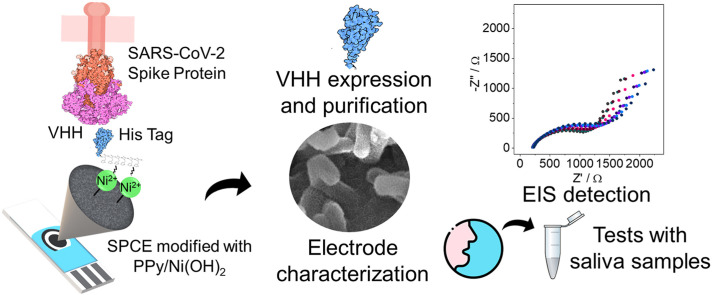
Abstract
SARS-CoV-2 rapid spread required urgent, accurate, and prompt diagnosis to control the virus dissemination and pandemic management. Several sensors were developed using different biorecognition elements to obtain high specificity and sensitivity. However, the task to achieve these parameters in combination with fast detection, simplicity, and portability to identify the biorecognition element even in low concentration remains a challenge. Therefore, we developed an electrochemical biosensor based on polypyrrole nanotubes coupled via Ni(OH)2 ligation to an engineered antigen-binding fragment of heavy chain-only antibodies (VHH) termed Sb#15. Herein we report Sb#15-His6 expression, purification, and characterization of its interaction with the receptor-binding domain (RBD) of SARS-CoV-2 in addition to the construction and validation of a biosensor. The recombinant Sb#15 is correctly folded and interacts with the RBD with a dissociation constant (KD) of 27.1 ± 6.4 nmol/L. The biosensing platform was developed using polypyrrole nanotubes and Ni(OH)2, which can properly orientate the immobilization of Sb#15-His6 at the electrode surface through His-tag interaction for the sensitive SARS-CoV-2 antigen detection. The quantification limit was determined as 0.01 pg/mL using recombinant RBD, which was expressively lower than commercial monoclonal antibodies. In pre-characterized saliva, both Omicron and Delta SARS-CoV-2 were accurately detected only in positive samples, meeting all the requirements recommended by the World Health Organization for in vitro diagnostics. A low sample volume of saliva is needed to perform the detection, providing results within 15 min without further sample preparations. In summary, a new perspective allying recombinant VHHs with biosensor development and real sample detection was explored, addressing the need for accurate, rapid, and sensitive biosensors.
Keywords: COVID-19, Impedimetric biosensor, Nanostructured electrode, Chemometrics, Saliva sample
Graphical abstract
1. Introduction
In December 2019, a group of patients diagnosed with severe febrile respiratory tract disease of unknown origin was reported in Wuhan (China). This disease outbreak was later associated with a previously unknown Coronavirus strain, subsequently named SARS-CoV-2, which quickly spread to all continents and the COVID-19 disease became world known [1]. After only three months, the World Health Organization (WHO) declared it a pandemic. Three years later (April/2023), more than 764,474,000 cases and 6,915,000 deaths have been confirmed worldwide (WHO, 2023). Despite high vaccination rates, COVID-19 still causes worldwide concern due to new variants emergence. The new variants, such as Omicron, can escape immunity acquired from the vaccine or previous infection [[2], [3], [4]] and cause further cases and deaths.
In addition to vaccination, one way to control the disease and the emergence of new variants is large-scale testing to identify and isolate infected patients, thus preventing virus spread. The real-time quantitative polymerase chain reaction (RT-qPCR) test is considered the gold standard assay for SARS-CoV-2 detection. However, the process complexity and equipment cost require centralized testing by highly trained technicians. In addition, the minimum time between test sample collection and result dissemination is approximately 3 h. This makes it difficult to perform tests in real-time and in locations far from diagnostic centers [[5], [6], [7]]. However, the type and quality of the patient specimen, the disease stage, and the degree of viral replication and/or clearance have an impact on test results. These factors are critical not only for PCR-based but also for other diagnostic test systems aiming to detect viral antigens [8].
The enzyme-linked immunosorbent assay and PCR techniques used in the diagnosis of viral diseases are at the forefront of well-established methods. However, these methods have deficiencies in terms of accessibility and rapidness. Concerning the technologies, there are currently more advantageous options available to be applied in the field of diagnostics [9]. Due to the limitations of traditional diagnostic techniques, antigen-detection rapid diagnostic tests have been widely used in the detection of SARS-CoV-2. Rapid tests are more practical as they can be processed outside the laboratory and provide a result within 15–20 min [10]. Although these tests are worldwide available, false positive and negative results are easily obtained, especially in the early days of infection when the viral load is small. Besides that, a nasopharyngeal sample is often collected with the use of a swab, which is a semi-invasive method with a correct collection technique causing discomfort in the testers [11]. In this context, electrochemical biosensors have the potential for the development of rapid tests for COVID-19 diagnosis. Biosensors have high sensitivity, selectivity, shorter response time, lower costs, and ease of application [[12], [13], [14]]. Several types of electrochemical biosensors have been studied to diagnose COVID-19. The mechanisms used for these detections include differential pulse voltammetry [15], square wave voltammetry [16,17], and electrochemical impedance spectroscopy (EIS) [18]. Among these detection techniques, EIS offers advantages compared to other electrochemical methods, such as high sensitivity, ease of signal quantification, minimal manipulation, and simple and rapid pretreatment [19]. In the EIS technique, biomolecule detection could be associated with differences between charge-transfer resistances, once it works by measuring the change in the whole impedance system when a specific binding event of receptors immobilized on a surface occurs. The most used biological recognition elements are antibodies, which bind specifically to an antigen to form an immune complex [20]. After the interaction between antigen and antibody, an increase in charge-transfer resistance occurs due to this interaction, which depends on the amount of immobilized antibodies and the antibody–antigen interaction [21].
Conducting polymers (CPs) are among the best materials for constructing biosensors [22]. CPs are unique materials because they are organic chains that have conjugated double bonds in the structure. In conjugation, the bonds between the carbon atoms alternate between single and double, giving the polymer semiconducting properties [23,24]. The use of CPs in biosensors has advantages such as greater sensitivity, improved conductivity, lower detection limits, cost-benefit, stability, and biocompatibility [23,24]. Polypyrrole (PPy) is one of the most used polymers in biosensor development due to its stability, biocompatibility, electrical conductivity, and good redox properties [25]. Furthermore, due to the important and attractive redox characteristics, the use of a redox probe in the electrolyte is unnecessary since the polymer presents Faradaic reactions [26]. Also, the electrochemical response can be enhanced by using nanostructures due to the larger sensor surface area, which increases sensitivity and selectivity and facilitates the charge transfer processes [27]. PPy is also promising for commercial applications due to greater environmental stability, ease of synthesis, and conductivity than other CPs [28].
Bioreceptor immobilization on the electrode is a crucial step in biosensor fabrication, success in this step ensures biosensor stability and sensitivity [22]. To create a new biosensor that can meet the current challenges, the points that must be considered include changing the surface chemistry for immobilization, improving detection limit and selectivity for better detection, and enabling data processing and analysis. Furthermore, many researchers have focused on increasing the sensitivity and sensing range of biosensors using semiconductor and metallic materials [29]. Immobilization of proteins via histidine-tag (His-tag) is an interesting and accessible method for immobilization since it focuses on protein orientation. The His-tag position on the protein can be controlled by genetic engineering, resulting in protein uniform orientation during immobilization [30,31]. His-tag proteins have a high affinity for bivalent transition metal ion complexes (e.g. Ni2+, Cu2+, Co2+) and can be immobilized on a surface through a coordination bond with the metal complex [32,33]. Nickel hydroxide is one of the materials used to construct biosensors [15]; however, the attachment of biomolecules through His-tag interaction with Ni(OH)2 was not explored so far, and this feature could be very interesting for the oriented biomolecules immobilization directly onto the material surface. In addition, Ni(OH)2 materials have some advantages, such as electrocatalytic properties, cost-effectiveness, high porosity, and electro-inactivity in physiological pH solutions [34,35].
Antibodies or immunoglobulins (Igs) are produced by vertebrate animals as part of their defensive immune response. They are heterodimeric proteins composed of two heavy and two light chains with an Fc region. The different antibody isotypes, including IgM, IgD, IgA, IgE, and IgG, are distinguished by structural differences in their heavy chain constant regions. Thanks to genetic engineering and recombinant antibody techniques, high-quality antibodies can be obtained in the laboratory, as monoclonal antibodies (mAbs) which are specific for one antigen and are produced in the laboratory by B cell hybridoma. Researchers have also created smaller antibody fragments through genetic engineering to enhance antibody performance and reduce assay costs [36,37].
Antigen-binding fragments of heavy chain-only antibodies or VHH are single-domain antibodies derived only from the heavy chain of camelid antibodies [38]. Due to the lack of glycosylation and their small size, the VHHs can be produced via Escherichia coli expression systems, being easier, faster, and with lower cost than other expression platforms, like mammalian cells [39]. Besides that, these antibodies have advantages when compared to conventional ones, such as solubility at higher concentrations, small size, greater resistance to denaturation, stability at high temperatures and high/low pH, and high specificity [[40], [41], [42], [43]]. Despite the advantages of VHHs over conventional antibodies, their use in the construction of electrochemical biosensors is still poorly reported.
In this work, we developed a biosensor based on a VHH as a biorecognition molecule for SARS-CoV-2 detection in saliva samples from SARS-CoV-2 infected patients. Saliva is also a preferable sample medium compared to nasopharyngeal samples, due to the possibility of self-collection without any discomfort, still with comparable sensitivity even with small sample quantities, which also presents high dilution capacity [44]. EIS was employed as the detection method for its advantages, such as high sensitivity, low detection limit, and rapid and non-destructive analysis, being one of the most used electrochemical techniques in biosensors [45,46]. Combining all these advantageous characteristics, in our approach, results could be readily obtained in 15 min, with a limit of quantification of 0.01 pg/mL for the proposed VHH Sb#15-His6. Accurate and sensitive detection was achieved according to WHO standards, in untreated real saliva samples, which is ideal for rapid diagnosis.
