Significance
We analyze the functional and spatiotemporal properties of 57 hard sweeps inferred in ancient human genomes to reconstruct human evolution during the poorly understood Out of Africa migration. Our analyses reveal a previously unsuspected extended period of genetic adaptation lasting ~30,000 y, potentially in Arabia or surrounding regions, prior to a rapid dispersal across the rest of Eurasia as far as Australia. Functional targets include multiple interacting loci involved in fat storage, neural development, skin physiology, and cilia function, with associations with multiple modern Western diseases. Similar adaptive signatures are also evident in introgressed archaic hominin loci and modern Arctic human groups, indicating that cold environments were a prominent historical selection pressure that potentially facilitated the successful peopling of Eurasia.
Keywords: adaptation, ancient DNA, hard sweeps, human migrations
Abstract
The evolutionarily recent dispersal of anatomically modern humans (AMH) out of Africa (OoA) and across Eurasia provides a unique opportunity to examine the impacts of genetic selection as humans adapted to multiple new environments. Analysis of ancient Eurasian genomic datasets (~1,000 to 45,000 y old) reveals signatures of strong selection, including at least 57 hard sweeps after the initial AMH movement OoA, which have been obscured in modern populations by extensive admixture during the Holocene. The spatiotemporal patterns of these hard sweeps provide a means to reconstruct early AMH population dispersals OoA. We identify a previously unsuspected extended period of genetic adaptation lasting ~30,000 y, potentially in the Arabian Peninsula area, prior to a major Neandertal genetic introgression and subsequent rapid dispersal across Eurasia as far as Australia. Consistent functional targets of selection initiated during this period, which we term the Arabian Standstill, include loci involved in the regulation of fat storage, neural development, skin physiology, and cilia function. Similar adaptive signatures are also evident in introgressed archaic hominin loci and modern Arctic human groups, and we suggest that this signal represents selection for cold adaptation. Surprisingly, many of the candidate selected loci across these groups appear to directly interact and coordinately regulate biological processes, with a number associated with major modern diseases including the ciliopathies, metabolic syndrome, and neurodegenerative disorders. This expands the potential for ancestral human adaptation to directly impact modern diseases, providing a platform for evolutionary medicine.
When anatomically modern human (AMH) populations moved out of Africa (OoA) and across periglacial Eurasia around 50 to 60,000 y ago (1), they encountered a range of environments that were markedly different from their African past. This is likely to have driven selection for new traits important for human survival, as has been observed for more recent episodes of human history with genes involved in malaria resistance (2) and lactose tolerance (3). While such important adaptations might be expected to include hard sweeps, where a new or rare beneficial allele is driven to high frequency by selection, modern human populations globally tend to show few classical genetic signatures of strong selection (4, 5). This has led to the suggestion that most recent human adaptation may have instead been behavioral or involved alternate modes of genetic selection that leave less pronounced signatures in genomes such as “soft” sweeps and polygenic selection (6, 7). However, recent analysis of >1,000 ancient western Eurasian genomes has shown that pronounced phases of population admixture can obscure earlier signatures of hard sweeps from modern populations (8). Here, we analyze ancient sweeps in non-African human groups spanning from the present to ~45 thousand years ago (kilo annum, ka) and compare published archaic hominin and modern human datasets to characterize the historic selection pressures and genetic loci underlying human adaptation to new environments. Understanding and identifying the influence of significant past selective events on the human genome is also an important means to reveal genetic factors driving sensitivity to disease in a modern environmental and cultural context (9).
Analysis of Ancient Human Genomes Support an OoA Origin for Hard Sweeps
To examine the molecular, functional, and temporal characteristics of human loci exhibiting signals of strong selection during the AMH migration OoA, we applied a range of analyses to a dataset comprising more than 1,500 ancient human genomes (including both shotgun-sequenced genomes and high-density single nucleotide polymorphism [SNP] scans) along with comparative modern datasets from global human populations (SI Appendix, Tables S1–S3 and Supplemental Materials 1). We have previously identified and validated a set of 57 high-confidence historical hard sweeps (SI Appendix, Table S4; study-wide false positive rate <11%, see ref. 8) in a dataset of 1,162 ancient west Eurasian genomes (mostly from the early Holocene and Bronze Age period, i.e., ~10–5 ka, SI Appendix, Table S1) grouped into 18 distinct ancient populations based on genetic and archaeological relationships (SI Appendix, Fig. S1, Table S1, and Supplemental Materials 1 and ref. 8). Genomic sequences within each of the 18 populations were aligned and scanned for evidence of hard selective sweeps (e.g., distorted allele frequency patterns and anomalously low diversity) using SweepFinder2 (SF2). Importantly, SF2 provides robust identification of hard sweeps using a dynamic sliding window approach to control for demographic history and population structure (10), and we were able to confirm that these properties also extend to ancient genomic datasets using extensive forward simulations that mimic Eurasian demographic history and ancient genome properties (i.e., missing data, SNP ascertainment, small sample sizes; SI Appendix, Fig. S2 and Supplemental Materials 1, ref. 8).
The 57 hard sweeps are limited to Eurasian populations and are absent from the Yoruba African population (8) and also exhibit no significant overlap with recently reported hard sweeps observed in multiple modern African populations (permutation test accounting for recombination rate: p ~ 0.4; SI Appendix, Supplemental Information 1). Accordingly, we used the SNP frequency differences between the ancient Eurasians and the Yoruba population to determine a set of divergent marker alleles that characterize each sweep haplotype [SI Appendix, Supplemental Materials 2, Datasets S1–S56 (https://doi.org/10.25909/22359865)]. After discarding one hard sweep (LINCO1153; see SI Appendix, Table S4 for sweep labels and metadata) with too few marker SNPs to make robust measurements, we could ascertain 56 sweep haplotypes in ancient and modern human genomes to examine the broad spatiotemporal dynamics of hard sweeps in recent human history (SI Appendix, Supplemental Materials 2).
The 56 ancient Eurasian sweep haplotypes are almost entirely absent from contemporary African populations, but many are present in the genomes of geographically distant individuals whose ancestors are thought to have separated from other OoA migrants shortly after the initial Eurasian dispersal [e.g., Andamanese Onge, Aboriginal Australians, Papuans; (11, 12)] (Fig. 1 and SI Appendix, Figs. S3 and S4 and Table S2). This suggests that the sweeps most likely arose in the founding OoA group following separation from other African populations [genetically estimated ~100 ka; (1)]. Although several sweeps appear to have been at high frequency, none were fixed prior to the subsequent widespread Eurasian dispersal (SI Appendix, Table S2). Further, the temporal haplotype patterns confirm the persistence of the sweep signals through the complex series of population replacement events in Europe between 45 and 12 ka (13, 14), before decreasing markedly in the Holocene during two well-known periods of extensive population admixture [i.e., the initial expansion of Near Eastern early Farmers [~8 to 7 ka] and the invasion of Steppe populations in the early Bronze Age [~5 ka] (15, 16); Fig. 1 and SI Appendix, Figs. S3 and S4]. A separate decrease in the sweep signals can be observed within Iberian data coinciding with the Moorish occupation around 700 AD (SI Appendix, Supplemental Materials 2). The subsequent rebound in sweep signals observed in some European regions (Fig. 1 and SI Appendix, Fig. S3) during this period may reflect local demographic changes rather than ongoing selection; for instance, through the well-documented Middle Holocene resurgence in WHG-related ancestry apparent throughout much of Europe (15, 17–21) (SI Appendix, Supplemental Materials 2).
Fig. 1.
