Significance
Proper repression of the floral homeotic genes AP3, PI, and AG in leaf primordia and activation of them in floral meristems is essential for reproductive fitness. Many studies have revealed molecular details about the activation of AP3, PI, and AG in floral meristems, but relatively less is known about how they are repressed in leaves and how this repression is lifted in floral meristems. Two genes encoding Arabidopsis ZINC FINGER PROTEIN (ZFP) transcription factors, ZP1 and ZFP8, are highly expressed in leaf primordia and down-regulated in floral primordia. Here, we show that ZP1 and ZFP8 repress these floral homeotic genes in leaves. LFY and AP1 down-regulate ZP1 and ZFP8 in floral meristems, lifting the repression on floral homeotic genes.
Keywords: ZP1, ZFP8, AP3, PI, AG
Abstract
Organ initiation from the shoot apical meristem first gives rise to leaves during vegetative development and then flowers during reproductive development. LEAFY (LFY) is activated after floral induction and together with other factors promotes the floral program. LFY functions redundantly with APETALA1 (AP1) to activate the class B genes APETALA3 (AP3) and PISTILLATA (PI), the class C gene AGAMOUS (AG), and the class E gene SEPALLATA3, which leads to the specification of stamens and carpels, the reproductive organs of flowers. Molecular and genetic networks that control the activation of AP3, PI, and AG in flowers have been well studied; however, much less is known about how these genes are repressed in leaves and how their repression is lifted in flowers. Here, we showed that two genes encoding Arabidopsis C2H2 ZINC FINGER PROTEIN (ZFP) transcription factors, ZP1 and ZFP8, act redundantly to directly repress AP3, PI, and AG in leaves. After LFY and AP1 are activated in floral meristems, they down-regulate ZP1 and ZFP8 directly to lift the repression on AP3, PI, and AG. Our results reveal a mechanism for how floral homeotic genes are repressed and derepressed before and after floral induction.
In most angiosperms, the shoot first produces leaves then flowers in their life cycle. The transition from vegetative development to reproductive development is tightly controlled by multiple pathways that respond to both endogenous and environmental cues (1, 2). These pathways converge on a set of floral integrators that activate floral meristem identity (FMI) genes such as LEAFY (LFY) and APETALA 1 (AP1). LFY and AP1 activate each other and a set of floral homeotic genes to specify floral organ identities in floral meristems (3–5). An Arabidopsis flower consists of four whorls of floral organs with four sepals in the first whorl, four petals in the second whorl, six stamens in the third whorl, and two fused carpels in the fourth whorl. These different floral organ identities are specified by different combinations of floral homeotic gene activities as specified in the ABCE model. The outer sepals are specified by class A+E activities and the petals specified by class A+B+E activities. The reproductive organs of a flower, stamen and carpel, are specified by class B+C+E and class C+E activities, respectively (6). AP1, a class A gene in addition to its role in FMI, the class B genes AP3 and PISTILLATA (PI), the class C gene AGAMOUS (AG), and the class E gene SEPALLATA 3 (SEP3) are all direct targets of LFY activation. SEP3 is not only an LFY direct target, but also a LFY cofactor in the direct activation of AP3, PI, and AG (7, 8).
While much work has focused on the activation of AP3, PI, and AG in the floral meristems, little is known about how these genes are silenced in leaves and how this silencing is released in FMs. Indeed, silencing of these genes is important for proper development of a plant (9, 10). AP3 and AG are silenced in vegetative leaves by polycomb repressive complex 2 (PRC2), which catalyzes trimethylation of histone 3 lysine 27 (H3K27me3) on their chromatin (11, 12). Whether transcription factors are involved in the silencing of AP3, PI, and AG and how the silencing is lifted remain unknown. Here, we showed that two zinc finger protein transcription factors, Arabidopsis ZINC FINGER PROTEIN 1 (AtZP1, ZP1) and ZFP8, are involved in the repression of AP3, PI, and AG in leaves. It was reported that ZP1 is involved in the suppression of root hair initiation and elongation, and ZFP8 is involved in epidermal trichome initiation (13–15). ZP1 has an Ethylene-responsive element binding factor-associated Amphiphilic Repression (EAR) motif that carries transcriptional repressor activities (13). Here, we found that ZP1 and ZFP8 repress AP3, PI, and AG in vegetative leaves and are down-regulated by LFY and AP1 in floral meristems to lift the repression on AP3, PI, and AG. Therefore, LFY activates the expression of AP3, PI, and AG both directly and indirectly to ensure robust production of flowers after floral induction (3, 7, 8).
Results
LFY and AP1 Repress AtZP1 and ZFP8 in Developing Flowers.
