Abstract
The emergence of fluoroquinolone and macrolide resistance in C. jejuni, a recognized zoonotic pathogen, has increased worldwide. This study aimed to investigate phenotypic resistance to ciprofloxacin and erythromycin, the molecular mechanisms involved, and the strain of C. jejuni isolated from broiler carcasses. Eighty C. jejuni isolates from broiler carcasses in southern Brazil were investigated for their susceptibility to ciprofloxacin and erythromycin at minimal inhibitory concentrations. Mismatch amplification mutation assay–polymerase chain reaction (MAMA-PCR) was performed to detect substitutions of Thr-86-Ile, A2074C, and A2075G of domain V in the 23S rRNA. The presence of ermB gene and CmeABC operon were investigated by PCR. DNA sequencing was used to detect substitutions in the L4 and L22 proteins of the erythromycin-resistant strains. The Short Variable Region (SVR) of flaA was used to type all the strains resistant to both antimicrobials. Ciprofloxacin and erythromycin resistance were detected in 81.25% and 30.00% of the strains, respectively, and minimal inhibitory concentration values ranged from ≤ 0.125 to 64 µg/mL for ciprofloxacin and 0.5 to > 128 µg/mL for erythromycin. The Thr-86-Ile mutation in gyrA was observed in 100% of the ciprofloxacin-resistant strains. Mutations in both the A2074C and A2075G positions of 23S rRNA were observed in 62.5% of the erythromycin-resistant strains, while 37.5% had only the mutation A2075G. None of the strains harbored CmeABC operon, and ermB was not detected. Using DNA sequencing, the amino acid substitution T177S was detected in L4, and substitutions I65V, A103V, and S109A were detected in L22. Twelve flaA-SVR alleles were identified among the strains, with the most common SVR-flaA allele, type 287, covering 31.03% of ciprofloxacin- and erythromycin-resistant isolates. The present study revealed a high incidence and high levels of resistance to ciprofloxacin and erythromycin, as well as broad molecular diversity in C. jejuni isolates from broiler carcasses.
Keywords: Antimicrobial resistance, Broiler, Campylobacter, flaA typing
Introduction
Campylobacteriosis is the most reported gastrointestinal bacterial infection in humans in the European Union [1] and the third in the USA [2] and Campylobacter jejuni is the species commonly associated with human cases. Chicken meat is a major source of Campylobacter infection in humans [3] and is recognized as a significant risk factor for acquiring this disease [4]. Human campylobacteriosis is a self-limiting illness that does not require antimicrobial treatment. However, in cases of invasive diseases and infections in immunosuppressed and young individuals, antibiotic therapy can be performed using erythromycin and ciprofloxacin as alternative drugs [5]. In Brazil, there is little information on human campylobacteriosis because this bacterium has not been routinely investigated in human diarrhea. However, an outbreak of Escherichia coli O:157 infection associated with C. jejuni that resulted in the death of two children has been reported in southern Brazil. The authors reported that the deaths were attributed to E. coli O157, but that co-infection with C. jejuni could contribute to the severity of the symptoms [6].
Since the 1980s, an increase in Campylobacter fluoroquinolone-resistant strains has been reported in many countries [7–9], coinciding with the introduction of fluoroquinolones in veterinary medicine [10]. Currently, fluoroquinolone-resistant Campylobacter is classified as a high-priority pathogen in the Global Priority Pathogens List of Antibiotic-Resistant Bacteria by the World Health Organization [11] and fluoroquinolone- and macrolide-resistant Campylobacter are listed as serious threats to public health by the Centers for Disease Control and Prevention [12]. The main mechanism of resistance to fluoroquinolones in Campylobacter is a mutation in the Quinolone Resistance Determinant Region (QRDR) of the gyrA gene [5]. Resistance to macrolides is related to mutations in positions 2074 and 2075 of domain V of 23 s rRNA. In both cases, the efflux pump CmeABC has been described as a secondary mechanism that acts synergistically by expelling toxic compounds such as antimicrobials, metals, and bile. Other mechanisms, such as ermB gene, which encodes a methylase that confers a high level of resistance to this class [13], and mutations in the L4 and L22 ribosomal proteins, have been reported in Campylobacter strains and are associated with low levels of resistance [14].
Several studies have reported high genetic diversity in C. jejuni [15–17]. Short variable regions of flaA gene sequence (SVR-flaA), Pulsed Field Gel Electrophoresis (PFGE), multilocus sequence typing (MLST), and whole genome sequencing (WGS) [18, 19] are typing methods commonly used to differentiate Campylobacter isolates. Although MLST and WGS are more recent typing techniques, SVR-flaA typing is considered a very useful tool for epidemiological studies of Campylobacter [20], in addition to being more affordable.
When compared to the other countries Brazil, has less data about Campylobacter jejuni. In this context, this study aimed to investigate the phenotypic resistance to ciprofloxacin and erythromycin, the molecular mechanisms involved, and the type of C. jejuni isolated from broiler carcasses.
Material and methods
Sample collection
A total of 80 C. jejuni strains isolated from broiler carcasses in slaughterhouses under federal inspection in three neighboring states (Parana = 28 strains; Santa Catarina and Rio Grande do Sul = 26 strains each) in southern Brazil between 2017 and 2018 [21] were included in this study. After species identification by Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) mass spectrometry, the strains were stored in skim milk at − 80 °C for further analysis.
