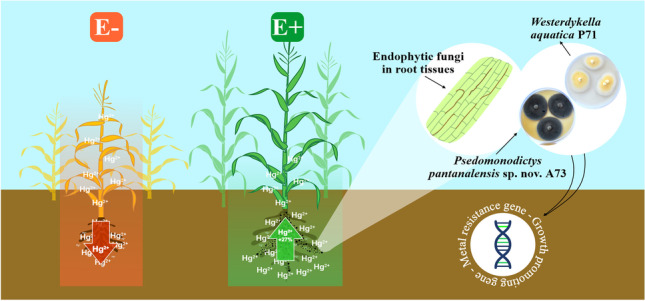
Abstract
Mercury is a non-essential and toxic metal that induces toxicity in most organisms, but endophytic fungi can develop survival strategies to tolerate and respond to metal contaminants and other environmental stressors. The present study demonstrated the potential of mercury-resistant endophytic fungi in phytoremediation. We examined the functional traits involved in plant growth promotion, phytotoxicity mitigation, and mercury phytoremediation in seven fungi strains. The endophytic isolates synthesized the phytohormone indole-3-acetic acid, secreted siderophores, and solubilized phosphate in vitro. Inoculation of maize (Zea mays) plants with endophytes increased plant growth attributes by up to 76.25%. The endophytic fungi stimulated mercury uptake from the substrate and promoted its accumulation in plant tissues (t test, p < 0.05), preferentially in the roots, which thereby mitigated the impacts of metal phytotoxicity. Westerdykella aquatica P71 and the newly identified species Pseudomonodictys pantanalensis nov. A73 were the isolates that presented the best phytoremediation potential. Assembling and annotation of P. pantanalensis A73 and W. aquatica P71 genomes resulted in genome sizes of 45.7 and 31.8 Mb that encoded 17,774 and 11,240 protein-coding genes, respectively. Some clusters of genes detected were involved in the synthesis of secondary metabolites such as dimethylcoprogen (NRPS) and melanin (T1PKS), which are metal chelators with antioxidant activity; mercury resistance (merA and merR1); oxidative stress (PRX1 and TRX1); and plant growth promotion (trpS and iscU). Therefore, both fungi species are potential tools for the bioremediation of mercury-contaminated soils due to their ability to reduce phytotoxicity and assist phytoremediation.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-023-00924-4.
Keywords: Endophytes, Genome draft, Secondary metabolites, Bioremediation
Introduction
Mercury (Hg) is a non-essential and toxic metal originated from natural sources and human activities [1–3]. The most significant natural sources of mercury are mineral weathering, erosion, and volcanic eruption [5, 6]. However, the anthropic sources have caused great concern because the exponential increase in the use of mercury in various sectors such as industry, agriculture, and mining has resulted in its excessive release into aquatic and terrestrial ecosystems [7–10]. The Pantanal is the world’s largest humid freshwater area and it is under increasing anthropogenic threats, including long-term regionally intensive gold mining practices [11] that are known to release harmful pollutants such as mercury to the surrounding environment [12].
Mercury causes a major global impact due to its toxicity, persistence, bioaccumulation, and biomagnification capacity that vary according to its chemical speciation [4, 13]. Methylated derivatives are the most toxic mercury species due to their affinity for sulfhydryl radicals of amino acids, which alter protein structure and cause loss of function [13, 14]. Bioaccumulation and biomagnification of mercury in living organisms can disturb different trophic levels of ecosystems, since mercury is easily transferred through the food chain [15, 16] and poses high risks to human health and animal in the short and long term [17, 18]. Mercury toxicity may aggravate neurological and neurodegenerative [19], renal [20], and cardiovascular diseases [21] in humans. The negative effects of mercury contamination can also strongly affect the ecosystem by limiting plant growth and altering the structure and composition of communities of microorganisms [22–25], which impair the maintenance of the structure and functioning of the ecosystem.
Several species of endophytic fungi are resistant to mercury toxicity, including Aspergillus sp. and Curvularia geniculata [26]. Endophytes attenuate the toxicity of metals to hosts in contaminated environments [22, 24–26], favor plant growth and phytoremediation, and enhance the host ability to extract soil contaminants [27, 28]. The use of ecological approaches to reduce soil mercury contamination has attracted increasing attention in the last 10 years. Bioremediation is a process that uses living organisms, including archaea, bacteria, fungi, and plants, to remove or immobilize contaminants [22, 23, 25, 27, 29]. It is an effective approach to treat soils contaminated with toxic metals, is cheaper than traditional physicochemical approaches, and has little or no environmental impact [28, 30].
Microorganisms have developed different mechanisms of resistance to toxic metals. The intracellular detoxification mechanism of some fungi, bacteria, and archaea may be mediated by enzymes encoded in the operon mer that are able to volatilize mercury [30–32]. Another extracellular mechanism of resistance to toxic metals is the ability to adsorb metal ions to cell wall components through binding to functional groups, such as amino, hydroxyl, carboxyl, and sulfhydryl groups [33, 34].
Polygonum acuminatum Kunth and Aeschynomene fluminensis Vell are abundant plant species in mercury-contaminated wetland soils, whose richness and composition of endophytic fungi are influenced by contamination [26]. These hosts harbor mercury-resistant endophytic species and thereby have the potential to assist phytoremediation [26, 27]. The high functional versatility of fungi is an important biotechnological characteristic applied to bioremediation that represent an advantage over the physicochemical removal of toxic metals in terms of cost and time efficiency [35]. However, the molecular mechanisms of tolerance and resistance to mercury in fungal cells are poorly known. There are some evidences that fungal cells have enzymes capable of volatilizing mercury and macromolecules that adsorb metals [27, 30, 34].
We hypothesize that mercury-resistant endophytic fungi are capable of promoting the growth of the host plant in mercury-contaminated soil, attenuating the effects of the contaminant on plants and assisting in phytoremediation. In this sense, the present study aims to (1) determine functional traits and growth promotion in maize (Zea mays L.—Poaceae) plants inoculated with endophytic fungi isolated from Aeschynomene fluminensis Vell (Fabaceae) and Polygonum acuminatum Kunth (Polygonaceae); (2) examine the role of endophytic fungi in mercury phytoextraction; and (3) assemble and annotate the genome of endophytic fungi in order to identify important factors for resistance to toxic metals.
Material and methods
Endophytic strains
In the present study, seven endophytic fungi isolates were randomly chosen among the mercury-resistant microorganisms (Table 1). The isolates were obtained from the Culture Collection of the Laboratory of Biotechnology and Microbial Ecology at Federal University of Mato Grosso (Cuiabá, Mato Grosso State, Brazil). Endophytic fungi were previously isolated from the root system of host species abundant in wet areas contaminated (3.24 mg.kg−1) (+ Hg site—S″16°15′42.7″ W″056°38′43.6″) or not contaminated with mercury (-Hg site—S″16°21′19.7″ W″056°20′13.9″), located in the city of Poconé [26]. The isolates were activated in potato dextrose agar culture medium for 7 days at 30 °C, and preserved in 20% glycerol, at − 20 °C.
Table 1.
Endophytic fungi used in maize growth promotion and mercury bioremediation assays
| Isolate | Identification | Host | TI (μg mL−1) | NCBI access number |
|---|---|---|---|---|
| A65 | Sordariomycetes2 | Aeschynomene fluminensis Vell (Fabaceae) | 0.82 | KY987119 |
| A73* | Pseudomonodictys pantanalensis sp. nov | Aeschynomene fluminensis Vell (Fabaceae) | 0.72 | KX381148 |
| P42** | Colletotrichum sp. | Polygonum acuminatum Kunth (Polygonaceae) | 1.03 | KX381174 |
| P71*,** | Westerdykella aquatica | Polygonum acuminatum Kunth (Polygonaceae) | 0.95 | KX381190 |
| P72 | Nemania sp. | Polygonum acuminatum Kunth (Polygonaceae) | 0.44 | KX381191 |
| P74 | Pleosporales 1 | Polygonum acuminatum Kunth (Polygonaceae) | 0.85 | KX381192 |
| P96 | Diaporthe miriciae | Polygonum acuminatum Kunth (Polygonaceae) | 0.94 | KX381202 |
TI, mercury tolerance index. TI was calculated from the growth rate of the strain exposed to mercury divided by the growth rate in the control plate (without mercury) [26]
*These isolates were previously identified as Lindgomycetaceae A73 and Westerdykella sp. P71 by Pietro-Souza et al. [26], and we propose changing the identification
**Sporulating fungi in potato dextrose agar medium
Characterization of functional traits of fungal isolates
The strains were inoculated in potato dextrose broth supplemented with 2.5 mg mL−1 tryptophan (pH 6.0) and incubated for 7 days, at 30 °C, under shaking at 100 rpm. The production of indole-3-acetic acid was determined using Salkowski’s reagent, according to Babu et al. [36].
Siderophores were quantified in the culture supernatant of strains grown in potato dextrose broth for 7 days, at 30 °C, under shaking at 100 rpm. The supernatant was centrifuged at 11,000 rpm for 5 min. One milliliter of the chromium azurol S indicator solution (1:1, v/v) [37] was mixed with 1 mL of the supernatant from different cultures, and absorbance was recorded in a spectrophotometer set at 630 nm [38].
Phosphate solubilization by fungal isolates was estimated using phosphate-supplemented growth medium, according to the protocol developed by Nautiyal [39]. Production of siderophores and phosphate solubilization by the strains were qualitatively evaluated and the results were expressed as positive ( +) and negative ( −) presence of the tested functional traits.
Promotion of maize plant growth by endophytic fungi under the effect of mercury
Maize seeds (Zea mays, Hybrid AG1051) were superficially disinfected with 70% ethanol for 1 min, 2.5% sodium hypochlorite for 5 min, and rinsed in sterile distilled water. Then, the seeds were placed in trays containing vermiculite and sand 1:1 (m:m), and kept in a greenhouse for 7 days, for germination. After this period, seedlings were transferred to pots (0.5 dm3) containing vermiculite and sand 1:1 (m:m) previously homogenized with 20 mg.Kg−1 of Hg+2 (HgCl2). The fungal strains were activated in potato dextrose agar medium for 7 days, at 30 °C. The seedling roots were inoculated with the fungal strains: 1 mL of spore suspension (106 conidia.mL−1) of sporulating strains or two discs (2 cm diameter) of medium containing mycelium of non-sporulating strains. After 24, 48, and 72 h of inoculation, 20 mg.Kg−1 of HgCl2 solution was applied, totaling 80 mg.Kg−1 Hg+2 (HgCl2) per pot [27].