The C2H2 zinc finger protein AtZP1 (AT4G17810, ZP1) is required for root hair initiation and elongation (13), but its roles in the aerial parts of a plant remain unknown. To analyze the role of ZP1 in Arabidopsis shoot development, we cloned a 5.5 kb-DNA fragment upstream of the ZP1 transcription start site (TSS) and fused it with β-glucuronidase (ZP1::GUS). A temporal analysis of ZP1 expression in the shoot by this construct showed that ZP1 is highly expressed in developing vegetative leaves and is down-regulated in flowers and fruits (Fig. 1A). Next, we examined vegetative and reproductive development in a zp1 T-DNA insertion line (zp1-3, SAIL_24_B09). qRT-PCR analysis on this mutant line showed that there was an approximately 75% reduction of ZP1 transcripts in this mutant, suggesting that this might be a strong allele (SI Appendix, Fig. S1 A and B). Further phenotypic analysis on the number of juvenile leaves, adult leaves, cauline leaves, and floral organs showed that there was no significant difference between this mutant and Col-0 (SI Appendix, Fig. S1 C–E). However, plants overexpressing ZP1 (35S::ZP1) never flowered in long-day (LD) conditions (SI Appendix, Fig. S1F). ZP1 is a member of the C2H2 ZFP transcription factor family that consists of 176 members, and the absence of a phenotype in the loss-of-function mutant might result from genetic redundancy. To identify potential C2H2 ZFPs that might act redundantly with ZP1, we performed RNA-seq analysis on 2-wk-old shoot apex and 4-wk-old inflorescence of Col-0. ZP1 has one zinc finger domain and belongs to the C1-1i subset with other 32 members (SI Appendix, Fig. S2). Our results showed that in the C1-1i subset family, ZP1, ZFP4, ZFP7, and ZFP8 were down-regulated more than two fold in 4-wk-old inflorescence, while SUPERMAN (SUP), KNUCKELS (KNU), and RABBIT EARS (RBE), three ZFPs that were known to function in floral organ development, were up-regulated in 4-wk-old inflorescence. ZFP8 was reported to function redundantly with GLABROUS INFLORESCENCE STEMS (GIS) and GIS2 to initiate epidermal trichomes in leaves, fruits, and stems (14). To confirm the RNA-seq results, we performed qRT-PCR analysis in LD-grown 2-wk-old shoot apex and 4-wk-old Col-0 inflorescence. Floral organs were not visible in 2-wk-old plants but were well developed in 4-wk-old plants, and the floral organ identity ABCE genes (AP1, AP2, AP3, PI, AG, and SEP3) were highly up-regulated in 4-wk-old tissues (Fig. 1B). ZP1, ZFP4, ZFP7, and ZFP8 were down-regulated about 60 to 80% from 2-wk to 4-wk-old tissues (Fig. 1B and (SI Appendix, Fig. S3A), while SUP, KNU, and RBE were up-regulated 5 to 100 folds (SI Appendix, Fig. S3A). We examined the relative expression levels of ZP1, ZFP4, ZFP7, and ZFP8 in cotyledons; young leaves (petiole have not developed yet); mature leaves; cauline leaves; the 1-wk-old shoot apex; and flowers at stages 4 to 6, stages 7 to 9, and stages 10 to 12 by qRT-PCR. Our results showed that ZP1 was expressed at higher levels in young leaves than that of cotyledons, it was down-regulated 30 to 40% in mature leaves, cauline leaves, 1-wk-old shoot apex, and it was down-regulated more than 90% in floral organs from stages 4 to 12 (Fig. 1C). Transcripts of ZFP4, ZFP7, and ZFP8 were not significantly changed in young leaves and mature leaves, nor shoot apex compared to their levels in cotyledons. However, they were down-regulated 90% in floral organs from stages 4 to 12 (Fig. 1C and SI Appendix, Fig. S3C). As LFY and AP1 promote FMI and floral organogenesis, we wondered whether downregulation of ZP1, ZFP4, ZFP7, and ZFP8 in Col-0 inflorescence might be regulated by LFY and AP1 (3, 4, 8, 16). To investigate the possible regulation of ZP1, ZFP4, ZFP7, and ZFP8 by LFY and AP1, we analyzed the abundance of ZP1, ZFP4, ZFP7, and ZFP8 transcripts in 4-wk-old Col-0, lfy-1, and ap1-10 inflorescence. AP1, AP3, PI, AG, and SEP3, direct targets of LFY (8, 16, 17), were down-regulated in lfy-1, while ZP1, ZFP4, and ZFP8 were up-regulated in lfy-1 (Fig. 1D and SI Appendix, Fig. S3D). Transcripts of AP3, PI, and SEP3 were not affected in ap1-10; however, ZP1, ZFP4, and ZFP8 were up-regulated in ap1-10 (Fig. 1E and SI Appendix, Fig. S3E).
Fig. 1.
LFY and AP1 repress ZP1 and ZFP8 during floral organogenesis. (A) Expression of ZP1::GUS in 1-wk-, 2-wk-, 3-wk-, and 4-wk-old plants grown in LD conditions. (Scale bars: 3 mm.) (B) qRT-PCR analysis of ABCE genes, ZP1 and ZFP8 in 2-wk-old Col shoot apex (including the SAM and leaf primordia smaller than 2 mm), and 4-wk-old Col inflorescence (excluding flowers at stage 12 and above). Levels of transcripts of each gene in 2-wk-old shoot apex were set to 1, and the values were log2 of their fold change. (C) qRT-PCR analysis of ZP1 and ZFP8 in different tissues of Col. COT: cotyledons; YL: young leaf; ML: mature leaf; CL: cauline leaf; SA: 1-wk-old shoot apex; FS4-6: flowers at stage 4 to 6; FS7-9: flowers at stage 7 to 9; FS10-12: flowers at stage 10 to 12. Transcripts from ZP1 or ZFP8 in cotyledons were set to 1. * Significantly different from the levels in COT, P < 0.05, t test. (D and E) qRT-PCR analysis of ABCE genes, ZP1 and ZFP8 in 4-wk-old lfy-1 (D), and ap1-10 (E). Transcripts from Col were set to 1, and the values in D were log2 of their fold change. Statistical analysis in B–E: *P < 0.05, **P < 0.01, ***P < 0.001, ns, p>0.05, one-way ANOVA. (F) In situ hybridization analysis of ZP1 and ZFP8 in 1-wk-old Col shoot apices, 4-wk-old Col, ap1-10, and lfy-1 inflorescence apices. Insets are ZP1 and ZFP8 in 1w zp1-3 and zfp8, respectively. (Scale bars: 50 µm.)