Antibiotic susceptibility test
The Minimum Inhibitory Concentrations (MIC) of erythromycin and ciprofloxacin (Sigma-Aldrich, Brazil) were determined according to the method reported by the Clinical and Laboratory Standards Institute [22]. MIC breakpoints for erythromycin and ciprofloxacin were ≥ 4 µg/mL and ≥ 32 µg/mL, respectively. The concentrations of erythromycin and ciprofloxacin ranged from 0.5 to 128 µg/mL and can be checked in Table 2. C. jejuni ATCC 33,560 was used as a quality control strain.
Table 2.
Distribution of the minimal inhibitory concentration values for 80 C. jejuni isolates from broiler carcasses
| Antimicrobial | State | MIC(μg/mL) | Susceptible isolates (%) | Resistant isolates (%) | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤ 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | > 128 | |||||
| Ciprofloxacin | Paraná | 12 | 0 | 1 | 0 | 0 | 0 | 1 | 10 | 4 | 0 | 0 | 13 (46.42) | 15 (53.58) | 28 (100) |
| Santa Catarina | 2 | 0 | 0 | 0 | 0 | 0 | 6 | 7 | 11 | 0 | 0 | 2 (7.70) | 24 (92.30) | 26 (100) | |
| Rio Grande do Sul | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 9 | 13 | 1 | 0 | 0 (0) | 26 (100) | 26 (100) | |
| Erythromycin | Paraná | 0 | 0 | 4 | 7 | 13 | 0 | 2 | 0 | 0 | 0 | 2 | 26 (92.85) | 2 (7.15) | 28 (100) |
| Santa Catarina | 0 | 0 | 2 | 5 | 9 | 2 | 0 | 0 | 0 | 0 | 8 | 18 (69.23) | 8 (30.77) | 26 (100) | |
| Rio Grande do Sul | 0 | 0 | 1 | 2 | 8 | 1 | 0 | 0 | 0 | 2 | 12 | 12 (46.15) | 14 (53.85) | 26 (100) | |
Molecular characterization of antimicrobial resistance
DNA extraction was performed using the Wizard® Genomic DNA Purification Kit (Promega, Brazil) following the manufacturer’s recommendations. The concentration and quality of genomic DNA were measured using a Nanodrop spectrophotometer (Biodrop®).
The strains were subjected to MAMA-PCR to detect gyrA mutation in codon 86 (Thr-Ile) [23] and A2075G/A2074C mutations in the 23S rRNA gene [24]. DNA sequencing to evaluate the presence of mutations in the rplD and rplV genes encoding the L4 and L22 ribosomal proteins was performed in erythromycin-resistant strains, according to Corcoran et al. (2006) [14]. The presence of the ermB gene was investigated as described by Wang et al. (2014) [13]. All strains were subjected to PCR to detect the genes cmeA, cmeB, and cmeC following the protocol described by Lin et al. (2002) [25]. The PCR reactions had a final volume of 25 µl containing:1X PCR buffer, 1.5 mM MgCl2, 5µL of DNA, 0.2 µM of each primer. All reactions were performed with GoTaq® G2 Hot Start Taq Polymerase (Promega, Brazil) in a T100 (Biorad) thermocycler. C. jejuni strains identified as carriers of the investigated genes and/or mutations were used as positive controls and ultrapure water was used as a negative control. The details of all primers used and their references are listed in Table 1. The obtained amplicons were loaded onto a 1.5% agarose gel (Lwt Biotec, Brazil) stained with ethidium bromide, submerged in Tris-Acetato-EDTA Buffer (Ludwig, Alvorada, Brazil), and subjected to electrophoresis in a horizontal electrophoresis system (Loccus, São Paulo, Brazil) at 90 V for 40 min.
Table 1.
