Abstract
OBJECTIVE
The Rare and Atypical Diabetes Network (RADIANT) will perform a study of individuals and, if deemed informative, a study of their family members with uncharacterized forms of diabetes.
RESEARCH DESIGN AND METHODS
The protocol includes genomic (whole-genome [WGS], RNA, and mitochondrial sequencing), phenotypic (vital signs, biometric measurements, questionnaires, and photography), metabolomics, and metabolic assessments.
RESULTS
Among 122 with WGS results of 878 enrolled individuals, a likely pathogenic variant in a known diabetes monogenic gene was found in 3 (2.5%), and six new monogenic variants have been identified in the SMAD5, PTPMT1, INS, NFKB1, IGF1R, and PAX6 genes. Frequent phenotypic clusters are lean type 2 diabetes, autoantibody-negative and insulin-deficient diabetes, lipodystrophic diabetes, and new forms of possible monogenic or oligogenic diabetes.
CONCLUSIONS
The analyses will lead to improved means of atypical diabetes identification. Genetic sequencing can identify new variants, and metabolomics and transcriptomics analysis can identify novel mechanisms and biomarkers for atypical disease.
Graphical Abstract
Introduction
Atypical diabetes comprises both uncommon genetic syndromes as well as clusters of phenotypically distinct forms of diabetes that fall within a spectrum between the two poorly defined poles of “type 1” and “type 2” diabetes (1–20). However, many more unrecognized or uncharacterized forms of diabetes exist. The Rare and Atypical Diabetes Network (RADIANT) is designed to characterize new diabetes subtypes and reveal novel mechanistic or causal pathways that can be leveraged for prevention or treatment.
Research Design and Methods
RADIANT, funded by the National Institutes of Health, consists of 14 clinical centers: Baylor College of Medicine (BCM), University of Chicago, University of Washington, Seattle Children’s Hospital, University of Colorado, SUNY Downstate Health Sciences University, Indiana University, Columbia University, Massachusetts General Hospital, University of Maryland, University of Michigan, University of North Carolina, Vanderbilt University, and Washington University in St. Louis. The Data Coordinating Center, University of South Florida, is responsible for the coordination and support of the study protocol and data collection and analysis.
The RADIANT Central Laboratory at the University of Florida is responsible for all biochemical analyses, autoantibody testing, and DNA and RNA extraction. Whole-genome sequencing (WGS) is done at the Broad Institute and RNA sequencing at BCM. Plasma metabolomics is performed at Duke University, and monocytes are preserved for pluripotent stem cell derivation at BCM. The study is overseen by the University of Utah Institutional Review Board.
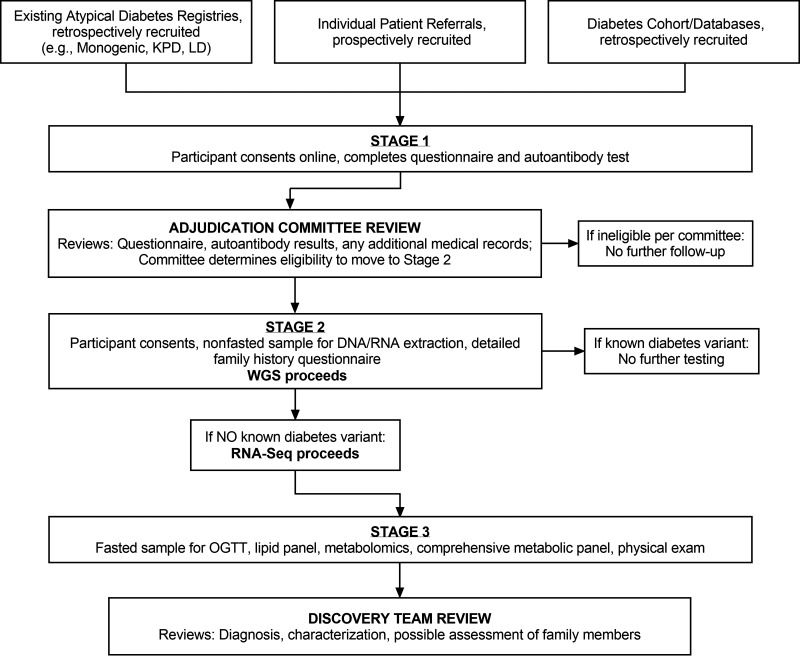
The RADIANT study includes three stages of participation (Fig. 1).
