Significance
Drop-in biohydrocarbons have the potential to directly replace fossil-based products due to their chemical and physical similarity, and are fully compatible with the existing petroleum processing, distribution, and components infrastructure. Herein, we report the structure and function of a cytochrome P450, a terminal (poly-unsaturated) olefin-producing decarboxylase, which enables the use of unsaturated fatty acids as feedstocks for alkene production. The enzyme exploits a distinct molecular mechanism for substrate binding and chemoselectivity comparing to the CYP-decarboxylases characterized thus far. The broad spectrum of substrates metabolized, and less-restrictive reaction conditions represent an advance toward the development of biological routes for the production of sustainable hydrocarbons, which is an urgent need for the mitigation of climate change and reduction of the dependence on petrochemicals.
Keywords: CYP152 peroxygenase, decarboxylation activity, molecular mechanism, alkene production, renewable hydrocarbons
Abstract
The enzymatic decarboxylation of fatty acids (FAs) represents an advance toward the development of biological routes to produce drop-in hydrocarbons. The current mechanism for the P450-catalyzed decarboxylation has been largely established from the bacterial cytochrome P450 OleTJE. Herein, we describe OleTPRN, a poly-unsaturated alkene-producing decarboxylase that outrivals the functional properties of the model enzyme and exploits a distinct molecular mechanism for substrate binding and chemoselectivity. In addition to the high conversion rates into alkenes from a broad range of saturated FAs without dependence on high salt concentrations, OleTPRN can also efficiently produce alkenes from unsaturated (oleic and linoleic) acids, the most abundant FAs found in nature. OleTPRN performs carbon–carbon cleavage by a catalytic itinerary that involves hydrogen-atom transfer by the heme-ferryl intermediate Compound I and features a hydrophobic cradle at the distal region of the substrate-binding pocket, not found in OleTJE, which is proposed to play a role in the productive binding of long-chain FAs and favors the rapid release of products from the metabolism of short-chain FAs. Moreover, it is shown that the dimeric configuration of OleTPRN is involved in the stabilization of the A-A’ helical motif, a second-coordination sphere of the substrate, which contributes to the proper accommodation of the aliphatic tail in the distal and medial active-site pocket. These findings provide an alternative molecular mechanism for alkene production by P450 peroxygenases, creating new opportunities for biological production of renewable hydrocarbons.
Renewable hydrocarbons have unparalleled potential to directly replace fossil fuels and chemicals due to their compatibility (e.g., chemical and physical properties) with the existing petroleum processing, distribution, and component infrastructure (1–6). The discovery of fatty acid (FA) decarboxylation by the enzyme OleTJE (terminal alkene forming from Jeotgalicoccus sp. 8,456) provided an attractive route for the biochemical conversion of FAs into renewable hydrocarbons (7). This enzyme generates terminal alkenes from saturated FAs under high salt concentrations and has since served as a model for the biocatalytic decarboxylation of FAs (8–10). OleTJE belongs to the CYP152 family of cytochrome P450 enzymes, often referred to as peroxygenases due to their ability to activate H2O2, bypassing the prototypical P450 catalytic itinerary of O2 activation, which needs assistance from redox protein(s) and NAD(P)H to form transient ferric-superoxo (11–13) and ferric-hydroperoxo species en route to production of the high-valent intermediate known as Compound I (14–17).
Notably, most of the well-characterized CYP152 family members are known to predominantly carry out hydroxylation instead of decarboxylation, including the cytochrome P450BSβ (15) and P450SPα (16). This suggests that CYP152 enzymes that can perform decarboxylation integrate subtle and intricate molecular adaptations to enable the circumnavigation of the oxygen-rebound step prevalent in most CYPs (17). The molecular basis for the partitioning of reaction pathways in CYP152s has been investigated and the current model points to a key role of active-site pocket architecture and a distal loop region (F-G loop) for modulating the chemoselectivity of CYP152 enzymes (18). Recent experimental and theoretical (19–23) studies of OleTJE, the only CYP152 decarboxylase that has been structurally characterized to date, indicate that the F-G loop affects the mobility of the FA in the substrate-binding pocket and increases the solvation of the active site that inhibits the Compound II oxygen rebound, favoring the decarboxylation reaction at the expense of hydroxylation (18, 24).
In this work, we describe an unprecedented mechanism for FA decarboxylation in which substrate binding and chemoselectivity are not associated with the presence of an extended F-G loop. Instead, this CYP152 member, termed OleTPRN [decarboxylase from Rothia nassimurium forming terminal (poly-unsaturated) olefins], features a dimeric configuration that is involved in the stabilization of the second-coordination sphere of the substrate, and a hydrophobic cradle that is proposed to be critical for the activity on long-chain and to favor product release of short-chain FAs. These distinguishing properties also confer the ability to efficiently convert both saturated and unsaturated FAs (such as oleic and linoleic acids) into alkenes. The expanded substrate range is instrumental, considering that unsaturated FAs are abundant feedstocks for alkene production and oleic acid is a known inhibitor for OleTJE. Taken together, our findings provide a new molecular basis for FA decarboxylation that can be leveraged for the development of biotechnological applications such as in vivo alkene production.
Results
Rothia Bacteria Harbor a Distant CYP152 Peroxygenase that Binds a Broad Range of Saturated FAs.
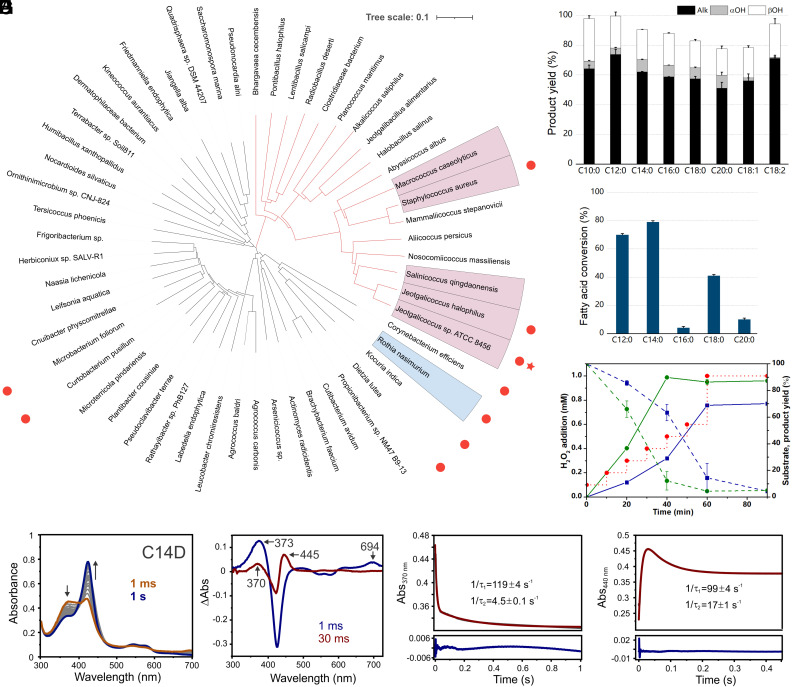
Several Rothia species have been shown to produce terminal alkenes (25). Genomic and phylogenetic analyses indicate that alkene-producing Rothia bacteria contain a CYP152-encoding gene (Uniprot: A7979_11370) that shares low sequence identity with the model decarboxylase OleTJE (9, 10) (32%) and other characterized OleTs (26) (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
(A) Phylogenetic tree of putative CYP-decarboxylases obtained from HMM analysis. Node highlighted in blue represents OleTPRN, and nodes highlighted in salmon display species with OleT already characterized. Red dots highlight bacterium genera with alkene production already reported. Red star highlights OleTJE. (B) Product yield of turnover reactions of OleTPRN with different FA substrates (experimental conditions: 2 µM enzyme, 500 µM substrate, 600 µM H2O2, 35 °C and 1 h). (C) Substrate preference assay of OleTPRN, performed with a mixture of even chain-length FAs ranging from C12 to C20. (D) Time course of myristic (blue) and oleic acid (green) simultaneous consumption (dashed line) and alkene production (straight line) through stepped H2O2 addition (red pointed line). (E) PDA spectra of the single turnover reaction between 10 µM C14D and 10 mM H2O2. Arrows indicate the direction of decreasing and increasing absorbances during the reactions. (F) Difference spectra generated by subtracting 1 s PDA trace from all other PDA traces collected during C14D single turnover. The blue spectrum (1 ms) was obtained by subtracting the 1 s PDA trace and shows the features of OleTPRN Compound I, which has a major Soret absorbance at 373 nm and a secondary absorption at 694 nm due to the π-cation radical. Wine red spectrum (30 ms) represents the split Soret absorbance at 370 and 445 nm attributed to Compound II. Two summed-exponential fits of 370 nm (G) and 440 nm time courses (H) PMT traces were collected during the single turnover reaction of C14D bound OleTPRN with 10 mM H2O2. The PMT data is shown in dark gray, with two-summed exponential fits in red, and the residuals below blue. Numbers in the figures represent the RRT constants associated with Compound I decay and Compound II formation and decay.
