 1.01 [95% CI 1.01–1.02]; P = 2.9 × 10−4) (Table 1). Data from the current study (DBP β ± SE 0.332 ± 0.153, P = 0.0297, SBP 0.595 ± 0.257, P = 0.0208, and hypertension OR 1.09 [95% CI 1.01–1.17]; P = 0.0174), the BBJ project (DBP β ± SE 0.013 ± 0.004, P = 2.1 × 10−3, in the study by Kanai et al. [30] and 0.013 ± 0.004, P = 6.5 × 10−4, in study by Sakaue et al. [20]), and the FinnGen project (essential hypertension OR 1.02 [95% CI 1.00–1.04]; P = 0.0481) supported these associations in patients with diabetes, East Asians and Europeans, albeit there was insufficient power to detect a genome-wide significant association (Supplementary Table 8). In the expression quantitative trait loci (eQTL) analysis, the C allele of rs10171703 was associated with increased expression of the PDE1A gene in subcutaneous adipose tissue (normalized effect size [NES] ± SE 0.110 ± 0.028, P = 1.1 × 10−4) (Supplementary Figs. 2 and 3). Results for other bioinformatic analyses are available in Supplementary Results (Supplementary Material).
Table 1.
Results of PheWAS for PDE1A rs10171703
| Sources | Related traits | Stage | Cohort (reference no.) | Population | Covariates | Total N (case subjects) | RAF | β ± SE/OR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|---|
| T2D Knowledge Portal | DBP | PheWAS | UK Biobank and ICBP BP GWAS (29) | Europeans | Sex, age, age2, BMI, and other study-specific covariates | 757,601 | 0.506 | 0.130 ± 0.017 | 4.1 × 10−14 |
| SBP | BP-related traits | UK Biobank and ICBP BP GWAS (29) | Europeans | Sex, age, age2, BMI, and other study-specific covariates | 738,170 | 0.505 | 0.098 ± 0.030 | 1.1 × 10−3 | |
| HT | BP-related traits | Heart rate and hypertension GWAS (with data from UK Biobank) (37) | Europeans | — | 458,554 (144,793) | 0.512 | 1.01(1.01–1.02) | 2.9 × 10−4 | |
| Body height | PheWAS | GIANT UK Biobank GWAS (38) | Europeans | Age, sex, recruitment center, genotyping batches, and PCs | 709,342 | 0.502 | −0.008 ± 0.001 | 4.2 × 10−9 | |
| PhenoScanner | TPM | PheWAS | UK Biobank (39,40) | Europeans | — | 330,995 | — | −0.009 ± 0.002 | 2.3 × 10−9 |
| PhenoScanner | TFM | PheWAS | UK Biobank (39,40) | Europeans | — | 331,030 | — | −0.009 ± 0.002 | 3.5 × 10−9 |
| Open Targets | ALM | PheWAS | UK Biobank (41) | Europeans | AFM, age, age2, PCs, assessment center, and genotyping array | 450,243 | 0.512 | −0.013 ± 0.002 | 1.2 × 10−11 |
ORs and 95% CIs were reported according to the CHD-related risk allele (C allele) of PDE1A rs10171703. AFM, apendicular fat mass; ALM, appendicular lean mass; GIANT, Genetic Investigation of ANthropometric Traits; HT, hypertension; RAF, risk allele frequency (C allele); TFM, trunk fat-free mass; TPM, trunk predicted mass.
