Significance
Bacteria persist under nutrient starvation by activating RelA/SpoT homolog (Rsh), which synthesizes a growth-regulating alarmone, ppGpp. Rsh is activated specifically upon recognizing a translation elongation complex with deacylated-tRNA at the A-site. It is however unclear how Rsh identifies such a complex inside the starving bacterial cell. We show here that conditions inducing ribosome hibernation in mycobacteria cause loss of intracellular Rsh, implying that association with translating ribosomes is necessary for intracellular stability of Rsh. Using structural analysis of Rsh-bound 70S translation initiation complex, we propose here that mycobacterial Rsh identifies a Rsh-activating ribosomal complex by constitutively surveilling the ribosome entering the translation cycle at the early elongation stage.
Keywords: RelA/SpoT, Mycobacterium, ribosome hibernation, Mpy, zinc
Abstract
The stringent response, which leads to persistence of nutrient-starved mycobacteria, is induced by activation of the RelA/SpoT homolog (Rsh) upon entry of a deacylated-tRNA in a translating ribosome. However, the mechanism by which Rsh identifies such ribosomes in vivo remains unclear. Here, we show that conditions inducing ribosome hibernation result in loss of intracellular Rsh in a Clp protease–dependent manner. This loss is also observed in nonstarved cells using mutations in Rsh that block its interaction with the ribosome, indicating that Rsh association with the ribosome is important for Rsh stability. The cryo-EM structure of the Rsh-bound 70S ribosome in a translation initiation complex reveals unknown interactions between the ACT domain of Rsh and components of the ribosomal L7/L12 stalk base, suggesting that the aminoacylation status of A-site tRNA is surveilled during the first cycle of elongation. Altogether, we propose a surveillance model of Rsh activation that originates from its constitutive interaction with the ribosomes entering the translation cycle.
The months-long treatment regimen for tuberculosis (TB) is widely attributed to the extraordinary persistence of the pathogen, Mycobacterium tuberculosis (Mtb), presumably in nutrient-deprived host environment. Molecular insights into the persistence mechanisms of nutrient-starved mycobacteria can lead to targets for a shorter TB therapy. Under nutrient starvation, Mtb and Mycobacterium smegmatis induce a highly conserved adaptation mechanism, called the stringent response, which promotes long-term survival and persistence of mycobacteria under various stresses including antibiotics (1–6). The stringent response in bacteria is a programmed adaptation to nutrient starvation facilitated by the secondary messengers, guanosine tetra- or penta-phosphates [collectively called (p)ppGpp] (7–10). (p)ppGpp reprograms the physiology of starving cells by binding to multiple cellular targets (9, 11–14).
The level of (p)ppGpp is primarily regulated by a guanosine di- or triphosphate (GDP/GTP) pyrophosphokinase and a GDP/GTP pyrophosphohydrolase, encoded either as two enzymes RelA and SpoT in β- and γ- proteobacteria (9), or as a single protein called Rel in other species (15). Further insights into the domain architectures of RelA/Rel and the mechanism of its activation have been obtained from the structures of either the free enzyme from Streptococcus dysgalactiae (subsp. equisimilis), or the enzyme from Escherichia coli or Bacillus subtilis in complex with the ribosome, messenger RNA (mRNA), and A- and P-site transfer RNAs (tRNAs) (16–20). RelA/Rel has five distinct domains (Fig. 1A): i) N-terminal domain (NTD) possessing the (p)ppGpp synthase and hydrolase (HYD) activities, ii) TGS (ThrRS, GTPase, SpoT) domain that interacts with 3′ CCA tail of deacylated-tRNA, iii) alpha helical domain (AHD), iv) zinc-binding domain (ZBD) that contacts the A-site finger (ASF) on the 23S ribosomal RNA (rRNA) and S19 at the intersubunit interface, and v) the ACT domain (aspartate kinase, chorismate, and TyrA) occupying a cavity in the large 50S ribosomal subunit (or LSU) (17–19). The interaction between the TGS domain and 3′ CCA tail of the deacylated-tRNA stabilizes the tRNA in a distorted state at the A-site, called the A/R state, while pinning the domain against the small 30S ribosomal subunit (or SSU) and extending the flexible catalytic NTD of RelA in an open and active conformation (17–19).
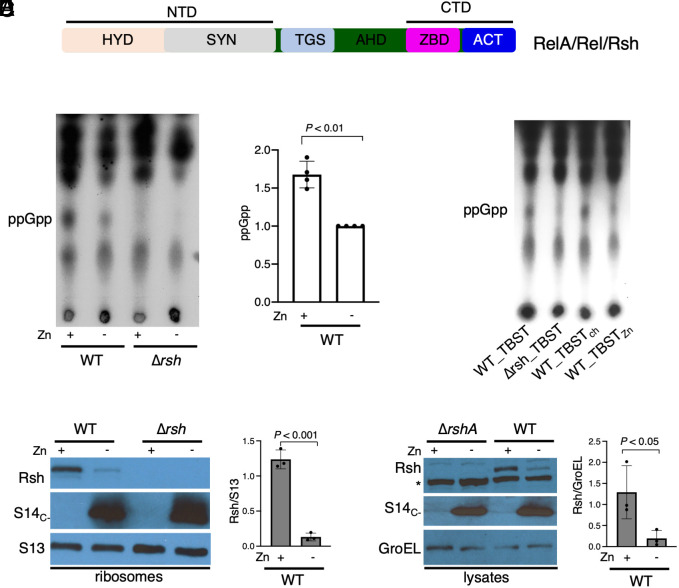
Fig. 1.
Attenuated stringent response and loss of Rsh in zinc-starved mycobacteria. (A) A conserved domain architecture in mycobacterial RelA/SpoT homolog (Rsh) compared to RelA and Rel in other species. The N-terminal domain (NTD) containing the hydrolase (HYD) and synthase (SYN) subdomains is distinguished from the C-terminal domain (CTD) by TGS (Thr-RS, GTPase, and SpoT) and alpha-helix domain (AHD). The CTD is comprised of ZBD (zinc-binding domain) and ACT (aspartokinase, chorismate mutase, and TyrA) subdomains. (B) Radio-TLC showing ppGpp levels in cultures of wild-type (WT) M. smegmatis isolated from Sauton’s medium with high zinc (1 mM ZnSO4, denoted as + Zn) and low zinc (1 µM TPEN, denoted as − Zn) after 96 h of growth. 32P-labeled sodium phosphate (100 µCi/mL) was added to the cultures 6 h prior to starvation in TBST for 3 h. After starvation, nucleotides were extracted in formic acid and amounts equivalent to 50,000 cpm were analyzed by TLC. Purified cold ppGpp was loaded as positional marker. The plot shows ppGpp levels in high-zinc WT samples relative to their low-zinc counterparts from four biologically independent experiments. (C) Radio-TLC showing ppGpp levels in WT cells cultured for 96 h in high-zinc Sauton’s medium, then labeled for 6 h, and resuspended in either TBST, or Chelex 100–treated TBST (TBSTch), or zinc-supplemented TBSTch (TBSTZn). A Δrsh mutant was used as control in panels B and C. (D) Immunoblot analysis of Rsh levels in 70S ribosomes purified from 96-h-old high- and low-zinc cultures of M. smegmatis strains described in panel B. S14c- was used as a marker for the remodeled C- ribosome, while S13 was used as the loading control for ribosomes. (E) Immunoblot analysis of Rsh in the cell lysates from the samples used for ribosome analysis in panel D. The asterisk indicates a nonspecific unknown protein cross-reactive to Rsh antibody. The plots to the right of the blots in panels D and E show the normalized average difference in Rsh between high- and low-zinc cultures from three independent experiments.
