Abstract
Background
Emerging evidence has revealed that tumor‐associated macrophages (TAMs) and exosomes play a crucial role in the microenvironment for tumor growth. However, the mechanisms through which exosomal miRNAs modulate TAMs and tumor development in breast cancer are not fully understood.
Methods
We constructed a macrophage model and an indirect coculture system consist of breast cancer cells and macrophages. Exosomes were isolated from BC cells culture supernatant and identified by transmission electron microscopy, Western blot and Nanosight LM10 system. The expression of miR‐148b‐3p in exosomes was determined by qRT‐PCR and the effect of exosomal miR‐148b‐3p on macrophage polarization was measured using qRT‐PCR and ELISA. The proliferation, migration and invasion of BC cells were estimated by EdU, wound healing assay and transwell assay. We employed bioinformatics, luciferase reporter assay and Western blot to identify the target gene of miR‐148b‐3p. Western blot was used to clarify the mechanism of exosomal miR‐148b‐3p mediated the crosstalk between BC cells and M2 macrophages.
Results
Cancer‐derived exosomes could induce M2 polarization of macrophages, which promoted the migration and invasion of breast cancer cells. We found that exosomal miR‐148b‐3p was overexpressed in breast cancer cell‐derived exosomes and correlated with lymph node metastasis, late tumor stage and worse prognosis. Upregulated miR‐148b‐3p expression in exosomes modulated macrophage polarization by targeting TSC2, which promoted the proliferation and might affect migration and invasion of breast cancer cells. Interestingly, we found that exosomal miR‐148b‐3p could induce M2 macrophage polarization via the TSC2/mTORC1 signaling pathway in breast cancer.
Conclusion
Overall, our study elucidated that miR‐148b‐3p could be transported by exosomes from breast cancer cells to surrounding macrophages and induced M2 polarization by targeting TSC2, providing novel insights for breast cancer therapy.
Keywords: breast cancer, exosomes, macrophage polarization, miR‐148b‐3p, TSC2
Breast cancer exosomes promote migration and invasion by inducing M2 macrophage polarization in breast cancer.
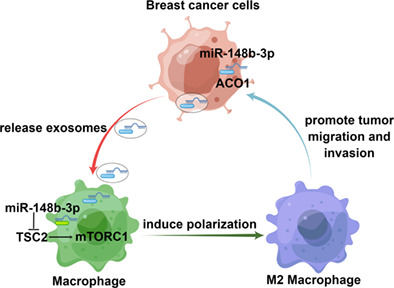
INTRODUCTION
Breast cancer (BC) is the most prevalent cancer in women and one of the leading causes of cancer deaths globally. 1 Triple‐negative breast cancer (TNBC) is the most severe molecular subtype, with a dismal five‐year overall survival rate of only 64.97%. 2 As a result, establishing efficient therapies in BC study requires a comprehensive understanding mechanisms of tumor progression.
Exosomes are 30–150 nm extracellular vesicles that can be released by several cell types and have been documented in numerous body fluids, such as blood, urine, saliva, and so on. 3 Exosomes include many signaling molecules, like proteins and nucleic acids, facilitating intercellular communication. 4 , 5 In addition, the genetic material in exosomes can be protected by cell membranes and shielded from enzyme degradation in the circulation, which is essential for transmitting important information to other cells. 6 There is an increasing consensus that exosomes play a key role in tumorigenesis and cancer progression in the tumor microenvironment and distant metastases, participating in immune escape, angiogenesis, spreading of cancer cells and invasion. 7 , 8 , 9 MicroRNAs (miRNAs) are a category of small endogenous noncoding RNAs, which can be packed into exosomes, conveying important information to neighboring or distant cells, thus subsequently modulating recipient cells. 10 As a negative regulator, miRNAs usually bind to the 3' UTR regions of target mRNAs, thus contributing to the progression of various cancers. 11 , 12 Therefore, exosomal miRNA secretion provides novel insights into communication between tumor‐associated macrophages and cancer cells. 13
Tumor‐associated macrophages (TAMs) are one type of immune cells that participate in the formation of the tumor microenvironment. It is well‐established that TAMs can promote tumor invasion and migration by immunosuppression, promoting tumor angiogenesis and remodeling the extracellular matrix. 14 Macrophages can be divided into two functional subtypes: classically activated M1 macrophages and alternatively activated M2 macrophages. 15 M1 macrophages are capable of promoting inflammation and exert antitumor effects, which are marked by cytokines expression, including TNF‐α, IL‐1β and the inducible type of nitric oxide synthase (iNOS). M2 macrophages exert anti‐inflammatory effects and promote tumorigenesis and development, featured by CD206, CD163, IL‐10 and arginase‐1 expression. 16 , 17 , 18 Meanwhile, tuberous sclerosis complex2 (TSC2) has been shown to negatively regulate the mTORC1 signaling pathway, which is widely thought to drive the polarization of M2 phenotype TAM that promotes cancer progression. 19 , 20 , 21 However, the regulation of exosomal miRNA induces the polarization of TAMs, and whether TSC2 affects the function of macrophage polarization in breast cancer, remains largely unclear.
Herein, we provide preliminary evidence that BC cell‐derived exosomal miR‐148b‐3p can stimulate cancer cell migration and invasion in vitro. In addition, miR‐148b‐3p can stimulate M2 macrophage polarization by targeting TSC2 and activating the mTORC1 pathway.
METHODS
Cell culture
Human BC cell line MDA‐MB‐231, human breast epithelial cell line MCF‐10A and human leukemic cell line THP‐1 were all acquired from American Type Culture Collection (ATCC). MDA‐MB‐231 and THP‐1 cells were cultivated in RPMI‐1640 medium containing 10% FBS. MCF‐10A cell was cultivated in MCF‐10A special medium (Procell). All cells were incubated over a humidified 5% CO2 atmosphere with 95% air at 37°C. THP‐1 cells (1 × 106) were supplemented with 100 ng/mL PMA (Sigma‐Aldrich) for 24 h to stimulate differentiation into M0 macrophages. As for exosome isolation, MDA‐MB‐231 and MCF‐10A cells were supplemented with exosome‐depleted FBS medium (SBI).
Cell coculture
THP‐1 cells (1 × 106) were supplemented with 100 ng/mL PMA for 24 h to stimulate differentiation into macrophages. PMA‐stimulated macrophages were cocultured with MDA‐MB‐231 cells‐derived exosomes or not for 24 h. Coculture of macrophages and breast cancer cells was performed in a 24‐well Boyden chamber (Corning). The upper chamber was seeded with MDA‐MB‐231 cells (5 × 104) suspended in serum‐free media, while the lower well was planted with preconditioned THP‐1 cells (1 × 104) in media containing 10% FBS at 37°C in a 5% CO2 humidified atmosphere for 24 h.
Exosome isolation and identification
Following instructions outlined in the exosome isolation and purification kit (cat. no: UR52121, Umibio), exosomes were isolated from the cell culture supernatant. The morphology of exosomes was examined by transmission electron microscope, stained with 1% uranyl‐oxalate, dried and imaged. Their size and concentration were estimated using the Nanosight LM10 system (Navato). The isolated exosomes were kept at −80°C for future studies. The obtained exosomes were verified by the expression of specific markers CD9, CD81 and TSG101 by western blotting.
Exosome labeling and tracking
After extraction and tagging with Vybrant DiL cell‐labeling solution (cat. no.: V‐22885, Molecular Probes), exosomes that had been separated from the culture medium were used according to the manufacturer's experimental protocols. THP‐1 cells (2 × 105) were supplemented with 100 ng/mL PMA for 24 h to stimulate differentiation into macrophages. Next, 20 μL tagged exosome sediment was resuspended and incubated with macrophages for the exosome uptake experiment. Cells were incubated for 30 min, 2 h, or 12 h at 37°C and were examined under fluorescence microscopy.
RNA extraction and real‐time quantitative PCR
The trizol kit was utilized to isolate total RNA (Invitrogen), and cDNA synthesis was carried out utilizing a reverse transcriptase qPCR RT kit (Toyobo) according to the manufacturer's instructions. miRNA cDNA was synthesized utilizing miRNA cDNA synthesis reagents (Cwbio). miRNA plus Poly(A) tail reaction solution was mixed and incubated at 37°C for 15 min, and the first strand of modified miRNA cDNA was synthesized at 42°C for 50 min and 85°C for 5 min. qRT‐PCR for mRNA was conducted using the SYBR Green rt‐PCR Master Mix (Toyobo), while RT‐ PCR for miRNA was performed using the miRNA qPCR assay (Cwbio). U6 and GAPDH were utilized as internalized controls for mRNA and miRNA expression data. Relative gene expressions were measured using the 2−ΔΔCt technique. 22 The primer sequences used in qPCR are displayed in Table S1.
Western blotting analysis
Total soluble proteins were extracted from cultivated cells using RIPA buffer, and the protein concentration was assessed by a BCA protein assay kit (P0012S, Beyotime Biotechnology). The proteins were boiled at 100°C for 15 min and then stored at −20°C. The proteins were separated utilizing SDS‐PAGE gel, which included 5% stacking gel and 10% or 15% separating gel, and 25 μg protein was loaded into each well of the gel. After running the electrophoresis, the gel bands were transferred to PVDF membranes. The membranes were blocked with 5% skim milk powder at room temperature for 1 h. After blocking, the membranes were incubated with various primary antibodies at 4°C overnight. The membranes were then washed three times with TBST at 90 rpm and incubated with corresponding secondary antibodies for 1 h at room temperature. After washing three times with TBST, ECL solutions were added to the membranes to observe the luminescence. Primary antibodies utilized in WB experiments were: CD9 (sc‐13 118, Santa Cruz), CD81 (sc‐166 029, Santa Cruz), CD206 (sc‐376 108, Santa Cruz), TSC2 (no. 4308, Cell Signaling Technology), p‐S6 (Ser235/236, no. 4858, Cell Signaling Technology), S6 (no. 2217, Cell Signaling Technology), TSG101 (ab125011, Abcam), ACO1 (ab183721, Abcam), β‐actin (A5441, Sigma), Histone H3 (no. 4499, Cell Signaling Technology), Alix (sc‐53 540, Santa Cruz), GAPDH (no. 5174, Cell Signaling Technology). The secondary antibodies used in this study were as follows: anti‐mouse IgG, HRP‐linked Antibody (no. 7076, Cell Signaling Technology) and anti‐rabbit IgG, HRP‐linked Antibody (no. 7074, Cell Signaling Technology).
