Abstract
Extended-spectrum β-lactamase (ESBL)-producing Gram negative bacteria are becoming increasingly important in veterinary and human medicine because they can hydrolyze the third generation β-lactams, penicillins, and monobactams. The aim of this study was to identify ESBL-producing Enterobacteriaceae in raw cow milk samples from northeast Brazil. Twenty-six bacterial isolates belonging to the Enterobacteriaceae family were obtained from milk samples from 257 cows with subclinical mastitis. Using microbiological tests, 53.85% (14/26) were identified as Escherichia coli, 15.38% (4/26) as Proteus mirabilis, 26.92% (7/26) as Klebsiella spp., and 3.85% (1/26) as Citrobacter spp. Of all the isolates, 61.54% (16/26) were positive in the ESBL screening test, of which 12.5% (2/16) were positive in the double-disc synergy test using three types of cephalosporins and amoxicillin/clavulanic acid. The two isolates were identified as Klebsiella spp. Among all the isolates, 53.85% (14/26) were positive for one or both ESBL-encoding genes, blaSHV and blaTEM; among these, 71.43% (10/14) were identified as E. coli. This study demonstrates that ESBL-producing bacteria can be found in raw cow milk from northeast Brazil. Cows with subclinical mastitis should be recognized as reservoirs of these strains, which can propagate to humans.
Keywords: Bacterial resistance, Food safety, Subclinical mastitis, Antimicrobials
Introduction
Antibiotic resistance is a major public health concern worldwide. The inappropriate use of antibiotics has led to the development of resistant bacteria for both human and veterinary medicine and, consequently, to an increase in the number of incurable infections. The environment and domestic animals are considered to be the biggest reservoirs of antimicrobial-resistant bacteria [1].
The use of antibiotics in veterinary medicine, human medicine, and agriculture, either as a prophylactic or as a treatment for infectious diseases, has a significant impact on public health because it stimulates the adaptation (mutation) and survival capacity (resistome) of the bacteria to toxic molecules such as antibiotics [2–4]. Among these microorganisms, the members of the Enterobacteriaceae family are considered reservoirs of resistance genes and are enlisted under group 1 of the list of priority pathogens for research and development of new antibiotics by the World Health Organization [5]; they can carry genes encoding extended-spectrum beta-lactamases (ESBL), which confer resistance against various cephalosporins, oxyimino cephalosporins (cefotaxime, ceftazidime, and ceftriaxone), and monobactams (azetreonam) [6].
Members from the Enterobacteriaceae family, mainly Escherichia coli and Klebsiella pneumoniae, carrying ESBL-encoding genes, have been isolated from animal food products, such as milk [7], chicken [8], beef, and pork [9]. Researchers have emphasized that food-producing animals should be considered in the epidemiological surveillance of resistant and multidrug-resistant strains.
In Brazil, ESBL-producing Enterobacteriaceae have been detected in raw milk and artisanal cheese from the southern region of the country [7]; however, similar surveillance in northeast Brazil is scarce.
The aim of this study was to identify ESBL-producing Enterobacteriaceae and strains carrying the blaSHV and blaTEM genes from raw milk of cows with subclinical mastitis in northeastern Brazil.
Material and methods
Sampling
Twenty-six isolates of enterobacteria, recovered from raw milk samples from 257 cows with subclinical mastitis, were analyzed. The sampling was performed in six dairy cattle farms in the state of Pernambuco during the first half of 2021.
Bacterial identification and ESBL screening test
The genus and species were identified through the characteristics of the colonies on agar base (enriched with 5% sheep blood), Gram staining, and biochemical tests, such as fermentation of glucose, fermentation of lactose, citrate utilization, lysine decarboxylation, and production of H2S, gas, indole, urease, and phenylalanine deaminase [10].