The mechanism by which RelA finds a stalled ribosome complex in a nutrient-starved cell is unclear. In the currently adopted “hopping model,” RelA samples translation elongation complexes and binds to a stalled ribosome with deacylated-tRNA in the A/R state, and after synthesizing (p)ppGpp, the enzyme “hops” to another stalled complex (21–23). The hopping model further hypothesizes that an inactive RelA in nonstarved cells acquires a ribosome-free, closed conformation, in which the catalytic activity is autoinhibited by the C-terminal domain (CTD), either through intermolecular (24–26) or intramolecular interactions (27–32). Winther et al. further proposed that free cytosolic RelA in E. coli binds to deacylated-tRNA before binding to the translating ribosome (22), although this model is challenged by the evidence of RelA bound to the ribosome without any tRNA at the A-site (18). Moreover, single-molecule tracking of RelA-fluorescent protein suggests constitutive association between RelA and ribosomes in nonstarved E. coli cells (33). Recognizing these discrepancies, Takada et al. offered a refined hopping model in B. subtilis in which activation of the stringent response appears to be initiated by a weak interaction between Rel and the ribosome before the complex is stabilized by the entry of deacylated-tRNA at the A-site, although they found RelA–ribosome interaction in E. coli to be too weak to universally validate their model (23). These studies, while revealing subtle differences in the properties of RelA homologues across bacterial species, also underscore the limitations of biochemical assays in modeling a highly dynamic in vivo interaction between the stringent factor and the translation machinery.
All annotated mycobacterial genomes encode a single dual-function enzyme, RelA/SpoT homolog (henceforth called Rsh), with both kinase and HYD activities located in distinct domains of the enzyme (25, 26, 34–36). The optimal kinase activity of mycobacterial Rsh is observed in an Rsh-activating complex comprising mRNA, deacylated-tRNA, and the ribosome (37). The ribosome in M. smegmatis and Mtb is remodeled under relatively moderate zinc starvation, and subsequently targeted for hibernation under severe zinc starvation (38, 39). Ribosome remodeling involves replacement of ribosomal proteins containing the CXXC motif (called C+ r- proteins) with their paralogs without the motif (called C- r-proteins) (38, 40). The C- ribosome is subsequently targeted for hibernation by mycobacterial protein Y (Mpy), with assistance from a zinc-sensing protein called Mrf (38, 39). A zinc-bound form of Mrf is sensitive to degradation by Clp protease, such that Mrf accumulation increases with decreasing concentration of zinc (39).
While investigating the interplay between the stringent response and ribosome hibernation in mycobacteria, we observed that stabilization of Mrf in zinc-starved M. smegmatis decreases the intracellular abundance of Rsh. We further discovered that interaction of Rsh with the ribosome is necessary for its intracellular stability in nonstarved cells, raising a hypothesis that Rsh constitutively interacts with translating ribosomes. Reconstitution of Rsh-bound 70S ribosome in a translation initiation complex and structural analysis of this complex allow us to propose a mechanism of starvation sensing by mycobacterial Rsh, in which its activation arises from its unique interaction with the ribosome entering the translation cycle.
Results
Mrf Attenuates the Stringent Response in Zinc-Starved Mycobacteria.
We tested how a zinc-limiting growth condition that induces ribosome hibernation in M. smegmatis would impact ppGpp synthesis. After 96-h of growth in high- or low-zinc Sauton’s medium, wild-type (WT) M. smegmatis cells were exposed for 3 h to macronutrient starvation in tris-buffered saline with Tween-80 (TBST) to induce the stringent response to a measurable level (2). Rsh-dependent accumulation of ppGpp upon TBST starvation of the high-zinc culture was significantly greater than that of the low-zinc culture (Fig. 1B). To distinguish the contribution of macronutrient depletion from zinc limitation in TBST, we examined the stringent response in cells from high-zinc cultures after exposure to metal-chelated or zinc-supplemented TBST. Metal chelation did not impact the ppGpp synthesis, indicating that macronutrient starvation in TBST was the primary inducer of the stringent response (Fig. 1C).
Interaction of RelA with ribosomes is critical for the stringent response (21). We therefore hypothesized that the attenuated stringent response in low-zinc cultures was likely due to zinc-responsive changes in the ribosome structure and/or function. We therefore analyzed the Rsh occupancy on the ribosome in high- and low-zinc cultures. Rsh occupancy on the ribosomes purified from low-zinc cells was significantly lower than that from high-zinc cells (Fig. 1D). Unexpectedly, the difference in Rsh abundance on the ribosome correlated tightly with the levels in the total cellular lysate (Fig. 1 D and E). Moreover, the lack of any other detectable smaller Rsh isoforms (SI Appendix, Fig. S1) suggests total loss of intracellular Rsh. Together, Fig. 1 B–E shows that physiological conditions developed during zinc starvation decrease the abundance of Rsh and thus its interaction with the ribosome, thereby diminishing the stringent response.
We next investigated the effect of two zinc-responsive physiological changes in the ribosome – remodeling and hibernation – in altered Rsh stability and Rsh–ribosome interaction. The Zur-mediated C+ to C− remodeling of the ribosome induced by zinc starvation did not impact the Rsh–ribosome interaction: Rsh occupancy on the ribosome from a high-zinc culture of wild type was comparable to Δzur and a strain constitutively synthesizing C- ribosomes (SI Appendix, Fig. S2). To determine the effect of zinc-responsive ribosome hibernation on Rsh–ribosome interaction, we analyzed the impact of Mpy recruitment factor (Mrf) on Rsh stability, activity, and its occupancy on the ribosome. Mrf binds to SSU and facilitates the inhibition of translation and growth retardation upstream to Mpy recruitment to the ribosome (39). In zinc-starved M. smegmatis, deletion of mrf restored the levels of intracellular and ribosome-bound Rsh similar to those in high-zinc WT cells, and the mutant phenotype was complemented by plasmid-borne mrf (Fig. 2 A and B). The increase in Rsh abundance in zinc-starved Δmrf also correlated with the increase in ppGpp levels upon exposure to TBST (Fig. 2C). Together, these data suggest that: i) the limiting intracellular free zinc concentration has little direct impact on the stringent response, and ii) accumulation of Mrf induced by zinc depletion and likely downregulation of translational activity decrease total Rsh abundance in the cell. Deletion of rsh did not impact Mpy–ribosome interaction (SI Appendix, Fig. S3), distinguishing mycobacteria from other species in which ppGpp transcriptionally induces factors necessary for ribosome hibernation (41–43).
Fig. 2.
Mrf accumulation causes loss of Rsh. (A and B) Analysis of Rsh in 70S ribosomes (A) and cell lysates (B) from high- (denoted as + Zn) or low-zinc (denoted as − Zn) cultures of Δmrf and complemented (Δmrfcomp) strains of M. smegmatis. High- and low-zinc cultures of WT were controls. S14c-, S13, and GroEL were probed as loading controls for ribosomes or cell lysates as indicated. The asterisk in panel B indicates a nonspecific protein cross-reactive to Rsh antibody. (C) Radio-TLC showing ppGpp levels in 96-h-old high- or low-zinc cultures of WT and low-zinc cultures of Δmrf and Δmrfcomp of M. smegmatis strains after 3 h of starvation in TBST. Purified cold ppGpp was loaded as positional marker. The plot shows relative ppGpp levels in each sample compared to the level in low-zinc culture of WT from two biologically independent experiments. (D) Ectopic Mrf expression causes loss of Rsh. Levels of Rsh in the cell lysates of a M. smegmatis strain (pYL222) expressing FLAG-tagged Mrf* under the acetamide-inducible promoter. Cells were cultured in high-zinc Sauton’s medium (1 mM ZnSO4) containing 0.2% succinate as the carbon source until OD 0.6, after which Mrf* expression was induced with 0.2% acetamide, and cells were grown for 96 h from inoculation. Uninduced cells were used as the reference. As controls, 96-h-old cells from the wild-type (WT) and Δrsh strains were cultured in either high- or low-zinc (1 µM TPEN) Sauton’s medium with succinate as the carbon source. The plot shows average normalized density of Rsh from two independent experiments. (E) Analysis of Rsh in ribosomes purified from 96-h-old induced or uninduced high-zinc cultures of pYL222 strain. (F) Simultaneous accumulation of Rsh and Mrf in YL9 cells (39) upon conditional depletion of ClpP1 protease. Cells were grown in high-zinc Sauton’s medium until OD of 0.6, after which anhydrotetracycline (ATc) was added to induce CRISPRi system to deplete ClpP1. Cell lysates were analyzed after 16 h of ClpP1 depletion. Parallel uninduced cells were used as control. YL9 cells grown for 96 h in high- or low-zinc Sauton’s medium without ATc were used as control. The plot shows average normalized density of Rsh from two independent experiments.