Cardiovascular Risk Reduction by BP Control Was Modified by PDE1A rs10171703
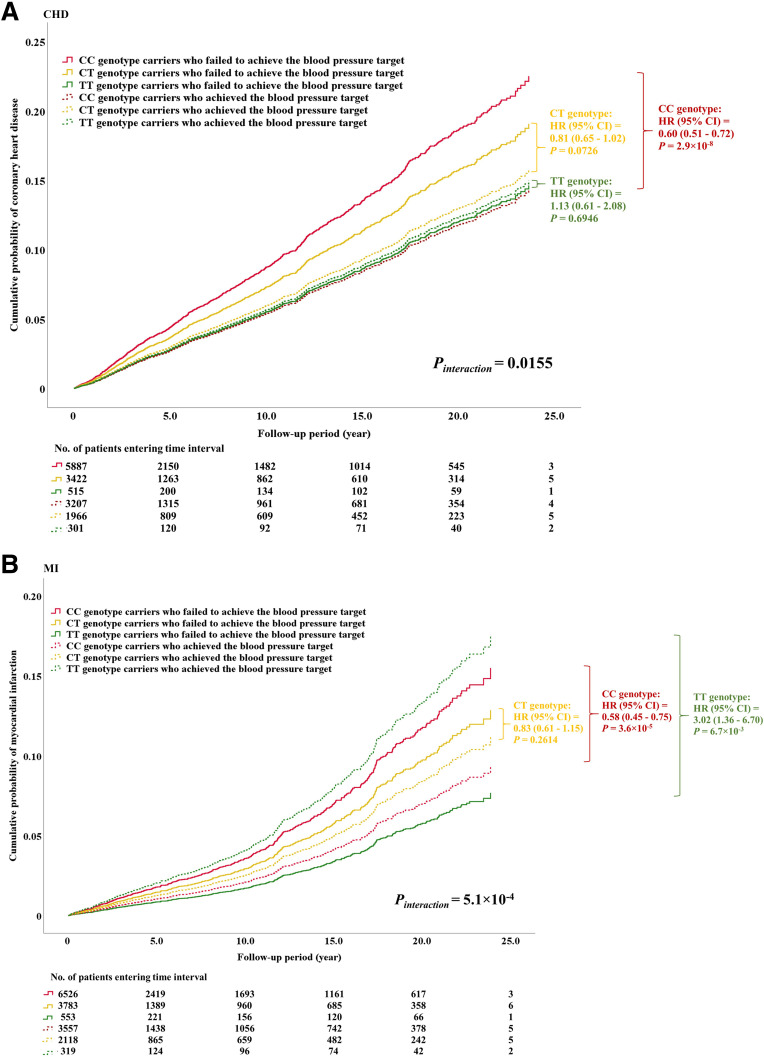
Given the consistent association of PDE1A loci with BP traits, we explored the interactive effect between this variant and each attainment of ABC goals defined according to the American Diabetes Association guidelines on CHD risk (i.e., HbA1c <7%, BP <130/80 mmHg, and LDL-C <2.6 mmol/L) in our combined prospective cohort of Chinese patients with T2D. In the Cox regression analysis, we found that the effect of BP goal attainment on cardiovascular outcomes varied among genotypes (Pinteraction = 0.0155 and 5.1 × 10−4 for incident CHD and MI, respectively) (Supplementary Table 9). Among the CC genotype carriers, BP goal attainment was associated with a 40% reduction in CHD (hazard ratio [HR] 0.60 [95% CI 0.51–0.72]; P = 2.9 × 10−8) in comparison with those not at goal. However, it did not have any significant effect among the carriers of CT genotype (0.81 [0.65–1.02]; P = 0.0726) and TT genotype (1.13 [0.61–2.08]; P = 0.6946) (Fig. 3A, Supplementary Fig 4A, and Supplementary Table 9). Likewise, BP goal attainment had a protective effect on incident MI in the CC genotype group (0.58 [0.45–0.75]; P = 3.6 × 10−5) but not in the CT genotype group (0.83 [0.61–1.15]; P = 0.2614) (Fig. 3B, Supplementary Fig. 4B, and Supplementary Table 9). We further found that attainment of BP goal was significantly associated with a threefold increased risk of MI in the TT genotype group (3.02 [1.36–6.70]; P = 6.7 × 10−3) (Fig. 3B, Supplementary Fig. 4B, and Supplementary Table 9). Adjustments for additional clinical risk factors for CVD (e.g., HbA1c and lipid levels) did not attenuate these interaction effects (CHD Pinteraction = 0.0424, MI Pinteraction = 1.7 × 10−3) (model II in Supplementary Table 10).
Figure 3.
Modulating influence of PDE1A rs10171703 on the association between BP control and new-onset cardiovascular complications among Chinese patients with T2D. These analyses were conducted in the combined cohort of the HKDR study and the HKDB phase 1 and 2 studies. Cumulative probability of new-onset CHD (A) and MI (B) is shown according to the combinations of PDE1A rs10171703 genotypes and the status of BP control. Pinteraction was the P value of the interaction term obtained from the Cox regression model including two main effects (PDE1A rs10171703 and achievement of BP target [i.e., SBP <130 mmHg and DBP <80 mmHg]), the interaction term of main effects, and the covariates (the study cohorts [HKDR study, HKDB phase 1 and 2 studies], enrollment year, sex, age, duration of diabetes, and principal components). P values and HRs and 95% CIs were obtained from the Cox regression model assessing the association between the achievement of BP target (“yes” [coded as 1] vs. “no” [coded as 0]) and diabetic cardiovascular complications, with adjustment for study cohorts, enrollment year, sex, age, duration of diabetes, and PCs, stratified by the genotypes of rs10171703. Numbers of patients entering various time intervals in each category are shown in the bottom panel of each plot.