To examine how LFY and AP1 repress ZP1 and ZFP8 spatially, we performed in situ hybridization analysis. ZP1 and ZFP8 could be detected in shoot apical meristems (SAM) and leaf primordia from 1-wk-old Col-0 (Fig. 1F). In reproductive tissues, ZP1 and ZFP8 mRNA could only be slightly detected in the inflorescence and stage 1 to stage 4 floral meristems. However, they could be detected at higher levels in lfy-1 and ap1-10 stage 1 to 4 floral meristems (Fig. 1F). Together, the qRT-PCR and in situ hybridization results showed that ZP1 and ZFP8 are highly expressed in vegetative tissues and were down-regulated in floral meristems by LFY and AP1 when floral organ identity genes are activated.
LFY and AP1 Bind to ZP1 and ZFP8 Directly.
The downregulation of ZP1 and ZFP8 by LFY and AP1 prompted us to examine whether LFY and AP1 directly regulate ZP1 and ZFP8 expression. We made a LFY::GFP-LFY construct (18), transformed it into lfy-1 heterozygotes, and selected LFY::GFP-LFY lfy-1 that complemented lfy-1 mutant (SI Appendix, Fig. S4). Flower development is synchronized in 35S::AP1-GR ap1 cal plant, and floral organ development could be activated by dexamethasone (Dex) treatment (17). We collected inflorescence for chromatin immunoprecipitation qPCR (ChIP-qPCR) analysis 24 h after mock or Dex treatment. Potential LFY or AP1 binding sites upstream of ZP1 and ZFP8 were selected for ChIP-qPCR analysis based on previous genome-wide ChIP seq analysis that showed peaks for LFY and AP1 association (17, 19). Our ChIP-qPCR analysis showed that LFY and AP1 bind to the same regions of the ZFP8 promoter, and LFY has a broader binding region than that of AP1 at ZP1 promoter (Fig. 2).
Fig. 2.
LFY and AP1 bind to ZP1 and ZFP8 directly. (A and B) Schematic structure of ZP1 (A) and ZFP8 (B) and localization of potential LFY binding sites. Arrow indicates TSS, and gray box represents the first exon. (C) ChIP-qPCR analysis of ZP1 and ZFP8 fragments in Col and LFY::GFP-LFY lfy-1. (D) ChIP-qPCR analysis of ZP1 and ZFP8 fragments in 35S::AP1-GR ap1 cal plant treated by mock or Dex. ns denotes not significantly different groups, P > 0.05. Stars denote significantly different groups, *P < 0.05, **P < 0.01, ***P < 0.001, ns, p > 0.05, one-way ANOVA. The TA3 retrotransposon (AT1G37110) served as a negative control locus for ChIP-qPCR.
Expression of ZP1 and ZFP8 under the Control of LFY and AP1 Promoters Prevents Activation of Class B and C Genes.
To examine the biological relevance of the downregulation of ZP1, ZFP4, ZFP7, and ZFP8 by LFY and AP1, we expressed the coding region of ZP1, ZFP4, ZFP7, and ZFP8 behind the promoters of LFY (18) and AP1 (3.5 kb). LFY::ZP1, LFY::ZFP4, LFY::ZFP7, LFY::ZFP8, AP1::ZP1, AP1::ZFP4, AP1::ZFP7, and AP1::ZFP8 were transformed into Col-0, and vegetative and reproductive development was analyzed in these plants. The number of rosette leaves was not significantly changed in these transgenic plants, except that there was a small reduction of rosette leaves in AP1::ZFP8 (SI Appendix, Fig. S5A). Inflorescences from LFY::ZP1 have many visible leaves, and there were large bracts subtending flowers as the inflorescence elongated (Fig. 3C). The basal flowers from LFY::ZP1 are fertile, with reduced number of petals and stamens (Fig. 3I). Many of the later-arising floral meristems from LFY:: ZP1 were arrested at early stages such that petals, stamens, and carpels were not visible (Fig. 3 I, Inset). LFY::ZFP8 plant produced more secondary inflorescences than those of Col-0 (SI Appendix, Fig. S5B), and flowers from LFY::ZFP8 were subtended with small bracts, had no petals, occasionally had 1 or 2 stamens (SI Appendix, Fig. S5C), and had carpels that were not properly fused (Fig. 3J). Flowers from AP1::ZP1 and AP1::ZFP8 were very much like flowers from LFY::ZFP8, and they all look like flowers lack of class B and C activities (Fig. 3 K and L). Floral organ identities in LFY::ZFP4, LFY::ZFP7, AP1::ZFP4, and AP1::ZFP7, however, were not affected (SI Appendix, Fig. S5 C and D).
Fig. 3.
