Significance
Regarding the risk factor for COVID-19, the significance of obesity differs among ethnicities. The Japanese population tends to become abdominal or visceral-dominant obesity, associated with subclinical inflammatory conditions. We identified that Japanese COVID-19 patients with abdominal obesity showed a lower survival rate than those with overweight; thus, the abdominal adipose burden was related to accelerated inflammatory responses. In the murine COVID-19 model, we demonstrated that C57BL/6 background obese mice with abdominal obesity died after the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) infection due to excessive inflammatory responses compared to lean mice. Inflammatory cytokine (IL-6) blockade and the prevention of obesity improved survival by reducing excessive immune responses. Our results suggest how abdominal obesity increases the risk of cytokine storm and death in patients with COVID-19.
Keywords: COVID-19, obesity, abdominal adiposity, cytokine storm, risk factor
Abstract
Obesity has been recognized as one of the most significant risk factors for the deterioration and mortality associated with COVID-19, but the significance of obesity itself differs among ethnicity. Multifactored analysis of our single institute–based retrospective cohort revealed that high visceral adipose tissue (VAT) burden, but not other obesity-associated markers, was related to accelerated inflammatory responses and the mortality of Japanese COVID-19 patients. To elucidate the mechanisms how VAT-dominant obesity induces severe inflammation after severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) infection, we infected two different strains of obese mice, C57BL/6JHamSlc-ob/ob (ob/ob), C57BLKS/J-db/db (db/db), genetically impaired in the leptin ligand and receptor, respectively, and control C57BL/6 mice with mouse-adapted SARS-CoV-2. Here, we revealed that VAT-dominant ob/ob mice were extremely more vulnerable to SARS-CoV-2 due to excessive inflammatory responses when compared to SAT-dominant db/db mice. In fact, SARS-CoV-2 genome and proteins were more abundant in the lungs of ob/ob mice, engulfed in macrophages, resulting in increased cytokine production including interleukin (IL)-6. Both an anti-IL-6 receptor antibody treatment and the prevention of obesity by leptin replenishment improved the survival of SARS-CoV-2-infected ob/ob mice by reducing the viral protein burden and excessive immune responses. Our results have proposed unique insights and clues on how obesity increases the risk of cytokine storm and death in patients with COVID-19. Moreover, earlier administration of antiinflammatory therapeutics including anti-IL-6R antibody to VAT-dominant patients might improve clinical outcome and stratification of the treatment for COVID-19, at least in Japanese patients.
The COVID-19, which is caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), continues to spread throughout the world and to threaten all human beings in spite of the best efforts to overcome this disease. A certain proportion of COVID-19 patients experience marked elevations of inflammatory mediators, termed “cytokine storm,” resulting in the deterioration of the respiratory condition (1, 2). Various comorbidities, including hypertension (3), diabetes (4), and chronic kidney disease (CKD) (5), have been implicated as risk factors predicting the deterioration or poor outcome of COVID-19 patients. Among such conditions, obesity is one of the most significant aggravating factors (6), although the underlying mechanisms have not been fully elucidated.
We have previously reported the clinical characteristics of Japanese COVID-19 patients focusing on the thrombotic complications (7). During inpatient management and data collection, we also noticed that the patients with abdominal fat dominant obesity tend to experience severer disease courses and worse outcomes. Therefore, we focused on the two types of obesity, namely visceral adipose tissue (VAT)–dominant and subcutaneous adipose tissue (SAT)–dominant obesity. The former is closely associated with metabolic disorders and immune-related diseases (8, 9), whereas the latter type of obesity does not preclude such association with specific diseases. Therefore, we hypothesized that the accumulation of VAT would fuel the systemic inflammatory responses in COVID-19 and become a more reliable marker to identify the patients with a potential risk than other obesity-associated markers, including body mass index (BMI).
In the present study, we elucidate that VAT accumulation rather than mere overweight is associated with a worse outcome in Japanese COVID-19 patients. To support our hypothesis and to explore a unique stratification of COVID-19 patients, we infected mouse-adapted SARS-CoV-2 with several obese mice, revealing that VAT-dominant C57BL/6JHamSlc-ob/ob (ob/ob) mice and diet-induced obesity mice died after infection with low-titer mouse-adapted SARS-CoV-2 virus due to the subsequent cytokine storm, whereas most of the nonobese wild-type mice or even similar obese C57BLKS/J-db/db (db/db) mice survived. The lethality of the ob/ob mice could be overcome with interleukin (IL)-6 signaling blockade and with reduction of adipose tissues before infection.
Results
Visceral, but Not Subcutaneous, Adipose Tissue Accumulation Was Associated with Death in Japanese COVID-19 Patients.
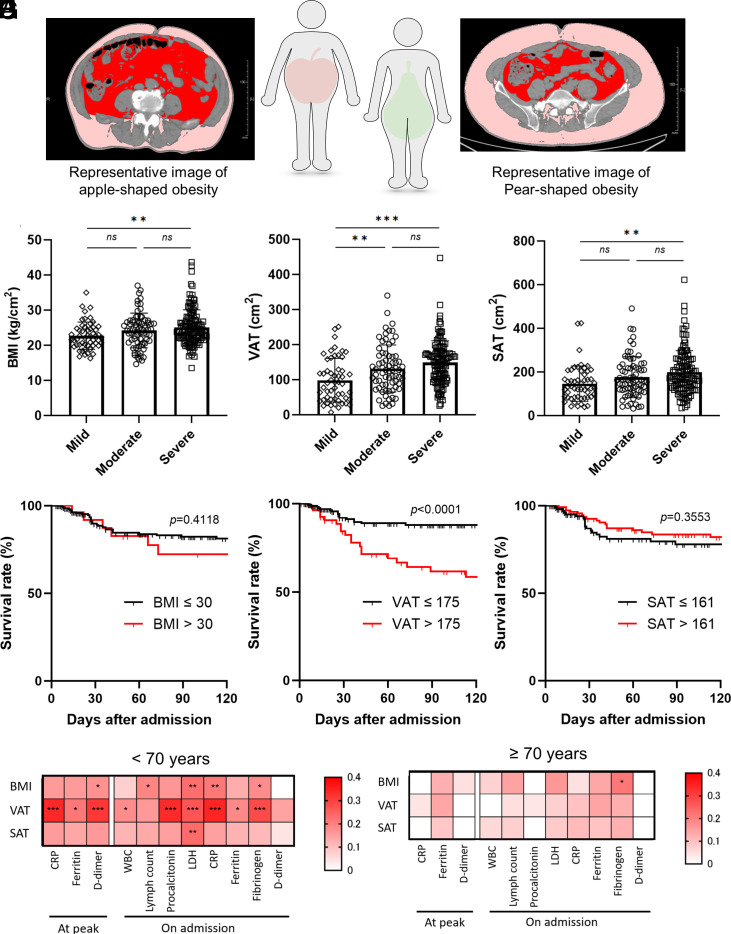
To evaluate whether body composition was associated with the outcome of COVID-19, we quantified the fat areas in the abdomen and in the subcutaneous tissues using abdominal computed tomography (CT) scan images (Fig. 1A) (10). These data were analyzed in correlation with disease severity and outcome. In addition to BMI, VAT area and SAT area were associated with the peak severity of COVID-19 (Fig. 1 B–D), consistent with the previous reports (11, 12). Interestingly, we could demonstrate that the VAT area was the single marker with a statistically significant association with death despite the size of our cohort (SI Appendix, Table S1). By drawing the receiver operating characteristic (ROC) curve, we designated the cutoff value of VAT area as 175 cm2 in order to maximize the sensitivity and specificity (81.0% and 55.9%, respectively). Indeed, multivariable analysis using a logistic regression model revealed that increased VAT area (P < 0.0001) was a robust independent risk factor for mortality due to COVID-19 besides other known risk factors (13–17) such as older age (P = 0.0389), CKD above grade 4 (P = 0.0382), and history of infarction (P = 0.0097), with high adjusted odds ratio (SI Appendix, Table S2). Kaplan–Meier survival analysis also revealed that the survival rate during hospitalization was worse in patients with high VAT burden but not in those with a high BMI or high SAT burden (Fig. 1 E–G). The Harrell’s c-statistic with optimism correction using the internal bootstrap method of the Cox regression model (18) including the known risk factors [gender male, hypertension, history of infarction, elevated HbA1c (>7.5%), and CKD (≥grade 4)] was 0.6632 (SI Appendix, Table S3). By adding increased VAT (>175 cm2) to this model, we could improve the value to 0.7229, whereas the addition of BMI (>30 kg/m2) or SAT (>160 kg/m2) decreased those values (0.6470 and 0.6157, respectively).