To shed light on the functional and mechanistic properties of this putative decarboxylase, which we refer to as OleTPRN, the recombinant enzyme was heterologously produced in Escherichia coli and purified to homogeneity (SI Appendix, Fig. S2A). The UV–visible spectrum of the OleTPRN exhibits a sharp heme Soret peak at 424 nm, and β- and α-bands at 543 and 573 nm, respectively, in the low-spin (LS) ferric resting state (SI Appendix, Fig. S2B). Moreover, the addition of carbon monoxide to the reduced OleTPRN resulted in a typical P450 heme spectrum (27), indicating the presence of a catalytically active thiolate-ligated heme-iron complex (SI Appendix, Fig. S2B).
The binding characteristics of OleTPRN were probed through monitoring the spin state conversion of the enzyme from the ferric low-spin (LS) to high-spin state (HS) upon FA binding. OleTPRN can bind a broad range of even-length saturated FAs (C10:0 to C20:0) and the LS Soret band shifts from 424 nm to 399 nm, with >70% spin state conversion for all substrates tested (SI Appendix, Table S1). Based on these results, it was possible to estimate the binding constants (KD) and determine that the highest affinity was for myristic acid (C14:0). However, the binding affinities for other tested FAs were very similar (SI Appendix, Fig. S3 and Table S2), indicating efficient binding of OleTPRN across a broad panel of FAs. This property is notably different from OleTJE that exhibits a strong correlation between KD values and substrate chain-length, resulting in the highest affinity towards long-chain (e.g., C20) substrates (9, 10).
OleTPRN operates across relatively broad pH and temperature ranges of 6.5 to 8.0 and 20 to 45 °C, respectively. Thermal stability assays demonstrated that OleTPRN retains more than 95% of its activity after 1-h incubation at 45 °C, in contrast to the loss of activity of OleTJE for temperatures approaching 40 °C (SI Appendix, Fig. S4). Moreover, thermal denaturation analyses showed that OleTPRN is significantly stabilized by the substrate (SI Appendix, Fig. S5 and Table S3), indicating that enzymatic processes could be compatible with even higher temperatures. OleTPRN exhibited tolerance to H2O2 at concentrations up to 10 mM, with steady stimulation of the activity (SI Appendix, Fig. S6A). Unlike OleTJE (9, 10) and other characterized OleTs (26, 28), OleTPRN does not require high salt concentrations for its activity (SI Appendix, Fig. S6 B and C). These combined features require less restrictive reaction conditions for downstream applications and make OleTPRN a more suitable enzyme for metabolic engineering for in vivo microbial alkene production, taking into consideration the low intracellular ion concentration in heterologous host organisms (29, 30).
OleTPRN Efficiently Converts Both Saturated and Unsaturated FAs into Alkenes.
To evaluate the substrate specificity and chemoselectivity for FA conversions, reactions were performed with saturated (C10:0 to C20:0) and unsaturated (C18:1 and C18:2) FAs. For the saturated ones, the overall activity increased with short- and medium-chain FAs, and C12:0 was the most readily converted, followed by C10:0 and C14:0 (Fig. 1B). Alkenes were the major reaction products with a yield of around 66 to 74% for all tested substrates, followed by minor β- and α-hydroxylated products (Fig. 1B and SI Appendix, Tables S4 and S5). The high Cβ regioselectivity of OleTPRN is a feature that likely favors high levels of decarboxylase activity (31).
The substrate promiscuity of OleTPRN was further investigated by competition assays using a mixture of even chain-length FAs ranging from C12 to C20 (Fig. 1C). Approximately 70% and 80% of C12 and C14 were converted, respectively. On the other hand, only <5% of C16, C18, and C20 FAs were converted by the enzyme in the presence of short-chain FAs, despite their high conversion rates (60%) when reacted with OleTPRN individually. These results reinforce the fact that OleTPRN has a significant preference for mid-chain FAs (C12 and C14) rather than long ones.
The oleic acid (C18:1) is one of the most abundant FAs available in nature, and its occurrence in most natural oils claims attention towards its use as a valuable substrate for alkene production (32). However, C18:1 conversion into alkene has not been demonstrated enzymatically in vitro by native OleT enzymes (7, 33–35). Moreover, this unsaturated substrate has been documented to have an inhibitory effect on the model enzyme OleTJE (36), which was corroborated by the drastic reduction in 1-tridecene production in a turnover assay conducted with both myristic and oleic acids simultaneously (SI Appendix, Fig. S7). Turnover assays for OleTPRN revealed a conversion rate of 79% and alkene yield of 58% for oleic acid (Fig. 1B). Furthermore, kinetic experiments performed with oleic and myristic acids, simultaneously, showed that oleic acid does not inhibit the conversion of other FAs by OleTPRN, unlike OleTJE (Fig. 1D). Instead, the mixture of saturated and unsaturated substrates favored a higher alkene yield by OleTPRN, compared to individually performed reactions. Interestingly, OleTPRN is not only able to convert oleic acid, but also linoleic acid (C18:2; Fig. 1B), which is another abundant FA in vegetable oils (37, 38) and microorganisms (39). Wang and co-authors engineered the CYP152 BSβ for higher turnover numbers for myristic acid, but the alkene production for longer chains (e.g, C16 and C18) was extremely inefficient (7 to 13%), and it also displayed low alkene yields for unsaturated substrates (40). In addition, the same P450BSβ-V74I/Q85H/F166M/G290V mutant was dependent on lower temperatures (between 0 and 10 °C), conditions unlikely feasible in an industrial environment. Otherwise, its chemoselectivity is compromised (40). Therefore, our findings create new opportunities for the use of the most abundant FAs in nature to produce renewable hydrocarbons by using a natively occurring enzyme that is fully functional under industrially relevant conditions.