The field capacity of the substrate was maintained at 70%, and the plants were irrigated weekly with 100% Hoagland solution [40]. Control treatments were carried out in pots containing non-inoculated plants grown in substrate with added Hg (positive control; C + Hg) or without added Hg (negative control; C − Hg). The pots were randomly distributed in the greenhouse, and each treatment contained 12 replicates.
Fifteen days after transplanting, the chlorophyll index was determined using a portable chlorophyll meter (SPAD-512, Minolta). Height, root length, and shoot and root dry weight data were recorded on the sixteenth day after inoculation. The growth promotion efficiency of the strains studied was determined using the parameters height, root length and dry mass, and shoot length and dry mass, which were determined after drying the plants in an oven at 65 °C until they reached constant mass [41]. Plant mass allocation was determined using plant dry mass data and was expressed as percentage, calculated from the ratio between organ and plant dry mass [42].
Data were analyzed by parametric analysis of variance (ANOVA) combined with Dunnett’s test (p < 0.05), and the p-values were adjusted by FDR (False Discovery Rate). Statistical analyzes were performed using the R v 3.6.3 software [43] with the multcomp package [44].
Quantification of mercury in plant tissues
The properly dried plant tissue samples were digested in a concentrated acidic solution of HNO3:H2O2 (5:3, v/v) in a microwave apparatus (Berghof), using the schedules recommended by the equipment manufacturer. The metal content was determined by inductively coupled plasma optical emission spectroscopy (ICP-OES), according to Mello et al. [31]. The ICP-OES system was calibrated using serial dilutions of mercury standards. The standard curve used Hg2 + quantification followed the equation y = 0.9469x + 1.7957 and R2 = 0.9972.
To determine the efficiency of mercury translocation from maize root to shoot, the translocation factor was calculated from the ratio between mercury concentration (in mg.Kg−1) in shoot and root [45]. Data were analyzed by parametric analysis of variance (ANOVA) and t test (p < 0.05), and p-values were adjusted by FDR (False Discovery Rate). Statistical analyses were performed using the R v 3.6.3 software [43] and the PAST V 3.25 software [46].
Total DNA extraction and genome sequencing
As the isolates A73 and P71 performed best in assisting phytoremediation, they were selected for genome sequencing. Endophytes were grown in potato dextrose broth and incubated for three days, at 30 °C, and under shaking at 100 rpm. Their mycelia were macerated with liquid nitrogen and DNA was extracted using the PowerSoil™ DNA extraction kit, according to the manufacturer’s instructions (Mobio Laboratories, Carlsbad, CA, USA). Genome was sequenced on the Illumina HiSeq 2500 platform using paired-end sequencing (2 × 150 bp). Libraries were prepared using Nextera™ XT Library Prep workflow, according to the Illumina Preparation Guide (http://www.illumina.com). DNA concentration was adjusted based on qPCR quantifications, using the KAPA Library Quantification Kit for Illumina® Platforms, according to the manufacturer’s specifications.
Genome assembly
The quality of reads was assessed using the FASTQC tool [47]. Snippets of adapters and poor-quality bases at the end of the sequences were removed using the Trimmomatic v0.38 software [33], with the parameters: cutting of adapters; LEADING:3; TRAILING:3; SLIDINGWINDOW:12:15; MINLEN:30 for the strain P71; and MINLEN:50 for the strain A73. Quality reads were assembled using the new SPAdes assemblers [48], Newbler [49], and the MeGAMerge post-assembler [50], using the standard parameters. The quality of the assembled reads was assessed using the QUAST tools [51]. Sequencing data are available on the NCBI Sequence Read Archive under accession numbers JAFHBV000000000 and JAFHBW000000000.
Prediction and functional gene annotation
To predict biosynthesis gene clusters, the assembled contigs were analyzed using the anti-SMASH v.5.0 version with predefined parameters by default [52]. Protein-coding genes were predicted using the AUGUSTUS software (Version: 3.2.1) with predefined parameters by default [53]. The completeness of the annotation was determined with BUSCO using the predicted proteins of the fungi_odb9 model [54]. The predicted and conceptually deduced genes in protein sequences were used for functional annotations.
Ortholog groups were identified using the eggNOG-Mapper 0.12.7 functional annotation tool [55] with DIAMOND v4.5.1 mapping mode [56], and protein functional signatures identified using the InterProScan v5 software [57]. The tRNA and rRNA regions were detected using the Aragorn and RNAmmer software, respectively [58, 59]. Based on the protein prediction results, the gene ontology terms (GO) were mapped into three groups: Biological Process, Molecular Function, and Cell Component. GO analysis was performed using the online platform AgBase (https://agbase.arizona.edu/). The new protein annotation files were generated with GOanna, GOanna2ga, and GOSlimViewer tools running with predefined default parameters [60].
DNA extraction, PCR amplification, and DNA barcoding sequencing for phylogenetic analysis
To identify Pseudomonodictys pantanalensis sp. nov., total DNA from the isolates P87, A73, and P100 was extracted using a DNA purification kit (Norgen Biotek Corp, Canada). The fragments of the regions corresponding to ITS, SSU, and LSU were amplified using the primers listed in Table S1. ITS1/F and ITS4/R were used to amplify the rDNA internal transcribed space region (ITS). LR0R/F and LR3/R were used to partially amplify the rDNA sequence of the 28S ribosomal large subunit (LSU). SCR1c/F and NS8/R were used to partially amplify the rDNA of ribosomal small subunit (SSU).
The PCR reactions were performed in a 25 μL reaction volume containing 50 ng of genomic DNA, 0.5 pmol of each primer, 1X PCR buffer with 50 mM MgCl2, 1 mM dNTPs, and 1 U Taq polymerase (EasyTaq-TranGen Biotech Co. LTD). The reaction program was initial denaturation at 95 °C for 3 min, 35 denaturation cycles at 95 °C for 30 s, annealing at 53 °C for 45 s, and elongation at 72 °C for 1 min. Final stretching was performed at 72 °C for 5 min. The amplicons were purified using Exosap (Thermo Fisher). The sequencing reactions were performed using the BigDye v3.1 kit (Thermo Fisher), according to the manufacturer’s recommendations. The sequences were analyzed on the 3500 Genetic Analyzer (Thermo Fisher) sequencer.
Phylogenetic analysis
Consensus sequences for each PCR-amplified barcoding were obtained based on alignment of forward and reverse sequences using a DNA baser assembly software (http://www.dnabaser.com/). Sequences obtained by amplification and from the complete genome for P. pantanalensis were deposited in GenBank (http://www.ncbi.nlm.nih) under the accession numbers KX381148, KX381153, and KX381197 (ITS), MZ766927 to MZ766928 (SSU), MZ766923 to MZ766925 (LSU), and MZ803210 (TEF). The isolates P87, A73, and P100 were phylogenetically identified using a collection of 32 type species belonging to the suborder Massarinae. Such collection included 29 isolates previously characterized, composed of members of the families Parabambusicolaceae (n = 16), Morosphariaceae (n = 6), Trematosphariaceae (n = 4), Bambusicolaceae (n = 2), Massarinaceae (n = 2), and Lindgomycetaceae (n = 2). Corynespora cassiicola and Corynespora smithii species were used as outgroups (Table S2).
For phylogenetic identification of W. aquatica P71, we used the dataset containing sequences of 25 isolates belonging to 14 type species, previously characterized from the genus Westerdykella, in addition to Preussia funiculata as outgroup (Table S3). The sequences obtained from the complete genome of W. aquatica P71 were deposited in GenBank, under the accession numbers MZ747709 (ITS), MZ766926 (LSU), and MZ803211 (tub2).
The sequences identified for each locus were individually aligned with the MAFFT tool in the UGENE software [61], and the alignments were plotted using the MEGA7 software [62]. Phylogenetic analyzes were conducted using concatenated sequences of the four loci for P. pantanalensis (isolates P87, A73, and P100) and three loci for Westerdykella isolate P71, using Bayesian Inference and Maximum Likelihood.
The Maximum Likelihood analysis included 1000 replicates (bootstrap) using all sites, General Time Reversible statistical model, and discrete gamma distribution with four categories. Bayesian Inference was based on the model adopted in PAUP*4 and Mrmodeltest2 v2 software [63]. All sites at the ITS, SSU, LSU, and TEF loci for the isolates P87, A73, and P100, as well as the sites at ITS, LSU, and tub2 for the isolate P71 were considered. Ten million generations were analyzed, with the first 25% of the trees discarded and burned using the MrBayes v3.7 tools available at CIPRES (https://www.phylo.org/). Posterior probability and tree topology were visualized with Figtree v 1.1.2 [64]. The consensus tree of the Maximum Likelihood and Bayesian Inference analyses was generated manually from the topology obtained by Figtree, derived from the Bayesian Inference analysis with the posterior probability values, plus the bootstrap values generated by the Maximum Likelihood analysis, using the CorelDraw editing package.
Phenotypic characterization of the new species Pseudomonodictys pantanalensis A73.
The fungal isolates were inoculated in Potato Dextrose Agar, 4% Sabouraud Glucose Agar (Fluka), and Oat Agar (50 g of oat flour, 15 g of agar, 1 L of distilled water), and grown for 7 days, at 30 °C. The endophytic fungi were characterized according to macromorphological aspects of the cultures, such as surface and reverse coloration of the culture medium, border, shape, relief, and mycelium coloration [65]. Permanent slides obtained from microcultures [66] in Potato Dextrose Agar, Sabouraud Glucose Agar, Oat Agar, and Rice Straw media [67] were mounted in order to analyze micromorphological characteristics of the isolates using an EVOS M5000 microscope (Invitrogen by Thermo Fisher Scientific).
Microscopic analysis of maize roots inoculated with Pseudomonodictys pantanalensis A73.
Maize seeds were superficially disinfected with 70% ethanol for 1 min, 2.5% sodium hypochlorite for 5 min, and three rinses in sterile distilled water. The disinfested seeds were inoculated in solid Murashige and Skoog basal medium [68] for germination. After 2 days, the radicles were inoculated with mycelium fragments and the seedlings were kept under 16–8 h photoperiod for 7 days. Then, the radicles were clarified with 10% KOH for 3 min and neutralized with 1% HCl for 5 min. The radicle fragments were stained with 0.05% trypan blue (prepared in 1:1:1 lactic acid, glycerol, and water) for 2 min, at 60 °C, in a water bath. Permanent slides were mounted with polyvinyl lactoglycerol [69] and the structures were analyzed in an EVOS M5000 microscope (Invitrogen by Thermo Fisher Scientific).