Despite smaller sample sizes in the individual cohorts, the interaction effect between rs10171703 and BP goal attainment remained consistent for both CHD and MI in the HKDR (CHD HRinteraction 0.84 [95% CI 0.65–1.08], MI 0.61 [0.43–0.87]) and HKDB (CHD 0.56 [0.36–0.88], MI 0.50 [0.27–0.91]) cohorts, suggesting that these were unlikely to be chance findings (Supplementary Table 10). Since BP control might vary over time and we only had baseline data without data on long-term BP exposure, we considered various follow-up periods (i.e., 1–10 years) for both outcomes in the interaction analysis. Results from these sensitivity analyses remained robust with respect to changes in follow-up period, indicating a persistent protective effect of good BP control on diabetes CVD among the CC genotype carriers but not among the CT and TT genotype carriers (Supplementary Table 10).
Conclusions
Here we report a two-stage GWAS for CHD in Chinese patients with T2D, followed by multiple replication studies in patients with T2D and general populations. We discovered a novel susceptibility locus at PDE1A (rs10171703) in T2D patients of Chinese ancestry and found an association in general populations of different ethnicities. This variant also interacted with BP goal attainment (130/80 mmHg) with reduced risk of MI in CC genotype carriers but increased risk in TT genotype carriers, demonstrating the potential utility of this variant for individualizing BP goals for optimizing benefit-to-harm ratio in prevention of CVD in T2D.
In two GWAS previously conducted in people with diabetes of mostly European ancestry, investigators did not detect any genetic variants specifically associated with CHD in diabetes (26,27). In one of these studies investigators concluded that people with or without diabetes had similar genetic architecture of CHD, although the study included individuals with type 1 diabetes and T2D known to have different genetic contributions (26). Although our sample size (n = 12,494) was modest compared with other CHD GWAS in the general population (5–7), we were able to identified a novel genetic locus rs10171703 at PDE1A for CHD among Chinese patients with T2D, with OR of 1.21. The effect size of this locus was attenuated in Japanese (OR 1.02–1.07) and European (OR 1.04) general populations (2.6 × 10−6 < P < 2.1 × 10−3 in the heterogeneity tests). Similarly, Qi et al. (9) used comprehensive genetic data and multiple cohorts of European population and identified a novel locus for CHD at the GLUL gene, which encodes glutamate-ammonia ligase. This locus was associated with CHD, with an OR of 1.36 (95% CI 1.22–1.51), in 4,188 patients with T2D but not in 2,374 individuals without diabetes (OR 0.99 [95% CI 0.87–1.13]), with significant gene-diabetes interaction on CHD risk (Pinteraction = 2.6 × 10−4). Given the importance of hyperglycemia in activating or amplifying multiple atherogenic pathways, GWAS performed in patients with T2D could facilitate the discovery of novel genetic loci as molecular targets for drug development (28).
A recent European meta-analysis (∼61,000 case and ∼578,000 control subjects) with use of data contributed by the UK Biobank and CARDIoGRAMplusC4D consortium identified eight novel loci for MI, including the PDE1A locus (22). The MI-related variant rs12693302 reported by the European study was independent of our CHD-related variant rs10171703, being at least 100 kbp apart. The linkage disequilibrium (LD) indicated by r2 was 0.202–0.245 in our Chinese samples. In our 12,494 Chinese patients with T2D, there was little change in the effect size of rs10171703 after conditioning on rs12693302 (ORunconditional 1.21 [95% CI 1.13–1.30], Punconditional = 2.4 × 10−8, vs. ORconditional 1.22 [1.13–1.32], Pconditional = 3.2 × 10−7) (Supplementary Table 11). Although the European variant, rs12693302, was also modestly associated with increased risk of CHD (ORunconditional 1.07 [1.01–1.14], Punconditional = 0.0227) in our Chinese patients with T2D this was attenuated after conditioning on rs10171703 (ORconditional 0.99 [0.92–1.06], Pconditional = 0.7069). Similar observations were made in the conditional analysis for MI outcome (Supplementary Table 11). Taken together, these findings implied that multiple common genetic variants within the PDE1A gene region might independently or synergistically increase risk for CHD albeit with heterogeneity in terms of effect sizes and locations due to differences in LD pattern and allele frequency (0.78 in East Asians vs. 0.46 in Europeans for rs10171703). This genetic heterogeneity can be further modified by differences in phenotypes, lifestyles, and treatments. Further investigations of this region in large-scale study of different ethnicities and disease populations as well as well-characterized prospective cohorts might discover population-specific causal variants relevant to CHD.