2. Materials and methods
2.1. Plasmid constructions
The amino acid sequence of the VHH termed sybody 15 (Sb#15), which recognizes the RBD of SARS-CoV-2, was obtained from the study reported by Walter et al. [47]. Initially, a synthetic gene encoding Sb#15 optimized for E. coli expression cloned into the BglII and XhoI restriction sites of plasmid pET32a in fusion with thioredoxin A (pET32a-Sb#15, Supplementary information) was purchased from the Biomatik Corporation (Kitchener, Ontario, Canada). For expression of Sb#15 without the thioredoxin fusion, the Sb#15 coding sequence was excised from plasmid pET32a-Sb#15 by digestion with the BglII and XhoI restriction enzymes and cloned into the BamHI and SalI sites of pETDuet-1. This cloning was possible since the BglII-BamHI and XhoI-SalI are compatible for ligation although they do not reconstitute any of the restriction sites. The resulting plasmid, pETDuet1-Sb#15, encodes Sb#15 with an N-terminal hexa-histidine tag followed by a tobacco etch virus (TEV) protease recognition site (Supplementary information). The VHH encoded by this plasmid was termed His6-Sb#15. For the expression of Sb#15 without an N-terminal fusion, the Sb#15 coding sequence was amplified by PCR using specific primers (Sb#15-Nco-F: GGCCGGCACCATGGCCCAGGTTCAGCTGGTTGAAAG and Sb#15-Xho-R: TATACTCGAGACCACCGGTTTCCGGCAGACCGCCGCTAACGGTAACCTGGGTAC) and inserted into the NcoI and XhoI restriction sites of plasmid pET28a. In the resulting plasmid (pET28a-Sb#15), the amino acid sequence of Sb#15 is expressed in fusion with a Sortase A recognition site and a C-terminal hexa-histidine tag (Supplementary information). The VHH encoded by this plasmid was termed Sb#15-His6.
A synthetic gene encoding the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein of the Wuhan strain (residues 319–533, accession number QHD43416.1) [48] was acquired from GenScript (Piscataway, NJ, USA) cloned into the HindIII e NotI restriction sites of plasmid pcDNA3.1(+). The coding sequence of the RBD contains a signal peptide at the N-terminal and a deca-histidine tag in the C-terminal. The deca-histidine tag is preceded by a TEV protease recognition site allowing for the removal of the tag if needed (See Supplementary information). For expression in HEK293 cells, the RBD coding sequence was subcloned into NheI and XhoI restriction sites of plasmid pIRES2-EGFP to create pIRES2-EGFP-RBD2.
2.2. Expression and purification of Sb#15
The E. coli strain BL21ΔslyD [49] transformed with pRARE (Novagen/Merck) was used throughout this study as a bacterial expression host. pETDuet1-Sb#15 and pET28a-Sb#15 were transformed into chemically competent BL21ΔslyDpRARE cells using a heat shock protocol as previously described [50] and plated on lysogenic broth (LB) plates supplemented with 33 μg/mL chloramphenicol and 100 μg/mL ampicillin (for pETDuet1-Sb#15) or 30 μg/mL kanamycin (pET28a-Sb#15). A single positive colony was inoculated into 10 mL of LB with the required antibiotics and incubated overnight at 37 °C with agitation of 200 rpm. This seed culture was used to inoculate a 1 L of LB culture with the required antibiotics and incubated up to an OD600nm between 0.6 and 0.8 at 37 °C with agitation of 200 rpm. Expression was induced with 0.25 mmol/L isopropyl-β-d-thiogalactopyranoside at 37 °C for 4 h. The cells were harvested by centrifugation at 6000 × g at 4 °C for 10 min, suspended in 20 mmol/L of buffer A (50 mmol/L Tris-HCl, 200 mmol/L NaCl, 20 mmol/L imidazole pH 8.0), and stored at −80 °C. Upon thawing, lysozyme and β-mercaptoethanol were added to final concentrations of 150 μg/mL and 10 mmol/L, respectively, and incubated for 30 min on ice with intermittent agitating, followed by mechanical lysis using a microfluidizer (Microfluidics, Newton, MA, USA). The lysate was cleared by centrifugation at 20,000 × g for 30 min at 4 °C. The recombinant His6-Sb#15 and Sb#15-His6 were initially purified by metal-chelating affinity chromatography. For this, the lysate was loaded onto a 1 mL HisTrap column (Cytiva, Marlborough, MA, USA), washed with 10 column volumes of buffer A and eluted with a gradient of 0-to-10% Buffer B (50 mmol/L Tris-HCl, 200 mmol/L NaCl, 500 mmol/L Imidazole pH 8.0), followed by a gradient of 10-to-100% of buffer B, which was collected in fractions of 0.5 mL. The peak fractions were pooled and concentrated to 2.5 mL and fractionated further on a size exclusion Superdex HiLoad 16/600 75 pg column (Cytiva, Marlborough, MA, USA) pre-equilibrated with 20 mmol/L Tris-HCl, pH 8.0, and 150 mmol/L NaCl. The purity of the purified Sb#15 was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), using 15% polyacrylamide gels. The samples were quantified on a Nanodrop 2000 spectrophotometer (ThermoFischer Scientific, Waltham, MA, USA) using, for His6-Sb#15 an extinction coefficient of 24,410 M−1/cm and a molecular weight of 16.38 kDa, and for Sb#15-His6, an extinction coefficient of 22,920 M−1/cm and a molecular weight of 14.45 kDa. The purified sample was supplemented with 10% glycerol and stored at −80 °C until use. The purified versions of Sb#15 were termed His6-Sb#15 or Sb#15-His6 according to the position of the hexa-histidine tag.
2.3. TEV protease digestion of His6-Sb#15
For removal of the hexa-histidine tag, purified His6-Sb#15 was dialyzed in TEV digestion buffer (50 mmol/L Tris.HCl pH 8.0, 200 mmol/L NaCl, 0.5 mmol/L ethylenediamine tetra acetic acid (EDTA) and 10 mmol/L β-mercaptoethanol) overnight at 4 °C. After dialysis, the sample was centrifuged at 20.000 × g for 30 min at 4 °C and quantified as described above and 5.7 mg of His6-Sb#15 was digested with 0.11 mg of TEV protease in final mass ratio of 50:1 for 3 h at 30 °C on a rotator shaker at 70 rpm. Subsequently, the sample was passed through a for removal of the hexa-histidine tag, and of residual undigested His6-Sb#15, the sample was passed through a 1 mL HisTrap column (Cytiva, Marlborough, MA, USA) using the same conditions as for the affinity chromatography described above to capture the hexa-histidine tag and any residual undigested His6-Sb#15. The flowthrough was collected, concentrated, and fractionated on a size exclusion Superdex HiLoad 16/600 75 pg column (Cytiva, Marlborough, MA, USA). The peak fractions of the size exclusion chromatography were pooled and concentrated to 1 mg/mL (71 μM) for the microscale thermophoresis (MST) assays. After removal of the hexa-histidine tag, the purified VHH was termed Sb#15.
2.4. Transfection of HEK293 cells and selection of clones expressing the RBD of the SARS-CoV-2 spike protein
Expression of the RBD domain of the SARS-CoV-2 spike protein was performed in HEK293 cells cultivated in a Forma Series II Water Jacketed CO2 incubator (Thermo Fischer Scientific, Waltham, MA, USA) at 37 °C with humidity and 5% CO2. Transfection of HEK293 cells with plasmid pIRES2-EGFP-RBD2 was performed according to the procedure described by Hawley–Nelson et al. [51]. Briefly, 6.255 × 105 cells in 5 mL of Opti-MEM supplemented with 2.5% of fetal bovine serum (FBS), 1× de GlutaMAX, and 1× penicillin/streptomycin (Thermo Fischer Scientific, Waltham, MA, USA) were seeded in 25 cm2 culture flask at a final cell density of 2.55 × 104 cells/cm2. In the following day, 6.25 μL (1 μg/μL) of linearized pIRES2-EGFP-RBD2 DNA and 37.5 μL of lipofectamine were first diluted in 312.5 μL of non-supplemented Opti-MEM in separate tubes. Subsequently, the content of the two tubes was mixed, and incubated at room temperature for 15 min. The medium was removed from the culture flask and replaced by 1.83 mL of Opti-MEM without FBS and the plasmid DNA-lipofectamine suspension gently distributed over the cells followed by gentle mixing. The final amounts of plasmid DNA and lipofectamine were 250 ng and 1.5 μL, respectively, per 2.5 × 104 cells/cm2. The 25 cm2 culture flask with the transfection reactions was placed back to the CO2 incubator and maintained at 37 °C for 5 h. At this time, 2.5 mL of Opti-MEM™ Reduced Serum Mediumsupplemented 5% FBS was added to obtain FBS at a final concentration of 2.5%. A parallel control transfection was performed using the same procedure except that no plasmid DNA was added. After four days, both the transfected and control cultures were subjected to antibiotic selection with G418 (600 μg/mL) (Sigma). Every 2–3 days, the medium was removed, the cells washed in Dulbecco′s Phosphate Buffered Saline (DBPS) 1× (2.6 mmol/L KCl, 1.47 mmol/L KH2PO4, 137.9 mmol/L NaCl, 8 mmol/L Na2HPO4 anhydrous) (Thermo Fischer Scientific, Waltham, MA, USA), new medium added, and the cells cultivated until the cell population expanded to reached about 35 × 106 cells.
After selection of the cell population resistant to G418, single cells expressing higher levels of green fluorescent proteins (GFP) were sorted by flow cytometry using a FACSAria II (BD Life Sciences, New Jersey, USA) with a 100 μm nozzle, excitation at 488 nm and a 530/30 filter, using a pressure limit of 20 psi at 4 °C. For sorting, the cells were diluted to 106 cells/mL in DPBS 1× containing 20% Opti-MEM and 2% FBS, and the flow cytometer adjusted to recover only the cells with the 10% highest fluorescence intensity. Isolated cells were collected in a 96-well plate in a 200 μL mix containing 50% DMEM, 20 mmol/L HEPES/NaOH (Sigma) pH 7.3, 10% FBS, 1× GlutaMAX and 1× penicillin/streptomycin, and 50% of conditioned DMEM plus 600 μg/mL of G418. Conditioned DMEM was obtained by cultivating HEK293 cells for 3–4 days in DMEM containing 10% FBS, 1× GlutaMAX, and 1× penicillin/streptomycin. The medium was collected, the debris removed by centrifugation for 5 min at 1000×g, transferred to a sterile flask, and stored at 4 °C until use for up to a week. Two weeks after sorting, the medium was replaced by Opti-MEM containing 2.5% FBS, 1× GlutaMAX, 1× penicillin/streptomycin, and 600 μg/mL of G418, and the culture expanded gradually first in 24-well plates and later in 300 cm2 bottles. Three clones originated from single cells were obtained with this procedure. For evaluation of RBD expression, the supernatant of 25 cm2 culture bottles was analyzed by Western blot using an anti-hexa-histidine antibody. All three clones presented similar levels of RBD expression and clone C4 was selected for expansion.