Figure 1.
RADIANT protocol flow. OGTT, oral glucose tolerance test; RNA-Seq, RNA sequencing; KPD, ketosis-prone diabetes; LD, lipodystrophy.
Stage 1: Recruitment
The following criteria or phenotypes are considered for suspecting “atypical” participants:
Type 2 diabetes diagnosed at a time when the individual was prepubertal or nonobese
Mendelian pattern, especially with early onset (<18 years old)
Syndromic (multiple systems involved)
Lipodystrophic
Extremes of BMI
“Mitochondrial” characteristics (e.g., myopathy, hearing deficits)
Nonprogressive
Rapidly progressive (“fulminant”)
Low insulin requirements (<0.5 units/kg/day)
Cyclical hyperglycemia with periods of remission
Lean individuals with polycystic ovarian syndrome
History of gestational diabetes mellitus when lean
Lean insulin resistant
If islet autoantibodies and β-cell function parameters have been measured (where A is islet cell autoantibodies and B is β-cell function): A−B− (i.e., lacking islet autoimmunity makers and lacking β-cell function) or A−B+ with unprovoked diabetic ketoacidosis at initial presentation (i.e., lacking islet autoimmune markers, with preserved β-cell function, but presenting with unprovoked diabetic ketoacidosis)
Exclusions from the study include the following:
Those with high likelihood of typical type 1, typical type 2, known monogenic, or other known secondary forms of diabetes
Refusal of consent for genetic testing
Islet autoantibody positive (participants who are islet autoantibody positive but present with additional atypical features, i.e., syndromic, strong linear family history of diabetes, may not be excluded)
Women who are currently pregnant
All adults or parents/legal guardians of children are directed to an online stage 1 screening consent form. Participants then complete questionnaires (Supplementary Table 1) for exclusion of autoimmune type 1 diabetes, typical type 2 diabetes, and secondary diabetes, as well as to reveal atypical features of diabetes. All questionnaires, available in Spanish and English, can be downloaded for manual completion. Participants are asked to sign a medical record release form for the RADIANT team. Where continuation in RADIANT is deemed appropriate, participants are sent a sample collection kit to draw a sample for diabetes autoantibody testing.
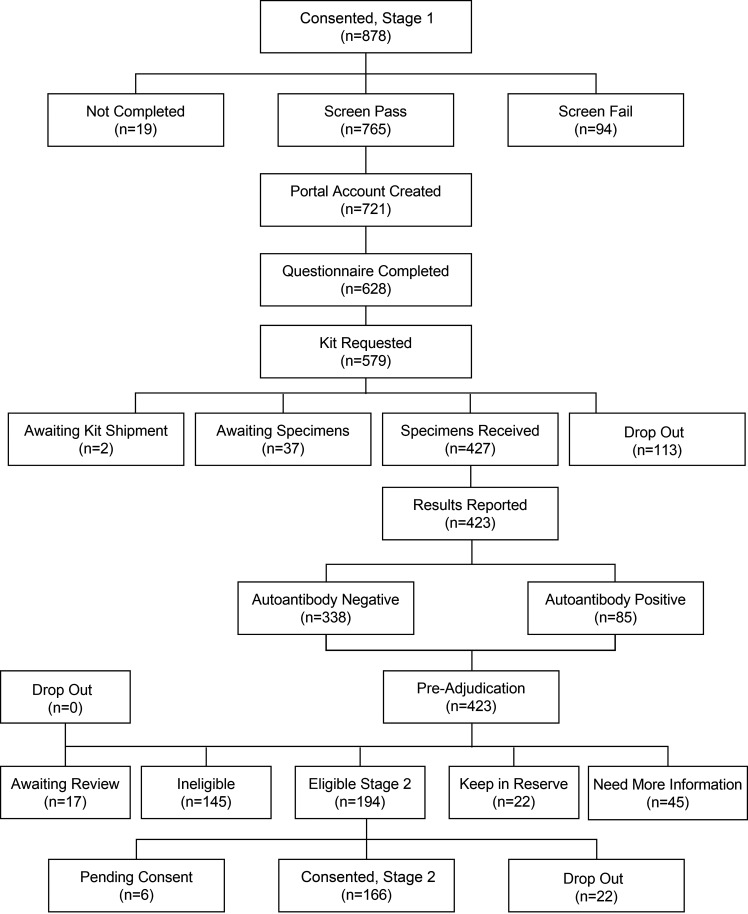
Samples are analyzed for GAD autoantibodies (GADA). If negative for GADA, the samples will be analyzed for insulinoma antigen 2 (IA-2) and zinc transporter 8 (ZnT8) autoantibodies. The RADIANT Adjudication Committee reviews the data collected in stage 1 and then selects candidates with potential atypical diabetes to proceed to stage 2 (Fig. 2).