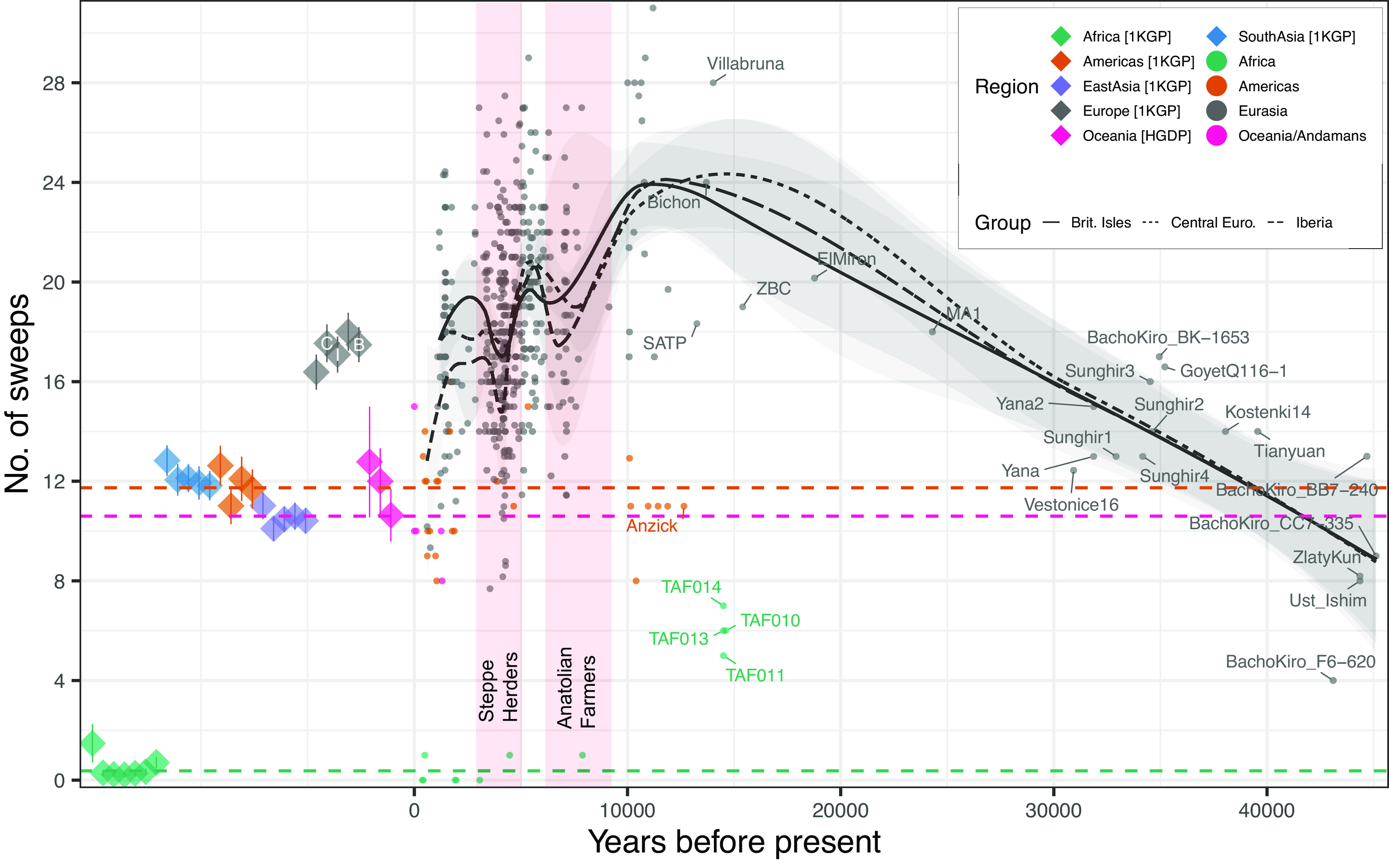
Detection of hard sweep haplotypes in ancient human specimens through time. Presence of each of the 56 sweep haplotypes in ancient samples (circles) and modern populations (diamonds; averages and SEs shown) obtained from the 1,000 Genomes Project (1KGP; with Northern European [CEU; C] British [GBR; B], and Iberian [IBS; I] populations labeled) and Human Genome Diversity Panel (HGDP). By fitting a local regression (LOESS) of sweep count (y axis) as a function of sample age (x axis; samples >12 ka are individually labeled) in ancient West Eurasians, we observe that the number of sweeps per specimen appears to steadily increase through the glacial conditions of the Upper Paleolithic until the start of the warm Holocene period (~12 ka). Population movements resulting from the rapidly warming Holocene temperatures are associated with sharp declines in sweep frequency across Central Europe, Iberia, and the British Isles (see legend), notably during known periods of extensive population admixture associated with the expansion of Early Farmers from the Near East (7 to 8 ka), and Steppe herders (~5 ka) (red shaded panels), with a further marked decline in Iberia around 1 ka associated with Moorish occupation (SI Appendix, Supplemental Materials 2). Mean sweep counts for ancient samples (dashed lines; with Moroccan Iberomaurusian samples [i.e., TAF10-14] omitted) are generally consistent with their modern regional counterparts, and counts are similar for the oldest Eurasians (~45 kya) and Oceanian individuals that likely separated from other Out of Africa migrants by 50 ka (SI Appendix, Supplemental Materials 2). This suggests that Oceanians should provide reasonable proxies for estimating ancestral sweep presence at the time of population separation from Main Eurasian lineages.
Sustained Adaptation to Cold Eurasian Environments
The observation that the majority of the 56 sweep haplotypes are currently segregating at intermediate frequencies in diverse modern non-African populations (SI Appendix, Fig. S5) implies ancient selection pressures acting during the early phase of human expansion beyond Africa at least 50,000 y ago. To explore this pivotal but poorly understood period of human evolution further, we performed a series of qualitative and quantitative analyses to investigate the possible biological functions and selection pressures that underlie the hard sweeps. We applied iSAFE, a statistical method for localizing the adaptive locus within a sweep region (22), to a set of modern European genomes (SI Appendix, Supplemental Materials 3). In 32 of the 56 sweep regions, we were able to identify single protein-encoding driver (i.e., candidate) genes as the putative target of selection, permitting functional analyses (SI Appendix, Tables S5 and S6 A–D and Supplemental Materials 3). Following recommendations for validating positively selected loci in the absence of clearly defined causal mutations (23), we inferred the core functionality of each candidate gene using phenotype information from human carriers of crippling or haploinsufficient alleles or from animal models with loss-of-function mutations in orthologous genes (SI Appendix, Table S6 A–D and Supplemental Materials 3). Surprisingly, the 32 ancient Eurasian candidate genes revealed a pattern of gene classes and biological functions strongly reminiscent of high-confidence candidate genes previously identified as being under population-specific selection in present-day Arctic human populations (58 functionally classified genes from refs. 24–28) or in archaic hominin adaptively introgressed (HAI) loci arising from past mixing events with Neandertal or Denisovan groups (54 functionally classified genes from refs. 29–31) which are also thought to have been cold-adapted (29) (SI Appendix, Tables S1 and S6 A–D and Supplemental Materials 3). This is consistent with suggestions that archaic hominin genetic adaptation to colder periglacial northern environments (29) may have provided beneficial alleles to the early AMH populations dispersing from Africa into those same Eurasian environments (SI Appendix, Supplemental Materials 3), such as the introgressed Denisovan locus impacting body fat and developmental traits now found at high frequencies in modern Greenland Inuit (30).
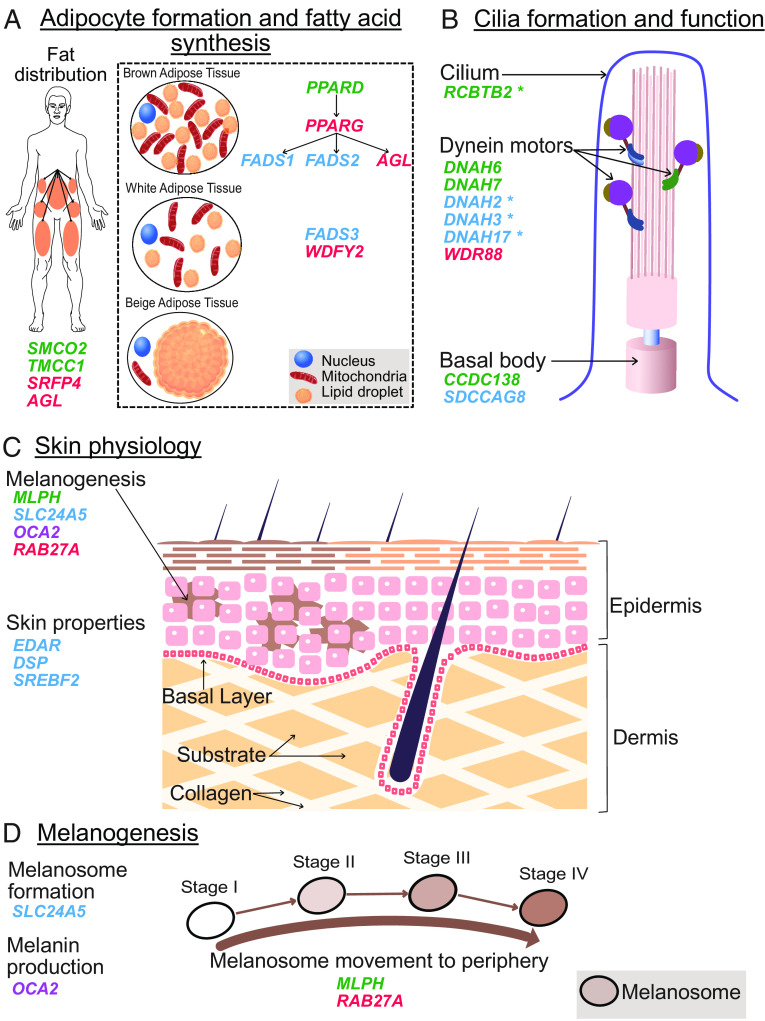
A closer examination of the functions across the candidate selected genes identified in the ancient Eurasian, Arctic human, and archaic hominin AI gene sets revealed surprising layers of concordant biological connectivity, including multiple biological processes known to be involved in human cold adaptation (31) (Fig. 2, Table 1, and SI Appendix, Table S6 A–D and Supplemental Materials 3). For example, roughly one-quarter of the genes in each candidate set are associated with metabolic processes, which include multiple genes that actively regulate fat metabolism, a key metabolic nexus for mammalian cold adaptation (32). This includes three ancient Eurasian genes: PPARD, a metabolically-sensitive transcription factor that regulates fatty acid oxidation for the generation of ATP or heat and is involved in adipogenesis (33); and SMCO and TMCC1 which have been linked to variation in body fat deposition (34, 35). Among the HAI genes, PPARG (a PPAR-family nuclear receptor like PPARD) and WDFY are required for the formation of white and brown adipocytes (33, 36, 37), which provide fuel storage as triglycerides or heat generation from oxidative phosphorylation, respectively, while AGL regulates glycogen metabolism (38), a major source of stored energy. Similarly, within the modern Arctic human candidate selected genes, FADS1, 2, and 3 also regulate fatty acid synthesis (39), while ANGPTL6 has profound impacts on the regulation of body adiposity and insulin sensitivity (40). Remarkably, metabolic loci across all three of the ancient Eurasian, Arctic human, and HAI candidate gene sets are also directly linked in regulatory networks (Fig. 2) as PPARD is a transcription factor regulating the expression of PPARG, which in turn is a transcription factor regulating the expression of FADS1 and FADS2, as well as the HAI metabolism gene AGL (41).
Fig. 2.
Functional concordance across genetic targets of selection (i.e., candidate genes) in ancient Eurasian sweeps (Green), adaptively introgressed archaic hominin [HAI] loci (Red), modern cold-adapted Arctic human groups (Blue), or both HAI loci and Arctic humans (Purple), in genes that: (A), regulate metabolism through adipogenesis, as well as fat synthesis and distribution [arrows indicate gene regulatory networks]; (B) are active in cilia function, particularly formation of the basal body complex and dynein motor complexes; or (C) control skin physiology including the “wooly” phenotype, wound healing, and skin formation, as well as (D) pigmentation through the formation of melanosomes, melanin synthesis within melanosomes, and melanosome transport to the cell periphery. The asterisk denotes putatively adaptive genes identified within the ancient Eurasian sweeps or reported for Arctic human groups that did not satisfy our conservative criteria for inclusion among the respective candidate gene sets. See SI Appendix, Table S6 A–D and Supplemental Materials 3 for detailed gene characteristics and functions.