RNA immunoprecipitation (RIP) experiment
RNA immunopreciptation (RIP) assay was performed using the Magna RIP kit (Millipore) according to the manufacturer's instructions. Cells were lysed by adding lysates containing protease inhibitors and RNase inhibitors. The lysates from cells or exosomes were saved as input. The cell extracts were incubated with RIP buffer containing protein A/G magnetic beads coated with anti‐ACO1 antibody (ab183721, Abcam) or anti‐IgG antibody (negative control) (no. 8726, Cell Signaling Technology). The RNA binding protein‐RNA complexes were then immunoprecipitated. After washing the beads with ice‐cold buffer, the RNA binding proteins were digested with proteinase K. The RNA in the coimmunoprecipitant complex was isolated using Trizol reagent, reverse transcribed and enrichment levels of miR‐148b‐3p was measured by qRT‐PCR, which was performed as described above. The enrichment levels of miR‐148b‐3p in the coimmunoprecipitant complex was presented as percent input sample (% input).
Luciferase reporter assay
The 3′UTR region (5′‐GGGGCCCUCCCUCCUGCACUGG‐3′) of TSC2 targeted by miR‐148b‐3p was predicted by TargetScan7.1 software (http://www.targetscan.org). The reporter genes containing 3'UTR terminal wildtype and mutant vector were established from GenePharma. 293 T cells were seeded in a 96‐well plate and then cotransfected with luciferase reporters and miR‐148b‐3p mimics or control as per lipofectamine 2000 instructions (Invitrogen). The cells were then grown for 48 h at 37°C. The reporter gene luciferase activities were measured upon transfection using a dual‐luciferase reporter assay system (Promega). It was normalized to Renilla luciferase activity.
Cell transfection
The Cy3 labeling miR‐148b‐3p mimics or inhibitors and the TSC2 siRNA or negative control were purchased from by RiboBio (Guangzhou). Lipofectamine 2000 (Invitrogen) was utilized to transfect macrophages with 100 nM miRNA mimics or inhibitors according to the manufacturer's instructions. The plasmid expressing TSC2 was a gift from Brendan Manning (Addgene plasmid no. 14129; http://n2t.net/addgene:14129; RRID: Addgene 14 129). 23 Cells were transfected with plasmids using lipofectamine 2000 according to the manufacturer's protocol. Transfection siRNA against human ACO1, SFRS13A and TSC2 were purchased from ThermoFisher.
Cell viability
The cell viability was determined by cell counting kit‐8 (CCK‐8) assay. Cancer cells (5 × 103) were seeded in the 96‐well plates and we waited for cell adherence. The medium was then changed to a different conditioned medium from macrophages for 1–3 days and refreshed every 2 days. When the culture was terminated, 10 μL CCK‐8 solution was added to each well for 2 h incubation at 37°C. The absorbance values were measured at 450 nm.
Cell proliferation, migration and invasion
An EdU assay kit (C10310, RiboBio) was utilized to evaluate the proliferation ability of BC cells based on the manufacturer's protocol. Briefly, cells (1 × 105) were plated into 24‐well plates and treated with 50 uM EdU solution at 37°C for 2 h. The cells were fixed with 4% paraformaldehyde for 30 min at room temperature and 50 μL of 2 mg/mL glycine for 5 min, 100 μL 0.5% TritonX‐100 for 10 min. After washing with PBS, the cells were incubated with 100 μL Apollo solution for 30 min and stained with Hoechst 33342 solution for 30 min. The images were taken using an Olympus microscope, and the positive cells were counted under dark conditions.
A wound healing assay was conducted to evaluate BC cell migration status. Cells (1 × 105) were plated into six‐well plates and allowed to reach 90% confluency. Notably, confluent cells were treated with 10 μg/mL mitomycin C for 2 h to block cell proliferation. Next, a mechanical wound was created by scratching the bottom of cells with a cell scraper (1.2 mm width). Cells were washed with PBS and the complete medium was replaced with low serum medium (FBS <2%) to continue the cell culture in order to reduce the influence of cell proliferation. The images were captured using a phase‐contrast microscope at 0, 48, and 72 h.
Transwell invasion assay was carried out to evaluate BC cell invasion utilizing 24 well‐plates and a Boyden chamber (8 μm pore size) covered with Matrigel (Corning). The upper chamber was seeded with MDA‐MB‐231 cells (5 × 104) suspended in serum‐free media, and the lower chamber was seeded with preconditioned THP‐1 cells (1 × 104) in media containing 10% FBS. After incubation at 37°C in a 5% CO2 humidified atmosphere for 24 h, the uninvaded cells were wiped off using a cotton swab from the upper surface of the chamber filter. Subsequently, the invaded cells on the bottom of the membrane were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 30 min. As for the transwell migration assay, the insert chambers were devoid of Matrigel before seeding the cells in the upper chamber. After incubation for 24 h at 37°C, migrated cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet dye. The number of migrated or invaded cells was counted under a microscope.
Bioinformatic analysis
To explore the differential miRNA expression profiles between MDA‐MB‐231‐exo and MCF‐10A‐exo, we downloaded the GEO DataSet (GSE50429) from the NCBI website (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50429). 24 Heatmaps were generated using R software. The target miRNAs of TSC2 were predicted by Targetscan7.1 software. A Venn plot was generated by Venny version 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/). Moreover, patient survival information analysis was performed using the UALCAN database, an online tool for performing gene expression profiling analyses in cancer and adjacent tissues based on TCGA. 25 Kaplan–Meier survival analysis was assessed by the online resource Kaplan–Meier plotter (http://kmplot.com/analysis/index.php?p=service&cancer=breast_mirna). 26 The RNA‐Binding Protein DataBase (RBPDB, http://rbpdb.ccbr.utoronto.ca/) was retrieved for specific interactions between miR‐148b‐3p sequence and motifs of RNA binding proteins (RBPs) with a threshold of 0.7. The correlation between TSC2 expression and macrophage cell abundance in BRCA was calculated using the TISIDB database (http://cis.hku.hk/TISIDB/). 27
Statistical analysis
The results are expressed as mean ± SD. The Student's t‐test or one‐way ANOVA test was adopted to assess the statistical significance of differences. All statistical analyses were analyzed and visualized using GraphPad Prism 8 software. A p‐value <0.05 was statistically significant.
RESULTS
Breast cancer‐derived exosomes induce polarization of M2 macrophages
After 48 h, exosomes were extracted from the MDA‐MB‐231 cell culture supernatant and were observed under electron microscopy. The isolated exosomes were spherical and had a diameter of approximately 30 to 150 nm, exhibiting typical exosome structures (Figure 1a,b). Western blotting (WB) was utilized to detect the presence of exosomal protein markers, including CD9, CD81 and tumor susceptibility gene 101 (TSG‐101), which confirmed the successful extraction of exosomes from BC cells (Figure 1c). Since it has been reported that tumor‐derived exosomes can directly affect M2 macrophage polarization, 28 we explored whether exosomes derived from BC cells account for this effect. As depicted in Figure 1d,e, we cultured human THP‐1 monocytes with phorbol 12‐myristate 13‐acetate (PMA) and THP‐1 cells were transformed into macrophages. PMA‐stimulated macrophages changed from suspended round cells to adherent and transparent cells, which is characterized by CD68 expression (Figure 1e). It has been reported that tumor cell‐derived exosomes induced macrophages to express M2 phenotype, resulting in a favorable niche for metastasis. 18 Next, the impact of BC cell‐derived exosomes on macrophage polarization was uncovered. MDA‐MB‐231 cell‐derived exosomes labeled with DiL could be internalized by the macrophages when cocultured with macrophages for 48 h, which was analyzed by immunofluorescence (Figure 1f). The expression of M2 macrophage‐related genes (CD163, CD206, IL‐10 and arginase‐1) was upregulated while M1 macrophage‐associated genes (IL‐1β, iNOS) were unaltered after coculture with MDA‐MB‐231 cell‐derived exosomes (Figure 1g). These findings suggested that BC exosomes can induce M2 macrophage polarization.
FIGURE 1.