Expression of ZP1 and ZFP8 driven by LFY and AP1 promoter, respectively, resulted in loss of floral organ identity. (A-F) Top view of inflorescences from Col (A), lfy-1 (B), LFY::ZP1 (C), LFY::ZFP8 (D), AP1::ZP1 (E), and AP1::ZFP8 (F). (G-L) Flowers from Col (G), lfy-1 (H),LFY::ZP1 (I), LFY::ZFP8 (J), AP1::ZP1 (K), and AP1::ZFP8 (L). In LFY::ZP1, basal flowers had reduced number of petals and stamens and apical flowers were arrested at earlier development stages that petals and stamens were not visible (Inset), and bracts developed with each flower (arrows). In LFY::ZFP8, AP1::ZP1, and AP1::ZFP8, petals and stamens were not properly developed, and carpels were not properly fused, resembling lfy-1 flowers. (M-R) Inflorescences from Col (M), lfy-1 (N),LFY::ZP1 (O), LFY::ZFP8 (P), AP1::ZP1 (Q), and AP1::ZFP8 (R). (Scale bars in top two rows: 1 mm, scale bars in bottom row: 5 mm.)
qRT-PCR analyses on these transgenic plants showed that LFY was slightly down-regulated (about 10 to 15%) in LFY::ZP1 and AP1::ZFP8 and was not changed in LFY::ZFP8 nor in AP1::ZP1. Interestingly, transcripts of AP1 were up-regulated (about 1.5-fold to twofold), and transcripts of AP2 were not changed or slightly down-regulated (Fig. 4 A–D). The class B genes AP3 and PI and the class C gene AG were down-regulated in all the backgrounds. In LFY::ZP1 which produces some fertile flowers, transcripts of AP3, PI, and AG were down-regulated 60 to 70%. In LFY::ZFP8, AP1::ZP1, and AP1::ZFP8, which have fewer fertile flowers than those of LFY::ZP1, transcripts of AP3, PI, and AG were down-regulated 70 to 90% (Fig. 4 A–D). Transcripts of SEP3 were not significantly changed in any of them (Fig. 4 A–D), suggesting that the downregulation of AP3, PI, and AG in the transgenic plants was not caused by reduced activities of LFY. Upregulation of AP1 may have resulted from reduced activities of AG, which acts antagonistically to AP1 (20, 21).
Fig. 4.
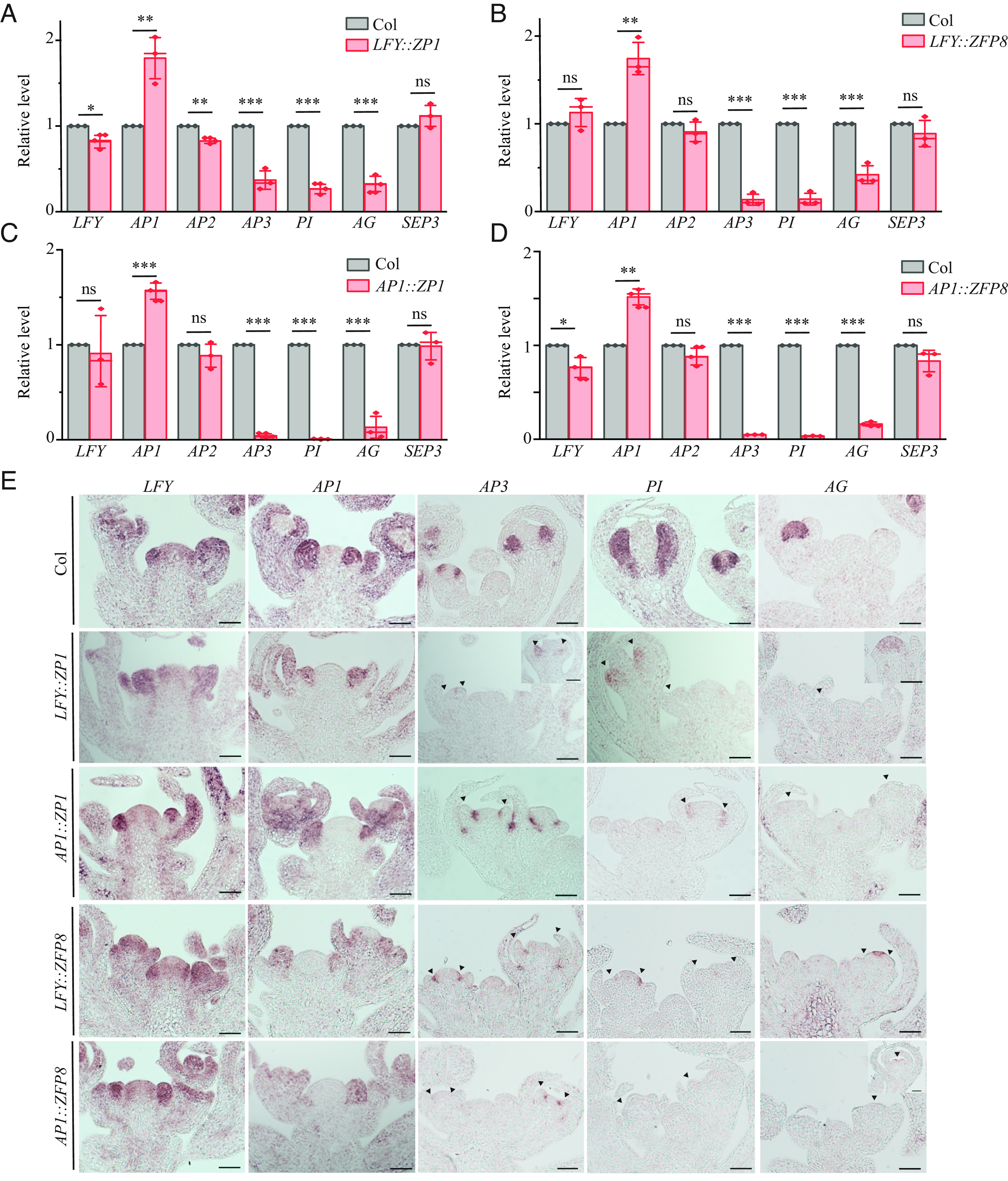
Expression of ZP1 and ZFP8 driven by LFY promoter and AP1 promoter resulted in downregulation of AP3, PI, and AG. (A–D) qRT-PCR analysis of LFY and ABCE genes in 4w-old inflorescence of Col, LFY::ZP1 (A), LFY::ZFP8 (B), AP1::ZP1(C), and AP1::ZFP8 (D). Transcripts of each gene in Col were set to 1, and values were relative fold to Col. Values represent the mean ± SE from three biological replicates (diamonds). ns denotes not significant, P > 0.05, *P< 0.05, **P< 0.01, ***P< 0.001, one-way ANOVA. (E) In situ hybridization analysis of LFY, AP1, AP3, PI, and AG in 4-wk-old inflorescence apices of Col, LFY::ZP1, AP1::ZP1, LFY::ZFP8, and AP1::ZFP8. (Scale bars: 50 µm.)