Figure 2.
Consolidated Standards of Reporting Trials (CONSORT) diagram: stage 1.
Stage 2 Procedures
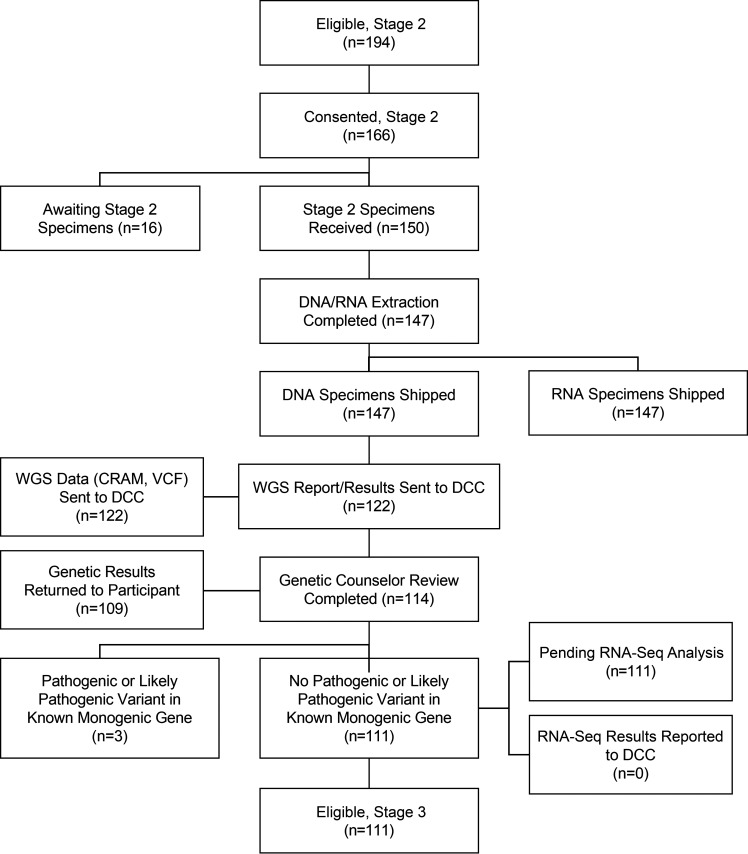
Participants deemed eligible for participation in stage 2 are contacted to provide informed consent/assent, including consent for genetic screening for known forms of monogenic diabetes, medically actionable variants as defined by the American College of Medical Genetics and Genomics (ACMG), and for blood collection for DNA and RNA extraction, storage, and sequencing (WGS and RNA sequencing). Participants are also asked to complete an online questionnaire about their family history to construct a pedigree (Fig. 3).
Figure 3.
Consolidated Standards of Reporting Trials (CONSORT) diagram: stage 2. DCC, Data Coordination Center, University of South Florida; RNA-Seq, RNA sequencing.
WGS is performed in a College of American Pathologists (CAP)/Clinical Laboratory Improvement Amendments (CLIA) laboratory. Identified variants are mapped to the human reference genome assembly. Analysis of identified nuclear and mitochondrial-encoded variants proceeds in two phases.
Clinical reporting phase: participants can elect to receive identification of known pathogenic variants in (a) known monogenic diabetes genes and (b) genes identified by the ACMG as medically actionable.
Discovery phase: identification of candidate causative variants for the atypical diabetes phenotype. This information is not shared with participants at this stage of the study.
DNA samples from participants with a suspected mitochondrial disease phenotype undergo mitochondrial genome sequencing, performed at BCM. Participants who have a causal genetic variant that is already known and consistent with the phenotype will not proceed to the next stage of the RADIANT study. If a participant elects to have the results of these analyses returned to them, results will be explained by a RADIANT genetic counselor. RNA sequencing of peripheral blood leukocytes is performed by the Human Genome Sequencing Center at BCM (Supplementary Fig. 1).
