Significance
There is an unmet need for strategies that uncover the roles of human lectins in modulating host–microbe interactions in complex physiological environments such as the mouth. We have developed a microbial glycan analysis probe (mGAP) from a predominant oral cavity lectin zymogen granule protein 16B (ZG16B) and applied the probe to interrogate the interactions of ZG16B. ZG16B binds to commensal bacteria including Streptococcus vestibularis through cell surface glycoconjugates and regulates its growth. ZG16B also recruits the salivary mucin MUC7 and induces the aggregation of S. vestibularis. Our findings suggest that ZG16B assists in maintaining homeostasis in the oral microbiome by capturing selective commensal bacteria and preventing overgrowth by a mucin-assisted clearance mechanism.
Keywords: human soluble lectins, oral microbiota, microbial glycans, glycan analysis probe, cell wall polysaccharides
Abstract
The oral microbiome is critical to human health and disease, yet the role that host salivary proteins play in maintaining oral health is unclear. A highly expressed gene in human salivary glands encodes the lectin zymogen granule protein 16 homolog B (ZG16B). Despite the abundance of this protein, its interaction partners in the oral microbiome are unknown. ZG16B possesses a lectin fold, but whether it binds carbohydrates is unclear. We postulated that ZG16B would bind microbial glycans to mediate recognition of oral microbes. To this end, we developed a microbial glycan analysis probe (mGAP) strategy based on conjugating the recombinant protein to fluorescent or biotin reporter functionality. Applying the ZG16B-mGAP to dental plaque isolates revealed that ZG16B predominantly binds to a limited set of oral microbes, including Streptococcus mitis, Gemella haemolysans, and, most prominently, Streptococcus vestibularis. S. vestibularis is a commensal bacterium widely distributed in healthy individuals. ZG16B binds to S. vestibularis through the cell wall polysaccharides attached to the peptidoglycan, indicating that the protein is a lectin. ZG16B slows the growth of S. vestibularis with no cytotoxicity, suggesting that it regulates S. vestibularis abundance. The mGAP probes also revealed that ZG16B interacts with the salivary mucin MUC7. Analysis of S. vestibularis and MUC7 with ZG16B using super-resolution microscopy supports ternary complex formation that can promote microbe clustering. Together, our data suggest that ZG16B influences the compositional balance of the oral microbiome by capturing commensal microbes and regulating their growth using a mucin-assisted clearance mechanism.
The oral cavity, a primary site for host–microbe interactions, engages numerous factors to promote the interplay between the host and the resident microbiome. Like the gut, the oral cavity harbors complex microbiota. The complexity of the oral microbiome composition in each individual arises from the unique ecological niches offered by each surface in the oral cavity. As the microbial population plays an active role in maintaining the physiology, nutrient availability, and defense mechanisms supporting oral health, it is important to understand the molecular details of the interactions in the oral cavity required for sorting and screening the complex oral microbiome (1, 2).
Soluble lectins, the carbohydrate-binding proteins produced by host cells, are essential components of innate immune defense against microorganisms (3–5). Lectins recognize and distinguish bacterial cell wall glycoconjugates uniquely associated with different microbial species. The majority of the studies on lectin–microbe interactions conducted to date have used commercially available plant lectins (6, 7). However, the microbe-binding properties of the human soluble lectins, including the C-type lectins, galectins, intelectins, and L-type lectins (3, 8, 9), have not been adequately investigated due to the paucity of suitable probes. Understanding the ligand specificity and recognition properties of human lectins is extremely challenging due to the complexity of microbial glycomes (10), the low binding affinity of small oligosaccharides and monosaccharides (Kd ~ mM to µM range) for the lectins (11, 12), and limited availability of the lectins.
An example of an understudied putative human lectin is zymogen granule protein 16 homolog B (ZG16B). ZG16B is a monomeric salivary protein. The gene encoding ZG16B is one of the most highly expressed in the oral cavity, with correspondingly high levels of protein secreted from the serous and seromucous acinar cells in the submandibular and sublingual glands (13, 14). The protein is reported to bind to the oral microbes Staphylococcus aureus (15) and Streptococcus mutans (16) and, via this binding, is proposed to regulate bacterial biofilm formation on oral surfaces. However, the mechanism by which ZG16B recognizes microbes and its microbial binding specificity is unknown. No lectin-like function has been described for this protein (13, 14). Still, ZG16B adopts a beta-prism fold structure (17) that is similar to the mannose-binding jacalin-related lectin BanLec (18). However, the putative sugar-binding peptide motif (GXXXD) in the binding loop of BanLec is altered to the QLLGIK sequence in ZG16B, replacing the conserved aspartic acid with Lys177 (17). ZG16B also features a positively charged patch comprising Lys87, Arg131, and Lys147 adjacent to the putative sugar-binding site. Thus, it is unclear whether ZG16B binds microbial glycans and, if so, what organisms it engages. We postulated that deciphering the recognition properties of ZG16B could provide valuable insights into its function.
Herein, we describe microbial glycan analysis probes (mGAPs) based on recombinant zymogen granule (ZG) proteins, ZG16B and the related zymogen granule protein 16 homolog P (ZG16P) (17, 19), the latter of which is found in the gastrointestinal (GI) tract. We converted the ZG proteins into mGAPs by modification with either biotin or a fluorophore. We used these probes to investigate the microbe-binding specificity of ZG16B by isolating bacteria from human oral biofilms. The data revealed that Streptococcus vestibularis is the predominant microbe bound by ZG16B. We then determined the bacterial cell wall components of the dental plaque microbes that bind ZG16B. We also found that ZG16B can mediate S. vestibularis aggregation and attenuate growth. Interaction of ZG16B with the host salivary mucin MUC7 amplifies this bacterial cell aggregation. Together, our data suggest that ZG16B can preferentially capture and regulate the growth of distinct species of oral bacteria. Such interactions could maintain homeostasis within the oral microbiome.
Results
Engineering ZG Lectins and Analysis of Interactions with Dental Plaque Bacteria.
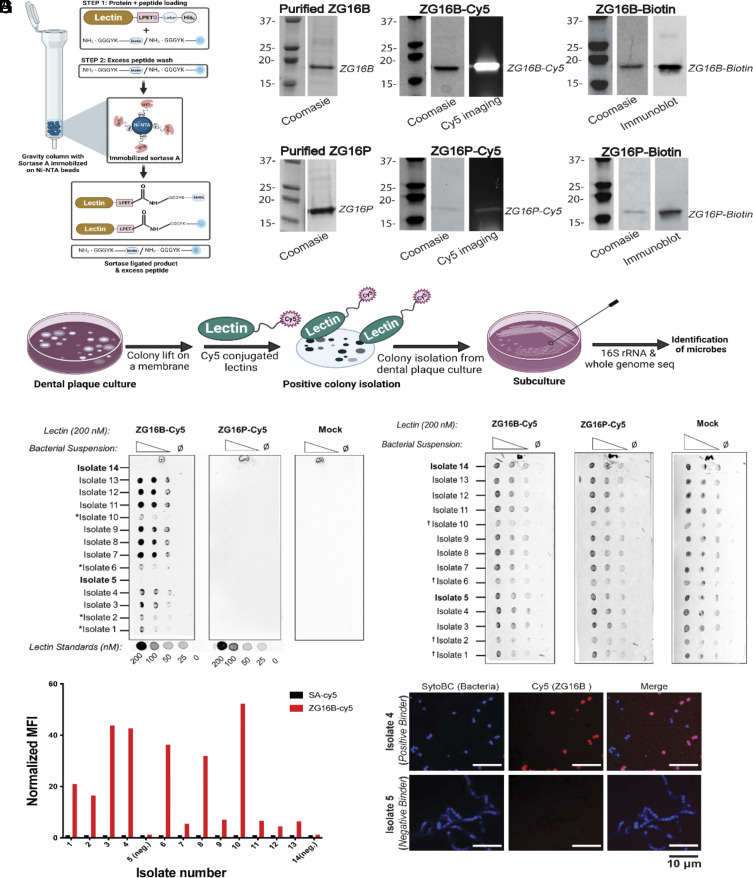
We developed an experimental pipeline for expression, purification, and sortase-mediated ligation (SML) to deploy ZG lectins as mGAPs (Fig. 1A). The probes were based on ZG16B and ZG16P to enable comparative analysis (Fig. 1B). These ZG lectins show distinct tissue-specific distributions: ZG16B in the oral cavity and ZG16P in the GI tract (19). Albeit genetically unrelated to ZG16B, ZG16P is an ideal control in these studies because of its similar secretory behavior (19, 20) and structural features (17, 21). The mGAPs represent the native lectin sequences, excluding the secretory signal sequences and including an additional C-terminal sortase motif (e.g. LPETG) and a His6 tag. The recombinant lectins (ZG16B and ZG16P) appear monomeric and folded based on dynamic light scattering (DLS) (SI Appendix, Fig. S1A) and differential scanning fluorimetry (DSF) (SI Appendix, Fig. S1B). Analysis of ZG16B (PDBID: 3AQG) and ZG16P (PDBID: 3APA) using the Protein Structure and Interaction Analyzer (PSAIA) supports a monomeric state in solution (SI Appendix, Materials and Methods section). The engineered ZG16B and ZG16P can be readily functionalized at the C-terminus with biotin- or sulfoCy5- (herein referred to as Cy5) conjugated polyglycine peptides using a SML technique, modified from the literature (Fig. 1 A and B) (22). SML provides a versatile method for conjugating reporter groups to the recombinant lectins.