2.5. Purification of the RBD of the SARS-CoV-2 spike protein
For purification, clone C4 was expanded to 9 × 106 cells in 300 cm2 culture bottles (3 × 104 cells/cm2) containing 50 mL of Opti-MEM containing 2.5% FBS, 1× GlutaMAX, 1× penicillin/streptomycin, and 600 μg/mL of G418. The culture reaches ∼90% confluence after three days. At this point, the medium was collected and stored at −80 °C for subsequent purification. The culture was treated with trypsin and diluted back to 3 × 104 cells/cm. This process was repeated several times and the culture medium was combined for purification. Purification of the RBD domain was typically performed with ∼300 mL of medium. Initially, the medium was defrosted overnight at 4 °C and aggregates were removed by centrifugation at 20,000 × g. The histidine-tagged RBD was purified by affinity chromatography using a HisTrapFF Crude 5 mL column using an ÄKTA Pure M25 chromatography system (Cytiva, Marlborough, USA) equipped with a sample pump using a flow rate of 2 mL/min. The column was pre-equilibrated in buffer A (20 mmol/L sodium phosphate pH7.5, 300 mmol/L NaCl; 20 mmol/L imidazole). After loading the samples, the column was washed with 5 column volumes and eluted with a gradient of 0-to-10% Buffer B (20 mmol/L sodium phosphate pH7.5, 300 mmol/L NaCl; 500 mmol/L imidazole) in 10 column volumes, followed by a gradient of 10-to-100% of buffer B. Fractions of 1.5 mL of the imidazole gradients were collected and analyzed by SDS-PAGE. Six peak fractions of the affinity chromatography were combined in 9 mL and concentrated to 2.5 mL by reverse dialysis using polyethylene-glycol 20000 (Sigma-Aldrich) and fractionated further by size exclusion chromatography using a HiLoad16/600 Superded 75 pg on a ÄKTA Pure M25 chromatography system (Cytiva, Marlborough, USA). The chromatography was performed in buffer containing 20 mmol/L sodium phosphate pH 7.5 and 150 mmol/L NaCl at flow rate of 1 mL/min. The elution peak was collected in fractions of 1 mL, analyzed by SDS-PAGE, and the RBD quantified on a Nanodrop 2000 spectrophotometer (ThermoFischer Scientific, Waltham, MA USA). The yield was approximately 1 mg of RBD per 300 mL of culture supernatant.
2.6. Stability analysis
Dynamic light scattering (DLS) analysis was performed to determine the hydrodynamic radius and polydispersity index (PDI) of the purified Sb#15 VHH versions and RBD using a Prometheus Panta device (NanoTemper Technologies, Munich, Germany). Briefly, the purified the Sb#15 versions in buffer containing 20 mmol/L Tris (pH 8.0), 150 mmol/L NaCl were concentrated by ultrafiltration with 3 kDa cut-off concentrators (Merk Millipore, Cork Ireland) up to 1 mg/mL for Sb#15 and 2 mg/mL for Sb#15-His6. The purified RBD in buffer 20 mmol/L sodium phosphate (pH 7.5) and 150 mmol/L NaCl was concentrated by reverse dialysis using polyethylene glycol 20000 (Sigma) up to 0.6 mg/mL, and the aggregates were removed by centrifugation at 20,000 × g, 4 °C for 15 min. 10 μL samples were loaded on standard capillaries in triplicates and placed on the device. Particle distribution was determined by measuring light scattering intensity using the size analysis function. Isothermal DLS scans (10 acquisitions, 30% light-emitting diode power, 100% DLS laser power) were performed at 20 °C. The average values of the hydrodynamic radius and PDI were recorded and analyzed using the manufacturer's software PR Panta Control. The same Prometheus Panta device was used to determine thermostability by nano-differential scanning fluorimetry (nanoDSF) using the thermal analysis function. The same samples used for DLS were heated from 20.0 to 90 °C at a ramp rate of 1.5 °C/min for acquisition of thermal unfolding parameters and were cooled down to 20 °C for acquisition of refolding parameters. For intrinsic tryptophan fluorescence measurements, 280 nm wavelength was used for excitation and emission measured at 330 nm and 350 nm to determine the 350 nm/330 nm ratio. Thermal transition midpoint (Tm) and aggregation curves were calculated using the Panta Analysis Software (NanoTemper Technologies, Munich, Germany) and first derivatives of experimental curves were computed to facilitate visualization.
2.7. Interaction analysis
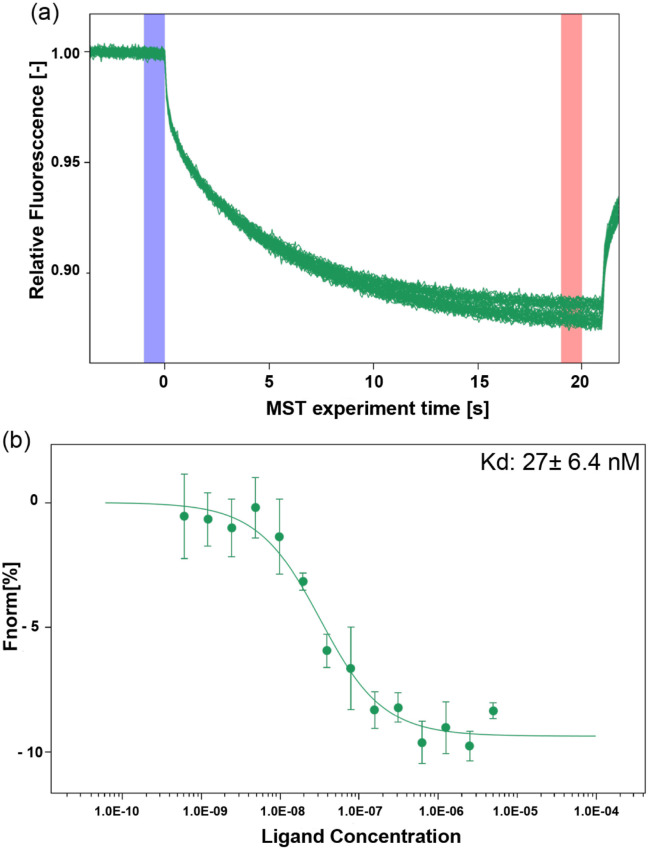
Evaluation of the Sb#15 ability to bind the RBD of the SARS-CoV-2 spike protein was performed by MST on a Monolith NT.115 instrument (NanoTemper Technologies, Germany). For the interaction assays, the purified RBD was dialyzed in buffer containing 20 mmol/L sodium phosphate (pH 7.3) and 100 m/mol−1/L NaCl and labeled using Monolith His-Tag Labeling Kit RED-tris-NTA (MO-L008, NanoTemper Technologies, Munich, Germany) at a concentration of 100 nmol/L. Initially, different dilutions of labeled RBD were tested to determine the best RBD concentration and MST power to obtain the appropriate range of fluorescence counts for the assays which was established in 20 nmol/L of labeled RBD. After removal of the hexa-histidine tag, Sb#15 was concentrated to 1 mg/mL (71 μM) in size exclusion chromatography buffer (20 mmol/L Tris-HCl, pH 8.0, and 150 mmol/L NaCl). Subsequent Sb#15 dilutions were performed using MST buffer (sodium phosphate pH 7.3, 100 mmol/L NaCl, 0.05% Tween-20). The experiments were carried out using 10 nmol/L of labeled RBD and 1:2 serial dilutions of unlabeled Sb#15 starting with 10 μmol/L down to 0.001 μmol/L. The binding reactions were loaded into premium capillaries (Monolith NT.115 Capillaries, NanoTemper Technologies) and thermophoresis measured with MST power set to 40%, excitation power set to 20%, and temperature control set to 25 °C. MST measurements were performed with four independent technical replicates. The dissociation constant (Kd) was calculated using the MO Affinity Analysis v1.5.3 software (NanoTemper Technologies).
2.8. Electrode modification
Electrochemical procedures were performed using IviumStat Potentiostat XRe equipment and a Metrohm DropSens STAT-I-400 portable potentiostat. Initially, the screen-printed carbon electrodes (SPCEs), containing a carbon working electrode, a carbon counter electrode, and a silver pseudo-reference electrode, Metrohm Brasil, were cleaned by polishing with aluminum oxide dispersion and then were electrochemically activated according to the method described by Rahmati et al. [15]. Briefly, the SPCEs were submerged in 1 mol/L H2SO4 (Synth) solution, and a voltage of 1.5 V was applied during 150s.
The electropolymerization of the monomer was carried out according to Hryniewicz et al. [52] in a solution containing 100 mmol/L of pyrrole monomer (98%, Sigma Aldrich), 5 mmol/L of methyl orange (Synth), and 8 mmol/L of KNO3 (Sigma Aldrich) as an electrolytic medium. The solution pH was adjusted with 1 mol/L HNO3 (Synth) to the final pH 2 at 25 °C. Electropolymerization was carried out by applying a constant potential of 0.8 V and a charge control of 500 mC/cm2, aiming to assure the same amount of polypyrrole nanotubes (PPy-NTs) in the different electrodes. Electrodeposition of nickel hydroxide on the polymer was carried out by adapting the method described by Rahmati et al. [15] The PPy-modified SPCE was submerged in a beaker containing 10 mL of 0.01 mol/L Ni(NO3)2 (Sigma Aldrich) and then Ni(OH)2 was deposited by cyclic voltammetry (CV, Fig. S1), varying the potential from 0 to −1.5 V, for 20 cycles with a scan rate of 50 mV/s.
Electrochemical characterization of PPy-NTs/Ni(OH)2-modified electrodes was performed by EIS and CV; all experiments were carried out in a phosphate buffer saline (PBS, tablet pH 7.4, Synth) solution. The EIS data were acquired by applying an alternating voltage of 10 mV, in open-circuit potential, in a frequency range from 10 kHz to 0.1 Hz, and by CV in a potential range from −0.5 to +0.5 V at a scan rate of 20 mV/s. The morphology of modified electrodes was analyzed using scanning electron microscopy in a TESCAN MIRA3 and transmission electron microscopy images in a JEOL JEM 1200EX-II.