Results and discussion
Functional characteristics of endophytic fungal strains
The functional profile of endophytic isolates was analyzed to determine functional traits important for plant growth. Three out of the seven strains analyzed—P. pantanalensis A73, W. aquatica P71, and Nemania sp. P72—secreted the phytohormone indole-3-acetic acid (55.77 to 69.72 µg.mL−1) and siderophores when cultured in vitro (Table 2). Endophytic fungi synthesize metabolites that stimulate plant growth and development under biotic and abiotic stresses [26, 70, 71], such as indole-3-acid acetic, siderophores, in addition to organic acids and phosphatases involved in solubilization and mineralization of phosphate [72–75].
Table 2.
Quantification of functional traits produced by endophytic fungi isolated from Polygonum acuminatum and Aeschynomene fluminensis
| Isolated name | IAA (µg. mL−1) | p | Siderophores |
|---|---|---|---|
| Sordariomycetes 2 A65 | - | − | − |
| Pseudomonodictys pantanalensis A73 | 55.77 | − | + |
| Colletotrichum sp. P42 | - | + | − |
| Westerdykella aquática P71 | 199.00 * | − * | + * |
| Nemania sp. P72 | 69.72 | − | + |
| Pleosporales 1 P74 | - | − | − |
| Diaporthe miriciae P96 | - | + | + |
IAA, indole-3-acetic acid; P, phosphate solubilization; ( +) positive test; ( −) negative test
*Pietro-Souza et al. [27]
Indole-3-acetic acid is a dominant type of auxin produced by plants and many endophytes that delays plant aging, regulates plant growth and development, and maintains endophytic symbiosis [73, 75–77]. This phytohormone stimulates root growth and thereby enhances the plant capacity to absorb nutrients and metals. Under stress conditions, endophytes can regulate the synthesis of phytohormones and promote physiological and metabolic changes in their hosts, which can indirectly favor removal or reduction of pollutants [25, 27, 76].
Fungi can produce one or different types of siderophores, depending on the species and environmental conditions [78]. Siderophores can facilitate soil demineralization and increase the solubilization of metals that are essential for the soil biota, such as iron [78, 79]. These small chelating molecules with high affinity for Fe2+ and other bivalent metal ions can help plants extract more metal ions from the soil, alleviate the stress caused by excess of metals, and even absorb more Fe2+, thus maintaining plant health and promoting plant growth [77, 78].
In this study, the isolated Colletotrichum sp. P42 and Diaporthe miriciae P96 solubilized phosphate, under the conditions assessed (Table 2). Phosphorus is an essential nutrient poorly bioavailable in soils that is usually found in insoluble organic and inorganic forms [74]. Soil microorganisms play an important role in increasing the phosphorus availability through inorganic phosphorus solubilization and organic phosphorus mineralization via processes that depend on the ability of microorganisms to secrete phosphatases, phytases, and organic acids [74, 80]. Microorganism-mediated solubilization of inorganic phosphate is one of the main mechanisms of plant growth promotion [74].
Mercury phytoremediation assisted by endophytic fungi
Addition of mercury (80 mg.Kg−1) to maize plants reduced the height (48.31%), dry biomass (28.13%), and chlorophyll index (11.31%) in non-inoculated plants treated with mercury (C + Hg), when compared with non-inoculated plants not treated with mercury (C − Hg) (Table 3). This result suggested that mercury contamination negatively affected the morphophysiological characteristics of maize, corroborating other studies that demonstrated mercury toxicity to maize plants [25, 81]. In addition to mercury, metals such as copper (Cu), lead (Pb) [81], cadmium (Cd) [82], and chromium (Cr) [83] are potentially toxic to maize.
Table 3.
Growth promotion of maize inoculated with endophytic fungal strains under the influence of mercury
| Treatment | Height (cm) | GPEA (%) | Root (cm) | GPER (%) | MSPA (g) | GPEMSPA (%) | A.MMSPA (%) | MSR (g) | GPEMSR (%) | A.MMSPA (%) | SPAD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sordariomycetes A65 | 34.22 ± 2.4* | 38.77 | 34.60 ± 6.5* | 61.23 | 0.274 ± 0.04* | 62.13 | 38.65 | 0.435 ± 0.08 | 17.57 | 61.35 | 33.64 ± 1.9 |
| P. pantanalensis A73 | 30.62 ± 1.8 | 24.17 | 22.82 ± 3.3 | 6.34 | 0.297 ± 0.03*** | 75.74 | 36.53 | 0.517 ± 0.12 | 39.46 | 63.47 | 30.42 ± 2.6 |
| Colletotrichum sp. P42 | 33.24 ± 2.5* | 34.79 | 28.82 ± 3.1 | 34.30 | 0.284 ± 0.02* | 68.05 | 42.97 | 0.377 ± 0.02 | 1.89 | 57.03 | 33.50 ± 2.2 |
| W. aquatica P71 | 37.48 ± 2.3*** | 51.99 | 35.08 ± 1.93* | 63.47 | 0.33 ± 0.02*** | 95.27 | 31.35 | 0.62 ± 0.03** | 67.57 | 68.65 | 36.98 ± 1.09* |
| Nemania sp. P72 | 31.08 ± 3.8 | 26.03 | 25.46 ± 2.3 | 18.64 | 0.202 ± 0.06 | 19.53 | 32.58 | 0.418 ± 0.01 | 12.97 | 67.42 | 32.28 ± 1.5 |
| Pleosporales P74 | 31.66 ± 3.0 | 28.39 | 32.30 ± 3.3 | 50.51 | 0.213 ± 0.03 | 26.04 | 29.18 | 0.517 ± 0.07 | 39.73 | 70.82 | 33.5 ± 2.2 |
| Diaporthe miriciae P96 | 28.36 ± 3.5 | 15.00 | 27.42 ± 7.6 | 27.77 | 0.234 ± 0.05 | 38.46 | 38.17 | 0.379 ± 0.02 | 2.43 | 61.83 | 37.12 ± 2.7** |
| C − Hg | 44.62 ± 3.0*** | 80.94 | 35.34 ± 4.3** | 64.68 | 0.442 ± 0.02*** | 161.54 | 58.93 | 0.308 ± 0.08 | -16.76 | 41.07 | 35.16 ± 1.4 |
| C + Hg | 24.66 ± 5.9 | 21.46 ± 8.2 | 0.169 ± 0.02 | 31.35 | 0.370 ± 0.03 | 68.65 | 31.18 ± 2.97 |
C − Hg uninoculated plants, without Hg and C + Hg uninoculated plants, with Hg; MSPA – aerial dry mass; MSR – dry root mass; GPE (%) – percentage of efficiency in growth promotion; A.M (%) – percentage of mass allocation; SPAD – chlorophyll index. Dunnett’s test, significance. codes: 0 “***” 0.001 “**” 0.01 “*” 0.05 “.” 0.1 “” 1 (p < 0.05). *Statistical difference compared to control (C + Hg). Data are expressed as means ± SD
Inoculation of maize plants with endophytes increased the plant height (15 to 51.98%), root elongation (6.34 to 63.47%), and total dry biomass (13.73 to 76.25%), in the presence and absence of mercury. Sordariomycetes A65 and W. aquatica P71 performed best to improve these parameters (Dunnett’s test, p < 0.05) (Table 3). Colonization by endophytic fungi can mitigate the impacts of metal toxicity in host plants, aid their adaptation, and improve their growth and tolerance to metals [26–28, 76].
The maize plant groups studied herein differed with respect to their chlorophyll content (Table 3). Plants treated with the endophytic strains W. aquatica P1 (36.98 ± 1.09) and D. miriciae P96 (37.12 ± 2.7) and mercury had significantly higher chlorophyll concentration, as compared with non-inoculated plants not treated with mercury (Dunnett’s test, p < 0.05). The other endophytic fungal strains did not alter the chlorophyll content when compared with the control groups. Other studies have reported that endophyte inoculation increases the chlorophyll content in maize even in the presence of toxic metals [27, 81]. The findings reported in this subsection suggested that endophytic fungi reduced metal toxicity, promoted host plant growth, and interfered with chlorophyll production in maize plants grown in metal-contaminated soils. Endophytes promote plant growth either directly by supplying nutrients or secreting plant hormones [84] or indirectly through mechanisms that control phytopathogens [85] or improve resistance to stressors such as salinity [86] and toxic metals [22, 25, 27, 31].
Quantification of mercury in maize plants
Mercury preferentially accumulated in the roots of maize plants, regardless of inoculation with endophytic fungi. However, compared with non-inoculated plants, inoculated plants accumulated less mercury content in the aerial parts, as evidenced by the translocation factor values that ranged from 0.02 to 0.06 (Table 4).
Table 4.
Mercury absorption and concentration profile in different tissues of Zea mays inoculated with endophytic fungal strains
| Concentration of Hg mg.Kg−1 | ||||
|---|---|---|---|---|
| Treatment | Shoot | Root | Total | TF S/R |
| Sordariomycetes A65 | 32.22 ± 0.15 *** | 1300 ± 10.74 *** | 1332 ± 10.76 *** | 0.02 |
| Pseudomonodictys pantanalensis A73 | 127.3 ± 1.74 *** | 1982 ± 4.23 *** | 2110 ± 4.51 *** | 0.06 |
| Colletotrichum sp. P42 | 48.45 ± 0.10 *** | 1611 ± 31.19 | 1659 ± 31.25 *** | 0.03 |
| Westerdykella aquatica P71 | 126.52 ± 5.01 *** | 1958,46 ± 23.70 *** | 2084,98 ± 28.71 *** | 0.06 |
| Nemania sp. P72 | 58.91 ± 0.28 *** | 1389 ± 0.32 *** | 1448 ± 0.31 *** | 0.04 |
| Pleosporales P74 | 94.19 ± 1.55 *** | 1950 ± 43.78 *** | 2044 ± 45.32 *** | 0.05 |
| Diaporthe miriciae P96 | 18.49 ± 0.10 *** | 1179 ± 9.62 *** | 1198 ± 9.72 *** | 0.02 |
| C + Hg | 282.9 ± 3.39 | 1562 ± 16.68 | 1845 ± 22.01 | 0.18 |
C + Hg – non-inoculated plants, with Hg; TF – Hg translocation factor; S/R – shoot/root. Results are expressed as mean ± SD. *control vs treatment; t test, p < 0.05
Plants inoculated with Sordariomycetes A65, Nemania sp. P72, and D. miriciae P96 had lower mercury concentrations in their roots and shoots (Table 4). In contrast, plants inoculated with P. pantanalensis A73, W. aquatica P71, and Pleosporales P74 had significantly higher (t test, p < 0.05) mercury concentrations in their roots and shoots, when compared with non-inoculated plants treated with mercury (C + Hg) (Table 4). Endophytic fungi can increase the bioaccumulation factor of mercury [27] and other metals such as cadmium [87], lead, and zinc [88] in plant tissues.