Fig. 1.
Engineering labeled ZG16B and interactions with dental plaque bacteria (A) Schematic representation of gravity flow–based sortase-mediated ligation (SML) used for conjugating biotin or fluorophore-labeled polyglycine peptide on the C-terminus of the recombinant lectins ZG16B and ZG16P. SrtA represents the His-tagged sortase immobilized on the Ni-NTA beads. (B) Polyacrylamide gel electrophoresis (PAGE) and western blotting analysis of mGAPs based on ZG16B and ZG16P, produced by SML shown in A. (C) Schematic representation of the colony lift and far-western (lectin) blotting assay used for identifying oral biofilm microbes bound by ZG16B-Cy5. (D) Dot blot analysis of the fourteen representative dental plaque isolates spotted at different OD600 dilutions and probed with 200 nM ZG16B-Cy5 or ZG16P-Cy5. Mock designates control treatment lacking lectins. (E) Membranes were counterstained with Ponceau S to confirm the presence of the bacteria immobilized on it. The (*) in panel D marks positive isolates with lower ZG16B-Cy5 fluorescence signal compared to the other isolates, which is due to the lower concentration of bacteria on the blot as shown by the † in the Ponceau S stain of the blots in panel E. (F) Relative mean fluorescence intensity (MFI) determined by flow cytometry is plotted as a bar graph, representing the signal obtained from ZG16B-Cy5 bound to dental plaque microbes in comparison to the control signal from the Cy5-labeled streptavidin (SA). (G) Confocal microscopic images of ZG16B-binding (Upper) and nonbinding (Lower) dental plaque microbes, stained with Syto BC dye and incubated with ZG16B-Cy5.
To test the microbe-binding of ZG16B, we screened dental plaques and tongue dorsal swabs from ten individuals using a colony lift (23), followed by far-western blotting with Cy5-conjugated ZG16B (ZG16B-Cy5) (Fig. 1C). We identified a total of 50 bacterial isolates from the oral biofilms of four volunteers that were bound by ZG16B-Cy5 but were not bound by the control lectin ZG16P-Cy5 (Fig. 1 D and E and SI Appendix, Tables S1 and S2). Among the bound isolates, most appeared to be facultative anaerobes and could be grown on blood agar plates, the original medium used for isolation, and cultured in brain–heart infusion (BHI) broth.
After culturing 12 representative ZG16B-binding and two nonbinding strains, we used flow cytometry to quantify ZG16B interactions (Fig. 1F). We confirmed bacterial isolate binding by ZG16B with the lectin exhibiting a range of affinities as suggested by mean fluorescence intensity (Fig. 1F). Interestingly, the forward scatter plots varied between some of the microbial isolates implying that ZG16B can bind different types of aggregates of dental plaque microbes (SI Appendix, Fig. S1 C–F). The selected negative control isolates failed to bind ZG16B-Cy5, indicating that ZG16B interacts selectively with certain microbial species within the oral microbiome (Fig. 1F).
We also used the ZG16B probe to visualize the bacterial isolates via confocal microscopy (Fig. 1G). The positively bound isolates appeared as individual cocci or small clusters of cocci (Fig. 1 G, Upper). In contrast, the nonbinding microbes appeared as chains of cocci (Fig. 1 G, Lower).
ZG16B Binds to Oral Commensals Including S. vestibularis.
The species and strain level identity of dental plaque isolates were determined by sequencing the V3-V4 region of the 16S rRNA, followed by whole genome shotgun sequencing. The 16S rRNA sequencing revealed that most of the ZG16B-bound microbes (a total of 50 bacterial isolates) were Streptococcus species in the salivarius group, including Streptococcus salivarius, S. vestibularis, and Streptococcus thermophilus (SI Appendix, Tables S1 and S2) (24). The remainder belonged to the genus Gemella (SI Appendix, Table S1). We chose 14 representative isolates from the first round of donor screening, including positive and negative binders, for whole genome shotgun sequencing to identify their strain level identity. Seven of the twelve positive dental plaque isolates were identified as three different strains of S. vestibularis (isolates 3–4, 7–9, and 11–12), whose genome sequences are closely related to strain NCTC12167. Three are distinct Gemella haemolysans strains related to the FDAARGOS 740 sequence (isolates 2, 6, and 10), and one is a strain with sequence similarity to the Streptococcus mitis group strain ChDC B345 (isolate 1). The isolates that do not bind to ZG16B include Streptococcus oralis strains (isolates 5 and 14) related to strain SF100 (SI Appendix, Tables S1 and S3).
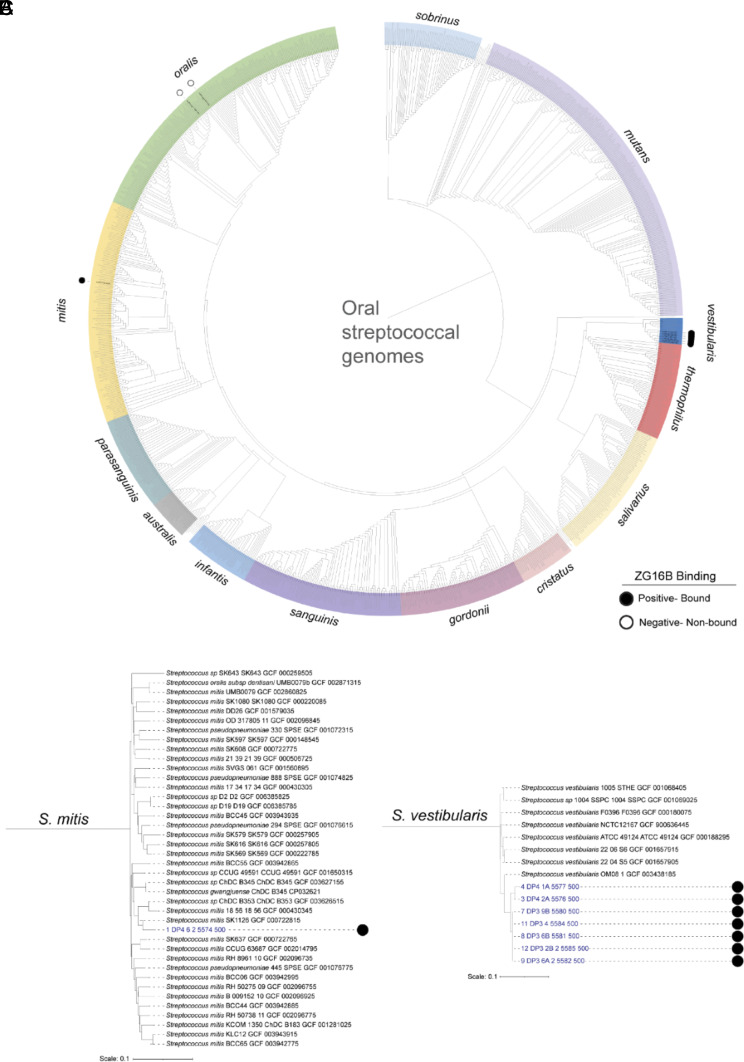
We used the genomic data of 13 representative isolates (excluding isolate 13, refer to SI Appendix, Materials and Methods section) to generate a phylogenetic comparison (SI Appendix, Table S1). Genomic analysis of the streptococcal bacteria (Fig. 2A) reveals that most of the S. vestibularis isolates bound by ZG16B are more closely related to each other than to other genome sequences of members of the same species (Fig. 2C). The isolated member of the S. mitis group (Fig. 2B), is phylogenetically different from S. vestibularis. The isolated G. haemolysans are individual strains, as evidenced by the differences in branch lengths in the tree (SI Appendix, Fig. S2A). The phylogenetic comparison suggests that ZG16B binds to specific species of bacteria, including S. vestibularis, S. mitis, and G. haemolysans (Fig. 2 B and C and SI Appendix, Fig. S2A) in the oral microbiome. The genomes of the two S. oralis isolates, which were not recognized by ZG16B, were distinct (SI Appendix, Fig. S2B).