2.9. Biosensor construction
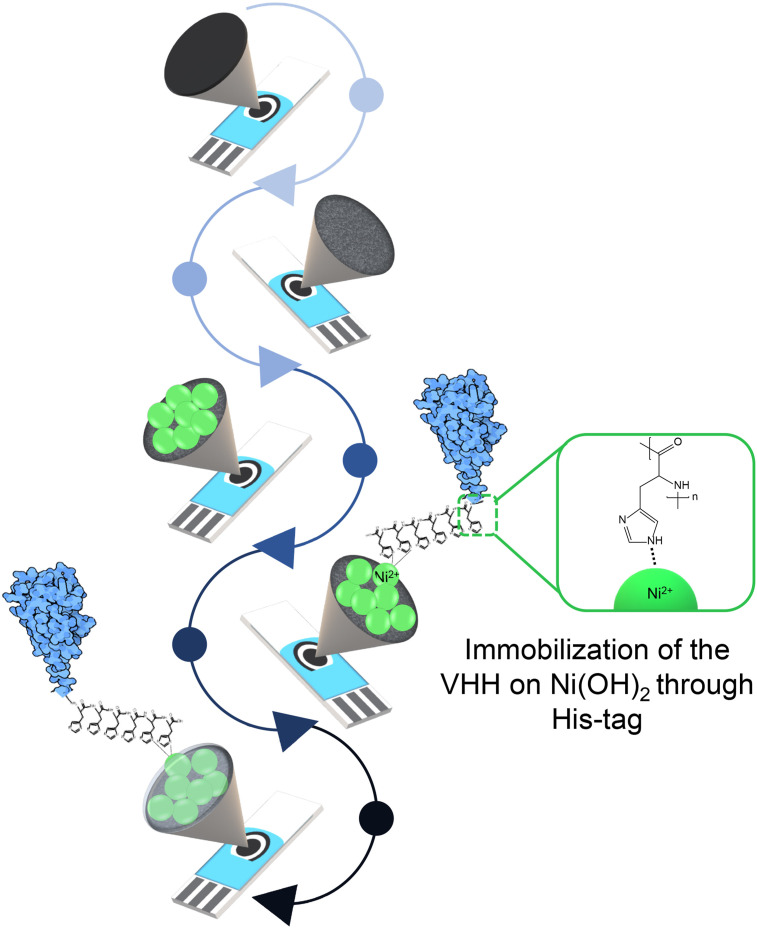
Antibody immobilization was performed by adding 15 μL of solution over the working electrode for 45 min of incubation. For commercial IgG antibodies, mAb10540 and mAb105802, (cat no. 10540 and 105802, R&D Systems) it was used at concentration of 500 ng/mL each, both diluted in PBS (pH 7.4); while for Sb#15-His6, it was used 100 ng/mL, prepared in 20 mM Tris-HCl, pH 8.0 and 150 mM NaCl buffer. Then, the electrodes were washed, by immersion, in PBS for 10 min to remove antibodies that did not bind to the Ni(OH)2. In sequence, 15 μL of bovine serum albumin (BSA) 1% prepared in PBS was added to the biosensor for 30 min to block the remaining active sites and minimize unspecific interactions. After this step, the electrode was washed, by immersion, in PBS for 10 min and it was ready to detect the spike protein RBD (cat no. AB275986, Abcam Plc) in solution and/or saliva samples. A scheme representing each step of biosensor construction is summarized in Fig. 1 .
Fig. 1.
Schematic representation of the biosensor development.
All the steps of biosensor construction were characterized by EIS and Fourier-transform infrared (FTIR) spectroscopy. The FTIR spectra were acquired in a Bruker Vertex 70 spectrometer, scanning from 400 to 4.000 cm−1, 32 scans, with a resolution of 4 cm−1. The samples were prepared with spectroscopic grade KBr powder.
2.10. Biosensors evaluation
To evaluate the ability to detect SARS-CoV-2 by the different antibodies, the biosensors were exposed to different concentrations of SARS-CoV-2 spike protein RBD diluted in PBS. The linear range of SARS-CoV-2 spike protein RBD was 0.05–10.0 ng/mL, 0.01–5.0 ng/mL, and 0.01–50.0 pg/mL for mAb10540, mAb105802, and Sb#15-His6, respectively. For detection, the biosensor was previously characterized by EIS. In sequence, 15 μL of spike protein RBD solution were added to the electrode and incubated for 15 min. After this time, the electrode was carefully washed in a PBS solution for 5 min, and EIS analysis was carried out.
2.11. Analysis of spike protein RBD in saliva samples
Saliva samples were collected at the SARS-CoV-2 detection service of the Laboratory of Immunogenetics and Histocompatibility of Universidade Federal do Paraná in Curitiba, Brazil. Signed informed consent forms were obtained from all participants, and the study was approved by the Ethics Committee of Universidade Federal do Paraná (CAAE: 43945121.4.0000.0102).
To analyze the ability of the biosensor to recognize the spike protein RBD in real samples, saliva samples pre-characterized with RT-PCR were used. A total of 22 saliva samples were used, including 7 negative and 15 positive samples for SARS-CoV-2. Among the positive samples, 5 were from the Delta variant and 10 were from the Omicron variant (7 from the BA.1.1 subvariant and 3 from the BA.2 subvariant). The samples were diluted in PBS (dilution factor 1:100) and then 15 μL were added to the biosensor and incubated for 15 min. Before EIS measurements, the electrode was washed in PBS for 5 min.
2.12. Analysis of RT-qPCR and genotyping of saliva samples
Saliva samples collected at the SARS-CoV-2 detection service of the Laboratory of Immunogenetics and Histocompatibility of UFPR, Curitiba, Brazil, from September 2021 to May 2022 were submitted to RT-qPCR to detect SARS-CoV-2 nucleic acids. When positive, they were genotyped. Briefly, samples were homogenized and centrifuged, and 100 μL of supernatant was used for RNA extraction by using an automated magnetic EXTRACTA–RNA and DNA Viral kit (Loccus Biotecnologia, São Paulo, Brazil) according to the manufacturer's instructions. 5 μL of extracted RNA as well from positive and negative controls (Kit BIOMOL OneStep/COVID-19 – IBMP, Curitiba, Brazil) were mixed with 15 μl of the solution provided on the kit into a PCR 96 well plate and analyzed using QuantiStudio5™ (Thermo Fisher Scientific Inc., Waltham, MA, USA). The parameters were as follows: reverse transcription 50 °C – 5 min, cDNA start denaturation 95 °C – 3 min, 40 cycles of denaturation 95 °C by 15 s, annealing 55 °C by 40 s, and 1 cycle to cool at 25 °C by 10 s. The test allows the detection of specific sequences in RNA extraction of target ORF-1ab (FAM), N gene (HEX-VIC), and intern control—human endogenous gene (ROX).
To assess whether different strains of SARS-CoV-2 can be differentially detected by the biosensor, we selected samples from the last quarter of 2021, when Delta was the main circulating variant, and from the first semester of 2022, when the Omicron variant prevailed in Curitiba [72].
For genotyping, two probe-based genotyping systems were used to identify the variants of concern (VOC) multiplexed RT-qPCR method [53], allows the detection of ORF1a Δ3675–3677, Spike Δ69–70 deletions, CDC-N1 (N gene) to discern the VOCs: Alpha (B.1.1.7), Beta (B.1.351), Gamma (P1), Delta (B.1.617.2), and Omicron (B.1.1.529 – BA.1).
Samples obtained from 2022 were analyzed in the quadruplex format (4-target detection), using TaqMan probes, by amplifying a target region in the N gene, and results were obtained (presence or absence) from the deletions (Del) S106, G107, and F108, in the gene ORF1a (nsp6) and Del. H69 and V70 in the Spike gene. This protocol allowed to detect and sort VOCs Alpha, Beta, Gamma, Delta, and Omicron. By the protocol, the criteria of interpretation from results were based on the PCR data, as shown in Table 1 .
Table 1.
Interpretation of target results.
| SC2-N (FAM) | Wt Del NSP6 (ROX) | Wt Del 69,70 (HEX/VIC) | RP (CY5) | Result |
|---|---|---|---|---|
| CT ≤ 40 | CT ≤ 40 | Undetected | CT ≤ 35 | Potentially B.1.351 (β), or P1 (γ), or BA.2 (ο) |
| CT ≤ 40 | Undetected | Undetected | CT ≤ 35 | Potentially B.1.1.7 (α), or BA.1(ο) |
| CT ≤ 40 | CT ≤ 40 | CT ≤ 40 | CT ≤ 35 | Potentially Wild, or B.1.617.2 (δ) |
Targets: SC2-N – gene N fragment from SARS-CoV-2, Wt Del NSP6 – gene ORF1a fragment without deletion in NSP6, Wt Del 69,70 – gene S fragment without deletion 69,70, RP – human constitutive gene fragment, RNAseP.
2.13. Statistical analysis
The statistical analyses and threshold determination to distinguish the samples between positive and negative for COVID-19 were performed based on in-house computational coded scripts written by authors in MATLAB R2018a (Mathworks, Natick, MA. USA). Every sample was classified based on the threshold value obtained by Bayes' Theorem, which aims to minimize the misclassification rate and allows evaluating the reject region for the class separation [54]. The Bayesian threshold assumes that the impedance values represent a populational distribution of future samples either for positive or negative samples. The threshold (i.e. the value that separates each class) is selected at the cross section of the two estimated distributions, at which the number of false positives and false negatives should be minimized for future predictions [55]. In addition, the proposed method was also evaluated by accuracy, sensitivity, and specificity. Accuracy corresponds to the global performance of the method, i.e. the ratio between the correct classified samples by the total number of samples; sensitivity (or true positive rate) corresponds to the ratio between the correct positive classified samples within the positive samples; specificity (or true negative rate) corresponds to the ratio between the correct negative classified samples within the negative samples [56].