Target genes/mutation, primer sequence, annealing temperature (°C), amplicon size, and reference of the primers used in this study
| Gene | Primer sequence (5′-3′) | Sense | Annealing temperature | Size (pb) | Reference |
|---|---|---|---|---|---|
| cmeA | TTTGGATCCTTGATGGCTAAGGCAACTTTC | Forward | 54° | 771 | [25] |
| CTCCAATTTCTTAAGCTTCGCTACCAA | Reverse | ||||
| cmeB | GGTACAGATCCTGATCAAGCC | Forward | 820 | ||
| AGGAATAAGTGTTGCACGGAAATT | Reverse | ||||
| cmeC | GCTTGGATCCTTATCTTGGGAAAAA | Forward | 624 | ||
| TTTTTAAAGCTTTAAGGTAATTTTCTT | Reverse | ||||
| ermB | GGGCATTTAACGACGAAACTGG | Forward | 55° | 421 | [13] |
| CTGTGGTATGGCGGGTAAGT | Reverse | ||||
| gyrA | TTTTTAGCAAAGATTCTGAT | Forward | 50° | 265 | [23] |
| CAAAGCATCATAAACTGCAA | Reverse | ||||
| 23sRNA | TTAGCTAATGTTGCCCGTACCG | Forward | 59° | 485 | [24] |
| TAGTAAAGGTCCACGGGGTCGC* | Reverse | ||||
| AGTAAAGGTCCACGGGGTCTGG** | Reverse | ||||
| L4 | TTATCCCTCTTTTGTAATAGATTCTAA | Forward | 51° | 614 | [14] |
| ATGAGTAAAGTAGTTGTTTTAAATGAT | Reverse | ||||
| L22 | TTAGCTTTCCTTTTTCACTGTTGCTTT | Forward | 55° | 425 | |
| ATGAGTAAAGCATTAATTAAATTCATAAG | Reverse | ||||
| TGAGAAGTTAAGTTTTGGAGAG | Reverse | ||||
| SVR-flaA | CTATGGATGAGCAATTWAAAAT | Forward | 60° | 402 | [26] |
| CAAGWCCTGTTCCWACTGAAG | Reverse |
*Mutation detection position 2074, **mutation detection position 207
Genetic diversity
Genetic diversity was assessed by DNA sequencing of the Short Variable Region (SVR) of the flaA gene according to Meinersmann et al. 1997 [26]. All erythromycin- and ciprofloxacin-resistant strains detected by the MIC test (n = 24) and five full susceptible strains were addressed. The amplicons were purified using a QIAquick PCR purification kit (Qiagen, USA) and sequenced with an ABI 3730 DNA sequencer (Applied Biosystems) using the RPT/Fiocruz Sequencing Platform. Sequences were manually edited and compared with those in current databases using the BLAST suite of programs. Nucleotide alignments were generated using ClustalW in the Unipro UGENE software [27]. For L4 and L22, the sequences were aligned to the corresponding sequence of the parent strain NCTC 11,168 using the same software to identify specific mutations, and the flaA-SVR nucleotide allele was obtained in the database found at PubMLST database (http://pubmlst.org/organisms/campylobacter-jejunicoli) [28]. The dendrogram was generated with the CLC GenomicsWorkbench 23.0.2 (Qiagen, The Netherlands), using the UPGMA method with the Jukes and Cantor distance correction model and bootstrap values calculated in 1000 replicates.
Statistical analysis
The chi-square test was performed using Epi Info, version 6.0, software (Centers for Disease Control and Prevention, Atlanta, Ga.) to evaluate the differences in resistance levels to erythromycin, ciprofloxacin, and their combination in strains from different states. A significant level of 0.05 was considered statistically significant.
Results
Of the 80 C. jejuni isolates, 81.25% (65/80) were resistant to ciprofloxacin, 30% (24/80) were resistant to erythromycin, and 18.75% (15/80) were susceptible to both the antimicrobials. The resistance observed in the Paraná state to ciprofloxacin and erythromycin was 53.57% (15/28) and 7.14% (2/28), respectively; in the Santa Catarina state 92.31% (24/26) and 30.77% (8/26); and 100% (26/26) and 53.85% (14/26) in the Rio Grande do Sul state, respectively. Strains from Paraná were more sensitive to both antimicrobials than those from Santa Catarina and Rio Grande do Sul (pciprofloxacin = 0.0000006; perythromycin = 0.00003). MIC values for ciprofloxacin ranged from ≤ 0.125 to 64 µg/mL and for erythromycin 0.5 to > 128 µg/mL. Most ciprofloxacin-resistant strains (55/80) showed MIC ≥ 16 µg/mL and almost all erythromycin-resistant strains (22/24) showed MIC > 128 µg/mL (Table 2).
All ciprofloxacin-resistant strains (65/80) had a mutation at codon 86 (Thr-86-Ile) in the QRDR of gyrA, whereas the sensitive strains had no substitution. Regarding the erythromycin-resistant strains, 62.5% (15/24) had a mutation in both the A2074C and A2075G positions of 23S rRNA, and 37.5% (9/24) had only the A2075G mutation. None of the sensitive strains harbored the A2074C or A2075G mutations. Six strains had amino acid substitutions in three codons (I65V, A103V, and S109A), and seven had two substitutions (I65V/S109A) in the L22 ribosomal protein. Seven strains harbored T177S substitutions in the L4 protein, and no strain had L4 and L22 substitutions concurrently. The ermB gene was not detected in the present study and all strains (80/80) harbored the cmeABC operon.
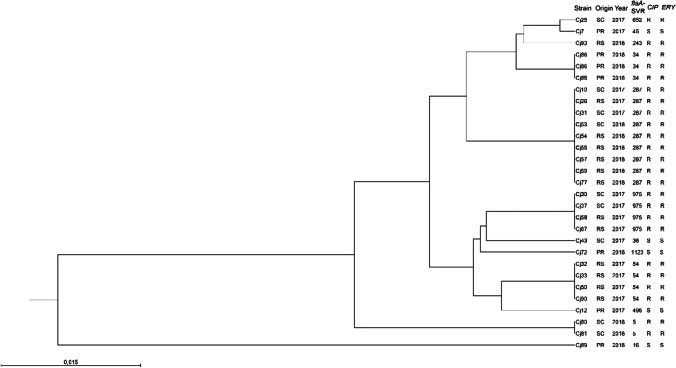
SVR-flaA typing revealed 12 different alleles in the 29 investigated strains. Allele 287 was the most predominant, covering 31.03% (9/29) of the isolates, followed by alleles 975 and 54 with four isolates each (4/29). The five SVR-flaA alleles detected in the five susceptible strains differed from those detected in resistant strains. Table 3 lists flaA alleles, antimicrobial resistances, and molecular markers associated with the 29 sequenced strains (Fig. 1). Two clusters were identified with a similarity greater than 84.11%. Only the Cj89 strain was grouped alone in one cluster and the other 28 strains evaluated were grouped in another cluster with a similarity ≥ 94.70%.