Table 1.
Biological role of genes inferred as under positive selection (i.e., candidate genes) in ancient Eurasians, cold-adapted modern human groups, or archaic hominin introgressed loci
| Biological impact of candidate gene | Ancient Eurasians n = 32, % | Adaptive archaic hominin introgression n = 54, % | Arctic humans n = 58, % |
|---|---|---|---|
| Neurological | 31 | 35 | 34 |
| Development | 34 | 31 | 28 |
| Metabolism | 28 | 24 | 22 |
| Immunity | 0 | 7 | 16 |
| Reproduction | 6 | 2 | 0 |
| Importance, % | |||
| Lethal phenotype | 25 | ||
| Constrained gene | 50 | ||
| Major physiological impact | 59 | ||
| Molecular function, % | |||
| Membrane proteins | 9 | ||
| Extracellular proteins | 3 | ||
| Intracellular proteins | 88 | ||
| Enzyme | 22 | ||
| Transcription regulator | 9 | ||
| Molecular motor | 6 | ||
| Other | 13 | ||
| Signaling | 38 | ||
| Adapters | 16 | ||
| Molecular complex | 9 | ||
| Guanidine exchange factor | 9 | ||
| Kinase | 3 | ||
Frequency [%] is calculated from the total number of candidate genes annotated for each respective data set (SI Appendix, Table S6 A–D). Key biological impacts (Importance) of the candidate genes identified in ancient Eurasians. Lethal phenotype defined by spontaneous embryonic lethality or premature lethality post-partum in humans or animals; constrained genes identified by LOUEF scores ≤ 0.5; major physiological impact defined as a loss-of-function mutation in human subjects and or from gene targeting studies in animal models which cause premature lethality, physical malformations, or developmental delay. Molecular function of candidate genes defined as membrane proteins (receptors, ion pumps, transporters, tethered proteins), extracellular proteins (secreted or otherwise released), or intracellular proteins (enzymes, transcription regulators, signaling molecules etc.). Signaling molecules were able to be further classified.
Functional Concordance Across Selected Loci Reveals Adaptation Targets Subcellular Machinery
Around a third of the genes in each candidate gene set are associated with developmental processes (Table 1 and SI Appendix, Table S6 A–D), including distinct subsets of genes involved in the developmental formation and function of cilia, skin properties, and DNA damage response (Fig. 2 and SI Appendix, Table S6 A–D and Supplemental Materials 3). Cilia are evolutionarily conserved hair-like cell structures that can function as cellular environmental sensors or provide locomotion (42) but are also important for lung and airway health in cold and arid environments (43). Cilia genes were also found to be overrepresented in a meta-analysis of genetic selection signals in Arctic mammals and multiple human populations from Greenland and Siberia (SI Appendix, Supplemental Materials 3; also see tables S8–S10 in ref. 32). The ancient Eurasian candidate genes DNAH6, DNAH7, and CCDC138—along with DNAH2, DNAH3, and DNAH17 identified in the Greenland Inuit study (25)—all encode molecular structural components of cilia [comprising specific components of the axonemal dynein motor complex that collectively provide the force generation for cilia movement (44)], while the Arctic human-selected gene SDCCAG8 controls cilia development (Fig. 2 and SI Appendix, Table S6 A–D). A further ancient Eurasian gene, RCBTB2, which fell just below the detection cut-off threshold (SI Appendix, Table S5 and Supplemental Materials 3), encodes an intracellular protein also essential for the formation of cilia (45, 46). In addition, the HAI gene WDR88 has a potential cilia function, as its close paralogue DAW1 is involved in building the cilia structures containing DNAH6 and DNAH7 (47).
Genes involved in skin physiological properties and pigmentation emerged as another functional concentration across the three candidate gene sets (Fig. 2). Skin pigmentation is dictated by melanosomes, which produce and transport melanin to intracellular and extracellular sites (48). Within this process, the ancient Eurasian gene MLPH interacts biochemically with the HAI gene RAB27A to shuttle melanin-containing melanosomes to the cellular periphery (49). Melanin synthesis itself requires the “P protein” encoded by the shared HAI and Arctic human gene OCA2 (50), while melanosome formation (i.e., melanogenesis) requires the Arctic-selected gene SLC24A5 (51). Mutations in either MLPH or RAB27A cause the hypopigmentation condition Griscelli syndrome (52, 53), while SLC24A5 and OCA2 variants are associated with albinism in humans (50, 54). In evolutionary terms, these “color” genes are functionally conserved across vertebrates, as polymorphisms in MLPH contribute to coat color in domestic cats (55) while mutations in SLC24A5 and OCA2 result in the “golden” phenotype in zebrafish and cichlids, respectively (53, 56, 57). Selective pressure on pigmentation during the OoA migration has been noted previously and suggested to be a response to UV and or sunlight levels (58). In this regard, it is notable that a number of the Eurasian sweep genes are involved in the DNA damage response (e.g., TP53BP1, GTSE1, and FANCD2) which may also relate to selection due to high UV levels (58).
Evidence for Neurological Adaptation in Eurasian Environments
A further one-third of the candidate genes are associated with neuronal functions (Table 1 and SI Appendix, Table S6 A–D). This was not necessarily expected, but neural tissues play a central role in coordinating environmental information into physiological and behavioral responses necessary to navigate new environments (59), while human cognitive performance is also impaired in cold conditions (60). Intriguingly, eight of the ten ancient Eurasian neuronal genes are associated with severe intellectual disabilities and developmental delay phenotypes in humans (Table 1 and SI Appendix, Table S6 A–D). This is provocative as it has been suggested that the rapid development of advanced sociality and behavior in late Pleistocene African AMH populations during the cold Marine Isotope Stage 4 (MIS4) climatic period from ~75 to 58 ka is likely to have involved selective pressure around cognition (61). Collectively, the neuronal candidate genes highlight fundamental neurological processes of vesicle trafficking, neurite growth, and cerebral cortex formation, suggesting that there may have been selection around the maintenance of environmental perception and cognitive functions in cold environments (SI Appendix, Supplemental Materials 3). In this regard, the ancient Eurasian gene MPP6 is required for nerve myelination (62), which changes in response to environmental cues throughout life and may represent a plastic neural response to environmental challenges, while TRPM2, a candidate selected gene identified in modern Arctic human populations, encodes a nerve receptor that relays temperature information to the central nervous system (63) (SI Appendix, Supplemental Materials 3).
Quantitative Validation of Functional Analyses
The surprising degree of functional concordance observed across the ancient Eurasian, modern Arctic human, and archaic hominin AI candidate loci (Fig. 2) suggests that sustained selection pressures have acted upon a common set of biological pathways over a long period of time, which minimally includes the founding AMH expansion into Eurasia and subsequent Holocene migrations into Arctic environments (24, 25). To further scrutinize the apparent elevated functional concordance across the three candidate gene sets, we used curated protein–protein interactions (PPIs) from the STRING database (64) to test if there are more PPIs between the candidate sets than expected among randomly sampled gene sets of the same size (SI Appendix, Supplemental Materials 3). These tests provide further support for high functional concordance across the three gene sets, with ~15 to ~170% more PPIs observed among genes from different candidate sets than expected (SI Appendix, Fig. S6, Table S7 A and B, and Supplemental Information 3).
A similar excess of PPIs was also apparent among the candidate genes when grouped according to our functional categories, while multiple functionally equivalent GO and biomedical annotations were overrepresented among candidate genes within each functional category (FDR < 0.05 for STRING enrichment tests; SI Appendix, Table S8 and Supplemental Information 3), indicating that these groupings formed biologically coherent units. In contrast, only neurological functions were significant when enrichment tests were performed on the combined set of candidate genes (SI Appendix, Table S8)—possibly because the large number of functionally coherent candidate loci within this category preserved the underlying statistical signal in the presence of additional functionally distinct genes. These results indicate that our analytical approach can discriminate robust fine-scale functional information that may not be detected when applying standard statistical tests to a multifunctional cohort of candidate genes.
The Ancient Hard Sweeps Do Not Appear to Arise from Introgressed Archaic Hominin Sequences
It is notable that 14 of the hard sweeps (~25%) overlap with introgressed archaic hominin DNA tracts that have previously been identified as putative targets of selection in AMH (SI Appendix, Figs. S7 and S8), raising the possibility that some of the 56 sweeps may have been driven by beneficial introgressed Neandertal or Denisovan variants. However, close inspection reveals that most of these overlapping introgressed sequences lie on the periphery of our sweep regions. Indeed, introgressed hominin regions inferred in ancient Eurasian genomes with the recently developed admixfrog method (65) were underrepresented near the peak sweep signal (Fig. 3 and SI Appendix, Figs. S7 and S8 and Supplementary Materials 4). Together, this suggests that the beneficial sweep variants were most likely AMH-derived, which removed introgressed hominin sequences lying near the beneficial variant while bringing linked introgressed sequences to higher frequencies through genetic hitchhiking. Such hitchhiking archaic hominin sequences may be the primary cause of the sizeable overlap between the Eurasian sweep regions and introgressed archaic hominin DNA tracts and could feasibly have produced false positive signals of adaptive introgression using current detection methods.
Fig. 3.