3. Results and discussion
3.1. VHH Sb#15 expression, purification, and stability analysis
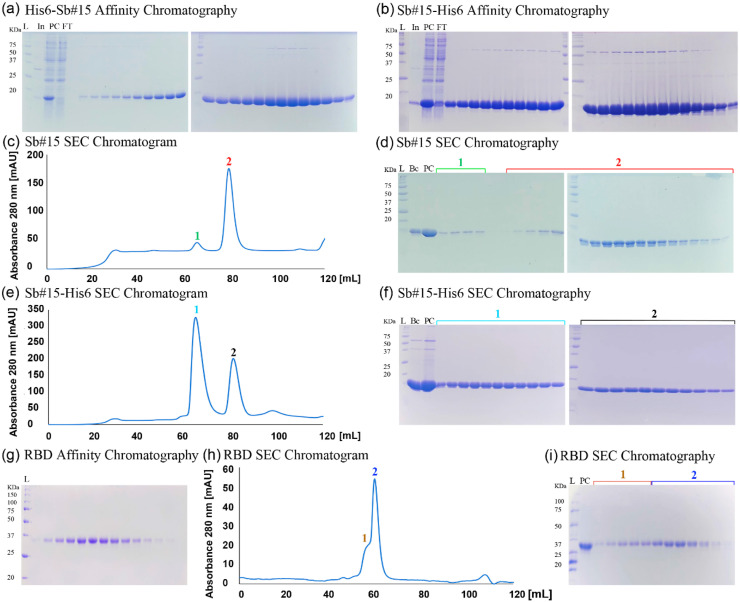
Two versions of Sb#15 were produced using plasmids pETDuet1-Sb#15 and pET28a-Sb#15 containing hexa-histidine tag either at the N-(His6-Sb#15) or C-Terminal (Sb#15-His6), respectively. Both were purified from soluble E. coli extracts on immobilized nickel columns followed by size exclusion chromatography. Representative results of the purification steps are shown in (Fig. 2 ).
Fig. 2.
Purification of His6-Sb#15, Sb#15-His6, and SARS-CoV-2 RBD. (a), (b), (g) SDS-PAGE analysis of the indicated proteins purified by affinity chromatography. (c), (e), (h) Size exclusion chromatography profiles of the indicated proteins. Sb#15 shows a major peak eluting at 78 mL, Sb#15-His6 elutes in two peaks eluting, respectively, at 64 and 81 mL, and SARS-CoV-2 RBD elutes in a major peak with 59 mL and a shoulder eluting earlier with 56.5 mL. (d), (f), (i) SDS-PAGE analysis of the indicated proteins after fractionation by size exclusion chromatography. L, molecular mass marker. In, input. PC, precolumn. FT, flow through. Bc, before concentrate. RBD, receptor-binding domain.
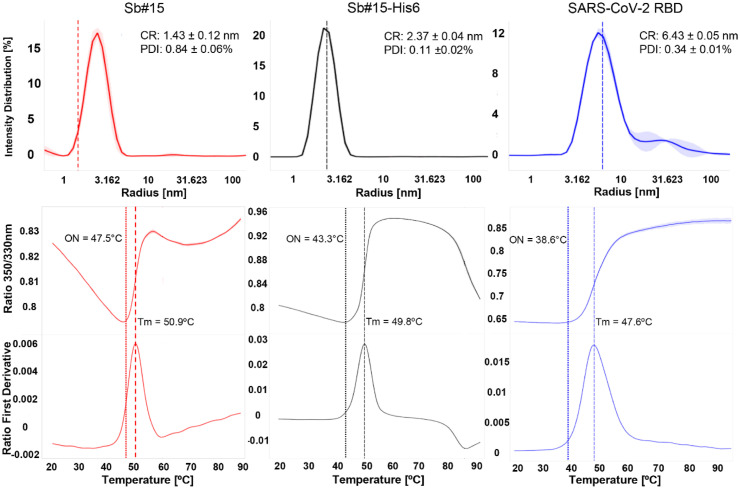
In the case of His6-Sb#15, the hexa-histidine tag was removed by TEV protease digestion before further analyses. In the size exclusion chromatography, Sb#15 without the hexa-histidine tag elutes in a major peak with 78 mL of buffer volume (Fig. 2(c)). Sb#15-His6, however, elutes from the size exclusion column in two peaks with volumes of 64 and 81 mL, respectively (Fig. 2(e)). Since both peaks contain pure Sb#15-His6 (Fig. 2(f)), the peak eluting in a smaller buffer volume should correspond to an artificial dimer or aggregate while the peak eluting in a higher buffer volume is expected to contain monomeric Sb#15-His6 and only the peak eluting with larger volume was used in subsequent experiments. Consistently, DLS analysis of the second peak revealed that Sb#15-His6 shows a cumulant hydrodynamic radius of 2.37 ± 0.04 nm with a PDI of 0.11 ± 0.02, and Sb#15 without the hexa-histidine tag showed a cumulant hydrodynamic radius of 1.43 ± 0.12 nm with a PDI of 0.84 ± 0.06. Hydrodynamic radii are consistent with a majoritarian monomeric conformation and a PDI lower than 0.2 indicates a uniform particle size for Sb#15-His6, while Sb#15, which showed a PDI of 0.84, shows a propensity to aggregation (Fig. 3 ).
Fig. 3.
Biophysical analysis of Sb#15, Sb#15-His6, and SARS-CoV-2 RBD. Top panels show DLS intensity distribution curves. Sb#15 shows a cumulant hydrodynamic radius (CR) of 1.43 ± 0.12 nm and polydispersity index (PDI) of 0.11 ± 0.02%, His6-Sb#15 shows CR of 2.37 ± 0.04 nm and PDI of 0.11 ± 0.02%, and SARS-CoV-2 RBD shows CR of 6.43 ± 0.05 nm and PDI of 0.34 ± 0.01%. Center panels show the 350nm/330nm fluorescence intensity ratio along the thermal denaturation curves of His6-Sb#15, Sb#15-His6, and SARS-CoV-2 RBD. Lower panels show the first derivative of the 350nm/330nm fluorescence intensity ratio to determine the transition midpoints (Tm) along the thermal denaturation curves for Sb#15 (Tm = 50.9 °C), Sb#15-His6 (Tm = 49.8 °C), and SARS-CoV-2 RBD (Tm = 47.6 °C). DLS, dynamic light scattering.
Thermal stability of Sb#15-His6 and Sb#15 was determined using nanoDSF with melting curves measured by the 350/330 nm fluorescence ratio turbidity followed by DLS. Sb#15 presented a melting onset at 47.5 °C and an inflection point or transition midpoint (Tm) of 50.9 °C. These results show that both Sb#15 and Sb#15-His6 eluting in the larger volume of size exclusion chromatography are folded in solution with no significant aggregation or higher order oligomerization present.
The primary goal of product and process development is to identify and limit the degradation pathways to enable safe and efficacious usage with commercially viable shelf life. Product liabilities are commonly identified during development via biochemical and biophysical characterization techniques. Knowledge of these properties helps to define parameters that can damage the product during transport and storage [57]. The VHH Sb#15-His6 sample consisted of high monomer content and had high protein melting temperatures, which are associated with long-term stability.
3.2. Expression, purification, and stability analysis of SARS-CoV-2 spike protein RBD
The deca-histidine-tagged RBD of SARS-CoV-2 spike protein expressed in adherent eukaryotic HEK293 cells was initially purified from the medium supernatant by affinity chromatography on immobilized nickel columns. Subsequent size exclusion chromatography revealed a major peak eluting with 59 mL with a shoulder eluting earlier (56.6 mL). Both the shoulder and the major peak contain purified RBD only, indicating that the RBD found in the shoulder may represent a different conformation. Therefore, only the fractions of the major peak were used for further analysis and in subsequent experiments. Representative results of RBD purification are shown in Fig. 2(g–i). The SARS-CoV-2 RBD of the major peak showed a cumulant hydrodynamic radius of 6.43 ± 0.05 nm with a PDI of 0.34 ± 0.01 which is compatible with a monomeric conformation but a PDI of 0.34 indicates that it shows some propensity to aggregation. Nevertheless, thermal denaturation monitored by nanoDSF showed that the SARS-CoV-2 RBD is quite stable, showing a transition midpoint (Tm) of 47.6 °C. Together, these analyses indicate that the SARS-CoV-2 RBD purified by this procedure is suitable for interaction assay to validate antibodies.
3.3. Characterization of Sb#15 interaction with SARS-CoV-2 RBD
The binding affinity (dissociation constant, KD) of recombinant VHH Sb#15 was determined using MST. MST time traces of Sb#15 titration with SARS-CoV-2 RBD as well as the corresponding thermophoresis signals showed a typical, sigmoidal shape. The curve presented as ΔFnorm is defined as the ratio between the average fluorescence of hot and cold regions. Raw normalized fluorescence (ΔFnorm) data were baseline-corrected by subtracting the mean of the normalized fluorescence values measured for the lowest compound concentration. The KD value obtained was 27.1 ± 6.4 nmol/L (Fig. 4 ), which is consistent with the data described by Walter et al. [47]. This result confirms VHH Sb#15 functionality and, therefore, we proceeded with biosensor development.
Fig. 4.