Table 3.
Strains, state, year of isolation, minimal inhibitory concentration (MIC) for ciprofloxacin (CIP) and erythromycin (ERY), molecular mechanisms associated with antimicrobial resistance and flaA alleles of 29 Campylobacter jejuni strains isolated from Broiler Carcasses in southern, Brazil
| Strain | State | Year of isolation | MIC CIP | Thr- 86-Ile | MIC ERY | 23S A2074C | 23S A2075G | ermB | L4 | L22 | cmeABC | flaA Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cj80 | SC | 2018 | 16 | Positive | > 128 | Negative | Positive | Negative | WT | WT | Positive | 5 |
| Cj81 | SC | 2018 | 16 | Positive | > 128 | Positive | Positive | Negative | WT | WT | Positive | 5 |
| Cj89* | PR | 2018 | ≤ 0.125 | Negative | 2 | Negative | Negative | Negative | WT | WT | Positive | 16 |
| Cj86 | PR | 2018 | 16 | Positive | > 128 | Positive | Positive | Negative | T177S | WT | Positive | 34 |
| Cj88 | PR | 2018 | 16 | Positive | > 128 | Positive | Positive | Negative | T177S | WT | Positive | 34 |
| Cj23 | RS | 2017 | 16 | Positive | > 128 | Negative | Positive | Negative | T177S | WT | Positive | 34 |
| Cj43* | SC | 2017 | ≤ 0.125 | Negative | 0.5 | Negative | Negative | Negative | NT | NT | Positive | 36 |
| Cj7* | PR | 2017 | ≤ 0.125 | Negative | 1 | Negative | Negative | Negative | NT | NT | Positive | 45 |
| Cj32 | RS | 2017 | 32 | Positive | > 128 | Positive | Positive | Negative | T177S | WT | Positive | 54 |
| Cj33 | RS | 2017 | 32 | Positive | > 128 | Positive | Positive | Negative | T177S | WT | Positive | 54 |
| Cj50 | RS | 2018 | 32 | Positive | > 128 | Positive | Positive | Negative | WT | I65V/S109A | Positive | 54 |
| Cj90 | RS | 2018 | 32 | Positive | > 128 | Negative | Positive | Negative | WT | I65V/S109A | Positive | 54 |
| Cj93 | RS | 2018 | 8 | Positive | > 128 | Positive | Positive | Negative | WT | WT | Positive | 243 |
| Cj10 | SC | 2017 | 8 | Positive | > 128 | Positive | Positive | Negative | WT | WT | Positive | 287 |
| Cj26 | RS | 2017 | 32 | Positive | > 128 | Negative | Positive | Negative | WT | WT | Positive | 287 |
| Cj31 | SC | 2017 | 16 | Positive | > 128 | Positive | Positive | Negative | WT | I65V/S109A | Positive | 287 |
| Cj53 | SC | 2018 | 8 | Positive | > 128 | Positive | Positive | Negative | T177S | WT | Positive | 287 |
| Cj54 | RS | 2018 | 16 | Positive | > 128 | Positive | Positive | Negative | WT |
I65V/ A103V/ S109A |
Positive | 287 |
| Cj55 | RS | 2018 | 32 | Positive | > 128 | Positive | Positive | Negative | WT | I65V/ A103V/ S109A | Positive | 287 |
| Cj57 | RS | 2018 | 32 | Positive | > 128 | Negative | Positive | Negative | WT | I65V/ A103V/ S109A | Positive | 287 |
| Cj59 | RS | 2018 | 16 | Positive | > 128 | Positive | Positive | Negative | WT | I65V/ A103V/ S109A | Positive | 287 |
| Cj77 | RS | 2018 | 8 | Positive | 64 | Positive | Positive | Negative | WT | I65V/ S109A | Positive | 287 |
| Cj12* | PR | 2017 | ≤ 0.125 | Negative | 2 | Negative | Negative | Negative | NT | NT | Positive | 496 |
| Cj25 | SC | 2017 | 32 | Positive | > 128 | Negative | Positive | Negative | WT | I65V/ A103V/ S109A | Positive | 652 |
| Cj30 | SC | 2017 | 32 | Positive | > 128 | Negative | Positive | Negative | WT | I65V/ S109A | Positive | 975 |
| Cj37 | SC | 2017 | 32 | Positive | > 128 | Negative | Positive | Negative | WT | I65V/ S109A | Positive | 975 |
| Cj58 | RS | 2018 | 16 | Positive | 64 | Negative | Positive | Negative | WT | I65V/ S109A | Positive | 975 |
| Cj87 | RS | 2018 | 32 | Positive | > 128 | Positive | Positive | Negative | T177S | WT | Positive | 975 |
| Cj72* | PR | 2018 | ≤ 0.125 | Negative | 1 | Negative | Negative | Negative | NT | NT | Positive | 1123 |
*Sensitive strains to both antimicrobials. NT, non-tested; WT, wild type; PR, Paraná; SC, Santa Catarina; RS, Rio Grande do Sul
Fig. 1.