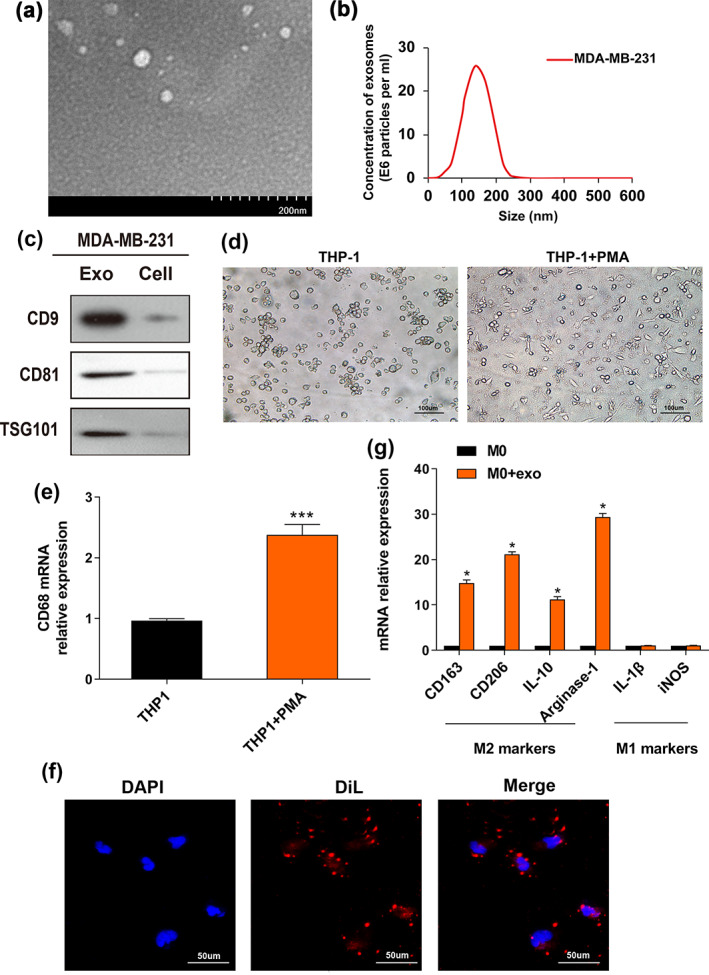
Breast cancer‐derived exosomes induce polarization of M2 macrophages. (a) Image of exosomes extracted from the conditioned medium of MDA‐MB‐231 cells was taken by electron microscopy. (b) The particle size distribution and concentration were estimated by the Nanosight LM10 system. (c) Exosomal markers (CD9, CD81 and TSG101) were evaluated by western blot (WB) assay in MDA‐MB‐231‐derived exosomes and cells. (d) THP‐1 cells were pretreated with PMA (100 ng/mL) for 24 h to generate M0 macrophages. Representative images of the two groups (THP‐1, THP‐1 + PMA) are shown. (e) qRT‐PCR was utilized to reveal macrophage marker CD68 expression. (f) Representative immunofluorescence images indicate the process of DiL‐labeled‐exosomes isolated from MDA‐MB‐231 internalized by macrophages. Scale bar, 50 μm. Original magnification ×200. (g) The M0 macrophages were incubated with PBS (control) or MDA‐MB‐231‐derived exosomes. qRT‐PCR was used to evaluate M2 markers (CD163, CD206, IL‐10, arginase‐1) and M1 markers (IL‐1β, iNOS) expression levels. The Student's t‐test was adopted to analyze the statistical significance of the difference between two groups, and one‐way ANOVA test was utilized to analyze multiple groups (*p < 0.05, **p < 0.01, ***p < 0.001).
Breast cancer‐derived exosomes promote breast cancer cell migration and invasion depending on M2 macrophage polarization
An increasing body of evidence suggests that the tumor‐associated M2 macrophage contributes to tumor growth and promotes tumor progression. 29 , 30 , 31 In this study, MDA‐MB‐231 cells were cultured with macrophages treated with cancer‐derived exosomes using the indirect coculture system in vitro (Figure 2a). We used the CCK‐8 assay to evaluate the cell viability. The results showed that macrophages pretreated with MDA‐MB‐231‐exo promoted cell viability of MDA‐MB‐231 cells compared to the control group at 48–72 h (Figure S1). ELISA assay was used to assess IL‐10 and TNF‐α secretion in macrophage supernatants (Figure 2b). Compared to the PBS group, the MDA‐MB‐231‐exo group displayed significantly increased IL‐10 secretion in macrophages and decreased TNF‐α secretion. BC migration and invasion were estimated by transwell assay, suggesting that MDA‐MB‐231‐exo treated macrophages significantly increased the migration and invasion in BC cells (Figure 2c,d). Collectively, these functional assays demonstrated that BC cell‐derived exosomes could trigger M2 macrophages polarization and promote BC cell migration and invasion in vitro.
FIGURE 2.
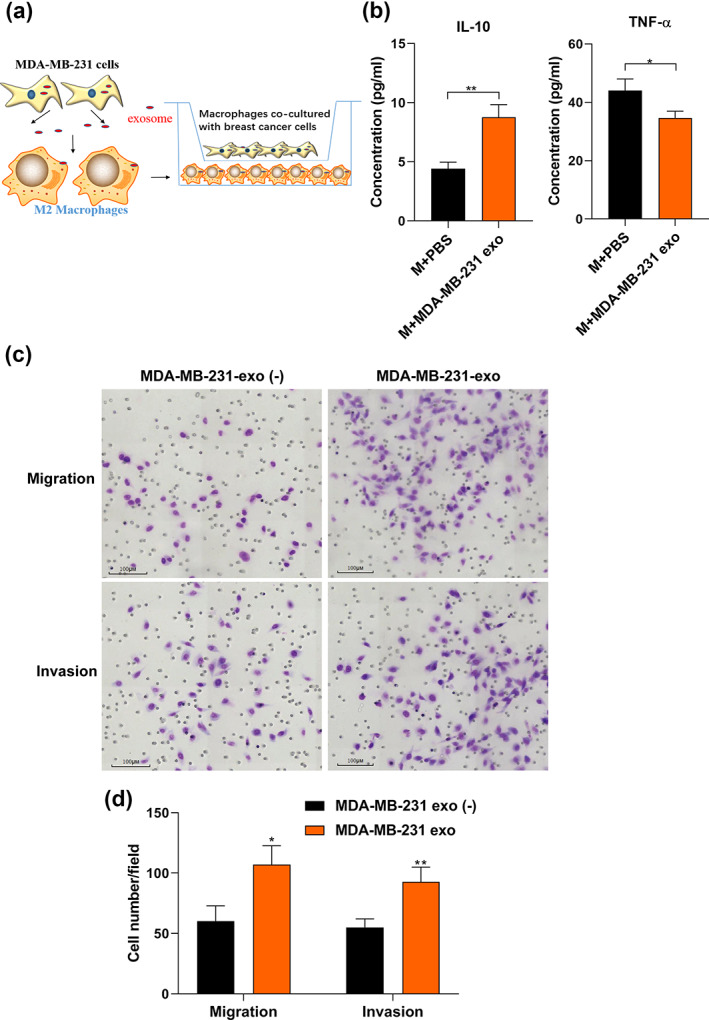
Breast cancer exosomes promote migration and invasion by inducing M2 macrophage polarization in breast cancer. (a) Schematic diagram of the indirect coculture system in vitro. (b) The M0 macrophages were incubated with PBS (control) or MDA‐MB‐231‐derived exosomes. After 72 h, supernatants were harvested to detect the release of IL‐10 and TNF‐α via ELISA. “M + MDA‐MB‐231 exo” refers to “macrophages treated with MDA‐MB‐231 exo”. (c) Migration and invasion abilities of breast cancer cells cocultured with macrophages treated with or without exosomes were estimated by transwell assay. Illustrative images of migrated or invasive cells are shown. Scale bar, 50 μm. Original magnification ×200. (d) The number of invaded or migrated cells on the membrane coated with or without Matrigel was counted. The Student's t‐test was adopted to analyze the statistical significance of the difference between two groups, and one‐way ANOVA test was utilized to analyze multiple groups (*p < 0.05, **p < 0.01).
miR‐148b‐3p is highly expressed in breast cancer cell‐derived exosomes and can be encapsulated in exosomes
Current evidence suggests that exosomal miRNAs mediate the intercellular connection between tumor cells and their microenvironments. 32 The tuberous sclerosis complex (TSC) gene encodes tumor suppressors TSC1 and TSC2, which can work synergistically to reduce TORC1 activity. 33 Previous studies have reported that the tuberous sclerosis complex2 (TSC2) is an mTOR upstream negative regulator, and mTORC1 could contribute to regulating macrophage polarization by inactivating TSC2. 19 , 34 , 35 , 36 Therefore, we assessed the significance of TSC2 in macrophage polarization. The differential analysis between BC cell‐derived exosomes (MDA‐MB‐231‐exo) and normal mammary epithelial cell‐derived exosomes (MCF‐10A‐exo) allowed the identification of dysregulated miRNAs based on the GEO Dataset (GSE50429). A heatmap was generated to visualize the distinct expression profiles of these miRNAs (Figure 3a). In total, 101 upregulated and 84 downregulated miRNAs were screened using the threshold of FDR corrected p‐value <0.05. The target miRNAs of TSC2 were predicted by Targetscan 7.1 software. After analysis by Venny V.2.1 (http://bioinfogp.cnb.csic.es/tools/venny/), four potential miRNAs were obtained (Figure 3b), and their expression in MDA‐MB‐231‐exo and MCF‐10A‐exo was validated by qRT‐PCR. It was discovered that miR‐148b‐3p expression in MDA‐MB‐231‐exo was the most upregulated miRNA compared to MCF‐10A‐exo (p < 0.05) (Figure 3c). According to TCGA dataset from UALCAN, miR‐148b‐3p expression was higher in BC relative to their corresponding nontumor tissues (Figure S2). 25 Moreover, the correlation between miR‐148b‐3p expression level and lymph node metastasis or tumor stage was statistically significant (Figure 3d,e). Most importantly, Kaplan–Meier survival analysis revealed significantly poorer overall survival (OS) in BC patients with miR‐148b overexpression (n = 1062, p = 0.0017) (Figure 3f). 26 Furthermore, miR‐148b‐3p expression treated with RNase A in conditioned medium (CM) of MDA‐MB‐231 cells remained unchanged, unlike with RNase A plus Triton X‐100 treatment (Figure 3g), imposing miR‐148b‐3p presence was encapsulated in a double‐layer membrane and could not be directly secreted. Further observation revealed that miR‐148b‐3p expression in MDA‐MB‐231 cell‐derived exosomes and CM was comparable, while the treatments with GW4869 (an exosome secretion inhibitor) or exosome‐depleted CM (ultracentrifugation depletion) could significantly reduce miR‐148b‐3p (Figure 3h,i). These results showed that the miR‐148b‐3p was significantly upregulated among BC cell‐derived exosomes and was encapsulated in BC cell‐derived exosomes.
FIGURE 3.