Ectopic Expression of Mrf Can Reduce Intracellular Abundance of Rsh.
To further establish the relationship between Rsh abundance and Mrf accumulation, we examined the impact of an ectopic expression of an Mrf variant (Mrf*) on Rsh levels in zinc-rich cells. Mrf* has reduced zinc affinity and is recalcitrant to regulation by the Clp protease system (39). Consequently, constitutive expression of Mrf* leads to its substantial accumulation in high-zinc mycobacteria (39), presumably in a conformation that is capable of binding to the ribosome and inhibiting both translation and cellular growth. The abundance of Rsh and its association with the ribosome were determined upon acetamide-inducible expression of Mrf* in high-zinc cultures of M. smegmatis. Fig. 2 D and E shows that the expression of Mrf* in high-zinc cells led to concomitant loss of Rsh, both in the lysates (Fig. 2D) and on the ribosomes (Fig. 2E).
Because SpoT in Salmonella enterica was identified as a potential substrate of the Clp protease system (44), we tested the possibility whether mycobacterial ClpP1P2 protease could be involved in regulating the abundance of free Rsh in cells. We used an M. smegmatis strain, which constitutively transcribed FLAG-tagged Mrf and harbored an anhydrotetracycline-inducible CRISPR-Ca9 system for targeting ClpP1 (39), to test whether Rsh loss during Mrf stabilization is Clp dependent. Depletion of ClpP1 in high-zinc cells resulted in concomitant accumulation of both Mrf and Rsh (Fig. 2F). Thus, loss of Rsh in cells during Mrf stabilization is likely achieved through a Clp-dependent mechanism, although whether Rsh is a direct substrate of Clp remains unknown.
Interaction with the Ribosome Is Necessary for Intracellular Stability of Rsh.
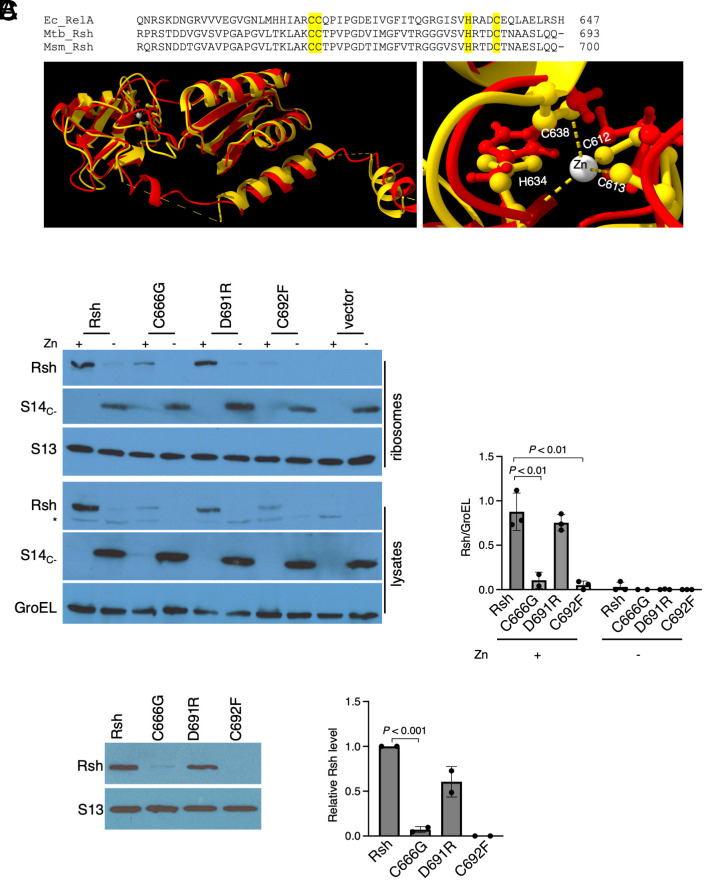
Above, we have established loss of Rsh in cells with progressively increasing amounts of Mrf prior to ribosome hibernation, suggesting that availability of translating ribosome might be important for Rsh stability. Moreover, similar levels of Rsh-bound ribosomes were observed in high-zinc cultures before and after nutrient starvation (SI Appendix, Fig. S4), suggesting that there is a constant level of Rsh–ribosome interaction regardless of the changes in the relative levels of (p)ppGpp. We therefore tested whether interaction with the ribosome impacts the intracellular stability of Rsh. E. coli RelA interacts with the ribosome primarily through its CTD containing a conserved CCHC-type zinc finger in the ZBD, which makes extensive contacts with the intersubunit region of the 70S ribosome, and ASF of the LSU (17, 45). Although the requirement of zinc in RelA–ribosome interaction remains unclear, the zinc-coordinating residues C612, C613, H634, and C638 (C666, C667, H688, and C692 in M. smegmatis Rsh) of ZBD are conserved in mycobacteria and likely acquire the same three-dimensional conformation as in E. coli RelA (Fig. 3A). Furthermore, mutation of C633 in Mtb Rsh (C692 in M. smegmatis) impairs the enzyme in vitro (25), supporting an important role of this residue in the Rsh activity. We therefore mutated the key ZBD residues and studied the impact on the intracellular abundance of Rsh. The mutation scheme closely mirrored the previous study in E. coli (24), which also showed that mutation in the neighboring D637 (D691 in M. smegmatis) had substantially less impact on protein function compared to C638 (C692 in M. smegmatis). Substitution of C666 and C692 residues resulted in substantial loss of Rsh in both the cell lysate and on the ribosome in actively growing M. smegmatis cells in zinc-rich conditions (Fig. 3B). The presence of WT Rsh on the ribosome was unambiguous (Fig. 3B), supporting the notion that Rsh–ribosome interaction in nonstarved cells is constitutive. Substitution in the adjacent D691 neither impacted the Rsh–ribosome interaction nor the intracellular abundance of the protein (Fig. 3B). Recombinant expression of these Rsh variants in E. coli did not significantly impact the protein yield (SI Appendix, Fig. S5), indicating that these mutations do not affect the intrinsic stability of the protein in vivo, and their properties are likely determined by the intracellular environment of mycobacteria. To further test whether the mutations specifically disrupted Rsh–ribosome interaction in mycobacteria, we mixed recombinant RshC666G and RshC692F variants with 70S ribosomes purified from Δrsh mutant of M. smegmatis and analyzed their occupancy on the ribosome. Ultracentrifugation on a 32% sucrose cushion was used to separate ribosome-bound from unbound Rsh. Compared to the WT Rsh, association with the ribosome was significantly compromised for both recombinant proteins, RshC666G and RshC692F (Fig. 3C). We thus conclude that Rsh constitutively interacts with the ribosome using its ZBD in nonstarved cells, and this interaction is necessary for the intracellular stability of the enzyme.