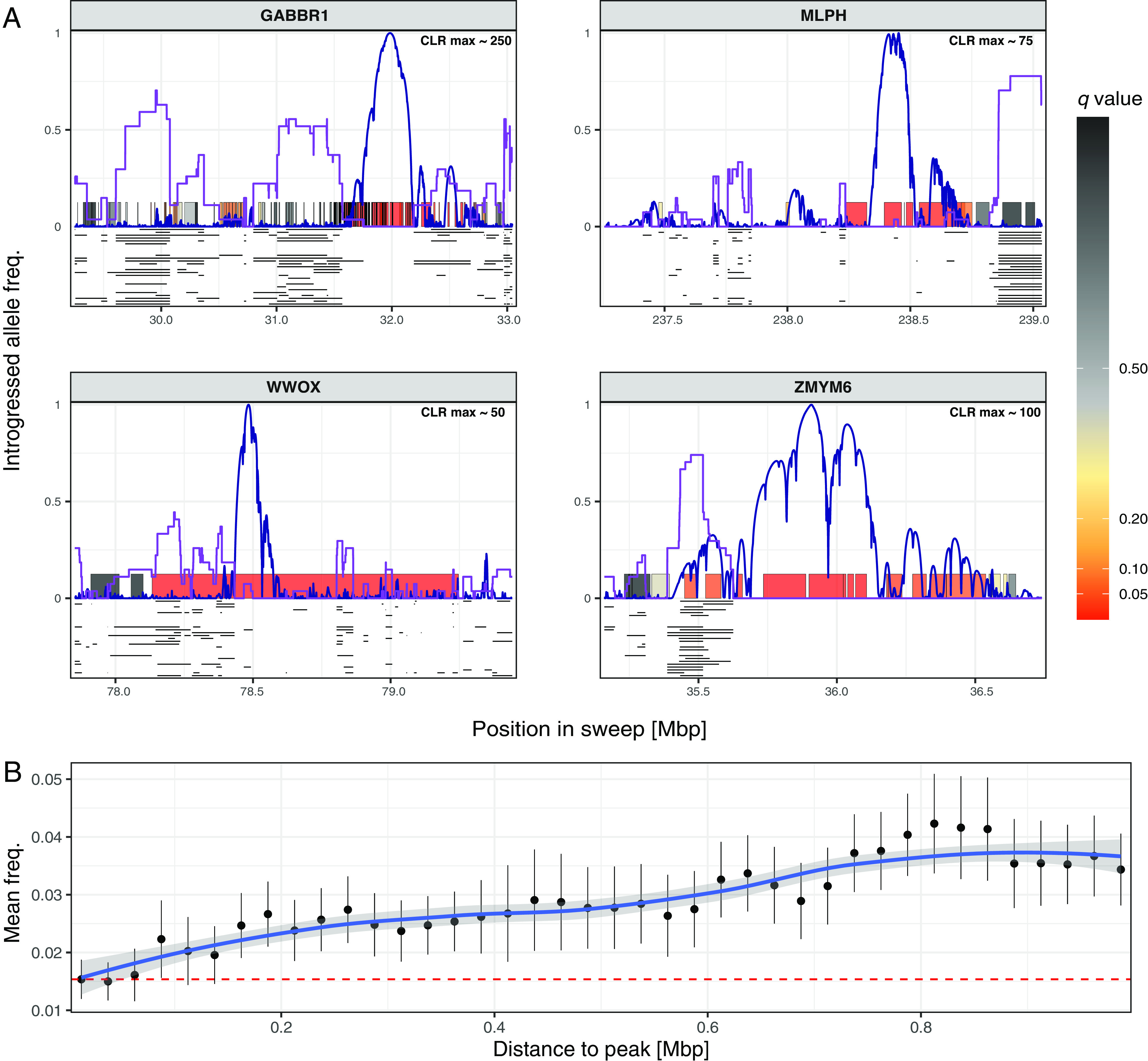
Introgressed archaic hominin loci in the vicinity of ancient hard sweeps. To determine the distribution of introgressed hominin loci around each sweep, we used admixfrog (65) to directly infer these loci in ancient genomes prior to the Holocene admixture events. (A) The inferred loci are shown for each of the 27 Anatolian Early Farmer individuals (black lines) for four sweeps (labeled panels), which includes the only candidate gene that is also found among the HAI candidate genes, WWOX. The frequencies of introgressed loci across each sweep region is indicated by the purple line. For comparison, we also depict the SweepFinder2 CLR scores (blue lines), with the maximum score indicating the most likely location of the underlying causal allele. Each gene in the region is shown as a colored rectangle, with the color indicating the gene score used to identify sweeps (see key). Notably, introgressed loci tend to occur at negligible frequencies beneath the peak CLR score and at higher frequencies when moving further away from the peak (peak CLR scores shown at Top-Right of each panel). (B) When averaging introgressed loci frequencies across all sweeps in successive 25 kb bins (black points, associated lines = mean ± two SEs), we find that these loci become significantly more common >150 kb from the peak than at the peak, consistent with introgressed loci hitchhiking to higher frequencies on a beneficial AMH-derived variant (red dashed line = mean frequency near peak, blue line and shading = best fitting LOESS curve ± two SEs, respectively).
Using Hard Sweep Signals to Reconstruct Paleolithic Human Migrations
The origins and geographic distribution of the hard sweeps provide a distinctive genetic marker to examine early AMH population movements OoA and across Eurasia, which remain poorly understood. The ~2% genomic signal of Neandertal admixture observed in modern non-African populations globally (1, 11, 66) has previously been used to trace the dispersal of a single ancestral OoA population across Eurasia and Island Southeast Asia (ISEA) as far as Australia around 60 to 50 ka (1, 67), but probably as recently as ~54 to 50 ka (SI Appendix, Supplemental Materials 2). The 54 to 50 ka date is in very close agreement with independent molecular clock dating of mitochondrial, Y chromosome, and autosomal DNA, all of which indicate the last common genetic ancestor of global non-African populations existed around 45 to 55 ka (1). In contrast, the genetic separation of the ancestral OoA population from other African populations is estimated to have occurred approximately 100 ka (1)—some 40 to 50 thousand years earlier. The early date of separation aligns with widespread archaeological evidence for the presence of early AMH groups throughout the Arabian Peninsula area (from the Levant to the Arabian Gulf, potentially including southern Iran) associated with discrete climatic wet phases around 125 ka, 100 ka, and 80 ka during MIS 5 (68–73), prior to the prolonged cold arid conditions of MIS 4 (71 to 57 ka). While some archaeological studies have supported the presence of AMH outside the African and Arabian areas before 55 ka, for example in India (74), much of the evidence is of uncertain AMH origin (1, 75) and the oldest uncontested records for AMH presence across Europe [~54 ka; (76)], and Asia and Australia [~50 ka; (75, 77)] closely match the genetic date estimates.
To examine genetic selection during the early stages of AMH dispersal from Africa, we reconstructed the spatiotemporal pattern of the underlying selection pressure(s) by quantifying the presence of the hard sweep haplotypes within moderate- to high-coverage genomic sequences of pre-Holocene Eurasian individuals up to ~45 ka in age, as well as indigenous Oceanic groups, such as Aboriginal Australians, whose genetic ancestry stems from the initial Main Eurasian dispersal and who have remained largely isolated since (SI Appendix, Supplemental Materials 2). Simulations indicate that our detection method is more prone to missing sweep haplotypes that are present in ancient samples (~26% detection rate in sweep heterozygotes) than making false detections (FPR ~6%) (SI Appendix, Figs. S9 and S10 and Supplemental Materials 2). Accordingly, we interpreted the oldest point at which the sweep haplotype was observed, or inferred within the genetically reconstructed Eurasian dispersal process (11), as evidence that the selection pressure was likely to have been present at that time point even if the locus was potentially not yet fixed in all individuals.
Around half of the hard sweeps (31/56) seem to have reached relatively high frequencies prior to the Eurasian dispersal as they are distributed widely across descendant European and Asian ancient and modern populations, as well as distant Oceanic populations in very different selective environments (Fig. 4 and SI Appendix, Table S2). The large number of sweeps inferred in the ancestral OoA group indicates that after separating from other African populations, it had experienced an extended period of both genetic isolation and selection prior to the Eurasian dispersal. Linear regression and population genomic analyses suggest this period of selection originated around 80,000 years ago (respective means ~83 and 79 ka, 95% CIs ~72 to 97 ka and 74 to 91 ka; SI Appendix, Fig. S11 and Supplemental Materials 2). This timing closely matches archaeological evidence of AMH activity across the Arabian Peninsula associated with the last wet climatic period in MIS 5 ~ 80 ka (68, 72, 78), immediately prior to the prolonged cold arid conditions of MIS 4 period (71 to 57 ka) (Fig. 5). From these observations, we infer that the ancestral OoA population underwent a prolonged period of genetic isolation and adaptation outside Africa, likely in the Arabian Peninsula area, potentially from around 100 ka but at least ~80 ka until the subsequent Eurasian dispersal ~54 to 50 ka (Figs. 4 and 5), and term this model the “Arabian Standstill” (SI Appendix, Supplemental Materials 2). The current lack of archaeological evidence for AMH presence in the Arabian Peninsula during the harsh conditions of MIS 4 has led to suggestions that the area was abandoned during this time (70–72). However, the genetic evidence indicates the continuous presence of at least a small AMH population, which may have been geographically constrained to a potential refugia such as the lower Arabian Gulf or Red Sea which are now largely submerged (68, 72) or adjacent areas such as southern Iran where Middle Paleolithic sites such as Boof Cave include MIS 4 dates (73) (SI Appendix, Supplemental Materials 2). Genomic studies have indicated that around the beginning of this period (~80 ka), the ancestral OoA population split into the now-extinct Basal Eurasians (1), which potentially occupied an area around the Arabian Gulf (79), and the Main Eurasian group which went on to admix with Neandertals at the end of the Standstill period, before dispersing globally (Figs. 4 and 5). The relatively large size of the resulting Neandertal genomic content (~2%) in the descendant populations indicates that the effective population size of the admixing Main Eurasian population was probably indeed very small, consistent with the presence of just two mitochondrial lineages (M and N) (SI Appendix, Supplemental Materials 2).
Fig. 4.