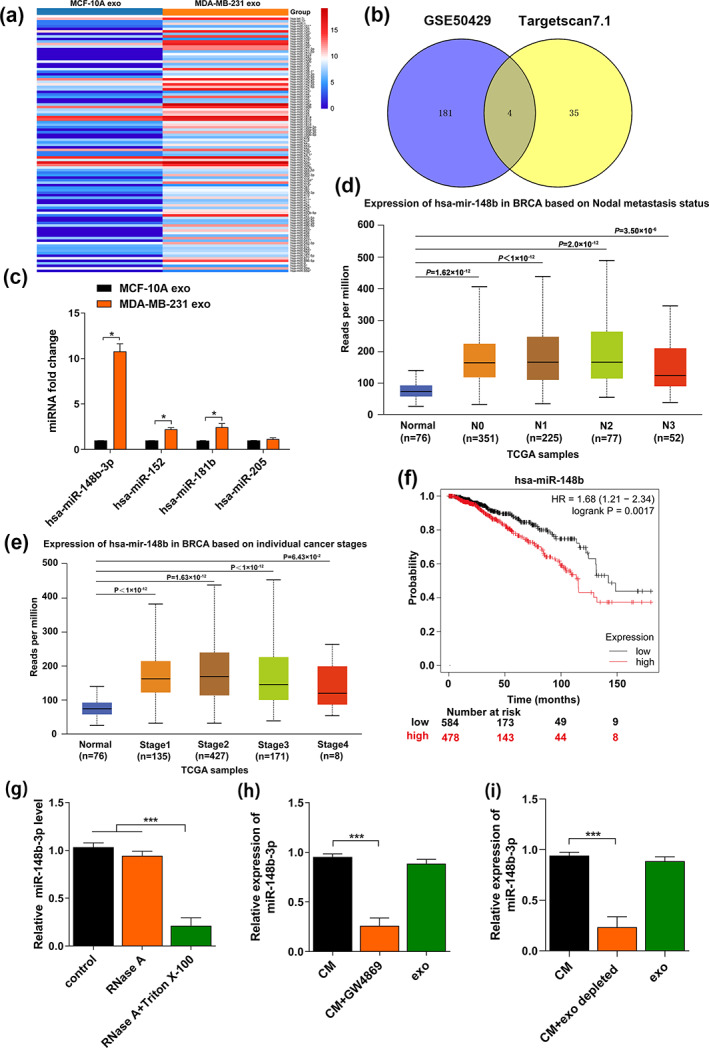
miR‐148b‐3p is upregulated in breast cancer cell‐derived exosomes. (a) Heatmap of differential miRNA expression between MDA‐MB‐231‐exo and MCF‐10A‐exo based on GEO Dataset (GSE50429, FDR corrected p‐value <0.05). (b) Venn diagram of upregulated miRNAs from GSE50429 and the target miRNAs of TSC2 predicted by Targetscan 7.1 software, which was calculated using Venny 2.1 software. (c) Expression levels of four potential upregulated miRNAs in exosomes derived from MDA‐MB‐231 were evaluated by qRT‐PCR. (d, e) Correlations between miR‐148b‐3p expression levels and lymph node metastasis and tumor stage in primary BRCA samples based on TCGA dataset from UALCAN. (f) The Kaplan–Meier survival curves of BC patients are shown according to the expression levels of serum exosomal miR‐148b‐3p (p = 0.0017 by log‐rank analysis). (g) qRT‐PCR was utilized to evaluate the miR‐148b‐3p expression in the conditioned medium of MDA‐MB‐231 cells pretreated with control medium or RNase A (2 mg/mL) alone or in combination with Triton X‐100 (0.1%) for 0.5 h. (h, i) qRT‐PCR was utilized to detect miR‐148b‐3p expression in the conditioned medium of MDA‐MB‐231 cells pretreated with GW4869 or depleted exosomes by ultracentrifugation. The Student's t‐test was adopted to analyze the statistical significance of the difference between two groups, and one‐way ANOVA test was utilized to analyze multiple groups (*p < 0.05, **p < 0.01, ***p < 0.001).
Overexpression of miR‐148b‐3p in exosomes modulates macrophage polarization and stimulates proliferation, migration and invasion of BC cells
It has been reported that miR‐148b‐3p could promote bladder and prostate cancer progression, while inhibiting gastric cancer, lung adenocarcinoma and retinoblastoma. 37 , 38 , 39 , 40 , 41 Nonetheless, the exact role of miR‐148b‐3p in BC remains obscure. We next examined the relationship between miR‐148b‐3p function and macrophage polarization. Macrophages were transfected with either a miR‐148b‐3p mimic, miR‐148b‐3p inhibitor, or miR‐148b‐3p‐nc. After 48 h, the stable phenotype was determined by qRT‐PCR (Figure S3). Interestingly, we found that the miR‐148b‐3p mimic upregulated M2 macrophage‐related genes, including CD163, CD206, IL‐10, and arginase‐1, while downregulating M1 macrophage‐related genes, like IL‐1β and iNOS. At the same time, the miR‐148b‐3p inhibitor yielded the opposite effect (Figure 4a). Furthermore, the ELISA assay indicated that miR‐148b‐3p mimics increased IL‐10 secretion and decreased TNF‐α secretion, while miR‐148b‐3p inhibitor yielded the opposite effect (Figure 4b). We further investigated the impact of M2 macrophages transfected with miR‐148b‐3p mimic on MDA‐MB‐231 cell proliferation, migration and invasion utilizing a coculture approach in vitro. The CCK‐8 assay showed that macrophages transfected with miR‐148b‐3p mimic promoted the viability of MDA‐MB‐231 cells compared to the miR‐nc group at 48‐72 h (Figure S4). Notably, we used mitomycin C to suppress cell proliferation before conducting the would‐healing assay. It was found that overexpression of miR‐148b‐3p increased cell migration at 48 h, and the scratched blank area healed faster than the control group (Figure 4c). In addition, transwell assays revealed that miR‐148b‐3p mimics significantly stimulated MDA‐MB‐231 cell migration and invasion capability (p < 0.05) (Figure 4d,e). Additionally, an EdU assay was performed to evaluate the proliferation of MDA‐MB‐231 cells. MDA‐MB‐231 cells were cocultured with the conditioned medium from macrophages for 48 h, and the results demonstrated that EdU‐positive cells were dramatically increased among miR‐148b‐3p‐overexpressing macrophages (Figure 4f,g). Overall, these results suggest that miR‐148b‐3p was involved in M2 macrophage polarization, which facilitated BC cell proliferation, and may affect migration and invasion.
FIGURE 4.
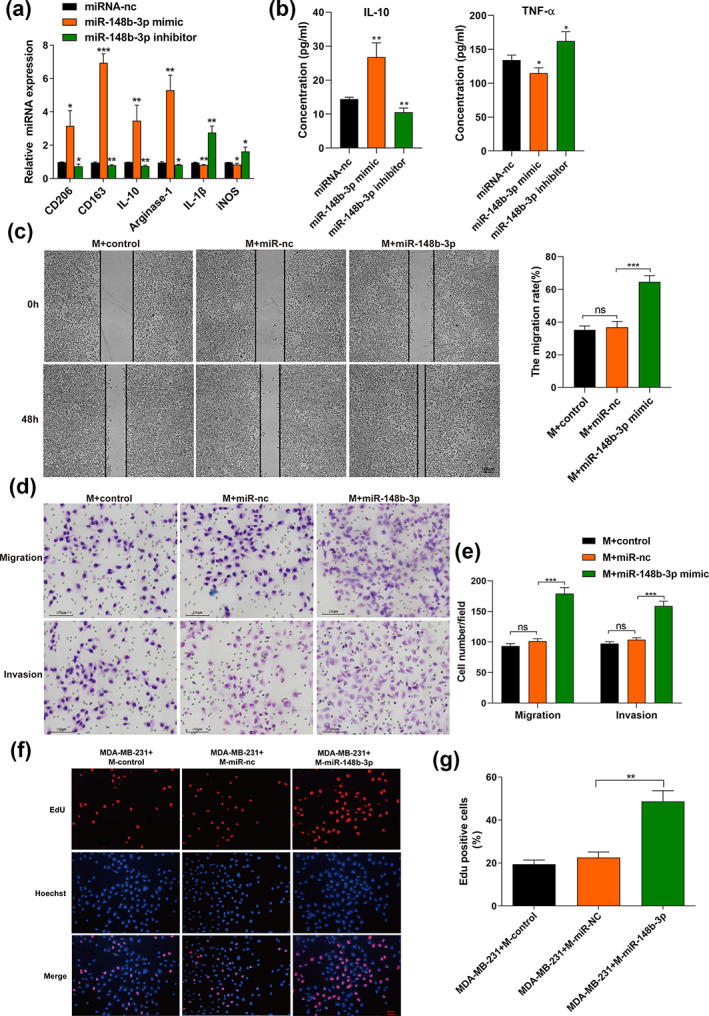
The overexpression of exosomal miR‐148b‐3p modulates macrophage polarization and promotes proliferation, migration and invasion of breast cancer cells. (a) Macrophages were transfected with miRNA‐nc, miR‐148b‐3p mimic or miR‐148b‐3p inhibitor. After 48 h, qRT‐PCR was applied to detect the expression of M2 markers (CD163, CD206, IL‐10, arginase‐1) and M1 markers (IL‐1β, iNOS). (b) The identical method used in (a) was applied to macrophages. The different conditioned macrophage supernatants were harvested to detect the release of IL‐10 and TNF‐α by ELISA. (c) Scratch experiment. Notably, we used mitomycin C (10 μg/mL) to treat MDA‐MB‐231 cells for 2 h before conducting the scratch assay. After scratching, the cells were washed with PBS and replaced with conditioned medium from macrophages transfected with miR‐nc, miR‐148b‐3p mimic or not. Images were taken at 0 and 48 h (scale bar, 100 μm). Original magnification ×100. “M+ miR‐148b‐3p” refers to “macrophages treated with miR‐148b‐3p mimic”. (d, e) Transwell assay was performed to evaluate the migration and invasion of MDA‐MB‐231 cells. Macrophages transfected with miR‐nc, miR‐148b‐3p mimic and then cultured with MDA‐MB‐231 cells in the indirect coculture system in vitro. The control group was not transfected with any exogenous miRNA. After incubation for 24 h, MDA‐MB‐231 cells were detached from the upper chamber, and the migration and invasion ability of MDA‐MB‐231 cells was detected. Illustrative images of migratory or invasive cells and quantifications are shown. Scale bar, 100 μm. Original magnification ×100. (f, g) EdU assay was performed to evaluate the proliferation of MDA‐MB‐231 cells. MDA‐MB‐231 cells were cocultured with conditioned macrophages as described in (a) for 48 h, and the positive cell ratio is shown. Scale bar, 50 μm. Original magnification ×200. The student's t‐test was adopted to analyze the statistical significance of the difference between two groups, and one‐way ANOVA test was utilized to analyze multiple groups (*p < 0.05, **p < 0.01, ***p < 0.001).