Fig. 2.
Identification of ZG16B-bound and nonbound strain identity by streptococcal genome sequence comparisons. The genome sequences of ZG16B-bound and nonbound bacterial isolates were compared to over six thousand publicly available (NCBI) and an additional set of 69 unpublished streptococcal genome sequences using a pairwise k-mer composition–based distance matrix method for identification of oral biofilm isolate strain identity. (A) Cladogram that displays the phylogeny of a subset of streptococcal genome sequences of species that commonly colonize the oral cavity. A pruned subtree of the (B) S. mitis and (C) S. vestibularis subclades from A that display the location of ZG16B-bound isolate genome sequences within the clade. The genome names of the isolates bound or not bound by ZG16B are marked by black-filled and open circles, respectively. The tree scales show the pairwise k-mer composition–based distance that is used for each branch length.
The ZG16B-Cy5 probe did not bind to any S. mutans strains tested (SI Appendix, Fig S1H), including strain UA159, which causes dental caries and disease. Previous studies had suggested an interaction between ZG16B and S. mutans strain UA159 (16). The authors found that a construct of the common salivary protein 1 (CSP-1) (a synonym for human ZG16B) failed to promote S. mutans aggregation and lacked significant bactericidal activity. However, in saliva with native CSP-1, S. mutans showed adherence to an experimental salivary pellicle on hydroxyapatite surfaces (16). S. mutans was absent in our screen of oral biofilm bacteria, and flow cytometry analysis showed that ZG16B-Cy5 probe did not bind to the typed S. mutans strains (SI Appendix, Fig S1H), including pathogenic strain UA159. Thus, the probes based on ZG16B show preferential binding to oral commensals.
Among the total 50 bacterial isolates identified as ZG16B binders, around 92% appeared to be in the S. salivarius group (SI Appendix, Tables S1 and S2). ZG16B also showed binding to the ATCC strain S. vestibularis NCTC 12166 (25) (SI Appendix, Fig. S1G), phylogenetically close to the identified dental plaque S. vestibularis strains (Fig. 2C), clearly indicating that ZG16B interacts with S. vestibularis species. We conducted 16S rRNA sequencing on 25 bacterial isolates identified in our screen that ZG16B did not bind (SI Appendix, Table S3). With one exception, these isolates did not belong to the S. salivarius group, supporting that the preference of ZG16B for S. vestibularis was unbiased. Due to the predominance of S. vestibularis over other species and the better growth in suspension, we pursued further analysis of this species.
The microbial species identified from the dental plaque isolates are primarily commensals that inhabit the oral cavities and upper respiratory tracts as early colonizers (1, 23). S. vestibularis is a commensal species, although it was reported to cause infective endocarditis in a patient with coexisting S. oralis endocarditis in a recent study (26, 27). The mGAP based on ZG16B binds S. vestibularis but not S. oralis, although both belong to the viridans group of streptococci that share several phylogenetic features (24). These findings suggest that molecular features of the cell surface of the microbes may underpin this lectin–microbe interaction.
ZG16B Binds to the Peptidoglycan Layer of the Cell Wall of Dental Plaque Microbes.
The bacterial cell wall features various glycoconjugates that could interact with ZG16B. Initial studies to screen the ligand specificity of the functionalized ZG16B using microbial glycan microarrays failed to yield a definitive ZG16B binding partner (SI Appendix, Fig. S3 A and B). This inability to find ligands may reflect the composition of the microarrays, which include isolated polysaccharides from mostly Gram-negative bacteria and capsular polysaccharides from some strains of Gram-positive bacteria (28). The current glycan arrays lack glycoconjugates from oral streptococci. Therefore, a top–down approach was taken to identify ZG16B binding partners from extracted cell wall glycans of the dental plaque bacteria S. vestibularis and S. oralis.
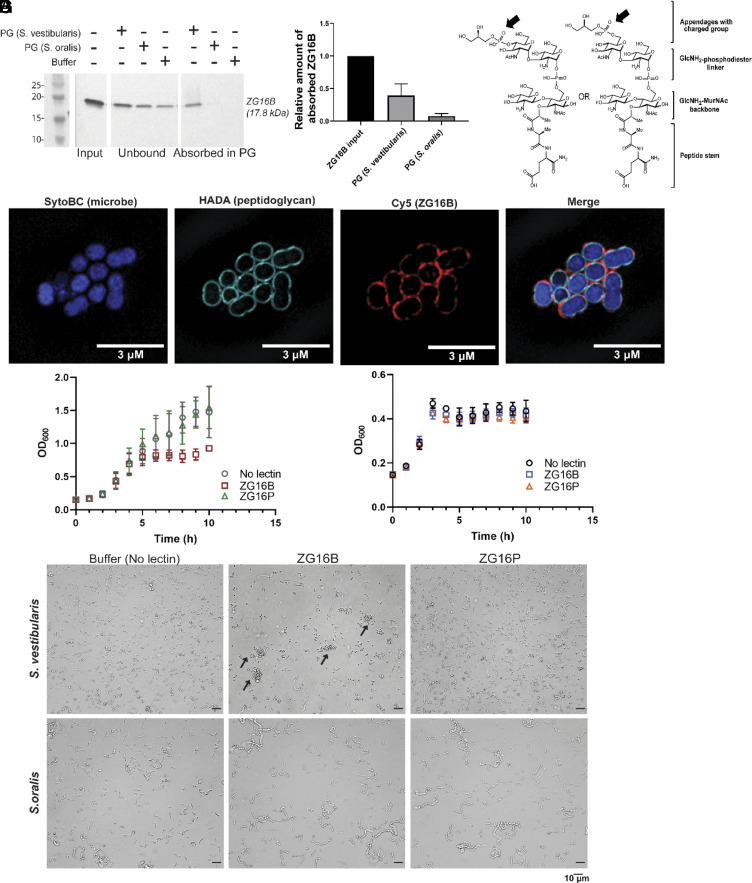
The cell wall extracts were separated into peptidoglycan and teichoic acid fractions (SI Appendix, Fig. S3C) using standard procedures (29). Dot blot analysis of the fractions immobilized on a nitrocellulose membrane probed with biotin-ZG16B qualitatively showed the binding of ZG16B to the peptidoglycan isolated from S. vestibularis (SI Appendix, Fig. S3D). However, the insolubility of the peptidoglycan led to weak signals in protein–ligand binding analyses, making quantitative analysis inconclusive. Therefore, to assess lectin absorption to the insoluble peptidoglycan, recombinant ZG16B was incubated with peptidoglycan isolated from S. vestibularis and S. oralis. Almost 40% of the input recombinant ZG16B was absorbed into the peptidoglycan extracted from S. vestibularis, approximately five times more than that absorbed into the peptidoglycan of S. oralis (Fig. 3 A and B). Digestion of the peptidoglycan with mutanolysin from Streptomyces globisporus abrogated the interaction between ZG16B and the peptidoglycan fraction from S. vestibularis (SI Appendix, Fig. S3 F and G), suggesting that particular modifications of the peptidoglycan might be a key feature of the lectin–microbe interaction. We also confirmed that the interaction of ZG16B with the microbes is not mediated by the cell surface adhesins; formaldehyde treatment of the cells, which denatures surface proteins, did not abrogate the lectin–microbe interactions (SI Appendix, Fig. S3H).