Fig. 1.
VAT area, but not the other obesity markers, was associated with COVID-19 mortality and inflammatory biomarkers. (A) Representative computed tomography images analyzed by fat scan. The Left and Right panels represent visceral fat–dominant obesity (apple-shaped) and subcutaneous-dominant obesity (pear-shaped), respectively. The visceral fat is indicated in red, and the subcutaneous fat is indicated in pink. (B–D) The association of the peak severity– and obesity-associated markers, VAT (B), BMI (C), and SAT (D). Data are mean ± SD. One-way ANOVA followed by Tukey’s post hoc test was performed to compare the three groups. (E–G) Kaplan–Meier plot of survival rate in COVID-19 patients during hospitalization until 120 d after admission. Graph shows survival curves when patients are divided into two groups based on the VAT (E), BMI (F), and SAT (G). The cutoff values were designated by drawing the receiver operating characteristic (ROC) curve to maximize sensitivity and specificity. (H and I) Heatmap showing the correlation between obesity-associated markers and biomarkers in patients less than 70 y old (H) and 70 y old or above (I). The correlations were evaluated by Spearman’s method. *P < 0.05; **P < 0.01; ***P < 0,001. VAT: Visceral adipose tissue, BMI: Body mass index, SAT: Subcutaneous adipose tissue.
Interestingly, increased adipose tissue has weak, but significant, correlations with several biomarkers, including C-related protein (CRP), D-dimer, and ferritin at both the early phase and the disease peak in the relatively younger group (Fig. 1 H and I and SI Appendix, Fig. S1 A–C). We considered that increased VAT burden also fueled the inflammatory responses during COVID-19 especially in the relatively younger patients and resulted in worsening of clinical outcomes in patients, since macrophages and adipocytes in VAT contributed to subclinical inflammation in relatively young patients whose immune-responding capacities are still intact.
Obese Mice with C57BL/6 Background Were Significantly More Susceptible to SARS-CoV-2 Infection.
Immune responses against mouse-adapted SARS-CoV-2, established from human isolates, reached peak levels at three days postinfection (dpi). After the infection, lung inflammation caused death in mature adult mice (16 wk or older) rather than young mice, as shown previously (19). To consider the influence of obesity on the production of inflammatory cytokines in the disease course of SARS-CoV-2 infection, we examined two obese mouse models aged 18 to 20 wk. The ob/ob mice and db/db mice have a genetic dysfunction of leptin ligand or receptor, respectively. Both mice had a similar level of obesity by hyperphagia (SI Appendix, Fig. S2A).
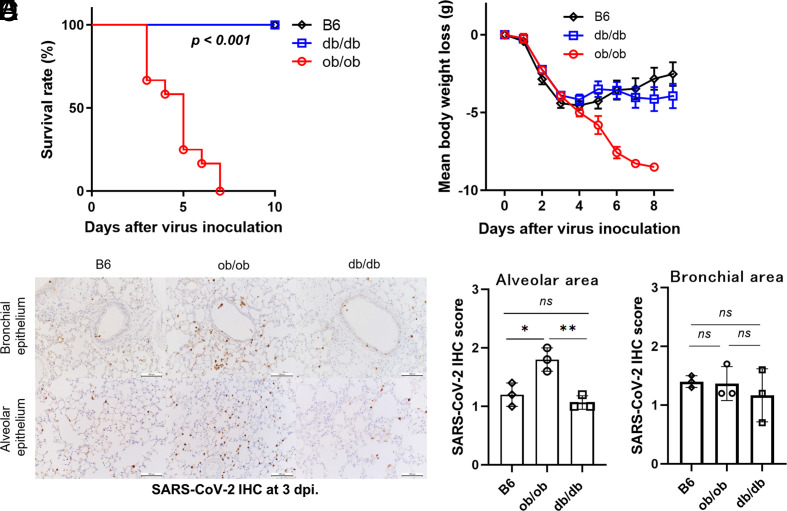
Intriguingly, all the ob/ob mice died around four to nine dpi after inoculation with mouse-adapted SARS-CoV-2 (Fig. 2 A and B). In contrast, all the db/db mice, as well as the control C57BL/6 (B6) mice, survived. The SARS-CoV-2-positive cells in the alveolar area were more abundant in ob/ob mice than those in the other two strains (Fig. 2 C–E), while those in the bronchial area were comparable among the three groups.
Fig. 2.
ob/ob mice were more vulnerable to SARS-CoV-2 infection with high infectivity. (A and B) Survival curve (A) and body weight (B) of ob/ob, db/db, and B6 mice (n = 11 to 12) after mouse-adapted SARS-CoV-2 inoculation. Data are pooled from two independent experiments showing similar results. Log-rank (Mantel–Cox) test was used to assess the survival curve. Data are mean ± SE. (C–E) Immunohistochemical staining using anti-SARS-CoV-2 N-specific antibody in the lungs of three strains at three days postinfection (dpi) in alveolar area (D) and bronchial area (E). (Scale bars were 100 µm.) Data are presented as the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. Data were analyzed by one-way ANOVA followed by Tukey’s post hoc test.
Considering the discrepancy of SARS-CoV-2 mortality between ob/ob mice and db/db mice, we need to take into account that db/db mice have a slightly different genetic background with ob/ob mice by accidental crossbreeding with DBA2, resulting in different major histocompatibility complex (MHC) class II haplotype (20) and insulin-producing capacity (21). Indeed, blood glucose levels were higher in db/db mice than those of ob/ob mice because of the sufficient insulin-producing capacity of ob/ob mice (SI Appendix, Fig. S2 B and C). Additionally, regarding the adipose tissue distribution, ob/ob mice showed relatively high weights of liver and perirenal adipose tissue, indicating VAT–dominant obesity rather than db/db mice (SI Appendix, Fig. S2 D and E), as shown previously (22, 23).
To expand our observations by avoiding leptin dysfunction as well as slightly different genetic background and insulin-producing capacity, we analyzed whether the susceptibility against SARS-CoV-2 increased in B6 wild-type obese mice. Compared to normal fat diet (NFD), high fat diet (HFD)–fed wild-type mice experienced increased body weight and elevated blood glucose and insulin levels after 10 wk of feeding (SI Appendix, Fig. S3 A–C). We noticed that HFD-induced obesity mice rapidly died after the infection, whereas NFD-fed mice survived (SI Appendix, Fig. S3D). Consistent with the results of the immunopathological analysis in ob/ob mice, the SARS-CoV-2-positive cells were more abundant in HFD-fed mice than those in the NFD-fed mice (SI Appendix, Fig. S3E).
Histological analysis revealed that relatively localized inflammation and attenuated lung injury were observed in db/db mice rather than ob/ob mice and B6 mice, suggesting the favorable outcome of db/db mice (SI Appendix, Fig. S4 A–C). However, regarding distribution of inflammation and acute lung injury, minor differences were observed between ob/ob mice and B6 mice at three dpi (SI Appendix, Fig. S4 B and C). Surprisingly, we revealed that both bronchial and alveolar cells of ob/ob mice had already showed greater score of SARS-CoV-2 immunohistochemical (IHC) at one dpi (SI Appendix, Fig. S4 D and E). Furthermore, the positive rate of Transmembrane protease, serine 2 (TMPRSS2) but not Angiotensin-converting enzyme 2 (ACE2) was higher in ob/ob mice than the other two groups under the noninfected condition (SI Appendix, Fig. S4 G–I). Blood glucose and insulin levels were lower under SARS-CoV-2 infection than those of the starved condition presumably because of the impaired food intake during infection. However, the blood glucose levels were still elevated in ob/ob and db/db mice than those of B6 mice (SI Appendix, Figs. S2 B and C and S3 B and C).