Whole blood, serum, plasma, and extracted DNA/RNA samples will be archived at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Central Repository. Additionally, WGS data with specific data-sharing restrictions may be submitted with the appropriate consent/data sharing information into one or more scientific databases, such as the database of Genotypes and Phenotypes (dbGaP); Analysis, Visualization, and Informatics Lab-space (AnVIL); and the clinical variant database ClinVar.
Stage 3 Procedures
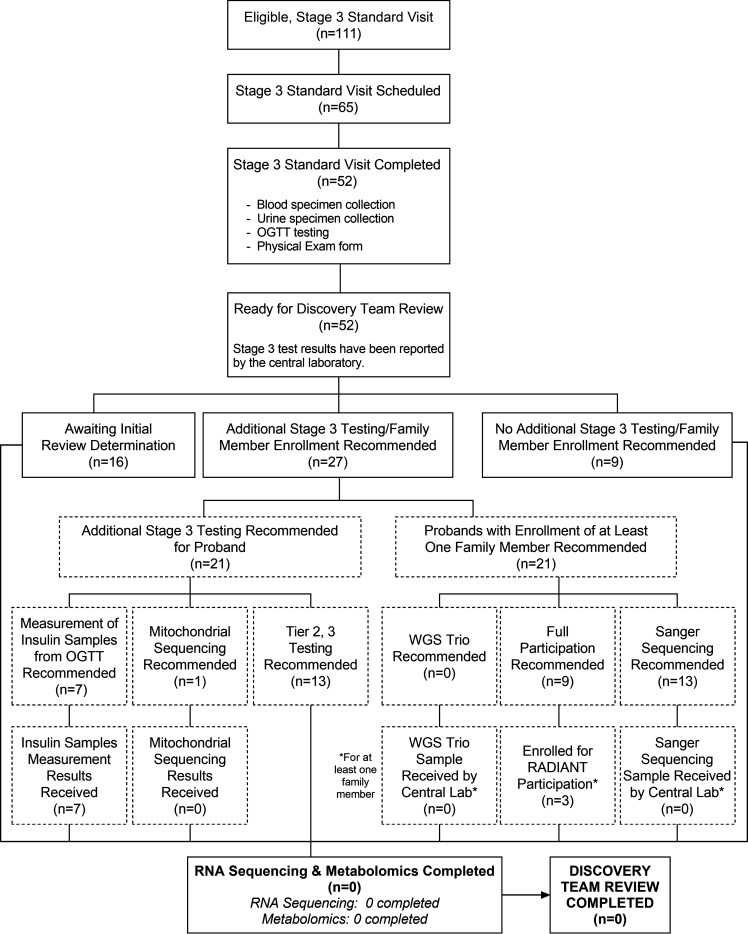
All participants eligible for stage 3 visit a study clinic and consent to a fasted blood draw for lipid profile, metabolic panel, metabolomics studies, and hemoglobin A1c measurement, followed by a standardized oral glucose tolerance test for glucose, insulin, and C-peptide measurements (Fig. 4). Quantitative estimates of insulin secretion and insulin sensitivity are derived (Supplementary Table 2). A comprehensive physical provides vital signs, biometric measurements, and optional photography.
Figure 4.
Consolidated Standards of Reporting Trials (CONSORT) diagram: stage 3. Boxes with dashed lines represent additional recommended testing that has not started as yet. OGTT, oral glucose tolerance test.
These data are considered by the RADIANT Discovery Team comprising genetics experts as well as clinical phenotyping and diabetes experts. Initially, a genome reviewer, a phenotyper/clinician reviewer, and an RNA sequencing reviewer will present the case for discussion and recommendations for deeper phenotyping (tiers 2 and 3 tests as described in Fig. 4 and Supplementary Material).
Enrollment of the proband’s family members may be recommended for additional genomic testing to determine if a candidate variant segregates with the identified phenotype in the family.
Data Analysis
Participants with known monogenic forms of diabetes are referred to experts who specialize in those forms of diabetes. Secondary findings (such as those recommended by ACMG) are reported back to the participant, if consent was given. For participants younger than 18 years of age, results returned include only information directly related to diseases and disorders with pediatric onset. When they reach 18 years of age, participants may request additional information. Genetic counseling is available for all RADIANT participants.
