Abstract
Endogenous viral elements (EVEs) offer insight into the evolutionary histories and hosts of contemporary viruses. This study leveraged DNA metagenomics and genomics to detect and infer the host of a non-retroviral dinoflagellate-infecting +ssRNA virus (dinoRNAV) common in coral reefs. As part of the Tara Pacific Expedition, this study surveyed 269 newly sequenced cnidarians and their resident symbiotic dinoflagellates (Symbiodiniaceae), associated metabarcodes, and publicly available metagenomes, revealing 178 dinoRNAV EVEs, predominantly among hydrocoral-dinoflagellate metagenomes. Putative associations between Symbiodiniaceae and dinoRNAV EVEs were corroborated by the characterization of dinoRNAV-like sequences in 17 of 18 scaffold-scale and one chromosome-scale dinoflagellate genome assembly, flanked by characteristically cellular sequences and in proximity to retroelements, suggesting potential mechanisms of integration. EVEs were not detected in dinoflagellate-free (aposymbiotic) cnidarian genome assemblies, including stony corals, hydrocorals, jellyfish, or seawater. The pervasive nature of dinoRNAV EVEs within dinoflagellate genomes (especially Symbiodinium), as well as their inconsistent within-genome distribution and fragmented nature, suggest ancestral or recurrent integration of this virus with variable conservation. Broadly, these findings illustrate how +ssRNA viruses may obscure their genomes as members of nested symbioses, with implications for host evolution, exaptation, and immunity in the context of reef health and disease.
Subject terms: Water microbiology, Microbial ecology, Virology
A study part of the Tara Pacific Expedition that surveyed newly sequenced and publicly available metagenomes and genomes revealed pervasive non-retroviral dinoflagellate-infecting endogenous +ssRNA viral elements within coral symbionts.
Introduction
Endogenous viral elements, or “EVEs,” arise when whole or fragmented viral genomes are incorporated into host cell germlines. Once integrated, EVEs may propagate across successive host generations, potentially becoming fixed in a population through natural selection or drift1,2. Therefore, the presence and content of EVEs can provide clues into the evolutionary relationships among host species and shed light on ancient and modern virus-host interactions3. To date, most EVEs described in metazoan and plant genomes are retroviral, as this viral group must integrate their genome (as a provirus) into the genome of the host to replicate. Retroviruses thus possess and encode all of the molecular machinery (e.g. reverse transcriptases, integrases) required to integrate autonomously4. Remarkably, however, sequences from viruses that do not encode reverse transcriptases or exploit integration as a component of an obligate replication strategy—even viruses with no DNA stage—have also recently been detected as EVEs in diverse eukaryotic genomes5–11. These non-retroviral RNA EVEs have been reported in hosts ranging from unicellular algae to chiropteran (bat) genomes12–18. Though the mechanisms behind non-retroviral integration continue to be explored, viral sequences may be introduced via nonhomologous recombination and repair, through interactions with host-provisioned integrases and reverse transcriptases supplied on mobile elements (e.g. retroelements), or by utilizing co-infecting viruses6,7.
Endogenization of any viral sequence (including non-retroviral EVEs) may have positive, neutral or negative effects on a host19–21. While many EVEs are functionally defective or deleterious and ultimately removed from a population via purifying selection, retained EVEs may remodel the genomic architecture of their hosts or introduce sources of genetic innovation later co-opted for host function (i.e. exaptation22,23). Such ‘domesticated’ EVEs can be co-opted by hosts and utilized as regulatory elements, transcription factors, or functional proteins with purposes ranging from organism development to synaptic plasticity in the mammalian brain24–28. In particular, non-retroviral EVEs potentially serve as antiviral prototypes that help hosts combat infection by exogenous viruses currently circulating in the population14,29–31. Mechanisms underpinning EVE-derived immunity can include cell receptor interference, nucleic acid sequence recognition (e.g., RNAi), or even replication sabotage through production of faulty virus proteins from EVEs32. If expressed, EVEs may have a significant influence on the health, physiology and/or behavior of their hosts in natural and experimental systems31,33,34.
Investigating the distribution, sequence identity, and function of EVEs can yield insight into virus-host interactions across generations. EVEs catalogue a subset of the viruses that a host lineage has encountered and can link homologous extant viruses to contemporary hosts or known disease states31. Because integrated elements may accrue mutations at a slower rate than exogenous viral genomes6,35, EVEs can fill gaps in virus-host networks and act as synapomorphies, indicating the minimum time that a virus may have interacted with a host. As ‘genomic fossils’, EVEs have helped paleovirologists date the minimum origin of Circoviridae36, Hepadnaviridae37, Bornaviridae38, Flaviviridae39, Lentiviridae40,41, and Spumaviridae42 infections within metazoans2,24,35,43,44 (reviewed by Barreat and Katzourakis in 202245).
Coral holobionts – the cnidarian animal and its resident microbial assemblage, including dinoflagellates in the family Symbiodiniaceae, bacteria, archaea, fungi, and viruses – are an ecologically and economically valuable, multipartite non-model system46,47. Symbiodiniaceae are key obligate nutritional symbionts of corals and support their hosts in the construction of reef frameworks48. However, environmental stress can break down coral-Symbiodiniaceae partnerships, resulting in bleaching – the mass loss of Symbiodiniaceae cells49. Some bleaching signs (paling of a coral colony) are hypothesized to also result from viral lysis of Symbiodiniaceae21,50–54, but direct evidence supporting this hypothesis remains limited. Overall, the role of viruses in coral colony health and disease requires further examination.
Non-retroviral +ssRNA dinoRNAV sequences were first reported in stony corals based on five metatranscriptomic sequences and corroborated by Symbiodiniaceae EST libraries55. Subsequent studies indicated that similar +ssRNA viruses are commonly detected in coral RNA viromes and metatranscriptomes, as well as via targeted amplicon assays54,56–58. These viruses exhibit synteny and significant homology to Heterocapsa circularisquama RNA virus (HcRNAV57), the sole recognized representative of the genus Dinornavirus and a known pathogen of free-living dinoflagellates59. Both HcRNAV and dinoRNAV sequences detected in coral holobiont tissues contain two ORFs – a Major Capsid Protein (MCP) and RNA dependent RNA polymerase (RdRp). Furthermore, icosahedral virus-like particle (VLP) arrays resembling HcRNAV (but with 40% smaller individual particle diameters) have been imaged in the Symbiodiniaceae-dense coral gastrodermis tissue and in Symbiodiniaceae themselves60. Levin et al. (2017)57 assembled the 5.2 kb genome of a putative dinoRNAV from a poly(A)-selected metatranscriptome generated from cultured Symbiodinium. The assembly contained a 5’ dinoflagellate spliced leader (“dinoSL”61) — a component of >95% of Symbiodiniaceae mRNAs, speculated to illustrate molecular mimicry — and exhibited >1000-fold higher expression in a thermosensitive Cladocopium C1 population relative to a thermotolerant population of this Symbiodiniaceae strain at ambient temperatures (27 °C48,57). Together, the findings from these studies suggest that Symbiodiniaceae are target hosts of reef-associated dinoRNAVs.
This study (1) systematically searched for putative endogenized dinoRNAVs in metagenomes from in situ (symbiotic) coral colonies and seawater, as well as in available genomes of Symbiodiniaceae and aposymbiotic (symbiont-free) cnidarians, (2) investigated the evolutionary relationship of putative dinoRNAV EVEs to exogenous reef-associated dinoRNAV sequences, and (3) made preliminary inferences regarding the distribution and possible function of these dinoRNAV EVEs based on their detection, prevalence, and genomic context.