Endophytic fungi reduced the translocation of mercury from roots to shoots. Plants inoculated with endophytes had lower mercury translocation factor (0.02–0.06) when compared with non-inoculated plants (0.18) (Table 4). Endophytes can act as biological filters and retain toxic metals in their cell wall or cytoplasm, which consequently reduces the metal translocation [30, 34].
Although the fungal isolates favored mercury accumulation in maize plants, they mitigated the negative effects of mercury toxicity and promoted plant growth. The isolates P. pantanalensis A73 and W. aquatica P71 stood out for their ability to induce maize plants to absorb greater amounts of mercury from the soil, retain most of the metal in their root system, and mitigate the metal phytotoxicity to the plant. Therefore, we selected the two endophyte species to analyze genome characteristics that may be related to molecular mechanisms of plant growth promotion and mercury detoxification/reduction.
Assembly and annotation of the P. pantanalensis A73 and W. aquatica P71 genomes
Data from sequencing, assembly, and gene annotation of filamentous fungi have helped to elucidate the mechanisms involved in the fungus-plant interaction, as well as the mechanisms of resistance to toxic metals, including mercury [30, 75]. Genome assembly data from P. pantanalensis A73 and W. aquatica P71 are reported in Table 5. The sequencing of P. pantanalensis A73 and W. aquatica P71 generated 2.4 and 4 million base pairs, respectively (Table S4). The use of MeGAMerge postmounter provided mounting with better parameters (Table S5). The assembly of P. pantanalensis A73 and W. aquatica P71 genomes generated, respectively: 45.70 Mb (4,791 contigs) and 31.80 Mb (1,993 contigs) with estimated mean depth sequencing coverage of 26.18-fold and 65.71-fold (Table 5).
Table 5.
Genome statistics and functional annotation of sequencing data for the isolates Pseudomonodictys pantanalensis A73 and Westerdykella aquatica P71
| Parameters | P. pantanalensis A73 | W. aquatica P71 |
|---|---|---|
| Genome assembly | ||
| Genome size (draft) (bp) | 45,700,434 | 31,805,049 |
| Contigs (bp) | 4791 | 1993 |
| Largest conting (bp) | 353,312 | 312,460 |
| N50 (bp) | 73,309 | 61,894 |
| L50 (bp) | 186 | 145 |
| Indels | 8 | 69 |
| GC (%) | 51.52 | 52.92 |
| Genome annotation | ||
| Coding DNA sequences (CDS) | 17,774 | 11,240 |
| Number of rRNA | 11 | 12 |
| Number of tRNA | 111 | 99 |
| BUSCO—complete (%) | 98.9 | 97.9 |
| BUSCO—complete and single-copy (%) | 97.9 | 97.2 |
| BUSCO—complete and duplicate (%) | 1 | 0.7 |
| BUSCO—fragmented (%) | 1 | 2.1 |
| BUSCO—missing (%) | 0.1 | 0 |
| Functional annotation | ||
| Genes with GO | 3910 | 4381 |
| Genes with Interproscan domain | 11,042 | 5090 |
| Genes with eggNOG | 11,239 | 4475 |
| GenBank accession number | JAFHBV000000000 | JAFHBW000000000 |
| BioSample | SAMN17374018 | SAMN17347913 |
Currently, there is only one species of the genus Westerdykella available at the NCBI WGS database: Westerdykella ornata—NCBI ID GCA_010094085.1. W. aquatica P71 (31.80 Mb) had a larger genome size than expected, which was 17.86% larger than the W. ornata genome. However, W. ornata and W. aquatica P71 had GC contents of 53% and 52.92%, respectively, as expected. As far as we know, the genome draft of P. pantanalensis (45.70 Mb) that we reported here was the first one identified for the genus Pseudomonodictys, and its size fitted the expected size for the order Pleosporales (25–79 Mb).
The prediction of protein-coding genes performed with AUGUSTUS identified 17,774 and 11,240 genes in the P. pantanalensis A73 and W. aquatica P71 genomes, respectively. The number of predicted proteins fitted the expected range for species of the order Pleosporales, such as Curvularia geniculata P1 (8692 genes) [89], Paraphaeosphaeria sporulosa (16,682 genes) [90], Laburnicola rhizohalophila (10,891 genes) [77], Corynespora cassiicola (18,487 genes), and Corynespora olivacea (13,501 genes) [91].
The tRNA and rRNA genes predicted from P. pantanalensis A73 and W. aquatica P71 genomes (Table 5) were distributed respectively into 111 and 99 tRNA genes (Table S6 and S7), 11 and 12 rRNA genes, including 5 and 9 8S rRNA, 3 and 1 18S rRNA, and 3 and 2 28S rRNA genes. Analysis of tRNA gene distribution can provide biologically important characteristics of intracellular tRNA and codon, and amino acid usage bias. rRNA, on the other hand, serves as a binding site for tRNA and a variety of protein synthesis factors, and often binds to mRNA in the extension of peptide chains [92].
The genome integrity of P. pantanalensis A73 and W. aquatica P71, based on BUSCO, using the fungi_odb9 dataset, is reported in Table 5. W. aquatica P71 genome had 97.9% completeness with 97.2% of complete and single copies, 0.7% duplicates, 2.1% fragmented, and 0% absent. P. pantanalensis A73 genome had 98.9% completeness with 97.9% complete and single copies, 1% duplicates, 1% fragmented, and 0.1% absent. These data demonstrated the good integrity of the assembled genomes, which made them suitable for further defined analyses.
GO analysis identified 3910 and 4381 genes in P. pantanalensis A73 and W. aquatica P71, respectively. They were distributed into three functional categories: 66, 30, and 38 GO terms in biological processes, cellular processes, and molecular function, respectively (Table S8). We highlighted genes with phosphatase activity (GO:0016791), oxidoreductase activity (GO:0016491), and ion binding (GO:0043167) that are involved in detoxification processes [30], as well as genes involved in DNA metabolism (GO:0006259) that are important to protect cells from metal-induced damage [30].
The anti-SMASH platform predicted gene clusters related to secondary metabolites. The P. pantanalensis A73 genome had 32 clusters of secondary metabolite biosynthetic genes (Table S9), which included polyketide synthase type 1 (T1PKS, n = 12), non-ribosomal peptide synthetase (NRPS, n = 7), similar non-ribosomal peptide synthetase (NRPS-like n = 6), hybrid clusters (NRPS-T1PKS n = 6), and terpenes (n = 4). The W. aquatica P71 genome had 42 clusters of genes related to secondary metabolite biosynthesis (Table S10), including T1PKS (n = 15), NRPS (n = 6), NRPS-like (n = 8), NRPS-T1PKS (n = 6), and terpenes (n = 4).
The gene clusters that shared 100% similarity with biosynthesis gene clusters stood out, such as those responsible for the synthesis of dimethylcoprogen (NRPS), melanin (T1PKS), and naphthopyrone (T1PKS) detected in the P. pantanalensis A73 genome; and dimethylcoprogen (NRPS), 6-methylsalicylic acid (T1PKS), melanin (T1PKS), and clavaric acid (terpene) detected in the W. aquatica P71 genome (Table S9 and S10). Melanin [93] and the siderophore dimethylcoprogen [94] are metal chelators and have antioxidant potential.
Fungi express genes involved in mechanisms of growth and resistance promotion and detoxification of toxic contaminants [30, 95, 96]. Even if some of these genes do not directly participate in detoxification processes of environmental pollutants, the products that they encode can play important roles in the interaction between endophytic fungi and plants or other microorganisms, and improve the tolerance or resistance of these organisms to different pollutants [77].
Functional annotation with the eggNOG-Mapper and analysis of functional protein signatures with InterProScan enabled identification of 13,791 and 6,052 genes encoding proteins in P. pantanalensis A73 and W. aquatica P71 genomes, with 80.07% and 84.12% of the proteins sharing similarity of protein domains, respectively (Table S11). We identified 24 genes involved in tolerance to different metals—including genes related to mercury resistance (merA and merR1)—, 15 genes involved in oxidative stress (katG, PRX1 and TRX1), 1 gene involved in DNA repair (rfc1), and 15 genes involved in plant growth promotion (trpS and iscU) (Table S9).
The genes PRX1, SOD4, and GPX2 involved in oxidative processes were predicted in the genomes of the isolates P. pantanalensis A73 and W. aquatica P71 (Table S11). Toxic metals are able to induce the generation of reactive oxygen species as cell defense mechanisms, resulting in lipid peroxidation, membrane damage, and enzyme inactivation [97, 98]. However, microorganisms that express catabolic genes can effectively detoxify or mineralize pollutants during their degradation/reduction, through the action of antioxidant enzymes such as glutathione peroxidase, catalase, and superoxide dismutase that metabolize reactive oxygen species [30, 95, 99–101].
In addition to genes involved in oxidative stress processes, fungi express genes that encode key enzymes for detoxification of toxic compounds, such as mer, cytochrome P450, and glutathione S-transferase [30, 102, 103]. The genes CYP46A1, ECM4, URE2, merA, and merR1 (Table S11) were detected in the P. pantanalensis A73 genome, suggesting that they participate in detoxification of toxic metals, including mercury.
The mer gene cluster is responsible for the reduction and consequent volatilization of mercury [30, 104]. The merA gene encodes a key enzyme for reduction of Hg (II) to volatile Hg (0) [30], while the merR gene encodes mercuric resistance operon regulatory proteins [104, 105]. Cytochrome P450 genes constitute a large superfamily of heme-thiolate proteins with a wide variety of functions, such as primary and secondary metabolism, including contaminant degradation and cell defense [103, 106]. Genes encoding glutathione S-transferase are important for detoxification of xenobiotics and response to oxidative stress [107]. In the future, the function of these genes in P. pantanalensis A73 and W. aquatica P71 can be validated using quantitative PCR or transcriptome sequencing.
