Abstract
The prickly nightshade Solanum rostratum, an annual malignant weed, is native to North America and has globally invaded 34 countries, causing serious threats to ecosystems, agriculture, animal husbandry, and human health. In this study, we constructed a chromosome-level genome assembly and annotation of S. rostratum. The contig-level genome was initially assembled in 898.42 Mb with a contig N50 of 62.00 Mb from PacBio high-fidelity reads. With Hi-C sequencing data scaffolding, 96.80% of the initially assembled sequences were anchored and orientated onto 12 pseudo-chromosomes, generating a genome of 869.69 Mb with a contig N50 of 72.15 Mb. We identified 649.92 Mb (72.26%) of repetitive sequences and 3,588 non-coding RNAs in the genome. A total of 29,694 protein-coding genes were predicted, with 28,154 (94.81%) functionally annotated genes. We found 99.5% and 91.3% complete embryophyta_odb10 genes in the pseudo-chromosomes genome and predicted gene datasets by BUSCO assessment. The present genomic resource provides essential information for subsequent research on the mechanisms of environmental adaptation of S. rostratum and host shift in Colorado potato beetles.
Subject terms: DNA sequencing, Sequence annotation, Comparative genomics, Genome duplication, Genome evolution
Background & Summary
The prickly nightshade, Solanum rostratum Dunal (Solanales: Solanaceae), an annual plant, is an invasive alien malignant weed which classified as an “agricultural weed”, an “environmental weed”, and a “noxious weed” in the Global Compendium of Weeds1. In China, it is listed as an entry quarantine pest and key management alien invasive species. This species has a fast growth rate and strong reproductive ability, whose seed production reaching 78,500 seeds per plant2. High competitiveness in light, water, nutrients, ecological niche, and other resources results in reduced agricultural land production, loss of native species’ competitive advantage, and decrease in biodiversity. In addition, the densely covered narrow and long prickles on the surface of the stem, leaf, calyx, and fruit can be mixed with fodder to hurt the oral cavity and gastrointestinal digestive tract of livestock. Moreover, the neurotoxin solanine present in whole plants can cause livestock poisoning3. It is also the host of the Colorado potato beetle Leptinotarsa decemlineata4, which is the most destructive pest on potatoes, the tomato golden mottle virus5, and the tomato severe leaf curl virus6. Thus, the invasion of S. rostratum seriously threatens the local ecological environment, agricultural production, grassland animal husbandry, biodiversity, and human health (Fig. 1).
Fig. 1.
Morphological characteristics of Solanum rostratum (a) habits in the grassland, (b) habits in the corn field, (c) infested by Colorado potato beetle in the field, (d) damage to livestock, (e) whole plant, (f) seedling, (g) flower, (h) root, (i) stem, (j) leaf, and (k) fruit.
The extremely strong ecological adaptability (could survive in the wasteland, grasslands, overgrazing pastures, roadside, garbage dumps, orchards, courtyards, irrigation ditches, and river beaches)7 and stress resistance (barren, drought, wet, and salt8) facilitate S. rostratum to spread and establish in a new environment as a dominant species. Native to North America9, S. rostratum is widely distributed in 34 countries and regions, including North America, Asia, Africa, Europe, and Oceania10. In China, since its first detection in 1981 in Chaoyang City, Liaoning Province11, it has spread to nine provinces and 54 counties within 30 years through water flow, wind, livestock trade, sand transportation, and other vectors.
Alien invasive plants usually can adapt to the new ecological environment and establish and expand populations within a short period12, which will seriously negatively impact the local ecosystem. High-quality reference genomes could help us profoundly comprehend and screen the genetic basis and variations associated with important traits and adaptation under different ecological and environmental conditions. Technological advances, including long-read sequencing by Pacific Biosciences (PacBio) or Oxford Nanopore Technologies (ONT), the chromosome conformation capture technique (Hi-C), and BioNano optical maps, have facilitated genome sequencing, assembly, and annotation, leading to the rapid expansion of the quantity and quality of public plant genomes in the past 20 years13,14. For the nightshade family, Solanaceae, which comprises approximately 90 genera and 3,000–4,000 species15, a total of 7 genera, 46 species, and 170 genomes have been reported. However, all previous genomic studies have focused on horticultural crops and their related wild species (for example, the cultivated tomato Solanum lycopersicum16 and the wild relative Solanum pimpinellifolium17, potato Solanum tuberosum18, hot pepper Capsicum annuum19, and eggplant Solanum melongena20), model plant organisms (tobacco Nicotiana tabacum21), ornamental flowers (Petunia inflata and Petunia axillaris22), and herbs (Datura stramonium23 and Lycium barbarum24). So far, the genome of the solanaceous malignant weed remains unsequenced. Therefore, a chromosome-level reference genome of S. rostratum is an essential resource to further elucidate the pathway and genes involved in ecological environment adaptation under different stresses, solanine biosynthesis, host shift from native host prickly nightshade to potato for Colorado potato beetle, etc., by integrating comparative genomics, functional genomics, metagenomics, and population genomics.
In this study, we constructed and annotated a high-quality chromosome-level reference genome using integrated sequencing data (Fig. 2). We performed an initial de novo assembly into a contig-level genome by Hifasm25 using PacBio High fidelity (HiFi) long-reads. Valid Illumina Hi-C paired-end reads were used to generate chromosome-level assemblies using the HiC-Pro pipeline26. After masked repeat sequences, three strategies were integrated to annotate the gene structure by EVidenceModeler (EVM)27, including homologous prediction against closely related species, transcriptome-based prediction using the transcripts generated from PacBio Isoform-Sequencing (Iso-seq) long-reads and Illumina Paired-end RNA-seq short-reads by Program to Assemble Spliced Alignments (PASA) pipeline28, and ab initio prediction based on the characteristics of genomic sequence data. After annotating protein-coding gene functional and protein domains against a related database, the completeness and quality of the genome assembly and annotation were evaluated by Benchmarking Universal Single-Copy Orthologs (BUSCO)29 analysis and genome mapping and coverage rates using Illumina Paired-end short-reads. These results indicate that the present genome assemblies and annotations are contiguous and accurate. Furthermore, comparative genomic analysis was conducted with other nineteen solanaceous species to provide insight into their phylogenetic relationship, divergence time, whole-genome duplication (WGD) events along the solanaceous speciation, and genomic evolutionary history. Thus, the present S. rostratum genomic resource will be a foundation for subsequent research on this weed.
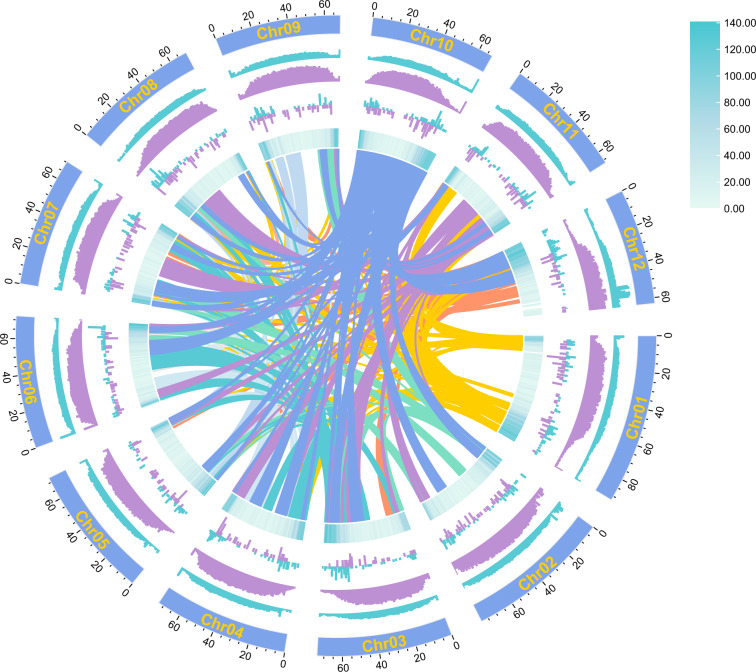
Fig. 2.