Fig. 3.
Ribosome-binding residues in Rsh are critical for its intracellular stability. (A, Top) Sequence alignment of the C-terminal region of Rsh from E. coli, M. smegmatis, and M. tuberculosis depicting the CCHC-like zinc finger (highlighted yellow) in the ZBD. (Bottom) Superimposition of the structure (PDBID: 5IQR) of the ZBD region from E. coli (yellow) and M. smegmatis (red). Zinc atom coordinated (shown with dashed lines) by the CCHC motif (shown as ball-and-stick model and numbered based on E. coli sequence) has been zoomed in the image to the Right. The corresponding residues from M. smegmatis (shown in red ball-and-stick model) predict similarity in spatial orientation with respect to their E. coli counterparts. (B) Immunoblot analysis of Rsh variants (C666G, D691R, and C692F) in cell lysates and 70S ribosomes purified from 96-h-old high- (denoted as + Zn) or low-zinc (denoted as − Zn) cultures of a Δrsh strain of M. smegmatis expressing each Rsh variant individually from an integrative plasmid (pYL238, pYL240, pYL241, and pYL242 for Rsh, RshC666G, RshD691R, and RshC692F, respectively). Cultures of Δrsh strain expressing WT Rsh were used as controls. Normalized average levels of Rsh and its variants in cell lysates from three independent experiments are shown in the plot. (C) In vitro ribosome binding of recombinant Rsh and its variants. 2.4 picomoles of 70S ribosomes purified from a 96-h-old high-zinc culture of Δrsh strain of M. smegmatis was mixed with 1.2 picomoles of Rsh or variant proteins (purified from recombinant E. coli harboring pYL243, pYL246, pYL247, and pY248 for Rsh, RshC666F, RshD691R, and RshC692F, respectively) and the mixture was incubated for 30 min at 37 °C. The complex was separated by ultracentrifugation through a 32% sucrose cushion. S13 was probed as the loading control. The plot shows quantitative analysis of Rsh from two biologically independent experiments.
Rsh Interacts with the Initiation Factor 2 (IF2)-Bound 70S Initiation Complex.
We next focused on understanding the stage of translation when Rsh would most likely interact with the ribosome in a nonstarved cell. We streamlined our approach based on the published structures (17–19), which delineate permissive and nonpermissive translation complexes for RelA binding. While RelA can bind to a late translation initiation intermediate (70S-fMet-tRNAiMet-mRNA with vacant A-site) (18), its binding to the ribosome during translation elongation is expected to be sterically hindered by intersubunit rotation (17–19). We therefore hypothesized that mycobacterial Rsh would enter the translation initiation complex through one of the free ribosomal subunits and likely remain on the complex until the start of the first cycle of elongation. This hypothesis also implies that Rsh must remain associated with the ribosome during its transition from initiation to preelongation stages, without affecting the overall canonical conformation of the translation complex.
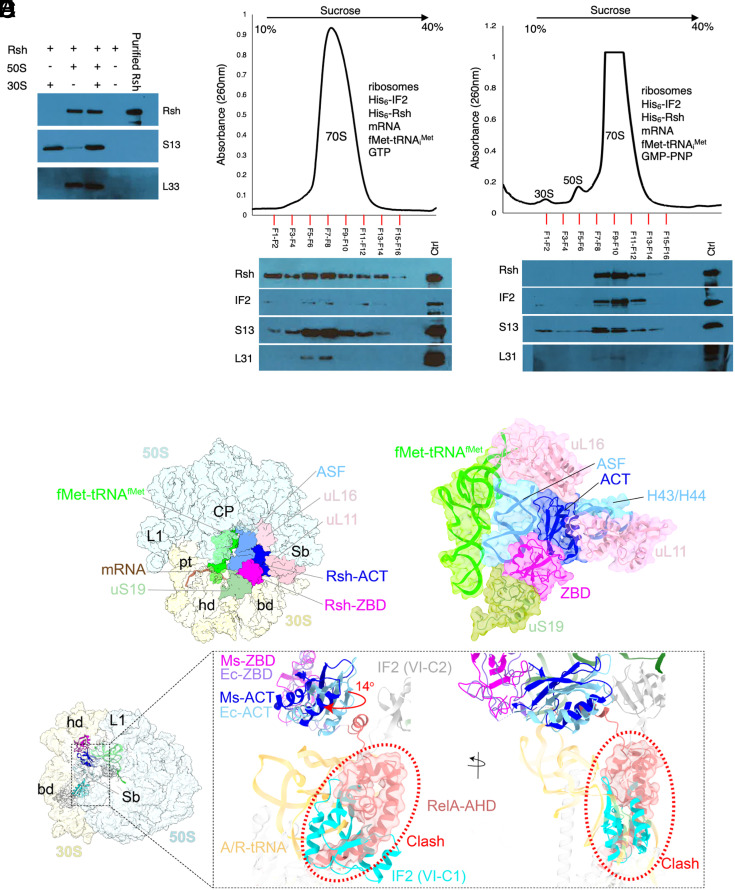
To investigate this possibility, we first determined the propensity of Rsh to interact with free ribosomal subunits. Purified recombinant Rsh was mixed individually with each of the two purified ribosomal subunits free of any canonical protein synthesis ligands, and the macromolecules were separated on a 32% sucrose cushion. Rsh appeared to preferentially cosediment with the LSU (Fig. 4A). We next asked whether Rsh would remain in the translation complex during its transitions through the various intermediate stages of initiation, especially given the potential of a steric clash between Rsh in active conformation and IF2. The contact points of IF2 on the 70S initiation complex significantly overlap with those of Rsh in active conformation (17–19, 46). We therefore tested the ability of Rsh to interact with the ribosome in the presence of IF2, fMet-tRNAiMet and mRNA in vitro. Dissociated ribosomes (SI Appendix, Fig. S6) were mixed with His6-IF2, His6-Rsh, fMet-tRNAiMet, leadered mRNA, and either GTP or a nonhydrolysable analog (GMP-PNP). Because GTP hydrolysis triggers the release of IF2 from the initiation complex (47), GMP-PNP allows us to address whether the presence of IF2 in the 70S initiation complex interferes with Rsh binding. The assembled 70S complex purified on 10 to 40% sucrose density gradient (SDG) was analyzed for the presence of IF2 and Rsh. The 70S ribosomes assembled in the presence of GTP contained Rsh but had undetectable levels of IF2 (Fig. 4B), demonstrating that Rsh can associate with the ribosome in the presence of the canonical ligands without interfering with GTP-dependent release of IF2. This property of mycobacterial Rsh contrasts with B. subtilis Rel, which does not form a stable complex with the 70S initiation complex reconstituted in the presence of GTP, unless an A-site deacylated-tRNA is included (23). Moreover, E. coli RelA has weaker association with starved ribosomal complex than B. subtilis Rel (23). In the light of these reports, our findings suggest that mycobacterial Rsh has a greater affinity for ribosomes than that of Rel/RelA of B. subtilis and E. coli. Unexpectedly, both Rsh and IF2 were detected in 70S initiation complex assembled in the presence of GMP-PNP (Fig. 4C), suggesting that Rsh and IF2 can co-occupy the same complex.
Fig. 4.