Then, we considered that the virus antigens were broadly distributed in the lungs of ob/ob mice and HFD-fed mice, suggesting that these findings were related to the lethality against SARS-CoV-2 infection.
SARS-CoV-2 Genome and Proteins Were More Abundant in ob/ob Mice, Engulfed in Lung Macrophages with Enhanced Immune Responses.
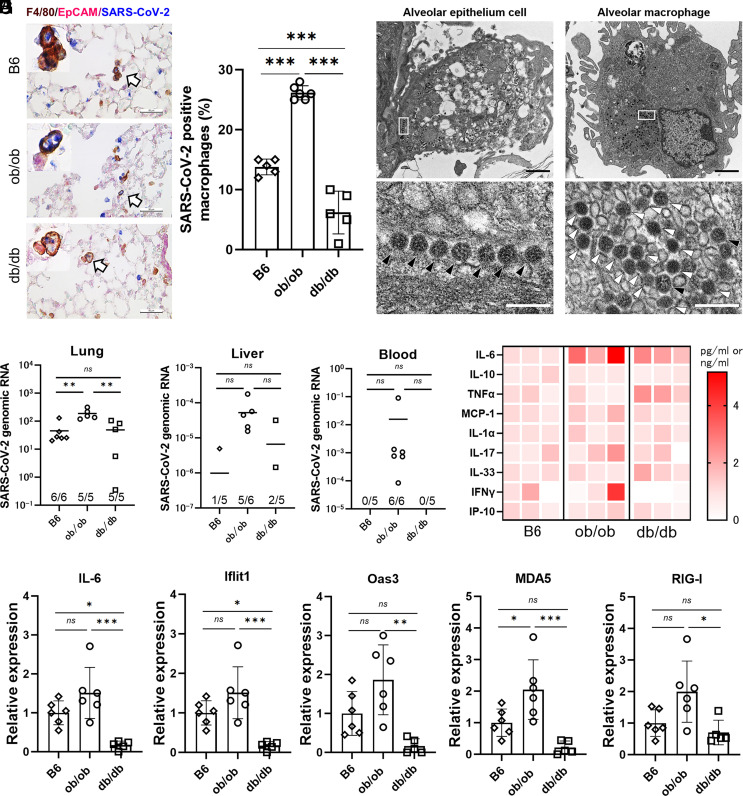
To identify the cell type of SARS-CoV-2-positive cells, we conducted IHC staining using anti-SARS-CoV-2 nucleocapsid antibody. Both lung epithelial cells and lung macrophages were stained in all the three strains (Fig. 3A), while the proportion of SARS-CoV-2 antigen–positive lung macrophages was significantly increased in ob/ob mice (Fig. 3B). Additionally, SARS-CoV-2 viral particles were observed using an electron microscope to be present in the lung macrophages and disrupted epithelial cells (Fig. 3C). However, no virus-derived ultrastructures, such as double-membrane vesicles and budding virions, during viral replication were found in the macrophage. The high density and irregular nucleocapsid arrangement of the viral particles in macrophage endosomes further indicated that they were distinct from infectious postbudding viral particles. These observations suggested that the lung macrophages in ob/ob mice carried disrupted viral particles during phagocytosis and did not support viral replication.
Fig. 3.
SARS-CoV-2-positive macrophages and genomes were the most abundant in ob/ob mice with enhanced inflammatory responses. (A) Representative image of triple IHC staining of F4/80 (brown), EpCAM (pink), and SARS-CoV-2 (blue) in the lungs of B6, ob/ob, and db/db. F4/80-positive macrophages (arrow) as well as EpCAM-positive lung epithelial cells (arrowhead) colocalized with SARS-CoV-2 antigens in all the three strains. (Scale bars were 50 µm.) (B) The proportions of SARS-CoV-2 and F4/80 double-positive cells in F4/80 single-positive cells. One-way ANOVA followed by Tukey’s post hoc test was performed to compare the three groups. Data are mean ± SD. (C) Electron microscopy images of viral particles in infected ob/ob mice. A higher-magnification image of the boxed area in the bottom. Nucleocapsid-visible viral particles and high-density viral particles were indicated with black arrowhead and white arrowhead, respectively. [Scale bars, 1.0 µm (Top); 0.2 µm (Bottom).] (D–F) The abundance of SARS-CoV-2 genomic RNA detected in the tissue samples derived from lung (D), peripheral blood (E), and liver (F) at three days postinfection (dpi). The results were normalized with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. (G) Heatmap of the concentrations of immune-related cytokines and chemokines in the lung homogenates at three dpi (n = 3 mice for each group), analyzed by Mouse Luminex Discovery Assay. The red color indicates higher concentrations of the biomarkers. (H and I) Quantitative analysis of representative immune-related, including IL-6 and interferon signature genes relative to B6 (H), and intracellular virus–sensing molecules (I). Data are presented as the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001. Data were analyzed by one-way ANOVA followed by Tukey’s post hoc test.
Indeed, more copies of SARS-CoV-2 genomic RNA-encoding spike (S) protein were detected in the liver and in blood, in addition to the lungs, particularly in ob/ob mice (Fig. 3 D–F). IL-6 and type I/II interferon responses were up-regulated in the lungs of ob/ob mice but not in db/db mice and wild-type mice at protein levels and RNA levels (Fig. 3 G–I). Additionally, we observed an impairment of type I interferon responses in db/db mice (Fig. 3 H and I).
Therefore, we considered that SARS-CoV-2 antigen–carrying phagocytes, such as lung macrophages, circulated systemically in ob/ob mice and might induce inflammasome-dependent activation. Our observation might support the results of several reports that SARS-CoV-2 antigens were sometimes detected in extrapulmonary tissues carried by circulating phagocytic cells (24, 25).
Innate Immune Activation and Cytokine Storm Occurred in the Lungs of ob/ob Mice.
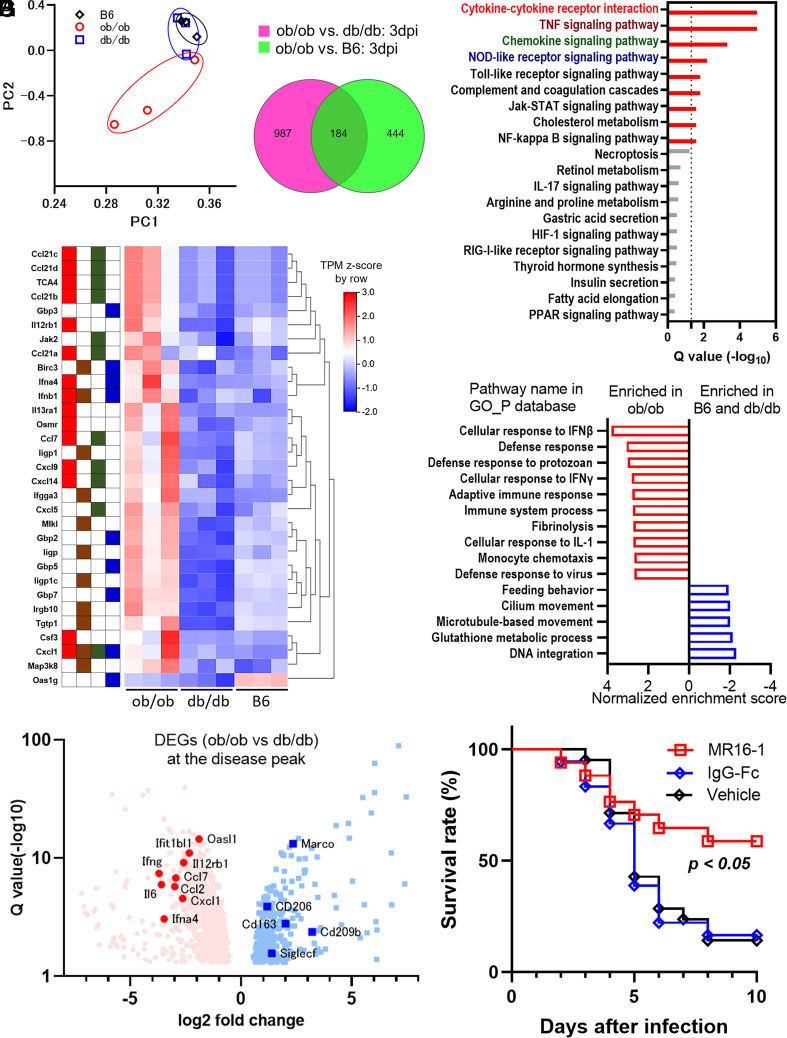
To assess our hypothesis using unbiased transcriptomic analysis, we performed RNA sequencing(RNA-Seq) for analyzing the transcripts of SARS-CoV-2-infected lungs at three dpi (E-MTAB-12617). The major principal components exhibited separated gene expression patterns between the three strains (Fig. 4A). Among 184 genes identified as shared differentially expressed genes (DEGs) (Fig. 4B), we identified using pathway or gene set enrichment analysis that global alterations of inflammation-associated pathways were enriched in the shared DEGs and were enhanced in ob/ob mice but not in the other mice (Fig. 4 C and E). The representative DEGs in the top four pathways of Fig. 4C were shown as a heatmap in Fig. 4D: cytokine–cytokine receptor interaction, tumor necrosis factor (TNF) signaling pathway, chemokine signaling pathway, and Nucleotide-binding and oligomerization domain (NOD)-like receptor pathway.