Fig. 3.
ZG16B binds to the peptidoglycan (PG) of S. vestibularis and regulates the growth of the bacteria. (A) Anti-His6 immunoblot for recombinant ZG16B, unbound or absorbed at the insoluble peptidoglycan, extracted from S. vestibularis or nonbinding S. oralis. (B) Bar graph representation of the amount of ZG16B absorbed in the peptidoglycan from three biological replicates of S. vestibularis or S. oralis, normalized to the lectin input amount. (C) Isobaric structures of peptidoglycan fragments identified from the high-resolution mass spectrometry analysis on the mutanolysin-digested peptidoglycan from ZG16B-bound dental plaque microbes. The negatively charged group indicated by the arrows is potentially involved in ZG16B binding. (D) Super-resolution images of S. vestibularis grown with peptidoglycan marker fluorescent D-amino acid (HADA) and incubated with ZG16B-Cy5, showing the colocalization of ZG16B with peptidoglycan. Growth curve of (E) S. vestibularis and (F) S. oralis in the presence and the absence of ZG16B or ZG16P at the MIC50 of ZG16B (1.25 μg/mL). (G) Brightfield images of S. vestibularis (Upper) and S. oralis (Lower) at 5 h of incubation with or without the recombinant lectins at 37 °C. The arrows highlight S. vestibularis aggregation in the presence of ZG16B.
Complementary confocal (SI Appendix, Fig. S3E) and super-resolution microscopy (Fig. 3D) show that ZG16B localizes to the cell wall of S. vestibularis. The microbes were grown with 3-[[(7-hydroxy-2-oxo-2H-1-benzopyran-3-yl)carbonyl]amino]-D-alanine hydrochloride (HADA), a fluorescent D-amino acid that is incorporated into the peptide linker during peptidoglycan biosynthesis (30). ZG16B-Cy5 (Fig. 3D) colocalized with the peptidoglycan marker HADA. No such interaction was observed with the fluorescent ZG16P (SI Appendix, Fig S3E). Additionally, ZG16B colocalization was not observed with the nonbinder S. oralis, validating the high binding selectivity of ZG16B with the S. vestibularis peptidoglycan.
The exact substructure of the S. vestibularis peptidoglycan that is responsible for selective ZG16B binding is not yet known. We observed that peptidoglycans extracted from S. oralis and S. vestibularis are both resistant to hen egg lysozyme but not to bacterial mutanolysin (SI Appendix, Fig. S3F), suggesting that the peptidoglycans from both oral streptococci include O-6-acylated N-acetylmuramic acid (MurNAc) or N-deacylated MurNAc or N-acetylglucosamine (GlcNAc) in the disaccharide repeat units (31). We then considered modifications of the peptidoglycan beyond the alternating GlcNAc/MurNAc glycan and peptide cross-linkers. The peptidoglycan of streptococci is often covalently modified with l-rhamnose (L-Rha)-rich cell wall polysaccharide (RhaCWPS), linked at the C6-OH of MurNAc via a bridging GlcNAc or glucosamine (GlcNH2) (32–34). The cell wall polysaccharide (CWPS) can serve as a functional replacement for wall teichoic acid and mediate virulence, host adhesion, immune evasion, and antimicrobial resistance (34). Strikingly, ovococcal bacteria, including streptococci, display a wide range of structural variations in their RhaCWPSs (32–34). The core repeating units with L-Rha building blocks often incorporate hexose sugars (e.g., Glc and Gal) and differ in length and glycosidic linkage (32, 35). Additionally, the side-chain substituents show extensive diversity in composition ranging from simple mono- or disaccharides to complex branched oligosaccharides comprising hexoses, HexNAcs, and charged modifications such as glycerol-1-phosphate (Gro-P) (36).
Currently, the details of the S. vestibularis RhaCWPS are unknown. However, characterization of the cell wall polysaccharides from strains of S. salivarius (37, 38) and S. thermophilus ST64987 (39, 40), the phylogenetically closest species in the salivarius group, shows the presence of L-Rha with Glc and Gal as the major components. Therefore, to assess the potential role of the CWPS, we investigated different experimental conditions in the ZG16B binding assays. ZG16B showed pH-dependent binding to S. vestibularis. Specifically, the interaction between ZG16B and S. vestibularis was impaired when immobilized microbes were probed with ZG16B-Cy5 or ZG16B-biotin and washed with buffers below pH 6 (SI Appendix, Fig. S3I). A similar reduction in binding was observed when whole cells, or extracted peptidoglycan, were probed with ZG16B-biotin and then washed with a buffer including 20 mM glycerol phosphate (Gro-P) (SI Appendix, Fig. S3 J and K). In contrast, when the wash buffers were supplemented with 20 mM L-lysine, binding was preserved, suggesting that the lectin might interact with the negatively charged Gro-P on the peptidoglycan of the microbes.
To investigate CWPS modifications on the peptidoglycan that might impact ZG16B binding, we performed high-resolution mass spectrometric (HRMS) analysis on the peptidoglycan fragments comparing the composition of ZG16B binder and nonbinder dental plaque microbes (Fig. 3C and SI Appendix, Table S4 and Fig. S4). We identified feasible isobaric fragments of the substructures of peptidoglycan of ZG16B binders (S. vestibularis isolates 3, 4, 8, and 12 and G. haemolysans isolate 10). These substructures were detected in both positive and negative electrospray ionization (ESI) modes (SI Appendix, Fig. S4) within a mass accuracy of 10 ppm. We also observed the presence of N-deacylated GlcNAc (GlcNH2) in the disaccharide repeat units of the peptidoglycan, consistent with mutanolysin digestion assays described above. Moreover, the identified masses indicate that a deacylated hexosamine, possibly a GlcNH2, is appended to the C6-OH of MurNAc through a phosphodiester linkage (SI Appendix, Fig. S4 Structures 1-3). Notably, we can detect the mass peaks corresponding to isobaric structures of fragments 4-7 (Fig. 3C and SI Appendix, Fig. S4 Structures 4-7) that indicate the presence of a Gro-P cap at a HexNAc sugar adjacent to the linker GlcNH2. The presence of the Gro-P moiety on the polysaccharide side chains appended on the peptidoglycan of the ZG16B binders is consistent with the findings from the dot-blot-based lectin blotting assays, implying that the charged group modification of the peptidoglycan plays a role in the binding of ZG16B to S. vestibularis.
As mentioned previously, the presence of the linker GlcNH2 is indicative of the existence of a polysaccharide chain on S. vestibularis peptidoglycan (32, 34). Under the current experimental conditions, we were not able to detect masses corresponding to complete CWPS attached to the peptidoglycan. The challenges of full characterization can arise because the GlcNH2-1-phosphate bond confers resistance to mild acid hydrolysis (41), the fragments from partial digestion of the peptidoglycan by mutanolysin have high molecular weight, or the resulting fragments are less efficiently ionized.
A parallel analysis was carried out on the ZG16B nonbinders (e.g., S. oralis isolate 5 and S. mutans UA159). In this case, a similar peptidoglycan fragment with the Gro-P modification was not detected under identical experimental conditions. The presence of Gro-P modifications in the group A carbohydrate in Streptococcus pyogenes (32, 34, 42, 43) and serotype c carbohydrate in S. mutans (32, 35, 36, 42) has been reported previously. We believe that the accessibility of the CWPS-modifying groups to ZG16B is one determining factor in its specificity toward particular microbial species. This analysis lays the groundwork for future characterization of the complete structure of the CWPS of S. vestibularis and the minimum glycan epitope necessary for ZG16B binding.