Chromosome-scale assembly genomic landscape of Solanum rostratum. Circos plot from the outer to the inner layers represents the following: (1) 12 pseudo-chromosomes length at the Mb scale; (2) GC content per Mb; (3) repeat density per Mb; (4) Copia (blue) and Gypsy (purple) LTR retroelement density per Mb; (5) gene density per Mb; and (6) center: intra-genomic syntenic blocks of S. rostratum.
Methods
Plant material collection and preparation
Healthy mature plants of S. rostratum were collected from the wasteland in Chaoyang City, Liaoning Province, China (120.504360° E, 41.604752° N) in August 2021. After washing with deionized water, the roots, stems, leaves, flowers, and fruits were harvested. All the tissues were put into liquid nitrogen immediately and preserved in an ultra-low temperature freezer until use.
DNA library construction and genome sequencing
High molecular weight genomic DNA was extracted from tender leaves with a modified 2 × cetyltrimethylammonium bromide (CTAB) method30. Approximately 200 mg of tender leaves were ground to powder using liquid nitrogen and then added to 800 μL of CTAB lysis buffer in a 2.0-mL tube. After incubation at 65 °C for 60 min, 800 μL of phenol/ chloroform/ isopentanol (25:24:1) was added and centrifuged at 12,000 rpm for 10 min. The supernatant was extracted into another 2.0-mL tube with an equal volume of chloroform/isopentanol (24:1). After mixing by gentle inversion, the tube was centrifuged at 12,000 rpm for 10 min. The supernatant was extracted to another tube with 0.6 times the volume of precooled (−20 °C) isopropanol. After being placed at −20 °C for over 2 h, the tube was centrifuged at 12,000 rpm for 10 min. The pellet was washed twice with 75% ethanol and dissolved in 50 µL of DNase and RNase-free Water for further study.
For the Illumina whole-genome shotgun raw sequencing, the genomic DNA was randomly fragmented, and a library with an average insert size of 350 bp was constructed using the Illumina TruSeq Nano DNA Library Prep Kit (Illumina, USA) following the manufacturer’s instructions. The library was sequenced on the Novaseq 6000 Platform set in the PE150 program, generating a total of 103.47 Gb of raw data. After filtering by fastp v0.12.431 with default to remove low quality and short reads and cut adapters and polyG, 102.14 Gb (113.69×) clean data were retained for the genome size estimation (Table 1).
Table 1.
Statistics of sequencing data for Solanum rostratum genome assembly and annotation.
| Platform | Type | sample | Molecule | Total clean data | Coverage | Usage | SRA accession number |
|---|---|---|---|---|---|---|---|
| Illumina NovaSeq | PE | Leaf | DNA | 102.14 Gb | 113.69× | correction | SRR23354532 |
| PacBio HiFi | CCS | Leaf | DNA | 25.83 Gb | 28.75× | de novo assembly | SRR23354533 |
| Illumina Hi-C | PE | Leaf | DNA | 102.09 Gb | 113.63× | chromosome-level assembly | SRR23354531 |
| PacBio Iso-seq | Subreads | Mixed | RNA | 19.81 Gb | 22.05× | gene structure annotation | SRR23354525 |
| Illumina NovaSeq | PE | Root | RNA | 6.00 Gb | 6.90× | gene structure annotation | SRR23354529 |
| Illumina NovaSeq | PE | Stem | RNA | 7.14 Gb | 8.20× | gene structure annotation | SRR23354528 |
| Illumina NovaSeq | PE | Leaf | RNA | 6.93 Gb | 7.97× | gene structure annotation | SRR23354530 |
| Illumina NovaSeq | PE | Flower | RNA | 6.92 Gb | 7.96× | gene structure annotation | SRR23354527 |
| Illumina NovaSeq | PE | Fruit | RNA | 6.94 Gb | 7.98× | gene structure annotation | SRR23354526 |
The PacBio Sequel II System, based on single-molecule real-time (SMRT) sequencing technology under the Circular Consensus Sequencing (CCS) model, was used for whole-genome sequencing. The DNA template was sheared by g-TUBE (Covaries, USA) to an average size of 15–20 kb, and the target DNA fragments were obtained using BluePippinTM Size-Selection System (Sage Science, USA). The library was constructed using SMRTbell Template Prep Kit 1.0 (Pacific Biosciences, USA) following the procedure and loaded onto PacBio Sequel™ Systems to read the sequence. Finally, approximately 366.02 Gb subreads were obtained with an average length of 13.59 kb and an N50 length of 15.25 kb after removing adaptors in polymerase reads (Table 1).
RNA library construction and transcriptome sequencing
Total RNA was isolated from the roots, stems, leaves, flowers, and fruits, respectively, using the standard TRIzol protocol (Invitrogen, USA)32. Approximately 100 mg of tissue was ground to powder using liquid nitrogen, and then 1000 μL of TRIzol was added in a 2.0-mL tube. After allowing the solution to stand for approximately 5 min, 200 μL of chloroform was added, shaken vigorously for 30 s, and allowed to stand for 3 min. After centrifugation at 12,000 rpm for 15 min at 4 °C, the upper aqueous phase was extracted to another 1.5-mL tube with 500 μL of isopropanol and then mixed by gently inverting. After standing for approximately 10 min, the tube was centrifuged at 12,000 rpm for 10 min. The supernatant was removed, and the pellet was washed twice with 75% ethanol and dissolved in 50 µL of DNase and RNase-free Water for further study.
For the Illumina paired-end reads sequencing, the mRNA was synthesized to cDNA, and five libraries were constructed with an insertion size of 350 bp using a TruSeq RNA library preparation kit (Illumina, USA) following the manufacturer’s instructions. Whole-genome shotgun raw sequencing was performed using the Novaseq 6000 Platform set in the PE150 program. In total, 32.91 Gb of clean data were generated from the RNA-seq library after filtering using fastp31 (Table 1).
For Iso-seq under the CCS model, the RNA samples extracted from root, stem, leaves, flowers, and fruits were equally mixed for sequencing. cDNA was synthesized using a Clontech SMARTer PCR cDNA Synthesis Kit (Takara Biotechnology, China). Then, the SMRTbell library (cDNAs length over 4 kb) was constructed using the Pacific Biosciences SMRTbell template prep kit (Pacific Biosciences, USA) and sequenced on the Pacific Bioscience Sequel II platform. A total of 19.81 Gb subreads were obtained with an average length of 2,562 bp and an N50 length of 3,005 bp after removing adaptors in polymerase reads (Table 1). The exported subreads were analyzed using packages of SMRT link v10.1, including highly accurate consensus sequence calling using package ccs v6.0.0 (https://github.com/PacificBiosciences/ccs), primer removal and demultiplexing using package lima v2.1.0 (https://github.com/pacificbiosciences/barcoding/), polyA tail and artificial concatemers removal using package isoseq3 v3.4.0 (https://github.com/PacificBiosciences/IsoSeq), and clustering and polishing using package isoseq3 v3.4.0. Finally, approximately 387.83 Mb high-quality consensus isoform sequences were generated with an average length of 3,843 bp.