Rsh interaction with a 70S translation initiation complex. (A) Cosedimentation of Rsh with the 50S ribosomal subunit. Pellets resulting from ultracentrifugation on a 32% sucrose cushion were analyzed for S13 to confirm SSU, L33C- to confirm LSU, and Rsh to test preferential subunit binding. Rsh in the absence of ribosomes was included as control to rule out sedimentation of free protein. Purified Rsh was loaded as marker. (B and C) Reconstitution of Rsh-bound 70S initiation complex in the presence of GTP (B) or GMP-PNP (C). Dissociated ribosomes (SI Appendix, Fig. S6) were mixed with indicated ligands in the presence of GTP or GMP-PNP. After incubation for 15 min in low-magnesium (1 mM MgCl2) buffer, recombinant Rsh was added, and magnesium was adjusted to 10 mM. The resulting mixture was incubated for additional 30 min. The complex was resolved in 10 to 40% SDG, and fractions corresponding to the 70S-peak and its margin were analyzed for Rsh, IF2, and indicated ribosomal proteins by immunoblotting using anti-Rsh, anti-His6 (for IF2), anti-S13, and anti-L31 antibodies. All reagents in the reactions in panels B and C were added in fivefold molar excess relative to the ribosomal subunits. (D) 2.7 Å resolution cryo-EM structure of the 70S-Rsh complex obtained for SDG in Fig. 4C. Besides the 30S and 50S subunits, which are shown in yellow and blue, respectively, other resolved ligands are fMet-tRNAiMet present in the P-site (green); mRNA (brown); and two C-terminal domains of Rsh, ZBD (magenta), and ACT (blue). Landmarks of the 50S subunit: L1, L1 stalk; CP, central protuberance; Sb, the L7/L12 stalk base; ASF, A-site finger; Landmarks of the 30S subunit: pt, platform; hd, head; and bd, body. (E) Ribosomal components interacting with Rsh ZBD and ACT domain, along with the fMet-tRNAfMet in the P-site (green), are shown. ZBD interacts with the components of intersubunit bridge B1a, uS19 (light green), and the A-site finger (ASF, sky blue). ACT interacts with the 50S components uL16 (sky blue), L11, and 23S rRNA helices H43/44 (sky blue). (F) Superimposition of M. smegmatis 70S-Rsh, E. coli 70S-RelA (PDB ID: 5KPV), and E. coli 70S-IF2 (PDB ID: 6HTQ) structures, comparing their binding sites and conformations on the ribosome. Position of IF2 (domain VI-C1, cyan) suggests a direct steric clash (highlighted by red dotted circle) with the E. coli RelA’s α-helical domain (RelA-AHD, light coral). M. smegmatis Rsh-ACT (Ms- ACT, blue) is rotated by ~14 degrees (indicated by a red arrow) relative to the E. coli RelA-ACT (Ec-ACT, light blue), while the ZBD domains (Ms-ZBD, magenta and Ec-ZBD in light purple) have the same overall position.
Cryogenic-Electron Microscopy (cryo-EM) Structure Reveals Unknown Ribosome–Rsh Interactions.
To gain further insight into the Rsh-bound M. smegmatis 70S ribosome in a translation initiation complex, we performed single-particle cryo-EM to determine the structure of the 70S ribosome assembled in the presence of IF2, Rsh, fMet-tRNAiMet, leadered mRNA, and GMP-PNP: the exact same complex used for SDG analysis in Fig. 4C. Three-dimensional classification of the cryo-EM dataset (SI Appendix, Figs. S7 and S8 and Table S3) revealed a unique class of Rsh-bound ribosome in a classical nonrotated state, but none of the classes showed any density for IF2, which might have fallen off the ribosome complex during the flash-freezing step of the cryo-EM grid preparation. The Rsh-bound 3D class refined to 2.7 Å resolution showed densities only for two CTDs of Rsh, ZBD, and ACT, along with an fMet-tRNAifMet bound at the ribosomal P site, while all other Rsh domains were found to be disordered (Fig. 4 D and E and SI Appendix, Figs. S7 and S9). The ZBD interacts with both the SSU and LSU: specifically, protein uS19 of the SSU and 23S rRNA helix H38 of the LSU (also referred to as ASF) (Fig. 4E). These interactions resemble those reported in the structures of E. coli RelA and B. subtilis Rel on the ribosome (18, 20). The Rsh-ACT domain occupies the intersubunit space and interacts with LSU proteins uL16, uL11, and the 23S rRNA helices H43-44 at the base of the protein L7/L12 stalk (Fig. 4E).
Previous structural studies suggest that IF2 and RelA binding to the ribosome would be mutually exclusive (18, 46). However, our data show coexistence of the two proteins in a reconstituted 70S translation initiation complex (Fig. 4C). To begin to resolve this issue, we superimposed either our mycobacterial structure or the E. coli 70S ribosome–RelA structure (18) with 70S-IF2-GMP-PNP initiation complexes (46). Indeed, RelA’s AHD, which was resolved only in the active conformation of RelA in the presence of deacylated-tRNA in the A/R state (18), would directly clash with domain VI-C1 of IF2 (Fig. 4F). In tertiary structure, the AHD of E. coli RelA lies adjacent to the ACT domain, while the ZBD is anchored to the ribosome via its interactions with uS19 and ASF (18), although the relative positions of ACT and ZBD domains were similar in structures with or without the deacylated-tRNA in the A/R state (18). In our structure, the rotational angle between the ACT and ZBD domains was ~14 degrees greater than that in E. coli RelA (Fig. 4F) and AHD was disordered, presumably displaced from its canonical position, thus avoiding a clash with domain VI-C1 of IF2 (Fig. 4F). The ACT domain is known to exhibit some conformational variations with respect to the ZBD domain in the previously published structures (18, 20), but an additional rotation by 14 degrees is unprecedented. It is possible that the adopted conformation of the ACT domain in our structure is to accommodate the exiting IF2 molecule upon completion of the translation initiation phase.
Compared to the E. coli RelA and B. subtilis Rel, mycobacterial Rsh shows an overall similarity in tertiary structures of ZBD and ACT domains, although the rotation of the ACT domain translates into its shift by ~6 Å toward the LSU’s L7/L12 stalk base (Fig. 5 A and B). This altered conformation of ACT in Rsh is stabilized through extensive interactions with the uL11 and the 23S rRNA helices H43/44 (Fig. 5C). Specifically, uL11 lysines, 89 and 96, interact with Y791, D789, and H776 of Rsh-ACT. Further, Rsh-ACT S779, N783, D789, and Y788 interact with the 23S rRNA helices H43-44 phosphate backbones. These interactions are in sharp contrast to the E. coli ribosome–RelA structures, where uL11 interacts with the A/R tRNA but not directly with RelA (18). Thus, our structure reveals a unique conformational state of the ribosome-bound Rsh. Multiple sequence alignment of the amino acid sequences does not reveal any apparent substitutions, insertions, or deletions that can explain the altered orientation of the ACT domain in M. smegmatis Rsh (SI Appendix, Fig. S10). Given that our structure was obtained in the presence of IF2-GMP-PNP, a key difference from the E. coli ribosome–RelA complex, a possible role of IF2 in inducing the rotation of ACT cannot be ruled out.
Fig. 5.
The Rsh-ACT domain interacts with the L7/L12 stalk base. (A) Superimposition of the Rsh/RelA/Rel interacting components from the 70S-Rsh/RelA/Rel complex structures from M. smegmatis (Ms) 70S-Rsh (ZBD, magenta; ACT, blue), B. subtilis (Bs) 70S-Rel (ZBD, light pink; ACT, gray), and E. coli (Ec) 70S-RelA (ZBD, light purple; ACT, steel blue). Only components interacting with RelA/Rsh, L7/L12 stalk base (H43/H44 in blue), A-site finger (ASF in blue), uL16 and uL11 (light pink), the A/R tRNA (light coral), and uS19 (sea green), along with the P-site tRNA (light gray), are displayed. (B) Superimposition of the C-terminal ZBD and ACT domains from Ms, Bs, and Ec (color-coded as in panel A) highlighting the rotation in Ms-ACT domain with respect to Ms-ZBD. (C) Interactions of Ms Rsh-ACT (blue) with L11 (light pink) and the 23S rRNA helices H43/44 (light blue) are shown from two viewing directions.