OleTPRN-Catalyzed Decarboxylation Involves the Intermediates Compounds I and II.
The decarboxylation of FAs by CYP152 peroxygenases begins with hydrogen atom transfer from the substrate Cβ by the Fe(IV)-oxo π-cation species Compound I, the first detectable intermediate in the reaction of the substrate bound to a CYP152 decarboxylase with H2O2 (31, 41–43). A second meta-stable Fe(IV)-OH species, known as Compound II (17, 18, 41, 44, 45), is then formed as a consequence of the hydrogen atom transfer.
Single turnover experiments carried out with a panel of protiated substrates did not result in the appreciable accumulation of Compound I (SI Appendix, Fig. S8), most likely due to facile C-H bond. However, by following the formation and decay of Compound II at 440 nm, it was possible to determine the rate constants for the decay of Compounds I and II (k ~ 400 s−1 and 12 s−1, respectively) (Table 1 and SI Appendix, Fig. S8C). These values are in close agreement with corresponding rate constants described for OleTJE (41). In an attempt to more assuredly monitor the formation and decay of Compound I in OleTPRN, the single turnover experiments were performed with perdeuterated FAs (41, 42). The analysis of the photodiode array spectra for C14D single turnover reactions revealed the accumulation of Compound I (Fig. 1E), as evidenced by a transient intermediate with a red-shifted Soret maximum at 373 nm and porphyrin cation radical absorbance at 694 nm that resembles the previously reported for P450 Compound I as well as analogous intermediates in synthetic thiolate-ligated porphyrins (41, 46–48) (Fig. 1F). A second intermediate is formed upon Compound I decay with a Soret maximum ~445 nm, akin to that observed for Compound II in OleTJE (41, 44, 45). Similar rate constants for the decay of Compounds I and II were observed for all chain-length FAs (Fig. 1 G and H and SI Appendix, Table S6). Notably, the value for the substrate 2H kinetic isotope effect (KIE) for OleTPRN was ~4, which is similar to that reported for the steady-state hydroxylation of myristic acid by P450SPα (49) and transient kinetics studies for eicosanoic acid decarboxylation by OleTJE (41). The KIE value suggests that, like OleTJE, alkene formation by OleTPRN is initiated by substrate C-H bond abstraction, a catalytic itinerary that is ubiquitous to CYP152 decarboxylases.
Table 1.
Compound I and Compound II decay rate constants for OleTPRN WT and mutants
| Variant | Fatty acid | Cpd-I decay (s−1) | Cpd-II decay (s−1) |
|---|---|---|---|
| WT | C14H | 389 ± 66 | 12 ± 0.5 |
| C14D | 99 ± 4 | 17 ± 1 | |
| S26F | C14H | ≥1,000 | 32.3 ± 0.8 |
| C14D | 153 ± 6 | 17 ± 3 | |
| E405F | C14H | ≥1,000 | 31 ± 5 |
| C14D | 150 ± 4 | 17 ± 1 |
A Heme Proximal Histidine Plays a Key Role in Regulating the Chemoselectivity of OleTPRN.
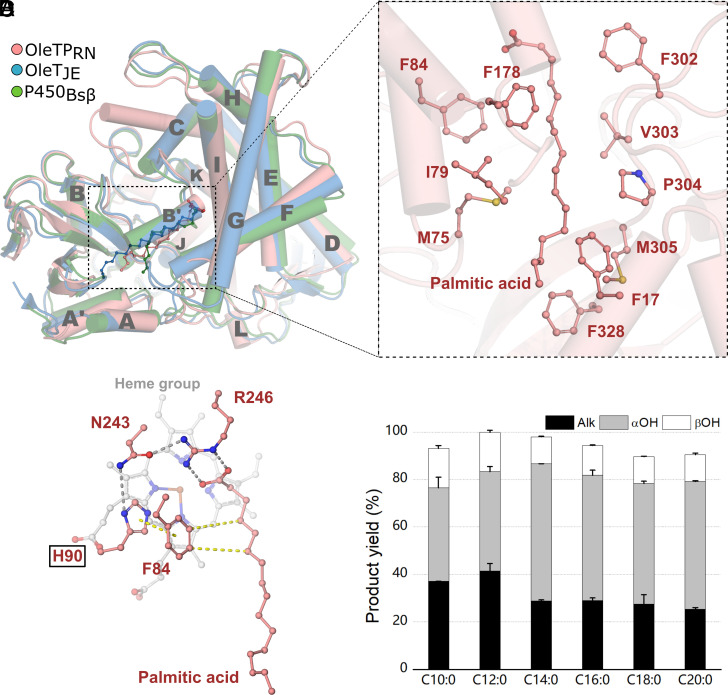
In order to explain the molecular basis for the decarboxylation activity of OleTPRN, the crystal structure of OleTPRN was determined in complex with palmitic acid (C16:0). The OleTPRN structure displays a typical P450 single-domain scaffold with a prosthetic heme group engraved within a long active site (Fig. 2 A and B and SI Appendix, Fig. S9). Regardless of the chemoselectivity, OleTPRN, OleTJE, and P450BSβ possess a wide positively charged surface around the catalytic tunnel that is thought to be required for the recognition of FAs (SI Appendix, Fig. S10). In the OleTPRN-C16:0 complex, the carboxylate moiety of palmitic acid forms a salt bridge with the guanidinium group of Arg246 (Arg245 in OleTJE, Arg242 in P450BSβ) with Nɛ and Nη2 atoms at 2.9 Å from O1 and O2 atoms, respectively (Fig. 2C). As observed in OleTJE and P450BSβ, Phe302 is strategically positioned to induce a kink in the acyl chain at the β-carbon, which favors salt bridge formation between the FA carboxylate and the Arg246 guanidinium group (Fig. 2B).
Fig. 2.
(A) Cartoon representation of superimposed OleTPRN, OleTJE, and P450Bsβ structure, showing substrates and helices names. (B) View of important amino acids for substrate positioning in OleTPRN pocket. (C) Catalytic site showing amino acid interactions, hydrogen bounds are displayed in gray and van der Waals forces in yellow. (D) Product yield of turnover reactions with OleTPRN H90Q (Experimental conditions: 2 µM of enzyme, 500 µM of substrate, 600 µM H2O2, 35 °C and 1 h of reaction).
The active site of hydroxylases and decarboxylases is generally well conserved including the catalytically relevant residues Phe79 and Arg242 in the hydroxylase P450BSβ that correspond to Phe79 and Arg245 in OleTJE and Phe84 and Arg246 in OleTPRN. Phe84 is critical not only for the activity of OleTPRN, as has been observed for OleTJE (10), but also for products distribution since its replacement by an alanine (F84A) shifted the enzyme towards hydroxylation for all saturated substrates (SI Appendix, Fig. S11). Other subtle differences are noted such as the Gln85 residue of P450BSβ is replaced by a histidine in the CYP152 decarboxylases (10) (His85 for OleTJE and His90 for OleTPRN). This led to the initial hypothesis that this adaptation would be instrumental for governing chemoselectivity among CYP152 peroxygenases. However, neither the Q85H mutation in P450BSβ nor the H85Q in OleTJE significantly alter the chemoselectivity of these enzymes, indicating that other molecular determinants are involved in functional differentiation (10). Intriguingly, the OleTPRN H90Q mutation drastically altered the chemoselectivity and compromised β-regioselectivity (Fig. 2D). Although the conversion rates remained unaltered, the alkene production for OleTPRN H90Q was substantially decreased at the expense of the formation of significant amounts of α-hydroxylated products for all tested FAs (Fig. 2D). Furthermore, the OleTPRN H90Q mutant did not present typical HS and spectral transitions (SI Appendix, Table S1), and more closely resembled the spectral profile (e.g., low spin-state conversion) observed for peroxygenases that contain a glutamine at this position, such as P450BSβ (15). These results reinforce that OleTPRN decarboxylation involves distinct molecular determinants compared to OleTJE and other characterized CYP152 members.