Results and discussion
Evidence of endogenized dinoRNAVs in coral holobiont metagenomes
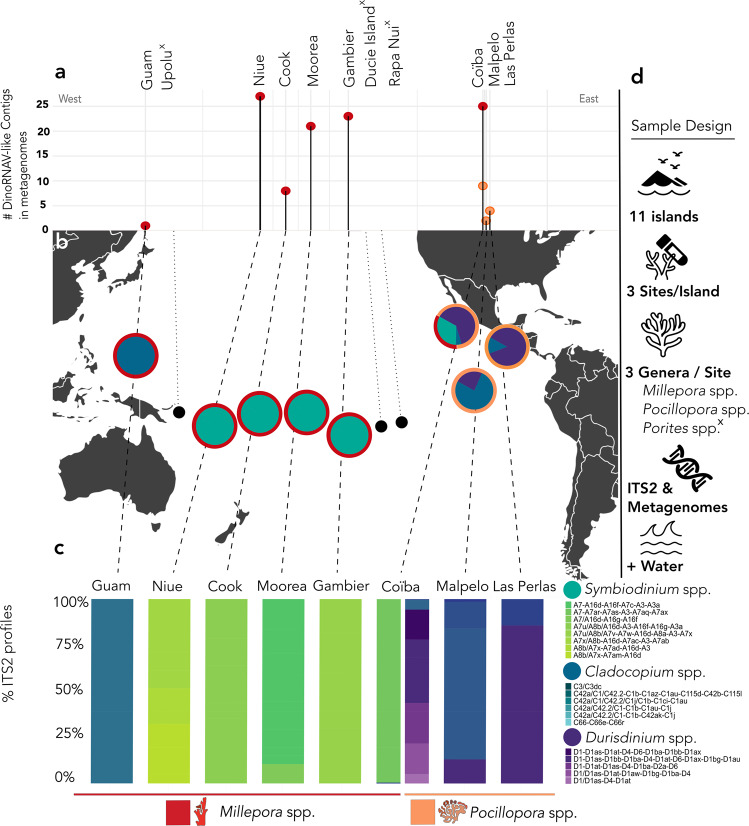
Putative dinoRNAV EVEs were detected in metagenomes generated from 42 cnidarian holobionts out of 269 sampled across the South Pacific Ocean (Supplementary Data 1). The majority of endogenized dinoRNAVs were identified in hydrocoral metagenomes (Millepora spp.; 70.5%, n = 105) which predominantly harbored Symbiodinium dinoflagellates but EVE-like sequences were also observed in scleractinian coral metagenomes (Pocillopora spp.; 29.5%, n = 15.) which predominantly harbored Cladocopium and Durusdinium dinoflagellates (Fig. 1a, c). No dinoRNAV-like sequences were detected among Porites spp. metagenomes (Figs. 1, 2). Hydrocoral metagenomes were sequenced at equivalent depths as scleractinian corals and had comparable levels of annotation (Supplementary Fig. 1, Supplementary Data 2); thus, higher dinoRNAV EVE prevalence in hydrocoral libraries was likely not a result of methodological bias. Of the 11 evaluated South Pacific islands, dinoRNAV EVEs were identified in samples from eight (Guam, Gambier, Moorea, Cook, Niue, Malpelo, Coïba, and Las Perlas), spanning 18 unique sites (Fig. 1b, d). Among Pocillopora spp. metagenomes, putative dinoRNAV EVEs were only identified on the Central American coast (CAMR, Coastal Pacific Longhurst Province) and were absent in Melanesia, Micronesia, and Polynesia; at these latter sites, dinoRNAVs were largely found in Millepora hydrocoral metagenomes. Importantly, endogenized dinoRNAV open reading frames (ORFs) appeared to be immediately adjacent to ORFs identified as dinoflagellate (typically Symbiodiniaceae) genes—they were not proximal to coral genes or those of other cellular organisms abundant in these metagenomes (Supplementary Data 3).
Fig. 1. Islands and species (cnidarian and dinoflagellate) correlating with dinoRNAV EVE-like sequence detection among Tara Pacific metagenomes.
a Count of scaffolds with putative endogenized dinoRNAV-like sequences among Tara Pacific metagenomes, grouped by island and spaced longitudinally by location sampled. b Sampling sites of Tara Pacific metagenomes explored for endogenized dinoRNAV-like sequences in this study. Internal circles indicate dominant Symbiodiniaceae genera based on ITS2 type profiles, outer ring denotes coral host(s) sampled at each island. c Symbiodiniaceae ITS2 type profile metabarcoding as delineated via Symportal119 within island and host. d Sample design of Tara Pacific libraries queried for dinoRNAV EVEs. [x] and black circles on map indicate island locations or species where no dinoRNAV-like sequences were detected. Icons derived from the Noun Project.
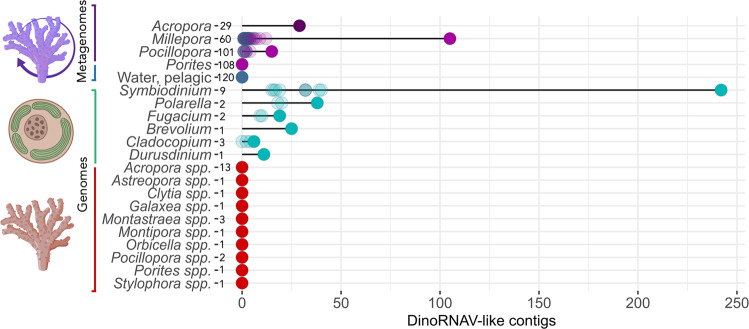
Fig. 2. Quantity of putative endogenized dinoRNAV per metagenome or genome.
Total quantity of putative endogenized dinoRNAV EVEs identified, broadly organized by sample source (metagenome or genome), and number of libraries or assemblies queried (after source name, Supplementary Data 6). Opaque circles denote the sum total of dinoRNAV EVE-like sequences identified from each source, while transparent circles denote individual counts of putative dinoRNAV EVEs per library. Icons created with BioRender.
We examined the Symbiodiniaceae ITS2 profiles associated with each metagenome and found that putative dinoRNAV EVEs were primarily associated with Symbiodinium, Cladocopium, and Durusdinium, which exhibited variation on both host and regional scales (Fig. 1c, Supplementary Data 4). DinoRNAV EVEs were more common in Symbiodinium-dominated cnidarians (F2,1044 = 25.8, p < 0.0001, nested ANOVA; Supplementary Fig. 2, Supplementary Data 5) relative to cnidarians hosting other Symbiodiniaceae genera, regardless of host. This suggested that dinoRNAV integration may be particularly recurrent or conserved within the genus Symbiodinium (Fig. 1).
To determine if these putative viral integrations were specific to cnidarian holobiont metagenomes and ensure that they were not artifacts of shared sample processing and sequencing procedures of the Tara Pacific pipeline, we also analyzed seawater metagenomes and publicly available metagenomes from the stony coral-dinoflagellate holobiont, Acropora spp. (Supplementary Data 1B,62–69). Examination of 120 Tara Oceans pelagic seawater metagenomes70 yielded no sequences sharing homology to dinoRNAVs. The concentration of Symbiodiniaceae cells within cnidarian tissues is considerably higher than that of the surrounding seawater71–74. On average, only 1.46 ± 0.08% of assembled contigs in seawater metagenomes were annotated as Symbiodiniaceae. Thus, lack of detection of dinoRNAV-like sequences from seawater metagenomes is likely due to reduced genomic signal of Symbiodiniaceae in the water column, rather than a lack of EVEs associated with Symbiodiniaceae lineages in seawater. However, it also must be noted that these Tara Oceans seawater metagenomes were not collected concurrently with coral samples75. Analysis of the 30 non-Tara Acropora holobiont metagenomes identified 29 more putative dinoRNAV EVEs (Fig. 2; Supplementary Data 6). These dinoRNAV EVEs were again neighboring dinoflagellate ORFs. While the Caribbean Acropora metagenomes analyzed contained too few reads to resolve the dominant Symbiodiniaceae present, earlier studies of the same coral colonies identified Symbiodinium spp. as the primary symbiont present76.
The identification of endogenized dinoRNAV-like sequences in cnidarian holobiont metagenomes, combined with the proximity of dinoRNAV-like ORFs to dinoflagellate-like sequences across metagenomes harboring diverse dinoflagellate consortia, collectively indicate that dinoRNAV EVEs are widespread among Symbiodiniaceae genera (Fig. 2 cyan dots).
Endogenized DinoRNAVs detected in Symbiodiniaceae genomes
To further test the hypothesis that dinoRNAVs on reefs infect dinoflagellate symbionts and not cnidarians, we examined 18 scaffold-scale genome assemblies representing the dinoflagellate families Symbiodiniaceae and Suessiaceae as well as 25 cnidarian genomes spanning 10 genera (Supplementary Data 1B62–69; Fig. 2; Table 1). Alignments revealed no evidence of endogenized dinoRNAVs in any of the 151,782 aposymbiotic (dinoflagellate-free) cnidarian scaffolds. In contrast, the same approach uncovered 351 (of 593,433) dinoflagellate scaffolds with evidence of endogenized dinoRNAVs (Fig. 2; Table 1). The identified 351 dinoRNAV EVE-containing scaffolds were observed across 17 of the 18 dinoflagellate genome assemblies (Table 1). DinoRNAV EVEs were also observed in two assemblies from the free-living dinoflagellate genus, Polarella (family Suessiaceae), which is closely related to the family Symbiodiniaceae, and served as an outgroup in this study77,78. Interestingly, assemblies belonging to Symbiodinium, the most ancestral Symbiodiniaceae genus48, contained a higher number of scaffolds with putative dinoRNAV EVEs (x̄=28.11, stdev=10.7) relative to assemblies of other Symbiodiniaceae genera (x̄=8.71, stdev=11; Fig. 2 cyan dots; Table 1). This result may clarify why observations of dinoRNAV-like ORFs were more common in metagenomes dominated by Symbiodinium (Fig. 1c). The dinoflagellate genome assembly with no detected dinoRNAV EVEs belonged to a relatively incomplete assembly of Cladocopium C15, which had the second lowest N50 and lowest BUSCO completeness score of all genomes examined (completeness 11.6%, relative to the average 24.54%; Table 1, Supplementary Data 7). The lower coverage/completeness of the Cladocopium C15 assembly indicates a reduced window into this genome. It is therefore possible that when a more complete assembly is generated, dinoRNAV EVE-like sequences will be detectable from this dinoflagellate. However, a linear model suggested that there was no relationship between dinoRNAV EVE detection and assembly statistics (i.e. query length, N50, or completeness; see Supplementary Data 8 for linear model output). Instead, dinoflagellate genus was the strongest predictor of dinoRNAV detection in a genome (LM results: Genus F = 5.74, p = 0.012) and dinoRNAV detections were significantly higher in Symbiodinium than Cladocopium genomes (pairwise estimated difference = −27.77 ± 5.91, p = 0.01; Supplementary Data 9). Furthermore, since we were unable to detect dinoRNAV EVEs in Porites metagenomes—a coral species primarily harboring Cladocopium C15 symbionts – we hypothesize that dinoRNAV endogenization was either less common in this lineage of Symbiodiniaceae or integrations have been lost over evolutionary time79,80.
Table 1.