Phylogenetic tree based on 29 SVR-flaA gene sequences of C. jejuni isolates from chicken carcasses in southern Brazil. The dendrogram was inferred by using the UPGMA method with Jukes and Cantor distance correction model and bootstrap values calculated in 1000 replicates. “PR” Paraná, “SC” Santa Catarina, “RS” Rio Grande do Sul, “CIP” Ciprofloxacin, “ERY” Erythromycin, “R” resistant strain, and “S” a susceptible strain
Discussion
High incidence and levels of resistance to both antimicrobials tested were detected in the C. jejuni strains tested in this study. The presence of these resistant strains represents an additional risk to public health, as infections may be more difficult to treat in cases where antimicrobial therapy is required. Strains from Paraná were more sensitive to both antimicrobials than those from Santa Catarina and Rio Grande do Sul. This may be related to the level of awareness of antimicrobial use by chicken producers, biosecurity in poultry farms leading to lower antimicrobial use, or even aspects related to differences in Campylobacter strains that circulate in each state. Several studies have detected fluoroquinolone-resistant strains isolated from chicken carcasses in Brazil and worldwide, with MIC exceeding 128 µg/mL [5, 29]. In our study, 81.25% (65/80) of the strains were resistant to ciprofloxacin with MIC reaching 64 µg/mL. Studies show that a single mutation at position Thr-86-Ile in the QRDR of gyrA is considered the main mechanism of resistance to fluoroquinolones and leads to the replacement of the amino acid threonine for isoleucine [5, 30, 31]. All resistant strains in our study harbored this mutation, which was not observed in the susceptible strains. This mutation confers a fitness benefit to Campylobacter in chickens by reducing the supercoiling activity of gyrA, which may help in the emergence, maintenance, and spread of fluoroquinolone-resistant isolates in poultry farms [32] and can overcome the colonization of susceptible strains [33]. Previous results obtained by our research group [29–31, 34] suggested the substitution of susceptible strains with resistant strains over time in the poultry production chain of Rio de Janeiro State, located in the southeast region of Brazil. The results obtained in the present study, including strains isolated between 2017 and 2018 from three other states, suggest that the replacement of susceptible strains with fluoroquinolone-resistant strains is widespread.
In our study, high levels (MIC ≥ 64 µg/mL) of erythromycin resistance were detected in 30% (24/80) of the strains. In previous studies in Brazil, erythromycin resistance in Campylobacter spp. isolated from poultry, ranged from 0 to 42.60% [35–38]. All resistant strains had the mutations A2074C/A2075G simultaneously or only A2075G. These mutations are considered to be the main mechanisms involved in the high levels of resistance to macrolides and do not represent any advantage for bacterial cells. Studies comparing colonization capacity demonstrated that strains without mutations supplanted mutant strains, indicating that in the absence of macrolides, these mutations decrease cell fitness [39]. ErmB encodes a ribosomal RNA methylase capable of methylating the macrolide-binding site and has also been linked to high levels of resistance in Campylobacter [13]. Since its first detection in China [13], reports of ermB-positive Campylobacter isolate have occurred in several countries [40–43]. Although one report was published in Latin America [44], this gene has not been detected in Campylobacter isolates from Brazil [36, 45, 46].
We detected the I65V, A103V, and S109A in L22 amino acid substitutions, and only the T177S substitution in the L4 ribosomal protein, as previously reported [14, 47, 48]. Mutations in L4 and L22 have been associated with a lower level of resistance to macrolides (erythromycin MIC = 32 μg/mL) in the absence of mutations in 23S rRNA genes [39]. In our study, it was difficult to estimate the real contribution of these substitutions to the level of resistance because the erythromycin-resistant strains had other mutations at positions 2074 and/or 2075 in the 23S rRNA, and low levels of resistance were not detected.
In addition to mutations in sites of antimicrobial action, such as gyrA and domain V of the 23S rRNA, the presence of efflux pumps in Campylobacter spp. is recognized as a secondary mechanism of resistance to ciprofloxacin and erythromycin and has been considered a factor that may potentiate resistance to these classes [49]. We detected genes related to the CmeABC efflux pump in 100% (80/80) of the strains, similar to the results obtained in other studies [50, 51]. The high prevalence of these genes is probably due to the known participation of efflux pumps in bacterial cell metabolism, mediation of resistance to bile salts in the intestinal tract, uptake of essential nutrients and ions, and excretion of bacterial metabolism products and toxic substances, which play a fundamental role in colonization by Campylobacter [49, 52].
Campylobacter is characterized by its high genetic variability, and the formation of new combinations of genetic alleles can be accelerated because this bacterium is naturally competent for DNA uptake and transformation [53]. Despite the wide use of MLST and PFGE, flaA typing is a commonly-employed technique offering discriminatory power, high reproducibility even if performed in different laboratories, and the possibility of comparison with strains from other countries deposited in the databases [18, 54]. However, SVR-flaA typing is a single-locus analysis method that does not assess the entire genome. Twelve different alleles of C. jejuni were identified. Although many strains clustered into the same allele, such as those belonging to the SVR-flaA alleles 287, 975, and 54, few differences in mutations and antimicrobial resistance levels to ciprofloxacin were observed. Frequent intra-and interspecies genetic mutations in C. jejuni result in different molecular variants, as determined by SVR-flaA typing [55]. The flaA allele type 287 was the most common, occurring in 31.03% (9/29) of the strains, although it was not detected in the Parana state. Other studies [56, 57] have detected this allele in samples from chickens and cases of human campylobacteriosis, reinforcing the role of chickens as a source of Campylobacter. Wieczorek et al. (2019) [55] found that this SVR-flaA type was the most common among numerous multidrug-resistant profiles, including resistance to ciprofloxacin in Campylobacter isolates from poultry chains. However, it was not clear whether there was a correlation between flaA-SVR 287 genotype and antimicrobial resistance. Other flaA allele types detected in this study, such as 34 and 45, have also been associated with human cases and chicken meat [57–59]. Additional research is necessary to investigate the connection between antimicrobial resistance and genotypes. Apart from the strains clustered into alleles 287, 975, and 54, we observed variability among alleles in the three different states. Since evolution is a dynamic process, monitoring the genotypes of Campylobacter is vital, as several anthropogenic factors such as intensive animal production and antibiotic use can act as selective pressures, changing epidemiological chains and how this microorganism interacts with its hosts.