All isolates were screened for the production of ESBL according to the method indicated in the guide of the European Committee on Antimicrobial Susceptibility Testing [11]. The test was performed on Mueller–Hinton agar with the following antibiotic discs: cefotaxime (30 µg), ceftazidime (30 µg), and ceftriaxone (30 µg). The isolates were suspended in sterile saline solution at a concentration of 0.5 on the McFarland standard for subsequent plate inoculation. After placing the antibiotic discs, the isolates were incubated at 37 °C for 18–24 h. All isolates that presented a diameter equal to or smaller than 21 mm for cefotaxime, 23 mm for ceftriaxone, and/or 22 mm for ceftazidime were selected for the ESBL confirmatory test.
ESBL confirmatory test
The confirmatory test was performed using the double-disc synergy test (DDST), following the guidelines for the Detection of Resistance Mechanisms and Specific Resistance of Clinical and/or Epidemiological Importance [11]. For the test, the following antibiotic discs were used: cephalosporins [cefotaxime (30 µg), ceftazidime (30 µg), and ceftriaxone (30 µg)] and amoxicillin with clavulanic acid (30 µg). The cephalosporin discs were placed 20 mm apart from the amoxicillin–clavulanic acid disc, on Mueller–Hinton agar (Fig. 1). After the incubation period (18–24 h), a strain was considered ESBL-positive if we observed an increase in the inhibition zone around any cephalosporin disc or an enlargement or “ghost zone” toward the clavulanic acid disc. The positive control for the confirmatory test was the K. pneumoniae ATCC 700,603 strain.
Fig. 1.
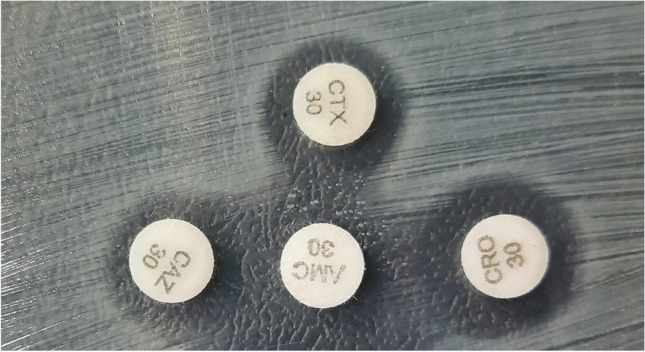
Positive isolate in the double-disc synergy test, where the enlargement or “ghost zone” toward the clavulanic acid disc can be observed. CTX, cefotaxime; CAZ, ceftazidime; CRO, ceftriaxone; AMC, clavulanic acid and amoxicillin
Genotypic identification of ESBL-encoding genes
For molecular analyses, DNA was extracted from all Enterobacteriaceae isolates, regardless of their DDST results, using the thermal extraction technique [12]. After extraction, all samples underwent qualitative and quantitative DNA analysis using a spectrophotometer (Thermo Scientific Multiskan Go) and DNA concentrations were adjusted to 50 ng/µL.
The identification of the ESBL-encoding genes (blaSHV and blaTEM) was carried out using conventional PCR. For amplifying the blaSHV gene, we used the primers SHV-F (5′-GCCGGGTTATTCTTATTTGTCGC-3′) and SHV-R (5′-ATGCCGCCGCCAGTCA-3′) [13]; for amplifying the blaTEM gene, we used the primers TEM-F (5′-TCGGGGAAATGTGCG-3′) and TEM-R (5′-TGCTTAATCAGTGAGGCACC-3′) [14].
The PCR reaction, with a final volume of 12.5 µL, contained the following: 2X GoTaq® G2 Green Master Mix, forward and reverse primers (SHV, 1 pmol/µL; TEM, 5 pmol/µL), ultrapure water, and 2.5 µl of DNA. The following thermal cycling conditions were used: initial denaturation at 95 °C for 5 min; 35 cycles of denaturation at 95 °C for 1 min, annealing at 60 °C for 30 s, extension at 72 °C for 40 s; and a final extension of 72 °C for 4 min. K. pneumoniae strain ATCC 700,603 was used as a positive control for blaSHV and E. coli strain ATCC 35,218 for blaTEM. E. coli strain ATCC 25,922 was used as a negative control.