Fig. 4.
Lethal level of excessive immune responses was induced in the lungs of ob/ob mice. (A–G) RNA sequencing analysis comparing the immune responses of ob/ob, db/db, and B6 mice at three days postinfection (dpi). (A) Principal component analysis of RNA-Sequencing data from nine murine lung samples in the three groups at three dpi. (B) Venn diagram showing differentially expressed genes (DEGs) in ob/ob mice vs. db/db mice (green) or B6 mice (magenta). (C) The KEGG enrichment analysis of the shared 184 DEGs. The pathway name of the top four pathways is shown in red, brown, dark green, and dark blue, respectively. (D) The heatmap of representative gene transcriptions in the top four pathways in Fig. 4C. The tiles next to the gene name indicate their categorized pathways: cytokine–cytokine receptor interaction pathway, TNF signaling pathway, chemokine signaling pathway, and NOD-like receptor signaling pathway are shown in red, brown, dark green, and dark blue, respectively. The Transcripts Per Kilobase Million (TPM) Z-scores were standardized by row direction. (E) The representative results of gene set enrichment analysis comparing ob/ob mice vs. B6 mice and db/db mice using the gene ontology (GO) database related to biological process (GO_P). (F) Volcano plot of DEGs in ob/ob vs. db/db mice. Red dot and blue dot indicate individual DEGs elevated in ob/ob mice or db/db mice, respectively. Representative gene names are shown, including inflammatory cytokines, chemokines, type I interferon downstream genes, and type 2 macrophage–associated genes. (G) The survival curve of vehicle (n = 22)-, control immunogloblin G (IgG) (n = 18)-, or MR16-1 (n = 17)-administrated mice is shown. MR16-1 was the mouse counterpart to tocilizumab, IL-6 receptor antibody. Data are pooled from three independent experiments showing similar results. The agents were administered intraperitoneally 1 h before infection and at three dpi. Log-rank (Mantel–Cox) test was used to assess the survival curve.
These findings suggested that ob/ob mice were more sensitive to SARS-CoV-2 infection rather than db/db mice, and excessive inflammatory responses were likely the cause of death in ob/ob mice. Among the DEGs in ob/ob vs. db/db mice, several inflammatory cytokines and chemokines were up-regulated in ob/ob mice as well as type I interferon downstream genes, whereas type 2 macrophage–associated genes were up-regulated in db/db mice (Fig. 4F). When we examined IL-6 blockade using IL-6 receptor antibody, MR 16-1, the mouse counterpart to tocilizumab (26), the survival rates were partially but significantly improved, indicating that excessive IL-6 production was at least one of the causes of death in ob/ob mice (Fig. 4G).
We next explored the differences between ob/ob mice and db/db mice by further RNA-Seq with or without SARS-CoV-2 infection (E-MTAB-12618). The pathway enrichment analysis of the 738 DEGs identified different gene expressions in several immune response–associated pathways, including the phagosome pathway (SI Appendix, Fig. S5 A–C). Based on the differences in genetic backgrounds mentioned above, different expressions of MHC class II molecules (H-2 molecules in mice) were identified (SI Appendix, Fig. S5D). Besides these differences, we observed a clear separation of macrophage phenotype–associated markers; specifically, inflammatory macrophage–associated markers were dominant in ob/ob mice, while pan-macrophage markers were up-regulated in both ob/ob mice and db/db mice after infection (SI Appendix, Fig. S5E).
Reducing the Adipose Tissue by Leptin Administration Prevented the Death of ob/ob Mice with Prompt Elimination of Virus Genome and Attenuation of Excessive Immune Responses.
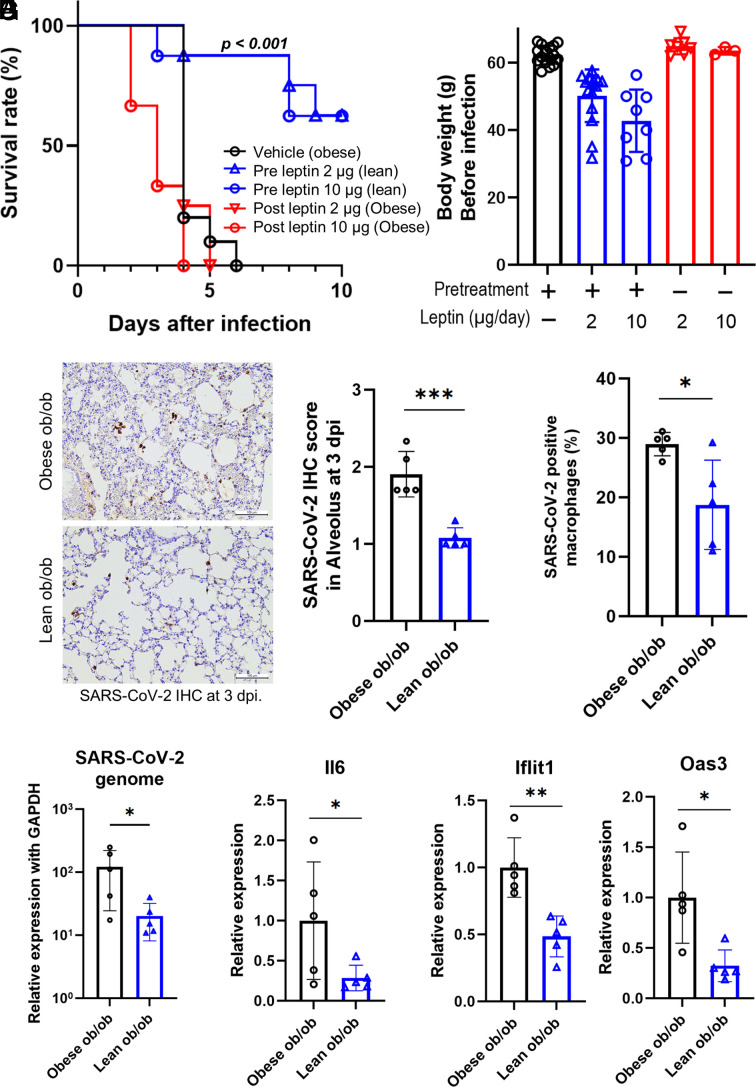
Then, we examined whether obesity was the essential factor of the susceptibility against SARS-CoV-2 in ob/ob mice. As one might expect, reduction of the body weight by preventive leptin administration to ob/ob mice (lean ob/ob) resulted in improved survival after infection, suggesting that excessive adipose accumulation altered the disease course in ob/ob mice (Fig. 5 A and B and SI Appendix, Fig. S6A). Indeed, therapeutic leptin administration with SARS-CoV-2 inoculation did not show any beneficial effects on the survival of obese ob/ob mice, suggesting that the susceptibility against SARS-CoV-2 was not due to immune-related function in leptin. In fact, preventive leptin administration could reduce the adipose tissue weight and improve the fatty liver with decreased number of crown-like structure around the portal veins (SI Appendix, Fig. S6 B–D). Since CLS are formed with inflammatory macrophages as a result of apoptotic adipocyte exposure, these findings suggested the reduction of inflammatory macrophages by prevention of obesity.