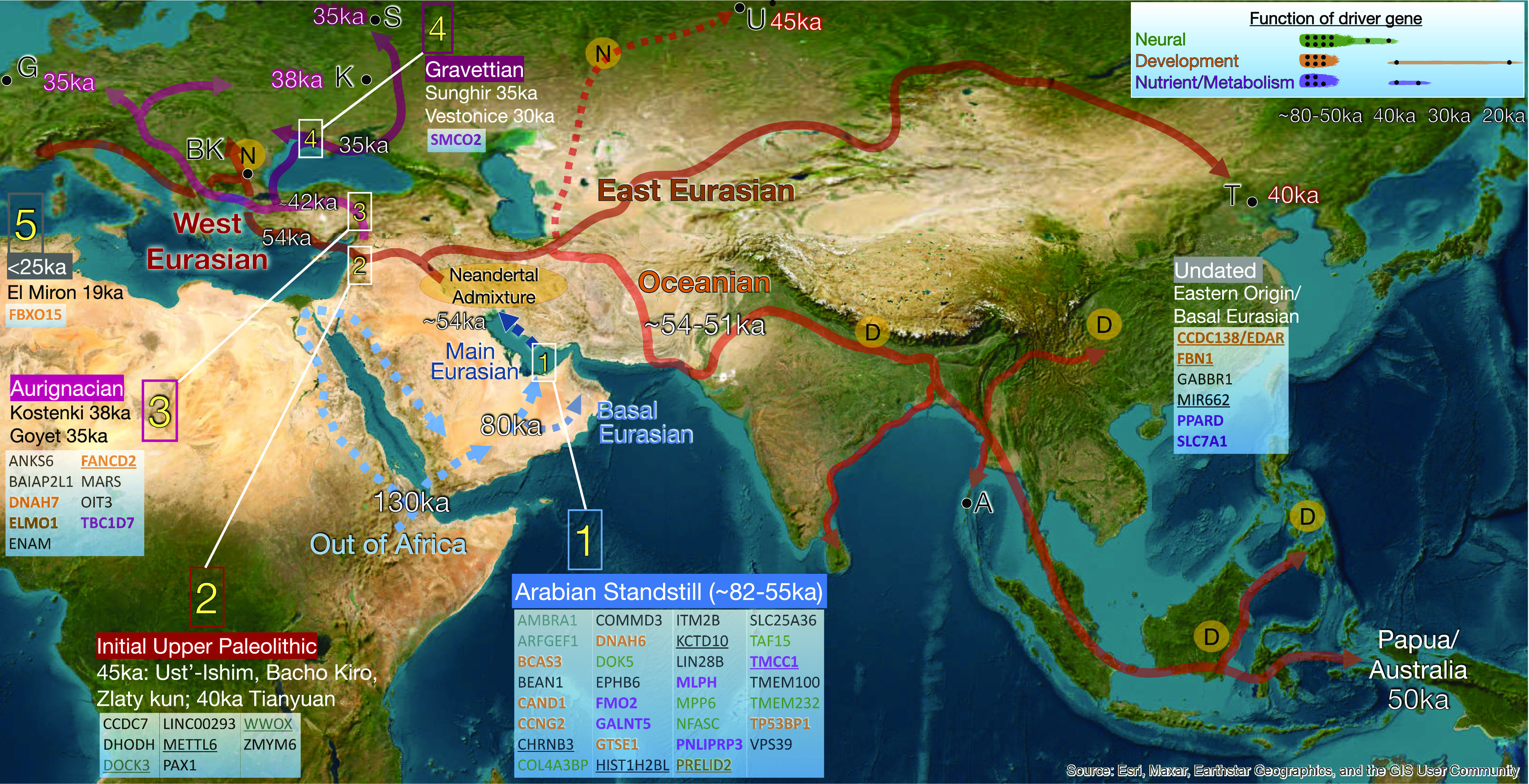
Dispersal of anatomically modern humans out of Africa. A simplified reconstruction of the AMH movement Out of Africa (130 to 80 ka) into the Arabian Peninsula area, and subsequent rapid expansion across the rest of Eurasia (~54 to 51 ka), based on the spatiotemporal distribution of the 56 hard sweeps, archaeological and genetic data, and savannah habitats (SI Appendix, Supplemental Materials 2). Archaeological evidence of AMH movements into the Arabian Peninsula area (potential southern and northern routes are shown, blue dashed lines) during discrete climatic wet phases from ~130 to 80 ka (Fig. 5) closely match the onset of an extended period of genetic isolation of the ancestral Out of Africa population, from around ~100 ka through to ~55 ka, which we term the Arabian Standstill phase. During this period the Main and Basal Eurasian lineages diverged (~80 ka), and the isolated Main Eurasian population experienced a significant phase of genetic selection starting around ~80ka, resulting in at least 31 hard sweeps (box 1). This is depicted in the potential Arabian Gulf Oasis/Iranian refugium (68, 72, 73). Around 54 to 51 ka, shortly after a major phase of Neandertal gene flow (oval), the Main Eurasian lineage rapidly dispersed across Eurasia, arriving in Europe by 54 ka (76), and as far as Australia by 50 ka (SI Appendix, Supplemental Materials 2). Discrete spatiotemporal groupings for the initial appearance of the hard sweeps are shown (boxes 1 to 5, with representative genomes), with an undated group appearing to originate outside the sampling range. Early European movements are simplified into three time bins (boxes 2 to 4) for clarity, marked as red, pink, and magenta. Areas of inferred admixture with archaic hominins are indicated (D; Denisovans: N; Neandertals). Key ancient specimens/sites: U = Ust’-Ishim, T = Tianyuan, K = Kostenki, S = Sunghir, G = Goyet, BK = Bacho Kiro, A = Andaman Islands. The function of identified candidate genes is indicated by color (key, along with an approximate timescale [brown = reproduction]). Underlining indicates sweeps identified as overlapping with adaptively introgressed archaic hominin loci.
Fig. 5.
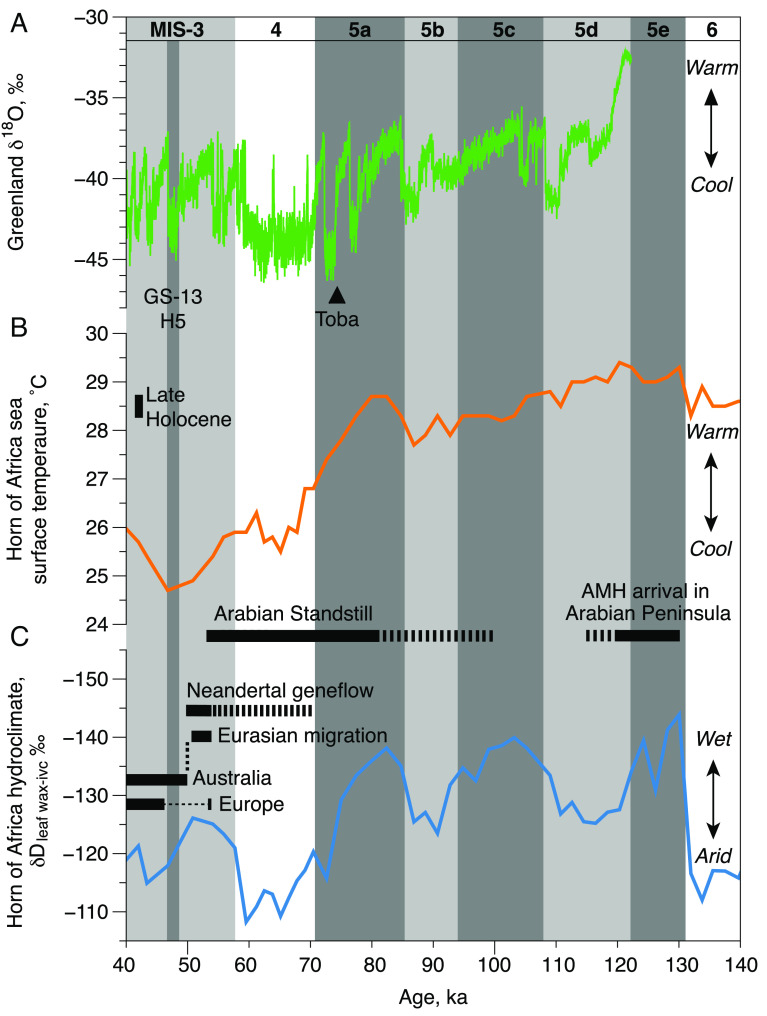
Environmental reconstruction for the Arabian Standstill. The estimated initial genetic isolation ~100 ka (dashed line), and subsequent onset of strong selection in the Arabian Standstill population ~80 ka (solid line) is shown in relation to the environmental conditions on the Arabian Peninsula during the final wet phase of MIS 5e, and severe cold arid conditions from the onset of MIS 4 (~71 ka) through to the warming of MIS 3 (~57 ka). The Arabian Standstill is contemporaneous with a marked arid cooling phase, potentially further exacerbated by events such as the Toba Eruption (~74 ka) and geographic isolation of the Arabian Peninsula area by desert and marine boundaries (SI Appendix, Supplemental Materials 2). The inability to migrate in response to the climatic deterioration is likely to have strengthened selective pressures for cold adaptation during the ~30 ka period of genetic selection. The subsequent expansion northwards, including admixture with local Neandertal populations, occurred with the return of moist, warmer conditions at the start of MIS 3 (~57 ka). (A) NGRIP δ18O record reported on the GICC05 timescale B.P. (CE 1950) (80); Greenland Stadial 13 (GS-13) and Heinrich 5 (H5), and Mt. Toba eruption (81) are shown. (B) Mean annual sea surface temperatures (SSTs) from the Gulf of Aden marine core RC09-166 (82). The Late Holocene (last 2.5 ka) temperature range is shown for comparison on the left-hand side. (C) Hydroclimate changes in northeast Africa reconstructed from stable hydrogen isotopic composition of leaf waxes corrected for ice volume contributions from RC09-166 (82). Horizontal dark gray bars define age ranges for key AMH events across the Arabian Peninsula and the Eurasia dispersal, including the apparently brief initial AMH presence in southern Europe around 54 ka (76), and potential for earlier Neandertal gene flow during the Arabian Standstill (SI Appendix, Supplemental Materials 2).