ACO1 mediates the transportation of exosomal miR‐148b‐3p to macrophages
Recent research has revealed that exosomal RNAs secretion is highly selective among different cell types requiring a specific RNA‐binding protein for transportation. 42 We next sought to investigate whether specific regulators mediated the transport of exosomal miR‐148b‐3p to macrophages. We retrieved the RBPDB database for specific interactions between miR‐148b‐3p sequence and motifs of RNA binding proteins (RBPs) with a threshold of 0.7. The results showed that aconitase 1 (ACO1) and serine and arginine rich splicing factor 10 (SFRS13A, also named SRSF10) motifs had putative miR‐148b‐3p binding sites (Figure 5a). To validate the targeting interactions between miR‐148b‐3p and their binding proteins, we knocked down ACO1 and SFRS13A with specific siRNAs in MDA‐MB‐231 cells (Figure S5, S6). The results revealed that ACO1 suppression significantly inhibited exosomal miR‐148b‐3p expression, whereas cellular miR‐148b‐3p level was generally unaffected (Figure 5b,c), suggesting that exosomal miR‐148b‐3p is regulated by ACO1 to some extent. ACO1 is a cytosolic, RNA‐binding protein that participates in the transportation and translation of mRNAs encoding proteins. 43 Furthermore, the miRNA pulldown assay showed that exosome marker Alix and cytoplasmic marker β‐actin were expressed, but not the nuclear marker Histone H3. These findings corroborated that the interactions between ACO1 and miR‐148b‐3p occurred in the cytoplasm and exosomes. Nevertheless, this binding ability might be compromised by modifying the miR‐148b‐3p sequence (CAGUGC) (Figure 5d). Additionally, RNA immunoprecipitation (RIP) experiments revealed miR‐148b‐3p was significantly enriched in anti‐ACO1 antibody group relative to the anti‐IgG group both in BC cells and their exosome lysates (p < 0.05) (Figure 5e). Overall, we provided preliminary evidence that ACO1 could assist in packaging miR‐148b‐3p into exosomes of BC cells by binding a specific motif (CAGUGC) of miR‐148b‐3p.
FIGURE 5.
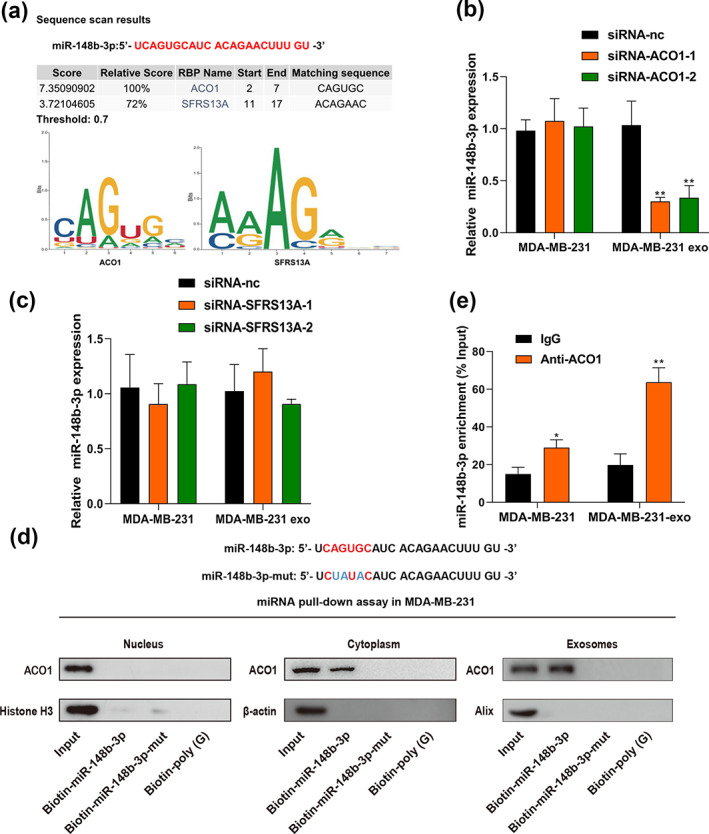
ACO1 is a mediator of exosomal miR‐148b‐3p delivery to macrophages. (a) RBPDB analysis was utilized to anticipate precise interaction between the miR‐148b‐3p sequence and RBP motifs (threshold 0.7). (b, c) ACO1 and SFRS13A were knocked down with specific siRNAs in MDA‐MB‐231 cells and exosomes respectively, and qRT‐PCR was used to evaluate miR‐148b‐3p expression level in cells and exosomes. (d) The miRNA pulldown assay of interactions between miR‐148b‐3p or mutated miR‐148b‐3p and ACO1 in the nucleus, cytoplasm and exosomes. Biotin‐poly (G) was used for negative control. (e) Anti‐ACO1 antibody and IgG as a negative control were used for the RIP assay. qRT‐PCR was used to detect miR‐148b‐3p levels in lysates from cells or exosomes of MDA‐MB‐231. The lysates from cells or exosomes were saved as input. The enrichment levels of miR‐148b‐3p in the coimmunoprecipitant complex are presented as percent input sample (% input). The Student's t‐test was adopted to analyze the statistical significance of the difference between two groups, and one‐way ANOVA test was utilized to analyze multiple groups (*p < 0.05, **p < 0.01).
TSC2 is the direct target of miR‐148b‐3p in macrophages
As an mTOR upstream negative regulator, TSC2 has been reported to be associated with macrophage polarization. 21 Based on the Targetscan7.1 software, the target miRNAs of TSC2 includes miR‐148b‐3p. Given that miRNAs can inversely regulate the expression of their target mRNAs, TSC2 was hypothesized to be a direct target of miR‐148b‐3p and responsible for M2 macrophage polarization. qRT‐PCR and WB assays were conducted to assess TSC2 expression throughout the macrophage‐like cell line THP‐1 following supplementation with ectopic miR‐148b‐3p mimic and inhibitor (Figure 6a,b). Results showed that TSC2 in macrophages was significantly downregulated by miR‐148b‐3p overexpression at both mRNA and protein levels. In contrast, the opposite results were observed when miR‐148b‐3p was inhibited. To further investigate the effects of miR‐148b‐3p on TSC2 expression, a mutant plasmid for upregulating TSC2 expression was transfected into macrophages. Likewise, TSC2 overexpression could be partially inhibited by miR‐148b‐3p mimic (Figure 6c,d). The luciferase reporter verified that the miR‐148b‐3p potential binding site across the 3'UTR sequence of TSC2 corresponded to the prediction made by the online bioinformatics tool TargetScan Human 7.2 database (Figure 6e,f). We further assessed the relationship between TSC2 expression and macrophage cell abundance in BRCA by the TISIDB database. In 1100 BRCA samples, TSC2 was negatively correlated with infiltration of macrophages (r = −0.249, p = 6.7e‐17, Figure S7). These data suggested that miR‐148b‐3p could specifically target TSC2 and suppress TSC2 expression at both protein and mRNA levels via binding to its 3'UTR region.
FIGURE 6.
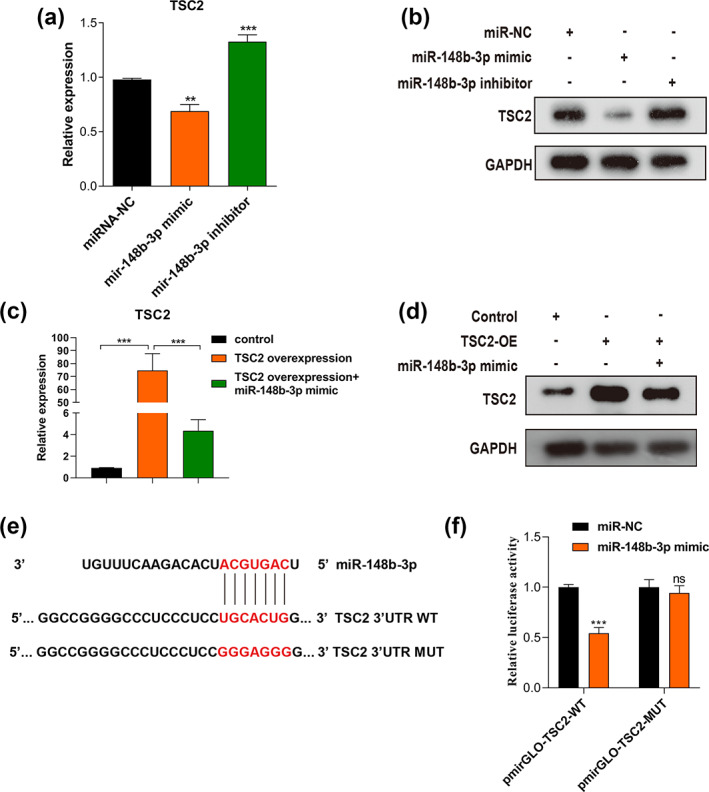
TSC2 is the direct target of miR‐148b‐3p in macrophages. (a, b) qRT‐PCR and WB assays were carried out to evaluate TSC2 expression in macrophages after incubated with ectopic miR‐148b‐3p mimic or inhibitor. (c, d) qRT‐PCR and western blot assays were done to determine TSC2 expression in macrophages after incubated with TSC2‐overexpression plasmids. “TSC2‐OE” refers to “macrophages transfected with TSC2‐overexpression plasmids”. (e) Sequencing of the wild‐type and mutant target sites in 3′UTR of TSC2 mRNA. (f) Luciferase reporter activity was measured to verify the target association between miR‐148b‐3p and TSC2. The student's t‐test was adopted to analyze the statistical significance of the difference between two groups, and one‐way ANOVA test was utilized to analyze multiple groups (*p < 0.05, **p < 0.01, ***p < 0.001).