DinoRNAV EVE-like open reading frames from representative Symbiodiniaceae and Suessiaceae dinoflagellate scaffold-level genome assemblies.
| Dinoflagellate Species (strain) | Total # Scaffolds | Host | Location | BUSCO score | dinoRNAV EVE ORFs on scaffolds | |||
|---|---|---|---|---|---|---|---|---|
| RdRp | MCP | Both | ||||||
| Symbiodiniaceae | Symbiodinium linucheae (CCMP2456)83 | 37,772 | Plexaura homamalla | Bermuda | 21.8% | 39 | 0 | 1 |
| Symbiodinium microadriaticum (04-503SCI.03)83 | 57,558 | Orbicella faveolata | Florida, USA | 41.6% | 30 | 1 | 3 | |
| Symbiodinium microadriaticum (CassKB8)83 | 67,937 | Cassiopea sp. | Hawaii, USA | 73.3% | 29 | 1 | 3 | |
| Symbiodinium microadriaticum (CCMP2467)113 | 9688 | Stylophora pistillata | Red Sea | 15.6% | 29 | 1 | 3 | |
| Symbiodinium natans (CCMP2548)83** | 2855 | N/A (Isolated from seawater) | Hawaii, USA | 15.5% | 14 | 1 | 3 | |
| Symbiodinium necroappetens (CCMP2469)83* | 104,583 | Condylactis gigantea | Jamaica | 22.8% | 37 | 4 | 2 | |
| Symbiodinium pilosum (CCMP2461)83** | 48,302 | Zoanthus sociatus | Jamaica | 19.8% | 15 | 0 | 0 | |
| Symbiodinium sp. A5 (formerly S. tridacnidorum)83 | 6245 | Heliofungia actiniformis | Australia | 21.1% | 17 | 1 | 0 | |
| Symbiodinium tridacnidorum (sh18 A3 Y106)114 | 16,176 | Tridacna crocea | Japan | 19.8% | 20 | 1 | 0 | |
| Brevolium minutum (Mf1.05b)115 | 21,899 | Orbicella faveolata | Florida, USA | 14.2% | 21 | 3 | 1 | |
| Cladocopium C15116 | 34,589 | Porites lutea | Australia | 11.6% | 0 | 0 | 0 | |
| Cladocopium sp. C1acro (formerly C. goreaui)89 | 41,289 | Acropora tenuis | Australia | 27.7% | 4 | 0 | 0 | |
| Cladocopium sp C92 (Y103)114 | 6686 | Fragum sp. | Japan | 19.5% | 2 | 0 | 0 | |
| Durusdinium trenchii117 | 19,593 | Favia speciosa | Japan | 28.7% | 10 | 1 | 0 | |
| Fugacium kawagutti (CS156 CCMP2468)89 | 16,959 | N/A (Free-living) | Hawaii, USA | 8.3% | 8 | 1 | 0 | |
| Fugacium kawagutti (CCMP2468)118 | 30,040 | N/A (Free-living) | Hawaii, USA | 17.9% | 9 | 1 | 0 | |
| n = 314 scaffolds with DinoRNAV EVE-like sequences | ||||||||
| Suessiaceae | Polarella glacialis (CCMP1383)78 ** | 33,494 | N/A (Free-living, isolated from seawater) | Antarctica | 20.8% | 20 | 0 | 0 |
| Polarella glacialis (CCMP2088)78 ** | 37,768 | N/A (Free-living, isolated from seawater) | Arctic | 21.8% | 18 | 0 | 0 | |
| n = 38 total scaffolds with DinoRNAV EVE-like sequences | ||||||||
Total counts of dinoflagellate scaffolds in genomes queried with individual endogenized dinoRNAV ORFs (RdRp, MCP) or both nearby each other, potentially indicating full viral genome integration. Table supplies associated dinoflagellate host species and location of isolation. RdRp = RNA-dependent RNA polymerase; MCP = major capsid protein. Assembly coverage and completeness are measured via BUSCO score (% completeness, or %C)102. * Indicates species with documented opportunistic life history; ** Indicates species with documented free-living life history per principal species description. Gonzalez-Pech et al.83, Aranda et al.113, Shoguchi et al.114, Shoguchi et al.115, Robbins et al.116, Liu et al.89, Shoguchi et al.117, Lin et al.118, Stephens et al.78. Further genome citations (including accession numbers) and BUSCO completion metrics can be found in Supplementary Data 7 and in superscript.
Incomplete ORFs and possible duplications indicate endogenization of DinoRNAVs
The repeated observation of putative dinoRNAV EVEs in dinoflagellate scaffolds and contigs from metagenomes and genomes suggests these sequences are either (1) conserved sequence artifacts of Symbiodiniaceae-dinoRNAV interactions, and/or (2) evidence of highly prevalent dinoflagellate viruses, commonly integrated and propagated via their single-celled hosts. If the observed dinoRNAV-like sequences represent active infections capable of generating virions during egress, we would, at minimum, expect essential ORFs associated with replication (RNA-dependent RNA polymerase, RdRp) and virion structure (Major Capsid Protein, MCP) to be endogenized on the same scaffold. We would additionally expect to observe overall conservation of ORF length/composition (with a lack of internal stop codons or substantial deletions) when aligning the dinoRNAV-like sequences detected here with known exogenous dinoRNAV sequences.
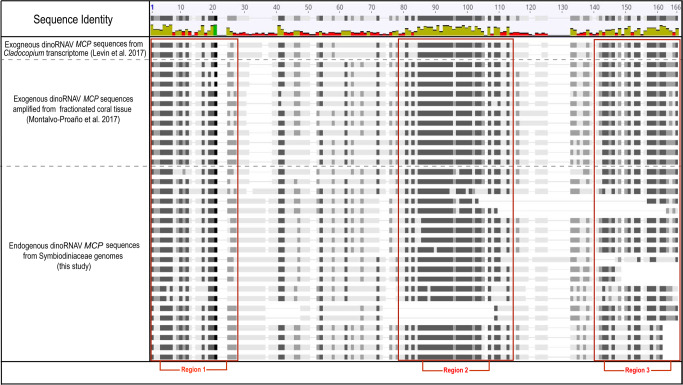
However, both DIAMOND and gene prediction analyses generally depicted dinoRNAV-like ORFs in isolation on separate scaffolds. While 28 MCP and 73 RdRp dinoRNAV ORFs were annotated, both ORFs were present on a Symbiodiniaceae scaffold – potentially representing whole dinoRNAV genome integrations – in only 14 instances. Thirteen of these 14 were from Symbiodinium genomes, whereas one scaffold was from Breviolum minutum, a member of the second most ancestral dinoflagellate genus (Table 1)48. To assess the conservation of putative dinoRNAV EVE sequence length/composition, we aligned the genomic and single ORF EVEs to reference exogenous dinoRNAV sequences. The reference genome for reef-associated dinoRNAVs is ~5 Kbp long and contains a 1,071 bp noncoding region between ORFs, with a 124-nucleotide internal ribosomal binding site57. In this study, for 13 of the scaffolds in which dinoRNAV ORFs were detected, the putative noncoding region between the MCP and RdRp EVEs ranged from ~200-800 bp (except for a scaffold belonging to S. linucheae CCMP2456, which contained a ~ 79 kbp noncoding region, and was excluded in further alignments). No internal ribosomal binding sites were detected within the putative dinoRNAV EVEs identified in dinoflagellate genomes. A nucleotide-based alignment to Levin et al.’s (2017)57 reference dinoRNAV genome indicated that the putative dinoRNAV EVEs presented here contained substantial insertions and/or deletions (Supplementary Fig. 3). Translated exogenous dinoRNAV MCP ORFs are reported to be ~358 aa in length57; Fig. 3 top sequences), but dinoRNAV-like MCP sequences recovered in this study ranged from 116-605aa in length. Furthermore, comparisons of these endogenous MCPs to exogenous reference sequences revealed internal stop codons and overall low similarity (Fig. 3). Amino acid-based alignment of endogenous dinoRNAV MCPs to metatranscriptome- and amplicon-generated exogenous reference sequences57,58 revealed indels and regions of low similarity between three conserved regions across both endogenous and exogenous MCP sequences (red boxes in Fig. 3).
Fig. 3. Amino acid alignment of putative endogenous and exogenous dinoRNAV-like +ssRNA virus Major Capsid Protein (MCP) sequences against transcriptome reference.
Putatively endogenous dinoRNAV Major Capsid Protein (MCP) amino acid sequences were aligned against exogenous references, including: (1) Symbiodiniaceae +ssRNA virus MCP ORFs recovered from a Cladocopium sp. transcriptome (Levin et al, 2017), and (2) dinoRNAV MCP amplicons from fractionated coral tissue (Montalvo-Proaño et al. 2017). Conserved regions were observed between exogenous and putatively endogenous viral sequences (labeled Regions 1–3).
Interestingly, multiple whole dinoRNAV integrations were sometimes observed in a single dinoflagellate genome. For example, genome assemblies of four different S. microadriacticum strains contained two or three whole dinoRNAV EVEs each (Table 1; Fig. 2). Pairwise alignments measuring shared nucleotide identity of whole dinoRNAV EVEs across Symbiodiniaceae scaffolds revealed that the S. microadriaticum genomes and the S. necroappetens genome share two whole genome dinoRNAV EVEs (provisionally dinoRNAV-A and dinoRNAV-B; Supplementary Fig. 3; Clustal-Omega)81. S. microadriaticum dinoRNAV-B was identical in all strains and shared 97% identity with the S. necroappetens dinoRNAV-B, yet proximal genes varied (Supplementary Data 10, 11). Importantly, the inconsistent composition and fragmented nature of both the genomic and single ORF dinoRNAV EVEs reported here supports the hypothesis that these sequences are not capable of generating replicative virions and are best interpreted as multiple integrations of dinoRNAVs into a host genome.