Fig. 5.
Lean ob/ob mice survived after SARS-CoV-2 infection by inducing an appropriate immune reaction. (A) Survival curve after mouse-adapted SARS-CoV-2 inoculation. The groups consisted of vehicle (n = 11), 2 µg/day of preventive leptin treatment (n = 8), 10 µg/day of preventive leptin treatment (n = 8), 2 µg/day of leptin after infection (n = 4), and 10 µg/day of leptin after infection (n = 3). Data are pooled from three independent experiments showing similar results. Log-rank (Mantel–Cox) test was used to assess the survival curve. (B) The body weight of both groups before infection. Data were mean ± SD. (C–G) Comparison of mice received vehicle (obese ob/ob, n = 5) or 2 µg/day of leptin pretreatment (lean ob/ob, n = 5) at three days postinfection (dpi). Immunohistochemical staining of the lung using anti-SARS-CoV-2 N-specific antibody (C). (Scale bars were 100 µm.) The proportions of SARS-CoV-2 and F4/80 double-positive cells in F4/80 single-positive cells (D). Quantitative analysis of SARS-CoV-2 genomic RNA (E), IL-6 (F), and interferon signature genes (G). A Student’s t test was used to compare two groups. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Regarding distribution of SARS-CoV-2, the IHC score, genomes, and the ratio of SARS-CoV-2-positive macrophages in lungs were lower in the lean ob/ob mice than those in the obese ob/ob mice (Fig. 5 C–E). Simultaneously, IL-6 induction and interferon-included responses were attenuated in lean ob/ob mice (Fig. 5 F and G). Glucose metabolism under SARS-CoV-2 infection was not significantly different between obese ob/ob mice and lean ob/ob mice (SI Appendix, Fig. S6 E and F).
Excessive Immune Responses Observed in the Obese ob/ob Mice under SARS-CoV-2 Infection Were Attenuated in Lean ob/ob Mice.
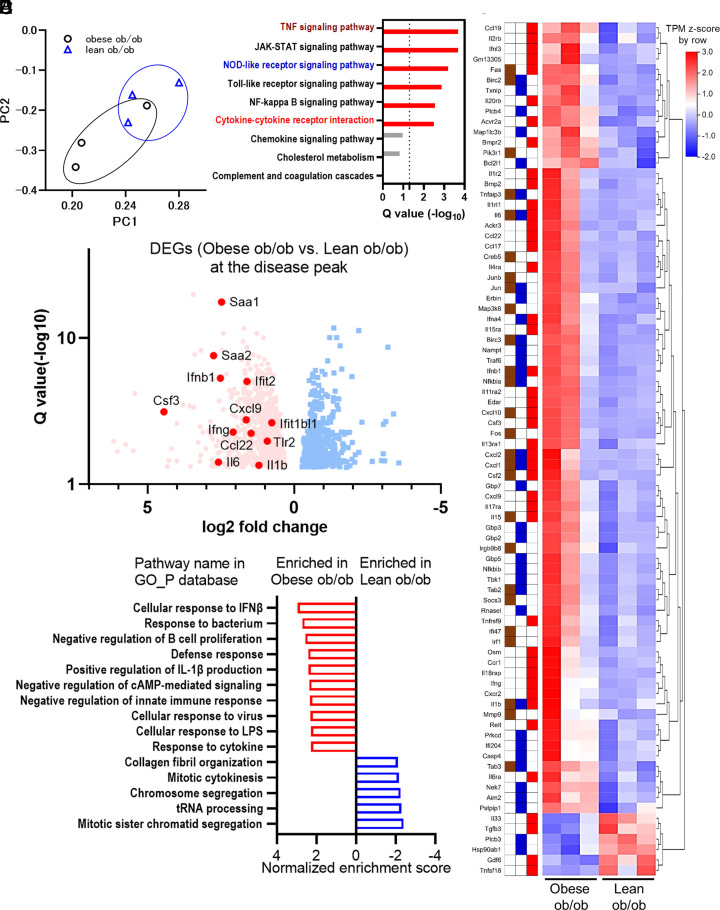
We performed RNA-Seq analyses to identify whether preventive leptin administration changed global gene expressions under SARS-CoV-2 infection (E-MTAB-12623). The major principal components exhibited separated gene expression patterns in obese and lean ob/ob mice (Fig. 6A). Among 1615 DEGs between lean ob/ob mice and obese ob/ob mice, 51 pathways were enriched using KEGG pathway enrichment analysis (SI Appendix, Table S4). Regarding the representative nine pathways enriched in obese ob/ob mice shown in Fig. 4C (Q value < 0.05), six downstream pathways of inflammation were still enriched in this analysis, while cholesterol metabolism, complement and coagulation cascades, and chemokine signaling pathways were not (Fig. 6B). The representative DEGs in the three pathways repeatedly enriched in Fig. 6B were shown as a heatmap in Fig. 6C: TNF signaling pathway, NOD-like receptor pathway, and cytokine–cytokine receptor interaction. Among the DEGs in lean ob/ob mice and obese ob/ob mice, effector molecules and acute-phase reactants were up-regulated in obese ob/ob mice rather than lean ob/ob mice (Fig. 6D). Similarly, gene set enrichment analysis revealed that several inflammation-associated pathways identified in Fig. 4E were attenuated in lean ob/ob mice rather than in obese ob/ob mice (Fig. 6E). Regarding alveolar macrophages, the proportions were fewer and lysosomal function was impaired in ob/ob and in db/db rather than those in B6 mice (SI Appendix, Fig. S7 A and C). However, these decrements/dysfunctions were rescued in the lean ob/ob mice, treated with preventive leptin administration (SI Appendix, Fig. S7 B and D).
Fig. 6.
Excessive immune responses observed in the obese ob/ob mice were attenuated in lean ob/ob mice under SARS-CoV-2 infection. (A–E) RNA sequencing analysis comparing the immune responses of obese ob/ob mice to lean ob/ob mice at three days postinfection (dpi). Obese ob/ob mice and lean ob/ob mice were continuously administrated with vehicle or 2 µg/day of leptin using ALZET pump for 6 wk. (A) Principal component analysis from six murine lung samples; obese ob/ob, n = 3 and lean ob/ob, n = 3. (B) KEGG enrichment analysis of the 1,615 DEGs in obese ob/ob vs. lean ob/ob mice. Nine representative pathways (Q < 0.05 in Fig. 4C) were shown. The colors of the pathway name were the same as those of Fig. 4C. (C) The heatmap of representative gene transcriptions categorized in TNF signaling pathway, NOD-like receptor signaling pathway, or cytokine–cytokine receptor interaction pathway in the 1,615 DEGs. The tiles next to the gene name indicate their categorized pathways: TNF signaling pathway, NOD-like receptor signaling pathway, and cytokine–cytokine receptor interaction pathway are shown in brown, dark blue, and red, respectively. The Transcripts Per Kilobase Million (TPM) Z-scores were standardized by row direction. (D) Volcano plot of DEGs in obese ob/ob vs. lean ob/ob mice. Red dot and blue dot indicate individual DEGs elevated in obese ob/ob mice or lean ob/ob mice, respectively. Representative gene names are shown, including inflammatory cytokines, chemokines, type I interferon downstream genes, and acute-phase reactants. (E) The representative results of gene set enrichment analysis comparing obese ob/ob mice vs. lean ob/ob mice using the gene ontology (GO) database related to biological process (GO_P).