After the initial 31 hard sweeps associated with the Arabian Standstill, new groups of sweeps appear in a sequence of ancient specimens that record the movement of Main Eurasian populations out of the Arabian area into the novel cold periglacial northern latitudes of Eurasia from ~54 to 51 ka (Fig. 4 and SI Appendix, Table S9 and Supplemental Materials 2). The earliest observations of eight sweeps occur in genomes from the first analyzed European and Asian AMH populations, which are associated with the Initial Upper Paleolithic culture, ~45 to 40 ka; (83). Following the IUP, early West Eurasian individuals dated between 38 and 18 ka subsequently recorded an additional 10 sweeps (Fig. 4 and SI Appendix, Table S9). Overall, the ancient West Eurasian specimens show four distinct temporal groupings of sweeps. Remarkably, each group correlates closely with a major early European archaeological culture (Fig. 4 and SI Appendix, Table S9 and Supplemental Materials 2). For example, after the Initial Upper Paleolithic specimens, nine sweeps are first detected in two specimens (Kostenki14, 38 ka, and GoyetQ116-1, 35 ka) associated with the Aurignacian Culture [~42 to 35 ka; (84)]. Further new sweeps then appear in individuals associated with the subsequent Gravettian Culture [35 to 25 ka; represented by Sunghir 1-4 (35 to 33 ka) and Věstonice16 (31 ka)] and the Magdalenian culture toward the end of the Last Glacial Maximum (represented by the El Mirón specimen, 19 ka) (84, 85). The patterns of temporally grouped sweep signals are consistent with previously recognized genetic population replacements between the IUP, Aurignacian, and Gravettian populations (13) (SI Appendix, Supplemental Materials 2 and Table S3). Interestingly, the currently unexplained replacement events between these three populations occur close in time to two major geomagnetic excursions (Laschamps ~41 ka and Mono Lake ~35 ka) recently suggested to have caused major environmental shifts (86). Lastly, individuals from late-glacial/Epigravettian cultures (e.g., Villabruna and the Azilian Bichon, 14 ka) contain a further six sweeps which appear to have originated earlier in populations to the east (SI Appendix, Supplemental Materials 2) but only spread geographically westward into view of the samples in this study around this time (Fig. 4 and SI Appendix, Table S9).
Early Selection Phase Coincides with Transition to Cold and Arid Climates
Together, the unexpected levels of biological connectivity and discrete temporal patterns of the sweep signals are consistent with a long-term selective pressure around cold adaptation, which appears to have started during the Arabian Standstill and continued through the Eurasian dispersal and archaic hominin introgression phase, into the Late Glacial (ca. 14 to 12 ka, Fig. 1). Candidate loci within each of the functional categories were widely distributed spatiotemporally (Fig. 4, Table 1, and SI Appendix, Tables S2 and S9) including modern Arctic populations, suggesting adaptations acquired during the Arabian Standstill continued to be important for subsequent expansion into new environments. While selective pressure for cold adaptation could be expected in the periglacial conditions across much of late Pleistocene Eurasia, the Arabian Standstill period was also characterized by a pronounced and sustained cooling phase ~71 to 57 ka associated with MIS 4 (Fig. 5), during which the mean annual temperatures in the Arabian Peninsula are estimated to have decreased ~4 °C (likely much greater during the boreal winter) (82). The onset of pronounced cooling and aridity in the Arabian area is close in age to the ~80 ka estimated date for the onset of the earliest sweeps, potentially reflecting the limited geographic ability of the isolated, small Arabian Standstill population to migrate in response to major climatic change, consistent with paleoenvironmental modeling (70) (SI Appendix, Supplemental Materials 2).
Discussion
The large amount of genomic information now available from ancient and modern AMH specimens, along with extensive databases of human functional and disease genetics, effectively makes humans a powerful model system to study evolution within the <0.1 Ma time frame, where the interplay of selective sweeps and population admixture can be observed. By identifying a set of high-confidence adaptive genes, our study has provided an initial view of key functional targets and environmental pressures during this crucial period. The ancient hard sweep signals persisted from the Arabian Standstill phase through the Eurasian dispersal as far as Oceania, indicating that there were no appreciable genetic contributions from any earlier AMH groups along this route, if they existed, similar to previous studies of the Neandertal genome content. The hard sweep signals were eventually diluted in western Eurasia by Holocene population admixture (Fig. 1), potentially involving groups with Basal Eurasian or African ancestry that did not possess the Arabian Standstill sweep haplotypes. The same admixture processes are thought to explain the reduced Neandertal genomic content of European versus Asian populations (1). The Holocene admixture has caused around 80% (41/57) of the hard sweeps we detect to instead be reported in modern European populations as partial sweeps of relatively recent origin (<30 ka) (8). This pattern suggests that hard sweeps may be more common in evolution than current studies have suggested.
The large number and broad function of the selected loci detected in ancient Eurasians raises the possibility that the speed of AMH movement OoA and across Eurasia may have been limited by both climatic cycles and the need for genetic adaptation to new environments (e.g., during the Arabian Standstill), rather than simply by the existing occupation of areas by archaic hominin groups. For example, the Eurasian dispersal moved very rapidly eastward through Asia and down as far as Australia by 50 ka, following a familiar (warmer) savannah ecozone (75, 87), despite the presence of multiple Denisovan and other ISEA hominin groups as well as significant marine barriers through ISEA (75, 87). The contrasting delay before AMH groups successfully colonized Europe around 47 ka, following an apparently unsuccessful initial foray around 54 ka (76), has often been explained by the presence of Neandertal populations in the area (88, 89). However, our data suggest that this delay may also have been associated with a distinct phase of genetic adaptation to the very cold northern environments, first seen as the set of sweeps in the Initial Upper Paleolithic individuals (Fig. 4 and SI Appendix, Tables S2 and S9) who appear in Europe immediately after the extreme cold conditions of Heinrich Event 5 (48 to 47 ka). Such an additional phase of cold adaptation is unlikely to have been necessary for the AMH expansion across the southern Asian savannah and tropical ISEA into Australia.
In comparison, the absence of ancient Eurasian sweep genes that are overtly involved in immune system functioning stands out, especially as introgressed archaic hominin immunogenetic variants appear to have been consistent targets of selection after Neandertal admixture ~54 to 51 ka (90–92). Indeed, recent epidemiological models predict that the successful expansion of AMH beyond the Levant likely required human migrants to adapt to the novel pathogen package carried by local Neandertal groups (89). While further testing is needed to reconcile this apparent discrepancy, these results align with recent research emphasizing the importance of genetic adaptation in facilitating range expansions into novel environments (93, 94) and suggest that multiple modes of selection, including soft sweeps and polygenic selection that were not directly evaluated in the present study, were involved in the peopling of Eurasia.
Many of the ancient Eurasian sweep haplotypes appear at moderate to high frequencies across contemporary non-African populations (Fig. 1 and SI Appendix, Fig. S5), where they might still contribute to differences in fitness and health. Remarkably, haploinsufficiency in over half of the Eurasian candidate genes causes Mendelian disease phenotypes, 25% are associated with premature lethality, while the majority exhibit low frequencies of loss-of-function mutations in human lineages (95) (Table 1 and SI Appendix, Fig. S12, Table S6 A–D, and Supplemental Materials 3). Further, a number of the Eurasian loci that coordinately regulate biological processes with those in the HAI and cold-adapted Arctic human gene sets are associated with major modern diseases including the ciliopathies (e.g., DNAH6), a recently recognized severe disease class that includes sensory, immunological, reproductive as well as developmental abnormalities (42); metabolic syndrome, including obesity and diabetes (e.g., PPARD); and neurodegenerative diseases including dementia and autism (e.g., TAF15, AMBRA1), all of which represent increasing or significant medical maladies in present-day populations (96, 97). This raises the potential that ancestral selection during the OoA expansion, focused around adaptation to novel cold environments, has established haplotypes that under modern conditions are associated with disease phenotypes.
The pattern of hard sweeps in recent human history provides an unexpected view of evolution, with the majority of gene targets concentrated around evolutionarily conserved intracellular machinery, dominated by enzymes, components of intracellular protein signaling complexes, and transcription regulators, rather than cell surface receptors and ligands, which might seem more obviously associated with sensing and responding to new environments (Table 1). The surprising prevalence of hard sweep signals may partly be explained by selection acting on the reduced haplotype diversity likely for these functionally important genes due to the prolonged small population sizes associated with the Arabian Standstill period (8, 98). Overall, these signals suggest that strong and persistent selective pressures have acted across sets of human genes that coordinate specific physiological functions, and can be observed from the early OoA phase through to modern Arctic populations.
Methods
Data Processing and Sweep Detection Pipeline (SI Appendix, Supplemental Materials 1).
A full description of the sweep detection pipeline—including ancient sample characterization, standardized genomic data processing, selection scan methodology, and various pipeline validation and performance measures—has been reported in ref. 8. We summarize key technical information for the population assignment (SI Appendix, section 1.1), data processing (SI Appendix, section 1.3), and the analytical pipeline (SI Appendix, sections 1.4–1.6) procedures in SI Appendix, section 1 and include new methodological information from tests on the sensitivity of sweep detection on SNP missingness (SI Appendix, section 1.7) and statistical comparisons with sweep signals from modern African populations (SI Appendix, section 1.8).
Inferring Selection Onset and Sweep Dynamics (SI Appendix, Supplemental Materials 2).
To track the occurence of the sweep haplotypes through time, we developed a heuristic method to determine a set of marker SNP alleles for each of the 57 sweeps and then used these markers to quantify the presence of the inferred sweep haplotype in modern and ancient human samples. Briefly, for each sweep, we evaluated SNP-wise Fst scores for all pairs of an African population (YRI) and ancient Eurasian populations with significant sweep signals. Up to 30 marker SNPs were determined for each sweep that satisfied threshold Fst scores obtained from background SNPs, with the most probable selected alleles at each marker SNP being combined to generate the putative underlying selected haplotype (SI Appendix, section 2.1). Sweep LINC01153 (SI Appendix, Table S4) only had 3 SNPs that passed all marker filtering criteria and was subsequently excluded from further marker-based analyses. Full discussion of the statistical properties of our sweep haplotype detection analyses is provided in SI Appendix, section 2.5 (also see SI Appendix, Figs. S9 and S10).