Results
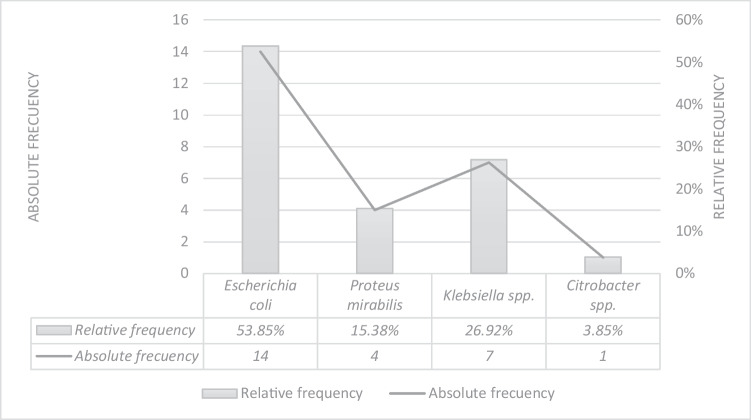
The identification of the genus and species of the enterobacteria isolates is shown in Fig. 2. The phenotypic and genotypic identification of ESBL-producing Enterobacteriaceae is presented in Table 1. Of all the isolates, 38.46% (10/26) showed an inhibition zone considered positive in the screening test for all three antibiotics, while 23.07% (6/26) of the isolates were positive for one or two antibiotics. All positive isolates in the ESBL screening test (61.54%, 16/26) were confirmed using the DDST, and 12.5% (2/16) tested positive in the ESBL confirmatory test and were identified as Klebsiella spp.
Fig. 2.
Absolute and relative frequency obtained in the phenotypic identification of the 26 isolates belonging to the Enterobacteriaceae family
Table 1.
Results of the screening test for the identification of ESBL-producing Enterobacteriaceae. Number of isolates that were positive to the confirmatory test, DDST. Isolates that presented the ESBL-encoding genes, blaSHV and blaTEM
| ID | No. of isolates | CTXa (≤ 21 mm) | CAZa (≤ 22 mm) | CROa (≤ 23 mm) | Positive ESBL screening test | ESBL* | blaSHV gene | blaTEM gene | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AR | RF (%) | AR | RF (%) | AR | RF (%) | AR | RF (%) | AR | RF (%) | AR | RF (%) | AR | RF (%) | ||
| Escherichia coli | 14 | 2 | 14.29% | 6 | 42.86% | 4 | 28.57 | 6 | 42.86% | 0 | 0% | 8 | 57.14% | 8 | 57.14% |
| Klebsiella spp. | 7 | 5 | 71.42% | 6 | 85.71% | 5 | 71.43 | 6 | 85.71% | 2 | 28.47 | 3 | 42.86% | 0 | 0% |
| Proteus mirabilis | 4 | 4 | 100% | 4 | 100% | 3 | 75 | 4 | 100% | 0 | 0% | 0 | 0% | 0 | 0% |
| Citrobacter spp. | 1 | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 100% |
| Total: 26 | 11 | 42.31%c | 16 | 61.54%c | 12 | 42.31%c | 16 | 61.54%c | 2 | 12.5%b | 11 | 79%d | 9 | 43%d | |
| Positive ESBL screening test: 16 (38.46%) | Positive PCR: 14 (53.85%) | ||||||||||||||
CTX cefotaxima, CAZ ceftazidima, CRO ceftriaxone, ESBL extended-spectrum β-lactamase, AR absolute frequency, RF relative frequency
*Confirmatory test—DDST (double-disc synergy test)
aPhenotypic test
bPercentage associated with the number of positive isolates in the screening test
cPercentage associated with the total number of isolates
dPercentage associated with the number of isolates positive for one or both genes
Of all the isolates, 53.85% (14/26) were positive for either one or both the ESBL-encoding genes (Table 1): 79% (11/14) had the blaSHV gene, 64% (9/14) had the blaTEM gene, and 43% (6/14) had both the genes. Of the two positive isolates from the DDST, one was positive for the blaSHV gene. Of the 14 isolates positive for one or two genes, 57% (8/14) were positive in the ESBL screening test while 43% (6/14) exhibited no resistance to the cephalosporins used in this study.