Interactions stabilizing Rsh in mycobacterial structure are primarily through LSU’s ASF, uL16, uL11, and rRNA helices H43/44, suggesting that Rsh associates primarily with the LSU, corroborating the results of our binding experiments that show the association of Rsh with free LSU. The rRNA helices H43/44 and uL11 together constitute the L7/L12 stalk base of LSU and are known to be mobile during elongation (48) and recycling (49) phases of translation. Comparison of structures of the L7/L12 stalk base regions in our M. smegmatis 70S-mRNA-fMet-tRNAifMet-Rsh complex with a 2.97 Å resolution Rsh–free initiation–like M. smegmatis 70S-mRNA-fMet-tRNAifMet complex (SI Appendix, Fig. S11) shows a ~3 Å shift in H43 and ~6 Å shift in a uL11 loop (amino acids 91 to 99 segment), likely to accommodate the ACT domain of Rsh. We thus propose that intensive contacts between the L7/L12 stalk base and the Rsh-ACT domain would transiently lock the ribosome in a stalled state before the start of the translation elongation cycle, as a mechanism to surveil the aminoacylation state of the incoming tRNA to the A site. A direct contact of Rsh with uL11 is distinct from E. coli RelA, which binds to the ribosome in uL11-independent manner, even though uL11 and a deacylated-tRNA at the A-site are required for (p)ppGpp synthesis (21, 50–52). While the structural changes in M. smegmatis complex here could be induced by the presence of IF2, we cannot exclude the possibility that these are intrinsic to M. smegmatis Rsh.
The superimposed structures also reveal some species-specific differences in Rsh/RelA/Rel, which could probably explain differences in their ribosome-binding affinities (SI Appendix, Fig. S10). The ZBD domains interact with uS19 and ASF in all the three cases; however, compared to M. smegmatis, B. subtilis Rel has additional interactions—two with uS19, one with ASF, and one with uS13 (SI Appendix, Fig. S10). The loss of the additional interactions with uS19 and ASF in M. smegmatis is likely due to substitution at corresponding positions with nonpolar amino acids (SI Appendix, Fig. S10). However, M. smegmatis Rsh-ZBD could potentially interact with uS13 since it has an Arg (R681) corresponding to K617 in B. subtilis. The E. coli RelA ZBD also has two additional interactions compared to M. smegmatis, one with uS19 and the other with ASF (SI Appendix, Fig. S10). However, E. coli RelA ZBD has a Gln (Q627) corresponding to K617 and R681 in B. subtilis and M. smegmatis, respectively, which might not interact with uS13 due to its shorter side chain. Since the density corresponding to ZBD is relatively strong in our structure, we designate it as the primary anchoring domain of mycobacterial Rsh.
Exclusion of Rsh from Mpy-Bound Ribosomes.
The requirement of the ribosomes for the intracellular stability of Rsh, combined with its loss in cells harboring Mrf and Mpy-bound hibernating ribosomes, suggests that Rsh is not able to bind to hibernating ribosomes. Mpy and Rsh bind at two physically distinct sites on the unrotated SSU of the ribosome (Fig. 6A). The Mpy NTD binds to the SSU spanning the decoding center by interacting extensively with the 16S rRNA components that converge from platform, body, and head regions at the SSU’s neck (38). By contrast, the Rsh ZBD interacts near the intersubunit bridge B1a (53)–forming components, including uS19 of the SSU and the LSU’s ASF (54) (Figs. 4E and 6A). Since the binding sites of the two factors do not overlap, we superimposed the structures of Rsh- and Mpy-bound M. smegmatis 70S ribosomes to understand the accessibility of Mpy-bound hibernating ribosome to Rsh binding. Both the ribosomes are in an unrotated state, but the SSU head domain in the Rsh-bound structure is tilted more toward the LSU in comparison to the Mpy-bound ribosome structure (Fig. 6B). We predict that the untilted SSU head in the MPY-bound state will prevent stable interactions between Rsh-ZBD and uS19 (Fig. 6C). ZBD is the primary anchoring domain of Rsh (18, 20); thus, any alteration in the bridge B1a configuration in the Mpy-bound state is likely to directly inhibit Rsh binding, thereby reducing the protection of labile Rsh provided by ribosome. Thus, reduction in the intracellular Rsh during ribosome hibernation could be a combined outcome of both a decrease in the synthesis of new Rsh and the occlusion of preexisting Rsh from the hibernating ribosome.
Fig. 6.
Structural basis of Rsh exclusion from an Mpy-bound 70S ribosome. (A) Superimposition of Ms 70S-Mpy (gray, PDB ID:6DZI) and Ms 70S-Rsh (gold) structures. Rsh (ZBD, magenta; ACT, blue) and Mpy (green) are shown in the space-filled rendering. (B) The 16S rRNAs corresponding to the head domain of the 30S subunit in panel A have been enlarged to highlight their relative position in the two functional states. (C) The position of uS19 (gray) in the Mpy-bound state would disrupt the observed interactions between Rsh-ZBD (magenta) and uS19 (orange).
Discussion
A significant knowledge gap exists in our understanding of how RelA/Rel/Rsh finds a stalled translation elongation complex with deacylated A-site tRNA in a nutrient-starved bacterium. Two models have emerged from recent studies: a) the hopping model in which a Rel–deacylated-tRNA complex is formed prior to its entry at the A-site of the elongation complex (22), and b) starvation-induced formation of a weak RelA–ribosome complex prior to its stabilization by deacylated-tRNA (23). Both models however preclude RelA–ribosome interaction in nonstarved cells, which is inconsistent with the evidence of constitutive RelA–ribosome interaction throughout the growth of E. coli and P. aeruginosa cells (33, 55). Here, we propose a model of Rsh recognition of a stalled translation complex in mycobacteria. In this model, which we call the surveillance model, Rsh constitutively interacts with ribosomes during the formation of the initiation complex, and its transition to the preelongation stage (Fig. 7A, step I). Consequently, entry of an amino acylated-tRNA would allow elongation, accompanied by intersubunit rotation that would displace Rsh from the complex (Fig. 7A, step II) (17–19). By contrast, entry of a deacylated-tRNA into the A-site would trigger Rsh activation and (p)ppGpp synthesis (Fig. 7A, step IV). Free Rsh appears to be labile (Fig. 7A, step VII). To maintain steady-state levels, Rsh must therefore associate with free LSU entering the translation cycle (Fig. 7A, step III), or another stalled elongation complex with deacylated-tRNA at the A-site (Fig. 7A, step V). The latter interaction is expected to be uncertain and less frequent given the low abundance of stalled elongation complexes relative to large abundance of free LSU. Given that Rsh abundance in an actively growing cell is ~200-fold less compared to the ribosome (21), a rapid rate of translation would likely maintain a sufficiently large pool of free LSU transitioning between termination and initiation cycles, thereby ensuring a stable population of Rsh–ribosome complex. The predictable fates of Rsh in the surveillance model render the model more stringent than either the original or the refined hopping model, both of which rely on random encounter of Rsh with a deacylated-tRNA and/or a stalled ribosome.
Fig. 7.