To evaluate sweep dynamics during the European Holocene period (SI Appendix, section 2.3), we quantified sweep presence in a dataset that combined 32 Paleolithic samples dated between 45 and 10 ka with a set of 424 Holocene-era (<10 ka) European samples sourced from three geographically distinct locations: the British Isles; Central Europe; and the Iberian Peninsula (SI Appendix, Table S3). Sweep haplotype dynamics were inferred by fitting a LOESS curve to the sweep presence data with respect to the radio-carbon dated sample age, with separate analyses performed for the three geographical regions after combining these samples with those from the Upper Paleolithic period (Fig. 1 and SI Appendix, Figs. S3 and S4 and section 2.3).
To ascertain the temporal origins of the sweeps (SI Appendix, section 2.4), we scanned for the presence of all 56 selected haplotypes among a set of 22 high-quality ancient and modern human genomes, which included multiple genomes from Eurasian Paleolithic samples and subsequent early Western HunterGatherers, along with genomes from the modern descendants of human lineages that are thought to have separated from the Eurasian dispersal at an early stage (SI Appendix, Fig. S11). Selection onset times were estimated using two separate statistical procedures: 1) the x-intercept from a standard linear model that regressed total sweep presence for each sample upon the reported specimen date and 2) the best fitting trajectory of aggregate sweep presence per sample through time inferred from population genetic simulations of hard selective sweeps under a plausible demographic model that accounts for sweep detection properties and the empirical characteristics of our samples.
Functional Analysis of Candidate Genes (SI Appendix, Supplemental Materials 3).
In addition to seven genes originating from sweeps that contained only a single gene, 25 putative target (i.e., candidate) genes were identified from the remaining sweep regions by running iSAFE on modern genomes from individuals with Northern and Western European ancestry (i.e., Utah residents from the Thousand Genomes Project [CEU]) and searching for cases where a single gene contained more than 50% of the top 20 iSAFE SNPs, and no other gene had more than 20% of these top 20 SNPs, in each sweep region (SI Appendix, section 3.1).
These genes were functionally classified by performing a systematic review of human disease literature, as well as animal and cell knockout phenotypic datasets available on public databases, and through utilization of available online bioinformatic functional annotation tools (SI Appendix, section 3.4). Similar classifications were also performed for comparative sets of candidate genes from modern Arctic human populations and HAI loci obtained from previously published studies (see SI Appendix, section 3.2 for full list of studies and candidate gene selection criteria). Discussion of the functional evaluations, including evidence for biological coordination across the three gene sets and all relevant literature, are provided in SI Appendix, sections 3.4–3.6.
We performed a series of quantitative analyses to assess the robustness of our functional classifications and levels of biological coordination across candidate gene sets. We assessed the essentiality of the candidate genes through statistical tests of multiple loss-of-function mutation metrics published in ref. 95 (SI Appendix, section 3.3). Evaluation of coherent biological functionality among the candidate gene sets was performed using standard enrichment tests of predefined functional and biomedical annotations on the STRING database (64) (SI Appendix, section 3.8). We also used PPI metrics from the STRING database to formally evaluate hypotheses concerning the degree of biological connectivity among the three candidate gene sets (SI Appendix, section 3.7).
Evaluating the Hominin Source of Sweep Driver Alleles (SI Appendix, Supplemental Materials 4).
To examine if the hard sweeps were driven by introgressed hominin alleles, we used admixfrog (65, 83) to infer introgressed hominin sequences within the combined set of genomes from the Anatolian EF population, which have the largest set of shotgun-sequenced genomes among the ancient Eurasian populations we had surveyed for hard sweeps. Hominin allele frequencies were calculated as the proportion of inferred introgressed sequences at each nucleotide position up to 1 Mb either side of the most likely target site at each sweep (taken as the position of the peak SF2 signal). For each sweep, hominin allele frequencies were averaged in successive 25 kb bins and SEs were compared to values at the putative target site (Fig. 3).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
The use of the term “Arabian Gulf” rather than the alternative “Persian Gulf” follows recent studies and does not reflect the political view of any author. We thank Murray Cox, Angad Johar, Jeremy Austin, Chris Auricht, and many colleagues who have provided valuable support and suggestions; This project was supported by the ARC; R.T. (DE190101069), Y.S. (DP190103705), A.C. (FL140100260), C.D.H. (DE180100883), N.B. (CE140100049), and NHMRC; S.T.G. (GNT1189235; GNT1140691).
Author contributions
R.T., Y.S., C.D.H., S.T.G., and A.C. designed research; R.T., Y.S., C.D.H., N.B., C.S.M.T., S.T.G., and A.C. performed research; R.T., Y.S., C.D.H., N.B., C.S.M.T., S.T.G., and A.C. analyzed data; and R.T., Y.S., C.D.H., S.T.G., and A.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Raymond Tobler, Email: ray.tobler@anu.edu.au.
Yassine Souilmi, Email: yassine.souilmi@adelaide.edu.au.
Shane T. Grey, Email: s.grey@garvan.org.au.
Alan Cooper, Email: alanjcooper42@gmail.com.
Data, Materials, and Software Availability
Datasets 1-56 and Dataset 57 have been deposited in adelaide.figshare.com and are accessible via https://doi.org/10.25909/22359865 (99) and https://doi.org/10.25909/22359874 (100), respectively. All genetic datasets used in this study are publicly available with summaries and links provided in the SI Appendix.
Supporting Information
References
- 1.Bergström A., Stringer C., Hajdinjak M., Scerri E. M. L., Skoglund P., Origins of modern human ancestry. Nature 590, 229–237 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Hamblin M. T., Thompson E. E., Di Rienzo A., Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 70, 369–383 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bersaglieri T., et al. , Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74, 1111–1120 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathieson I., Human adaptation over the past 40,000 years. Curr. Opin. Genet. Dev. 62, 97–104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez R. D., et al. , Classic selective sweeps were rare in recent human evolution. Science 331, 920–924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrider D. R., Kern A. D., Soft sweeps are the dominant mode of adaptation in the human genome. Mol. Biol. Evol. 34, 1863–1877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritchard J. K., Pickrell J. K., Coop G., The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20, R208–R215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Souilmi Y., et al. , Admixture has obscured signals of historical hard sweeps in humans. Nat. Ecol. Evol. 6, 2003–2015 (2022), 10.1038/s41559-022-01914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benton M. L., et al. , The influence of evolutionary history on human health and disease. Nat. Rev. Genet. 22, 269–283 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber C. D., DeGiorgio M., Hellmann I., Nielsen R., Detecting recent selective sweeps while controlling for mutation rate and background selection. Mol. Ecol. 25, 142–156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipson M., Reich D., A working model of the deep relationships of diverse modern human genetic lineages outside of Africa. Mol. Biol. Evol. 34, 889–902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang M. A., A genetic history of migration, diversification, and admixture in Asia. Hum Popul Genet Genom. 2, 0001 (2022). [Google Scholar]
- 13.Fu Q., et al. , The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazaridis I., The evolutionary history of human populations in Europe. Curr. Opin. Genet. Dev. 53, 21–27 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Haak W., et al. , Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazaridis I., et al. , Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olalde I., et al. , The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittnik A., et al. , The genetic prehistory of the Baltic Sea region. Nat. Commun. 9, 442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Günther T., et al. , Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. U.S.A. 112, 11917–11922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes D. M., et al. , A genomic neolithic time transect of hunter-farmer admixture in central Poland. Sci. Rep. 8, 14879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathieson I., et al. , The genomic history of southeastern Europe. Nature 555, 197–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbari A., et al. , Identifying the favored mutation in a positive selective sweep. Nat. Methods 15, 279–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szpak M., Xue Y., Ayub Q., Tyler-Smith C., How well do we understand the basis of classic selective sweeps in humans? FEBS Lett. 593, 1431–1448 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Cardona A., et al. , Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS One 9, e98076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fumagalli M., et al. , Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349, 1343–1347 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Zhou S., et al. , Genetic architecture and adaptations of Nunavik Inuit. Proc. Natl. Acad. Sci. U.S.A. 116, 16012–16017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds A. W., et al. , Comparing signals of natural selection between three Indigenous North American populations. Proc. Natl. Acad. Sci. U.S.A. 116, 9312–9317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khrunin A. V., Khvorykh G. V., Fedorov A. N., Limborska S. A., Genomic landscape of the signals of positive natural selection in populations of Northern Eurasia: A view from Northern Russia. PLoS One 15, e0228778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gower G., I. Picazo P., Fumagalli M., Racimo F., Detecting adaptive introgression in human evolution using convolutional neural networks eLife 10, e64669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gittelman R. M., et al., Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr. Biol 26, 3375–3382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Setter D., et al., VolcanoFinder: Genomic scans for adaptive introgression. PLOS Genetics 16, e1008867 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yudin N. S., Larkin D. M., Ignatieva E. V., A compendium and functional characterization of mammalian genes involved in adaptation to Arctic or Antarctic environments. BMC Genet. 18, 111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans R. M., Barish G. D., Wang Y.-X., PPARs and the complex journey to obesity. Nat. Med. 10, 355–361 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Yoneyama S., et al. , Gene-centric meta-analyses for central adiposity traits in up to 57 412 individuals of European descent confirm known loci and reveal several novel associations. Hum. Mol. Genet. 23, 2498–2510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma H., et al. , Identifying selection signatures for backfat thickness in Yorkshire Pigs highlights new regions affecting fat metabolism. Genes 10, 254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barak Y., et al. , PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell 4, 585–595 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Fritzius T., Moelling K., Akt- and Foxo1-interacting WD-repeat-FYVE protein promotes adipogenesis. EMBO J. 27, 1399–1410 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen J., et al. , Mutations in exon 3 of the glycogen debranching enzyme gene are associated with glycogen storage disease type III that is differentially expressed in liver and muscle. J. Clin. Invest. 