Conclusion
A high incidence and level of resistance to ciprofloxacin and erythromycin and related point mutations were detected in C. jejuni. These results are of public health concern when antibiotic therapy is required for human campylobacteriosis caused by poultry strains. We also detected variability in the SVR-flaA alleles among the resistant strains, corroborating the high diversity reported in several Campylobacter studies. Owing to the high plasticity and genetic diversity of Campylobacter jejuni, whole-genome sequencing should be performed on strains circulating in the Brazilian poultry industry to investigate the possible relationship between certain genotypes and antimicrobial resistance.
Funding
This work was supported by the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (E-26/010.001349/2019 and E-26/202.348/2022), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, code 001), and Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Mariana X Byndloss
The original online version of this article was revised: In Conclusion section, the text "whole-genome sequencing was performed on strains circulating in the Brazilian poultry" needs to be corrected to "whole-genome sequencing should be performed on strains circulating in the Brazilian poultry"
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/28/2023
A Correction to this paper has been published: 10.1007/s42770-023-00983-7
References
- 1.European Food Safety Authority (EFSA) (2021) The European Union One Health 2020 Zoonoses Report. EFSA J 19:. 10.2903/j.efsa.2021.6971 [DOI] [PMC free article] [PubMed]
- 2.CDC (2019) Surveillance for foodborne disease outbreaks United States, 2017: annual report
- 3.EFSA & ECDC (2021) The European Union One Health 2019 Zoonoses Report. EFSA Journal Eur Food Saf Auth 19:e06406. 10.2903/j.efsa.2021.6406 [DOI] [PMC free article] [PubMed]
- 4.Rosner BM, Schielke A, Didelot X, et al. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014 /692/308/174 /692/499 article. Sci Rep. 2017;7:1–12. doi: 10.1038/s41598-017-05227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elhadidy M, Ali MM, El-Shibiny A, et al. Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases. PLoS One. 2020;15:1–16. doi: 10.1371/journal.pone.0227833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartz FW, Teixeira LB, Schroder R, et al. First fatal cases due to Escherichia coli O157 and Campylobacter jejuni subsp. jejuni outbreak occurred in southern Brazil. Foodborne Pathog Dis. 2022;19:241–247. doi: 10.1089/fpd.2021.0075. [DOI] [PubMed] [Google Scholar]
- 7.Reina J, Borrell N, Serra A. Emergence of resistance to erythromycin and fluoroquinolones in thermotolerant Campylobacter strains isolated from feces 1987–1991. Eur J Clin Microbiol Infect Dis. 1992;11:1163–1166. doi: 10.1007/BF01961137. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez R, Fernandez-Baca V, Diaz MD, et al. Evolution of susceptibilities of Campylobacter spp. to quinolones and macrolides. Antimicrob Agents Chemother. 1994;38:1879–1882. doi: 10.1128/AAC.38.9.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cody AJ, Clarke L, Bowler IC, Dingle KE. Ciprofloxacin-resistant campylobacteriosis in the UK. Lancet. 2010;376:1987. doi: 10.1016/S0140-6736(10)62261-1. [DOI] [PubMed] [Google Scholar]
- 10.Endtz HP, Ruijs GJ, van Klingeren B, et al. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J Antimicrob Chemother. 1991;27:199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization (WHO) (2018) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics
- 12.CDC (2019) Antibiotic resistance threats in the United States, 2019. Atlanta, Georgia
- 13.Wang Y, Zhang M, Deng F, et al. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob Agents Chemother. 2014;58:5405–5412. doi: 10.1128/AAC.03039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corcoran D, Quinn T, Cotter L, Fanning S. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int J Antimicrob Agents. 2006;27:40–45. doi: 10.1016/j.ijantimicag.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Aquino MHC, Filgueiras ALL, Matos R, et al. Diversity of Campylobacter jejuni and Campylobacter coli genotypes from human and animal sources from Rio de Janeiro, Brazil. Res Vet Sci. 2010;88:214–217. doi: 10.1016/j.rvsc.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Elhadidy M, Arguello H, Álvarez-Ordóñez A, et al. Orthogonal typing methods identify genetic diversity among Belgian Campylobacter jejuni strains isolated over a decade from poultry and cases of sporadic human illness. Int J Food Microbiol. 2018;275:66–75. doi: 10.1016/j.ijfoodmicro.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Rapp D, Ross C, Hea SY, Brightwell G. Importance of the farm environment and wildlife for transmission of campylobacter jejuni in a pasture-based dairy herd. Microorganisms. 