Contig-level genome assembly
The in-built High-Quality Region Finder (HQRF) was used to identify the longest high-quality regain for each read of exported subreads according to the signal noise ratio (SNR). HiFi reads were then generated from filtered subreads using the CCS model of SMRT link v10.1 with the following parameters: --maxLength = 50000, --minPasses = 3, and --minPredictedAccuracy = 0.99. The sequences in fastq.gz were converted from the BAM file using bam2fastx v1.3.1 (https://github.com/pacificbiosciences/bam2fastx/). 25.83 Gb (28.75×) of CCS reads were obtained with an average length of 15.34 kb and an N50 length of 15.78 kb (Table 1). Then, Hifiasm v0.16.025 was used to assemble the genome into contigs with default parameters. To check for the potential contaminant sequences, assembled contigs were classified using Kraken2 against the custom database33. Four contigs were identified as bacteria (904,041 bp, 0.10%), which were flagged and removed from the final assembly. After removal, the final contig-level assembly was submitted to the NCBI independent contamination check to confirm the result, resulting in an 898.42 Mb contig-level genome consisting of 113 contigs and an N50 length of 62.00 Mb (Table 2).
Table 2.
Statistics of the Solanum rostratum genome assembly.
| Feature | Metric |
|---|---|
| Hifiasm-derived contigs | |
| Number of contigs | 113 |
| Total length of contigs | 898,418,632 bp |
| Longest contig | 92,793,329 bp |
| contig N50 | 62,003,212 bp (contig number = 7) |
| Contig N60 | 51,582,526 bp (contig number = 8) |
| Contig N70 | 44,575,705 bp (contig number = 10) |
| Contig N80 | 31,624,497 bp (contig number = 13) |
| Contig N90 | 12,481,291 bp (contig number = 17) |
| Hi-C scaffolded assembly | |
| Number of scaffolds | 224 |
| Total length of scaffolds | 869,692,437 bp |
| Longest scaffold | 92,283,834 bp |
| Scaffold N50 | 72,149,870 bp (scaffold number = 6) |
| Scaffold N60 | 69,863,894 bp (scaffold number = 8) |
| Scaffold N70 | 68,611,519 bp (scaffold number = 9) |
| Scaffold N80 | 67,684,033 bp (scaffold number = 10) |
| Scaffold N90 | 63,146,500 bp (scaffold number = 12) |
| GC content | 36.99% |
Hi-C library construction and pseudo-chromosome anchoring
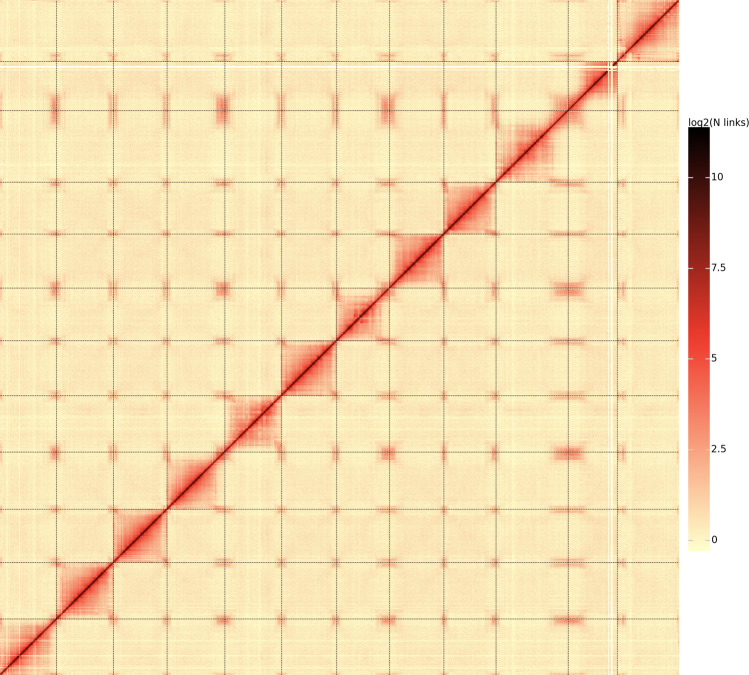
Tender leaves were cut into approximately 2-cm2 pieces for cellular protein cross-linking in 2% formaldehyde. The isolated DNA was purified, digested with Dpnii restriction enzyme, tagged with biotin-14-dCTP, sheared into 300–600 bp fragments, and blunt-end-repaired. Then, the Hi-C library was sequenced using the Illumina NovaSeq platform, which generated 100.16 Gb filtered clean data (113.63×) to anchor contigs into pseudo-chromosomes (Table 1). The cleaned Hi-C sequencing data were aligned on the contig assembly using bowtie2 v2.2.534 to obtain the unique mapped paired-end reads using the following parameters:--very-sensitive -L 20--score-min L, -0.6, --0.2 --end-to-end --reorder --rg-id BMG --phred33-quals -p 5. Quality control of read alignment and pairing was conducted using HiC-Pro v2.7.826 to discard low-quality alignment, singleton, multiple hits, and invalid pairs. A total of 156,223,644 valid paired-end reads were used to build the interaction matrices and scale up the primary genome assembly in contigs to chromosome-scale scaffolds (pseudo-chromosomes). A total of 869.69 Mb of the contig-level assembled sequences (96.80% anchored rate) were anchored and orientated onto 12 pseudo-chromosomes, which was consistent with the karyotype (2n = 24) analysis35, with lengths ranging from 63.15 to 92.28 Mb (Table 3). In summary, the size of the pseudochromosome-level S. rostratum genome that was obtained was 869.69 Mb with 212 unanchored contigs (total length 28.73 Mb), with a contig N50 of 72.15 Mb (Table 2). To validate the correction of the pseudo-chromosome anchoring result, the pseudo-chromosomes were divided into bins of equal size in 50 kb to construct genome-wide interaction matrices based on the interaction signals between each pair of bins. The interaction matrix heatmap was visualized using HiCPlotter v0.6.636 (Fig. 3).
Table 3.
Statistics of Solanum rostratum genome assembly result by Hi-C.
| Chromosome | Total length | ATCG base number | N base number | Gap ratio |
|---|---|---|---|---|
| Chr01 | 92,283,834 | 92,280,834 | 3,000 | 0.003251% |
| Chr02 | 79,062,878 | 79,056,378 | 6,500 | 0.008221% |
| Chr03 | 73,759,959 | 73,758,959 | 1,000 | 0.001356% |
| Chr04 | 73,025,073 | 73,024,073 | 1,000 | 0.001369% |
| Chr05 | 72,660,254 | 72,653,754 | 6,500 | 0.008946% |
| Chr06 | 72,149,870 | 72,137,370 | 12,500 | 0.017325% |
| Chr07 | 70,619,802 | 70,617,802 | 2,000 | 0.002832% |
| Chr08 | 69,863,894 | 69,863,894 | 0 | 0.000000% |
| Chr09 | 68,611,519 | 68,611,019 | 500 | 0.000729% |
| Chr10 | 67,684,033 | 67,678,033 | 6,000 | 0.008865% |
| Chr11 | 66,824,821 | 66,821,821 | 3,000 | 0.004489% |
| Chr12 | 63,146,500 | 63,144,000 | 2,500 | 0.003959% |
| Total anchored | 869,692,437 (96.80%) | 869,647,937 | 44,500 | 0.005117% |
| Unanchored | 28,726,195 (3.20%) |
Fig. 3.