The most prevalent gene was blaSHV with 11 positive isolates, of which 73% (8/11) were identified as E. coli and 27% (3/11) as Klebsiella spp. The blaTEM gene was found in nine isolates, of which 89% (8/9) were E. coli and 11% (1/9) were Citrobacter spp. Both the genes were found in 66% (6/9) of the positive isolates, and all were identified as E. coli.
Discussion
In food-producing animals, Enterobacteriaceae family members, such as E. coli and Klebsiella spp., are a part of the intestinal commensal microbiota and possess the ability to horizontally receive and transfer resistance genes from/to several bacterial species [1]. Members of the Enterobacteriaceae family have been identified as the etiological agents for bovine mastitis. They are considered opportunistic pathogens because they readily colonize the mammary glands of their hosts when the animals experience stress, caused by animal exploitation and overcrowding [15].
In our study, two isolates of ESBL-producing Klebsiella spp. were identified in the state of Pernambuco, using the DDST. In Brazil, ESBL-producing Enterobacteriaceae have already been identified in the agricultural system [16], but the incidence of such bacteria in bovine and goat milk has not been reported in the northeastern region of the country. De Campos et al. [17] analyzed two samples of artisanal cheese from the state of Bahia and reported negative results for ESBL-producing bacteria. Palmeira et al. [18] identified 40 strains of ESBL-producing E. coli in fecal samples from cattle farms, of which six were obtained from farms in Pernambuco, implying that cattle should be considered an important reservoir of ESBL-producing bacteria.
ESBLs, mainly encoded on plasmids, can hydrolyze different types of cephalosporins. These enzymes are identified as sulfhydryl variable (SHV), temoneira (TEM), or cefotaximases (CTX-M) [1]. In our study, of the two ESBL-producing Klebsiella spp., only one was positive for the blaSHV gene, while the other was negative for both the genes tested. It may harbor genes encoding CTX-M-like β-lactamases, which were not investigated in this study.
Most of the isolates carrying the blaSHV and/or blaTEM genes were identified as E. coli, which is in concordance with similar studies carried out in other countries such as Turkey [19], Switzerland [20], Egypt [21], and France [22]. The blaSHV gene was found to be more frequent than the blaTEM gene. In Brazil, while studying raw milk and/or artisanal cheese, E. coli has been determined as the most frequent bacteria from the Enterobacteriaceae family in cases of subclinical mastitis of environmental origin. However, the blaTEM gene was more frequently detected in these studies [7], contrasting with the results obtained in the present study. In a more recent study, Palmeira et al. [18] identified the CTX-M gene as the most incident, followed by the blaSHV gene and, in lesser amount, the blaTEM gene in E. coli isolates from bovine feces samples in northeastern Brazil. Although the CTX-M gene was not investigated in this study, our results are compatible with Palmeira et al. [18], since the blaSHV gene was detected at a higher frequency than the blaTEM gene in isolates of enterobacteria from raw bovine milk from Pernambuco.
Of the 14 isolates positive for ESBL-producing genes, one was positive in the DDST, seven were positive in the ESBL screening test, and six were negative in all phenotypic tests. This discrepancy between phenotypic and genotypic characteristics has already been observed by Son et al. [23], who identified non-ESBL-producing E. coli isolates (in the phenotypic test) carrying the CTX-M and blaTEM genes. Hughes, Andersson [24] affirmed that the presence of a gene might not predict a given phenotypic trait; it may be related to environmental conditions that can modify phenotypic expressions and/or to other genetic factors, such as combinations of several resistance genes, mutations that can change the phenotypic expression, and the presence of insertion sequences that can influence the antibiotic resistance mechanisms, such as drug inactivation, alteration of the target, or production of specific efflux pumps.