Exosomal miR‐148b‐3p regulates macrophage polarization by inhibiting TSC2 and promotes breast cancer by activating the mTORC1 signaling pathway
To explore the mechanisms associated with TSC2 in macrophage polarization, small interfering RNAs were transfected into macrophages to downregulate TSC2 expression (si‐TSC2). As shown in Figure 7a, knockdown of TSC2 significantly enhanced the expression of M2 macrophage markers (CD163, IL‐10 and arginase‐1) (Figure 7a). Consistent results were observed during ELISA assay, knockdown of TSC2 significantly increased IL‐10 secretion and decreased TNF‐α secretion (Figure 7b). Overall, our findings revealed that miR‐148b‐3p induces polarization of M2 macrophages by inhibiting TSC2. Next, we explored whether knocking down TSC2 in macrophages could enhance BC progression in the coculture system. When we treated the macrophages with si‐TSC2, BC proliferation was enhanced using the EdU assay (Figure 7c,d). Similarly, the transwell assay showed that TSC2 knockdown could significantly enhance BC migration and invasion after treated with si‐TSC2 in the coculture system (Figure 7e,f). Taken together, these results suggested that knockdown of TSC2 in macrophages was significantly associated with BC proliferation, migration and invasion.
FIGURE 7.
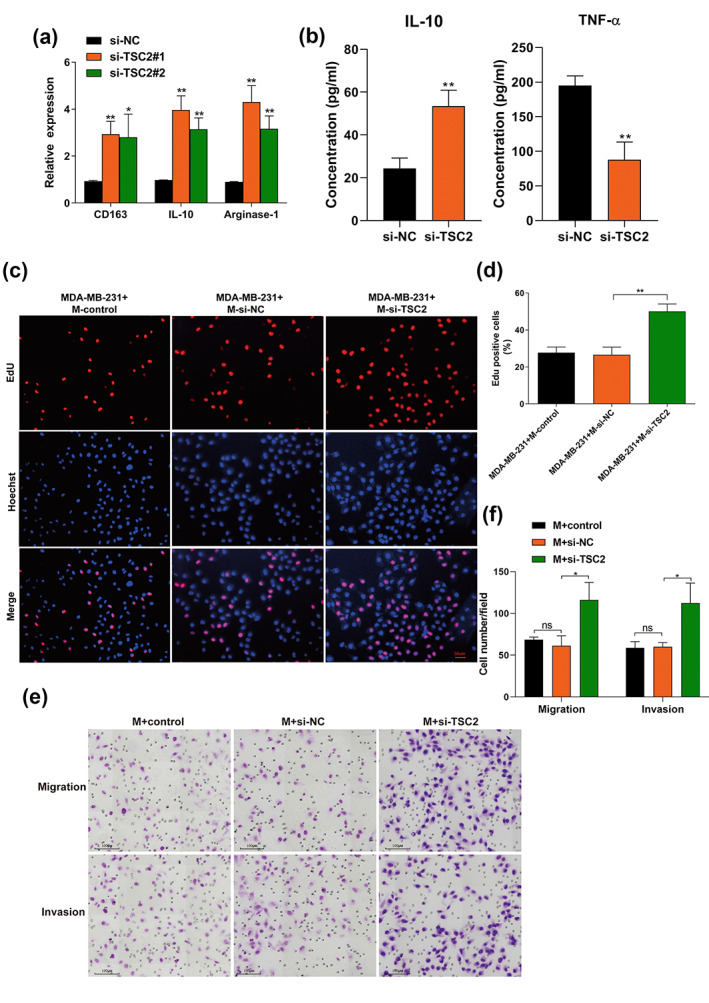
Exosomal miR‐148b‐3p regulates macrophage polarization by inhibiting TSC2, thus promoting breast cancer migration and invasion. (a) qRT‐PCR was used to evaluate M2 markers (CD163, IL‐10 and arginase‐1) expression in macrophages transfected with small interfering RNAs (si‐TSC2) for downregulating TSC2 expression. “M + si‐TSC2” refers to “macrophages transfected with si‐TSC2”. (b) The identical method used in (a) was applied to macrophages. The different conditioned macrophage supernatants were harvested to detect the release of IL‐10 and TNF‐α by ELISA. (c, d) The identical method used in (a) was applied to macrophages, and an EdU assay was performed to evaluate the proliferation of MDA‐MB‐231 cells. MDA‐MB‐231 cells were cocultured with the conditioned medium from macrophages for 48 h, and the positive cell ratio is shown. Scale bar, 50 μm. Original magnification ×200. (e, f) The identical method used in A was applied to macrophages, which were subsequently cultured with MDA‐MB‐231 cells in the indirect coculture system in vitro. After incubation for 24 h, MDA‐MB‐231 cells were detached from the upper chamber, and the migration and invasion ability of MDA‐MB‐231 cells was detected by transwell aasay. Representative pictures of migratory or invasive cells and quantifications are shown. Scale bar, 100 μm. Original magnification ×100. The Student's t‐test was adopted to analyze the statistical significance of the difference between two groups, and one‐way ANOVA test was utilized to analyze multiple groups (*p < 0.05, **p < 0.01).
An increasing body of evidence suggests that TSC2 is an mTOR‐negative upstreaming regulator, and mTORC1 may be crucial in regulating macrophage polarization by the inactivation of TSC2. 19 , 34 , 35 , 36 Thus, we hypothesized that exosomal miR‐148b‐3p could induce M2 macrophage polarization through the TSC2/mTORC1 signaling pathway. Considering that mTORC1 activity in macrophages can be determined through S6 ribosomal protein phosphorylation (p‐S6), 20 mTORC1 activity was evaluated under different macrophage‐polarizing conditions. We found that BC cell‐derived exosomes upregulated the expression of phosphorylated S6 and M2‐related gene CD206 (Figure 8a). In addition, miR‐148b‐3p overexpression or TSC2 knockdown enhanced phosphorylated S6 and CD206 expression while miR‐148b‐3p inhibitor exhibited the opposite effect (Figure 8b,c). Meanwhile, TSC2 overexpression suppressed phosphorylated S6 and CD206 expression. In particular, WB assay indicated that TSC2 overexpression could be partially mitigated by overexpression of miR‐148b‐3p (Figure 8d). Taken together, our data indicated that BC cell‐derived exosomal miR‐148b‐3p could induce M2 macrophage polarization by targeting TSC2‐mTORC1 signaling pathway in macrophages (Figure 8e).
FIGURE 8.
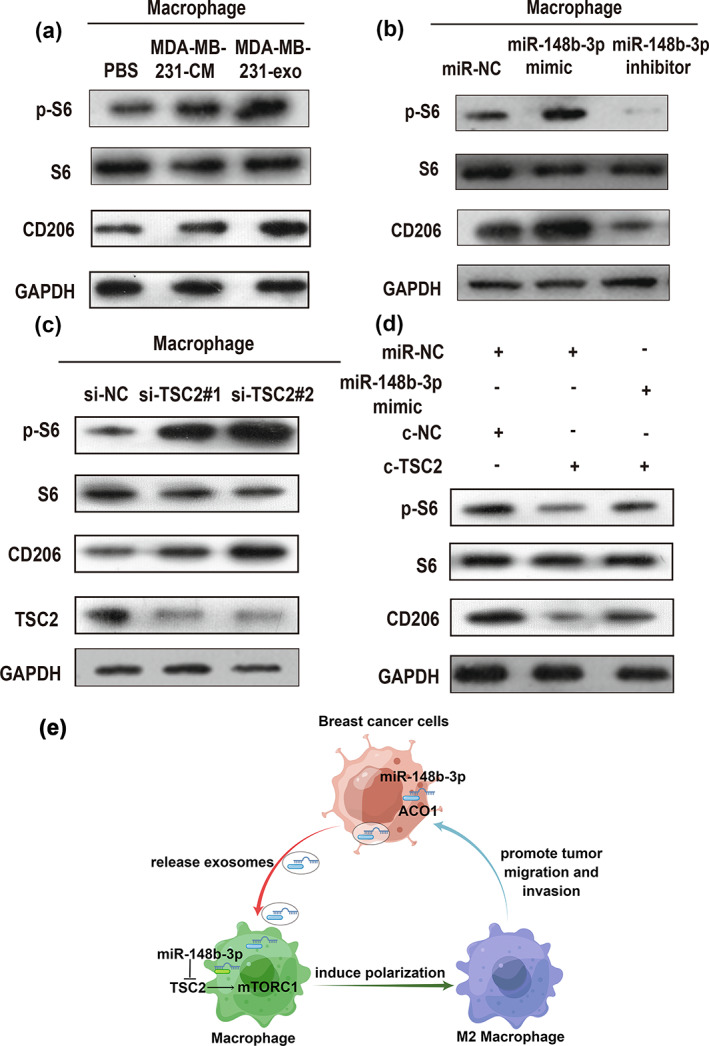
Exosomal miR‐148b‐3p regulates macrophage polarization by inhibiting TSC2 and activating the mTORC1 signaling pathway. (a) Western blot (WB) analysis of mTORC1 activity markers (p‐S6, S6) and M2‐related gene CD206 expression in M0 macrophages treated with PBS, MDA‐MB‐231‐CM, MDA‐MB‐231‐exo or (b) miR‐NC, miR‐148b‐3p mimic, miR‐148b‐3p inhibitor. (c) WB assay of mTORC1 activity markers (p‐S6, S6), TSC2 and M2‐related gene CD206 expression in M0 macrophages treated with si‐NC as control, si‐TSC2#1 and si‐TSC2#2 for downregulating TSC2 expression. (d) WB analysis of mTORC1 activity markers (p‐S6, S6) and M2‐related gene CD206 expression in M0 macrophages treated with the indicated treatment. Each experiment was conducted at least three times. (e) Schematic diagram of exosomal miR‐148b‐3p promoted M2 macrophage polarization via the TSC2/mTORC1 signaling pathway was drawn using Figdraw.