Heat map of genome-wide Hi-C intra-chromosome interactions in Solanum rostratum. The interaction density is measured by the number of supporting Hi-C reads and illustrated by the color bar from dark red (high density) to light pink (low density).
Genome annotation and functional prediction
Identifying repeat sequences
The repeat sequences in the genome were identified using a combination of homologous sequence prediction and ab initio prediction. For homologous sequence prediction, RepeatMasker v1.32337 and RepeatProteinMask v1.3638 were used to predict the homology sequences against known repeat sequences in the database RepBase39. For ab initio prediction, RepeatModeler open-1.0.840 was used to establish a de novo repeat sequence database, and RepeatMasker v1.32337 was used for prediction. Tandem Repeats Finder (TRF) v4.07b41 was used to find tandem repeat sequences in the genome. Combined with the results, 649.21 Mb repeat sequences were identified, accounting for 72.26% of the S. rostratum genome. The four predominant categories were long terminal repeats (LTR) (accounting for 46.06% of genome size), long interspersed nuclear elements (LINE) (3.62%), DNA elements (3.14%), and short interspersed nuclear elements (SINE) (0.22%) (Table 4).
Table 4.
Statistics of repeat elements in the genome of Solanum rostratum.
| Class | RepeatMasker TEs | RepeatProteinMask TEs | RepeatModeler TEs | Combined TEs | ||||
|---|---|---|---|---|---|---|---|---|
| Type | Length (bp) | % in genome | Length (bp) | % in genome | Length (bp) | % in genome | Length (bp) | % in genome |
| DNA | 20,569,654 | 2.29 | 769,635 | 0.09 | 11,659,600 | 1.3 | 28,196,585 | 3.14 |
| LINE | 22,128,879 | 2.46 | 19,023,801 | 2.12 | 28,490,398 | 3.17 | 32,487,489 | 3.62 |
| SINE | 1,961,417 | 0.22 | 0 | 0 | 0 | 0 | 1,961,417 | 0.22 |
| LTR | 231,529,759 | 25.77 | 163,968,428 | 18.25 | 363,534,714 | 40.46 | 413,866,571 | 46.06 |
| Unknown | 34,056 | 0 | 0 | 0 | 234,827,738 | 26.14 | 234,857,657 | 26.14 |
| Other | 1,462,968 | 0.16 | 16,942,527 | 1.89 | 9,137,996 | 1.02 | 25,927,555 | 2.89 |
| Total | 277,121,683 | 30.84 | 200,396,923 | 22.3 | 624,136,628 | 69.47 | 649,214,848 | 72.26 |
Abbreviations: LINE, long interspersed nuclear element; SINE, short interspersed nuclear element; LTR, long terminal repeat.
Identifying non-coding RNA (ncRNA) gene
Rfam42 was used to predict ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), and micro RNAs (miRNAs) by comparison with known non-coding RNA libraries. Transfer RNAs (tRNAs) were predicted using tRNAscan-SE v1.3.143. In total, 3,588 ncRNAs were annotated in the S. rostratum genome, including 547 miRNAs, 1,288 tRNAs, 1,110 rRNAs, and 643 snRNAs (Table 5).
Table 5.
Statistics for non-coding RNA genes in the genome of Solanum rostratum.
| Class | Type | Numbers | Average length (bp) | Total length (bp) | Percentage in genome (%) |
|---|---|---|---|---|---|
| miRNA | 547 | 96.73 | 52,910 | 0.00589 | |
| tRNA | 1,288 | 75.25 | 96,925 | 0.01079 | |
| rRNA | 18 S | 84 | 603.13 | 50,663 | 0.00564 |
| 28 S | 82 | 132.06 | 10,829 | 0.00121 | |
| 5.8 S | 30 | 134.53 | 4,036 | 0.00045 | |
| 5 S | 914 | 119.64 | 109,349 | 0.01217 | |
| snRNA | CD-box | 359 | 101.62 | 36,481 | 0.00406 |
| HACA-box | 58 | 123.33 | 7,153 | 0.00080 | |
| splicing | 226 | 135.37 | 30,593 | 0.00341 |
Gene structure prediction
Three strategies were applied to predict the gene structure from the repeat-masked genome. The first strategy was homologous prediction. BLAST v2.2.2844 with an E-value cutoff of 1e-5 and GeMoMa v1.645 were used to predict gene structure by comparing with seven closely related species (C. annuum19, Solanum chilense46, Solanum commersonii47, S. lycopersicum16, S. melongena20, Solanum pennellii48, and S. tuberosum18). The second strategy was based on transcriptome data. The filtered Illumina RNA-seq sequences from five libraries were assembled into transcripts using Trinity v2.11.049 with default parameters. Then, the Trinity RNA-Seq assemblies and full-length cDNAs were aligned and mapped to the soft-masked genome assembly using GMAP v2014-10-250 and BLAT Src3551. Candidate gene structures were extracted from the PASA v2.128 pipeline based on the open reading frame (ORF). The third strategy included using ab initio prediction based on the characteristics of the genomic sequence data. Using Augustus v3.352, SNAP v3892653, and GeneMark v4.3354, 29,485, 33,190, and 26,142 protein-coding genes were identified, respectively. Finally, EVM v1.1127 integrated the above three strategies, resulting in a non-redundant gene set, with weighting as default. Overall, 29,694 protein-coding genes were obtained, with an average gene length of 4,308 bp, cds length of 1,172 bp, exon length of 237 bp, and intron length of 795 bp (Table 6).
Table 6.
Summary of gene structure prediction by three strategies of Solanum rostratum.