Dynamic interaction of Rsh with the ribosome in mycobacteria, influenced by the abundance of deacylated-tRNA and hibernating ribosome in a zinc-responsive manner. (A) Rsh interaction with the ribosome through various steps of translation. In a nutrient-rich condition supporting high translation activity, labile Rsh predominantly remains associated with the free 50S subunit, which confers stability to the protein. Rsh-50S complex joins with a 30S preinitiation complex (pre-IC). Following the release of initiation factors, Rsh likely remains in the preelongation intermediate (PEI), which in an unrotated conformation is ready to elongate upon entry of the first A-site tRNA (step I). Biochemical evidence of the coexistence of IF2 and Rsh in PEI in the presence of GMP-PNP and the structure of the complex (indicated by a brown arrow pointing downward) are presented in this study. Rsh is likely sterically displaced during intersubunit rotation associated with the first cycle of elongation (step II). Free Rsh associates with the 50S subunit (step III) and the cycle continues until a deacylated-tRNA enters the A-site at the second codon (type I pause), which leads to activation of Rsh (step IV). Rsh could also potentially be activated if free Rsh interacts with a stalled ribosome downstream of the second codon (type II pause) (step V), although its frequency could be limited by relatively low abundance of such stalled complexes compared to the free 50S subunit. Conditions inducing hibernating ribosomes, which are likely less accessible to Rsh, would sequester ribosomes from the translation cycle and displace Rsh (step VI), thereby rendering Rsh labile in a Clp protease–dependent manner (step VII). (B) Depiction of the relationship between zinc, ribosome hibernation, and Rsh activation. Zinc limitation can potentially diminish the activity of zinc-dependent tRNA synthetases like threonyl-tRNA synthetase, thereby generating stalled ribosomes for Rsh activation (step IV in panel A). Acute zinc limitation leads to ribosome hibernation, which in turn reduces intracellular Rsh (and thus inhibiting its activation) in two ways: by limiting Rsh synthesis and by promoting its loss due to ribosome inaccessibility.
The lack of Rsh under conditions such as those inducing ribosome hibernation further supports the model that optimum protein synthesis and accessibility to LSU entering the translation cycle is critical for the intracellular stability of Rsh (Fig. 7A, step VI). This loss of Rsh further differentiates the physiological conditions for ribosome hibernation and the stringent response. The stringent response is an upstream response before ribosome hibernation (Fig. 7B); a logical order given that the stringent response reprograms gene expression to fine-tune metabolism, while ribosome hibernation directly shuts down global protein synthesis. Moreover, restoration of (p)ppGpp synthesis in a zinc-depleted Δmrf strain implies that Rsh–ribosome interaction and Rsh activation are relatively less sensitive to intracellular depletion of free zinc. We hypothesize that the coordinating zinc residues of Rsh have high affinity for zinc, supporting continued interaction with the ribosome even during the declining intracellular level of free zinc. This would allow activation of Rsh under low-zinc conditions before the induction of Mpy-dependent ribosome hibernation.
Zinc is an essential nutrient that binds to 6 to 10% of all proteins in bacteria (56). Notably, several amino acyl tRNA synthetases are zinc-binding proteins (56). A lack of zinc in the active site of threonyl-tRNA synthetase inactivates the aminoacylation activity of the enzyme (57). Moreover, after alanine (26%), threonine (17%) is the second-most frequent amino acid to follow the initiator Met in the top 183 most translated genes in M. smegmatis. Limitation in zinc therefore will directly increase the level of deacylated-Thr-tRNA, activating the stringent response through the surveillance model. Thus, the intracellular availability of zinc in mycobacteria could potentially be a shared signal for the sequential onset of the stringent response and ribosome hibernation, mediated directly through sensing the translation activity of the ribosome (Fig. 7B). Beyond zinc starvation as the documented condition leading to mycobacterial ribosome hibernation, it is likely that other conditions inducing ribosome hibernation would also attenuate the stringent response in a similar manner.
The Clp-dependent loss of ribosome-free intracellular Rsh, while unveiling a unique mechanism of regulation of intracellular Rsh activity, does not entirely rule out the CTD-dependent autoinhibition of the synthase activity through oligomerization: a model primarily proposed from in vitro characterization of the enzyme (24–26). However, the evidence of ribosome-bound Rsh as the dominant intracellular species suggests that Rsh oligomers, if any, would be a relatively minor molecular species in the cell. The predominance of ribosome-bound intracellular Rsh further suggests a reversible transition between the synthase and the HYD activities that may occur through differential allosteric effects of ppGpp and GDP/ATP binding to the HYD and the synthase domains, respectively (28).
In summary, we propose a unique structure-based mechanism of mycobacterial Rsh recognition of translating ribosomes stalled with deacylated A-site tRNA, in which the enzyme predominantly surveils the amino acylation status of the tRNA during the first cycle of elongation to activate the stringent response. This mechanism of Rsh activation is programmed to slow down upon induction of ribosome hibernation. These findings will serve as a molecular frame of reference for further investigation of the relationship between the stringent response and ribosome hibernation in mycobacteria.
Materials and Methods
Detailed and fully referenced methods including the lists of strains, plasmids, oligonucleotides, specimen preparation, cryo-EM data collection, image processing, as well as the statistical parameters for cryo-EM model are provided as SI Appendix. Briefly, bacteria were cultured in either standard medium or customized high- and low-zinc medium for indicated period as previously described (39). Ribosomes were purified using previously described method (39). For reconstitution of the 70S initiation complex in the presence of Rsh, recombinant His6-Rsh and His6-IF2 were purified using Ni-NTA resins (Qiagen), fMet-tRNAiMet was enzymatically prepared from tRNAiMet, whereas mRNA and nucleotides were commercially sourced. For cryo-EM analysis of the complex, samples were placed on carbon-coated copper grids, flash frozen, and data were collected on Titan Krios electron microscope at 300 keV using a K3 direct electron–detecting camera (Gatan). After motion correction of micrograph frames using MotionCor2 (58), images were processed using the pipeline of RELION (59). Cryo-EM maps were analyzed using Chimera (60), Coot (61), and PHENIX (62).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by grants to A.K.O. (NIH: AI132422, NIH: AI163599), C.L.S. (AI111696), R.K.A. (NIH:GM61576), and Y.-M.H. (NIH: GM134931). R.K.A. also acknowledges support to his lab through NIH R01 grants AI132422, GM139277, and AI155473. Support from the Wadsworth Center core facilities—Applied Genomic Technologies and the media core facility—is acknowledged. We also acknowledge Wadsworth Center’s and New York Structural Biology Center’s (NYSBC’s) three-dimensional Electron Microscopy (3D-EM) facilities. NYSBC EM facilities are supported by grants from the Simons Foundation (349247), New York State Office of Science, Technology and Academic Research (NYSTAR) (presently Empire State Development's Division of Science, Technology and Innovation), the NIH (GM103310), and the Agouron Institute (F00316). We acknowledge technical support from Ms. Kelley-Hurst and discussions with Drs. Pallavi Ghosh, Keith Derbyshire, Todd Gray, and Erica Lasek-Nesselquist.
Author contributions
Y.L., S.M., R.T., R.K.A., and A.K.O. designed research; Y.L., S.M., R.T., M.R.S., J.C., R.K.A., and A.K.O. performed research; J.C., H.B.G., S.R.M., J.P., C.L.S., Y.-M.H., and A.K.O. contributed new reagents/analytic tools; Y.L., S.M., R.T., M.R.S., N.K.B., R.K.A., and A.K.O. analyzed data; and Y.L., S.M., R.T., M.R.S., H.B.G., S.R.M., J.P., N.K.B., C.L.S., Y.-M.H., R.K.A., and A.K.O. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Rajendra K. Agrawal, Email: rajendra.agrawal@health.ny.gov.
Anil K. Ojha, Email: anil.ojha@health.ny.gov.