Interaction analysis between Sb#15 and SARS-CoV-2 RBD. (a) Representative MST time-trace of labeled RBD tested with serial dilutions of Sb#15. Cold (blue) and hot (red) regions as defined in central data analysis to calculate ΔFnorm. (b) Representative curve for the interaction of SARS-CoV-2 RBD at 10 nmol/L with Sb#15 using serial dilutions of Sb#15 starting at 10 μmol/L. Each curve was measured in quadruplicates. The binding constant (KD) was 27 ± 6.4 nmol/L. RBD, receptor-binding domain. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Biosensor construction and characterization
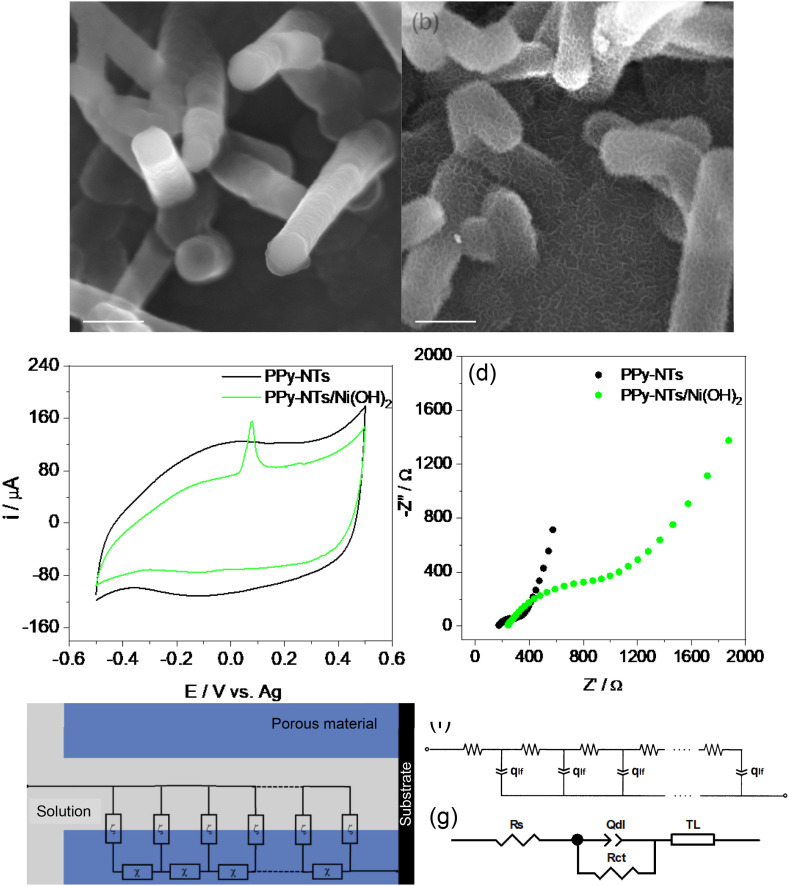
PPy-NTs and PPy-NTs/Ni(OH)2 were electrochemically deposited onto SPCEs and characterized using SEM images, as shown in Fig. 4. PPy-NTs modified electrode (Fig. 5 (a)) presents a nanotubular structure, characteristic of this CP when synthesized in the presence of methyl orange aggregates [52]. In the presence of Ni(OH)2 (Fig. 5(b)), the nanotubes are covered with a narrow film of nanoflakes, characteristic of Ni(OH)2 [58,59], indicating that it was successfully deposited on the electrode. The transmission electron microscopy images for PPy-NTs (Fig. S2(a) and PPy-NTs/Ni(OH)2 (Fig. S2(b)) are very similar, presenting a hollow structure and a rough surface, so Ni(OH)2 is deposited as a very thin film. The PPy-NTs/Ni(OH)2 presents nanotubes with a mean diameter of 135 ± 93 nm.
Fig. 5.
SEM images of (a) PPy-NTs and (b) PPy-NTs/Ni(OH)2 modified SPCEs (accelerating voltage = 20 kV; Detector: In-Bean SE). Electrochemical characterization of the modified electrodes using (c) CV at a scan rate of 20 mV/s and (d) EIS in ocp. Electrolyte = PBS pH 7.4. (e) Single-channel TL with distributed elements of impedance representing the solid network (χ) and the charge accumulation in the polymeric matrix (ζ). (f) TL and (g) equivalent circuit models used to adjust the EIS data. CV, cyclic voltammetry; EIS, electrochemical impedance spectroscopy; PPy-NT, polypyrrole nanotube.
The PPy-NTs-modified electrodes were characterized by CV as shown in Fig. 5(c). They show a CV profile with a large capacitive current and a broad redox peak, characteristic of this material, while in the presence of Ni(OH)2, a small oxidation peak at 80 mV appears, related to NiII(OH)2/NiIIIOOH pair. The reduction peak is not visible probably due to the large capacitive current of PPy-NTs. The well-defined oxidation peak, far from the oxygen evolution, indicates an α-phase of Ni(OH)2 material [60,61].
The materials were also characterized by EIS, presented in Fig. 5(d). It is very common to find an experimental Nyquist plot of a CP as the one present in Fig. 5(a), consisting of a charge-transfer semicircle and semi-infinite/finite diffusional processes, as described by Skinner and Hall [62]. In our case, the semi-infinite diffusion is not a limiting process and did not appear in the diagram, as can be seen by the absence of a straight line of 45° after the semicircle (lower frequencies). Instead, EIS data show two straight lines in the Nyquist diagram. The first one, approaching 45° at high frequencies, is related to the electronic transport in the polymeric film and it appears in the presence of a potential drop inside of the porous material, as the PPy-NTs [63]. The second straight line approaching 90° at low frequencies is related to the finite diffusional process and assigned to the charge accumulation in the polymer matrix to maintain the charge neutrality during the oxidation process [64]. The semicircle between the two lines is related to processes taking place at the electrode/electrolyte interface, as the charge-transfer resistance and double layer capacitance [64,65]. Considering the nature of the porous materials, where an ohmic drop leading to a current distribution in the polymeric material can be assigned, an approach based on transmission line (TL) modeling described by de Levie can be used to fit the EIS data [[66], [67], [68], [69]]. It must be emphasized that the behavior leading to a TL modeling is characterized by a linear relation (near 45° slope straight line) between the imaginary and real branches of the complex impedance in the high frequencies region [70]. A single-channel model was used to fit the data and calculate the parameters, as shown in Fig. 5(e). The χ distributed element is impedance per unit length (Ω/m), being transverse to the outer surface and representing the potential drop at the CP. The ζ distributed element is an impedance length (Ω m), which is parallel to the outer surface and represents the charge accumulation process in the polymeric material to maintain the electroneutrality of the PPy during the oxidation process.
Taking advantage of the charge diffusion/accumulation model used by Bisquert's group [71], a resistance accounting for the electronic transport in the polymer (rpol) was considered as the χ distributed element, and a constant-phase element (qlf) was considered as a ζ distributed element, related to the charge accumulation at the polymeric matrix (Fig. 5(f)) [71]. Considering the non-ideality of the system, all capacitances considered in the study will be replaced by a constant-phase element [71]. A series resistance (Rs), which represents the resistance of the electrolyte, cables, and current collectors, is placed in series to the TL, in addition to a resistance in parallel to a CPE, related to the charge-transfer resistance (Rct) and a double-layer capacitance (Qdl) at the polymer/electrolyte interface (Fig. 5(g)). It is important to mention that the results could be fitted using a simpler approximation, for example, using the equivalent circuit modeling only; however, it would give less information about the changes in the system during the modifications. The calculated parameters through fitting the EIS data are shown in Table 2 .
Table 2.
Calculated parameters through fitting the EIS data
| Parameter | PPy-NTs | PPy-NTs/Ni(OH)2 |
|---|---|---|
| RS/Ω | 168.3 | 224 |
| Qdl/mF sn−1 | 0.10 | 0.06 |
| ndl | 0.82 | 0.83 |
| Rct/Ω | 73.38 | 345.6 |
| rpol/Ω | 419.7 | 2606 |
| qlf/mF sn−1 | 1.93 | 1.10 |
| nlf | 0.81 | 0.82 |
Rs values variations can be assigned to changes in the constant cell each time that a new electrode was used, and a new measurement was carried out. After Ni(OH)2 deposition, a slight decrease in the Qdl value and a huge increment in Rct can be observed, indicating that the electrode/electrolyte interface became less electroactive and with a slightly smaller electrode/electrolyte interface area. The rpol value increased significantly after Ni(OH)2 deposition, indicating an increase in the electronic transport resistance in the material containing Ni(OH)2. Rct and rpol increment is expected since the EIS was performed in ocp (around 10 mV) where the insulating Ni(OH)2 is present [72] and can not only difficult the electronic transport but also block some polymeric electroactive sites at the electrode/electrolyte interface. As commented in the CV, Ni(OH)2 can be oxidized to a highly conductive and electroactive material (NiOOH) at 80 mV. The qlf parameter decreased in the presence of Ni(OH)2, which can also be related to polymeric sites blockage, which can difficult the ionic intercalation in the PPy matrix. Besides that, Ni(OH)2 particles have an overall negative surface charge [60], being possible to contribute to the charge neutrality during the oxidation of PPy, needing fewer ions to make the counterbalance.