| Gene set | Number of proteins | Average gene length (bp) | Average cds length (bp) | Average exons per gene | Average exon length (bp) | Average intron length (bp) | |
|---|---|---|---|---|---|---|---|
| ab initio prediction | Augustus | 29,485 | 4,661.13 | 1,284.52 | 4.89 | 262.70 | 869.09 |
| GeneMark | 33,190 | 4,931.98 | 1,127.19 | 5.35 | 210.83 | 876.39 | |
| SNAP | 26,142 | 17,230.72 | 596.34 | 3.93 | 151.56 | 5669.30 | |
| Transcriptome-based prediction | cDNA | 27,408 | 7,687.74 | 1,493.68 | 6.93 | 387.13 | 845.39 |
| Homology-based prediction | Capsicum annuum | 32,842 | 3,902.16 | 1,137.54 | 4.76 | 239.12 | 736.83 |
| Solanum chilense | 27,145 | 3,902.19 | 1,150.88 | 4.79 | 240.17 | 726.58 | |
| Solanum commersonii | 41,616 | 3,270.56 | 905.08 | 4.35 | 208.08 | 707.19 | |
| Solanum lycopersicum | 28,703 | 4,264.34 | 1,255.14 | 5.04 | 248.87 | 745.25 | |
| Solanum melongena | 32,600 | 3,497.00 | 1,018.97 | 4.47 | 227.85 | 714.71 | |
| Solanum pennellii | 29,777 | 4,275.29 | 1,252.46 | 5.03 | 249.07 | 751.35 | |
| Solanum tuberosum | 29,750 | 4,256.09 | 1,245.78 | 5.03 | 247.55 | 747.54 | |
| Total | EVM | 29,694 | 4,308.30 | 1,171.80 | 4.95 | 236.83 | 795.49 |
Gene function annotation
Functional annotation of the protein-coding genes was carried out via BLAST44, with an E-value cutoff of 1e-5, against the public protein databases, including the Non-redundant protein database (NR) (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz), the nucleotide sequence database (NT) (https://www.ncbi.nlm.nih.gov/nucleotide/), SwissProt protein database (SwissProt)55, Kyoto Encyclopedia of Genes and Genomes (KEGG)56, Eukaryotic Orthologous Groups of proteins (KOG)57, and eggNOG-mapper v2.1.0-158. Protein domains were predicted by searching against the Protein Families Database (Pfam)59 using Hmmer v3.1b160 with default settings. Gene Ontology (GO)61 terms were obtained based on the corresponding InterPro62 or Pfam59 entry. A total of 28,154 genes (94.81%) were annotated using at least one public database (Table 7).
Table 7.
Statistics for the Solanum rostratum functionally annotated protein-coding genes.
| Database | Number | Percentage in genome (%) | |
|---|---|---|---|
| Protein-coding genes | 29,694 | 100 | |
| Annotated genes | 28,154 | 94.81 | |
| BLASTP | 21,756 | 73.27 | |
| BLASTX | 21,542 | 72.55 | |
| GO | 21,987 | 74.05 | |
| KEGG ID | 8,047 | 27.10 | |
| KEGG Pathway | 5,062 | 17.05 | |
| NR | 27,830 | 93.72 | |
| NT | 26,451 | 89.08 | |
| PFAM | 21,703 | 73.09 | |
| eggNOG | 18,076 | 60.87 | |
| Unannotated genes | 1,540 | 5.19 | |
Solanaceous orthology identification, phylogenetic tree construction, and divergence time estimation
Twenty solanaceous species were selected for comparative genomic analysis, with Ipomoea trifida as the outgroup. The longest transcripts, which were extracted using TBtools v1.10663, were used as the gene set for the following analysis. The orthogroups and orthologs classification were identified using Orthofinder v2.5.464 with parameters -S diamond, -M msa, and -T fasttree. As a result, 824,030 genes (93.10% of total genes) were assigned to 56,426 orthogroups among 21 species, with 7,963 orthogroups shared in all the species and 799 shared single-copy orthogroups. Among the 29,694 genes in S. rostratum, 28,514 were clustered into 17,237 orthogroups, with 12,096 genes in single-copy orthologs, 16,418 genes in multiple-copy orthologs, and 1,065 genes in 298 species-specific orthogroups.
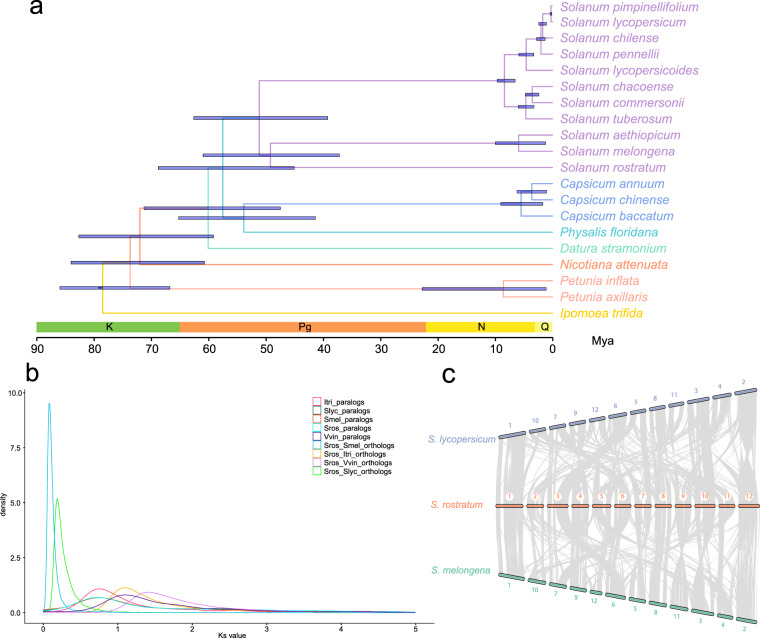
A phylogenetic tree was constructed using the concatenated 799 single-copy orthogroup gene alignment generated using Orthofinder64. The maximum-likelihood method software raxmlHPC v8.2.1265 was implemented with the parameters -m PROTGAMMAJTT, -f a, and -# 100. The solanaceous tree recovered the monophyly of 3 subfamilies, 5 tribes, and 6 genera with 100 support values at all nodes, revealing a sister group relationship between S. rostratum and S. melongena + S. aethiopicum (Fig. 4a).
Fig. 4.
Comparative genomic and evolution analysis of solanaceous species. (a) Phylogenetic topology constructed based on shared single-copy genes, and divergence times estimation of solanaceous species with Ipomoea trifida as an outgroup. All the nodes supported bootstrap values are 100. The blue bars on the nodes represent the divergence time range with 95% confidence intervals (million years ago, Mya). The below scale represents the geologic time divisions, covering Cretaceous (K), Paleogene (Pg), Neogene (N), and Quaternary (Q). (b) Whole Genome Duplication events revealed by synonymous substitution rate (Ks) analysis. The Ks frequency density distributions of syntenic orthologous or paralogous block pairwise within and between genomes of Solanum rostratum (Sros) and Vitis vinifera (Vvin), Ipomoea trifida (Itri), Solanum lycopersicum (Slyc), and Solanum melongena (Smel). (c) Whole-genome synteny between S. rostratum and two other closely related Solanum species (S. lycopersicum and S. melongena). Conserved syntenic blocks are highlighted with grey color corresponding to the twelve pseudo-chromosomes, indicating visible genome rearrangements occurred during evolution among Solanum species.
Four-fold Degenerate Synonymous Site (4DTv) was extracted from single-copy orthogroup genes to estimate the divergence time among Solanaceae using MCMCTree in PAML v4.10.366 with the following parameters: clock = correlated rates, model = H85KY, alpha = 0.5, burn in = 100,000, sample frequency = 2, and sample number = 1,000,000. Two calibrations were set, which were obtained from Timetree67: the divergence time between Solanaceae and Convolvulaceae (59.1–83.9 million years ago [Mya]), and the divergence time between S. lycopersicum and S. tuberosum (6.1–9.0 Mya). The results revealed that S. rostratum split from the common ancestor ca. 49.26 Mya (Fig. 4a).