Conclusion
The data obtained in this study confirm the occurrence and circulation of ESBL-producing Enterobacteriaceae in cows with subclinical mastitis in northeastern Brazilian; this can be a public health concern because the raw milk from these cows is used for the consumption and production of artisanal “coalho” cheese. Epidemiological studies on ESBL-producing bacteria in the agricultural environment are necessary to develop epidemiological, surveillance, and health policies.
Acknowledgements
We thank the FACEPE (Fundação de Amparo a Ciência e Tecnologia de Pernambuco) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior) for granting a scholarship doctorate level.
Author contribution
Tania Alexandra Ortega Sierra and Rinaldo Aparecido Mota contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Tania Alexandra Ortega Sierra, Atzel Candido Acosta, Renata Pimentel Bandeira de Melo, Pollyanne Raysa Fernandes de Oliveira, and José Wilton Pinheiro Junior. The first draft of the manuscript was written by Tania Alexandra Ortega Sierra. The first version of the manuscript was edited by Rinaldo Aparecido Mota. The revision and edition of all the following versions of the manuscript were in charge of Rodolfo de Morais Peixoto, Erika Fernanda Torres Samico Fernandes, and José Wilton Pinheiro Junior. All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval
This research was approved by the Ethics Committee on the Use of Animals (license number 4848110121).
Consent to participate
Not applicable.
Consent for publication
All authors read and approved the manuscript.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Argudín MA, Deplano A, Meghraoui A, et al. Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics. 2017;6(2):1–38. doi: 10.3390/antibiotics6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA. Global trends in antimicrobial use in food animals. PNAS. 2015;112(18):5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockwell VO, Duffy B. Use of antibiotics in plant agriculture. OIE Rev Sci Tech. 2012;31(1):199–210. doi: 10.20506/rst.31.1.2104. [DOI] [PubMed] [Google Scholar]
- 4.Wright GD. The antibiotic resistome. Expert Opin Drug Discov. 2010;5(8):779–788. doi: 10.1517/17460441.2010.497535. [DOI] [PubMed] [Google Scholar]
- 5.WHO. WHO. Global priority list of antibiotic-resistant batceria to guide research, discovery, and development of new antibiotics. Published 2017. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
- 6.Bergšpica I, Kaprou G, Alexa EA, Prieto M, Alvarez-Ordóñez A. Extended spectrum β-lactamase (ESBL) producing Escherichia coli in pigs and pork meat in the European Union. Antibiotics. 2020;9(10):1–23. doi: 10.3390/antibiotics9100678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parussolo L, Antônio R, Sfaciotte P, et al. Detection of virulence genes and antimicrobial resistance profiles of Escherichia coli isolates from raw milk and artisanal cheese in Southern Brazil. Semin Ciências Agrárias. 2019;40(1):163–178. doi: 10.5433/1679-0359.2019v40n1p163. [DOI] [Google Scholar]
- 8.Ortega-Paredes D, de Janon S, Villavicencio F, et al (2020) Broiler farms and carcasses are an important reservoir of multi-drug resistant Escherichia coli in Ecuador. Front Vet Sci. 7(November). 10.3389/fvets.2020.547843 [DOI] [PMC free article] [PubMed]
- 9.Guo S, Aung KT, Leekitcharoenphon P, et al. Prevalence and genomic analysis of ESBL-producing Escherichia coli in retail raw meats in Singapore. J Antimicrob Chemother. 2021;76(3):601–605. doi: 10.1093/jac/dkaa461. [DOI] [PubMed] [Google Scholar]
- 10.Koneman EW, Procop GW, Church DL, et al (2018) Diagnóstico Microbiológico - Texto e Atlas. 7a edição. Grupo GEN