Next, we examined the spatial expression of LFY, AP1, AP3, PI, and AG in these transgenic plants by in situ hybridization. LFY and AP1 were detected in stage 1 and stage 2 floral meristems in the transgenic plants, comparable to their expression in Col-0 (Fig. 4E). In Col-0, AP3 and PI were easily detected in stage 3 and stage 4 floral meristems in the second and third whorls where petal and stamen would be initiated. AP3 and PI were absent or much reduced in the second and third whorls of LFY::ZP1, LFY::ZFP8, AP1::ZP1, and AP1::ZFP8 or only detected at the base of the third whorl close to its boundary with the fourth whorl (Fig. 4E). In Col-0, AG was expressed in the center of the floral meristem that would give rise to stamens and carpels. However, AG expression was dramatically reduced or present in a narrower domain in these transgenic plants (Fig. 4E). Together, the qRT-PCR and in situ hybridization analysis showed that AP3, PI, and AG were significantly down-regulated in these transgenic plants, while LFY and AP1 were maintained at similar levels as they were in Col-0. Our results suggest that downregulation of ZP1 and ZFP8 in the LFY and AP1 expression domain is essential for upregulation of AP3, PI, AG, and proper development of petal, stamen, and carpel identities.
Rescue of Petal Development in ap1-10 and lfy-1 by Loss-of-Function zp1 zfp8.
LFY and AP1 activate each other, and they function redundantly to activate the expression of AP3 and PI (3–5, 16, 17). To examine whether downregulation of ZP1 and ZFP8 is required for floral organ development, we crossed zp1-3 and zfp8 (14) (SI Appendix, Fig. S6I) to ap1-10 and lfy-1. We first analyzed floral organ development in zp1-3 and zfp8 single mutants and the zp1-3 zfp8 double mutant. Our results showed that mutations in ZP1 and ZFP8 did not affect flowering time, FMI, and floral organ development (SI Appendix, Fig. S6). The ap1-10 mutant produces secondary flowers in the axils of the first whorl leaf-like organs. The primary flower usually does not contain petals, although secondary flowers may produce some petals. ap1-10 zp1-3 and ap1-10 zfp8 double mutants sometimes produced three petals per flower in their secondary flowers. The ap1-10 zp1-3 zfp8 triple mutant produced two, three, or four petals per flower in their secondary flowers more often, significantly more than the average number of petals per flower in the double mutants (Fig. 5 A and C). Most lfy-1 flowers do not produce petals either, with one to two petals being produced occasionally. More lfy-1 zp1-3 flowers had two petals, but the average number of petals per flower is not significantly different from lfy-1. More lfy-1 zp1-3 zfp8 triple mutant flowers produced two petals, and the average number of petals per flower was significantly higher than that of the double mutants (Fig. 5 A and D). The occurrence of petals was significantly increased in ap1-10 zp1-3 zfp8 and lfy-1 zp1-3 zfp8 triple mutants (Fig. 5 A, C, and D), suggesting that class B floral organ identity genes might be up-regulated in these backgrounds. We then examined the transcripts of AP3, PI, and AG by in situ hybridization. Our results showed that AP3, PI, and AG were easily detected in Col-0 and zp1-3 zfp8 stage 3 and stage 4 floral meristems, while they could hardly be detected in lfy-1. AP3 and PI expression was partially restored, while AG was only slightly restored in lfy-1 zp1-3 zfp8 stage 3 and stage 4 floral meristems (Fig. 5B). Together, our results suggest that LFY and AP1 promote the activation of AP3, PI, and AG both directly and indirectly, and the indirect pathway involves redundant actions of ZP1, ZFP8, and other factors.
Fig. 5.
Loss-of-function zp1-3 zfp8 partially rescues petal development in ap1-10 and lfy-1. (A) Flowers from Col and mutants. (Scale bars: 1 mm.) (B) In situ hybridization analysis of AP3, PI, and AG in Col, zp1-3 zfp8, lfy-1, and lfy-1 zp1-3 zfp8 floral meristems. (Scale bars: 50 µm.) (C and D) Statistical analysis on the number of petals per flower in ap1-10 zfp mutants (C) and lfy-1 zfp mutants (D). Shared letters above each group (C and D) indicate not significantly different groups, p > 0.05. Different letters above each group indicate significantly different groups. P < 0.05, one-way ANOVA.