WGD analysis
To investigate the WGD event history of Solanaceae, the synonymous substitution rate (Ks) frequency density distributions of syntenic orthologous block pairwise between genomes and syntenic paralogous block pairwise within genomes were calculated by wgd v1.1.268, including S. rostratum (Sros), Vitis vinifera69 (Vvin), Ipomoea trifida70 (Itri), S. lycopersicum16 (Slyc), and S. melongena20 (Smel). For one-versus-one orthologs Ks distributions calculation, the module dmd was implemented to extract orthologs by all-versus-all blastp using the diamond71 algorithm with the parameters–nostrictcds -e 1e-10. The module ksd66 was then used to construct one-versus-one ortholog Ks distributions. For whole-paranome Ks distribution calculation, the module dmd was used to extract paralogs and cluster gene families using the Markov cluster (MCL)72 algorithm. Then, the module ksd66 was used to construct whole-paranome Ks distributions with the parameter -mp 1000. Finally, the module syn identified and extracted paralogs in intra-genomic colinear blocks using i-ADHoRe v3.073. A shared peak was detected within Solanaceae at approximately 0.68, which occurred after the divergence peak with V. vinifera, and before the Solanaceae speciation peak, indicating that an an ancient WGD occurred in the ancestor of the Solanaceae. However, there was no subsequent WGD after species differentiation within the Solanaceae. Within Solanaceae, S. rostratum first diverged from S. lycopersicum at 0.14, and S. melongena at 0.03 (Fig. 4b).
Whole-genome synteny
To understand the extend of genomic rearrangement of S. rostratum during evolution, whole-genome synteny analysis was conducted between S. rostratum (Sros) and S. lycopersicum16 (Slyc), and between S. rostratum and S. melongena20 (Smel). The protein sequences of Sros and Slyc, and Sros and Smel were blasted using blastp with parameter -evalue 1e-5. The multiple alignments of syntenic blocks were identified by MCScanX74 with the parameter -s 15 (number of genes required to call a collinear block) and visualized by jcvi v1.2.875 with the parameter–minspan = 30. The complicated conserved syntenic blocks among the twelve pseudo-chromosomes, indicate that visible genome rearrangements occurred during evolution among Solanum (Fig. 4c).
Data Records
All raw sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) (Table 1) under Bioproject number PRJNA932047, including the genomic Illumina sequencing data (SRR23354532)76, genomic PacBio HiFi sequencing data (SRR23354533)77, transcriptome Illumina sequencing data (SRR23354526-SRR23354530)78–82, Hi-C sequencing data (SRR23354531)83, and transcriptome Pacbio-Sequel II sequencing data (SRR23354525)84.
The final chromosome-level assembled genome sequences were deposited in the NCBI Assembly database under Accession Number JARACL00000000085.
The genome annotation results, including repeated sequences, gene structure, and functional predictions were deposited in the Figshare database (10.6084/m9.figshare.22016024)86.
Technical Validation
Evaluation of the quality of genomic DNA and RNA
The purification, concentration, and integrity of the DNA template were quantitatively determined using a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific, USA), Qubit Fluorometers (Thermo Fisher Scientific, USA), and Agilent 4200 Bioanalyzer (Agilent Technologies, USA), respectively. The evaluation results require the 15 kb insert library of the PacBio Sequel sequencing platform to meet the following criteria: including (1) the DNA content ≥ 10 μg, (2) the DNA concentration ≥ 80 ng/μL, (3) the DNA peak size was 32.59 kb which was over than 20 kb, (4) the DNA absorbance was 1.8 ≤ OD260/280 ≤ 2.0 and 1.6 ≤ OD260/230 ≤ 2.5.
The purification, concentration, and integrity of the RNA template were quantitatively determined using a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific, USA), an Agilent 2100 Bioanalyzer (Agilent Technologies, USA), and an Agilent RNA 6000 Nano Kit (Agilent Technologies, USA), respectively. The evaluation results required all the meet for the Iso-seq library construction, including (1) the RNA content ≥ 4 μg, (2) the RNA concentration ≥ 250 ng/μL, (3) RNA Integrity Number (RIN) value ≥ 6.0, (4) the RNA absorbance was 2.0 ≤ OD260/280 ≤ 2.2, and 1.6 ≤ OD260/230 ≤ 2.1.
Evaluating the completeness and quality of the genome assembly and annotation
Flow cytometry analysis
FACScalibur Flow cytometry (BD Biosciences, USA) analysis87 was conducted to estimate the S. rostratum genome size with three replicates, and ModFit software v5.0 (Yerity SoftwareHouse, USA) was used to analyze the results. The genome size of the internal reference standard Glycine max is 978.4 Mb (1 pg DNA = 0.978 G)88. The 2 C DNA content in pg of S. rostratum was calculated according to the following formula89: . The peak values of G. max were 104.29, 106.54, and 103.70, respectively. The corresponding peak values for S. rostratum were 94.32, 97.10, and 94.28, respectively. The genome size of S. rostratum was estimated to be approximately 885.36–892.20 Mb, which was very close to the genome size of the pseudo-chromosome-level assembly in 898.42 Mb.
Mapped to the genome using Illumina data
Illumina paired-end reads were mapped back to the draft genome using Burrows-Wheeler Aligner (BWA) v0.7.9a90. Then, depth, mapping rates, and coverage at each position were calculated using samtools v0.1.1991. The results showed that 99.04% of read pairs were mapped to the genome with an average depth of 105.24 and a coverage rate of 97.54%, indicating high single-base concordance.
BUSCO assessment
The completeness of the contig-level genome, Hi-C pseudo-chromosome-level genome, and predicted gene datasets were further evaluated with BUSCO (default parameters) v5.1.229 based on the ortholog database embryophyta_odb10 (1,614 genes). The results were visualized by the python script generate_plot.py of BUSCO, showing a high completeness level with 99.4%, 99.5%, and 91.3% complete genes found in the contig-level genome, Hi-C pseudo-chromosome-level genome, and predicted gene datasets, respectively (Fig. 5).
Fig. 5.
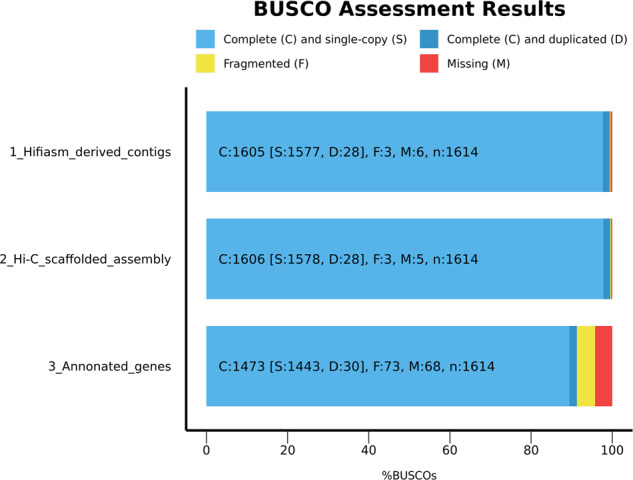
Benchmarking of genome completeness of Solanum rostratum genome assembly and annotation, evaluated by BUSCO based on embryophyta_odb10 database which includes 1,614 genes. C: the number of complete genes, S: the number of complete and single-copy genes, D: the number of complete and duplicated genes, F: the number of incomplete genes, M: the number of missing genes.