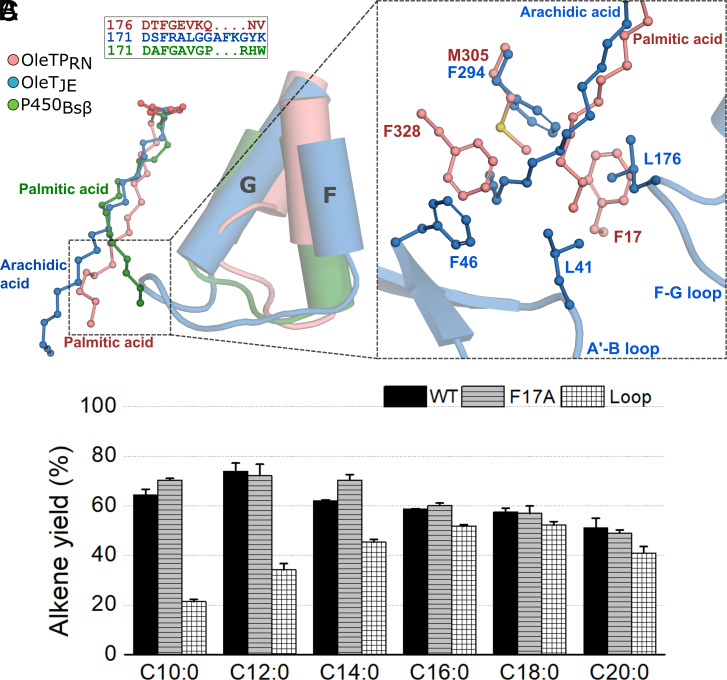
OleTPRN Lacks the F-G Loop Considered Essential for Decarboxylation Reaction.
The unaltered chemoselectivity of OleTJE upon H85Q mutation led to the exploration of other non-conserved regions, which culminated in the discovery of the F-G loop as another structural motif that is critical for the retention of decarboxylase activity (18). It was demonstrated that the extended F-G loop in OleTJE consists of a structurally disordered region responsible for the entry, anchoring and reduced mobility of FAs in the active-site pocket and also to participate in the product release (18). In particular, the Leu176 residue in OleTJE creates a hydrophobic microenvironment at the entrance of the distal pocket of the substrate channel, establishing a hydrophobic interaction with the FA tail, which is considered to be a key determinant for chemoselectivity (18). Unexpectedly, the F-G loop is significantly shortened in OleTPRN (by four residues) and more closely resembles the hydroxylase P450BSβ (18) (Fig. 3A), which lacks the corresponding Leu176. However, the shorter F-G loop seems to be compensated by a Phe residue (Phe17) protruding from the N-terminal α-helix that occupies an equivalent region in OleTPRN, suggesting a similar role (Fig. 3B). Thus, Phe17 was mutated to alanine (F17A) to test this hypothesis, but surprisingly, it did not significantly affect the decarboxylation activity (Fig. 3C).
Fig. 3.
(A) Representation of longer F-G loop present in OleTJE in contrast to the shorter loop of OleTPRN and P450Bsβ. (B) Phe17 present in OleTPRN in substitution of the longer F-G loop and A’-B loop interactions of OleTJE. (C) Comparison of alkene yield from turnover reactions with OleTPRN WT and mutants (Experimental conditions: 2 µM of enzyme, 500 µM of substrate, 600 µM H2O2, 35 °C and 1 h of reaction).
We also redesigned the F-G loop of OleTPRN to resemble OleTJE by insertion of an amino acid segment consisting of the sequence FRALGGAFK, which did not result in any positive effect on substrate conversion. On the contrary, the introduction of the extended F-G loop was detrimental for the conversion of short-chain FAs, C10:0 and C12:0, with substantial effects observed for both substrate conversion, and more notably, a threefold lower yield of alkene compared to the wild-type enzyme (Fig. 3C and SI Appendix, Fig. S12). Interestingly, the loop exchange in OleTPRN was also detrimental to the conversion rate of the unsaturated oleic acid into alkadiene (SI Appendix, Fig. S13), suggesting that the extended F-G loop in OleTJE could be one of the factors limiting its activity in this substrate. These findings again reinforce that the OleTPRN decarboxylation reaction is governed by structural elements that differ from the model enzyme OleTJE.
A Hydrophobic Cradle Is Critical for OleTPRN Activity.
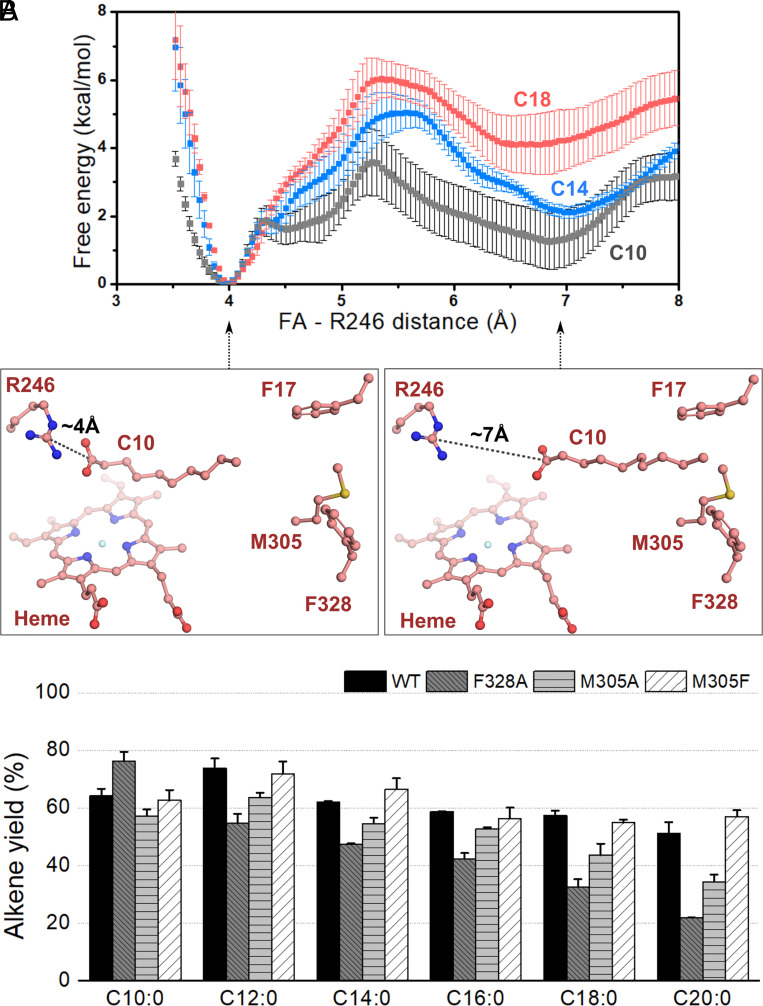
To gain more insights into structural determinants associated with FA decarboxylation by OleTPRN, molecular dynamics (MD) simulations were performed with different FAs and the heme iron in the penta-coordinated HS state. The simulations showed that short FAs (C10 to C14) could frequently visit two states according to the Arg246-COO- interaction distances (SI Appendix, Figs. S14 and S15). The bound state as in the crystal structure and a pseudo-bound state in which Arg246-COO- interaction is weakened and the FA acyl tail then interacts with a highly hydrophobic region located approximately 15 Å apart from the heme group. This region, termed the hydrophobic cradle, is populated by the residues Phe17, Phe24, Met305, and Phe328 (Fig. 3B and SI Appendix, Fig. S14). For long FAs, like C16-C20, the Arg246-COO- interaction is more persistent, in agreement with the fact that the chain length allows the simultaneous interactions of the FA with Arg246 and the hydrophobic cradle (SI Appendix, Fig. S15). Additionally, we observed that C16, C18, and C20 are less solvated than the short ones (C10, C12, and C14) (SI Appendix, Fig. S16), reflecting lower frequency of long-chains in the pseudo-bound state.