Our PheWAS analysis revealed important pleiotropic effects of PDE1A locus on BP. The CHD-related risk allele of rs10171703 was reported to be associated with elevated BP in European and East Asian populations (20,29,30), which was also confirmed in our Chinese patients with T2D (Supplementary Table 8). More recently, associations between BP levels and two common variants, rs16823124 and rs12693302 at PDE1A, were reported in the general population (20,29). However, our top variant rs10171703 had low correlation with these variants (r2 <0.25 in Chinese population) and we did not detect their associations with BP levels in our cohorts (P > 0.05 for both SBP and DBP). Apart from ethnic differences, it remained possible that the causal variants for BP in the PDE1A region might not be the same in general populations and patients with T2D. In our prospective cohort with documentation of other risk factors and outcomes, adjustments for BP levels and use of antihypertensive drugs partially attenuated the association between rs10171703 and CHD risk (Supplementary Table 5).
Given the importance of BP as a major risk factor for CHD in patients with T2D, the risk associations of PDE1A locus with BP and CHD were biologically plausible (31). To this end, the Ca2+-calmodulin–stimulated phosphodiesterases (PDEs) encoded by the PDE1A gene preferentially hydrolyze the second messenger cGMP, which regulates many cellular functions. In the media of vascular smooth muscle cells (VSMCs) of normal carotid arteries, PDE1A was expressed mainly in the cytoplasm (32,33). However, in experimental injury models (e.g., rat aorta with tolerance to nitrate-induced vasorelaxation) (34) or passage-induced senescent human VSMCs (35), there was increased expression of PDE1A with subcellular localization. In other experimental models such as neointimal VSMCs of rat balloon injury model and mouse carotid ligation model, as well as human coronary arteries with restenosis lesions, PDE1A was detected in the nuclei with cellular dedifferentiation. These findings implicated a role of PDE1A in VSMC growth and survival and might contribute to the neointima formation during atherosclerosis and restenosis.
Against this background, it is noteworthy that in our Chinese patients with T2D, we were able to demonstrate how a single genetic variant (e.g., rs10171703) might be used as a stratification tool for prioritizing patients with T2D. Among the CC genotype carriers at risk, who represented 60% of the Chinese patients with T2D, those who attained BP goal had 40% reduction in risk of MI compared with those who were not at goal, while neutral effect was observed in the 35% of patients with intermediate-risk CT genotype. More importantly, in the remaining 5% of patients with the low-risk TT genotype, those at BP goal had three-times-increased risk in MI in comparison with those not at goal. These modulating effects of rs10171703 were independent of other risk factors related to CVD such as HbA1c and LDL-C, suggesting that they were not due to a confounding effect of adverse glycemic or lipid controls among patients with high BP levels. These findings remained consistent in our subanalysis including patients recruited in different periods where BP control had improved over time. In the Action to Control Cardiovascular Risk in Diabetes (ACCORD) substudy with comparison of intensive SBP lowering to <120 mmHg vs. the recommended goal of 140 mmHg, there was no further reduction in cardiovascular events in patients with T2D, with a numerically higher death rate in the 120 mmHg group versus the 140 mmHg group (3.3% vs. 1.3%) (14). In a recent systematic review by the Blood Pressure Lowering Treatment Trialists’ Collaboration, investigators suggested that similar BP targets can be adopted for people with or without diabetes for cardiovascular protection (36). Taken together, our findings suggested that the benefit-to-harm ratio of intensive BP control might depend on the genetic background of an individual, making rs10171703 a potential biomarker for stratified medicine.
This study has several limitations. First, the sample size of our genome-wide scan analysis was modest, resulting in a small number of new loci and limited power to detect variants with small genetic effects. Second, our discovery study was limited to Southern Han Chinese patients with T2D. Third, some patients in the control group might have subclinical CHD or might have developed CHD outside of our study period who were not captured by our definitions. This misclassification might attenuate the genetic effects estimated in the current study with false-negative results. Furthermore, we did not further validate the modulatory effect of PDE1A rs10171703 on the association between BP control and diabetes cardiovascular complications in an independent cohort. However, analysis conducted in individual cohorts of HKDR and HKDB showed consistent results with respect to the main analysis, suggesting that our finding is unlikely due to chance. Further investigation is warranted to confirm our findings, elucidate the underlying mechanisms, and determine whether this interaction effect can be translated into new strategies to prevent cardiovascular complications in T2D.