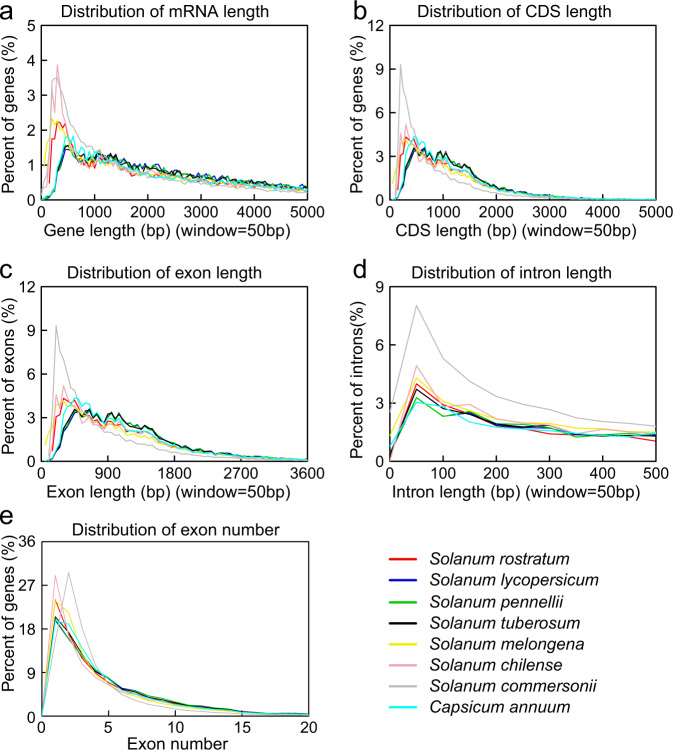
Protein coding genes comparison with close species
To determine the prediction accuracy and reliability, the distribution of mRNA length, CDS length, exon length, intron length, and exon number in S. rostratum and other closely related species (C. annuum19, S. chilense46, S. commersonii47, S. lycopersicum16, S. melongena20, S. pennellii48, and S. tuberosum18) were determined. The consistent distribution tendency among all species further supported an ideal annotated gene dataset in S. rostratum (Fig. 6).
Fig. 6.
Annotated genes comparison of the distribution of (a) mRNA length (b) CDS length (c) exon length (d) intron length (e) exon number in Solanum rostratum with other closely related species. The x-axis represents the length or number and the y-axis represents the density of genes.
Hence, a high-quality completeness and accuracy S. rostratum genome was assembled and annotated in the present study.
Supplementary information
Acknowledgements
The work was supported by the National Key R&D Program of China (2021YFD1400300), China Postdoctoral Science Foundation (2022M723444), National Key R&D Program of China (2022YFC2601400), and Central Public-interest Scientific Institution Basal Research Fund (BSRF202216).
Author contributions
G.L.Z. and W.D.F. conceived and designed the study; Z.H.W. and Z.L.Y. collected the samples and extracted the genomic DNA for sequencing; Y.Z. and Z.S. assembled the genome; Y.Z. and Z.L.Y. performed Hi-C scaffolding and genome annotation; W.C.G., Z.S. and J.H.G. performed technical validation; Y.Z. wrote the draft manuscript. W.C.G. and Y.Z. modified the manuscript. All authors have read, revised, and approved the final manuscript for submission.
Code availability
No specific custom codes were developed in this study. All commands and pipelines used for data analyses were conducted according to the manuals or protocols provided by the corresponding software development team, which are described in detail in the Methods section. Default parameters were employed if no detailed parameters were mentioned for the software used in this study. Supplementary Table 1 lists the versions, settings, and parameters of the relevant software used in this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Weidong Fu, Email: fuweidong@caas.cn.
Guoliang Zhang, Email: zhangguoliang@caas.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-023-02247-3.
References
- 1.Randall, R. P. A Global Compendium of Weeds. (Department of Agriculture and Food Western Australia, 2012).
- 2.Lin Y, Tan DY. The potential and exotic invasive plant: Solanum rostratum. Acta Phytotaxonomiea Sinica. 2007;45:675–685. doi: 10.1360/aps07010. [DOI] [Google Scholar]
- 3.Bowers KA. The pollination ecology of Solanum rostratum (Solanaceae) Am. J. Bot. 1975;62(6):633–638. doi: 10.1002/j.1537-2197.1975.tb14094.x. [DOI] [Google Scholar]
- 4.Weber DC, Drummond FA, Ferro DN. Recruitment of Colorado potato beetles (Coleoptera: Chrysomelidae) to solanaceous hosts in the field. Environ. Entomol. 1995;24(3):608–622. doi: 10.1093/ee/24.3.608. [DOI] [Google Scholar]
- 5.Mauricio-Castillo JA, Argüello-Astorga GR, Ambriz-Granados S, Alpuche-Solís AG, Monreal-Vargas CT. First Report of Tomato golden mottle virus on Lycopersicon esculentum and Solanum rostratum in Mexico. Plant Dis. 2007;91(11):1513–1513. doi: 10.1094/PDIS-91-11-1513B. [DOI] [PubMed] [Google Scholar]
- 6.Singh RP, Bagnall RH. Solanum rostratum Dunal., a new test plant for the potato spindle tuber virus. Am. J. Potato Res. 1968;45(9):335–336. doi: 10.1007/BF02849770. [DOI] [Google Scholar]
- 7.Bassett IJ, Munro DB. The biology of Canadian weeds.: 78. Solanum carolinense L. and Solanum rostratum Dunal. Can. J. Plant Sci. 1986;66(4):977–991. doi: 10.4141/cjps86-120. [DOI] [Google Scholar]
- 8.Wei S, et al. Factors affecting buffalobur (Solanum rostratum) seed germination and seedling emergence. Weed Sci. 2009;57(5):521–525. doi: 10.1614/WE-09-054.1. [DOI] [Google Scholar]
- 9.USDA-NRCS. The PLANTS Database. National Plant Data Centerhttp://plants.usda.gov/ (2014).
- 10.2022. GBIF Backbone Taxonomy. GBIF Secretariat. [DOI]
- 11.Guan G, et al. Solanum rostratum - A quarantine weed. Plant Quarantine. 1984;11:359–426. [Google Scholar]
- 12.Kane NC, Rieseberg LH. Genetics and evolution of weedy Helianthus annuus populations: adaptation of an agricultural weed. Mol. Ecol. 2008;17(1):384–394. doi: 10.1111/j.1365-294X.2007.03467.x. [DOI] [PubMed] [Google Scholar]
- 13.Marks RA, Hotaling S, Frandsen PB, VanBuren R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants. 2021;7(12):1571–1578. doi: 10.1038/s41477-021-01031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Shang L, Zhu QH, Fan L, Guo L. Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci. 2021;27(4):391–401. doi: 10.1016/j.tplants.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 15.PBI Solanum Project. Solanaceae Source website. USA: Planetary Biodiversity Inventories (PBI), National Science Foundationhttp://www.solanaceaesource.org/ (2014).
- 16.Hosmani, P.S. et al. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. BioRxiv, 767764 (2019).