DISCUSSION
Triple‐negative breast cancer is well‐established as the most aggressive molecular subtype of BC. It is widely thought that the interactions between BC cells and the microenvironment could reprogram macrophages into different phenotypes, enabling tumor cells to maintain tumor progression and distant metastasis. 44 In addition, exosomes play a crucial role in activating M2‐type macrophages, which are well‐recognized as important participants in cancer progression. 28 In this study, we discovered that cancer‐derived exosomes induced M2 macrophage polarization, which promote the migration and invasion of breast cancer cells. Interestingly, exosomal miR‐148b‐3p was overexpressed in breast cancer cell‐derived exosomes and was positively correlated with lymph node metastasis, late tumor stage and worse prognosis, which suggested that exosomal miR‐148b‐3p may function as an oncogene in BC and might be harnessed as a biomarker in the future.
Exosomes are microscopic vesicles released by numerous cell types, especially tumor cells, which have been documented in cancer patients' blood. 3 Emerging evidence suggests that exosomes can act as transportation cargos, facilitating the connection between cells and allowing tumor cells to send signals to surrounding stromal cells, which promotes carcinogenesis. 8 , 9 Our results showed BC cell‐derived exosomes could be internalized by macrophages when cocultured with macrophages, thus stimulating M2 macrophage polarization and further inducing BC cell migration and invasion in vitro. Our results verified that the exosomes act as an important messenger between tumor cells and the microenvironment.
MicroRNAs are an endogenous type of short noncoding RNAs found in exosomes and can be taken by neighboring or distant cells, thereby regulating recipient cells. 10 As a negative regulator, miRNAs usually bind to 3'UTR regions of target mRNAs, thus contributing to the progression of various cancers. 11 , 12 It has been reported that miR‐148b‐3p could promote tumor progression in bladder cancer, prostate cancer and inhibit gastric cancer, lung adenocarcinoma and retinoblastoma. 37 , 38 , 39 , 40 , 41 In early stages of BC, plasma expression level of miR‐148b‐3p was significantly higher in breast cancer cases than the healthy group, and miR‐148b was secreted from breast cancer cell lines. 45 In an in vitro experiment, miR‐148b‐3p mimics significantly promoted the proliferation and migration of breast cancer cell lines, 46 , 47 , 48 suggesting the tumor‐promoting role of miR‐148b‐3p in breast cancer. In contrast, miR‐148b‐3p could increase adriamycin sensitivity by directly targeting SPIN1 in breast cancer cells in vitro. 49 In addition, human umbilical cord mesenchymal stem cells (HUCMSCs) derived exosomal miR‐148b‐3p could inhibit cell proliferation, invasion and migration in breast cancer by downregulating TRIM59. 50 Therefore, the impact of miR‐148b‐3p on BC cells remains unclear. Our study aimed to explore the role of exosomal miR‐148b‐3p from BC cells on macrophage polarization. Additionally, we demonstrated that miR‐148b‐3p was highly concentrated in BC cell exosomes and could be transported from BC cells to macrophages by binding particular regulator ACO1, thus inducing M2 polarization. Subsequently, polarized M2 macrophages could influence the biological behaviors of MDA‐MB‐231 cells. miR‐148b‐3p overexpressed in exosomes induced BC cell proliferation and may affect migration and invasion. Furthermore, miR‐148b‐3p overexpression was significantly related to a worse prognosis in BC patients. These results suggested that exosomal miR‐148b‐3p may play an important role in tumor cells and the microenvironment.
Tuberous sclerosis complex 2 (TSC2) is a tumor suppressor gene that is mutated in the tuberous sclerosis complex (TSC) tumor syndrome and is associated with TSC1 forming a functional complex to reduce TORC1 activity specifically. 33 There are several studies available substantiating that TSC2 is an mTOR upstream negative regulator, and mTORC1 could play an important function in regulating macrophage polarization by the inactivation of TSC2. 19 , 34 , 35 , 36 Although Chen et al. confirmed that the TSC2‐mTOR pathway could induce monocyte differentiation into M2 macrophages, contributing to tumor angiogenesis, 21 its specific roles in breast cancer remain largely unknown. Our study corroborated that TSC2 expression was decreased by miR‐148b‐3p overexpression at both mRNA and protein levels in macrophages. Utilizing bioinformatic analysis and luciferase reporter, TSC2 was found to be a miR‐148b‐3p target gene. Moreover, high expression of TSC2 was positively correlated with overall survival in BC patients and negatively with HER2‐positive and TNBC subclasses of BRCA samples (Figure S8). Additionally, we revealed that miR‐148b‐3p could induce macrophage polarization by suppressing TSC2 and stimulating the mTORC1 signaling pathway, thereby facilitating BC progression. Moreover, we demonstrated that TSC2 knockdown promoted M2 macrophage polarization. Accordingly, knockdown of TSC2 could promote tumor cell proliferation, invasion, and migration abilities. Since phosphorylation of S6 ribosomal protein (p‐S6) is a mTORC1 activation hallmark, 20 mTORC1 activity was evaluated in different macrophage‐polarizing conditions. We uncovered that exosomes derived from BC cells promoted p‐S6 and M2‐related gene CD206 expression. miR‐148b‐3p overexpression or TSC2 knockdown also enhanced p‐S6 and CD206 expression whereas miR‐148b‐3p inhibitor yielded the opposite effect. Our findings validated that exosomal miR‐148b‐3p could induce M2 macrophage polarization by targeting the TSC2‐mTORC1 signaling pathway in macrophages. These findings lay the groundwork for future research on breast cancer therapy. However, a limitation of the present study is that we did not conduct more experiments to validate the function of this miRNA in vivo and in clinical samples. Therefore, further studies are required to elucidate the effect of this miRNA on the progression of breast cancer.
When comparing our results with previous reports, there are similarities as well as distinct differences. Multiple studies have shown that exosomal miRNAs play direct and indirect roles between breast cancer cells and TAMs. Overexpression of miR‐130 and miR‐33 in exosomes of MDA‐MB‐231 cells increased the expression of M1 macrophages related genes. The conditioned medium from miRNA mimic‐treated macrophage was coincubated with tumor cells, which reduced the invasion and migration ability of tumor cells. It is suggested that upregulated miR‐130 and miR‐33 in breast cancer cells can transform macrophage polarization from M2 type to M1 type, which is crucial for inhibiting the occurrence and metastasis of breast cancer. 51 Our study found that exogenic miR‐148b‐3p can induce M2‐type polarization of macrophages and enhance the invasion and migration ability of breast cancer. Studies have found that TAMs promote the invasion and metastasis of breast cancer possibly through macrophage‐secreted exosomes, which could deliver invasive miRNAs to breast cancer cells. They cocultured TAMs with breast cancer cells and found that miR‐233 was elevated in exosomes released by macrophages, and the expression of this miRNA was also significantly upregulated in cocultured MDA‐MB‐231 cells. After inhibiting miR‐233 expression in TAMs, the invasiveness of cocultured breast cancer cells was also reduced. It is suggested that macrophages enhance the invasive ability of breast cancer through miR‐233, 52 whereas we focused on the effect of exosomes secreted from breast cancer cells on tumor microenvironment, especially TAMs. The breast cancer‐derived exosomes could induce the polarization of TAMs and enhance tumor invasion and migration ability. Moreover, miR‐183‐5p can be transported from mouse breast cancer cells 4 T1 to TAMs via exosomes and promote the secretion of proinflammatory cytokines by inhibiting the expression of PPP2CA protein, thus promoting the progression of breast cancer. 53 That study was aimed at mouse breast cancer cells, while we studied human triple negative breast cancer cells.
In summary, we explored the molecular mechanism by which tumor‐derived exosomal miRNA regulated the crosstalk between MDA‐MB‐231 cells and M2 macrophages. We revealed that miR‐148b‐3p was upregulated in BC cell‐derived exosomes and could stimulate M2 macrophage polarization through inhibition of TSC2 and stimulation of the mTORC1 signaling pathway, which promoted BC proliferation, migration and invasion. According to these findings, miR‐148b‐3p may serve as a possible diagnostic marker and therapeutic target for BC therapy.
AUTHOR CONTRIBUTIONS
Baogang Zhang contributed to the study design. Chong Hao and Zhimei Sheng performed the experiment. Wenhao Wang performed the statistical analysis. Ruijun Feng, Qinpei Xiao and Yuanhang Zheng carried out cell culture and molecular biology studies. Baogang Zhang engaged in the conception, design and coordination of the study. Chong Hao drafted the manuscript, which all authors then reviewed. All authors read and approved the final manuscript.
FUNDING INFORMATION
This project was established by the National Natural Science Foundation of China (grant nos. 81672631 and 81872163).
CONFLICT OF INTEREST STATEMENT
The authors state that they have no competing interests that might have influenced their study.
Supporting information
Figure S1. The cell viability of MDA‐MB‐231 cells cocultured with conditioned medium from macrophages pretreated with or without MDA‐MB‐231‐exo.
Figure S2. miR‐148b‐3p expression levels in primary BRCA samples compared with their corresponding normal tissues based on TCGA dataset from UALCAN.
Figure S3. PMA‐stimulated macrophages were transfected with various miRNAs. The stable phenotype was determined by qRT‐PCR.
Figure S4. The cell viability of MDA‐MB‐231 cells co‐cultured with conditioned medium from macrophages transfected with miR‐nc, miR‐148b‐3p mimic or not.
Figure S5. SFRS13A expression of the MDA‐MB‐231 cells transfected with si‐SFRS13A or si‐nc was assessed by qRT‐PCR.
Figure S6. ACO1 expression of the MDA‐MB‐231 cells transfected with si‐ACO1 or si‐nc was assessed by qRT‐PCR.
Figure S7. TSC2 expression was significantly negatively related with infiltration of macrophage cell in BRCA.
Figure S8. High expression of TSC2 was positively correlated with overall survival in BC patients and negatively with HER2‐positive and TNBC subclasses of BRCA samples.
Table S1. Sequences of RT‐PCR oligonucleotide primers.