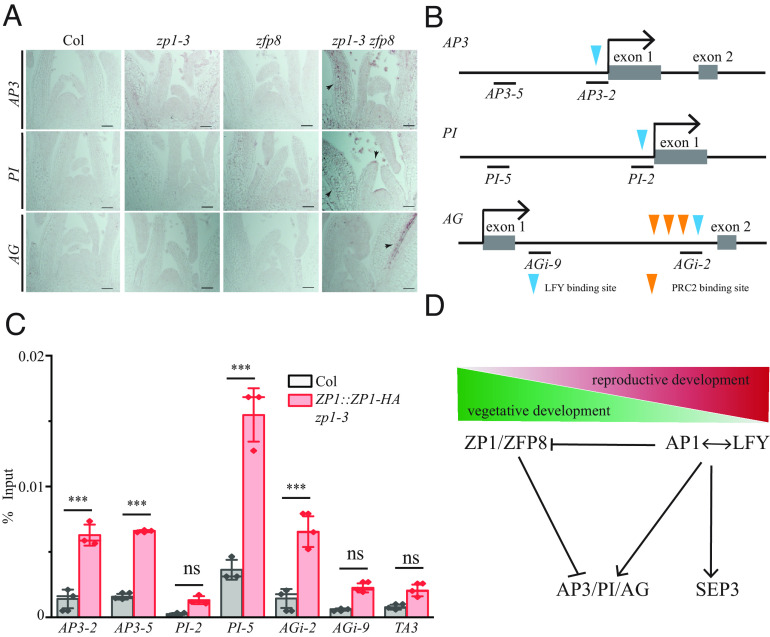
ZP1 Binds to AP3, PI, and AG Directly to Repress Their Expression in Vegetative Tissues.
ZP1 and ZFP8 are expressed at relatively high levels in vegetative tissues compared to their levels in flowers at stages 4 to 12, while AP3, PI, and AG are expressed at low levels in 2-wk-old shoot apex when flowers are not visible and are expressed at high levels in 4-wk-old inflorescence when flowers are well developed (Fig. 1). To examine whether ZP1 and ZFP8 repress AP3, PI, and AG in vegetative tissues, we fixed 11-d-old LD-grown shoot apices from Col-0, zp1-3, zfp8, and zp1-3 zfp8 for in situ hybridization analysis. AP3, PI, and AG could not be detected in Col-0 and zp1-3, nor zfp8, but could be detected in a few cells in zp1-3 zfp8 leaf primordia (Fig. 6A). To examine whether ZP1 represses AP3, PI, and AG directly, we transformed ZP1::ZP1-HA into zp1-3 mutant and selected transgenic lines which expressed ZP1 at a higher level than Col-0 (SI Appendix, Fig. S6H). Our ChIP-qPCR results showed that ZP1 binds to the region AP3-2 (close to the TSS, a LFY binding site) (8, 22) and AP3-5 (~400 bp upstream of the TSS). ZP1 binds to PI-5 (~1,100 bp upstream of the TSS), but not to PI-2 (close to TSS, a LFY binding site) (Fig. 6 B and C) (8). These results showed that ZP1 could bind to AP3 and PI independent of LFY. The ChIP-qPCR results are consistent with the in situ hybridization results that AP3 and PI could be easily detected in lfy-1 zp1-3 zfp8, though in a narrower domain, as well as the phenotypic analysis showing that petal development could be partially rescued in lfy-1 zp1-3 zfp8. Because many regulatory elements for AG lie in the second intron of AG (23), we examined the binding of ZP1 to the second intron of AG. Our mapping of the ZP1 binding to AG regulatory sequences showed that ZP1 associates with the 3′end of its second intron, which is also bound by LFY and CURLY LEAF (CLF, a methyltransferase in PRC2) (12, 22), but not to the beginning of the second intron (AGi-9) (Fig. 6 B and C)). This suggests that ZP1 and LFY may compete for binding to AG, which is consistent with the in situ hybridization analysis that AG could hardly be up-regulated in lfy-1 zp1-3 zfp8, and that carpel development could not be rescued in lfy-1 zp1-3 zfp8 (SI Appendix, Fig. S6J). Together, our results showed that ZP1 binds to AP3, PI, and AG directly in vegetative tissues to repress their expression.
Fig. 6.
ZP1 binds to AG, AP3, and PI directly. (A) In situ hybridization analysis of AP3, PI, and AG in LD-grown 11-d old shoot apices from Col, zp1-3, zfp8, and zp1-3 zfp8 double mutant. (B) Schematic structure of AP3, PI, and AG and DNA fragments tested for ZP1 binding. Arrow indicates TSS, and gray box represents exons. LFY and PRC2 binding sites are represented as blue and orange triangles, respectively. (C) ChIP-qPCR analysis of AP3, PI, and AG fragments in Col and ZP1::ZP1-HA. ns denotes not significantly different groups, P > 0.05. Stars denote significantly different groups, ***P< 0.001, two-way ANOVA. (D) Model for activation of AP3, PI, and AG. In vegetative tissues where ZP1 and ZFP8 are highly expressed, they function redundantly (probably together with other zinc finger proteins) to repress AP3, PI, and AG. Once LFY and AP1 are activated in floral meristems, they down-regulate ZP1 and ZFP8 to release the repression of AP3, PI, and AG. LFY and AP1 activate AP3, PI, and AG directly in parallel to the derepression of AP3, PI, and AG. SEP3 is activated by LFY directly and is not repressed by ZP1 and ZFP8.