In summary, we identified a novel association of CHD with the BP-raising C allele of rs10171703 at PDE1A locus in Chinese patients with T2D. Its modest effect on CHD in general populations emphasized the modifying effects of diabetes on genetic association with CHD. Apart from its ability to discriminate patients with T2D at high risk for developing CHD, the effect of BP control on risk of MI varied by genotypes. These findings support the potential utility of using genetic information to individualize treatment goals and strategies to optimize benefit-to-risk ratio in pursuit of precision care. Validation in independent cohorts is required to confirm our findings.
Article Information
Acknowledgments. The authors are grateful for all study participants for their contribution. Special thanks are extended to all physicians and nurses at all participating diabetes centers for their dedication and professionalism. The authors also thank all team members for their kind assistance and their efforts in the recruitment of patients and data collection. The authors acknowledge the participants and investigators of the BBJ project, the FinnGen study, the CARDIoGRAMplusC4D consortium, the UK Biobank, the MEGASTROKE project, the SUMMIT consortium, CKDGen Consortium, and the Genotype-Tissue Expression (GTEx) project for making their data available for analysis. The GTEx project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by the National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. The data used for the analyses described in this manuscript were obtained from GTEx Portal (https://gtexportal.org/home/). The authors also acknowledge help from Sandun Sanjiv, Nanda Aryal, and Rachel O’Connell from the NHMRC Clinical Trials Centre, University of Sydney, who assisted with data lookup.
Funding. This work was funded by the Research Grants Council of the Hong Kong Special Administrative Region (CU R4012-18), the Research Grants Council Theme-based Research Scheme (T12-402/13N), the Focused Innovation Scheme, Vice-Chancellor One-off Discretionary Fund, University Grants Committee Research Grants Matching Scheme, the Postdoctoral Fellowship Scheme of The Chinese University of Hong Kong, The Chinese University of Hong Kong–Shanghai Jiao Tong University Joint Research Fund, Natural Science Foundation of China–National Health and Medical Science Council, Australia Joint Research Scheme (no. 81561128017), the Hong Kong Foundation for Research and Development in Diabetes, The Chinese University of Hong Kong, the Croucher Foundation Senior Medical Research Fellowship, and Research Grants Council Senior Research Fellow Scheme (SRFS2021-4S04).
The funding sources did not have any role in the design, interpretation of the study, or the decision to publish the results.
Duality of Interest. J.C.N.C. reported receiving consultancy fees from AstraZeneca, Bayer, Boehringer Ingelheim, Celltrion, Merck Sharp & Dohme, Pfizer, Sanofi, and Viatris; speaker fees from AstraZeneca, Bayer, Boehringer Ingelheim, Merck Sharp & Dohme, Merck, Sanofi, and Servier; and research grants through her institutions from Applied Therapeutics, AstraZeneca, Hua Medicine, Lee Powder, Lilly, Merck, and Servier. A.P.S.K. reported receiving research grants from Abbott, AstraZeneca, Bayer, Boehringer Ingelheim, Eli Lilly, Kyowa Kirin, and Merck Serono and honoraria for consultancy or giving lectures from Nestle, Novo Nordisk, Pfizer, and Sanofi. E.Y.K.C reported receiving speaker fees from Sanofi and Novartis and institutional research funding from Sanofi, Medtronic Diabetes, and Powder Pharmaceuticals. R.C.W.M reported receiving research funding from AstraZeneca, Bayer, Merck Sharp & Dohme, Novo Nordisk, Pfizer, and Tricida for carrying out clinical trials and speaker honoraria or consultancy in advisory boards from AstraZeneca, Bayer, and Boehringer Ingelheim. All proceeds were donated to The Chinese University of Hong Kong to support diabetes research. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. C.H.T.T., C.K.P.L., A.O.Y.L., J.C.N.C., and R.C.W.M. were involved in study design and conception. C.K.P.L., A.O.Y.L., A.P.S.K., E.Y.K.C., W.-y.S., J.C.N.C., and R.C.W.M. conceived and coordinated the investigation. C.K.P.L., A.O.Y.L., A.P.S.K., R.O., E.Y.K.C., K.F.L., S.C.S., G.H., C.C.T., K.P.L., J.Y.Y.L., E.Y.N.C., M.W.T., G.K., I.T.L., J.K.Y.L., V.T.F.Y., E.L., S.L., S.F., Y.L.C., C.C.C., W.-y.S., J.C.N.C., and R.C.W.M. contributed to data acquisition. A.C.W.N. and H.-m.L. performed the biochemical measurements and DNA extraction from blood samples. C.H.T.T., M.S., E.S.H.L., B.F., and G.J. carried out data processing and quality control, as well as advised on the analytical methods. C.H.T.T., M.S., FIELD Study Investigators, and R.C.W.M. conducted the statistical analyses. C.H.T.T., M.S., X.F., T.F.C., K.Y.L.Y., S.L., W.Y., S.K.W.T., Y.H., H.-y.L., C.C.S., N.L.S.T., B.T., Y.H., A.J.J., A.K., J.C.N.C., and R.C.W.M. were involved in the interpretation of the results. C.H.T.T., M.S., H.M.C., J.C.N.C., and R.C.W.M. wrote the first draft of the manuscript. All authors edited, reviewed, and approved the final version of the manuscript. R.C.W.M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.22734467.