Hao C, Sheng Z, Wang W, Feng R, Zheng Y, Xiao Q, et al. Tumor‐derived exosomal miR‐148b‐3p mediates M2 macrophage polarization via TSC2/mTORC1 to promote breast cancer migration and invasion. Thorac Cancer. 2023;14(16):1477–1491. 10.1111/1759-7714.14891
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Wang QX, Cai YF, Chen YY, Zhang W, Jin WX, Chen ED, et al. Additional prognostic value of lymph node ratio (LNR) and number of negative lymph nodes (NLNs) in Chinese patients with triple negative breast cancer. Ann Clin Lab Sci. 2017;47:68–75. [PubMed] [Google Scholar]
- 3. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–8. [DOI] [PubMed] [Google Scholar]
- 4. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 5. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silverman JM, Clos J, de'Oliveira CC, Shirvani O, Fang Y, Wang C, et al. An exosome‐based secretion pathway is responsible for protein export from leishmania and communication with macrophages. J Cell Sci. 2010;123:842–52. [DOI] [PubMed] [Google Scholar]
- 7. Jia Y, Chen Y, Wang Q, Jayasinghe U, Luo X, Wei Q, et al. Exosome: emerging biomarker in breast cancer. Oncotarget. 2017;8:41717–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, et al. Tumour‐released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15:80–8. [DOI] [PubMed] [Google Scholar]
- 10. Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. [DOI] [PubMed] [Google Scholar]
- 12. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 13. Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y, et al. Exosomal miRNAs and miRNA dysregulation in cancer‐associated fibroblasts. Mol Cancer. 2017;16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noy R, Pollard JW. Tumor‐associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan Y, Yu Y, Wang X, Zhang T. Tumor‐associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh A, Talekar M, Raikar A, Amiji M. Macrophage‐targeted delivery systems for nucleic acid therapy of inflammatory diseases. J Control Release. 2014;190:515–30. [DOI] [PubMed] [Google Scholar]
- 17. Wang J, Cao Z, Zhang XM, Nakamura M, Sun M, Hartman J, et al. Novel mechanism of macrophage‐mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res. 2015;75:306–15. [DOI] [PubMed] [Google Scholar]
- 18. Ham S, Lima LG, Chai EPZ, Muller A, Lobb RJ, Krumeich S, et al. Breast cancer‐derived exosomes alter macrophage polarization via gp130/STAT3 signaling. Front Immunol. 2018;9:871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson JL, Nagele T, Linke M, Demel F, Fritsch SD, Mayr HK, et al. Inverse data‐driven modeling and multiomics analysis reveals Phgdh as a metabolic checkpoint of macrophage polarization and proliferation. Cell Rep. 2020;30:1542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kimura T, Nada S, Takegahara N, Okuno T, Nojima S, Kang S, et al. Polarization of M2 macrophages requires Lamtor1 that integrates cytokine and amino‐acid signals. Nat Commun. 2016;7:13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen W, Ma T, Shen XN, Xia XF, Xu GD, Bai XL, et al. Macrophage‐induced tumor angiogenesis is regulated by the TSC2‐mTOR pathway. Cancer Res. 2012;72:1363–72. [DOI] [PubMed] [Google Scholar]
- 22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 23. Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex‐2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3‐kinase/akt pathway. Mol Cell. 2002;10:151–62. [DOI] [PubMed] [Google Scholar]
- 24. Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer‐secreted miR‐105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, et al. miRpower: a web‐tool to validate survival‐associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–46. [DOI] [PubMed] [Google Scholar]
- 27. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor‐immune system interactions. Bioinformatics. 2019;35:4200–2. [DOI] [PubMed] [Google Scholar]
- 28. Baig MS, Roy A, Rajpoot S, Liu D, Savai R, Banerjee S, et al. Tumor‐derived exosomes in the regulation of macrophage polarization. Inflamm Res. 2020;69:435–51. [DOI] [PubMed] [Google Scholar]
- 29. Hagemann T, Biswas SK, Lawrence T, Sica A, Lewis CE. Regulation of macrophage function in tumors: the multifaceted role of NF‐kappaB. Blood. 2009;113:3139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan B, Shi X, Zhang J, Qin J, Zhang N, Ren H, et al. Inhibition of Rspo‐Lgr4 facilitates checkpoint blockade therapy by switching macrophage polarization. Cancer Res. 2018;78:4929–42. [DOI] [PubMed] [Google Scholar]
- 31. Xiang W, Shi R, Kang X, Zhang X, Chen P, Zhang L, et al. Monoacylglycerol lipase regulates cannabinoid receptor 2‐dependent macrophage activation and cancer progression. Nat Commun. 2018;9:2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inoki K. Role of TSC‐mTOR pathway in diabetic nephropathy. Diabetes Res Clin Pract. 2008;82(Suppl 1):S59–62. [DOI] [PubMed] [Google Scholar]
- 34. Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. [DOI] [PubMed] [Google Scholar]
- 35. Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57. [DOI] [PubMed] [Google Scholar]
- 36. Byles V, Covarrubias AJ, Ben‐Sahra I, Lamming DW, Sabatini DM, Manning BD, et al. The TSC‐mTOR pathway regulates macrophage polarization. Nat Commun. 2013;4:2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shan G, Zhou X, Gu J, Zhou D, Cheng W, Wu H, et al. Downregulated exosomal microRNA‐148b‐3p in cancer associated fibroblasts enhance chemosensitivity of bladder cancer cells by downregulating the Wnt/beta‐catenin pathway and upregulating PTEN. Cell Oncol. 2021;44:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arambula‐Meraz E, Bergez‐Hernandez F, Leal‐Leon E, Romo‐Martinez E, Picos‐Cardenas V, Luque‐Ortega F, et al. Expression of miR‐148b‐3p is correlated with overexpression of biomarkers in prostate cancer. Genet Mol Biol. 2020;43:e20180330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X, Jiang M, Chen D, Xu B, Wang R, Chu Y, et al. miR‐148b‐3p inhibits gastric cancer metastasis by inhibiting the Dock6/Rac1/Cdc42 axis. J Exp Clin Cancer Res. 2018;37:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang Z, Ma Y, Zhang P, Si J, Xiong Y, Yang Y. Long non‐coding RNA H19 confers resistance to gefitinib via miR‐148b‐3p/DDAH1 axis in lung adenocarcinoma. Anticancer Drugs. 2020;31:44–54. [DOI] [PubMed] [Google Scholar]
- 41. Lu H, Zhang Z, Lu Y, Xiu W, Cui J. LncRNA NEAT1 acts as an miR‐148b‐3p sponge to regulate ROCK1 inhibition of retinoblastoma growth. Cancer Manag Res. 2021;13:5587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA‐binding specificities. Nucleic Acids Res. 2011;39:D301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oskarsson GR, Oddsson A, Magnusson MK, Kristjansson RP, Halldorsson GH, Ferkingstad E, et al. Predicted loss and gain of function mutations in ACO1 are associated with erythropoiesis. Commun Biol. 2020;3:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nasrollahzadeh E, Razi S, Keshavarz‐Fathi M, Mazzone M, Rezaei N. Pro‐tumorigenic functions of macrophages at the primary, invasive and metastatic tumor site. Cancer Immunol Immunother. 2020;69:1673–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen J, Hu Q, Schrauder M, Yan L, Wang D, Medico L, et al. Circulating miR‐148b and miR‐133a as biomarkers for breast cancer detection. Oncotarget. 2014;5:5284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aure MR, Leivonen SK, Fleischer T, Zhu Q, Overgaard J, Alsner J, et al. Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biol. 2013;14:R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dai W, He J, Zheng L, Bi M, Hu F, Chen M, et al. miR‐148b‐3p, miR‐190b, and miR‐429 regulate cell progression and act as potential biomarkers for breast cancer. J Breast Cancer. 2019;22:219–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang XY, Mao L. Circular RNA Circ_0000442 acts as a sponge of MiR‐148b‐3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene. 2021;766:145113. [DOI] [PubMed] [Google Scholar]
- 49. Chen X, Wang YW, Gao P. SPIN1, negatively regulated by miR‐148/152, enhances Adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J Exp Clin Cancer Res. 2018;37:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan L, Liu Y, Qu Y, Liu L, Li H. Exosomes derived from MicroRNA‐148b‐3p‐overexpressing human umbilical cord mesenchymal stem cells restrain breast cancer progression. Front Oncol. 2019;9:1076. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51. Moradi‐Chaleshtori M, Bandehpour M, Soudi S, Mohammadi‐Yeganeh S, Hashemi SM. In vitro and in vivo evaluation of anti‐tumoral effect of M1 phenotype induction in macrophages by miR‐130 and miR‐33 containing exosomes. Cancer Immunol Immunother. 2021;70:1323–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion‐potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guo J, Duan Z, Zhang C, Wang W, He H, Liu Y, et al. Mouse 4T1 breast cancer cell‐derived exosomes induce proinflammatory cytokine production in macrophages via miR‐183. J Immunol. 2020;205:2916–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The cell viability of MDA‐MB‐231 cells cocultured with conditioned medium from macrophages pretreated with or without MDA‐MB‐231‐exo.
Figure S2. miR‐148b‐3p expression levels in primary BRCA samples compared with their corresponding normal tissues based on TCGA dataset from UALCAN.
Figure S3. PMA‐stimulated macrophages were transfected with various miRNAs. The stable phenotype was determined by qRT‐PCR.
Figure S4. The cell viability of MDA‐MB‐231 cells co‐cultured with conditioned medium from macrophages transfected with miR‐nc, miR‐148b‐3p mimic or not.
Figure S5. SFRS13A expression of the MDA‐MB‐231 cells transfected with si‐SFRS13A or si‐nc was assessed by qRT‐PCR.
Figure S6. ACO1 expression of the MDA‐MB‐231 cells transfected with si‐ACO1 or si‐nc was assessed by qRT‐PCR.
Figure S7. TSC2 expression was significantly negatively related with infiltration of macrophage cell in BRCA.
Figure S8. High expression of TSC2 was positively correlated with overall survival in BC patients and negatively with HER2‐positive and TNBC subclasses of BRCA samples.
Table S1. Sequences of RT‐PCR oligonucleotide primers.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.