Discussion
Vegetative development precedes reproductive development in most flowering plants. The homeotic genes AP3 and AG are repressed by PRC2 in vegetative tissues (11, 12). However, transcription factors mediating the repression of AP3, PI, and AG were unknown. Here, we showed that two ZFP transcription factors, ZP1 and ZFP8, are highly expressed in vegetative tissues and act redundantly to repress AP3, PI, and AG in these tissues. Activation of LFY and AP1 after floral induction leads directly to the downregulation of ZP1 and ZFP8. Thus, LFY and AP1 activate AP3, PI, and AG both directly and indirectly (Fig. 6D). Our results suggest a mechanism for the temporal regulation of AP3, PI and AG, which ensures fitness and robust activation of floral program.
Ectopic expression of ZP1 or ZFP8 in floral meristems is sufficient to down-regulate AP3, PI, and AG in floral meristems. However, inactivating ZP1 and ZFP8 simultaneously could only slightly derepress AP3, PI, and AG in leaves. This suggests the presence of additional factors that function redundantly with ZP1 and ZFP8. SUP encodes a related C2H2 zinc finger protein with an EAR repression domain. SUP functions to define the boundary between whorl 3 and whorl 4 to restrict the expression of AP3 from whorl 4 (24). Mutations in sup resulted in an expansion of AP3 expression into whorl 4 and consequently a change of the cell fate from female to male with additional stamens replacing carpels in the fourth whorl (24–28). RBE, encoding another related C2H2 zinc finger protein, is expressed in cells giving rise to petal primordia, and is required for proper sepal and petal development through repressing AG in the outer whorls (29, 30). Though SUP and RBE repress AP3, PI, and AG in floral meristems, it is unlikely that they function redundantly with ZP1 and ZFP8 to maintain the repression of AP3, PI, and AG in vegetative leaves. SUP and RBE are expressed at low levels in vegetative tissues and high in floral meristems, which is opposite to the expression pattern of ZP1 and ZFP8 (SI Appendix, Fig. S3 A and B). Finding zinc finger proteins that function redundantly with ZP1 and ZFP8 in repressing homeotic gene expression in leaves is essential for understanding the temporal regulation of homeotic genes in leaves and flowers.
Another C2H2 zinc finger protein KNUCKLES (KNU), that functions in regulating floral determinacy, shares a conserved C2H2 zinc finger domain and an EAR domain with SUP, ZP1, and ZFP8 (13) (SI Appendix, Fig. S7). Genetic and molecular analyses of KNU, SUP, and ZP1 showed that they are transcriptional repressors (13, 27, 31). The transcriptional repressor activities in them might be carried out through the interaction between the EAR domain and TOPLESS (TPL), which is a transcriptional repressor that recruits histone deacetylase19 (32, 33). Several lines of evidence suggest that their repression activities could also be carried out through interaction with PRC2. SUP represses the auxin biosynthesis genes YUCCA1/4 (YUC1/4) by recruiting PRC2 to YUC1/4 to fine-tune local auxin signaling to determine floral meristem determinacy (27). KNU recruits PRC2 to initiate and maintain downregulation of WUSCHEL (WUS) to regulate floral meristem determinacy (31). Our ChIP-qPCR analysis of the ZP1 binding sites at AG showed that ZP1 binds to the 3′end of the second intron of AG. This region is also bound by PRC2, suggesting that ZP1 might be involved in recruiting PRC2 to AG. In vivo protein–protein interaction analysis showed that both the zinc finger domain and EAR domain are required for KNU and FIE (a protein in the PRC2 complex) interaction and SUP and CLF interaction. However, motif analysis among transcription factors that recruit PRC2 to target loci showed that they share an EAR domain (34), suggesting that the EAR domain may recruit PRC2. The EAR domain in KNU is important for H3ac levels and H3K27me3 levels in vivo, indicating that the EAR domain is important for recruiting histone deacetylases and PRC2 to target loci (34). It remains to be determined whether both the zinc finger domain and EAR domain in ZP1 and ZFP8 are required and how they are required for recruiting PRC2 to AP3 and AG in vegetative leaves.
C2H2 zinc finger proteins contain two cysteine residues and two histidine residues that hold one zinc ion. There are 176 C2H2 zinc finger proteins in Arabidopsis, which are divided into three subfamilies (A, B, and C). Among them, 64 are classified in the C1 subclass (35). A lot of the C1 subclass members are involved in stress responses, but relatively little is known about how they function in plant development (13, 35). This study provides insights into how ZP1 and ZFP8 act to repress floral homeotic gene expression in inappropriate developmental times and tissues. Protein sequence alignment analysis of ZP1 and ZFP8 among Arabidopsis, wild cabbage, tomato, and soybean showed that the zinc finger domain and the EAR domain are conserved among them, suggesting conserved mechanisms and function in dicots (SI Appendix, Fig. S8).