A Potential Mechanism for dinoRNAV Endogenization: Host-Provisioned Retroelements
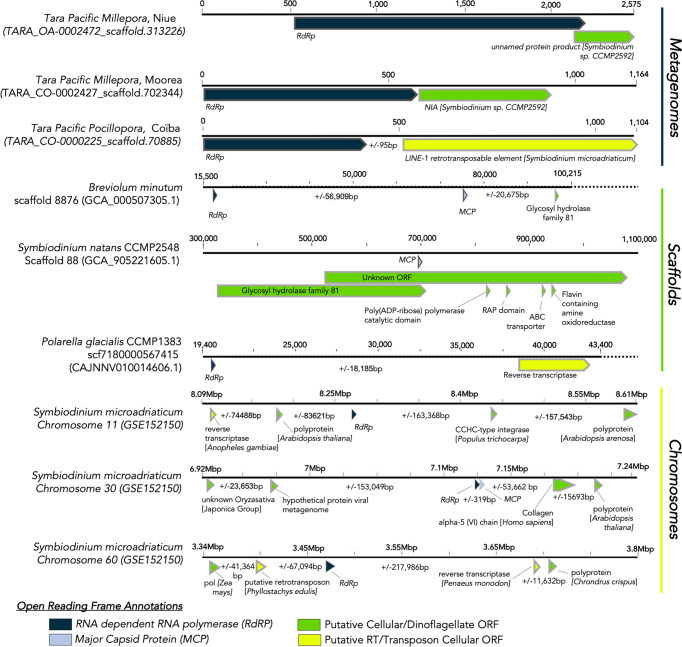
To assess if general genomic “neighborhoods” are conserved across dinoRNAV integrations (e.g. site location and synteny) and to better understand the genes proximal to EVEs on Symbiodiniaceae genomes, a chromosome-scale Symbiodinium microadriaticum genome assembly was evaluated (Fig. 4). The highest quality dinoflagellate genome assembly currently available revealed dinoRNAV-like ORFs on 18 of 94 chromosomes, with at least one RdRp on each, and some with multiple (two with n = 2 RdRps, three with n = 3 RdRps). On three of the chromosomes (# 30, 35, and 74), there were predicted ORFs annotated as dinoRNAV MCPs in close proximity to a RdRp ORF (separated by noncoding regions 319-656nt), indicative of a potential full-length dinoRNAV genome integration. These results corroborate detections of multiple genomic dinoRNAV EVEs in scaffold-scale assemblies of Symbiodinium microadriaticum genomes (Supplementary Fig. 3). The higher-resolution S. microadriaticum chromosome-level assembly facilitated the identification of an additional dinoRNAV genomic EVE (n = 4 for chromosome-level vs. n = 3 for scaffold-level, Supplementary Fig. 3), two of which were identified on Chromosome 74 and were separated by 2501 nucleotides. Of note, Nand et al. (2021)82 reported a decreasing abundance and expression of genes towards the center of chromosomes (past ~2Mpb of a telomere), where there was an increase in repetitive elements; this is where 26 of 29 putative dinoRNAV EVEs were identified in the chromosome-level assembly. Furthermore, ORFs neighboring integrations often varied widely, both in proximity and predicted function, from collagen and RNA binding protein to reverse transcriptase and non-LTR retrotransposable elements. These ORFs potentially contributed to the endogenization of dinoRNAV via mechanisms such as retrotransposition (Fig. 4, Supplementary Data 10).
Fig. 4. Representative scaffolds and chromosome fragments containing putative dinoRNAV EVEs.
Scaffolds annotation: MCP ORFs are indicated by light blue, RdRp indicated by navy blue ORFs (with complete description in Supplementary Data 10). Open reading frame (ORF) color broadly indicates cellular versus putative +ssRNA viral homology; yellow and some green (e.g., integrases, polyproteins) ORFs may be exploited mechanisms for viral integration. (+/−) base pair values represent sequence lengths between ORFs.
Retroposition through host-provisioned retroelements is one proposed mechanism of non-retroviral RNA virus integration into eukaryotic genomes6,7. An indicator of this form of integration is the nearby presence of a relict dinoflagellate spliced leader (“dinoSL”), a 22nt sequence located at the 5’ end of mRNAs83–86. Such a sequence flanks the RdRp gene on some extant dinoRNAVs57. We detected dinoSLs within 500 bp of 23.1% (six of 26) endogenized RdRp ORFs on S. micoradriaticum chromosomes, providing support for retroposition of these viral elements into Symbiodiniaceae genomes (Supplementary Data 11, 12). DinoRNAV gene integration may be facilitated by any of three major orders of retroelements associated with Symbiodiniaceae, including long terminal repeat (LTR) retrotransposons, short interspersed nuclear elements (SINEs), and long interspersed nuclear elements (LINEs83,87,88). Evidence suggests that these LINEs are common and non-active remnants of an ancient proliferation of LINEs that preceded the diversification of Suessiales78,83,89. Symbiodinium contains more LINEs relative to other Symbiodiniaceae genera, comprising 74.10-171.31 Mbp of Symbiodinium genomes, relative to an average of 7.48 Mbp of the genomes of in other genera, indicating the loss of these retroelements across speciation events82,83. The loss of LINEs in more recently derived Symbiodiniaceae genera coincides with a decrease in dinoRNAV EVE detection in these genomes (Table 1). Conversely, the genomes of Polarella, the psychrophilic and free-living outgroup from which Symbiodiniaceae diversified ~160 million years ago, are LINE-rich and generally have comparable numbers of dinoRNAV EVEs to Symbiodinium (Table 148,77,78,83). Together, this suggests that LINE activity during speciation may have facilitated dinoRNAV integration and the resulting EVEs may constitute dinornavirus “fossils.” This may explain their degree of sequence fragmentation and relatively low sequence similarity to modern extant dinoRNAVs (Fig. 3).
LINE-mediated retroposition is further supported by the observation of a LINE reverse transcriptase homolog ~17 kbp upstream of a RdRp EVE with a relict dinoSL on chromosome 45 (Supplementary Data 12) and a LINE retroelement 95 bp downstream of an EVE recovered from a Pocillopora metagenome (Fig. 4). Additionally, ~40% of annotated ORFs (35 of 88 annotated proteins) proximal to dinoRNAV ORFs on S. microadriaticum chromosomes were similar to non-LTR elements seen in other eukaryotic genomes sometimes <300 bp 5’ upstream (Supplementary Data 12, Supplementary Fig. 4). Collectively, these findings implicate host provisioned retroelements, such as LINEs, as facilitators of dinoRNAV gene integration.
DinoRNAV EVEs show homology to extant exogenous viruses
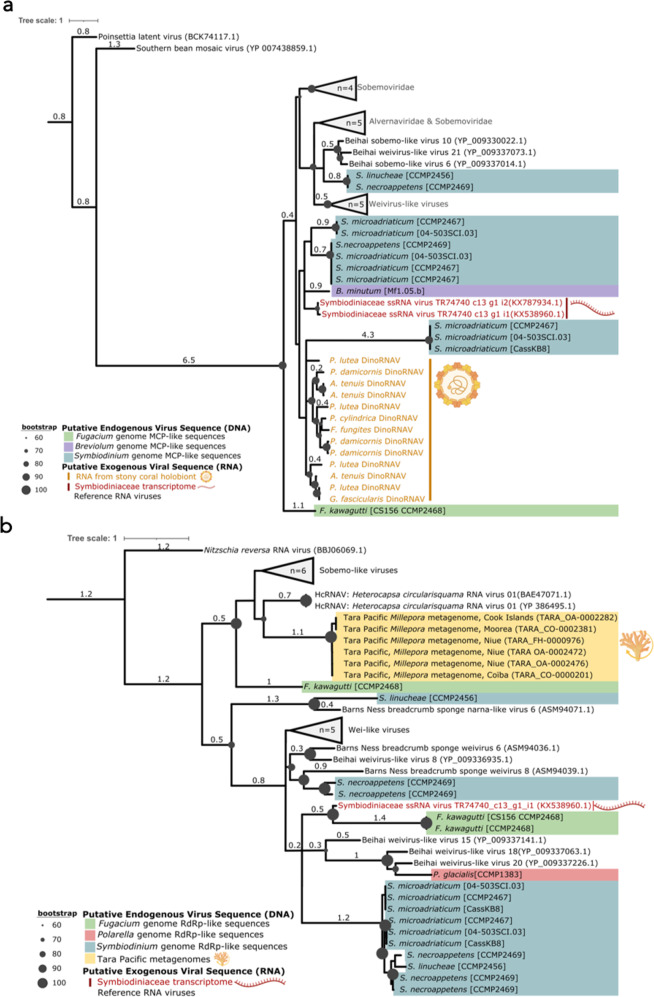
Modern, exogenous dinoRNAVs (Order: Sobelivirales) are highly divergent and hypothesized to form chronic infections within dinoflagellate hosts54,55,57,58. This chronic infection strategy likely provides opportunities for retroelement-driven endogenization into host genomes. Because many EVEs evolve at the rate of the host genome, rather than at the much faster rate of exogenous +ssRNA viral genomes, EVEs can serve as a snapshot of viral ancestry90. We compared translated dinoRNAV EVEs to exogenous dinoRNAVs and other Dinornavirus taxa to assess the conservation of EVEs, the potential for host utilization of these elements, and their relatedness to contemporary dinoRNAVs. We found that amino acid translations of endogenous dinoRNAV MCP sequences contained conserved motifs observed in the exogenous MCP sequences (e.g. Regions 1–3 in Fig. 3), yet the associated phylogeny was highly polyphyletic along inferred ancestral nodes (Fig. 5a). Endogenous MCP ORFs also appear to be evolving under neutral selection (dN/dS=0.958).