2020;8:1–11. doi: 10.3390/microorganisms8121877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazão MR, de Souza RA, Medeiros MIC, et al. Molecular typing of Campylobacter jejuni strains: comparison among four different techniques. Brazilian J Microbiol. 2020;51:519–525. doi: 10.1007/s42770-019-00218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark CG, Taboada E, Grant CCR, et al. Comparison of molecular typing methods useful for detecting clusters of Campylobacter jejuni and C. coli isolates through routine surveillance. J Clin Microbiol. 2012;50:798–809. doi: 10.1128/JCM.05733-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Adawy H, Hotzel H, Tomaso H, et al. Detection of genetic diversity in Campylobacter jejuni isolated from a commercial Turkey flock using flaA typing, MLST analysis and microarray assay. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0051582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues CS, Armendaris PM, de Sá CVGC, et al. Prevalence of Campylobacter spp. in chicken carcasses in slaughterhouses from South of Brazil. Curr Microbiol. 2021;78:2242–2250. doi: 10.1007/s00284-021-02478-w. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute (CLSI) (2015) M45. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria [DOI] [PubMed]
- 23.Zirnstein G, Li Y, Swaminathan B, Angulo F. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J Clin Microbiol. 1999;37:3276–3280. doi: 10.1128/jcm.37.10.3276-3280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonso R, Mateo E, Churruca E, et al. MAMA-PCR assay for the detection of point mutations associated with high-level erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains. J Microbiol Methods. 2005;63:99–103. doi: 10.1016/j.mimet.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Overbye Michel L, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meinersmann RJ, Helsel LO, Fields PI, Hiett KL. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J Clin Microbiol. 1997;35:2810–2814. doi: 10.1128/jcm.35.11.2810-2814.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 28.Jolley KA, Bray JE, Maiden MCJ (2018) Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed]
- 29.Panzenhagen PHN, Aguiar WS, da Silva FB, et al. Prevalence and fluoroquinolones resistance of Campylobacter and Salmonella isolates from poultry carcasses in Rio de Janeiro, Brazil. Food Control. 2016;61:243–247. doi: 10.1016/j.foodcont.2015.10.002. [DOI] [Google Scholar]
- 30.Frasão BS, Côrtes LR, Nascimento ER, et al. Detecção de resistência às fluoroquinolonas em Campylobacter isolados de frangos de criação orgânica. Pesqui Vet Bras. 2015;35:613–619. doi: 10.1590/s0100-736x2015000700003. [DOI] [Google Scholar]
- 31.da Silva Frasao B, Medeiros V, Barbosa AV, et al. Detection of fluoroquinolone resistance by mutation in gyrA gene of Campylobacter spp. isolates from broiler and laying (Gallus gallus domesticus) hens, from Rio de Janeiro State. Brazil Cienc Rural. 2015;45:2013–2018. doi: 10.1590/0103-8478cr20141712. [DOI] [Google Scholar]
- 32.Han J, Wang Y, Sahin O, et al. A fluoroquinolone resistance associated mutation in gyrA affects DNA supercoiling in Campylobacter jejuni. Front Cell Infect Microbiol. 2012;2:21. doi: 10.3389/fcimb.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo N, Pereira S, Sahin O, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci. 2005;102:541–546. doi: 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dias TS, Nascimento RJ, Machado LS, et al. Comparison of antimicrobial resistance in thermophilic Campylobacter strains isolated from conventional production and backyard poultry flocks. Br Poult Sci. 2021;62:188–192. doi: 10.1080/00071668.2020.1833302. [DOI] [PubMed] [Google Scholar]
- 35.Melo RT, Grazziotin AL, Júnior ECV, et al. Evolution of Campylobacter jejuni of poultry origin in Brazil. Food Microbiol. 2019;82:489–496. doi: 10.1016/j.fm.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Dias TS, Machado LS, Vignoli JA, et al. Phenotypic and molecular characterization of erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains isolated from swine and broiler chickens. Pesqui Vet Bras. 2020;40:598–603. doi: 10.1590/1678-5150-PVB-6466. [DOI] [Google Scholar]
- 37.Hungaro HM, Mendonça RCS, Rosa VO, et al. Low contamination of Campylobacter spp. on chicken carcasses in Minas Gerais state, Brazil: molecular characterization and antimicrobial resistance. Food Control. 2015;51:15–22. doi: 10.1016/j.foodcont.2014.11.001. [DOI] [Google Scholar]
- 38.Paravisi M, Laviniki V, Bassani J, et al (2020) Antimicrobial resistance in Campylobacter jejuni Isolated from Brazilian poultry slaughterhouses. Brazilian J Poult Sci 22:. 10.1590/1806-9061-2020-1262
- 39.Bolinger H, Kathariou S (2017) The current state of macrolide resistance in Campylobacter spp.: trends and impacts of resistance mechanisms. Appl Environ Microbiol 83:. 10.1128/AEM.00416-17 [DOI] [PMC free article] [PubMed]