Phylogenetic analysis of Westerdykella aquatica P71
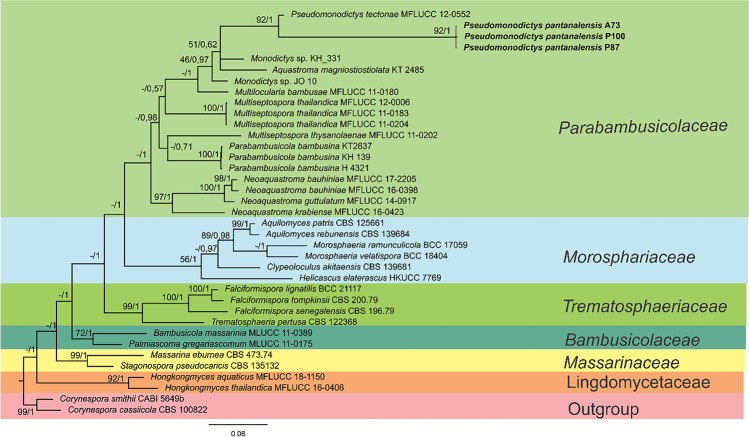
Multigene analysis of the isolate P71 identified 3626 characters, including gaps. The ITS region corresponded to 692 characters, while the LSU and tub2 regions provided 1398 and 1628 characters, respectively. The most adequate evolutionary model to explain the Bayesian Inference was General Time Reversible + I + G for all regions. The isolate P71 had high bootstrap support and posterior probability (100 and 1, respectively) and thereby grouped with the W. aquatica (JAUCC1788) type species (Fig. 1).
Fig. 1.
Phylogram generated from ML (maximum likelihood) analysis based on combined data from ITS, LSU, and tub2 sequences of type species of the genus Westerdykella. The numbers indicate branch support, ML (maximum likelihood), and PP (posterior probability) (ML/PP)
The isolate P71 was previously identified as belonging to the genus Westerdykella, using similarity of sequences from the ITS region [26]. However, the polyphasic analysis of the LSU and tub2 regions, associated with the ITS region, led us to infer that the isolate belonged to the species W. aquatica. Song et al. [108] first isolated this species that produces phytases as an endophyte of Acorus calamus grown in rice field mud in Jiangxi province, China.
Phylogenetic analysis Pseudomonodictys pantanalensis sp. nov
The alignment of genes amplified from the isolates A73, P87, and P100 generated 3227 characters, including gaps. The ITS region corresponded to 775 characters, while the SSU, LSU, and TEF regions provided 595, 873, and 984 characters, respectively. The most adequate evolutionary model to explain the Bayesian Inference was General Time Reversible + I + G for all regions. Five families structured in monophyletic clades stood out from the set analyzed. The three isolates obtained from Aeschynomene fluminensis roots formed a sister clade with the species Pseudomonodictys tectonae, with high posterior probability and bootstrap support; this finding revealed the second species for the genus that we named Pseudomonodictys pantanalensis (Fig. 2).
Fig. 2.
Phylogram generated from Bayesian analysis based on combined data from ITS, SSU, LSU, and tef sequences of type species of the Parabambusicolaceae family and related family. The numbers indicate branch support, ML (maximum likelihood), and PP (posterior probability) (ML/PP)
The isolates A73, P87, and P100 were previously identified as belonging to the Lindgomycetaceae family, using similarity of sequences from the ITS region [26]. The polyphasic analysis of LSU, SSU, ITS, and tef1 sequence data enabled us to infer that the isolates belonged to the genus Pseudomonodictys. To date, there is no ITS sequence for this genus deposited in the NCBI database, which made its initial identification difficult [26]. The morphological and microstructural characteristics confirmed the classification of the A73, P87, and P100 strains into the genus Pseudomonodictys, including hyphomycete asexual morph, semi-macronematous conidiophores that produce several conidia with granular content [109, 110].
Analysis of endophytism in vitro
The fact that P. pantanalensis A73 colonized maize roots without causing apparent damage to the plant confirmed that it was an endophyte. This species infected maize root cells by differentiating microsclerotia and septate melanized hyphae, which are characteristic of dark septate fungi (Fig. S1). Our findings corroborated the report of the presence of microsclerotia and melanized hyphae in the root cortex of A. fluminensis and maize colonized by the isolate P87, previously identified as Lindgomycetaceae [27].
Taxonomy
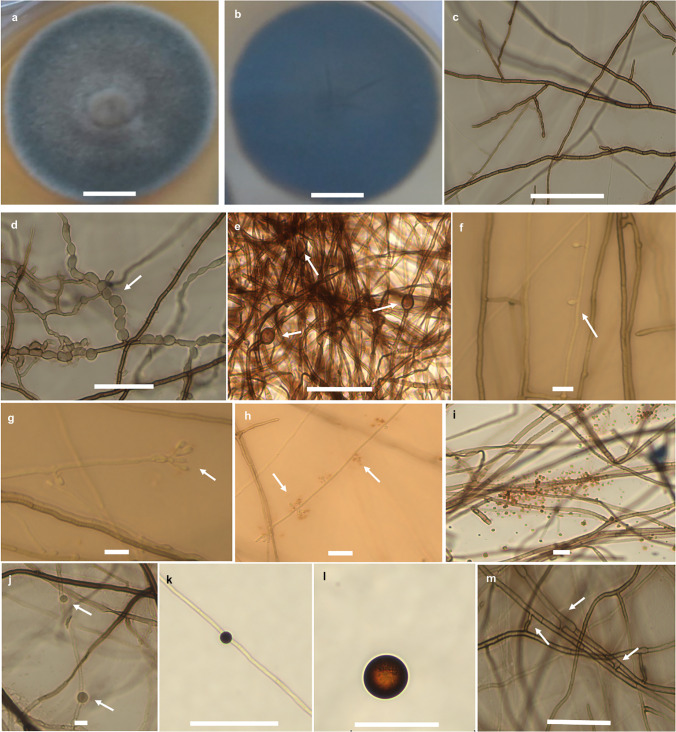
Pseudomonodictys pantanalensis Soares, Senabio, Silva, Sousa sp. nov. (Fig. 3).
Fig. 3.
Pseudomonodictys pantanalensis (holotype). a Seven-day colony (from above) in the BDA; b seven-day colony (reverse view) at the BDA; c asexual, branched, and septate hyphae with brown coloration; d chlamydoconidia in chains, intercalary; e intercalary chlamydoconidia; f conidiophores producing immature conidia; g immature non-uniform ellipsoidal aseptated subhyaline immature conidia; h conidia producing orange pigments; i orange pigment; j–k intercalated chlamydospore; l chlamydospore; m anastomosis of vegetative hyphae. Arrows indicate structures. Bars, 10 µm (f–j), 50 µm (d, e, k, l, and m), 100 µm (c), and 10 mm (a and b)
MYCOBANK: MB842113
Etymology: Name refers to the Brazilian wetland biome named Pantanal, from which the strain was isolated.
Holotype: A73 is an isolate maintained in the Culture Collection of the Laboratory of Biotechnology and Microbial Ecology at Federal University of Mato Grosso (UFMT, Cuiabá, Mato Grosso State, Brazil) and INPA—National Institute for Amazonian Research (holotype = INPA 1073), preserved in 20% glycerol at − 20 °C. Mycelium was grown in Potato Dextrose Agar medium and covered with mineral oil, while mycelium discs placed in tubes with water, stored at 4 °C, were designated as P. pantanalensis holotype. It was isolated from roots of Aeschynomene fluminensis Vell, collected in Poconé (Mato Grosso State, Brazil), in October 2014, by Pietro-Souza, W and Soares, MA. Other isolates of this type include P87 (P87 = Laboratory of Biotechnology and Microbial Ecology, UFMT, Cuiabá, Mato Grosso State, Brazil) and P100 (P100 = Laboratory of Biotechnology and Microbial Ecology, UFMT, Cuiabá, Mato Grosso State, Brazil).
Diagnosis: Pseudomonodictys pantanalensis sp. nov. cultured in Potato Dextrose Agar medium at 25 °C differs from the species Pseudomonodictys tectonae in that it does not produce conidia and red pigments in the mycelium. In Sabouraud Glucose Agar, Oat Agar, and Rice Straw media, P. pantanalensis produces chain, intercalary, and terminal chlamydoconidia. It only produces conidia in Rice Straw medium, with 6–18.5 µm length and 5–12 µm width (x̄ = 10.7 × 8.6 μm, n = 30), and mature conidia release orange pigmentation. Polyphasic analysis of the regions (SSU, LSU, and tef) identified the formation of a sister clade with the species P. tectonae, with high posterior probability and bootstrap support.
Description: Sexual morphology: Undetermined. Asexual morphology: Colonies on Rice Straw medium after 20 days in the dark at 25 °C, conidiophores up to 165 μm length and 2.4–3.7 width, semi-macronematous conidiophores, sometimes macronematous, erect to slightly curved, septate, branched, slightly contracted in septa, subhyaline, conidiophores producing immature conidia. Immature conidia, non-uniform conidia, ellipsoidal, aseptate and truncated base, subhyaline, 1.5–6.8 µm length and 1–2.5 µm width (x̄ = 3.87 × 1.8 µm, n = 20), conidia producing orange pigments. Intercalated and terminal chlamydoconidia, chained, intercalated and terminal chlamydoconidia, initially subhyaline, becoming brown to dark brown, 6–18.5 × 5–12 µm (x̄ = 10.7 × 8.6 μm, n = 30). Chlamydospores initially hyaline, turning brown to dark brown with maturity, subglobular, thick-walled, smooth, 5.2–27.3 × 5–26.6 µm (x̄ = 13.5 × 13.3 μm, n = 10). Colony diameters reached 34.5–44 mm after 7 days of growth, in the dark, at 25 °C, in Potato Dextrose Agar, Sabouraud Glucose Agar, and Oat Agar media, with the entire border, raised, circular, superficial in the center, velvety, moderately dense, olive gray in the center, white on edge, reverse black with brown mycelium irradiation (Fig. 3). Hyphae 1.5–4.5 μm wide, partially superficial, verrucous, demacean ranging from light brown to dark brown, septate and branched. Colonies detected after 30 days of growth, in the dark, at 25 °C, in Sabouraud Glucose Agar and Oat Agar, with hyphae of 1.5–4.5 μm wide, branched and irregular maceous septate hyphae, intercalated and terminal chlamydoconidia, chained, intercalated and terminal chlamydoconidia.