To evaluate this molecular event more quantitatively, umbrella sampling MD simulations were employed to obtain the free energy profile along the order parameter corresponding to the Arg246-FA distance for C10, C14, and C18 (SI Appendix, Fig. S17). The results show that for all substrates considered, the bound state, with the Arg246-FA interaction established, is the most stable, corresponding to an Arg246-FA carbon-carbon distance of 4 Å. As the Arg246-FA distance increases up to ~5 to 6 Å, the free energy also increases and reaches a secondary minimum at about 6 to 7 Å (Fig. 4A). Furthermore, we observed that both the free energy barrier associated with the disruption of the Arg246–FA interaction (therefore the rate of this event) and the free energy of the secondary minimum (therefore the equilibrium between bound and pseudo-bound states) are strongly dependent on the chain-length. The general trend is the larger the hydrophobic chain, the higher the free energy barrier for disrupting the Arg246-FA interaction and the higher the free energy difference between the bound and pseudo-bound states. These results indicate that all tested FA chains adopt the configuration in which the Arg246–FA interaction is established, but shorter chains are less tightly bound and prone to visit the pseudo-bound state more frequently.
Fig. 4.
(A) Free energy profile along the Arg246-FA distance, measured between the Arg246 Cζ atom and the FA carboxylate C atom. Snapshots corresponding to the first minimum (bound state) and second minimum (pseudo-bound state) are shown for C10. The free energy profiles show the existence of bound-pseudo-bound equilibrium which shifts to the bound state with an increase in the chain-length. Error bars correspond to the SD associated with four free energy profiles, computed for blocks of 10 ns of sampling per window. (B) Alkene yield from turnover reactions with OleTPRN mutants of the hydrophobic cradle amino acid (Experimental conditions: 2 µM of enzyme, 500 µM of substrate, 600 µM H2O2, 35 °C and 1 h of reaction).
Despite the fact that short FAs are more likely to sample the pseudo-bound state, it is not detrimental to the conversion rates nor chemoselectivity. On the contrary, biochemical assays revealed high conversion rates for short FAs. Previous studies of OleTJE showed that product release limits the enzyme kinetics (18) and based on this evidence, one might suggest that the pulling effect promoted by the hydrophobic cradle could contribute to detaching the alkene from the heme pocket, thus favoring product release. Our MD simulations showed that short FAs exhibit high mobility in the catalytic tunnel featuring high RMSF values (up to 6 Å) compared to long FAs (SI Appendix, Fig. S18). This is further corroborated by Molecular Mechanics/Poisson–Boltzmann Surface Area MM/PBSA calculations, in which binding free energies are more negative for longer acyl chains (SI Appendix, Fig. S19). Additionally, we also observed that alkene products, such as C9 and C13, exhibit notable mobility and higher binding free energies compared to the parental substrate that could facilitate the migration to the hydrophobic cradle (SI Appendix, Fig. S20). In sharp contrast to the C13 alkene, simulations of the C14 β-hydroxylated product showed that it maintains an interaction with Arg246, like the substrate. Furthermore, a comparison of the mobility of the F-G loop of OleTPRN in the presence of different alkene products with OleTJE, revealed that this region in OleTPRN is nearly twofold more flexible (SI Appendix, Fig. S21). Such analyses indicate that short-chain alkenes may have a more favorable product release kinetics in OleTPRN, supporting the biochemical assays.
Given the relevance of the hydrophobic cradle for OleTPRN activity as indicated by structural analysis and MD simulations, the residues comprising this region were mutated to better understand the role of the hydrophobicity at the end of the binding pocket in the enzyme’s functionality. The deleterious mutation of the hydrophobic cradle (quadruple mutant—F17A, F24A, M305A, and F328A) or the single OleTPRN F24A mutant resulted in a loss of protein stability, impairing activity assays. The OleTPRN F328A mutation had a pronounced negative impact on long FA conversion rates, while increasing the activity over short C10 FA (Fig. 4B and SI Appendix, Fig. S12). The same trend was observed for the OleTPRN M305A mutation, whereas the OleTPRN M305F mutation, which enhances the hydrophobicity of the hydrophobic cradle, had a beneficial effect on the conversion rates of long-chain FAs (Fig. 4B and SI Appendix, Fig. S12). These mutation studies demonstrate the importance of the hydrophobic cradle for OleTPRN activity over long-chain FAs.
Taking these results together, we could infer that the hydrophobic cradle plays distinct roles depending on the substrate chain-length. For long-chain FAs, the high hydrophobicity of this region is critical for the productive binding, but probably due to the shortened FG-loop in OleTPRN, the alkene production is more effective compared to OleTJE. On the other hand, for short-chain FAs, the pulling effect promoted by the hydrophobic cradle seems to be associated with high substrate conversion rates into alkenes.
Dimerization Facilitates Productive Substrate Binding.