Complete member lists for the Hong Kong Diabetes Biobank Study Group and the TRansomics ANalysis of Complications and ENdpoints in Diabetes (TRANSCEND) Consortium can be found in the supplementary material online.
Contributor Information
Hong Kong Diabetes Biobank Study Group, FIELD Study Investigators:
Ronald C.W. Ma, Juliana C.N. Chan, Risa Ozaki, Andrea O.Y. Luk, Wingyee So, Ka-fai Lee, Shing-chung Siu, Grace Hui, Chiu-chi Tsang, Kam-piu Lau, Jenny Y.Y. Leung, Man-wo Tsang, Grace Kam, Elaine Cheung, Ip-tim Lau, June K.Y. Li, Vincent T.F. Yeung, Jo Jo Kwan, Samuel Fung, Stanley Lo, Emmy Lau, Yuk-lun Cheng, Stephen K.W. Tsui, Yu Huang, Huiyao Lan, Weichuan Yu, Brian Tomlinson, Si Lok, Ting-fung Chan, Kevin Y.L. Yip, Cheuk-chun Szeto, Xiaodan Fan, Nelson L.S. Tang, Xiaoyu Tian, Claudia H.T. Tam, Guozhi Jiang, Shi Mai, Baoqi Fan, Fei Xie, Sen Zhang, Pu Yu, Meng Wang, Heung-man Lee, Cadmon K.P. Lim, Fangying Xie, Alex C.W. Ng, Grace P.Y. Cheung, Alice P.S. Kong, Elaine Y.K. Chow, Ming-wai Yeung, Chun-chung Chow, Kitty K.T. Cheung, Rebecca Y.M. Wong, Honcheong So, Katie K.H. Chan, Chin-san Law, Anthea K.Y. Lock, Ingrid K.Y. Tsang, Susanna C.P. Chan, Yin-wah Chan, Cherry Chiu, Chi-sang Hung, Cheuk-wah Ho, Ivy H.Y. Ng, Maria W.H. Mak, Kai-man Lee, Candy H.S. Leung, Ka-wah Lee, Hui-ming Chan, Winnie Wat, Tracy Lau, Cheuk-yiu Law, Ryan H.Y. Chan, Candice Lau, Pearl Tsang, Vince Chan, Lap-ying Ho, Eva Wong, Josephine Chan, Sau-fung Lam, Jessy Pang, and Yee-mui Lee
TRansomics ANalysis of Complications and ENdpoints in Diabetes (TRANSCEND) Consortium:
Ronald C.W. Ma, Juliana C.N. Chan, Yu Huang, Hui-yao Lan, Si Lok, Brian Tomlinson, Stephen K.W. Tsui, Weichuan Yu, Kevin Y.L. Yip, Ting-fung Chan, Xiaodan Fan, Wing-yee So, Cheuk-chun Szeto, Nelson L.S. Tang, Andrea O.Y. Luk, Xiaoyu Tian, Claudia H.T. Tam, Guozhi Jiang, Heung Man Lee, Cadmon K.P. Lim, Katie K.H. Chan, Fangying Xie, Alex C.W. Ng, Grace P.Y. Cheung, Ming-wai Yeung, Shi Mai, Fei Xie, Wei Jiang, Sen Zhang, Pu Yu, Meng Weng, Kelly Y. Li, Chuiguo Huang, and Gechang Yu
References
- 1. Leon BM, Maddox TM. Diabetes and cardiovascular disease: epidemiology, biological mechanisms, treatment recommendations and future research. World J Diabetes 2015;6:1246–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Einarson TR, Acs A, Ludwig C, Panton UH. Prevalence of cardiovascular disease in type 2 diabetes: a systematic literature review of scientific evidence from across the world in 2007-2017. Cardiovasc Diabetol 2018;17:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peters SAE, Huxley RR, Woodward M. Diabetes as risk factor for incident coronary heart disease in women compared with men: a systematic review and meta-analysis of 64 cohorts including 858,507 individuals and 28,203 coronary events. Diabetologia 2014;57:1542–1551 [DOI] [PubMed] [Google Scholar]
- 4. Zhao W, Rasheed A, Tikkanen E, et al. CHD Exome+ Consortium; EPIC-CVD Consortium; EPIC-Interact Consortium; Michigan Biobank . Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat Genet 2017;49:1450–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ishigaki K, Akiyama M, Kanai M, et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet 2020;52:669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koyama S, Ito K, Terao C, et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet 2020;52:1169–1177 [DOI] [PubMed] [Google Scholar]
- 8. Matsunaga H, Ito K, Akiyama M, et al. Transethnic meta-analysis of genome-wide association studies identifies three new loci and characterizes population-specific differences for coronary artery disease. Circ Genom Precis Med 2020;13:e002670. [DOI] [PubMed] [Google Scholar]