- 40.Florez-Cuadrado D, Ugarte-Ruiz M, Quesada A, et al. Description of an erm(B)-carrying Campylobacter coli isolate in Europe. J Antimicrob Chemother. 2016;71:841–843. doi: 10.1093/jac/dkv383. [DOI] [PubMed] [Google Scholar]
- 41.Jehanne Q, Bénéjat L, Ducournau A, et al. Emergence of erythromycin resistance methyltransferases in Campylobacter coli strains in France. Antimicrob Agents Chemother. 2021;64:810. doi: 10.1128/AAC.01124-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramatla T, Mileng K, Ndou R et al (2022) Campylobacter jejuni from slaughter age broiler chickens : genetic characterization, virulence, and antimicrobial resistance genes. 2022: [DOI] [PMC free article] [PubMed]
- 43.Wallace RL, Bulach D, Valcanis M, et al. Identification of the first erm(B)-positive Campylobacter jejuni and Campylobacter coli associated with novel multidrug resistance genomic islands in Australia. J Glob Antimicrob Resist. 2020;23:311–314. doi: 10.1016/j.jgar.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Anampa D, Benites C, Lázaro C et al (2020) Detección del gen ermB asociado a la resistencia a macrólidos en cepas de Campylobacter aisladas de pollos comercializados en Lima , Perú. Pan Am J Public Heal 1–7 [DOI] [PMC free article] [PubMed]
- 45.Rauber N, Ramires T, F S De et al (2021) Antimicrobial resistance genes and plasmids in Campylobacter jejuni from broiler production chain in Southern Brazil. 144:. 10.1016/j.lwt.2021.111202
- 46.Gomes CN, Fraza MR, Passaglia J et al (2019) Molecular epidemiology and resistance profile of Campylobacter jejuni and Campylobacter coli strains isolated from different sources in Brazil. 00: 10.1089/mdr.2019.0266 [DOI] [PubMed]
- 47.García-Sánchez L, Melero B, Jaime I, et al. Biofilm formation, virulence and antimicrobial resistance of different Campylobacter jejuni isolates from a poultry slaughterhouse. Food Microbiol. 2019;83:193–199. doi: 10.1016/j.fm.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 48.Wei B, Kang M (2018) Molecular basis of macrolide resistance in Campylobacter strains isolated from poultry in South Korea. Biomed Res Int 2018:. 10.1155/2018/4526576 [DOI] [PMC free article] [PubMed]
- 49.Vieira A, Ramesh A, Seddon AM, Karlyshev AV. CmeABC multidrug efflux pump promotes Campylobacter jejuni survival. Appl Environ Microbiol. 2017;83:1–13. doi: 10.1128/AEM.01600-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nascimento RJ, Frasão BS, Dias TS, et al. Detection of efflux pump CmeABC in enrofloxacin resistant Campylobacter spp. strains isolated from broiler chickens (Gallus gallus domesticus) in the state of Rio de Janeiro. Brazil Pesqui Vet Bras. 2019;39:728–733. doi: 10.1590/1678-5150-pvb-6004. [DOI] [Google Scholar]
- 51.Rokney A, Valinsky L, Vranckx K, et al. WGS-based prediction and analysis of antimicrobial resistance in Campylobacter jejuni isolates from Israel. Front Cell Infect Microbiol. 2020;10:1–13. doi: 10.3389/fcimb.2020.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J, Martinez A. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J Antimicrob Chemother. 2006;58:966–972. doi: 10.1093/jac/dkl374. [DOI] [PubMed] [Google Scholar]
- 53.Burnham PM, Hendrixson DR. Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat Rev Microbiol. 2018;16:551–565. doi: 10.1038/s41579-018-0037-9. [DOI] [PubMed] [Google Scholar]
- 54.Gomes CN, Souza RA, Passaglia J, et al. Genotyping of Campylobacter coli strains isolated in Brazil suggests possible contamination amongst environmental, human, animal and food sources. J Med Microbiol. 2016;65:80–90. doi: 10.1099/jmm.0.000201. [DOI] [PubMed] [Google Scholar]
- 55.Wieczorek K, Wolkowicz T, Osek J (2019) FlaA-SVR based genetic diversity of multiresistant campylobacter jejuni isolated from chickens and humans. Front Microbiol 10:. 10.3389/fmicb.2019.01176 [DOI] [PMC free article] [PubMed]
- 56.Giacomelli M, Andrighetto C, Rossi F, et al. Molecular characterization and genotypic antimicrobial resistance analysis of Campylobacter jejuni and Campylobacter coli isolated from broiler flocks in northern Italy. Avian Pathol. 2012;41:579–588. doi: 10.1080/03079457.2012.734915. [DOI] [PubMed] [Google Scholar]
- 57.Marotta F, Garofolo G, Di Donato G et al (2015) Population diversity of Campylobacter jejuni in poultry and its dynamic of contamination in chicken meat. Biomed Res Int 2015:. 10.1155/2015/859845 [DOI] [PMC free article] [PubMed]
- 58.Wieczorek K, Osek J. Genetic diversity of Campylobacter jejuni isolated from the poultry food chain. J Vet Res. 2019;63:35–40. doi: 10.2478/jvetres-2019-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giacomelli M, Andrighetto C, Lombardi A, et al. A longitudinal study on thermophilic Campylobacter spp. in commercial Turkey flocks in Northern Italy: occurrence and genetic diversity. Avian Dis. 2012;56:693–700. doi: 10.1637/10141-032312-Reg.1. [DOI] [PubMed] [Google Scholar]