Fig. 5. Phylogenies of dinoRNAV major capsid protein (MCP) and RNA-dependent RNA polymerase (RdRp) ORFs.
a dinoRNAV Major Capsid Protein (MCP) ORF phylogeny. Maximum-likelihood tree of MCP amino acid sequences generated with a LG + F + G4 substitution model and 50,000 parametric bootstraps, illustrating the similarity of putative dinoRNAV EVEs (this study) to extant dinoRNAVs from stony coral colonies. b RNA-dependent RNA polymerase (RdRp) ORF phylogeny. Maximum-likelihood tree of RdRp amino acid sequences generated with a Blosum62 + G4 substitution model and 50,000 parametric bootstraps, demonstrating the similarity of metagenomic dinoRNAV EVE RdRps to RdRps of the sole recognized Dinornavirus, Heterocapsa circularisquama RNA virus (HcRNAV), as well as alignment to each other. ORFs were recovered from host metagenomes, transcriptomes, genomes, and extant +ssRNA reference viruses from amplicon libraries (a only). Both trees include Dinornavirus reference sequences and visualized in iTOL.
Endogenized dinoRNAV MCP form their own clades within the MCP tree, each closely related to specific clades consisting of extant dinoRNAVs or environmental (i.e. unclassified) sobeliviruses with similar conserved motifs. The majority of dinoRNAV MCP EVEs shared similarity to extant MCPs identified from unfractionated stony coral holobionts via amplicon sequencing58; these sequences formed an independent, disorganized clade (Fig. 5a clade containing yellow and blue sequences), relative to those recovered from dinoflagellate transcriptomes or those of other invertebrate hosts. Likewise, dinoRNAV RdRp EVEs identified via metagenomics appear most similar to HcRNAV, the defining member of family Alvernaviridae and a protist pathogen, further supporting the affiliation of this EVE with a dinoflagellate host. MCP and RdRp ORFs putatively derived from the same dinoflagellate genomes often shared clades (clades containing multiple blue or green sequences in Fig. 5a, b), perhaps indicative of duplications within genomes or multiple integration events of particular dinoRNAV lineages within host genera. The detection of putative dinoRNAV RdRp ORFs within Polarella genomes is therefore indicative of either the antiquity of dinoRNAV-dinoflagellate interactions and/or a propensity for recent dinoRNAV integration across Dinophyceae families. However, the exclusion of the P. glacialis dinoRNAV-RdRp from RdRps of other dinoflagellate clades (pink, Fig. 5B) further illustrates the congruence between EVEs and their host genomes. Overall, the evident homology to contemporary Dinornaviruses support these integrations as Alvernaviridae within order Sobelivirales.
The expression and functional potential of endogenized dinoRNAV elements (if any) remains unclear. With no isolated Symbiodiniaceae-infecting dinoRNAV strains available, investigation into EVE functionality is limited to in silico approaches. Sequence data mining efforts identified RNA sequences either sharing sequence similarity with dinoRNAVs, or containing whole dinoRNAV-like ORFs that also annotated as dinoflagellate transcripts (i.e. with cellular ORFs or sequence similarity) in seven out of nine publicly accessible dinoflagellate transcriptomes (Supplementary Data 13). Additionally, two transcripts from an exogenous dinoRNAV infection identified in Cladocopium transcriptomes carried MCP ORFs of +ssRNA viral sequences (‘TR74740_c13-g1_i1’ and ‘TR74740_c13-g1_i2’57, red text in Fig. 5a) and form a clade with putative Symbiodinium dinoRNAV EVEs (Fig. 5). Likewise, the RdRp ORF of ‘TR74740_c13-g1_i1’ and the RdRp of ‘GAKY01194223.1’— a transcript derived from a cultured Symbiodinium microadriaticum A1 transcriptome—shared some areas of similarity to putative endogenous dinoRNAVs (Fig. 5b57,91. Importantly, both RNA transcripts also shared features characteristic of dinoflagellates, such as a 5’ dinoSL61 or dinoflagellate sequence space flanking the dinoRNAV itself91. Furthermore, ‘TR74740_c13-g1_i1’ appeared to be in the top 0.03% of expressed transcripts at under certain thermal conditions, and GAKY01194223.1 appeared to exhibit moderately differential expression at the extremes of temperature and ionic stress in a cultured host57,91.
While viral RdRps have been leveraged by eukaryotes in multiple pathways92, the apparent fragmentation of the putative dinoRNAV EVEs in silico may indicate a role in triggering antiviral mechanisms within their hosts31,93. Given that the Symbiodinium genome contains all core RNAi protein machinery, including Argonaute and Dicer, and that GAKY01194223.1 folds into several hairpins (ΔG = −142.5 kcal/mol; Supplementary Fig. 5 examples), Symbiodiniaceae may use the putative EVE ncRNA identified here to develop host immunity against extant, exogenous dinoRNAVs. Furthermore, Symbiodiniaceae harboring dinoRNAV EVEs also contained numerous non-retroviral EVEs of other viral families (Supplementary Data 11, Fig. 7) in close proximity, such as Herpesviridae, Baculoviridae, Poxviridae, Iridoviridae, Phycodnaviridae, Pandoraviridae and Pithoviridae, ssDNA viruses of the family Shotokuvirae, -ssRNA viruses from the family Rhabdoviridae and +ssRNA viruses from the family Coronaviridae (Supplementary Fig. 6). Metagenomes corroborate findings of similar RdRps from these viral families (Supplementary Fig. 6). This provides support for host-mediated integration (e.g. retroposition) as a means of defense for single celled organisms, though further research is needed94.
Conclusions
Over recent decades, endogenous viral elements (EVEs) have enabled investigators to better understand the evolutionary history of viruses (“paleovirology”) in diverse terrestrial systems, uncovering ancient and modern virus-host interactions. Our study further demonstrates how in silico identification of EVEs can provide ecological context for enigmatic viral genomes in non-model, multipartite systems such as coral holobionts, impacting how we study coral reefs and their viral consortia. Here, we detected heritable integrations of multiple putative dinoRNAV genes in Symbiodiniaceae scaffolds from cnidarian metagenomes, as well as in diverse genomes of cultured Symbiodiniaceae; no integrations were detected from seawater metagenomes nor diverse aposymbiotic cnidarian genomes. The apparent pervasive nature of dinoRNAV-like sequences among dinoflagellate genomes (especially the genus Symbiodinium) suggests widespread and recurrent/ancestral integration of these EVEs. We propose that host-provisioned mechanisms drive dinoRNAV integration into single-celled dinoflagellate genomes as EVEs. The findings presented in this study further validate the dinoRNAV-Symbiodiniaceae virus-host pair, enhancing our understanding of ecologically and economically important cnidarian holobionts and opening the door to examining the role of EVEs in reef health.
Methods
Identification and computational validation of dinoRNAV EVEs leveraging meta’omics
The Tara Pacific Expedition (2016-2018) sampled coral reefs to investigate reef health and ecology using multiple methods, including amplicon sequencing and metagenomics (see Pesant et al. 202095 and 10.5281/zenodo.4068293 for coral reef sampling and processing methods). In this study, we explored metagenomes generated from hydrocorals (n = 60 Millepora), stony corals (n = 108 Porites, n = 101 Pocillopora) sampled from 11 islands (three replicate sites per island) across the South Pacific Ocean during the Tara Pacific Expedition for dinoRNAV EVEs (Fig. 1, Supplementary Data 1A, 1B95). Amplicon libraries of the dinoflagellate Internal Transcribed Spacer 2 (ITS2) gene fragment were sequenced in tandem with the metagenomes, to characterize the dominant Symbiodiniaceae harbored by hydrozoan and stony coral colonies95.
To confirm that these dinoRNAV EVE sequences were affiliated with coral holobionts and reduce the possibility that they are technical artifacts, publicly available metagenome libraries were analyzed (Supplementary Data 1B). These additional libraries included 120 assembled pelagic water samples presumed to include pelagic dinoflagellate sequences from the Tara Oceans dataset (2009-201370) and 30 MiSeq metagenomes from unfractionated samples of the stony coral genus Acropora, which were processed and sequenced via a different pipeline (Supplementary Data 1B, Supplementary Fig. 7). Publicly accessible transcriptomes from nine Symbiodiniaceae assemblies (Supplementary Data 1B) were also queried to determine if dinoRNAV-like sequences were present in poly(A)-selected dinoflagellate transcriptomes and resembled EVEs in terms of proximal gene composition and presence of a characteristic pre-mRNA spliced leader (dinoSL) sequence (as in Levin et al, 201757). Details regarding the collection of samples, generation of metagenomes and associated Symbiodiniaceae amplicon libraries, and associated bioinformatic analyses are provided in Supplementary Fig. 7).