ITS Barcode: KX381148 (alternative markers: SSU = MZ766928; LSU = MZ766923; tef1 = MZ803210).
Other isolates examined: P87 (markers: ITS = KX381197; SSU = MZ766927; LSU = MZ766924) and P100 (markers: ITS = KX381153; LSU = MZ766925), isolated from roots of Aeschynomene fluminensis Vell, collected in Poconé, Mato Grosso State, Brazil.
Conclusion
Two out of seven mercury-resistant endophytic fungi effectively interacted with Zea mays plants to improve mercury fixation and mitigate phytotoxicity, assist phytoremediation, and promote growth of the host under mercury stress condition: Westerdykella aquatica P71 and the newly identified species Pseudomonodictys pantanalensis sp. nov. A73. Both species harbored genes involved in plant growth promotion, antioxidant processes, and metal remediation, including mercury, as identified after genome assembling and annotation. Transcriptome studies are required to elucidate the mechanisms by which (i) P. pantanalensis A73 and W. aquatica P71 promote plant growth and mercury phytoremediation, and (ii) the differential expression of genes affects mercury toxicity.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
J.A.S. designed and conducted the research and analyzed the data. J.A.S, F.C.P. and W.P.S. conducted the experiments and collected the data. M.A.S. designed the research, analyzed the data, contributed with tools and reagents, and reviewed the final version. J.A.S. wrote the manuscript. T.F.S. and G.F.S. analyzed the phylogeny.
Funding
This work was supported by grants from National Council for Scientific and Technological Development (CNPq, grant # 409062/2018–9), The Mato Grosso State Research Foundation (FAPEMAT, grant # 568258/2014), and Coordination for the Improvement of Higher Education Personnel (CAPES)—Financial Code 001.
Data availability
All data are available in the article and deposited in the NCBI database.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jaqueline Alves Senabio, Email: jaquelinesenabio@hotmail.com.
Felipe de Campos Pereira, Email: felipebsbcampos@gmail.com.
William Pietro-Souza, Email: wpietro.souza@gmail.com.
Thiago Fernandes Sousa, Email: thiago.f.sousa@ufv.br.
Gilvan Ferreira Silva, Email: gilvan.silva@embrapa.br.
Marcos Antônio Soares, Email: drmasoares@gmail.com.
References
- 1.Marrugo-Negrete J, Durango-Hernández J, Pinedo-Hernández J, Olivero-Verbel J, Díez S. Phytoremediation of mercury-contaminated soils by Jatropha curcas. Chemosphere. 2015;127:58–63. doi: 10.1016/j.chemosphere.2014.12.073. [DOI] [PubMed] [Google Scholar]
- 2.Obrist D, Kirk JL, Zhang L, Sunderland EM, Jiskra M, Selin NE. A review of global environmental mercury processes in response to human and natural perturbations: Changes of emissions, climate, and land use. Ambio. 2018;47(2):116–140. doi: 10.1007/s13280-017-1004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connor D, Hou D, Ok YS, Mulder J, Duan L, Wu Q, Rinklebe J. Mercury speciation, transformation, and transportation in soils, atmospheric flux, and implications for risk management: a critical review. Environ Int. 2019;126:747–761. doi: 10.1016/j.envint.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Li F, Zhang J, Jiang W, Liu C, Zhang Z, Zhang C, Zeng G. Spatial health risk assessment and hierarchical risk management for mercury in soils from a typical contaminated site, China. Environ Geochem Health. 2017;39(4):923–934. doi: 10.1007/s10653-016-9864-7. [DOI] [PubMed] [Google Scholar]
- 5.Dixit R, Malaviya D, Pandiyan K, Singh UB, Sahu A, Shukla R, Paul D. Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustainability. 2015;7(2):2189–2212. doi: 10.3390/su7022189. [DOI] [Google Scholar]
- 6.Ermolin MS, Fedotov PS, Malik NA, Karandashev VK. Nanoparticles of volcanic ash as a carrier for toxic elements on the global scale. Chemosphere. 2018;200:16–22. doi: 10.1016/j.chemosphere.2018.02.089. [DOI] [PubMed] [Google Scholar]
- 7.Samal AC, Chakraborty S, Mallick A, Santra SC. Mercury contamination in urban ecosystem–a case study in and around Kolkata metropolis, West Bengal. India Int J Exp Res Rev. 2017;13:38–43. [Google Scholar]
- 8.Castro MS, Hilderbrand RH, Kaumeyer M. Mercury concentrations in northern two-lined salamanders from stream ecosystems in Garrett County, Maryland. Arch Environ Contam Toxicol. 2018;75(1):17–24. doi: 10.1007/s00244-017-0496-4. [DOI] [PubMed] [Google Scholar]
- 9.Schudel G, Miserendino RA, Veiga MM, Velasquez-López PC, Lees PS, Winland-Gaetz S, Bergquist BA. An investigation of mercury sources in the Puyango-Tumbes River: using stable Hg isotopes to characterize transboundary Hg pollution. Chemosphere. 2018;202:777–787. doi: 10.1016/j.chemosphere.2018.03.081. [DOI] [PubMed] [Google Scholar]
- 10.Tang Z, Fan F, Deng S, Wang D. Mercury in rice paddy fields and how does some agricultural activities affect the translocation and transformation of mercury-a critical review. Ecotoxicol Environ Safety. 2020;202:110950. doi: 10.1016/j.ecoenv.2020.110950. [DOI] [PubMed] [Google Scholar]
- 11.Ceccatto AP, Testoni MC, Ignácio AR, Santos-Filho M, Malm O, Díez S. Mercury distribution in organs of fish species and the associated risk in traditional subsistence villagers of the Pantanal wetland. Environ Geochem Health. 2016;38(3):713–722. doi: 10.1007/s10653-015-9754-4. [DOI] [PubMed] [Google Scholar]
- 12.Pestana IA, de Rezende CE, Almeida R, de Lacerda LD, Bastos WR. Let's talk about mercury contamination in the Amazon (again): the case of the floating gold miners’ village on the Madeira River. Ext Ind Soc. 2022;11:101122. doi: 10.1016/j.exis.2022.101122. [DOI] [Google Scholar]
- 13.Wang X, He Z, Luo H, Zhang M, Zhang D, Pan X, Gadd GM. Multiple-pathway remediation of mercury contamination by a versatile selenite-reducing bacterium. Sci Total Environ. 2018;615:615–623. doi: 10.1016/j.scitotenv.2017.09.336. [DOI] [PubMed] [Google Scholar]
- 14.Frossard A, Donhauser J, Mestrot A, Gygax S, Bååth E, Frey B. Long-and short-term effects of mercury pollution on the soil microbiome. Soil Biol Biochem. 2018;120:191–199. doi: 10.1016/j.soilbio.2018.01.028. [DOI] [Google Scholar]
- 15.Feng X, Meng B, Yan H, Fu X, Yao H, Shang L (2018) Bioaccumulation of mercury in aquatic food chains. In: Biogeochemical cycle of mercury in reservoir systems in Wujiang River Basin, Southwest China. Springer, Singapore, pp 339–389. 10.1007/978-981-10-6719-8
- 16.Luo H, Wang Q, Liu Z, Wang S, Long A, Yang Y. Potential bioremediation effects of seaweed Gracilaria lemaneiformis on heavy metals in coastal sediment from a typical mariculture zone. Chemosphere. 2020;245:125636. doi: 10.1016/j.chemosphere.2019.125636. [DOI] [PubMed] [Google Scholar]
- 17.Skaldina O, Sorvari J (2019) Ecotoxicological effects of heavy metal pollution on economically important terrestrial insects. In Networking of mutagens in environmental toxicology 137–144. Springer, Cham 10.1007/978-3-319-96511-6_7
- 18.Khan F, Momtaz S, Abdollahi M. The relationship between mercury exposure and epigenetic alterations regarding human health, risk assessment and diagnostic strategies. J Trace Elem Med Biol. 2019;52:37–47. doi: 10.1016/j.jtemb.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Carocci A, Rovito N, Sinicropi MS, Genchi G (2014) Mercury toxicity and neurodegenerative effects. In: Reviews of environmental contamination and toxicology. Springer, pp 1–18. 10.1007/978-3-319-03777-6_1 [DOI] [PubMed]
- 20.Han B, Lv Z, Han X, Li S, Han B, Yang Q, Zhang Z. Harmful effects of inorganic mercury exposure on kidney cells: mitochondrial dynamics disorder and excessive oxidative stress. Biol Trace Elem Res. 2022;200(4):1591–1597. doi: 10.1007/s12011-021-02766-3. [DOI] [PubMed] [Google Scholar]
- 21.Hu XF, Lowe M, Chan HM. Mercury exposure, cardiovascular disease, and mortality: a systematic review and dose-response meta-analysis. Environ Res. 2021;193:110538. doi: 10.1016/j.envres.2020.110538. [DOI] [PubMed] [Google Scholar]
- 22.Mariano C, Mello IS, Barros BM, da Silva GF, Terezo AJ, Soares MA. Mercury alters the rhizobacterial community in Brazilian wetlands and it can be bioremediated by the plant-bacteria association. Environ Sci Pollut Res. 2020;27(12):13550–13564. doi: 10.1007/s11356-020-07913-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Wang LA, Zhan X, Huang Y, Wang J, Wang X. Response mechanism of microbial community to the environmental stress caused by the different mercury concentration in soils. Ecotoxicol Environ Safety. 2020;188:109906. doi: 10.1016/j.ecoenv.2019.109906. [DOI] [PubMed] [Google Scholar]
- 24.da Silva Maciel JH, Mello IS, Vendrusculo SJ, Senabio JA, da Silva RC, de Siqueira AB, Soares MA (2021) Endophytic and rhizospheric bacterial communities are affected differently by the host plant species and environmental contamination. Symbiosis 85:191–206. 10.1007/s13199-021-00804-1
- 25.Mello IS, Pietro-Souza W, Barros BM, da Silva GF, Campos ML, Soares MA. Endophytic bacteria mitigate mercury toxicity to host plants. Symbiosis. 2019;79(3):251–262. doi: 10.1007/s13199-019-00644-0. [DOI] [Google Scholar]