- 9. Qi L, Qi Q, Prudente S, et al. Association between a genetic variant related to glutamic acid metabolism and coronary heart disease in individuals with type 2 diabetes. JAMA 2013;310:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doria A, Wojcik J, Xu R, et al. Interaction between poor glycemic control and 9p21 locus on risk of coronary artery disease in type 2 diabetes. JAMA 2008;300:2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morieri ML, Gao H, Pigeyre M, et al. Genetic tools for coronary risk assessment in type 2 diabetes: a cohort study from the ACCORD clinical trial. Diabetes Care 2018;41:2404–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elliott J, Bodinier B, Bond TA, et al. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA 2020;323:636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Action to Control Cardiovascular Risk in Diabetes Study Group; Gerstein HC, Miller ME, Byington RP, et al. Effects of intensive glucose lowering in type 2 diabetes. N Engl J Med 2008;358:2545–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cushman WC, Evans GW, Byington RP, et al. ACCORD Study Group . Effects of intensive blood-pressure control in type 2 diabetes mellitus. N Engl J Med 2010;362:1575–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ginsberg HN, Elam MB, Lovato LC, et al. ACCORD Study Group . Effects of combination lipid therapy in type 2 diabetes mellitus. N Engl J Med 2010;362:1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tam CHT, Lim CKP, Luk AOY, et al. Hong Kong Diabetes Register TRS Study Group; Hong Kong Diabetes Biobank Study Group . Development of genome-wide polygenic risk scores for lipid traits and clinical applications for dyslipidemia, subclinical atherosclerosis, and diabetes cardiovascular complications among East Asians. Genome Med 2021;13:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. FIELD Study Investigators . The need for a large-scale trial of fibrate therapy in diabetes: the rationale and design of the Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study. [ISRCTN64783481]. Cardiovasc Diabetol 2004;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurki MI, Karjalainen J, Palta P, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. 6 March 2022 [preprint]. medRxiv:2022.03.03.22271360 [Google Scholar]
- 19. Matsushita K, van der Velde M, Astor BC, et al. Chronic Kidney Disease Prognosis Consortium . Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: a collaborative meta-analysis. Lancet 2010;375:2073–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakaue S, Kanai M, Tanigawa Y, et al. FinnGen . A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet 2021;53:1415–1424 [DOI] [PubMed] [Google Scholar]
- 21. Malik R, Chauhan G, Traylor M, et al. AFGen Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium; International Genomics of Blood Pressure (iGEN-BP) Consortium; INVENT Consortium; STARNET; BioBank Japan Cooperative Hospital Group; COMPASS Consortium; EPIC-CVD Consortium; EPIC-InterAct Consortium; International Stroke Genetics Consortium (ISGC); METASTROKE Consortium; Neurology Working Group of the CHARGE Consortium; NINDS Stroke Genetics Network (SiGN); UK Young Lacunar DNA Study; MEGASTROKE Consortium . Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hartiala JA, Han Y, Jia Q, et al. INVENT Consortium; CHARGE Consortium Hemostasis Working Group; GENIUS-CHD Consortium; Biobank Japan . Genome-wide analysis identifies novel susceptibility loci for myocardial infarction. Eur Heart J 2021;42:919–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sandholm N, Van Zuydam N, Ahlqvist E, et al. The FinnDiane Study Group; The DCCT/EDIC Study Group; GENIE Consortium; SUMMIT Consortium . The genetic landscape of renal complications in type 1 diabetes. J Am Soc Nephrol 2017;28:557–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Zuydam NR, Ahlqvist E, Sandholm N, et al. Finnish Diabetic Nephropathy Study (FinnDiane); Hong Kong Diabetes Registry Theme-based Research Scheme Project Group; Warren 3 and Genetics of Kidneys in Diabetes (GoKinD) Study Group; GENIE (GEnetics of Nephropathy an International Effort) Consortium; Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC) Research Group; SUrrogate markers for Micro- and Macrovascular hard endpoints for Innovative diabetes Tools (SUMMIT) Consortium . A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 2018;67:1414–142729703844 [Google Scholar]