ZG16B Suppresses S. vestibularis Growth.
Plant lectins can exhibit antimicrobial activity through their interactions with various glycoconjugates; antimicrobial roles for animal lectins are less well characterized (44–46). To determine the effect of ZG16B on S. vestibularis growth, the bacteria were cultured in the presence of recombinant ZG16B or the nonbinding lectin ZG16P. ZG16B prevented the outgrowth of S. vestibularis (Fig. 3E and SI Appendix Fig. S5A) with MIC50 values of 2.5 to 1.25 µg/mL (SI Appendix Fig. S5A). In contrast, the lectin had no influence S. oralis growth (Fig. 3F and SI Appendix Fig S5B). Consistent with our previous observations, the related lectin ZG16P had no influence on S. vestibularis (Fig. 3E and SI Appendix Fig. S5A) or S. oralis (Fig. 3F and SI Appendix Fig. S5B) growth. We did not observe any significant growth suppression of the microbes at higher concentrations (~20 µg/mL) of ZG16B, presumably due to aggregation of the lectin upon incubation at 37 °C for 12 h, as supported by dynamic light scattering (DLS) (SI Appendix, Fig. S5C).
The observed bacteriostatic effect of ZG16B may be due to its ability to aggregate cells. To test this hypothesis, we used brightfield imaging to visualize the impact of the lectin on bacterial cells. S. vestibularis grown for 5 h (near the end of the log phase) aggregated in the presence of ZG16B (Fig. 3G), while neither ZG16B nor ZG16P aggregated S. oralis (Fig. 3G).
ZG16B Recruits Salivary Mucin MUC7 onto S. vestibularis.
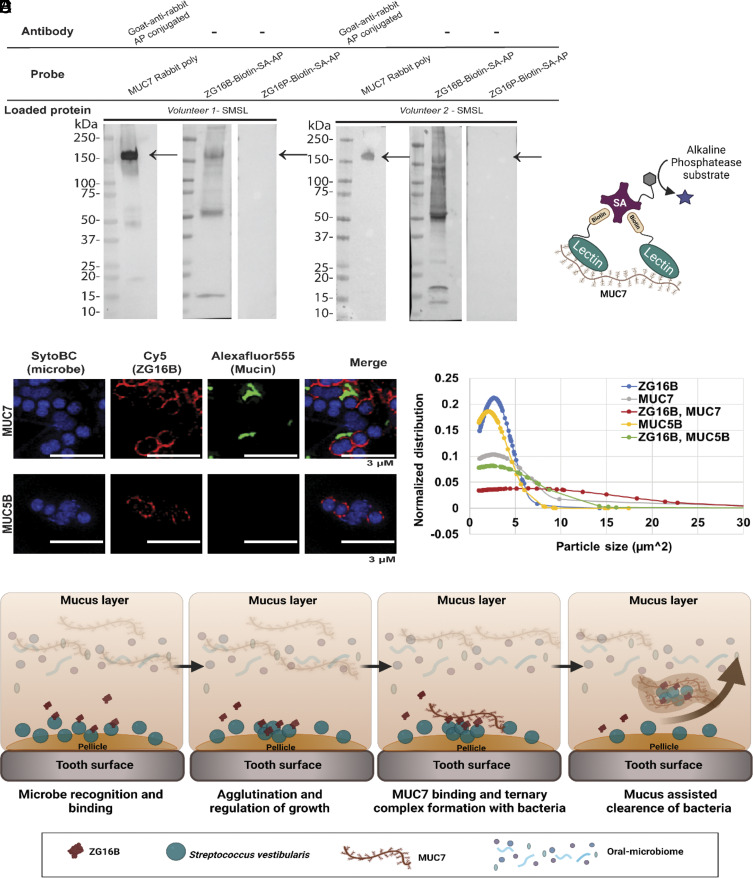
The binding of lectins to microbial cell surface glycoconjugates often arises from multivalent interactions; however, ZG16B appears to be a monomeric lectin in solution. We postulated that in a native environment, such as the oral cavity, ZG16B might function as part of a multicomponent salivary agglutination complex. To test this hypothesis, we assessed the interaction of ZG16B with other salivary proteins by lectin blot analysis. Two donor saliva samples from submandibular/sublingual (SMSL) glands were separated by gel electrophoresis (SI Appendix, Fig. S6A), transferred to a nitrocellulose membrane, and probed with mGAPs based on ZG16B. ZG16B-Cy5 showed weak binding (SI Appendix, Fig. S6B, lane 1) to a target at 150 kDa, which corresponds to the low Mr salivary mucin MUC7 (Fig. 4A, lanes 1 and 4) (47). Interestingly, a multivalent arrangement of the lectin generated from biotinylated ZG16B precomplexed with streptavidin-conjugated alkaline phosphatase (Fig. 4A, lanes 2 and 5, Fig. 4B) or Cy5 (SI Appendix, Fig. S6B, lane 2), bound more avidly to MUC7, suggesting that multivalent interactions promote ZG16B to MUC7. Control experiments where the salivary protein blots were probed with precomplexed ZG16P-biotin (Fig. 4A, lanes 3 and 6 and SI Appendix, Fig. S6B, lane 3) did not show a lectin–mucin interaction in a monovalent or multivalent presentation. Furthermore, ZG16B maintained its binding specificity to MUC7 that was extracted and enriched from saliva samples (SI Appendix, Figs. S6 C and D). No binding to the other major salivary mucin MUC5B was detected (SI Appendix, Fig. S6D).
Fig. 4.
ZG16B interactions with MUC7 and S. vestibularis. (A) Interaction of ZG16B with salivary mucin MUC7 detected by far-western blot analysis of two salivary samples probed for biotinylated lectins that were precomplexed with alkaline phosphatase–conjugated streptavidin (SA-AP). The presence of MUC7 in the saliva samples was detected by probing with an anti-MUC7 antibody. (B) Schematic representation of the molecular level interaction between the MUC7 and biotinylated ZG16B, complexed with SA-AP for colorimetric detection. (C) Super-resolution images of Syto BC stained S. vestibularis in the presence of ZG16B-Cy5 and mucin (MUC7 or MUC5B)-enriched samples probed with Alexafluor 555. (D) The average size distribution of the microbial clusters of S. vestibularis in the presence and absence of MUC7- or MUC5B-rich samples was calculated from the image analysis using Fiji software. On average, 100 particles were chosen from six regions of interest from each sample shown in SI Appendix, Fig S4C. (E) Schematic representation of the potential biological role of ZG16B in the oral cavity. ZG16B can capture and aggregate the S. vestibularis on the pellicle and influence homeostasis by this mechanism and also through mucus-assisted clearance by recruiting oral mucin MUC7.
The binding specificity of ZG16B for MUC7 relative to MUC5B may arise from the differences in the glycosylation of these two mucins. The abundance of sialic acid glycans is higher on MUC7 than on MUC5B with extended branched sialylation on core glycan structures (SI Appendix, Fig. S6E) (47–51). Moreover, the two mucins share only a few oligosaccharide sequences. The glycan profile on MUC7 is more conserved than MUC5B in individuals, while MUC5B shows intra-individual variation in glycosylation (52, 53). To assess whether the interaction between ZG16B and MUC7 is mediated by the O-linked glycans, the SMSL saliva fractions from two individuals were treated with sodium periodate to oxidize cis-diol of the MUC7 glycans (54). The samples were probed with biotinylated lectins, precomplexed with streptavidin-Cy5 on a nitrocellulose membrane. For both saliva samples, periodate treatment completely abrogated the interaction between ZG16B and MUC7 (SI Appendix, Fig. S6F), suggesting that the lectin–mucin interaction is mediated by mucin carbohydrate residues sensitive to sodium periodate (e.g., sialic acids). Furthermore, the binding of ZG16B and MUC7 was partially lost upon treatment of the saliva sample with α2-3,6,8 neuraminidase or with both neuraminidase and O-glycosidase (SI Appendix, Fig. S6G), supporting that the terminal sialic acids of the O-glycans of MUC7 may act as the binding ligands of ZG16B in the lectin–mucin interaction.