- 26.Pietro-Souza W, Mello IS, Vendruscullo SJ, da Silva GF, da Cunha CN, White JF, Soares MA. Endophytic fungal communities of Polygonum acuminatum and Aeschynomene fluminensis are influenced by soil mercury contamination. PloS one. 2017;12(7):e0182017. doi: 10.1371/journal.pone.0182017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pietro-Souza W, de Campos Pereira F, Mello IS, Stachack FFF, Terezo AJ, da Cunha CN, Soares MA. Mercury resistance and bioremediation mediated by endophytic fungi. Chemosphere. 2020;240:124874. doi: 10.1016/j.chemosphere.2019.124874. [DOI] [PubMed] [Google Scholar]
- 28.Lindblom SD, Wangeline AL, Valdez Barillas JR, Devilbiss B, Fakra SC, Pilon-Smits EA (2018) Fungal endophyte Alternaria tenuissima can affect growth and selenium accumulation in its hyperaccumulator host Astragalus bisulcatus. Front Plant Sci 9:1213. 10.3389/fpls.2018.01213 [DOI] [PMC free article] [PubMed]
- 29.Ullah R, Muhammad S (2020) Heavy metals contamination in soils and plants along with the mafic-ultramafic complex (Ophiolites), Baluchistan, Pakistan: evaluation for risk and phytoremediation potential. Environ Technol Innov 100931 10.1016/j.eti.2020.100931
- 30.Chang J, Shi Y, Si G, Yang Q, Dong J, Chen J (2020) The bioremediation potentials and mercury (II)-resistant mechanisms of a novel fungus Penicillium spp. DC-F11 isolated from contaminated soil. J Hazard Mater 122638 1016/j.jhazmat.2020.122638 [DOI] [PubMed]
- 31.Mello IS, Targanski S, Pietro-Souza W, Stachack FFF, Terezo AJ, Soares MA. Endophytic bacteria stimulate mercury phytoremediation by modulating its bioaccumulation and volatilization. Ecotoxicol Environ Safety. 2020;202:110818. doi: 10.1016/j.ecoenv.2020.110818. [DOI] [PubMed] [Google Scholar]
- 32.Norambuena J, Miller M, Boyd JM, Barkay T. Expression and regulation of the mer operon in Thermus thermophilus. Environ Microbiol. 2020;22(4):1619–1634. doi: 10.1111/1462-2920.14953. [DOI] [PubMed] [Google Scholar]
- 33.Chen SH, Cheow YL, Ng SL, Ting ASY. Mechanisms for metal removal established via electron microscopy and spectroscopy: a case study on metal tolerant fungi Penicillium simplicissimum. J Hazard Mater. 2019;362:394–402. doi: 10.1016/j.jhazmat.2018.08.077. [DOI] [PubMed] [Google Scholar]
- 34.Hoque E, Fritscher J. Multimetal bioremediation and biomining by a combination of new aquatic strains of Mucor hiemalis. Sci Rep. 2019;9(1):1–16. doi: 10.1038/s41598-019-46560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein HP, Navajas-Pérez R, Aranda E (2018) Potential for CRISPR genetic engineering to increase xenobiotic degradation capacities in model fungi. In Approaches in bioremediation 61–78. Springer, Cham 10.1007/978-3-030-02369-0_4
- 36.Babu AG, Shim J, Bang KS, Shea PJ, Oh BT. Trichoderma virens PDR-28: a heavy metal-tolerant and plant growth-promoting fungus for remediation and bioenergy crop production on mine tailing soil. J Environ Manage. 2014;132:129–134. doi: 10.1016/j.jenvman.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Arora NK, Verma M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech. 2017;7(6):381. doi: 10.1007/s13205-017-1008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giovanella P, Cabral L, Costa AP, de Oliveira Camargo FA, Gianello C, Bento FM. Metal resistance mechanisms in Gram-negative bacteria and their potential to remove Hg in the presence of other metals. Ecotoxicol Environ Saf. 2017;140:162–169. doi: 10.1016/j.ecoenv.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170(1):265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 40.Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Circular. California agricultural experiment station, 347(2nd edit)
- 41.Almoneafy AA, Kakar KU, Nawaz Z, Li B, Saand MA, Chunlan Y, Xie GL. Tomato plant growth promotion and antibacterial related-mechanisms of four rhizobacterial Bacillus strains against Ralstonia solanacearum. Symbiosis. 2014;63:59–70. doi: 10.1007/s13199-014-0288-9. [DOI] [Google Scholar]
- 42.Arnone JA, Gordon JC. Effect of nodulation, nitrogen fixation and CO2 enrichment on the physiology, growth and dry mass allocation of seedlings ofAlnus rubraBong. New Phytol. 1990;116(1):55–66. doi: 10.1111/j.1469-8137.1990.tb00510.x. [DOI] [Google Scholar]
- 43.R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
- 44.Hothorn T, Bretz F, Westfall P, Heiberger RM, Schuetzenmeister A, Scheibe S, Hothorn MT (2016) Package ‘multcomp’. Simultaneous inference in general parametric models. Project for Statistical Computing, Vienna, Austria. L https://multcomp.R-forge.R-project.org
- 45.Khan AR, Ullah I, Waqas M, Park GS, Khan AL, Hong SJ, Lee IJ. Host plant growth promotion and cadmium detoxification in Solanum nigrum, mediated by endophytic fungi. Ecotoxicol Environ Saf. 2017;136:180–188. doi: 10.1016/j.ecoenv.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 46.Hammer Ø, Harper DA, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia electronica, 4(1), 9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm
- 47.Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. In: Bioinformatics. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 29 Sept 2019
- 48.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protocol Bioinforma. 2020;70(1):e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 49.Forth LF, Höper D, Beer M, Eschbaumer M (2020) High-resolution composition analysis of an inactivated polyvalent foot-and-mouth disease vaccine. Pathogens 9(1):63. 10.3390/pathogens9010063 [DOI] [PMC free article] [PubMed]
- 50.Scholz M, Lo CC, Chain PS. Improved assemblies using a source-agnostic pipeline for MetaGenomic Assembly by Merging (MeGAMerge) of contigs. Sci Rep. 2014;4:6480. doi: 10.1038/srep06480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47(W1):W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32(suppl 2):W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 55.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, Bork P. Fast genome-wide functional annotation through orthology assignment by uclea-mapper. Mol Biol Evol. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Bork P. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44(D1):D286–D193. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, Pesseat S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lagesen K, Hallin PF, Rødland E, Stærfeldt HH, Rognes T, Ussery DW. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCarthy FM, Gresham CR, Buza TJ, Chouvarine P, Pillai LR, Kumar R, Ozkan S, Wang H, Manda P, Arick T, Bridges SM, Burgess SC (2010) "AgBase: supporting functional modeling in agricultural organisms." Nucleic Acids Res 2011 39(Database issue):D497–506 [DOI] [PMC free article] [PubMed]
- 61.Okonechnikov K, Golosova O, Fursov M, Ugene Team Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 62.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mole Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nylander JAA (2004) MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
- 64.Rambaut A (2009) FigTree v1.3.1 http://tree.bio.ed.ac.uk/software/figtree
- 65.Pimentel IC, Figura G, Auer CG. Fungos endofíticos associados a acículas de Pinus taeda. Summa phytopathologica. 2010;36:85–88. doi: 10.1590/S0100-54052010000100016. [DOI] [Google Scholar]
- 66.Kern ME, Blevins KS (1999) Medical mycology - text and atlas, 2nd edn. São Paulo
- 67.Tanaka K, Harada Y. Pleosporales in Japan (3) The genus Massarina. Mycoscience. 2003;44(3):173–185. doi: 10.1007/S10267-003-0102-7. [DOI] [Google Scholar]
- 68.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 69.Koske RE, Tessier B (1983) A convenient, permanent slide mounting medium. Mycol Soc Amer Newsl 34:59
- 70.Berthelot C, Leyval C, Foulon J, Chalot M, Blaudez D (2016) Plant growth promotion, metabolite production and metal tolerance of dark septate endophytes isolated from metal-polluted poplar phytomanagement sites. FEMS Microbiol Ecol 92(10):fiw144. 10.1093/femsec/fiw144 [DOI] [PubMed]
- 71.Contreras-Maizeejo HA, Macías-Rodríguez L, del-Val E, Larsen J. The root endophytic fungus Trichoderma atroviride induces foliar herbivory resistance in maize plants. Appl Soil Ecol. 2018;124:45–53. doi: 10.1016/j.apsoil.2017.10.004. [DOI] [Google Scholar]
- 72.Bilal L, Asaf S, Hamayun M, Gul H, Iqbal A, Ullah I, Hussain A. Plant growth promoting endophytic fungi Asprgillus fumigatus TS1 and Fusarium proliferatum BRL1 produce gibberellins and regulates plant endogenous hormones. Symbiosis. 2018;76(2):117–127. doi: 10.1007/s13199-018-0545-4. [DOI] [Google Scholar]
- 73.Numponsak T, Kumla J, Suwannarach N, Matsui K, Lumyong S. Biosynthetic pathway and optimal conditions for the production of indole-3-acetic acid by an endophytic fungus, Colletotrichum fructicola CMU-A109. PloS one. 2018;13(10):e0205070. doi: 10.1371/journal.pone.0205070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adhikari P, Pandey A. Phosphate solubilization potential of endophytic fungi isolated from Taxus wallichiana Zucc. Roots Rhizosphere. 2019;9:2–9. doi: 10.1016/j.rhisph.2018.11.002. [DOI] [Google Scholar]
- 75.Jahn L, Hofmann U, Ludwig-Müller J. Indole-3-acetic acid is synthesized by the endophyte Cyanodermella asteris via a tryptophan-dependent and-independent way and mediates the interaction with a non-host plant. Int J Mol Sci. 2021;22(5):2651. doi: 10.3390/ijms22052651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mehmood A, Hussain A, Irshad M, Hamayun M, Iqbal A, Khan N. In vitro production of IAA by endophytic fungus Aspergillus awamori and its growth promoting activities in Zea mays. Symbiosis. 2019;77(3):225–235. doi: 10.1007/s13199-018-0583-y. [DOI] [Google Scholar]