- 25. Wuttke M, Li Y, Li M, et al. Lifelines Cohort Study; V. A. Million Veteran Program . A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 2019;51:957–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fall T, Gustafsson S, Orho-Melander M, Ingelsson E. Genome-wide association study of coronary artery disease among individuals with diabetes: the UK Biobank. Diabetologia 2018;61:2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Zuydam NR, Ladenvall C, Voight BF, et al. SUMMIT Steering Committee; CARDIOGRAMplusC4D Steering Committee . Genetic predisposition to coronary artery disease in type 2 diabetes mellitus. Circ Genom Precis Med 2020;13:e002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Low Wang CC, Hess CN, Hiatt WR, Goldfine AB. Clinical update: cardiovascular disease in diabetes mellitus: atherosclerotic cardiovascular disease and heart failure in type 2 diabetes mellitus - mechanisms, management, and clinical considerations. Circulation 2016;133:2459–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Evangelou E, Warren HR, Mosen-Ansorena D, et al. Million Veteran Program . Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet 2018;50:1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanai M, Akiyama M, Takahashi A, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet 2018;50:390–400 [DOI] [PubMed] [Google Scholar]
- 31. Ray KK, Seshasai SR, Wijesuriya S, et al. Effect of intensive control of glucose on cardiovascular outcomes and death in patients with diabetes mellitus: a meta-analysis of randomised controlled trials. Lancet 2009;373:1765–1772 [DOI] [PubMed] [Google Scholar]
- 32. Nagel DJ, Aizawa T, Jeon KI, et al. Role of nuclear Ca2+/calmodulin-stimulated phosphodiesterase 1A in vascular smooth muscle cell growth and survival. Circ Res 2006;98:777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khammy MM, Dalsgaard T, Larsen PH, et al. PDE1A inhibition elicits cGMP-dependent relaxation of rat mesenteric arteries. Br J Pharmacol 2017;174:4186–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim D, Rybalkin SD, Pi X, et al. Upregulation of phosphodiesterase 1A1 expression is associated with the development of nitrate tolerance. Circulation 2001;104:2338–2343 [DOI] [PubMed] [Google Scholar]
- 35. Bautista Niño PK, Durik M, Danser AHJ, et al. Phosphodiesterase 1 regulation is a key mechanism in vascular aging. Clin Sci (Lond) 2015;129:1061–1075 [DOI] [PubMed] [Google Scholar]
- 36. Nazarzadeh M, Bidel Z, Canoy D, et al. Blood Pressure Lowering Treatment Trialists’ Collaboration. Blood pressure-lowering treatment for prevention of major cardiovascular diseases in people with and without type 2 diabetes: an individual participant-level data meta-analysis. Lancet Diabetes Endocrinol 2022;10:645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Z, Wang X, Li X, et al. International COPD Genetics Consortium . Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir Res 2019;20:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yengo L, Sidorenko J, Kemper KE, et al. GIANT Consortium . Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Staley JR, Blackshaw J, Kamat MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016;32:3207–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019;35:4851–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pei YF, Liu YZ, Yang XL, et al. The genetic architecture of appendicular lean mass characterized by association analysis in the UK Biobank study. Commun Biol 2020;3:608. [DOI] [PMC free article] [PubMed] [Google Scholar]