Data, Materials, and Software Availability
The cryo-EM map and atomic coordinates of the M. smegmatis 70S-Rsh-fMet-tRNAiMet complex are deposited in the Electron Microscopy and PDB Data Bank (wwPDB.org). Accession codes are EMD-29397 and 8FR8, respectively (63). All other relevant data of this study and methods used are presented within the manuscript and SI Appendix.
Supporting Information
References
- 1.Dahl J. L., et al. , The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 10026–10031 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stallings C. L., et al. , CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138, 146–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss L. A., Stallings C. L., Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J. Bacteriol. 195, 5629–5638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards J. P., Cai W., Zill N. A., Zhang W., Ojha A. K., Adaptation of Mycobacterium tuberculosis to biofilm growth is genetically linked to drug tolerance. Antimicrob. Agents Chemother. 63, e01213-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vilcheze C., Jacobs W. R. Jr., The isoniazid paradigm of killing, resistance, and persistence in Mycobacterium tuberculosis. J. Mol. Biol. 431, 3450–3461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutta N. K., et al. , Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci. Adv. 5, eaav2104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cashel M., Gallant J., Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841 (1969). [DOI] [PubMed] [Google Scholar]
- 8.Chatterji D., Ojha A. K., Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4, 160–165 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Potrykus K., Cashel M., (p)ppGpp: Still magical? Annu. Rev. Microbiol. 62, 35–51 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Prusa J., Zhu D. X., Stallings C. L., The stringent response and Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 76, fty054 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross W., Vrentas C. E., Sanchez-Vazquez P., Gaal T., Gourse R. L., The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalebroux Z. D., Swanson M. S., ppGpp: Magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Diez S., Ryu J., Caban K., Gonzalez R. L. Jr., Dworkin J., The alarmones (p)ppGpp directly regulate translation initiation during entry into quiescence. Proc. Natl. Acad. Sci. U.S.A. 117, 15565–15572 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irving S. E., Choudhury N. R., Corrigan R. M., The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Atkinson G. C., Tenson T., Hauryliuk V., The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6, e23479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R., Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117, 57–68 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Brown A., Fernandez I. S., Gordiyenko Y., Ramakrishnan V., Ribosome-dependent activation of stringent control. Nature 534, 277–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loveland A. B., et al. , Ribosome*RelA structures reveal the mechanism of stringent response activation. Elife 5, e17029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arenz S., et al. , The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 44, 6471–6481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pausch P., et al. , Structural basis for regulation of the opposing (p)ppGpp synthetase and hydrolase within the stringent response orchestrator rel. Cell Rep. 32, 108157 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Wendrich T. M., Blaha G., Wilson D. N., Marahiel M. A., Nierhaus K. H., Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Winther K. S., Roghanian M., Gerdes K., Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-site. Mol. Cell 70, 95–105.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Takada H., et al. , Ribosome association primes the stringent factor Rel for tRNA-dependent locking in the A-site and activation of (p)ppGpp synthesis. Nucleic Acids Res. 49, 444–457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gropp M., Strausz Y., Gross M., Glaser G., Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J. Bacteriol. 183, 570–579 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avarbock A., et al. , Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44, 9913–9923 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Jain V., Saleem-Batcha R., China A., Chatterji D., Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 15, 1449–1464 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takada H., et al. , The C-terminal RRM/ACT domain is crucial for fine-tuning the activation of "long" RelA-SpoT homolog enzymes by ribosomal complexes. Front. Microbiol. 11, 277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamman H., et al. , A nucleotide-switch mechanism mediates opposing catalytic activities of Rel enzymes. Nat. Chem. Biol. 16, 834–840 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Turnbull K. J., Dzhygyr I., Lindemose S., Hauryliuk V., Roghanian M., Intramolecular interactions dominate the autoregulation of Escherichia coli stringent factor RelA. Front. Microbiol. 10, 1966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mechold U., Murphy H., Brown L., Cashel M., Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184, 2878–2888 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronneau S., et al. , Regulation of (p)ppGpp hydrolysis by a conserved archetypal regulatory domain. Nucleic Acids Res. 47, 843–854 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gratani F. L., et al. , Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet. 14, e1007514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.English B. P., et al. , Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 108, E365–E373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avarbock D., Salem J., Li L. S., Wang Z. M., Rubin H., Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233, 261–269 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Primm T. P., et al. , The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182, 4889–4898 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ojha A. K., Mukherjee T. K., Chatterji D., High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect. Immun. 68, 4084–4091 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avarbock D., Avarbock A., Rubin H., Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry 39, 11640–11648 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Li Y., et al. , Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 8191–8196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Corro J. H., Palmer C. D., Ojha A. K., Progression from remodeling to hibernation of ribosomes in zinc-starved mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 117, 19528–19537 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prisic S., et al. , Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis. Mol. Microbiol. 97, 263–280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izutsu K., Wada A., Wada C., Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells 6, 665–676 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Hood R. D., Higgins S. A., Flamholz A., Nichols R. J., Savage D. F., The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 113, E4867–E4876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schafer H., et al. , The alarmones (p)ppGpp are part of the heat shock response of Bacillus subtilis. PLoS Genet. 16, e1008275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao X., Yeom J., Groisman E. A., The expanded specificity and physiological role of a widespread N-degron recognin. Proc. Natl. Acad. Sci. U.S.A. 116, 18629–18637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudrin P., et al. , The ribosomal A-site finger is crucial for binding and activation of the stringent factor RelA. Nucleic Acids Res. 46, 1973–1983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sprink T., et al. , Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci. Adv. 2, e1501502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall R. A., Aitken C. E., Puglisi J. D., GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol. Cell 35, 37–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datta P. P., Sharma M. R., Qi L., Frank J., Agrawal R. K., Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol. Cell 20, 723–731 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Yokoyama T., et al. , Structural insights into initial and intermediate steps of the ribosome-recycling process. EMBO J. 31, 1836–1846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friesen J. D., Fiil N. P., Parker J. M., Haseltine W. A., A new relaxed mutant of Escherichia coli with an altered 50S ribosomal subunit. Proc. Natl. Acad. Sci. U.S.A. 71, 3465–3469 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker J., Watson R. J., Friesen J. D., A relaxed mutant with an altered ribosomal protein L11. Mol. Gen. Genet. 144, 111–114 (1976). [DOI] [PubMed] [Google Scholar]
- 52.Yang X., Ishiguro E. E., Involvement of the N terminus of ribosomal protein L11 in regulation of the RelA protein of Escherichia coli. J. Bacteriol. 183, 6532–6537 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabashvili I. S., et al. , Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell 100, 537–549 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Gao H., et al. , Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell 113, 789–801 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Pletzer D., et al. , The stringent stress response controls proteases and global regulators under optimal growth conditions in pseudomonas aeruginosa. mSystems 5, e00495-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreini C., Banci L., Bertini I., Rosato A., Zinc through the three domains of life. J. Proteome Res. 5, 3173–3178 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Sankaranarayanan R., et al. , The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell 97, 371–381 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen E. F., et al. , UCSF chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta. Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Majumdar S., Sharma M. R., Manjari S. R., Banavali N. K., Agrawal R. K., Structure of Mycobacterium smegmatis Rsh bound to a 70S translation initiation complex. Protein Data Bank. https://www.rcsb.org/structure/8fr8. Deposited 6 January 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The cryo-EM map and atomic coordinates of the M. smegmatis 70S-Rsh-fMet-tRNAiMet complex are deposited in the Electron Microscopy and PDB Data Bank (wwPDB.org). Accession codes are EMD-29397 and 8FR8, respectively (63). All other relevant data of this study and methods used are presented within the manuscript and SI Appendix.