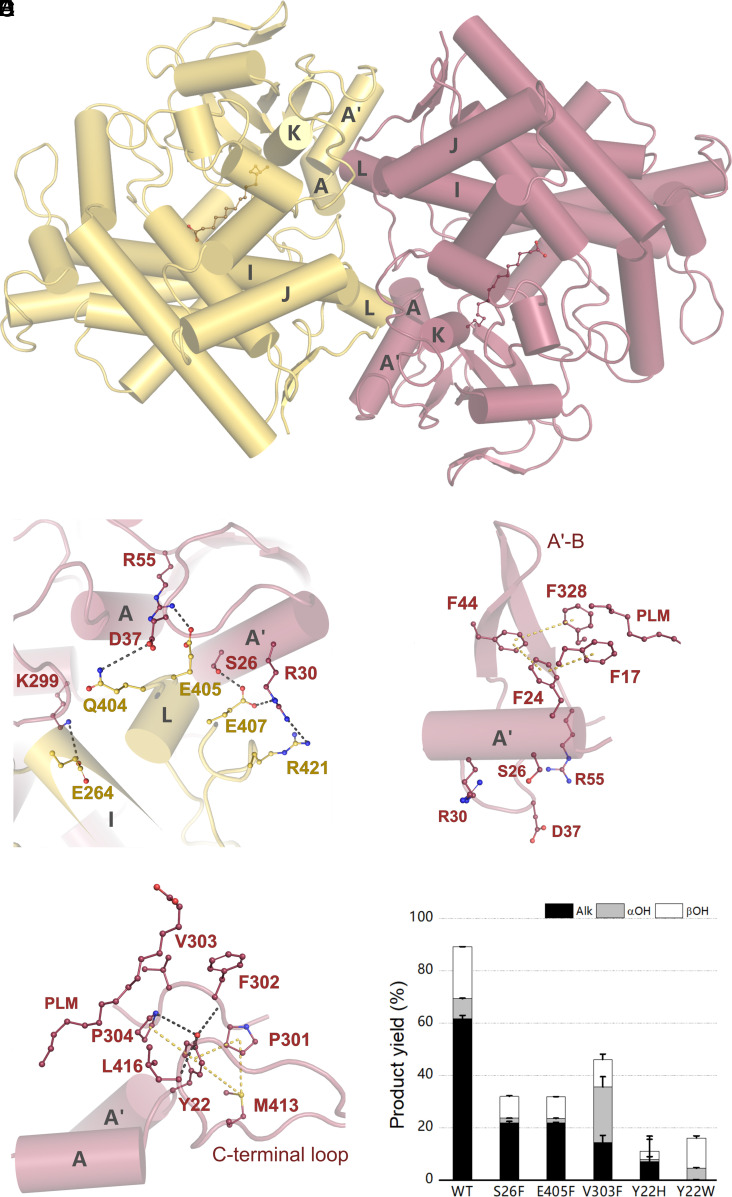
Although the structural organization of P450 superfamily is diverse (50), characterized members of the CYP152 family have been described as monomeric in nature (9, 15, 16). The analysis of OleTPRN crystalline interfaces indicated that a dimeric assembly is energetically favorable (Fig. 5A). Multiple analytical techniques showed that OleTPRN behaves as a dimer in solution, supporting a biological relevance for dimerization (SI Appendix, Figs. S22 and S23). To further support this, we evaluated the OleTPRN elution from analytical size exclusion chromatography in the presence of C14 or hydrogen peroxide. For all these conditions, the enzyme was found as a dimer (SI Appendix, Fig. S22C).
Fig. 5.
(A) Cartoon representation of OleTPRN dimerization, highlighting helices involved in the dimer interface. One protomer is shown in yellow and the other in salmon. (B) Highlight of important amino acids involved in the dimer interface contact. Hydrogen bonds are shown in gray. (C) Ideal placement of Phe24 established by the dimerization, anchoring Phe17 and Phe328, via Phe44, which are both important for substrate positioning. Van der Waals forces are displayed in yellow. (D) Tyr22 interactions that hold important amino acids for substrate positioning in the binding pocket. (E) Product yield of turnover reactions of myristic acid with OleTPRN mutants that destabilize either protein dimerization or substrate positioning.
The dimeric interface is mainly stabilized by electrostatic (Lys299-Glu264; Arg55-Glu405 and Arg30-Glu407) and polar (Ser26-Glu407 and Asp37-Gln404) contacts between the N-terminal A-A’ helical motif of one subunit and the long loop from the C-terminal β-hairpin of another protomer (Fig. 5B). These residues are not conserved in OleTJE nor P450BSβ, which is expected since both are monomeric (SI Appendix, Fig. S24). To interrogate the relevance of this oligomeric state for the enzyme function, the OleTPRN mutants S26F and E405F were designed with the aim of preventing the formation of the dimerization interface. The mutation strategy was to replace polar and charged residues by voluminous hydrophobic residues, which would promote steric clashes and prevent dimer stabilization. As envisaged, both S26F and E405F mutants eluted mainly as monomers (SI Appendix, Fig. S22B).
Both mutants displayed lower protein stability (SI Appendix, Fig. S30) and were drastically less active compared to the wild-type enzyme, with a reduction in FA conversion of more than 50% (Fig. 5E). However, in transient kinetic studies with C14, the mutants did not exhibit a significant alteration in the decay kinetics of Compounds I and II (SI Appendix, Figs. S25–S28 and Table 1). We further investigated the activation of H2O2 for wild-type and monomeric mutants with C14 and the proteins exhibited very similar apparent KD values for H2O2 (SI Appendix, Fig. S29). These results collectively reinforce that catalytic steps from substrate binding to Compound II decay are largely unaffected by the oligomerization state.
Structural analysis showed that the second-coordination sphere of the substrate, the A-A′ helical motif adjacent to the hydrophobic cradle, is stabilized by the dimeric interface and the monomerization would perturb this region. Specifically, the A-A’ loop contains the Tyr22 residue, which interacts and stabilizes the conformation of the loop containing the residues Phe302 and Val303 (Fig. 5C). Val303 directly interacts with the substrate, and Phe302, as aforementioned, is involved in the kink of the acyl chain at the β-carbon, favoring the Arg246–COO- interaction (Fig. 5D). Furthermore, the Phe24 residue from the A’ helix is interacting with the aromatic residues forming the hydrophobic cradle (Phe17, Phe44 and Phe328) (Fig. 5C). To experimentally demonstrate the role of these stabilizing interactions at the second-coordination sphere in the OleTPRN activity, the mutants Y22H and Y22W were designed and, as envisaged, the perturbation of the interactions between the A-A’ loop and the loop containing Phe302 and Val303 resulted in a significant decrease in the enzyme activity (Fig. 5E). It is worth noting that any perturbation at the aliphatic tail-interacting residues of the (distal and medial) active-site pocket seems to be highly detrimental to the activity of OleTPRN since even the replacement of hydrophobic residues with smaller side-chain volume (e.g., V303F) impaired the catalytic activity of OleTPRN (Fig. 5E). These results endorse the interpretation that upon the disruption of the dimer, the conformational stability of the A-A′ helical motif is compromised, affecting the configuration of aliphatic tail-interacting motifs including the hydrophobic cradle, which leads to a non-optimal substrate binding. This finding is in agreement with the reported importance of proper substrate accommodation in the active-site pocket for catalytic activity (18, 22, 23).
Discussion
The investigation of the biochemical basis for alkene production by the Rothia genus led to the discovery of a novel CYP152 decarboxylase, which is not salt-dependent, peroxide tolerant, and predominantly generates alkenes from a broad range of saturated and unsaturated FAs.
The current model for decarboxylation reaction by CYP152 peroxygenases is based on the mechanistic study of the enzyme OleTJE, which postulates the importance of the F-G loop for decarboxylase activity instead of the polymorphic residue (Gln→His substitution) from the catalytic triad of CYP152 peroxygenases. The literature has shown that the F-G loop in OleTJE, mainly because of its extended length and the presence of Leu176, contribute to FA anchoring, acting as a modulator of chemoselectivity (18).
Our work revealed that OleTPRN, despite forming the intermediates Compounds I and II during the catalytic cycle, exploits a distinct molecular mechanism for driving decarboxylation and chemoselectivity compared to other characterized CYP152 decarboxylases. Primarily, the H90Q substitution has an important role, not altering the total FA conversion capacity, but by modulating the enzyme chemoselectivity, which is not observed for the model enzyme OleTJE. Furthermore, OleTPRN possesses a shortened F-G loop similar to the hydroxylase P450BSβ, lacking the 4-residue extension and the residue Leu176 observed in OleTJE. The redesign of OleTPRN sequence mimicking the OleTJE F-G loop did not confer any positive effect in alkene production nor result in higher conversion rates. It highlights that CYP152 members can exploit distinct molecular bases to promote the decarboxylation of FAs and those from OleTPRN confer a high capacity for producing alkenes from the broad range of saturated and unsaturated FAs.