Bioinformatic and statistical analysis will include several clustering methods, including partitioning around medoids recursive partitioning analysis and Bayesian nonnegative matrix factorization (21–24).
Data and Resource Availability
By signing the study consent forms, participants agree to have their samples and data stored for future research and that data may be deposited into controlled-access databases such as dbGaP. They may change their mind up until the end of the study, at which point information that identifies them to the sample will be destroyed. Any data collected prior to the date the participant withdraws will be maintained within the RADIANT database. When the study is completed, access to study data and/or biospecimen samples will be provided through the NIDDK Central Repository.
Results
Stage 1 opened on 30 September 2020, 878 participants enrolled through 19 July 2022 (Fig. 2). A pathogenic or likely pathogenic variant in a known diabetes monogenic gene was found in 3 of 122 (2.5%) individuals for whom WGS results have been received. To date, six new monogenic variants have been identified: SMAD5 [c.844C>T (p.Arg282Cys)], PTPMT1 [NM_175732.2 c.349T>A p.Phe117Ile], INS [NM_000207.2 c.188-11C>A], NFKB1 [NFKB1(NM_003998.4):c.1753-1G>C], IGF1R [c.1447G>A], and PAX6 NC_000011.9:g.31822258_31822284delins [GGTCG;NC_012920.1:m.(10929_?);NC_012920.1:m.(?_10674)].
The most frequent phenotypic clusters are lean type 2 diabetes, autoantibody-negative and insulin-deficient diabetes, and new forms of possible monogenic, oligogenic, or lipodystrophic diabetes. Special interest groups have been formed to study each in more depth.
Conclusions
RADIANT has far-reaching implications. The genotypic and phenotypic analyses will lead to improved means of identifying cases with atypical diabetes. Genetic sequencing could help identify new variants and metabolomics and transcriptomics analysis novel pathogenic mechanisms and biomarkers for atypical forms of the disease. Ultimately these findings may permit a more precise, etiologically based clinical classification of diabetes.
Article Information
Acknowledgments. The authors acknowledge the many providers who helped identify cases of suspected atypical diabetes and referred the patients to RADIANT and acknowledge the patients themselves who have joined this project.
Funding. The RADIANT Study is funded by NIDDK grants U54 DK118638 and U54 DK118612.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. All RADIANT investigators contributed to the design of the RADIANT study and reviewed and approved the manuscript. Jeffrey Krischer (Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa, FL) also contributed to the design of the RADIANT study, wrote the initial draft, and reviewed and approved the final manuscript. Jeffrey Krischer is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Clinical trial reg. no. NCT05544266, clinicaltrials.gov
This article contains supplementary material online at https://doi.org/10.2337/figshare.22367230.
A complete list of members of the RADIANT Study Group can be found in supplementary material online.
Contributor Information
RADIANT Study Group:
Ashok Balasubramanyam, Maria J. Redondo, William Craigen, Hongzheng Dai, Ansley Davis, Dimpi Desai, Monica Dussan, Jordana Faruqi, Ruchi Gaba, Iliana Gonzalez, Shalini Jhangiani, Elizabeth Kubota-Mishra, Pengfei Liu, David Murdock, Jennifer Posey, Nalini Ram, Aniko Sabo, Stephanie Sisley, Mustafa Tosur, Eric Venner, Marcela Astudillo, Adriana Cardenas, Mary Ann Fang, Erica Hattery, Adrienne Ideouzu, Julizza Jimenez, Nupur Kikani, Graciela Montes, Nikalina G. O’Brien, Lee-Jun Wong, Robin Goland, Wendy K. Chung, Anabel Evans, Rachelle Gandica, Rudolph Leibel, Kaisha Mofford, James Pring, Carmella