- 77.He W, Megharaj M, Wu CY, Subashchandrabose SR, Dai CC. Endophyte-assisted phytoremediation: mechanisms and current application strategies for soil mixed pollutants. Crit Rev Biotechnol. 2020;40(1):31–45. doi: 10.1080/07388551.2019.1675582. [DOI] [PubMed] [Google Scholar]
- 78.Forester NT, Lane GA, Steringa M, Lamont IL, Johnson LJ. Contrasting roles of fungal siderophores in maintaining iron homeostasis in Epichloë festucae. Fungal Genet Biol. 2018;111:60–72. doi: 10.1016/j.fgb.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Al Shaer D, Al Musaimi O, Beatriz G, Albericio F (2020) Hydroxamate siderophores: natural occurrence, chemical synthesis, iron binding affinity and use as Trojan horses against pathogens. Euro J Med Chem 112791 10.1016/j.ejmech.2020.112791 [DOI] [PubMed]
- 80.Della Mónica IF, Godoy MS, Godeas AM, Scervino JM. Fungal extracellular phosphatases: their role in P cycling under different pH and P sources availability. J Appl Microbiol. 2018;124(1):155–165. doi: 10.1111/jam.13620. [DOI] [PubMed] [Google Scholar]
- 81.Rizvi A, Khan MS. Heavy metal induced oxidative damage and root morphology alterations of maize (Zea mays L.) plants and stress mitigation by metal tolerant nitrogen fixing Azotobacter chroococcum. Ecotoxicol Environ Saf. 2018;157:9–20. doi: 10.1016/j.ecoenv.2018.03.063. [DOI] [PubMed] [Google Scholar]
- 82.Shafiq S, Adeel M, Raza H, Iqbal R, Ahmad Z, Naeem M, Azmi UR. Effects of foliar application of selenium in maize (Zea Mays L.) under cadmium toxicity. In Biol Forum-An Int J. 2019;11(2):27–37. [Google Scholar]
- 83.Gopi K, Jinal HN, Prittesh P, Kartik VP, Amaresan N. Effect of copper-resistant Stenotrophomonas maltophilia on maize (Zea mays) growth, physiological properties, and copper accumulation: potential for phytoremediation into biofortification. Int J Phytorem. 2020;22(6):662–668. doi: 10.1080/15226514.2019.1707161. [DOI] [PubMed] [Google Scholar]
- 84.Soares MA, Li HY, Bergen M, Da Silva JM, Kowalski KP, White JF. Functional role of an endophytic Bacillus amyloliquefaciens in enhancing growth and disease protection of invasive English ivy (Hedera helix L.) Plant Soil. 2016;405(1):107–123. doi: 10.1007/s11104-015-2638-7. [DOI] [Google Scholar]
- 85.Liotti RG, da Silva Figueiredo MI, Soares MA. Streptomyces griseocarneus R132 controls phytopathogens and promotes growth of pepper (Capsicum annuum) Biol Control. 2019;138:104065. doi: 10.1016/j.biocontrol.2019.104065. [DOI] [Google Scholar]
- 86.Soares MA, Li HY, Kowalski KP, Bergen M, Torres MS, White JF. Evaluation of the functional roles of fungal endophytes of Phragmites australis from high saline and low saline habitats. Biol Invasions. 2016;18(9):2689–2702. doi: 10.1007/s10530-016-1160-z. [DOI] [Google Scholar]
- 87.Shahabivand S, Parvaneh A, Aliloo AA. Different response of Alyssum montanum and Helianthus annuus to cadmium bioaccumulation mediated by the endophyte fungus Serendipita indica. Acta Ecol Sin. 2020;40(4):315–322. doi: 10.1016/j.chnaes.2019.09.002. [DOI] [Google Scholar]
- 88.Sharma VK, Li XY, Wu GL, Bai WX, Parmar S, White JF, Jr, Li HY. Endophytic community of Pb-Zn hyperaccumulator Arabis alpina and its role in host plants metal tolerance. Plant Soil. 2019;437(1):397–411. doi: 10.1007/s11104-019-03988-0. [DOI] [Google Scholar]
- 89.de Siqueira KA, Senabio JA, Pietro-Souza W, de Oliveira Mendes TA, Soares MA. Aspergillus sp A31 and Curvularia geniculata P1 mitigate mercury toxicity to Oryza sativa L. Arch Microbiol. 2021;203(9):5345–5361. doi: 10.1007/s00203-021-02481-6. [DOI] [PubMed] [Google Scholar]
- 90.Baroncelli R, Da Lio D, Vannacci G, Sarrocco S. Genome resources for the endophytic fungus Paraphaeosphaeria sporulosa. Mol Plant Microbe Interact. 2020;33(9):1098–1099. doi: 10.1094/MPMI-04-20-0097-A. [DOI] [PubMed] [Google Scholar]
- 91.Dal’Sasso TCDS, Rody HVS, Grijalba PE, de Oliveira LO (2021) Genome sequences and in silico effector mining of Corynespora cassiicola CC_29 and Corynespora olivacea CBS 114450. Arch Microbiol 20(8):5257–5265. 10.1007/s00203-021-02456-7 [DOI] [PubMed]
- 92.Chi BB, Lu YN, Yin PC, Liu HY, Chen HY, Shan Y. Sequencing and comparative genomic analysis of a highly metal-tolerant Penicillium janthinellum P1 provide insights into its metal tolerance. Front Microbiol. 2021;12:1475. doi: 10.3389/fmicb.2021.663217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tran-Ly AN, Reyes C, Schwarze FW, Ribera J. Microbial production of melanin and its various applications. World J Microbiol Biotechnol. 2020;36(11):1–9. doi: 10.1007/s11274-020-02941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chung KR, Wu PC, Chen YK, Yago JI. The siderophore repressor SreA maintains growth, hydrogen peroxide resistance, and cell wall integrity in the phytopathogenic fungus Alternaria alternata. Fungal Genet Biol. 2020;139:103384. doi: 10.1016/j.fgb.2020.103384. [DOI] [PubMed] [Google Scholar]
- 95.Phale PS, Sharma A, Gautam K (2019) Microbial degradation of xenobiotics like aromatic pollutants from the terrestrial environments. In: Pharmaceuticals and personal care products: waste management and treatment technology. Butterworth-Heinemann, pp 259–278. 10.1016/B978-0-12-816189-0.00011-1
- 96.Leontovyčová H, Trdá L, Dobrev PI, Šašek V, Gay E, Balesdent MH, Burketová L. Auxin biosynthesis in the phytopathogenic fungus Leptosphaeria maculans is associated with enhanced transcription of indole-3-pyruvate decarboxylase LmIPDC2 and tryptophan aminotransferase LmTAM1. Res Microbiol. 2020 doi: 10.1016/j.resmic.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 97.Huang C, Lai C, Xu P, Zeng G, Huang D, Zhang J, Wang R. Lead-induced oxidative stress and antioxidant response provide insight into the tolerance of Phanerochaete chrysosporium to lead exposure. Chemosphere. 2017;187:70–77. doi: 10.1016/j.chemosphere.2017.08.104. [DOI] [PubMed] [Google Scholar]
- 98.Kumar V, Dwivedi SK. Multimetal tolerant fungus Aspergillus flavus CR500 with remarkable stress response, simultaneous multiple metal/loid removal ability and bioremediation potential of wastewater. Environ Technol Innov. 2020;20:101075. doi: 10.1016/j.eti.2020.101075. [DOI] [Google Scholar]
- 99.Asemoloye MD, Ahmad R, Jonathan SG. Transcriptomic responses of catalase, peroxidase and laccase encoding genes and enzymatic activities of oil spill inhabiting rhizospheric fungal strains. Environ Pollut. 2018;235:55–64. doi: 10.1016/j.envpol.2017.12.042. [DOI] [PubMed] [Google Scholar]
- 100.Cheng YT, Zhang L, He SY. Plant-microbe interactions facing environmental challenge. Cell Host Microbe. 2019;26(2):183–192. doi: 10.1016/j.chom.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yun BR, Malik A, Kim SB. Genome based characterization of Kitasatospora sp MMS16-BH015, a multiple heavy metal resistant soil actinobacterium with high antimicrobial potential. Gene. 2020;733:144379. doi: 10.1016/j.gene.2020.144379. [DOI] [PubMed] [Google Scholar]
- 102.Ravintheran SK, Sivaprakasam S, Loke S, Lee SY, Manickam R, Yahya A, Rajandas H. Complete genome sequence of Sphingomonas paucimobilis AIMST S2, a xenobiotic-degrading bacterium. Scientific Data. 2019;6(1):1–6. doi: 10.1038/s41597-019-0289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goutam J, Sharma J, Singh R, Sharma D (2021) Fungal-mediated bioremediation of heavy metal–polluted environment. In: Microbial rejuvenation of polluted environment. Springer, pp 51–76. 10.1007/978-981-15-7455-9_3
- 104.Villar E, Cabrol L, Heimbürger-Boavida LE. Widespread microbial mercury methylation genes in the global ocean. Environ Microbiol Rep. 2020;12(3):277–287. doi: 10.1111/1758-2229.12829. [DOI] [PubMed] [Google Scholar]
- 105.Wei Q, Yan J, Chen Y, Zhang L, Wu X, Shang S, Zhang H. Cell surface display of MerR on Saccharomyces cerevisiae for biosorption of mercury. Mol Biotechnol. 2018;60(1):12–20. doi: 10.1007/s12033-017-0039-2. [DOI] [PubMed] [Google Scholar]
- 106.Chadha S, Mehetre ST, Bansal R, Kuo A, Aerts A, Grigoriev IV, Mukherjee PK. Genome-wide analysis of cytochrome P450s of Trichoderma spp: annotation and evolutionary relationships. Fungal Biol Biotechnol. 2018;5(1):12. doi: 10.1186/s40694-018-0056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen M, Zhao DK, Qiao Q, Liu L, Wang JL, Cao GH, Zhao ZW. Identification of glutathione S-transferase (GST) genes from a dark septate endophytic fungus (Exophiala pisciphila) and their expression patterns under varied metals stress. PloS one. 2015;10(4):e0123418. doi: 10.1371/journal.pone.0123418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Song HY, El Sheikha AF, Zhong PA, Liao JL, Wang ZH, Huang YJ, Hu DM. Westerdykella aquatica sp. nov., producing phytase. Mycotaxon. 2020;135(2):281–292. doi: 10.5248/135.281. [DOI] [Google Scholar]
- 109.Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KT, Dai DQ, Spirin V. Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal diversity. 2015;75(1):27–274. doi: 10.1007/s13225-015-0346-5. [DOI] [Google Scholar]
- 110.Phukhamsakda C, Bhat DJ, Hongsanan S, Xu JC, Stadler M, Hyde KD. Two novel species of Neoaquastroma (Parabambusicolaceae, Pleosporales) with their phoma-like asexual morphs. MycoKeys. 2018;34:47. doi: 10.3897/mycokeys.34.25124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the article and deposited in the NCBI database.