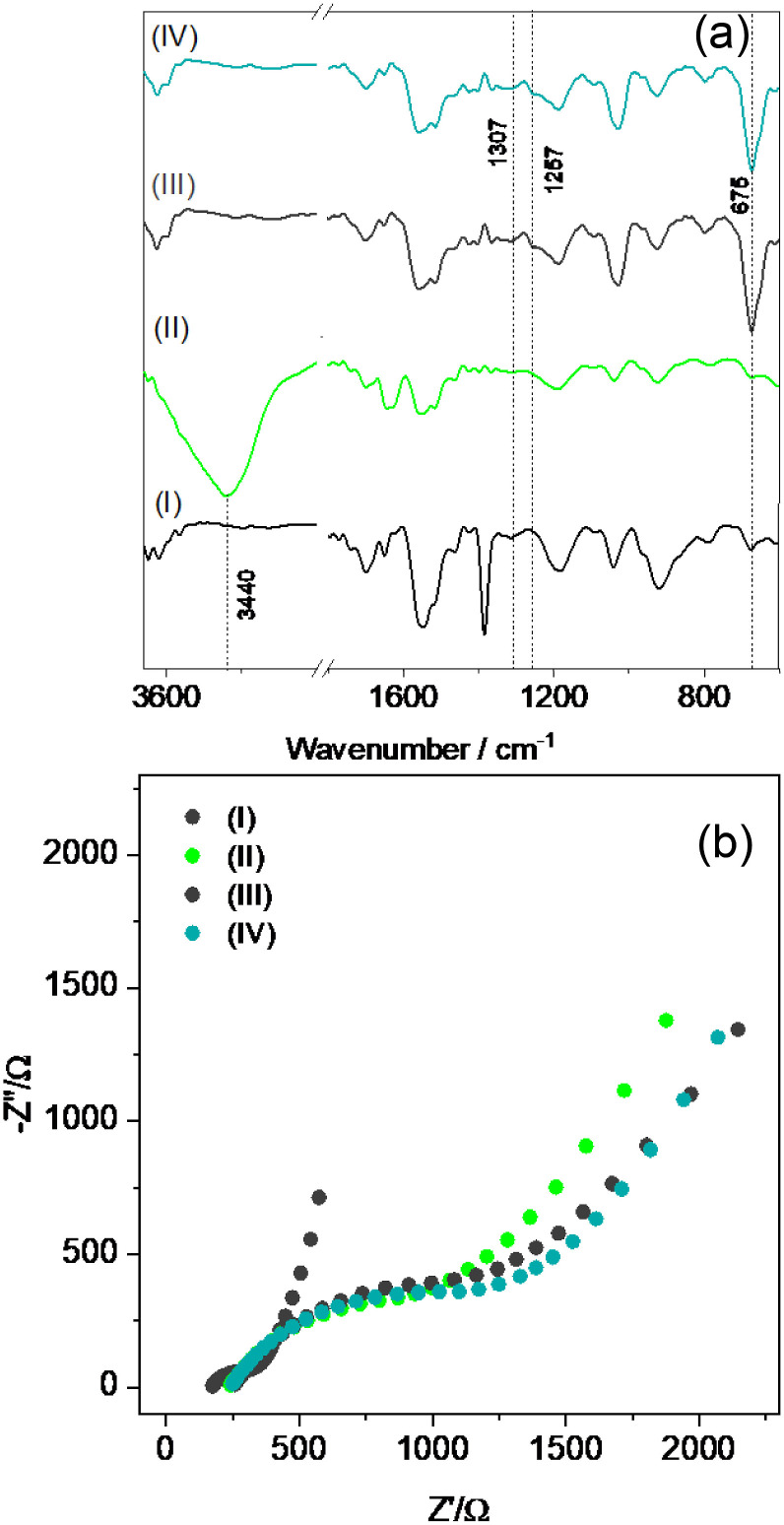
Each step of biosensor construction was characterized by FTIR (Fig. 6 (a)). PPy spectrum shows characteristic bands, as in 920 and 1186 cm−1 indicating the doping state of PPy [73], the band at 1042 cm−1 related to the C–H in-plane vibration and ring vibrations at 1552 and 1472 cm−1 [74]. After Ni(OH)2 deposition, a new band at 3440 cm−1 appeared, related to the hydrogen bonding O–H vibration of the water molecules present in the interlamellar space of Ni(OH)2 and to –OH groups that are non-hydrogen bonded [75,76]. After immobilization of Sb#15-His6 (Fig. 5(a)), mAb10540 (Fig. S3(a)), and mAb105802 (Fig. S3(b)), the bands at 1307 cm−1 (1320 cm−1 for mAb105802) and 1257 cm−1 (1242 cm−1 for mAb10540) appeared, which are related to the α-helix and β-sheet of the amide III region, respectively [77], and 675 cm−1, assigned to the –NH2 and –NH wagging [78], indicating that the proteins were successfully attached at the electrode surface. Also, the band at 3440 cm−2 disappeared after the antibodies and VHH immobilization, showing that the –OH vibrations of Ni(OH)2 were hampered. The absence of the bands associated with N–H stretching vibration between 3100 and 3030 cm−1 after the antibodies’ deposition can be related to the fact that the amide A band absorbs weakly in this region once it is usually part of Fermi resonance doublet with amide B [79]. It was not possible to verify changes after BSA attachment due to signals overlapping with bands from the antibodies.
Fig. 6.
Step-by-step characterization of the biosensor construction by (a) FTIR and (b) EIS measurements. I: PPy-NTs, II: PPy-NTs/Ni(OH)2, III: PPy-NTs/Ni(OH)2/Sb#15-His6, IV: PPy-NTs/Ni(OH)2/Sb#15-His6/BSA. EIS, electrochemical impedance spectroscopy; FTIR, Fourier transform infrared.
The biosensor construction was also monitored by EIS, as shown in Fig. 6(b). The obtained data were fitted according to the equivalent circuit present in Fig. 5(g), and the results are presented in Table 3 .
Table 3.
Calculated parameters through fitting the EIS data
| Parameter | PPy-NTs/Ni(OH)2 | PPy-NTs/Ni(OH)2/Sb#15-His6 | PPy-NTs/Ni(OH)2/ Sb#15-His6/BSA |
|---|---|---|---|
| RS/Ω | 224 | 238.1 | 228.3 |
| Qdl/mF sn−1 | 0.06 | 0.06 | 0.04 |
| ndl | 0.83 | 0.79 | 0.73 |
| Rct/Ω | 345.6 | 496.6 | 612.0 |
| rpol/Ω | 2606 | 2979 | 2392 |
| qlf/mF sn−1 | 1.10 | 1.18 | 1.10 |
| nlf | 0.82 | 0.81 | 0.79 |
After each step of biosensor construction, some parameters showed relevant changes. The main change was observed in Rct values, which increased significantly after the addition of VHH Sb#15-His6 (Fig. 6(b)), mAb10540 (Fig. S3(c)), mAb105802 (Fig. S3(d)), and BSA. This indicates that biomolecules are being successfully immobilized onto the electrode surface, blocking the material electroactive sites, which leads to Rct increment. Besides that, a decrease in Qdl and ndl can be observed, indicating that the electrode/electrolyte interface area is decreasing in the presence of the biomolecules and the surface is becoming less homogeneous. Finally, rpol, qlf, and nlf parameters did not present significant changes, suggesting that the electronic transport and charge intercalation processes are not affected by the biomolecules.
3.5. Biosensors analytical evaluation
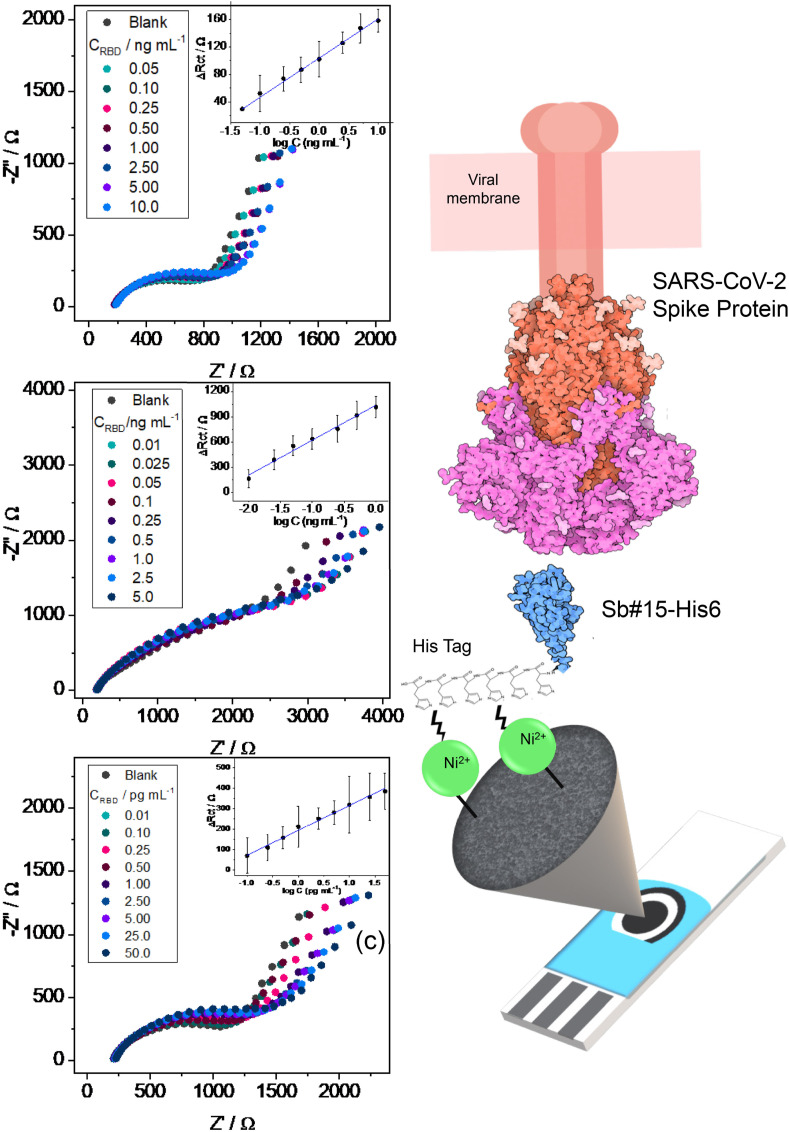
Different RBD concentrations were used for detection by mAb10540 (Fig. 7 (a)), mAb105820 (Fig. 7(b)), and Sb#15-His6 (Fig. 7(c)) biosensors. The Nyquist diagrams evidence a tendency of increasing the semicircle diameter with increasing RBD concentrations, indicating that EIS is a good technique to detect different amounts of RBD by monitoring the changes in Rct. The EIS data were fitted with the equivalent circuit present in Fig. 7 and the ΔRct values, which is the difference between the Rct of a certain RBD concentration and the blank Rct, were plotted against the log concentration for each antibody in quadruplicate (inset in Fig. 7) to construct an analytical curve.
Fig. 7.
EIS response to different spike protein RBD concentrations in PBS for (a) mAb10540, (b) mAb105802, and (c) Sb#15-His6 biosensors. The corresponding analytical curves are shown inset in the figures. (d) Representation of the fabricated biosensor using Sb#15-His6 (not-to-scale). PBS, phosphate buffer saline; RBD, receptor-binding domain.
The linear range for RBD detection by mAb10540 biosensor was 0.05–10 ng/mL, with mAb105802 biosensor was 0.01–5.0 ng/mL and with Sb#15-His6 0.01–50 pg/mL, showing the lower LOQ for Sb#15-His6 in comparison to commercial IgG antibodies, being possible to quantify 10 fg/mL of RBD. The regression equation for mAb10540 was attained as: ΔRct (Ω) = 15.18 log [RBD] (ng/mL) + 103.74 (R2 = 0.9989), for mAb105802 as: ΔRct (Ω) = 414.77 log [RBD] (ng/mL) + 1035.99 (R2 = 0.9853) and for Sb#15-His6 as: ΔRct (Ω) = 120.79 log [RBD] (ng/mL) + 556.71 (R2 = 0.9911), which evidences a higher sensitivity of Sb#15-His6 in comparison to mAb10540 and a lower sensitivity in comparison to mAb105802. Besides that, the lower LOQ found for the Sb#15-His6 biosensor shows the great potential for using this VHH to detection of small RBD concentrations, which means SARS-CoV-2 early infection. In Fig. 7(d), a representation of the developed biosensor and its interaction with SARS-CoV-2 Spike RBD is shown.