98, 352–357 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H., Park W. J., Unsaturated fatty acids, desaturases, and human health. J. Med. Food 17, 189–197 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Oike Y., et al. , Angiopoietin-related growth factor antagonizes obesity and insulin resistance. Nat. Med. 11, 400–408 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Fang L., et al. , PPARgene: A database of experimentally verified and computationally predicted PPAR target genes. PPAR Res. 2016, 6042162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiter J. F., Leroux M. R., Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milla C. E., The evolving spectrum of ciliopathies and respiratory disease. Curr. Opin. Pediatr. 28, 339–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canty J. T., Tan R., Kusakci E., Fernandes J., Yildiz A., Structure and mechanics of dynein motors. Annu. Rev. Biophys. 50, 549–574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L., et al. , RC/BTB2 is essential for formation of primary cilia in mammalian cells. Cytoskeleton 72, 171–181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stubbs J. L., Vladar E. K., Axelrod J. D., Kintner C., Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 14, 140–147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lesko S. L., Rouhana L., Dynein assembly factor with WD repeat domains 1 (DAW1) is required for the function of motile cilia in the planarian Schmidtea mediterranea. Dev. Growth Differ. 62, 423–437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohbayashi N., Fukuda M., Recent advances in understanding the molecular basis of melanogenesis in melanocytes. F1000Res. 9, 608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hume A. N., Ushakov D. S., Tarafder A. K., Ferenczi M. A., Seabra M. C., Rab27a and MyoVa are the primary Mlph interactors regulating melanosome transport in melanocytes. J. Cell Sci. 120, 3111–3122 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Crawford N. G., et al. , Loci associated with skin pigmentation identified in African populations. Science 358, eaan8433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yousaf S., et al. , Molecular characterization of SLC24A5 variants and evaluation of Nitisinone treatment efficacy in a zebrafish model of OCA6. Pigment Cell Melanoma Res. 33, 556–565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ménasché G., et al. , Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1). J. Clin. Invest. 112, 450–456 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aksu G., Kütükçüler N., Genel F., Vergin C., Omowaire B., Griscelli syndrome without hemophagocytosis in an eleven-year-old girl: Expanding the phenotypic spectrum of Rab27A mutations in humans. Am. J. Med. Genet. A 116A, 329–333 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Lee S. T., et al. , Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader-Willi syndrome plus albinism. N. Engl. J. Med. 330, 529–534 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Ishida Y., et al. , A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics 88, 698–705 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Lamason R. L., et al. , SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Kratochwil C. F., Urban S., Meyer A., Genome of the Malawi golden cichlid fish (Melanochromis auratus) reveals exon loss of oca2 in an amelanistic morph. Pigment Cell Melanoma Res. 32, 719–723 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Jablonski N. G., Chaplin G., Colloquium paper: Human skin pigmentation as an adaptation to UV radiation. Proc. Natl. Acad. Sci. U.S.A. 107 Suppl 2, 8962–8968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corbetta M., Patel G., Shulman G. L., The reorienting system of the human brain: From environment to theory of mind. Neuron 58, 306–324 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mäkinen T. M., Human cold exposure, adaptation, and performance in high latitude environments. Am. J. Hum. Biol. 19, 155–164 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Ambrose S. H., "Chronological calibration of Late Pleistocene modern human dispersals, climate change and archaeology with geochemical isochrons" in Modern Human Origins and Dispersal, Yonatan Sahle, et al., Eds. (Kerns Verlag, Tübingen, 2017), pp. 171–213. [Google Scholar]
- 62.Saitoh Y., Kamijo A., Yamauchi J., Sakamoto T., Terada N., The membrane palmitoylated protein, MPP6, is involved in myelin formation in the mouse peripheral nervous system. Histochem Cell Biol 151, 385–394 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Tan C. -H., McNaughton P. A., The TRPM2 ion channel is required for sensitivity to warmth. Nature 536, 460–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szklarczyk D., et al. , The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peter B. M., 100,000 years of gene flow between Neandertals and Denisovans in the Altai mountains. bioRxiv [Preprint] (2020). 10.1101/2020.03.13.990523 (Accessed 1 April 2021). [DOI]
- 66.Bergström A., et al. , Insights into human genetic variation and population history from 929 diverse genomes. Science 367, 674986 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skoglund P., Mathieson I., Ancient genomics of modern humans: The first decade. Annu. Rev. Genomics Hum. Genet. 19, 381–404 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Armitage S. J., et al. , The Southern route “Out of Africa”: Evidence for an early expansion of modern humans into Arabia. Science 331, 453–456 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Stewart M., et al. , Human footprints provide snapshot of last interglacial ecology in the Arabian interior. Sci. Adv. 6, eaba8940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beyer R. M., Krapp M., Eriksson A., Manica A., Climatic windows for human migration out of Africa in the past 300,000 years. Nat. Commun. 12, 4889 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Groucutt H. S., et al. , Multiple hominin dispersals into Southwest Asia over the past 400,000 years. Nature 597, 376–380 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rose J. I., An Introduction to Human Prehistory in Arabia: The Lost World of the Southern Crescent (Springer International Publishing, 2022), (December 1, 2022). [Google Scholar]
- 73.Shoaee M. J., Vahdati Nasab H., Petraglia M. D., The Paleolithic of the Iranian Plateau: Hominin occupation history and implications for human dispersals across southern Asia. J. Anthropol. Archaeol. 62, 101292 (2021). [Google Scholar]
- 74.Clarkson C., et al. , Human occupation of northern India spans the Toba super-eruption ~74,000 years ago. Nat. Commun. 11, 961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Connell J. F., et al. , When did Homo sapiens first reach Southeast Asia and Sahul? Proc. Natl. Acad. Sci. U.S.A. 115, 8482–8490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slimak L., et al. , Modern human incursion into Neanderthal territories 54,000 years ago at Mandrin, France. Sci. Adv. 8, eabj9496 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun X.-F., et al. , Ancient DNA and multimethod dating confirm the late arrival of anatomically modern humans in southern China. Proc. Natl. Acad. Sci. U.S.A. 118, e2019158118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petraglia M. D., Breeze P. S., Groucutt H. S., "Blue Arabia, Green Arabia: Examining human colonisation and dispersal models" in Geological Setting, Palaeoenvironment and Archaeology of the Red Sea, Rasul N. M. A., Stewart I. C. F., Eds. (Springer International Publishing, 2019), pp. 675–683. [Google Scholar]
- 79.Ferreira J. C., et al. , Projecting ancient ancestry in modern-day Arabians and Iranians: A key role of the past exposed Arabo-Persian Gulf on human migrations. Genome Biol. Evol. 13, evab194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rasmussen S. O., et al. , A stratigraphic framework for abrupt climatic changes during the Last Glacial period based on three synchronized Greenland ice-core records: Refining and extending the INTIMATE event stratigraphy. Quat. Sci. Rev. 106, 14–28 (2014). [Google Scholar]
- 81.Williams M. A. J., et al. , Reply to the comment on “Environmental impact of the 73ka Toba super-eruption in South Asia” by Williams M. A. J., Ambrose S. H., van der Kaars S., Ruehlemann C., U. Chattopadhyaya, Pal J., Chauhan P. R. [Palaeogeography, Palaeoclimatology, Palaeoecology 284 (2009) 295–314]. Palaeogeogr. Palaeoclimatol. Palaeoecol. 296, 204–211 (2010). [Google Scholar]
- 82.Tierney J. E., deMenocal P. B., Zander P. D., A climatic context for the out-of-Africa migration. Geology 45, 1023–1026 (2017). [Google Scholar]
- 83.Hajdinjak M., et al. , Initial upper palaeolithic humans in Europe had recent neanderthal ancestry. Nature 592, 253–257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Higham T., et al. , Precision dating of the Palaeolithic: A new radiocarbon chronology for the Abri Pataud (France), a key Aurignacian sequence. J. Hum. Evol. 61, 549–563 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Banks W. E., et al. , An application of hierarchical Bayesian modeling to better constrain the chronologies of Upper Paleolithic archaeological cultures in France between ca. 32000–21000 calibrated years before present. Quat. Sci. Rev. 220, 188–214 (2019). [Google Scholar]
- 86.Cooper A., et al. , A global environmental crisis 42,000 years ago. Science 371, 811–818 (2021). [DOI] [PubMed] [Google Scholar]
- 87.Teixeira J. C., Cooper A., Using hominin introgression to trace modern human dispersals. Proc. Natl. Acad. Sci. U.S.A. 116, 15327–15332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hublin J.-J., et al. , Initial upper palaeolithic homo sapiens from Bacho Kiro Cave, Bulgaria. Nature 581, 299–302 (2020). [DOI] [PubMed] [Google Scholar]
- 89.Greenbaum G., et al. , Disease transmission and introgression can explain the long-lasting contact zone of modern humans and Neanderthals. Nat. Commun. 10, 5003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zammit N. W., et al. , Denisovan, modern human and mouse TNFAIP3 alleles tune A20 phosphorylation and immunity. Nat. Immunol. 20, 1299–1310 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Gittelman R. M., et al. , Archaic hominin admixture facilitated adaptation to Out-of-Africa environments. Curr. Biol. 26, 3375–3382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Enard D., Petrov D. A., Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell 175, 360–371.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Szűcs M., et al. , Rapid adaptive evolution in novel environments acts as an architect of population range expansion. Proc. Natl. Acad. Sci. U.S.A. 114, 13501–13506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miller T. E. X., et al. , Eco-evolutionary dynamics of range expansion. Ecology 101, e03139 (2020). [DOI] [PubMed] [Google Scholar]
- 95.Karczewski K. J., et al. , The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saklayen M. G., The global epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 20, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hebert L. E., Weuve J., Scherr P. A., Evans D. A., Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 80, 1778–1783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hermisson J., Pennings P. S., Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics 169, 2335–2352 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tobler R., Souilmi Y., D. Huber C., Supplemental Datasets S1-S56. Figshare. 10.25909/22359865. Deposited 30 March 2023. [DOI]
- 100.Tobler R., Souilmi Y., D. Huber C., Supplemental Dataset S57. Figshare. 10.25909/22359874. Deposited 30 March 2023. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Datasets 1-56 and Dataset 57 have been deposited in adelaide.figshare.com and are accessible via https://doi.org/10.25909/22359865 (99) and https://doi.org/10.25909/22359874 (100), respectively. All genetic datasets used in this study are publicly available with summaries and links provided in the SI Appendix.