Metagenomic and transcriptomic scaffolds were annotated against a curated database of dinoRNAV-like sequences (Supplementary Data 14) via BLASTx (e-value < 1 × 10−5; see Supplementary Fig. 7 for workflow96). Alignments to the custom database with a bit score <50 and percent shared amino acid identity <30% were excluded from further analysis. A length penalty was not imposed during this step due to the limited length of assembled scaffolds (average N50 = 3341 ± 127 nt across all queried libraries). Open reading frames (ORFs) from selected scaffolds were called via Prodigal (v.2.6.397) and annotated against the NCBI-nr database (e-value < 0.001; DIAMOND v.2.0.698) to confirm homology to dinoRNAVs and to identify adjacent dinoflagellate sequences (e-value < 1 × 10-5, bit≥50). In the absence of complete ORFs (potentially due to the limited size of scaffolds, partial integrations, etc.), homology was confirmed through comparison of the initial alignments to the curated database and 300nt of upstream/downstream flanking sequences (bedtools v.2.30.099) against the NCBI-nr database (e-value < 0.001; DIAMOND v.2.0.698). This served as further curation and verification, as EVEs can exist in fragmented or degraded states. Non-normalized quality-controlled reads were mapped via bbmap (v.38.84100), and putative EVEs were assessed for uniform read coverage across scaffolds, reducing the probability of chimeric assembly. RNA secondary structure was predicted via mfold (v.3.5101).
dinoRNAV EVEs in dinoflagellate and aposymbiotic cnidarian genomes
Publicly available dinoflagellate and aposymbiotic (dinoflagellate-free) cnidarian genome assemblies were queried to resolve the putative host(s) of dinoRNAVs, to assess homology among detected dinoRNAVs within coral holobionts, and to compare genes proximal to dinoRNAV EVEs in different host species/strains. A chromosome-scale dinoflagellate genome assembly generated from a Symbiodinium microadriaticum culture (Accession: GSE152150)82, and scaffold-scale genome assemblies were examined for dinoRNAV EVEs (Supplementary Data 1B, Supplementary Fig. 7). Scaffold-scale genome assemblies were from the closely related families Symbiodiniaceae and Suessiaceae, and included representatives from the genera Symbiodinium (n = 9), Breviolum (n = 1), Cladocopium (n = 3), Durusdinium (n = 1), Fugacium (n = 2), and Polarella (n = 2), as well as 25 aposymbiotic cnidarian genome assemblies, including the stony coral genera Acropora (n = 13), Astreopora (n = 1), Galaxea (n = 1), Montastraea (n = 1), Montipora (n = 3), Orbicella (n = 1), Pocillopora (n = 2), Porites (n = 1), and Stylophora (n = 1), and the jellyfish Clytia (n = 1; Fig. 2, Supplementary Data 1B). All publicly available genome assemblies had undergone a form of microbial decontamination, trimming, and quality control prior to assembly, minimizing risk of microbial contamination. Genome completeness and quality further were assessed via BUSCO (v3)102 with the Eukaryota dataset and QUAST (v5.0.2103), respectively. Scaffolds/chromosomes containing putative dinoRNAV EVEs were identified by aligning sequences to the protein version of the Reference Viral DataBase (RVDB v.19104) using DIAMOND BLASTx (v0.9.30)98. The same exclusion criteria were maintained for alignments of metagenomic scaffolds, also omitting alignments <100 amino acids. Regions of dinoflagellate genomes exhibiting similarity to the MCP or RdRp of reef-associated dinoRNAV reference genomes57 or other closely related +ssRNA viruses (Supplementary Data 14) were extracted and re-aligned to the NCBI-nr database to further confirm viral homology.
We tested the relationship between the number of identified dinoRNAV EVE-containing scaffolds, dinoflagellate genera, and genome quality metrics using a linear model. Model selection was performed with an F-test (package car, v.3.0-12) and assumptions were visually checked. Pairwise comparisons between genera were conducted using the package emmeans (v.1.7.2). Putative whole dinoRNAV-like genomes within scaffolds were identified based on the presence of MCP and RdRp-like sequences on the same scaffold no further than 1.5 Kbp apart (Table 1; Supplementary Fig. 3). IRESPred105 was utilized to identify internal ribosomal entry sites (IRES) with default parameters on putative dinoRNAV EVE with whole sequence integrations.
ORFs were predicted and annotated from dinoRNAV EVE-containing scaffolds and all dinoflagellate chromosomes using Prodigal97 and MAKER2 annotation pipeline106 with the AUGUSTUS gene prediction software107. Translated ORFs were then aligned to a hybrid database containing the UniProt/Swiss-Prot database and protein version of RVDB (v.19; DIAMOND-BLASTp). ORFs on putative dinoRNAV EVE-containing scaffolds and chromosomes were further annotated using InterProScan (v5.48-83.0, Pfam analysis with default parameters) to identify sequences proximal to putative dinoRNAV integrations. The presence of dinoflagellate spliced leaders (“dinoSLs”) were examined within 500nt of dinoRNAV EVEs using BLASTn with default parameters (except word size=9, excluding two ambiguous positions as specified in Gonzalez-Pech et al. 202183).
Phylogenetic analysis of dinoRNAV EVEs
Amino acid-based phylogenetic trees were generated with dinoRNAV EVE ORFs (MCP and RdRp) from scaffold-scale genomic assemblies, metagenomes, transcriptomes, and sequences from exogenous and closely related +ssRNA reference viruses (Supplementary Data 1 A, B, Supplementary Data 14). Sequences were aligned using the best fit algorithm determined by MAFFT (v7.464)108 and reviewed and trimmed manually in MEGA (v7)109. Maximum-likelihood trees were generated with IQTREE2110 using the model determined by ModelFinder111 and 50,000 parametric bootstraps112 with nearest neighbor interchange optimization. ORFs from the chromosome-level assembly for S. microadriaticum culture CCMP2467 were not included in the phylogeny in order to avoid redundancy with those from the analogous scaffold-level assembly. To calculate dN/dS, ORFs were aligned in Clustal Omega (v.1.2.4), refined in MUSCLE (v.3.6), before using pal2nal (v.14) for codon-based nucleic acid alignment. Evolutionary trajectory was then assessed via CODEML (PAML package, v.4.10.5).
Statistics and reproducibility
As indicated throughout the article, metagenomes (n = 269) and genomes (n = 18) served as technical replicates, and ORFs or full EVE sequences served as comparative ecoevolutionary units (replicates described in Table 1) when available. Negative controls (seawater metagenomes, coral host, etc) were also evaluated. All statistical packages are reported in methods or Supplementary Fig. 7.
Reporting summary
Further information on research design and collection permits are available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
Special thanks to the Tara Ocean Foundation, the R/V Tara crew and the Tara Pacific Expedition Participants (10.5281/zenodo.3777760). Thank you also to Carsten G.B. Grupstra and Clark Hamor for their input regarding analyses. We are keen to thank the commitment of the following institutions for their financial and scientific support that made this unique Tara Pacific Expedition possible: CNRS, PSL, CSM, EPHE, Genoscope, CEA, Inserm, Université Côte d’Azur, ANR, agnès b., UNESCO-IOC, the Veolia Foundation, the Prince Albert II de Monaco Foundation, Région Bretagne, Billerudkorsnas, AmerisourceBergen Company, Lorient Agglomération, Oceans by Disney, L’Oréal, Biotherm, France Collectivités, Fonds Français pour l’Environnement Mondial (FFEM), Etienne Bourgois, FRANCE GENOMIQUE (#ANR-10-INBS-09 to P.W.), and the Tara Ocean Foundation teams. Tara Pacific would not exist without the continuous support of the participating institutes. This research is further supported by NSF OCE #2145472 to AMSC, NSF DOB Grant 2025457 to RLVT, and with additional support from NSF PRFB #1907184 to KSIB.
Author contributions
A.J.V. and K.S.I.B. contributed equally to this study. K.S.I.B. identified endogenization in metagenomes with support from RVT; A.J.V. confirmed the presence in genomes, with support from AMSC. All other authors (C.R.V., B.C.C.H., H.J.R., S.P, D.A., E.B., C.M., G.B., P.W., J.P., G.I., S.R., S.A., B.B., E.B., C.B., C.d.V., E.D., M.F., D.F., P.F., P.G., E.G., F.L., S.P., S.R., S.S., O.T., R.T., D.Z.) are contributing members of the Tara Pacific Expedition, collecting biosamples for metagenomics used in this study or guiding the expedition, with essential bioinformatic support in metagenomic assembly from HJR/SS, sequencing support from PW and JP, and ITS2 analysis from CRV/BCCH. A.V., K.S.I.B., A.M.S.C., and R.V.T. wrote the manuscript. This is publication number #26 of the Tara Pacific Consortium.
Peer review
Peer review information
Communications Biology thanks Guan-Zhu Han, Raúl González-Pech and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Zhijuan Qiu and Gene Chong.
Data availability
Metadata are accessible in zenodo: https://zenodo.org/record/6299409#.Y-ClwuzMKml. Metagenomes are available via 10.5281/zenodo.7839794. Seawater metagenomes are available through the European bioinformatics institute (Tara Oceans; ERP001736) and NCBI (PRJEB1787). NCBI accession numbers for individual holobiont species metagenomes, genome assemblies and reference sequences can be found in Supplementary Data 1B, 3 and 14, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alex J. Veglia, Kalia S.I. Bistolas.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-04917-9.