Evans-Molina, Farrah Anwar, Gabriela Monaco, Anna Neyman, Zeb Saeed, Emily Sims, Maria Spall, Marimar Hernandez-Perez, Kieren Mather, Kelly Moors, Miriam S. Udler, Jose C. Florez, Melissa Calverley, Victoria Chen, Kathy Chu, Sara Cromer, Aaron Deutsch, Mariella Faciebene, Evelyn Greaux, Dorit Koren, Raymond Kreienkamp, Mary Larkin, William Marshall, Pam Ricevuto, Amy Sabean, Nopporn Thangthaeng, Christopher Han, Jordan Sherwood, Liana K. Billings, Mary Ann Banerji, Kylnt Bally, Necole Brown, Beisi Ji, Lina Soni, Melissa Lee, Jennifer Abrams, Lorraine Thomas, Jennifer Abrams, Samara Skiwiersky, Louis H. Philipson, Siri Atma W. Greeley, Graeme Bell, Shanna Banogon, Jui Desai, David Ehrmann, Lisa R. Letourneau-Freiberg, Rochelle N. Naylor, Erin Papciak, Lainie Friedman Ross, Manu Sundaresan, Colleen Bender, Persephone Tian, Neda Rasouli, Mohsen Bahmani Kashkouli, Chelsea Baker, Andrew Her, Courtney King, Avinash Pyreddy, Vatsala Singh, Jules Barklow, Noosha Farhat, Rebecca Lorch, Carter Odean, Gregory Schleis, Chantal Underkofler, Toni I. Pollin, Hadley Bryan, Kristin Maloney, Ryan Miller, Paula Newton, Maria Eleni Nikita, Devon Nwaba, Kristi Silver, Jessica Tiner, Hilary Whitlatch, Kathleen Palmer, Stephanie Riley, Elizabeth Streeten, Elif A. Oral, David Broome, Anabela Dill Gomes, Maria Foss de Freitas, Brigid Gregg, Seda Grigoryan, Salman Imam, Melda Sonmez Ince, Adam Neidert, Carman Richison, Baris Akinci, Rita Hench, John Buse, Chase Armstrong, Chad Christensen, Jamie Diner, Rachael Fraser, Karla Fulghum, Tahereh Ghorbani, Alex Kass, Klara Klein, M. Sue Kirkman, Irl B. Hirsch, Jesica Baran, Xiaofu Dong, Steven E. Kahn, Dori Khakpour, Patali Mandava, Lori Sameshima, Thanmai Kalerus, Catherine Pihoker, Beth Loots, Kathleen Santarelli, Cisco Pascual, Kevin Niswender, Norma Edwards, Justin Gregory, Alvin Powers, Andrea Ramirez, Jennifer Scott, Jordan Smith, Fumihiko Urano, Jing Hughes, Stacy Hurst, Janet McGill, Stephen Stone, Jennifer May, Jeffrey P. Krischer, Rajesh Adusumalli, Bruce Albritton, Analia Aquino, Paul Bransford, Nicholas Cadigan, Laura Gandolfo, Jennifer Garmeson, Joseph Gomes, Robert Gowing, Christina Karges, Callyn Kirk, Sarah Muller, Jean Morissette, Hemang M. Parikh, Francisco Perez-Laras, Cassandra L. Remedios, Pablo Ruiz, Noah Sulman, Michael Toth, Lili Wurmser, Christopher Eberhard, Steven Fiske, Brandy Hutchinson, Sidhvi Nekkanti, Rebecca Wood, Jose C. Florez, Ahmed Alkanaq, MacKenzie Brandes, Nöel Burtt, Jason Flannick, Phebe Olorunfemi, Miriam S. Udler, Lizz Caulkins, Clive Wasserfall, William Winter, David Pittman, Beena Akolkar, Christine Lee, David J. Carey, Daniel Hood, Santica M. Marcovina, and Christopher B. Newgard
References
- 1. Froguel P, Vaxillaire M, Sun F, et al. Close linkage of glucokinase locus on chromosome 7p to early-onset non-insulin-dependent diabetes mellitus. Nature 1992;356:162–164 [DOI] [PubMed] [Google Scholar]
- 2. Froguel P, Zouali H, Vionnet N, et al. Familial hyperglycemia due to mutations in glucokinase. Definition of a subtype of diabetes mellitus. N Engl J Med 1993;328:697–702 [DOI] [PubMed] [Google Scholar]
- 3. Hattersley AT, Turner RC, Permutt MA, et al. Linkage of type 2 diabetes to the glucokinase gene. Lancet 1992;339:1307–1310 [DOI] [PubMed] [Google Scholar]
- 4. Gloyn AL, Pearson ER, Antcliff JF, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med 2004;350:1838–1849 [DOI] [PubMed] [Google Scholar]
- 5. van den Ouweland JM, Lemkes HH, Ruitenbeek W, et al. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet 1992;1:368–371 [DOI] [PubMed] [Google Scholar]