The ability of ZG16B to bind different glycan sequences in MUC7 and bacterial cell wall polysaccharides can be rationalized based on the structural features (17). ZG16B has a basic patch comprising Lys 87, Arg131, and Lys147, which could serve as the binding site for the sialic acid–rich O-glycans of MUC7, leaving the sugar-binding loop available for engaging the microbial CWPS. In this model, lysine 177 is poised to neutralize any negative charge in the CWPS. To support the two-binding-site model, we tested for ternary complex formation between ZG16B, MUC7, and S. vestibularis. We used confocal laser scanning (SI Appendix, Fig. S6 H–J) and super-resolution microscopy (Fig. 4C) to assess different complexes. S. vestibularis was incubated with ZG16B-Cy5, a MUC7-enriched sample, or both. MUC7 localized to S. vestibularis in the presence of ZG16B only, while ZG16B maintained binding to the microbes in the presence and absence of MUC7. Both ZG16B and MUC7 colocalized to the surface of S. vestibularis (Fig. 4C and SI Appendix, Table S5 and Fig. S6 H and J), consistent with the formation of a ternary complex between the lectin, the mucin, and the microbes. In the future, the molecular details of the two proposed sugar-binding sites of ZG16B can be studied in detail. Ternary complex formation was not observed with nonbinding S. oralis (SI Appendix, Fig. S6I) or the nonbinding mucin MUC5B (Fig. 4C and SI Appendix, Fig. S6 H and J).
The ZG16B-MUC7 interaction is likely to be physiologically important as the low Mr mucin MUC7 lacks a terminal cysteine-rich domain, making it unable to form a polymeric gel-like structure that shapes the protective layer on the oral cavity surfaces (55). Indeed, S. vestibularis exposed to ZG16B and MUC7 formed larger clusters of bacteria with an average cluster size three-fold larger than that of the lectin or the mucin alone (Fig. 4D and SI Appendix, Table S6 and Fig. S6H). In contrast, the presence of MUC5B with ZG16B failed to increase microbe clustering, indicating that enhanced microbial clustering occurs only during the recruitment of MUC7 on S. vestibularis by ZG16B. Although we may have anticipated ZG16B to precomplex with MUC7 to provide a multivalent scaffold to bind to bacteria, what we observed was that ZG16B binding to bacteria results in a multivalent interaction to which MUC7 then binds and provides a cross-linking scaffold for further agglutination.
Discussion
The oral cavity is a “front line” interface for host–microbe interactions. With its diverse microbiota, the host oral cavity must maintain the diversity of commensal species and limit overgrowth resulting in dysbiosis and disease (1, 14). Currently, limited information is available on the roles of ZG16B, an abundant protein in the oral proteome with a lectin fold. That ZG16B is highly expressed in mature salivary glands and not in the fetal glands suggests its significance in shaping the oral immune responses to the microbial environment during the developmental differentiation of salivary glands (14). Our goal was to investigate the functional significance of ZG16B.
In the present study, by converting ZG16B into a functional mGAP, we identified target bacteria in the oral microbiota. Our data indicate that ZG16B can distinguish between the oral viridans group streptococci and binds mainly to S. vestibularis (25) and G. haemolysans (56), but not S. oralis (57). Moreover, we did not observe binding to an S. mutans strain (16), indicating that this oral protein recognizes a subset of species in the microbiome. Although belonging to the same genus, the streptococci recognized are in different phylogenic groups (24), and these groups exhibit some biochemical differences in cell wall composition (24, 58).
We analyzed the cell envelope components that ZG16B binds to and found it interacts with CWPS appended to the peptidoglycan. The diversity in the chemical composition of CWPS widens the serotype variations among different groups of streptococci. The specificity of ZG16B for S. vestibularis appears to be mediated mainly by substructures of the CWPS, which are covalently linked to the peptidoglycan. That ZG16B distinguishes chemical differences in peptidoglycan constitution from different oral streptococci makes it an excellent peptidoglycan-binding probe to rapidly identify specific microbes. The presence of the Gro-P modifications on CWPS has been reported previously (36) for some ZG16B nonbinder streptococci, e.g., S. mutans, but not for S. vestibularis. Therefore, the position and orientation of the negatively charged group on the CWPS may have a role in the lectin specificity for a particular microbe. Our research lays the foundation and the rationale for further assessment of the complete structure of the CWPS of S. vestibularis to understand the precise features that drive lectin binding.
Our data suggest that ZG16B might act as “crowd control” for the oral microbes it recognizes. In vitro, ZG16B is bacteriostatic toward the oral commensal S. vestibularis, a feature that would allow the maintenance of this species in the oral cavity while controlling its growth. The observed aggregation among S. vestibularis in the presence of ZG16B may arise due to the charge neutralization on the microbial cell surface upon lectin binding. It is also possible that the presence of ZG16B could induce stress among the microbes and cause the bacteria to autoaggregate by expressing proteins or other glycans that result in aggregation.
The identification of ZG16B as a lectin led us to examine its interactions with oral mucins. We found that ZG16B binds to the salivary mucin MUC7 and forms a ternary complex with S. vestibularis. It is intriguing that the monomeric ZG16B can form a ternary complex with microbes and the mucin glycoprotein. These data support the hypothesis that the binding sites for microbial glycan determinant and mucin O-glycans on ZG16B are different. That ZG16B possesses basic amino acid residues in both primary and secondary binding sites (17) implies that it may use the lysine in the sugar-binding site for the negatively charged groups on the CWPS and the basic patch for binding to the sialic acids on the mucin glycopolymers.
The binding of ZG16B to the salivary mucin MUC7 gives information on the biological role of the lectin in its native environment. Enhanced S. vestibularis aggregation through ZG16B–MUC7 interactions may influence bacterial load on oral cavity surfaces (Fig. 4E). As MUC7 was reported to bind a diverse range of oral streptococci and non-Streptococcus microbes in the oral cavity (55), ZG16B may serve as the specificity handle for targeting commensals such as S. vestibularis. The recruitment of commensals to mucins may also assist in a mucus-assisted clearance system, a known mechanism for the removal of bacteria from the body (59). Finally, the ZG16B–MUC7 findings provide an intriguing model of host factor synergy in host–microbe interactions. We show that ternary complex formation is strengthened by multivalent interactions between the mucin and lectin, and these multivalent interactions are enabled by the immobilization of ZG16B through binding to the microbial cell surface.
By converting the oral lectin ZG16B into an mGAP, we successfully confirm the formation of a relevant ZG16B complex by identifying its target microbe and binding protein partner. The strategy presented here provides the proof of concept for applying human lectin–based probes to discover host–microbe interactions in complex environments. Such probes can also provide insight into the roles of other mediators in the environment, in this case, MUC7, which actively participates in the lectin–microbe interactions. Finally, the remarkable specificity of the ZG16B mGAP for a small subset of organisms promises to be the foundation for a class of probes with unique glycan-binding specificities for bacterial sugar profiling. Future studies using the ZG16B-mGAP with a broader set of both healthy and diseased subjects will provide insights into the range of microbes bound by ZG16B, potentially revealing the significance of the lectin in oral health and disease. Based on this foundation and the strategy presented, it is likely that the development of additional mGAPs will provide valuable insights into the common features of the human soluble lectins regarding their binding specificities to different microbes or glycoproteins and shaping host–microbe interactions.