- 17.Takei H, et al. De novo genome assembly of two tomato ancestors, Solanum pimpinellifolium and Solanum lycopersicum var. cerasiforme, by long-read sequencing. DNA Res. 2021;28(1):dsaa029. doi: 10.1093/dnares/dsaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham GM, et al. Construction of a chromosome-scale long-read reference genome assembly for potato. GigaScience. 2020;9(9):giaa100. doi: 10.1093/gigascience/giaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin C, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA. 2014;111(14):5135–5140. doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barchi L, et al. Improved genome assembly and pan-genome provide key insights into eggplant domestication and breeding. Plant J. 2021;107(2):579–596. doi: 10.1111/tpj.15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sierro N, et al. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014;5(1):3833. doi: 10.1038/ncomms4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, et al. The Physalis floridana genome provides insights into the biochemical and morphological evolution of Physalis fruits. Hortic. Res. 2021;8(1):244. doi: 10.1038/s41438-021-00705-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajewski A, Carter-House D, Stajich J, Litt A. Datura genome reveals duplications of psychoactive alkaloid biosynthetic genes and high mutation rate following tissue culture. BMC Genomics. 2021;22(1):1–19. doi: 10.1186/s12864-021-07489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao YL, et al. Wolfberry genomes and the evolution of Lycium (Solanaceae) Commun. Biol. 2021;4(1):671. doi: 10.1038/s42003-021-02152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Servant N, et al. HiC-Pro: An optimized and flexible pipeline for Hi-C processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:1–22. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seppey, M., Manni, M. & Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness. In Gene prediction, M. Kollmar, ed. (New York, USA: Springer), pp. 227-245 (2019). [DOI] [PubMed]
- 30.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 31.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rio DC, Ares M, Hannon GJ, Nilsen TW. Purifcation of RNA using TRIzol (TRI reagent) Cold Spring Harbor Protocols. 2010;6:pdb–prot5439. doi: 10.1101/pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 33.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:1–13. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice A, et al. The Chromosome Counts Database (CCDB) - a community resource of plant chromosome numbers. New Phytol. 2015;206:19–26. doi: 10.1111/nph.13191. [DOI] [PubMed] [Google Scholar]
- 36.Akdemir KC, Chin L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 2015;16:1–8. doi: 10.1186/s13059-015-0767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tempel S. Using and Understanding RepeatMasker. Methods Mol. Biol. 2012;859:29–51. doi: 10.1007/978-1-61779-603-6_2. [DOI] [PubMed] [Google Scholar]
- 38.Tarailo‐Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009;25:4–10. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 39.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flynn JM, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S, et al. Rfam: an RNA family database. Nucleic Acids Res. 2003;31(1):439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(suppl_2):W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keilwagen, J., Hartung, F. & Grau, J. GeMoMa: Homology-based gene prediction utilizing intron position conservation and RNA-seq data. In Gene prediction, Kollmar, M. ed. (New York, USA: Springer), pp. 161-177 (2019). [DOI] [PubMed]
- 46.Stam R, et al. The de novo reference genome and transcriptome assemblies of the wild tomato species Solanum chilense highlights birth and death of NLR genes between tomato species. G3. 2019;9(12):3933–3941. doi: 10.1534/g3.119.400529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.2023. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/assembly/GCA_018258275.1/
- 48.Bolger A, et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014;46(9):1034–1038. doi: 10.1038/ng.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21(9):1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 51.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32(suppl_2):W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leskovec J, Sosič R. Snap: A general-purpose network analysis and graph-mining library. ACM T. Intel. Syst. Tec. 2016;8(1):1–20. doi: 10.1145/2898361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18(12):1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4(1):41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huerta-Cepas J, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn, R.D. et al. Pfam: the protein families database. Nucleic Acids Res. 42(Database issue), 222-30 (2014). [DOI] [PMC free article] [PubMed]
- 60.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paysan-Lafosse T, et al. InterPro in 2022. Nucleic Acids Res. 2023;51(D1):D418–D427. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:1–14. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang ZH. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 67.Benton MJ, Donoghue PCJ, Asher RJ. Calibrating and constraining molecular clocks. The Timetree of Life. 2009;35:86. [Google Scholar]
- 68.Zwaenepoel A, Van de Peer Y. wgd - simple command line tools for the analysis of ancient whole genome duplications. Bioinformatics. 2019;35(12):2153–2155. doi: 10.1093/bioinformatics/bty915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.The French-Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449(7161):463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 70.Hirakawa H, et al. Survey of genome sequences in a wild sweet potato, Ipomoea trifida (HBK) G. Don. DNA Res. 2015;22(2):171–179. doi: 10.1093/dnares/dsv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using diamond. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 72.van Dongen S.M. Graph Clustering by Flow Simulation. PhD Thesis, University of Utrecht, Utrecht, The Netherlands (2000).
- 73.Proost S, et al. i-ADHoRe 3.0: fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012;40(2):e11. doi: 10.1093/nar/gkr955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang YP, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49–e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang H, et al. Synteny and collinearity in plant genomes. Science. 2008;320(5875):486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 76.2023. NCBI Sequence Read Archive. SRR23354532
- 77.2023. NCBI Sequence Read Archive. SRR23354533
- 78.2023. NCBI Sequence Read Archive. SRR23354526
- 79.2023. NCBI Sequence Read Archive. SRR23354527
- 80.2023. NCBI Sequence Read Archive. SRR23354528
- 81.2023. NCBI Sequence Read Archive. SRR23354529
- 82.2023. NCBI Sequence Read Archive. SRR23354530
- 83.2023. NCBI Sequence Read Archive. SRR23354531
- 84.2023. NCBI Sequence Read Archive. SRR23354525
- 85.Zhang Y, 2023. Solanum rostratum isolate YZ_2023a, whole genome shotgun sequencing project. GenBank. JARACL000000000
- 86.Zhang Y, 2023. Chromosome-level genome assembly and annotation of the prickly nightshade Solanum rostratum Dunal. figshare. [DOI] [PMC free article] [PubMed]
- 87.Duda M, Gasińska A, Gregoraszczuk E. Flow cytometric cell cycle analysis of two subpopulations of porcine granulosa cells. Exp. Clin. Endocrinol. Diabetes. 1999;107(03):203–207. doi: 10.1055/s-0029-1212099. [DOI] [PubMed] [Google Scholar]
- 88.Valliyodan B, et al. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 2019;100(5):1066–1082. doi: 10.1111/tpj.14500. [DOI] [PubMed] [Google Scholar]
- 89.Doležel J, Bartoš JAN. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot-London. 2005;95(1):99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv e-prints (2013).
- 91.Danecek P, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2022. GBIF Backbone Taxonomy. GBIF Secretariat. [DOI]
- 2023. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/assembly/GCA_018258275.1/
- 2023. NCBI Sequence Read Archive. SRR23354532
- 2023. NCBI Sequence Read Archive. SRR23354533
- 2023. NCBI Sequence Read Archive. SRR23354526
- 2023. NCBI Sequence Read Archive. SRR23354527
- 2023. NCBI Sequence Read Archive. SRR23354528
- 2023. NCBI Sequence Read Archive. SRR23354529
- 2023. NCBI Sequence Read Archive. SRR23354530
- 2023. NCBI Sequence Read Archive. SRR23354531
- 2023. NCBI Sequence Read Archive. SRR23354525
- Zhang Y, 2023. Solanum rostratum isolate YZ_2023a, whole genome shotgun sequencing project. GenBank. JARACL000000000
- Zhang Y, 2023. Chromosome-level genome assembly and annotation of the prickly nightshade Solanum rostratum Dunal. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
No specific custom codes were developed in this study. All commands and pipelines used for data analyses were conducted according to the manuals or protocols provided by the corresponding software development team, which are described in detail in the Methods section. Default parameters were employed if no detailed parameters were mentioned for the software used in this study. Supplementary Table 1 lists the versions, settings, and parameters of the relevant software used in this study.