- 11.UECAST (2017) EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and / or epidemiological importance. Published 2017. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf
- 12.Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Gheldre Y, Avesani V, Berhin C, Delmée M, Glupczynski Y. Evaluation of oxoid combination discs for detection ofextended-spectrum -lactamases. J Antimicrob Chemother. 2003;52(4):591–597. doi: 10.1093/jac/dkg415. [DOI] [PubMed] [Google Scholar]
- 14.Vercauteren E, Descheemaeker P, Ieven M, Sanders CC, Goossens H. Comparison of screening methods for detection of extended-spectrum beta-lactamases and their prevalence among blood isolates of Escherichia coli and Klebsiella spp in a Belgian teaching hospital. J Clin Microbiol. 1997;35(9):2191–2197. doi: 10.1128/jcm.35.9.2191-2197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal M, Regasa A, Gizaw F. Etiology, pathogenesis, risk factors, diagnosis and management of bovine mastitis : a comprehensive review etiology, Pathogenesis, Risk Factors, Diagnosis and Management of Bovine Mastitis : A Comprehensive Review. Int J Anim Vet Sci. 2020;60:40–55. [Google Scholar]
- 16.Santiago GS, Coelho IS, Bronzato GF, et al. Short communication : extended-spectrum AmpC – producing Escherichia coli from milk and feces in dairy farms in Brazil. J Dairy Sci. 2018;101(9):7808–7811. doi: 10.3168/jds.2017-13658. [DOI] [PubMed] [Google Scholar]
- 17.de Campos A, Puño-Sarmiento J, Medeiros L, et al. Virulence genes and antimicrobial resistance in Escherichia coli from cheese made. Foodborne Pathog Dis. 2017;20(20):1–7. doi: 10.1089/fpd.2017.2345. [DOI] [PubMed] [Google Scholar]
- 18.Palmeira JD, Haenni M, Metayer V, Madec JY, Ferreira HMN. Epidemic spread of IncI1/pST113 plasmid carrying the Extended-Spectrum Beta-Lactamase (ESBL) blaCTX-M-8 gene in Escherichia coli of Brazilian cattle. Vet Microbiol. 2020;243:1–5. doi: 10.1016/j.vetmic.2020.108629. [DOI] [PubMed] [Google Scholar]
- 19.Tekiner IH, Özpınar H. Occurrence and characteristics of extended spectrum beta-lactamases-producing Enterobacteriaceae from foods of animal origin. Brazilian J Microbiol. 2016;47:444–451. doi: 10.1016/j.bjm.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geser N, Stephan R, Hächler H. Occurrence and characteristics of extended- spectrum b -lactamase ( ESBL ) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. Vet Res. 2012;8(21):1–9. doi: 10.1186/1746-6148-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalifa SM, Abd El-Aziz AM, Hassan R, Abdelmegeed ES. β-lactam resistance associated with β-lactamase production and porin alteration in clinical isolates of E. coli and K. pneumoniae. PLoS One. 2021;16(5 May):1–22. doi: 10.1371/journal.pone.0251594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dao TL, Hoang VT, Magmoun A, et al. Acquisition of multidrug-resistant bacteria and colistin resistance genes in French medical students on internships abroad. Travel Med Infect Dis. 2020;2021(39):19–21. doi: 10.1016/j.tmaid.2020.101940. [DOI] [PubMed] [Google Scholar]
- 23.Son TV, Manh ND, Trung NT, et al. Molecular detection of bla CTX - M gene to predict phenotypic cephalosporin resistance and clinical outcome of Escherichia coli bloodstream infections in Vietnam. Ann Clin Microbiol Antimicrob. 2021;20(60):1–9. doi: 10.1186/s12941-021-00466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes D, Andersson DI. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev. 2017;fux004(41):374–391. doi: 10.1093/femsre/fux004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.