Materials and Methods
Detailed descriptions of the following standard procedures are provided in the SI Appendix: Construction of strains and plasmids for expression and purification of ZG16B and ZG16P; procedures for SML of ZG16B and ZG16P to generate Cy5 and biotin ligated mGAPs; collection of human oral biofilms according to the study approved by the University at Buffalo Human Subjects IRB (study # 030–505616) with informed consent obtained from all human participants; culture and isolation of bacteria from mixed-species oral biofilms and screening for bacteria bound to ZG16B-Cy5 by replica plating and flow cytometry (SI Appendix, Fig. S1 and Tables S1 and S2); 16S rRNA (SI Appendix, Tables S1–S3) and whole genome sequencing (SI Appendix, Table S1, data from whole genome sequencing results submitted under the BioProject number PRJNA880893 (60); bacterial isolate species and strain identification based on whole genome comparisons (SI Appendix, Fig. S2); confocal microscopy of dental plaque microbes in the presence of ZG16B and MUC7; determination of MIC50 of ZG16B and brightfield imaging of live cells; lectin absorption to peptidoglycan assay; lectin blotting on salivary mucins; extraction and fractionation of the cell wall capsule from dental plaque isolates; peptidoglycan digestion and turbidity assay; dot blot analysis and biolayer interferometry; DLS and nDSF of ZG16B and ZG16P; screening of precomplexed ZG16B on microbial glycan array; and PSAIA analysis of ZG proteins.
Super-Resolution Microscopy.
The cells were grown overnight in BHI medium with 200 μM HADA (R&D Systems) at 37 °C, without shaking. Cells were then fixed and stained with ZG16B-Cy5 following the same protocol used in the confocal microscopy of the dental plaque microbes in the presence of ZG16B- and MUC7-rich saliva samples. For analysis by microscopy, stained cells were pelleted and suspended in PBS supplemented with 1% w/v DABCO (1,4-diazabicyclo[2.2.2]octane) (Sigma-Aldrich) as an antifading reagent. Each sample was spotted onto a glass-bottomed microwell dish (MatTek corporation # P35G-1.5-14-C), allowed to settle overnight, and covered with a precooled 1% (w/v) agarose pad. Images were collected on an Applied Precision DeltaVision-OMXv4 Super-Resolution Microscope (60×/1.42 NA oil immersion lens, sequential imaging on two sCMOS cameras). Structured illumination microscopy (SIM) reconstruction and warp-based image alignments were performed with Applied Precision softWoRx. Multicolor Image alignments were calculated from an Applied Precision grid test slide, checked with TetraSpeck beads (0.1 μm, Molecular Probes), and verified with an Argo-SIM test slide (Axiom Optics) before data collection. Brightness and contrast were identically adjusted with the open-source Fiji distribution of ImageJ. Images were then converted to an RGB format to preserve normalization and then assembled into panels.
High-Resolution Mass Spectrometry of Soluble Peptidoglycan Fragments.
The soluble peptidoglycan fragments from S. vestibularis (isolates 3, 4, 8, and 12), G. haemolysans (isolate 10), S. oralis (isolate 5), and S. mutans (UA159) were processed using a 3-kDa MWCO spin filter to separate soluble fragments from nonsoluble peptidoglycan and lyophilized. The liquid chromatography method involved a 0.5 mL min−1 linear gradient starting from 0% A (0.1% formic acid in water) to 50% B (0.1% formic acid in acetonitrile) over 4 min. The eluting peaks were subjected to high-resolution mass analysis on the Q-Exactive Orbitrap (Acquity UPLC BEH C18 column 2.1 × 50 mm (Waters) using a Dionex ultrahigh performance liquid chromatography (UHPLC) coupled to a Q-Exactive Orbitrap (Thermo Fisher Scientific)). Thermo Xcalibur Qual Browser was used to process and analyze the data generated. All species showed the expected mass with the correct m/z ratio within ±10 ppm. HRMS spectra of identified peptidoglycan fragments in ZG16B-bound dental plaque bacteria are shown in the SI Appendix, Fig. S4 and Table S4.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Jeffrey R. Kuhn of the Microscopy Core Facility at the Koch Institute Robert A. Swanson Biotechnology Center for technical support with super-resolution microscopy. We thank Cassandra Roger of the WM Keck imaging facility at the Whitehead Institute for ANDOR confocal microscopy. We thank Stuart Levine and Huiming Ding of the BioMicro Center at MIT for technical and analytical support to obtain the whole genome sequencing. We thank Katharina Ribbeck, Dept. of Biological Engineering, MIT, for generously providing us with the MUC5B-enriched sample and S. mutans strains. We are grateful to Karen Allen, Dept. of Chemistry, Boston University, for help with PSAIA analysis of ZG proteins. We thank Suzanne Walker, Microbiology and Molecular Genetics, Harvard University, for kindly sharing the protocol for extraction and fractionation of cell wall capsules of Gram-positive bacteria. We thank Teddy Warner, Dept. of Chemistry, MIT, for his help in analyzing mass spectrometry data. This work is supported by the NIH U01 CA231079 (L.L.K. and B.I.), R01 AI055258 (L.L.K.), U01 CA221244, 5R01DE019807, and 2T32DE023526 (S.R.), and 1F32DE031963-01A1 (C.P.A.).
Author contributions
S.G., C.P.A., C.R.I., V.M.M., G.J.D., H.B., S.R., L.L.K., and B.I. designed research; S.G., C.P.A., C.R.I., V.M.M., G.J.D., H.B., R.L.M., A.E.D., S.J., L.N., and R.P. performed research; S.G., C.P.A., R.P., and C.L.G. contributed new reagents/analytic tools; S.G., C.P.A., C.R.I., V.M.M., G.J.D., H.B., H.T., R.P., S.R., L.L.K., and B.I. analyzed data; and S.G., C.P.A., V.M.M., S.R., L.L.K., and B.I. wrote the paper.
Competing interests
B.I. is cofounder and on the Board of Directors of AssayQuant Technologies.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Laura L. Kiessling, Email: kiesslin@mit.edu.
Barbara Imperiali, Email: imper@mit.edu.
Data, Materials, and Software Availability
Bacterial whole genome sequence and assembly data have been deposited in GenBank (BioProject accession number: PRJNA880893; https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA880893) (60). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Cross B. W., Ruhl S., Glycan recognition at the saliva–Oral microbiome interface. Cell. Immunol. 333, 19–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heo S.-M., Ruhl S., Scannapieco F. A., Implications of salivary protein binding to commensal and pathogenic bacteria. J. Oral Biosci. 55, 169–174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmskov U., Thiel S., Jensenius J. C., Collectins and ficolins: Humoral lectins of the innate immune defense. Annu. Rev. Immunol. 21, 547–578 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Garlatti V., et al. , Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J. 26, 623–633 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drickamer K., Taylor M. E., Biology of animal lectins. Annu. Rev. Cell. Biol. 9, 237–264 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Manimala J. C., Roach T. A., Li Z., Gildersleeve J. C., High-throughput carbohydrate microarray analysis of 24 lectins. Angew. Chem. Int. Ed. Engl. 45, 3607–3610 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Li Q., Rodriguez L. G., Gildersleeve J. C., An array-based method to identify multivalent inhibitors. J. Am. Chem. Soc. 132, 9653–9662 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasta G. R., Roles of galectins in infection. Nat. Rev. Microbiol. 7, 424–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wesener D. A., Dugan A., Kiessling L. L., Recognition of microbial glycans by soluble human lectins. Curr. Opin. Struct. Biol. 44, 168–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imperiali B., Bacterial carbohydrate diversity—A brave new world. Curr. Opin. Chem. Biol. 53, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kussrow A., et al. , Measurement of monovalent and polyvalent carbohydrate−lectin binding by back-scattering interferometry. Anal. Chem. 81, 4889–4897 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee R. T., Lee Y. C., Affinity enhancement by multivalent lectin–carbohydrate interaction. Glycoconjugate J. 17, 543–551 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Costa-da-Silva A. C., et al. , Salivary ZG16B expression loss follows exocrine gland dysfunction related to oral chronic graft-versus-host disease. iScience 25, 103592 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou M., et al. , Functional specialization of human salivary glands and origins of proteins intrinsic to human saliva. Cell Rep. 33, 108402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo S.-M., et al. , Host defense proteins derived from human saliva bind to Staphylococcus aureus. Infect Immun. 81, 1364–1373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambatipudi K. S., et al. , Human common salivary protein 1 (CSP-1) promotes binding of Streptococcus mutans to experimental salivary pellicle and glucans formed on hydroxyapatite surface. J. Proteome Res. 9, 6605–6614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanagawa M., et al. , Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related β-prism fold. Biochem. Biophys. Res. Commun. 