Through a series of computational and experimental approaches, we further demonstrated the existence of a novel motif, named as hydrophobic cradle, at the distal region of the substrate-binding pocket, which is critical for OleTPRN activity on long-chain FAs and plays a role in facilitating the product release of short-chain FAs. Computational simulations suggest a putative pulling effect of the hydrophobic cradle, which might favor the release of short-chain products from the heme pocket, increasing the conversion rates. Another molecular event that likely benefits the high conversion rates of short-chain FAs is the high solvation of the catalytic tunnel compared to long-chain FAs, which might facilitate the peroxide diffusion to the heme pocket, consequently contributing to enhanced activity. The high solvation of the catalytic pocket would also play a role in favoring decarboxylation reaction instead of hydroxylation since the presence of water molecules near to heme group might prevent the oxygen rebound required for hydroxylation (24).
To our knowledge, OleTPRN represents the first CYP152 family member to adopt a dimeric configuration. Such quaternary conformation seems to play a role in proper functioning of the enzyme via stabilizing the hydrophobic cradle and the loop containing the residues Phe302 and Val303. Both structural motifs were shown to be instrumental for enzyme function and therefore by non-direct interactions, the dimerization confers structural stability to the second-coordination sphere, specifically to the A-A’ helical motif, ensuring a productive substrate binding.
Together, the spectroscopic, structural, biochemical, and computational data presented herein uncovered unique molecular mechanisms governing the decarboxylation reaction of FA, which include dimerization, hydrophobic cradle, and Q→H substitution for chemoselectivity. Such results provide a novel mechanistic model for decarboxylase activity by CYP152 peroxygenases, with great biotechnological potential. This enzyme associated with tailored microbial chassis opens a new horizon for the biosynthesis of hydrocarbon from renewables feedstocks, including unsaturated ones.
Materials and Methods
Mutagenesis, Heterologous Expression, Enzyme Assays, and Biophysical Characterization.
Detailed materials and methods are provided in SI Appendix. OleTPRN mutants were generated by inverse PCR. The wild-type enzyme and mutants were recombinantly produced in E. coli BL21(DE3) harboring the pG-TF2 plasmid (Takara Bio, Kusatsu, JPN) and further purified to homogeneity. Enzyme assays were carried out by a range of spectroscopic and gas chromatographic methods for determination of dissociation constants, transient kinetics, steady-state turnovers, FA quantification, and substrate competitive assays. Biophysical and hydrodynamic properties were assessed by circular dichroism, small-angle X-ray scattering, and size-exclusion chromatography coupled with multi-angle light scattering.
X-Ray Crystallography.
OleTPRN samples were concentrated to 60 mg mL−1 and submitted to crystallization trials, using the vapor diffusion method in sitting drops. Single crystals were obtained from a solution containing 25% (w/v) PEG3500, 15% (w/v) 2-methyl-2,4-pentanediol, 0.1 M imidazole (pH 6.5), and 0.2 M lithium sulfate. Data collection and refinement procedures are described in SI Appendix. The refined atomic coordinates and structure factors have been deposited to the Protein Data Bank under the accession code 8D8P.
Computational Simulations.
MD simulations were performed with OleTPRN with the heme group in the pentacoordinate high-spin state in the presence of different FAs and products. Binding free energies were obtained with the MM/PBSA method. Potentials of mean forces were obtained with the Umbrella Sampling method. Detailed procedures are in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We gratefully thank the Brazilian Synchrotron Light Laboratory (LNLS, CNPEM, Campinas, Brazil) for the use of Manacá and D01A-SAXS2 beamlines; the Biosciences National Laboratory (LNBio, CNPEM, Campinas, Brazil) for the automated crystallization facility (Robolab), the Biophysics of Macromolecules and Metabolomics of LNBR (CNPEM, Campinas, Brazil) for the use of facilities; the National Laboratory for Scientific Computing (LNCC/MCTI, Brazil) for providing HPC resources of the SDumont supercomputer; the Center for Computing in Engineering & Science for the computational resources. This work was supported by grants from the São Paulo Research Foundation (#2018/04897-9; #2019/08855-1; #2019/12599-0; #2020/01967-6). This work was partially supported by grants from the NIH (1R01GM135315 to T.M.M.), NSF (CHE155066 to T.M.M.) and by LNBR – CNPEM (Campinas, Brazil).
Author contributions
L.L.R., W.C.G., S.D., A.S.S., T.M.M., and L.M.Z. designed research; L.L.R., W.C.G., S.D., A.S.S., R.L.S., M.C.A., P.S.V., R.Y.M., A.B.B.L., J.A.A., R.R.d.M., N.M., G.F.P., M.T.M., T.M.M., and L.M.Z. performed research; J.A.A. contributed new reagents/analytic tools; L.L.R., W.C.G., S.D., A.S.S., R.L.S., M.C.A., P.S.V., R.Y.M., A.B.B.L., J.A.A., R.R.d.M., N.M., G.F.P., A.M.F.L.J.B., M.T.M., T.M.M., and L.M.Z. analyzed data; and L.L.R., W.C.G., S.D., R.L.S., T.M.M., and L.M.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. S.P.d.V. is a guest editor invited by the Editorial Board.
Contributor Information
Thomas M. Makris, Email: tmmakris@ncsu.edu.
Leticia M. Zanphorlin, Email: leticia.zanphorlin@lnbr.cnpem.br.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. The protein structure has been deposited in Protein Data Bank with accession number 8D8P (51).