References
- 1.Johnson WE. Endogenous retroviruses in the genomics era. Annu. Rev. Virol. 2015;2:135–159. doi: 10.1146/annurev-virology-100114-054945. [DOI] [PubMed] [Google Scholar]
- 2.Johnson WE. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019;17:355–370. doi: 10.1038/s41579-019-0189-2. [DOI] [PubMed] [Google Scholar]
- 3.Johnson WE. Endless forms most viral. PLoS Genet. 2010;6:e1001210. doi: 10.1371/journal.pgen.1001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 5.Gallot-Lavallée L, Blanc G. A glimpse of nucleo-cytoplasmic large DNA virus biodiversity through the eukaryotic genomics window. Viruses. 2017;9:17. doi: 10.3390/v9010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn PJ, Moreau CS. Assessing the diversity of endogenous viruses throughout ant genomes. Front Microbiol. 2019;10:1139. doi: 10.3389/fmicb.2019.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horie M, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLOS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba S, et al. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLOS Pathog. 2011;7:e1002146. doi: 10.1371/journal.ppat.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu H, Jo Y, Cho WK. Evolution of endogenous non-retroviral genes integrated into plant genomes. Curr. Plant Biol. 2014;1:55–59. doi: 10.1016/j.cpb.2014.07.002. [DOI] [Google Scholar]
- 11.Kojima, S. et al Virus-like insertions with sequence signatures similar to those of endogenous nonretroviral RNA viruses in the human genome. Proc Natl Acad Sci. 118, 10.1073/pnas.2010758118 (2021). [DOI] [PMC free article] [PubMed]
- 12.Ballinger MJ, Bruenn JA, Taylor DJ. Phylogeny, integration and expression of sigma virus-like genes in. Drosoph. Mol. Phylogenet Evol. 2012;65:251–258. doi: 10.1016/j.ympev.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Tromas N, Zwart MP, Forment J, Elena SF. Shrinkage of genome size in a plant RNA virus upon transfer of an essential viral gene into the host genome. GBE. 2014;6:538–550. doi: 10.1093/gbe/evu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palatini U, et al. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics. 2017;18:512. doi: 10.1186/s12864-017-3903-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, et al. Endogenous viral elements in algal genomes. Acta Ocean. Sin. 2014;33:102–107. [Google Scholar]
- 16.Jebb D, et al. Six reference-quality genomes reveal evolution of bat adaptations. Nature. 2020;583:578–584. doi: 10.1038/s41586-020-2486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moniruzzaman M, Weinheimer AR, Martinez-Gutierrez CA, Aylward FO. Widespread endogenization of giant viruses shapes genomes of green algae. Nature. 2020;588:141–145. doi: 10.1038/s41586-020-2924-2. [DOI] [PubMed] [Google Scholar]
- 18.Skirmuntt EC, Escalera-Zamudio M, Teeling EC, Smith A, Katzourakis A. The potential role of Endogenous Viral Elements in the evolution of bats as reservoirs for zoonotic viruses. Annu. Rev. Virol. 2020;7:103–119. doi: 10.1146/annurev-virology-092818-015613. [DOI] [PubMed] [Google Scholar]
- 19.Roossinck MJ. The good viruses: viral mutualistic symbioses. Nat. Rev. Microbiol. 2011;9:99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 20.Harrison E, Brockhurst MA. Ecological and evolutionary benefits of temperate phage: what does or doesn’t kill you makes you stronger. BioEssays. 2017;39:1700112. doi: 10.1002/bies.201700112. [DOI] [PubMed] [Google Scholar]
- 21.Correa AMS, et al. Revisiting the rules of life for viruses of microorganisms. Nat. Rev. Microbiol. 2021;19:501–513. doi: 10.1038/s41579-021-00530-x. [DOI] [PubMed] [Google Scholar]
- 22.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev. Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. Changes in viral protein function that accompany retroviral endogenization. Proc. Natl Acad. Sci. 2007;104:17506–17511. doi: 10.1073/pnas.0704313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 25.Frank JA, Feschotte C. Co-option of endogenous viral sequences for host cell function. COVIRO. 2017;25:81–89. doi: 10.1016/j.coviro.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortelmans K, Wang-Johanning F, Johanning GL. The role of human endogenous retroviruses in brain development and function. APMIS. 2016;124:105–115. doi: 10.1111/apm.12495. [DOI] [PubMed] [Google Scholar]
- 27.Sofuku, K., & Honda, T. Influence of endogenous viral sequences on gene expression. IntechOpen. 10.5772/intechopen.71864 (2018)
- 28.Takahashi, H., Fukuhara, T., Kitazawa, H., & Kormelink, R. Virus latency and the impact on plants. Front. Microbiol. 1010.3389/fmicb.2019.02764 (2019). [DOI] [PMC free article] [PubMed]
- 29.Whitfield ZJ, et al. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr. Biol. 2017;27:3511–3519.e7. doi: 10.1016/j.cub.2017.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ter Horst AM, Nigg JC, Dekker FM, Falk BW. Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWI-interacting RNAs. J. Virol. 2019;93:e02124–18. doi: 10.1128/JVI.02124-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki Y, et al. Non-retroviral Endogenous Viral Element limits cognate virus replication in Aedes aegypti ovaries. Curr. Biol. 2020;30:3495–3506.e6. doi: 10.1016/j.cub.2020.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aswad S, Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol. Evol. 2012;27:627–36. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Parker BJ, Brisson JA. A laterally transferred viral gene modifies aphid wing plasticity. Curr. Biol. 2019;29:2098–2103.e5. doi: 10.1016/j.cub.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson W, Francis I, Ryan K, Davy S. Temperature induction of viruses in symbiotic dinoflagellates. Aquat. Micro. Ecol. 2001;25:99–102. doi: 10.3354/ame025099. [DOI] [Google Scholar]
- 35.Aiewsakun P, Katzourakis A. Endogenous viruses: connecting recent and ancient viral evolution. Virology. 2015;479–480:26–37. doi: 10.1016/j.virol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Belyi VA, Levine AJ, Skalka AM. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the parvoviridae and circoviridae are more than 40 to 50 million years old. J. Virol. 2010;84:12458–62. doi: 10.1128/JVI.01789-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert C, Feschotte C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010;8:e1000495. doi: 10.1371/journal.pbio.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawasaki J, Kojima S, Mukai Y, Tomonaga K, Horie M. 100-My history of bornavirus infections hidden in vertebrate genomes. Proc. Natl Acad. Sci. 2021;118:e2026235118. doi: 10.1073/pnas.2026235118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Y. Q. et al. Discovery of Flaviviridae-derived endogenous viral elements in shrew genomes provide novel insights into Pestivirus ancient history. bioRxiv. 02.11.480044. 10.1101/2022.02.11.480044 (2022) [DOI] [PMC free article] [PubMed]
- 40.Cui J, Holmes EC. Endogenous lentiviruses in the ferret genome. J. Virol. 2012;86:3383–5. doi: 10.1128/JVI.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;5:7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katzourakis A, Gifford RJ, Tristem M, Gilbert MT, Pybus OG. Macroevolution of complex retroviruses. Science. 2009;18:1512. doi: 10.1126/science.1174149. [DOI] [PubMed] [Google Scholar]
- 43.Katzourakis A. Paleovirology: inferring viral evolution from host genome sequence data. Philos. Trans. R. Soc. 2013;368:20120493. doi: 10.1098/rstb.2012.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel MR, Emerman M, Malik HS. Paleovirology—ghosts and gifts of viruses past. COVIRO. 2011;1:304–309. doi: 10.1016/j.coviro.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barreat JGN, Katzourakis A. Paleovirology of the DNA viruses of eukaryotes. Trends Microbiol. 2022;30:281–292. doi: 10.1016/j.tim.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Knowlton N, Rohwer F. Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am. Nat. 2003;162:S51–S62. doi: 10.1086/378684. [DOI] [PubMed] [Google Scholar]
- 47.Matthews JL, et al. Symbiodiniaceae‐bacteria interactions: rethinking metabolite exchange in reef‐building corals as multi‐partner metabolic networks. Environ. Microbiol. 2020;22:1675–1687. doi: 10.1111/1462-2920.14918. [DOI] [PubMed] [Google Scholar]
- 48.LaJeunesse TC, et al. Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018;28:2570–2580.e6.. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 49.Glynn PW. Coral reef bleaching: facts, hypotheses and implications. Glob. Change Biol. 1996;2:495–509. doi: 10.1111/j.1365-2486.1996.tb00063.x. [DOI] [Google Scholar]
- 50.van Oppen MJH, Leong J-A, Gates RD. Coral-virus interactions: a double-edged sword? Symbiosis. 2009;47:1–8. doi: 10.1007/BF03179964. [DOI] [Google Scholar]
- 51.Correa AMS, et al. Viral outbreak in corals associated with an in situ bleaching event: atypical herpes- like viruses and a new Megavirus infecting. Symbiodinium. Front Microbiol. 2016;7:127. doi: 10.3389/fmicb.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vega Thurber R, Payet JP, Thurber AR, Correa AMS. Virus–host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 2017;15:205–216. doi: 10.1038/nrmicro.2016.176. [DOI] [PubMed] [Google Scholar]
- 53.Messyasz A, et al. Coral bleaching phenotypes associated with differential abundances of Nucleocytoplasmic Large DNA Viruses. Front Mar. Sci. 2020;7:555474. doi: 10.3389/fmars.2020.555474. [DOI] [Google Scholar]
- 54.Grupstra CGB, et al. Thermal stress triggers productive viral infection of a key coral reef symbiont. ISME J. 2022;16:1430–1441. doi: 10.1038/s41396-022-01194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Correa AMS, Welsh RM, Vega Thurber RL. Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 2013;7:13–27. doi: 10.1038/ismej.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weynberg KD, Wood-Charlson EM, Suttle CA, van Oppen MJH. Generating viral metagenomes from the coral holobiont. Front Microbiol. 2014;5:206. doi: 10.3389/fmicb.2014.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levin RA, Voolstra CR, Weynberg KD, van Oppen MJH. Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J. 2017;11:808–812. doi: 10.1038/ismej.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montalvo-Proaño J, Buerger P, Weynberg KD, van Oppen MJH. A PCR-Based Assay targeting the major capsid protein gene of a Dinorna-Like ssRNA virus that infects coral photosymbionts. Front. Microbiol. 2017;8:1665. doi: 10.3389/fmicb.2017.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagasaki K, et al. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 2005;71:8888–8894. doi: 10.1128/AEM.71.12.8888-8894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence SA, Davy JE, Aeby GS, Wilson WH, Davy SK. Quantification of virus-like particles suggests viral infection in corals affected by Porites tissue loss. Coral Reefs. 2014;33:687–691. doi: 10.1007/s00338-014-1168-8. [DOI] [Google Scholar]
- 61.Zhang H, Zhuang Y, Gill J, Lin S. Proof that dinoflagellate Spliced Leader (DinoSL) is a useful hook for fishing dinoflagellate transcripts from mixed microbial samples: Symbiodinium kawagutii as a case study. Protist. 2013;164:510–527. doi: 10.1016/j.protis.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Voolstra CR, et al. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci. Rep. 2017;7:17583. doi: 10.1038/s41598-017-17484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buitrago-López C, Mariappan KG, Cárdenas A, Gegner HM, Voolstra CR. The Genome of the Cauliflower Coral Pocillopora verrucosa. Genome Biol. Evol. 1. 2020;12:1911–1917. doi: 10.1093/gbe/evaa184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci. Rep. 2018;8:16134. doi: 10.1038/s41598-018-34459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helmkampf M, Bellinger MR, Geib SM, Sim SB, Takabayashi M. Draft Genome of the Rice Coral Montipora capitata Obtained from Linked-Read Sequencing. Genome Biol. Evol. 1. 2019;11:2045–2054. doi: 10.1093/gbe/evz135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kitchen SA, et al. Genomic variants among threatened acropora corals. G3 (Bethesda) 2019;9:1633–1646. doi: 10.1534/g3.119.400125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.ReFuGe 2020 Consortium. The ReFuGe 2020 Consortium—using “omics” approaches to explore the adaptability and resilience of coral holobionts to environmental change. Front. Mar. Sci. 2. 10.3389/fmars.2015.00068 (2015).