Materials and Methods
Plants were in the Col-0 background and were grown under 16-h light/8-h dark at 22 °C. Either vegetative apices (1-wk-old or 11-d-old shoot apices) or inflorescences of plants were harvested for gene expression analysis. RNA isolation and quantitative RT-PCR were performed as previously described (36). qRT-PCR and ChIP qPCR results are mean ± SEM from three biological replicates. Detailed information on plant material, construction of plasmids for transformation, qRT-PCR analysis, GUS staining analysis, in situ hybridization, and coimmunoprecipitation followed by qPCR analysis is provided in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by a grant from the NSF (IOS 1947274) and start-up funds from the University of South Carolina to M.X. X.L. was supported by start-up funds awarded to Chunhua Zhang from Purdue University and a research assistantship from the Purdue Center for Plant Biology. We thank Dr. Beth Krizek for critical reading of the manuscript.
Author contributions
T.H. and M.X. designed research; T.H., L.D., D.M., and M.X. performed research; T.H., X.L., and M.X. analyzed data; and M.X. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The raw RNA-Seq data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) and are publicly accessible under the BioProject ID PRJNA968041 (https://www.ncbi.nlm.nih.gov/bioproject/968041) (37). The associated metadata, including sample information, experimental design, and sequencing details, can be found within the BioProject records. All other study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Amasino R., Seasonal and developmental timing of flowering. Plant J. Cell Mol. Biol. 61, 1001–1013 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Baurle I., Dean C., The timing of developmental transitions in plants. Cell 125, 655–664 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Wagner D., Sablowski R. W., Meyerowitz E. M., Transcriptional activation of APETALA1 by LEAFY. Science. 285, 582–584 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Weigel D., Meyerowitz E. M., Activation of floral homeotic genes in Arabidopsis. Science. 261, 1723–1726 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Ng M., Yanofsky M. F., Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13, 739–753 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krizek B. A., Fletcher J. C., Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 6, 688–698 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Liu C., Xi W., Shen L., Tan C., Yu H., Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Winter C. M., et al. , LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20, 430–443 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Mizukami Y., Ma H., Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 9, 393–408 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krizek B. A., Meyerowitz E. M., The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development. 122, 11–22 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Goodrich J., et al. , A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Schubert D., et al. , Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25, 4638–4649 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han G., et al. , Arabidopsis ZINC FINGER PROTEIN1 Acts Downstream of GL2 to Repress Root Hair Initiation and Elongation by Directly Suppressing bHLH Genes. Plant Cell 32, 206–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan Y., Liu C., Yu H., Broun P., Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development. 134, 2073–2081 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Zhou Z., et al. , Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 157, 673–682 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb R. S., Hill T. A., Tan Q. K., Irish V. F., Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development. 129, 2079–2086 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann K., et al. , Orchestration of floral initiation by APETALA1. Science., 328, 85–89 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y., et al. , TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 11, 5118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goslin K., et al. , Transcription Factor Interplay between LEAFY and APETALA1/CAULIFLOWER during Floral Initiation. Plant Physiol. 174, 1097–1109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustafson-Brown C., Savidge B., Yanofsky M. F., Regulation of the arabidopsis floral homeotic gene APETALA1. Cell 76, 131–143 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Irish V. F., Sussex I. M., Function of the apetala-1 gene during Arabidopsis floral development. The Plant cell 2, 741–753 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu M. F., et al. , SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 109, 3576–3581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busch M. A., Bomblies K., Weigel D., Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Prunet N., Yang W., Das P., Meyerowitz E. M., Jack T. P., SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc. Natl. Acad. Sci. U.S.A. 114, 7166–7171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goto K., Meyerowitz E. M., Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes. Dev. 8, 1548–1560 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Ito T., Sakai H., Meyerowitz E. M., Whorl-specific expression of the SUPERMAN gene of Arabidopsis is mediated by cis elements in the transcribed region. Curr. Biol. CB 13, 1524–1530 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Xu Y., et al. , SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37, e97499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai H., Krizek B. A., Jacobsen S. E., Meyerowitz E. M., Regulation of SUP expression identifies multiple regulators involved in arabidopsis floral meristem development. Plant Cell 12, 1607–1618 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krizek B. A., Lewis M. W., Fletcher J. C., RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J. Cell Mol. Biol. 45, 369–383 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Tian C., et al. , A gene expression map of shoot domains reveals regulatory mechanisms. Nat. Commun. 10, 141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun B., et al. , Integration of transcriptional repression and polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell 31, 1488–1505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Causier B., Ashworth M., Guo W., Davies B., The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagale S., Rozwadowski K., EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baile F., Merini W., Hidalgo I., Calonje M., EAR domain-containing transcription factors trigger PRC2-mediated chromatin marking in Arabidopsis. Plant Cell 33, 2701–2715 (2021), 10.1093/plcell/koab139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie M., Sun J., Gong D., Kong Y., The roles of Arabidopsis C1–2i subclass of C2H2-type zinc-finger transcription factors. Genes (Basel) 10, 653 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu M., Hu T., Smith M. R., Poethig R. S., Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28, 28–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu T., Li X., Du L., Manuela D., Xu M., RNA-seq of 2-week-old shoot apex and 4-week-old inflorescence of Arabidopsis thaliana (Col-0). NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/968041. Deposited 6 May 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The raw RNA-Seq data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) and are publicly accessible under the BioProject ID PRJNA968041 (https://www.ncbi.nlm.nih.gov/bioproject/968041) (37). The associated metadata, including sample information, experimental design, and sequencing details, can be found within the BioProject records. All other study data are included in the article and/or SI Appendix.