Supporting Information
References
- 1.Zargar A., et al. , Leveraging microbial biosynthetic pathways for the generation of “drop-in” biofuels. Curr. Opin. Biotechnol. 45, 156–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keasling J., et al. , Microbial production of advanced biofuels. Nat. Rev. Microbiol. 19, 701–715 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y. J., Kerkhoven E. J., Nielsen J., Barriers and opportunities in bio-based production of hydrocarbons. Nat. Energy 3, 925–935 (2018). [Google Scholar]
- 4.Liao J. C., Mi L., Pontrelli S., Luo S., Fuelling the future: Microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 14, 288–304 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Keasling J., et al. , Microbial production of advanced biofuels. Nat. Rev. Microbiol. 19, 701–715 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Peralta-Yahya P. P., Zhang F., del Cardayre S. B., Keasling J. D., Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Rude M., Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microb. 77, 1718–1727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthews S., et al. , Production of alkenes and novel secondary products by P450 OleTJE using novel H2O2 -generating fusion protein systems. FEBS Lett. 591, 737–750 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Belcher J., et al. , Structure and biochemical properties of the alkene producing cytochrome p450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 bacterium. J. Biol. Chem. 289, 6535–6550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews S., et al. , Catalytic determinants of alkene production by the cytochrome P450 peroxygenase OleTJE. J. Biol. Chem. 292, 5128–5143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davydov R., et al. , Hydroxylation of camphor by reduced oxy-cytochrome p450cam: Mechanistic implications of EPR and ENDOR studies of catalytic intermediates in native and mutant enzymes. J. Am. Chem. Soc. 123, 1403–1415 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Denisov I. G., Makris T. M., Sligar S. G., Cryotrapped reaction intermediates of cytochrome p450 studied by radiolytic reduction with phosphorus-32. J. Biol. Chem. 276, 11648–11652 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Kimata Y., Shimada H., Hirose T., Ishimura Y., Role of THR-252 in cytochrome P450CAM: A study with unnatural amino acid mutagenesis. Biochem. Biophys. Res. Commun. 208, 96–102 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Vidakovic M., Sligar S. G., Li H., Poulos T. L., Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect. Biochemistry 37, 9211–9219 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Lee D. S., et al. , Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis. Crystallographic, spectroscopic, and mutational studies. J. Biol. Chem. 278, 9761–9767 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Matsunaga I., Sumimoto T., Ueda A., Kusunose E., Ichihara K., Fatty acid-specific, regiospecific, and stereospecific hydroxylation by cytochrome P450 (CYP152B1) from Sphingomonas paucimobilis: Substrate structure required for alpha-hydroxylation. Lipids 35, 365–371 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Hsieh C. H., et al. , The enigmatic P450 decarboxylase OleT is capable of, but evolved to frustrate. Oxygen rebound chemistry. Biochemistry 56, 3347–3357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaya J. A., Rutland C. D., Leschinsky N., Makris T. M., A distal loop controls product release and chemo- and regioselectivity in cytochrome P450 decarboxylases. Biochemistry 57, 344–353 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Faponle A. S., Quesne M. G., De Visser S. P., Origin of the regioselective fatty-acid hydroxylation versus decarboxylation by a cytochrome P450 peroxygenase: What drives the reaction to biofuel production? Chem. Eur. J. 22, 5478–5483 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Du J., Liu L., Guo L. Z., Yao X. J., Yang J. M., Molecular basis of P450 OleTJE: An investigation of substrate binding mechanism and major pathways. J. Comput. Aided Mol. Des. 31, 483–495 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Pickl M., et al. , Mechanistic studies of fatty acid activation by CYP152 peroxygenases reveal unexpected desaturase activity. ACS Catal. 9, 565–577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantú Reinhard F. G., Lin Y. T., Stańczak A., de Visser S. P., Bioengineering of cytochrome P450 OleT JE: How does substrate positioning affect the product distributions? Molecules 25, 2675 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y. T., de Visser S. P., Product distributions of cytochrome P450 OleTJE with phenyl-substituted fatty acids: A computational study. Int. J. Mol. Sci. 22, 7172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bharadwaj V. S., Kim S., Guarnieri M. T., Crowley M. F., Different behaviors of a substrate in P450 decarboxylase and hydroxylase reveal reactivity-enabling actors. Sci. Rep. 8, 12826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surger M., Angelov A., Liebl W., Distribution and diversity of olefins and olefin-biosynthesis genes in Gram-positive bacteria. Biotechnol. Biofuels 13, 70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y., et al. , Biochemical characterization of three new α-olefin-producing P450 fatty acid decarboxylases with a halophilic property. Biotechnol. Biofuels 12, 79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loew G. H., Harris D. L., Role of the heme active site and protein environment in structure, spectra, and function of the cytochrome p450s. Chem. Rev. 100, 407–419 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Xu H., et al. , In vitro oxidative decarboxylation of free fatty acids to terminal alkenes by two new P450 peroxygenases. Biotechnol. Biofuels 10, 208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke R., Ingram P. J., Haynes K., An integrative model of ion regulation in yeast. PLoS Comput. Biol. 9, 1002879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szatmári D., Intracellular ion concentrations and cation-dependent remodelling of bacterial MreB assemblies. Sci. Rep. 10, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munro A. W., McLean K. J., Grant J. L., Makris T. M., Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem. Soc. Trans. 46, 183–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ntambi J. M., Miyazaki M., Recent insights into stearoyl-CoA desaturase-1. Curr. Opin. Lipidol. 14, 255–261 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y., et al. , Unexpected reactions of α, β-unsaturated fatty acids provide insight into the mechanisms of CYP152 peroxygenases. Angew. Chem. Int. Ed. Engl. 60, 24694–24701 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Armbruster J., et al. , P450Jα: A new, robust and α-selective fatty acid hydroxylase displaying unexpected 1-alkene formation. Chem. Eur. J. 26, 15910–15921 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., et al. , Hydrogen peroxide-independent production of α-alkenes by OleTJE P450 fatty acid decarboxylase. Biotechnol. Biofuels 7, 28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zachos I., et al. , Photobiocatalytic decarboxylation for olefin synthesis. Chem. Commun. 51, 1918–1921 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Li F., et al. , A genetically-encoded synthetic self-assembled multienzyme complex of lipase and P450 fatty acid decarboxylase for efficient bioproduction of fatty alkenes. Bioresour. Technol. 272, 451–457 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Yan J., Liu Y., Wang C., Han B., Li S., Assembly of lipase and P450 fatty acid decarboxylase to constitute a novel biosynthetic pathway for production of 1-alkenes from renewable triacylglycerols and oils. Biotechnol. Biofuels 8, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michalik B., Biel W., Lubowicki R., Jacyno E., Chemical composition and biological value of proteins of the yeast Yarrowia lipolytica growing on industrial glycerol. Can. J. Anim. Sci. 94, 99–104 (2014). [Google Scholar]
- 40.Wang S., Jiang S., Chen H., Bai W. J., Wang X., Directed evolution of a hydroxylase into a decarboxylase for synthesis of 1-alkenes from fatty acids. ACS Catal. 10, 14375–14379 (2020). [Google Scholar]
- 41.Grant J. L., Mitchell M. E., Makris T. M., Catalytic strategy for carbon-carbon bond scission by the cytochrome p450 oleT. Proc. Natl. Acad. Sci. U.S.A. 113, 10049–10054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant J. L., Hsieh C. H., Makris T. M., Decarboxylation of fatty acids to terminal alkenes by cytochrome P450 Compound I. J. Am. Chem. Soc. 137, 4940–4943 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Hsieh C. H., et al. , The enigmatic P450 decarboxylase OleT is capable of, but evolved to frustrate. Oxygen rebound chemistry. Biochemistry 56, 3347–3357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Ullrich R., Hofrichter M., Groves J. T., Heme-thiolate ferryl of aromatic peroxygenase is basic and reactive. Proc. Natl. Acad. Sci. U.S.A. 112, 3686–3691 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yosca T. H., et al. , Iron(IV)hydroxide pK(a) and the role of thiolate ligation in C-H bond activation by cytochrome P450. Science 342, 825–829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kellner D. G., Hung S. C., Weiss K. E., Sligar S. G., Kinetic characterization of compound I formation in the thermostable cytochrome P450 CYP119. J. Biol. Chem. 277, 9641–9644 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Rittle J., Green M. T., Cytochrome P450 compound I: Capture, characterization, and C-H bond activation kinetics. Science 330, 933–937 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Bukowski M. R., et al. , A thiolate-ligated nonheme oxoiron(IV) complex relevant to cytochrome P450. Science 310, 1000–1002 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Matsunaga I., et al. , Enzymatic reaction of hydrogen peroxide-dependent peroxygenase cytochrome P450s: Kinetic deuterium isotope effects and analyses by resonance raman spectroscopy. Biochemistry 41, 1886–1892 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Mallinson S. J. B., et al. , A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion. Nat. Commun. 9, 2487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vieira P. S., Murakami M. T., Zanphorlin L. M., Crystal structure of a novel fatty acid decarboxylase from Rothia nasimurium. RCSB Protein Data Bank (RCSB PDB). 10.2210/pdb8D8P/pdb. Deposited 8 June 2022. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix. The protein structure has been deposited in Protein Data Bank with accession number 8D8P (51).