- 6. Steenkamp DW, Alexanian SM, Sternthal E. Approach to the patient with atypical diabetes. CMAJ 2014;186:678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balasubramanyam A, Tandon N, Yajnik CS. Non-traditional forms of diabetes worldwide: implications for translational investigation. In Translational Endocrinology & Metabolism: Type 2 Diabetes Update. Robertson RP, Ed. The Endocrine Society, 2011, p. 43–67 [Google Scholar]
- 8. Stenström G, Gottsäter A, Bakhtadze E, Berger B, Sundkvist G. Latent autoimmune diabetes in adults: definition, prevalence, beta-cell function, and treatment. Diabetes 2005;54(Suppl. 2):S68–S72 [DOI] [PubMed] [Google Scholar]
- 9. Andersen MK, Lundgren V, Turunen JA, et al. Latent autoimmune diabetes in adults differs genetically from classical type 1 diabetes diagnosed after the age of 35 years. Diabetes Care 2010;33:2062–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maldonado M, Hampe CS, Gaur LK, et al. Ketosis-prone diabetes: dissection of a heterogeneous syndrome using an immunogenetic and beta-cell functional classification, prospective analysis, and clinical outcomes. J Clin Endocrinol Metab 2003;88:5090–5098 [DOI] [PubMed] [Google Scholar]
- 11. Otiniano ME, Balasubramanyam A, Maldonado M. Presence of the metabolic syndrome distinguishes patients with ketosis-prone diabetes who have a type 2 diabetic phenotype. J Diabetes Complications 2005;19:313–318 [DOI] [PubMed] [Google Scholar]
- 12. Balasubramanyam A, Garza G, Rodriguez L, et al. Accuracy and predictive value of classification schemes for ketosis-prone diabetes. Diabetes Care 2006;29:2575–2579 [DOI] [PubMed] [Google Scholar]
- 13. Balasubramanyam A, Nalini R, Hampe CS, Maldonado M. Syndromes of ketosis-prone diabetes mellitus. Endocr Rev 2008;29:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haaland WC, Scaduto DI, Maldonado MR, et al. A−β− subtype of ketosis-prone diabetes is not predominantly a monogenic diabetic syndrome. Diabetes Care 2009;32:873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patel SG, Hsu JW, Jahoor F, et al. Pathogenesis of A−β+ ketosis-prone diabetes. Diabetes 2013;62:912–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernandez R, Misra R, Nalini R, Hampe CS, Ozer K, Balasubramanyam A. Characteristics of patients with ketosis-prone diabetes (KPD) presenting with acute pancreatitis: implications for the natural history and etiology of a KPD subgroup. Endocr Pract 2013;19:243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brooks-Worrell BM, Iyer D, Coraza I, et al. Islet-specific T-cell responses and proinflammatory monocytes define subtypes of autoantibody-negative ketosis-prone diabetes. Diabetes Care 2013;36:4098–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mulukutla SN, Tersey SA, Hampe CS, Mirmira RG, Balasubramanyam A. Elevated unmethylated and methylated insulin DNA are unique markers of A+β+ ketosis prone diabetes. J Diabetes Complications 2018;32:193–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mulukutla SN, Hsu JW, Gaba R, et al. Arginine metabolism is altered in A-β+ ketosis prone diabetes (KPD). J Nutr 2018;148:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gonzaga-Jauregui C, Harel T, Gambin T, et al.; Baylor-Hopkins Center for Mendelian Genomics . Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep 2015;12:1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons, 2009 [Google Scholar]
- 22. Strobl C, Malley J, Tutz G. An introduction to recursive partitioning: rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol Methods 2009;14:323–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan VY, Févotte C. Automatic relevance determination in nonnegative matrix factorization with the β-divergence. IEEE Trans Pattern Anal Mach Intell 2013;35:1592–1605 [DOI] [PubMed] [Google Scholar]
- 24. Deng P, Li T, Wang H, Horng S-J, Yu Z, Wang X. Tri-regularized nonnegative matrix tri-factorization for co-clustering. Knowl Base Syst 2021;226:107101 [Google Scholar]