From these findings, we concluded that the excessive adipose accumulation was related to delayed elimination of the virus and to the activation of inflammatory macrophage and cytokine storm, leading to high mortality in SARS-CoV-2-infected obese mice.
Discussion
Here, we have shown that VAT burden is a risk factor contributing to the deterioration and death of Japanese COVID-19 patients, and that excessive adipose tissue is a more accurate predictor of mortality (SI Appendix, Fig. S8), which has been previously suggested in several studies (11, 12). Additionally, the levels of inflammatory biomarkers were correlated with the visceral adipose burden, especially in relatively younger population. Interestingly, the contribution of overweight to COVID-19 mortality was more pronounced in Asian and Black populations than in White, even with a similar level of obesity (27, 28). Since VAT-dominant obesity is more common in Asian population and closely associated with metabolic disorders (8, 29), VAT accumulation might accurately reflect the discrepancy of epidemiological or ethnic risks of COVID-19 mortality in high BMI patients, at least in the Japanese population. We should consider increased VAT as a risk factor for the deterioration and mortality seen in obese patients, especially in younger population.
In animal experiments, we have demonstrated that obesity increased the susceptibility against SARS-CoV-2 infection in two obese mouse models with C57BL/6 background (ob/ob and HFD-fed mice), whereas C57BLKS background obese mice (db/db) were resistant even with the similar level of obesity and with the leptin pathway deficiency. Additionally, we identified that SARS-CoV-2 infection induced excessive inflammatory responses in obese ob/ob mice, while the responses were attenuated in lean ob/ob mice (SI Appendix, Fig. S8). Despite the immunological function of leptin, the vulnerability of ob/ob mice against SARS-CoV-2 was not merely due to such effects because the therapeutic administration of leptin to the obese ob/ob mice did not improve their disease course. We demonstrated an apparent discrepancy in two leptin deficiency obese mice. Ob/ob mice and db/db mice are considered similarly immunocompromised (30–32), except for the report demonstrating greater sensitivity to endotoxin in ob/ob mice (33). However, in addition to our observations of the differences in MHC class II haplotype, residual macrophage phenotype, and adipose tissue distributions, recent reports demonstrated their different microbiomes and metabolites (22, 23). It is also intriguing that insulin levels were elevated in ob/ob mice and HFD-fed mice under noninfected condition. A continuous insulin signaling might impair the polarization to M2 macrophages and production of antiinflammatory cytokines (34). As shown in the human survey, the influence of obesity on infectious diseases might differ depending on various factors including genetic backgrounds. We found VAT accumulation as a mortality risk at least in Japanese COVID-19 patients as well as mice with C57BL/6 background.
Recently, various reports demonstrated that macrophage activation is the essential factor of the cytokine storm complicated in severe COVID-19 patients and animal models of SARS-CoV-2 infection (24, 35–37). Several studies revealed that adipose tissues serve as a target and reservoir for SARS-CoV-2, with evidence of infection in adipocytes (38, 39) and infiltrating monocytes (36). Although the susceptibility of adipocytes to the SARS-CoV-2 infection is controversial, Saccon et al. revealed that human visceral fat–derived cells were more susceptible to SARS-CoV-2 infection and higher ACE2 expression than their subcutaneous counterparts (40). In addition, Salina et al. revealed that the engulfment of SARS-CoV-2-infected proapoptotic cells by macrophages resulted in the production of inflammatory cytokines and impaired efferocytosis, namely, the failure in phenotype conversion to antiinflammatory macrophages (41). Impaired efferocytosis was considered as a major mechanism of metabolic disorders induced by obesity (42) and was observed in ob/ob mice (43). In this study, we showed that SARS-CoV-2 transcripts were abundant systemically and impaired lysosomal function and skewed expressions of M1 macrophage markers in ob/ob mice. Therefore, we considered that lung macrophages were activated by engulfing the SARS-CoV-2 antigens, and failed to cease unleashed inflammatory responses, resulting in the production of lethal cytokines in VAT-dominant obesity mice.
In addition to antiviral agents, antiinflammatory therapies, including steroids, IL-6 blockers, and a JAK inhibitor baricitinib, are being administered to patients with moderate-to-severe COVID-19. Such anticytokine strategies attenuate excessive immune responses and provide benefits to certain subgroups of patients, whereas these interventions would delay viral elimination and induce opportunistic infections. It is problematic at this moment that we cannot predict the efficacy of the antiinflammatory/immunosuppressive treatments in the real-world clinics. Since patients prone to cytokine storms are more likely to respond to these therapies, our current data are expected to be helpful in the stratification of the patients who would benefit from anticytokine therapeutics in the earlier phase of COVID-19.
We must admit several limitations in this study. Because clinical questions were raised from our retrospective cohort of COVID-19 patients, we could not avoid the residual confounding and lack of data regarding some biomarkers or follow-up data. We should further identify the mechanisms how visceral adipose accumulation recruited or converted macrophages into inflammatory phenotype in the lungs and VAT.
In conclusion, our findings indicated that VAT-dominant patients would be at risk for cytokine storm due to COVID-19 and might be good candidates to receive cytokine-suppressing therapy during the disease course.
Materials and Methods
The Patient Cohort of COVID-19 in Tokyo Medical and Dental University (TMDU) Hospital.
In the present study, we analyzed 250 patients who underwent abdominal CT out of 594 consecutive COVID-19 patients admitted to TMDU Hospital during April 1, 2020, to August 31, 2021. Clinical characteristics of the patients are shown in SI Appendix, Table S5. Among them, 34 patients died in hospital, while 216 survived. Based on the “guideline of the medical treatment of the noble coronavirus” presented by Japanese Ministry of Health, Labour and Welfare (44), disease severity was defined as follows: mild, for patients who do not need oxygen therapy; moderate, for patients who need oxygen of equal or less than 5 L/min; and severe, for patients who need oxygen of more than 5 L/min or mechanical ventilation. Peak disease severity was determined at the most severe clinical condition during hospitalization. The diagnostic criteria and data collection were described previously (7).
The study protocols and procedures complied with the Declaration of Helsinki and followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline. This study was approved by the ethics committees of TMDU as M2020-027. Informed consent was obtained in the opt-out approach on the website of the ethics committees.
Quantification of Fat Tissues from CT Images.
The abdominal fat distribution was measured on the umbilicus level of abdominal CT images using Fat Scan version 2.0 software (N2 System Co., Osaka, Japan) (10). Briefly, the regions of interest (ROI) of the fat areas were defined by tracing their contour and the outline of the abdominal wall. The visceral and subcutaneous fat areas were calculated automatically from ROI following the caliper of each CT image.
Animals.
Male C57BL/6JHamSlc-ob/ob (ob/ob), C57BLKS/J-db/db (db/db), and C57BL/6J (B6) mice were purchased from Japan SLC, Inc. (Hamamatsu, Japan) or CLEA Japan, Inc. (Tokyo, Japan) and were maintained in specific pathogen-free facilities in National institute of infectious diseases. All animals were housed until 18 to 20 wk old with free access to water and diet and provided with 12 h of light/dark cycle. Unexpected death was not observed.
SARS-CoV-2 Challenge Experiments.
Mouse-adapted SARS-CoV-2 strain was established in the previous study (19). Mice were anesthetized and then inoculated viral solution intranasally. Viral solutions for inoculation were diluted with phosphate-buffered saline (PBS) and 30 µL of solution contained twice of the 50% lethal dose (2LD50) determined in 24-wk-old BALB/c mice. Body weight was measured, and abnormal behaviors were assessed daily for 10 d. The humane endpoint was defined as the appearance of clinically diagnostic signs of respiratory stress or more than 25% weight loss. The mice were killed with an overdose of isoflurane if severe disease symptoms or weight loss was observed.
In Vivo Anti-IL-6R Antibody (MR16-1) Treatment.