404, 201–205 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Meagher J. L., Winter H. C., Ezell P., Goldstein I. J., Stuckey J. A., Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology 15, 1033–1042 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Bergström J. H., et al. , Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc. Natl. Acad. Sci. U.S.A. 113, 13833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleene R., Dartsch H., Kern H.-F., The secretory lectin ZG16p mediates sorting of enzyme proteins to the zymogen granule membrane in pancreatic acinar cells. Eur. J. Cell Biol. 78, 79–90 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Kumazawa-Inoue K., et al. , ZG16p, an animal homolog of β-prism fold plant lectins, interacts with heparan sulfate proteoglycans in pancreatic zymogen granules. Glycobiology 22, 258–266 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Policarpo R. L., et al. , Flow-based enzymatic ligation by sortase A. Angew. Chem. Int. Ed. Engl. 53, 9203–9208 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Ruhl S., Eidt A., Melzl H., Reischl U., Cisar J. O., Probing of microbial biofilm communities for coadhesion partners. Appl. Environ. Microbiol. 80, 6583–6590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whiley R. A., Beighton D., Current classification of the oral streptococci. Oral. Microbiol. Immunol. 13, 195–216 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Whiley R. A., Hardie J. M., Streptococcus vestibularis sp. nov. from the human oral cavity. Int. J. Syst. Bacteriol. 38, 335–339 (1988). [DOI] [PubMed] [Google Scholar]
- 26.Doyuk E., Ormerod O. J., Bowler I. C. J. W., Native valve endocarditis due to Streptococcus vestibularis and Streptococcus oralis. J. Infect. 45, 39–41 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Tufan M. A., et al. , Spondylodiscitis and endocarditis caused by S. vestibularis. Braz. J. Infect. Dis. 14, 377–379 (2010). [PubMed] [Google Scholar]
- 28.Stowell S. R., et al. , Microbial glycan microarrays define key features of host-microbial interactions. Nat. Chem. Biol. 10, 470–476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown S., Meredith T., Swoboda J., Walker S., Staphylococcus aureus and Bacillus subtilis W23 make polyribitol wall teichoic acids using different enzymatic pathways. Chem. Biol. 17, 1101–1110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuru E., Tekkam S., Hall E., Brun Y. V., Van Nieuwenhze M. S., Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc. 10, 33–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vollmer W., Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32, 287–306 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Guérin H., Kulakauskas S., Chapot-Chartier M.-P., Structural variations and roles of rhamnose-rich cell wall polysaccharides in Gram-positive bacteria. J. Biol. Chem. 298, 102488 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavelle K., Sinderen D. V., Mahony J., Cell wall polysaccharides of Gram positive ovococcoid bacteria and their role as bacteriophage receptors. Comput. Struct. Biotechnol. J. 19, 4018–4031 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mistou M.-Y., Sutcliffe I. C., van Sorge N. M., Bacterial glycobiology: Rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol. Rev. 40, 464–479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovacs C. J., Faustoferri R. C., Bischer A. P., Quivey R. G. Jr., Streptococcus mutans requires mature rhamnose-glucose polysaccharides for proper pathophysiology, morphogenesis and cellular division. Mol. Microbiol. 112, 944–959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar R. J., et al. , Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides. Nat. Chem. Biol. 15, 463–471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montague E. A., Knox K. W., Antigenic components of the cell wall of Streptococcus salivarius. J. Gen. Microbiol. 54, 237–246 (1968). [DOI] [PubMed] [Google Scholar]
- 38.Weerkamp A. H., McBride B. C., Identification of a Streptococcus salivarius cell wall component mediating coaggregation with Veillonella alcalescens VI. Infect. Immun. 32, 723–730 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonnell B., et al. , A cell wall-associated polysaccharide is required for bacteriophage adsorption to the Streptococcus thermophilus cell surface. Mol. Microbiol. 114, 31–45 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Szymczak P., et al. , Cell wall glycans mediate recognition of the dairy bacterium Streptococcus thermophilus by bacteriophages. Appl. Environ. Microbiol. 84, e01847-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rush J. S., et al. , PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides. Nat. Comm. 13, 590 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Beek S. L., et al. , Streptococcal dTDP-L-rhamnose biosynthesis enzymes: Functional characterization and lead compound identification. Mol. Microbiol. 111, 951–964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rush J. S., et al. , The molecular mechanism of N-acetylglucosamine side-chain attachment to the Lancefield group A carbohydrate in Streptococcus pyogenes. J. Biol. Chem. 292, 19441–19457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breitenbach Barroso Coelho L. C., et al. , Lectins as antimicrobial agents. J. Appl. Microbiol. 125, 1238–1252 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Vaishnava S., et al. , The Antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 334, 255–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blenda A. V., et al. , Galectin-9 recognizes and exhibits antimicrobial activity toward microbes expressing blood group-like antigens. J. Biol. Chem. 298, 1–18 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen P. A., Mandel U., Therkildsen M. H., Clausen H., Differential expression of human high-molecular-weight salivary mucin (MG1) and low-molecular-weight salivary mucin (MG2). J. Dent. Res. 75, 1820–1826 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Takehara S., Yanagishita M., Podyma-Inoue K. A., Kawaguchi Y., Degradation of MUC7 and MUC5B in human saliva. PLoS One 8, e69059 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhury N. M. A., Proctor G. B., Karlsson N. G., Carpenter G. H., Flowers S. A., Reduced Mucin-7 (Muc7) sialylation and altered saliva rheology in Sjögren’s Syndrome associated oral dryness. Mol. Cell. Proteom. 15, 1048–1059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karlsson N. G., Thomsson K. A., Salivary MUC7 is a major carrier of blood group I type O-linked oligosaccharides serving as the scaffold for sialyl Lewis x. Glycobiology 19, 288–300 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Zalewska A., Zwierz K., Zółkowski K., Gindzieński A. J. A. B. P., Structure and biosynthesis of human salivary mucins. Acta. Biochim. Pol. 47, 1067–1079 (2000). [PubMed] [Google Scholar]
- 52.Thomsson K. A., et al. , The salivary mucin MG1 (MUC5B) carries a repertoire of unique oligosaccharides that is large and diverse. Glycobiology 12, 1–14 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Thomsson K. A., Schulz B. L., Packer N. H., Karlsson N. G., MUC5B glycosylation in human saliva reflects blood group and secretor status. Glycobiology 15, 791–804 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Klement E., Lipinszki Z., Kupihár Z., Udvardy A., Medzihradszky K. F., Enrichment of O-GlcNAc modified proteins by the periodate oxidation-hydrazide resin capture approach. J. Proteome Res. 9, 2200–2206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frenkel E. S., Ribbeck K., Salivary mucins in host defense and disease prevention. J. Oral Microbiol. 7, 29759–29759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins M. D., “The Genus Gemella” in The Prokaryotes: Volume 4: Bacteria: Firmicutes, Cyanobacteria, Dworkin M., et al., Eds. (Springer, 2006), pp. 511–518. [Google Scholar]
- 57.Gobbetti M., Calasso M., “STREPTOCOCCUS | Introduction” in Encyclopedia of Food Microbiology, Batt C. A., Tortorello M. L., Eds. (Academic Press, Oxford, ed. 2, 2014), pp. 535–553. [Google Scholar]
- 58.Hogg S. D., Whiley R. A., De soet J. J., Occurrence of lipoteichoic acid in oral streptococci. Int. J. Syst. Bacteriol. 47, 62–66 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Johansson M. E. V., Sjövall H., Hansson G. C., The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 10, 352–361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh S., Streptococcus vestibularis, Gemella haemolysans, Streptococcus gwangjuense whole genome shotgun sequencing project. GenBank. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA880893. Deposited 15 September 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Bacterial whole genome sequence and assembly data have been deposited in GenBank (BioProject accession number: PRJNA880893; https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA880893) (60). All study data are included in the article and/or SI Appendix.