- 68.Shinzato C, et al. Eighteen coral genomes reveal the evolutionary origin of acropora strategies to accommodate environmental changes. Mol. Biol. Evol. 2021;4:16–30. doi: 10.1093/molbev/msaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ying H, et al. Comparative genomics reveals the distinct evolutionary trajectories of the robust and complex coral lineages. Genome Biol. 2018;2:175. doi: 10.1186/s13059-018-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tara Oceans Consortium Coordinators. et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data. 2015;2:150023. doi: 10.1038/sdata.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Littman RA, van Oppen MJH, Willis BL. Methods for sampling free-living Symbiodinium (zooxanthellae) and their distribution and abundance at Lizard Island (Great Barrier Reef) J. Exp. Mar. Biol. Ecol. 2008;364:48–53. doi: 10.1016/j.jembe.2008.06.034. [DOI] [Google Scholar]
- 72.Scheufen T, Iglesias-Prieto R, Enríquez S. Changes in the number of symbionts and Symbiodinium cell pigmentation modulate differentially coral light absorption and photosynthetic performance. Front. Mar. Sci. 2017;4:309. doi: 10.3389/fmars.2017.00309. [DOI] [Google Scholar]
- 73.Fujise L, et al. Unlocking the phylogenetic diversity, primary habitats, and abundances of free‐living Symbiodiniaceae on a coral reef. Mol. Ecol. 2021;30:343–360. doi: 10.1111/mec.15719. [DOI] [PubMed] [Google Scholar]
- 74.Grupstra CGB, Rabbitt KM, Howe-Kerr LI, Correa AMS. Fish predation on corals promotes the dispersal of coral symbionts. Anim. Microbiome. 2021;3:25. doi: 10.1186/s42523-021-00086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sunagawa S, et al. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 76.Muller EM, Bartels E, Baums IB. Bleaching causes loss of disease resistance within the threatened coral species Acropora cervicornis. eLife. 2018;7:e35066. doi: 10.7554/eLife.35066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Janouškovec J, et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl Acad. Sci. 2017;114:E171–E180. doi: 10.1073/pnas.1614842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stephens TG, et al. Genomes of the dinoflagellate Polarella glacialis encode tandemly repeated single-exon genes with adaptive functions. BMC Biol. 2020;18:56. doi: 10.1186/s12915-020-00782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan YTR, et al. Endosymbiont diversity and community structure in Porites lutea from Southeast Asia are driven by a suite of environmental variables. Symbiosis. 2020;80:269–277. doi: 10.1007/s13199-020-00671-2. [DOI] [Google Scholar]
- 80.Qin Z, et al. Diversity of Symbiodiniaceae in 15 coral species from the Southern South China Sea: potential relationship with coral thermal adaptability. Front Microbiol. 2019;10:2343. doi: 10.3389/fmicb.2019.02343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sievers F, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nand A, et al. Genetic and spatial organization of the unusual chromosomes of the dinoflagellate Symbiodinium microadriaticum. Nat. Genet. 2021;53:618–629. doi: 10.1038/s41588-021-00841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.González-Pech RA, et al. Comparison of 15 dinoflagellate genomes reveals extensive sequence and structural divergence in family Symbiodiniaceae and genus Symbiodinium. BMC Biol. 2021;19:73. doi: 10.1186/s12915-021-00994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang H, Hou Y, Miranda L, Lin S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl Acad. Sci. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lidie KB, van Dolah FM. Spliced leader RNA-mediated trans-splicing in a dinoflagellate, Karenia brevis. J. Eukaryot. Microbiol. 2007;54:427–35. doi: 10.1111/j.1550-7408.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- 86.Song B, Chen S, Chen W. Dinoflagellates, a unique lineage for retrogene research. Front. Microbiol. 2018;9:1556. doi: 10.3389/fmicb.2018.01556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elbarbary, R. A., Lucas, B. A., Maquat, L. E. Retrotransposons as regulators of gene expression. Science. 12, 351(6274):aac7247 (2016) [DOI] [PMC free article] [PubMed]
- 88.Mita P, Boeke JD. How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 2016;37:90–100. doi: 10.1016/j.gde.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu H, et al. Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 2018;1:95. doi: 10.1038/s42003-018-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holmes EC. The evolution of endogenous viral elements. Cell Host Microbe. 2011;10:368–377. doi: 10.1016/j.chom.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baumgarten S, et al. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics. 2013;14:704. doi: 10.1186/1471-2164-14-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lipardi C, Paterson BM. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc. Natl Acad. Sci. 2009;106:15645–15650. doi: 10.1073/pnas.0904984106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Blair, C. D., Olson, K. E., & Bonizzoni, M. The Widespread occurrence and potential biological roles of endogenous viral elements in insect genomes. Curr. Issues Mol. Biol. 34, 13–30 (2020). [DOI] [PubMed]
- 94.Yan N, Chen Z. Intrinsic antiviral immunity. Nat. Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pesant, S. et al. (2020). Tara Pacific samples provenance and environmental context—version 2. Zenodo. 10.5281/zenodo.4068293 (2020).
- 96.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 97.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 99.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bushnell B, Rood J, Singer E. BBMerge—accurate paired shotgun read merging via overlap. PLOS ONE. 2017;12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210––3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 103.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bigot T, Temmam S, Pérot P, Eloit M. RVDB-prot, a reference viral protein database and its HMM profiles. F1000 Res. 2020;8:530. doi: 10.12688/f1000research.18776.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kolekar, P. et al. IRESPred: web server for prediction of cellular and viral internal ribosome entry site. IRES. Sci. Rep.6, 27436 (2016). [DOI] [PMC free article] [PubMed]
- 106.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Minh BQ, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aranda M, et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016;6:39734. doi: 10.1038/srep39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shoguchi E, et al. Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics. 2018;19:458. doi: 10.1186/s12864-018-4857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shoguchi E, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 116.Robbins SJ, et al. A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat. Microbiol. 2019;4:2090–2100. doi: 10.1038/s41564-019-0532-4. [DOI] [PubMed] [Google Scholar]
- 117.Shoguchi E, et al. A new dinoflagellate genome illuminates a conserved gene cluster involved in sunscreen biosynthesis. GBE. 2021;13:evaa235. doi: 10.1093/gbe/evaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin S, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350:691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 119.Hume BCC, et al. SymPortal: A novel analytical framework and platform for coral algal symbiont next‐generation sequencing ITS2 profiling. Mol. Ecol. Resour. 2019;19:1063–1080. doi: 10.1111/1755-0998.13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Metadata are accessible in zenodo: https://zenodo.org/record/6299409#.Y-ClwuzMKml. Metagenomes are available via 10.5281/zenodo.7839794. Seawater metagenomes are available through the European bioinformatics institute (Tara Oceans; ERP001736) and NCBI (PRJEB1787). NCBI accession numbers for individual holobiont species metagenomes, genome assemblies and reference sequences can be found in Supplementary Data 1B, 3 and 14, respectively.