A rat anti-mouse IL-6 receptor monoclonal Ab (clone MR16-1, rat IgG1) was provided by Chugai Pharmaceutical (Tokyo, Japan). Purified rat IgG1 (isotype-matched control Ab) (Sigma-Aldrich) was administered as an isotype-matched control. Two milligrams of antibodies and vehicle was administered intraperitoneally 1 h before the virus inoculation and on three dpi. The body change and abnormal behaviors were monitored daily up to 10 dpi to consider humane endpoints.
The Administration of Leptin.
For the continuous administration of leptin, we used ALZET pumps (DURECT Corp., Cupertino, CA, USA) with model 2004 for 6-wk treatment and model 2001 for 1-wk treatment. These pumps delivered 2 or 10 μg of mouse recombinant leptin (498-OB, R&D Systems, Inc., Minneapolis, MN, USA) per day and were inoculated in the back of 9-wk-old ob/ob mice for the 6-wk administration and 15-wk-old ob/ob mice for the administration just before the SARS-CoV-2 infection, respectively. Body weights were measured every week after pump inoculation until the SARS-CoV-2 infection.
Diet-Induced Obesity Model.
To develop visceral dominant obesity, 8-to-10-wk-old male B6 mice were fed with HFD (D12492, Research Diet Inc., New Brunswick, NJ, USA), which contained 60% of fat and ad libitum access to food and water for 10 wk. Body weights were measured every week during HFD feeding until the SARS-CoV-2 infection.
Measurement of Plasma Glucose and Insulin Levels.
Blood was collected from a submandibular vein after 6 h of fasting. Plasma glucose level was measured using a Rabo Glucometer (ForaCare Japan, Tokyo, Japan). Plasma insulin level was measured using Ultra-Sensitive Mouse Insulin ELISA Kit (M1104, Morinaga Institute of Biological Science, Yokohama, Japan) according to the manufacturer’s instructions.
Ethics Statements for Animal Experiments.
All infected experiments were handled in BSL3 animal facilities and noninfected experiments were in TMDU according to the guidelines of the committee. Experiments using pathogens were approved by the Committee for Pathogens at the National Institute of Infectious Diseases, Tokyo, Japan. Animal experiments were approved by the Animal Care and Use Committee of the National Institute of Infectious Diseases in Japan (120148 and 121093) and the ethics committees of TMDU (G2020-023C5).
Immunohistopathological Analysis.
Tissue samples were analyzed with histopathology and immunohistochemistry. The detailed protocols are described in SI Appendix.
Electron Microscopy Analysis of SARS-CoV-2 Infection.
Tissue samples were prefixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer for 3 d at 4 °C and then postfixed in 2% osmium tetroxide, dehydrated with a graded series of alcohols and propylene oxide, and embedded in epoxy resin. After trimming the epoxy resin–embedded tissue, ultrathin sections (70 nm thick) were cut using an ultramicrotome (Diatome), mounted on grids, and stained with 4% uranyl acetate and lead citrate for Transmission Electron Microscope analysis using a HT7700 (Hitachi Ltd., Japan) at 80 kV.
Multiplex Assay for Cytokines and Chemokines.
Homogenized lung samples from 19-wk-old ob/ob, db/db, and B6 mice were analyzed using mouse Luminex Discovery Assay (R&D Systems) according to the manufacturer’s protocol, which detects 15 cytokines and chemokines: IP-10, IFN-γ, IL-6, IL-17/IL-17A, TNF-α, MCP-1, IL-1α/IL-1F1, IL-10, and IL-33. The assay samples were analyzed on a Luminex 200 instrument with xPONENT software described by the manufacturer. The unit concentrations of IL-6, IL-10, TNF-α, IL-1-α, and IFN-γ were pg/mL and those of MCP-1, IL-33, and IP-10 were ng/mL.
Flow Cytometric Analysis.
The lung-derived cells were incubated with anti-mouse CD16/32 (553142, BD Biosciences) at room temperature (RT) for 15 min to block nonspecific bindings, and then incubated with mixtures of antibodies at RT °C for 15 min. Antibodies were purchased from commercial sources as follows: anti-CD45 (103138, BD Biosciences), anti-CD11b (101228, BD Biosciences), anti-CD11c (117318, BD Biosciences), and anti-Siglec-F (S17007L, BD Biosciences). Data were acquired on a FACS Lyric (BD Biosciences) and analyzed using FlowJo software version X10.0.7r2 (Tree Star Inc., Ashland, OR).
Ex Vivo Phagocytosis Assay.
CD45-positive cells were isolated from lung-derived cells using MACS beads (130-052-301, Miltenyi Biotec) according to the manufacturer’s protocol and harvested in 24-well plates at 5 × 105 cells/well. After overnight incubation, the plated cells were washed with PBS and pHrodo Green Zymosan BioParticles Conjugate (P35365, Invitrogen) was added at the indicated final concentration. After 150-min incubation, adhered cells were detached using Accumax (AM105, Innovative Cell Technologies, Inc. San Diego, CA, USA). The suspended cells were prepared as described above using anti-CD45 and anti-F4/80 (565411, BD Biosciences).
Quantified PCR and RNA-Seq.
Extracted RNA was assessed by real-time quantitative PCR (qPCR) and RNA-Seq analysis. The detailed protocols and primers are described in SI Appendix.
Statistics.
We compared the distributions of continuous and categorical variables between the two groups using the Mann–Whitney U test and Chi-squared test (or Fisher’s exact test), respectively. The cutoff values were determined by drawing the ROC curve about VAT, SAT, and BMI to maximize sensitivity and specificity. Univariate and multivariate logistic regression analyses were employed to explore the impacts of known risk factors on mortality during admission. The variables were selected based on the previous reports and our research interests. Multicollinearity was then checked using the variance inflation factor for the final predictor variables. When developing the prediction model of the time to death based on the Cox regression analysis, we followed the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) 2015 guideline (45). For each model, we calculated the Harrell’s c-statistic with optimism correction using the internal bootstrap method (18) along with apparent c-statistic. The bootstrap was replicated 1,000 times. In the animal experiments, statistical significances in mean values among the groups were analyzed by ANOVA and Student’s t tests. The survival rates were analyzed by log-rank test. All the statistical analyses were performed using GraphPad Prism software version 8.0 (GraphPad Software, San Diego, CA), EZR software version 1.54, a free software for using R on graphical user interface (46), and the SAS (version 9.4; SAS Institute Inc., Cary, NC, USA). All statistical analyses were two sided, and statistical significance was considered as P < 0.05.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Katsuko Yamasaki for her expert technical assistance with histological analysis. We also thank Lucinda Beck for carefully proofreading the manuscript. We would like to thank all the participants in our institute for the management of patients with COVID-19. S.Y. was supported by the Japan Agency for Medical Research and Development (AMED) under grant number 21ek0410083h0002. T. Suzuki was supported by AMED under grants number JP22fk0108637 and JP22wm0125008. T.H. was supported by a research grant from Kobayashi Foundation. M.K. was supported by GSK Japan Research Grant 2021 from GlaxoSmithKline.
Author contributions
T.H., H.H., T. Suzuki, and S.Y. designed research; T.H., S.O., Y.K., D.K., H.I., S.M., M. Kataoka, M.T., T.K., A.A., K.W., Y.O., and Y. Miyazaki performed research; T.H., S.O., Y.K., D.K., M. Kamiya, H.I., S.M., M. Kataoka, M.T., T.K., A.A., H.S., A.H., Y. Mitsui, T. Satoh, K.W., T.Y., Y.O., Y. Miyazaki, and T. Suzuki analyzed data; and T.H., S.O., M. Kamiya, T. Satoh, K.W., Y. Miyazaki, H.H., T. Suzuki, and S.Y. wrote the paper.
Competing interests
S.Y. received research funding from AbbVie, Asahi Kasei Pharma, Chugai Pharmaceutical, CSL Behring, Eisai, ImmunoForge, Mitsubishi Tanabe, Pharma, and Ono pharmaceutical and speaking fees from AbbVie, Asahi Kasei Pharma, Chugai Pharmaceutical, Eisai, Eli Lilly, GlaxoSmithKline, Mitsubishi Tanabe Pharma, Ono pharmaceutical, and Pfizer. Y. Miyazaki received a research grant and an honorarium from Chugai Pharmaceutical Co., Ltd. M. Kamiya received research grant from GlaxoSmithKline. The other authors have declared no conflicts of interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
RNA-Seq data are available in ArrayExpress (E-MTAB-12618 (47), E-MTAB-12617 (48), and E-MTAB-12623 (49)). The other data that support the findings of this study are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Huang C., et al. , Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucas C., et al. , Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark C. E., McDonagh S. T. J., McManus R. J., Martin U., COVID-19 and hypertension: Risks and management. A scientific statement on behalf of the British and Irish Hypertension Society. J. Hum. Hypertens. 354, 304–307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega E., et al. , Risk factors for severe outcomes in people with diabetes hospitalised for COVID-19: A cross-sectional database study. BMJ Open 11, e051237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortiz A., et al. , Chronic kidney disease is a key risk factor for severe COVID-19: A call to action by the ERA-EDTA. Nephrol. Dial. Transplant. 36, 87–94 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwok S., et al. , Obesity: A critical risk factor in the COVID-19 pandemic. Clin. Obes. 10, e12403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oba S., et al. , Arterial and venous thrombosis complicated in COVID-19: A retrospective single center analysis in Japan. Front. Cardiovasc. Med. 8, 767074 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazare J. A., et al. , Ethnic influences on the relations between abdominal subcutaneous and visceral adiposity, liver fat, and cardiometabolic risk profile: The international study of prediction of intra-abdominal adiposity and its relationship with cardiometabolic risk/intra-abdominal adiposity. Am. J. Clin. Nutr. 96, 714–726 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Peters M. C., et al. , Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: A cross-sectional analysis of two cohorts. Lancet. Respir. Med. 4, 574–584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshizumi T., et al. , Abdominal fat: Standardized technique for measurement at CT. Radiology 1, 283–286 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Favre G., et al. , Visceral fat is associated to the severity of COVID-19. Metabolism 115, 154440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheffler M., et al. , Prognostic role of subcutaneous and visceral adiposity in hospitalized octogenarians with COVID-19. J. Clin. Med. 10, 5500 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appelman B., et al. , Mortality and readmission rates among hospitalized COVID-19 patients with varying stages of chronic kidney disease: A multicenter retrospective cohort. Sci. Rep. 121, 1–8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mubarik S., et al. , The association of hypertension with the severity of and mortality from the COVID-19 in the early stage of the epidemic in Wuhan, China: A multicenter retrospective cohort study. Front. Med. 8, 623608 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peckham H., et al. , Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 111, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu T., et al. , History of coronary heart disease increased the mortality rate of patients with COVID-19: A nested case–control study. BMJ Open 10, e038976 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holman N., et al. , Risk factors for COVID-19-related mortality in people with type 1 and type 2 diabetes in England: A population-based cohort study. Lancet Diabetes Endocrinol. 8, 823–833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrell F. E., Lee K. L., Mark D. B., Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 15, 361–387 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Iwata-Yoshikawa N., et al. , A lethal mouse model for evaluating vaccine-associated enhanced respiratory disease during SARS-CoV-2 infection. Sci. Adv. 8, eabh3827 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freitas J. M., et al. , The MHC gene region of murine hosts influences the differential tissue tropism of infecting Trypanosoma cruzi Strains. PLoS One 4, e5113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis R. C., et al. , Early hepatic insulin resistance precedes the onset of diabetes in obese C57BLKS-db/db mice. Diabetes 59, 1616–1625 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suriano F., et al. , Novel insights into the genetically obese (ob/ob) and diabetic (db/db) mice: Two sides of the same coin. Microbiome 9, 1–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giesbertz P., et al. , Metabolite profiling in plasma and tissues of ob/ob and db/db mice identifies novel markers of obesity and type 2 diabetes. Diabetologia 58, 2133–2143 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Sefik E., et al. , Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature. 606, 585–593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junqueira C., et al. , FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 606, 576–584 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okazaki M., Yamada Y., Nishimoto N., Yoshizaki K., Mihara M., Characterization of anti-mouse interleukin-6 receptor antibody. Immunol. Lett. 84, 231–240 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Yates T., et al. , A population-based cohort study of obesity, ethnicity and COVID-19 mortality in 12.6 million adults in England. Nat. Commun. 13, 1–9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M., et al. , Associations between body-mass index and COVID-19 severity in 6·9 million people in England: A prospective, community-based, cohort study. Lancet Diabetes Endocrinol. 9, 350–359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadowaki T., et al. , Japanese men have larger areas of visceral adipose tissue than Caucasian men in the same levels of waist circumference in a population-based study. Int. J. Obes. 307, 1163–1165 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Thaiss C. A., et al. , Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science. 359, 1376–1383 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Ikejima S., et al. , Impairment of host resistance to Listeria monocytogenes Infection in Liver of db/db and ob/ob Mice. Diabetes 54, 182–189 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Honce R., Schultz-Cherry S., Impact of obesity on influenza A virus pathogenesis, immune response, and evolution. Front. Immunol. 10, 1071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faggioni R., et al. , Leptin deficiency enhances sensitivity to endotoxin-induced lethality. Am. J. Physiol. 276, R136–R142 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Kubota T., et al. , Downregulation of macrophage Irs2 by hyperinsulinemia impairs IL-4-indeuced M2a-subtype macrophage activation in obesity. Nat. Commun. 9, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant R. A., et al. , Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 590, 635–641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez-Colón G. J., et al. , SARS-CoV-2 infection drives an inflammatory response in human adipose tissue through infection of adipocytes and macrophages. Sci. Transl. Med. 14, eabm9151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitsui Y., et al. , CiDRE+ M2c macrophages hijacked by SARS-CoV-2 cause COVID-19 severity. bioRxiv [Preprint] (2022). 10.1101/2022.09.30.510331 (Accessed 31 March 2023). [DOI]
- 38.Zickler M., et al. , Replication of SARS-CoV-2 in adipose tissue determines organ and systemic lipid metabolism in hamsters and humans. Cell Metab. 34, 1–2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiterer M., et al. , Hyperglycemia in acute COVID-19 is characterized by insulin resistance and adipose tissue infectivity by SARS-CoV-2. Cell Metab. 33, 2174–2188.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saccon T. D., et al. , SARS-CoV-2 infects adipose tissue in a fat depot- and viral lineage-dependent manner. Nat. Commun. 13, 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salina A. C., et al. , Efferocytosis of SARS-CoV-2-infected dying cells impairs macrophage anti-inflammatory functions and clearance of apoptotic cells. Elife 11, e74443 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tajbakhsh A., et al. , The regulation of efferocytosis signaling pathways and adipose tissue homeostasis in physiological conditions and obesity: Current understanding and treatment options. Obes. Rev. 10, e13487 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Li S., et al. , Defective phagocytosis of apoptotic cells by macrophages in atherosclerotic lesions of ob/ob mice and reversal by a fish oil diet. Circ. Res. 105, 1072–1082 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Japanese ministry of Health Labour and Welfare, Guide of the Medical Treatment of the Noble Coronavirus. ed. 5. https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/0000121431_00111.html (Accessed 30 August 2021).
- 45.Moons K. G. M., et al. , Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 162, W1–W73 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Kanda Y., Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 48, 452–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosoya T., Lung samples from obese mice with or without mouse-adapted SARS-CoV-2 infection. Annotare 2.0. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12618/. Deposited 12 January 2023.
- 48.Hosoya T., Mouse-adapted SARS-CoV-2 infected Lungs from wild type or genetically obese mice. Annotare 2.0. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12617/. Deposited 17 January 2023.
- 49.Hosoya T., Mouse-adapted SARS-CoV-2 infected lungs from lean or obese ob/ob mice. Annotare 2.0. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12623/. Deposited 17 January 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
RNA-Seq data are available in ArrayExpress (E-MTAB-12618 (47), E-MTAB-12617 (48), and E-MTAB-12623 (49)). The other data that support the findings of this study are included in the article and/or SI Appendix.