The proposed biosensor performance was compared to other EIS biosensors reported in the literature concerning the detection of different SARS-CoV-2 antigens, as shown in Table 4 . The Sb#15-His6 biosensor showed impressive results when compared to other studies, especially in relation to the biorecognition element concentration and time of detection. For example, Ehsan et al. [80] constructed a biosensor based on graphene/carbon modified electrodes and IgG antibodies to detect the spike protein RBD. The obtained LOQ is 0.25 fg/mL, which is expressively lower than the LOQ obtained in this work; however, the IgG antibody incubation was made with 30 mg/mL, much higher than 100 ng/mL used in this work. The same can be argued with the work by Rahmati et al. [81], who obtained a linear range for spike protein detection from 0.25 fg/mL – 1 μg/mL, but with an IgG antibody immobilization in a concentration of 30 μg/mL. Aydın et al. proposed a biosensor based on gold nanoparticles modified ITO electrodes for the detection of SARS-CoV-2 spike protein RBD, using a small concentration of the biorecognition element (4.4 ng/mL) [82]. The results showed a linear range from 0.002 to 100 pg/mL, which is slightly better than the biosensor proposed herein; however, the detection time is 60 min, in contrast to the 15 min of this work. In addition, the present work uses an SPCE, making it possible the portability and miniaturization of the system.
Table 4.
Comparison of EIS biosensors for SARS-CoV-2 antigens detection between this study and previous works from the literature.
| Electrode | Biorecognition element (conc./ng/mL) | Sample | Detection time/min | Linear range/pg/mL | LOQ/pg/mL | Ref |
|---|---|---|---|---|---|---|
| SPCE1/graphene/carbon | IgG antibody/(30 × 109) Spike S1 IgG antibody + Protein A (30 × 109) |
RBD spiked in diluted nasopharyngeal sample and [Fe(CN)6]3-/4 | 5 | 25 × 10−5 – 1 × 103 25 × 10−5 – 1 × 106 |
25 × 10−5 | [80] |
| SPCE/Cu2O nanocube | IgG antibody (30 × 106) | Spike protein in PB and [Fe(CN)6]3-/4- | 20 | 25 × 10−5 – 1 × 106 | – | [81] |
| SPCE | ACE2 (0.32 × 106) | Spike protein in PBS or saliva and [Fe(CN)6]3-/4 | 4 | 20 × 10−5 – 1 × 105 (PBS) 0.1 – 1 × 105 (saliva) |
7.26 10−3 (PBS) 4.63 (saliva) |
[83] |
| SPCE | SARS-CoV-2 Nucleocapsid Antibody (50 × 106) | SARS-CoV-2 N-protein in PBS and [Fe(CN)6]3-/4- | 30 | 1 – 1 × 104 | – | [84] |
| SPCE/gold nanoparticle | SARS-CoV-2 Nucleocapsid Antibody (2 × 106) |
N protein in PBS and [Fe(CN)6]3-/4- | 40 | 10 – 1 × 105 | – | [11] |
| Glassy carbon electrode/rGO4 | SARS-CoV-2 Spike S1 antibody (2.5 × 106) | SARS-CoV-2 spike protein RBD in PBS and [Fe(CN)6]3-/4- |
– | 1.6 × 105 – 1.25 × 106 2.5 × 106 – 40 × 106 |
– | [85] |
| ITO/ gold nanoparticles |
Anti-RBD antibody (4.4 × 103) |
SARS-CoV-2 spike protein RBD in KCl and [Fe(CN)6]3-/4 |
60 | 0.002 – 100 | – | [82] |
| GCE/Bi2WO6/Bi2S3 | SARS-CoV-2 Nucleocapsid Antibody (20 × 106) | SARS-CoV-2 N protein in saliva and H2O2 | 30 | 0.01–1.00 | – | [86] |
| SPCE/PPy-NTs/Ni(OH)2 | SARS-CoV-2 Spike VHH Sb#15-His6 (100) |
SARS-CoV-2 spike protein RBD in PBS |
15 | 0.01–50 | 0.01 | This work |
VHH has a molecular weight 10 times lower than IgG, and therefore, a larger number of molecules in solution are needed for immobilization on the biosensor. However, this smaller size is advantageous because it is possible to immobilize more molecules in the same area, thus increasing the surface of interaction with the antigen and, consequently, the sensitivity. To obtain greater sensitivity, it is necessary to use a higher concentration of IgG, which leads to an increase in costs; due to post-translational modifications, the IgG-type antibody must be produced in more complex and expensive expression systems. On the other hand, the production of VHH Sb#15-His6 has a high yield and lower cost production. Overall, the proposed biosensor is constructed with a small mass of VHH and without a redox probe for fast, sensitive, selective, and portable detection of spike protein RBD small concentrations.
A control version of the biosensor was also constructed following the same procedure but with Sb#15 without the His-tag to evaluate if Ni(OH)2 could effectively bind to Sb#15 via –NH2 covalent bond, as related before for other bioreceptors [15,87]. EIS data of three different biosensors constructed with 1000 ng/mL Sb#15 are shown in Fig. S4 in the presence of different spike protein RBD concentrations. The results indicate that the biosensor could not detect RBD even in high concentrations (320 ng/mL) and using a high concentration of the bioreceptor (VHH), evidencing the His-tag importance for attaching more and oriented VHH unities, resulting in higher sensitivity and lower limits of quantification.
3.6. Detection of SARS-CoV-2 spike protein RBD in saliva samples
The analysis of SARS-CoV-2 spike protein RBD in saliva samples was performed to evaluate the accuracy, sensitivity, and specificity of biosensors for different SARS-CoV-2 variants. For this purpose, 22 saliva samples pre-characterized by RT-qPCR were used, including 7 negative and 15 positive samples (5 Delta variant samples and 10 Omicron variant samples). The selected samples were characterized according to diagnosis and genotyping analysis (Supplementary Tables S1 and S2).
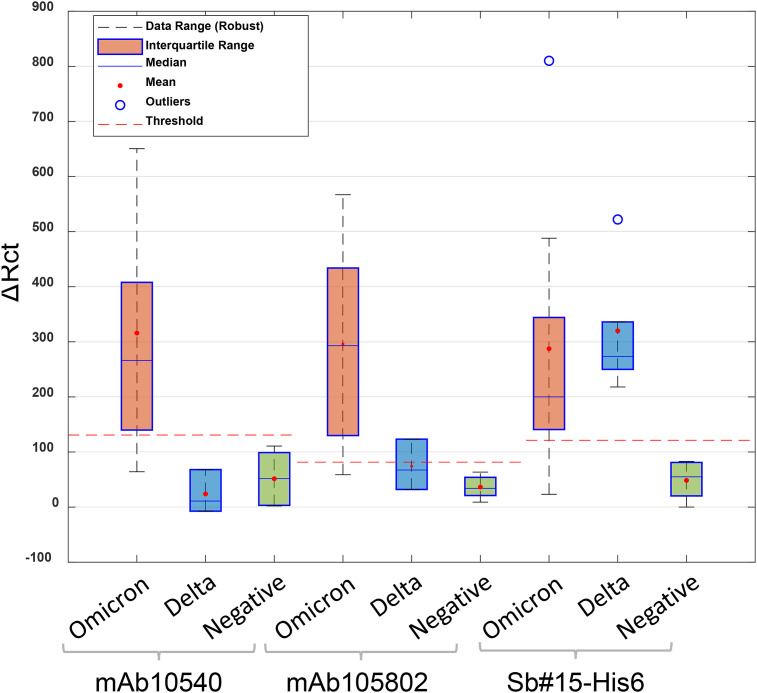
Rct values measured after exposure with positive samples increased significantly compared to negative samples (Figs. S5 and S6), indicating the interaction of spike protein with the antibodies immobilized on the electrode surface. The Rct values obtained for positive–negative samples were also submitted to statistical analysis and the threshold was calculated (Fig. 8 ). Threshold values obtained were 131 Ω, 81.5 Ω, and 130 Ω for mAb10540, mAb105802, and Sb#15-His6, respectively. For commercial antibodies, only three samples of Delta variant were tested. According to the manufacturer (R & D Systems), these antibodies do not detect the Delta variant, which justifies ΔRct values below the threshold. For Sb#15-His6, all Delta variant samples were successfully detected, with ΔRct values above the threshold. For Omicron variant samples, the same result was obtained for all biosensors, of the 10 positive samples, 9 were detected. The positive sample that was not detected by the biosensors was the subvariant BA.1.1.
Fig. 8.
The statistical analyses and threshold determination obtained for biosensors in the detection of SARS-CoV-2 variants (Delta and Omicron) and negative saliva samples.
When choosing which rapid test to use, sensitivity and specificity are parameters that must be considered, as they provide information about the diagnostic test accuracy [10]. According to WHO, rapid tests must meet minimum performance requirements of ≥80% sensitivity and ≥97% specificity compared with a molecular test [88].
The values for accuracy, sensitivity, and specificity are shown in Table 5 . For the Omicron variant, the biosensors showed values higher than 89% for all parameters; however, for the Delta variant, only Sb#15-His6 biosensor had values above the one recommended by WHO. When comparing all positive samples, Sb#15-His6 was the only biosensor that met the requirements recommended by WHO with accuracy of 95%. Besides that, the authors who first reported Sb#15 do not describe the cross-reaction of this VHH with other coronaviruses [47]. WHO recognizes that despite lower sensitivity than molecular tests, rapid diagnostic antigen detection tests may offer a faster and cheaper way to diagnose active SARS-CoV-2 infection, helping to prevent virus transmission [88].
Table 5.
Values obtained for accuracy, sensitivity, and specificity for biosensors in tests with saliva samples diluted in PBS.
| Omicron variant | |||
|---|---|---|---|
| mAb10540 | mAb105802 | Sb#15-His6 | |
| Accuracy | 94% | 94% | 94% |
| Sensitivity | 90% | 90% | 89% |
| Specificity |
100% |
100% |
100% |
| Delta variant | |||
| mAb10540 |
mAb105802 |
Sb#15-His6 |
|
| Accuracy | 22% | 90% | 100% |
| Sensitivity | 67% | 67% | 100% |
| Specificity |
0% |
100% |
100% |
| All positive samples | |||
| mAb10540 |
mAb105802 |
Sb#15-His6 |
|
| Accuracy | 85% | 85% | 95% |
| Sensitivity | 77% | 77% | 93% |
| Specificity | 100% | 100% | 100% |
The results obtained clearly demonstrate that the developed biosensor with Sb#15-His6 was more sensitive to detect SARS-CoV-2 in saliva samples than the biosensors developed with conventional IgG antibodies. In addition, it is essential to note that the Sb#15-His6 mass used in immobilization was five times lower than the others, which demonstrates the high specificity and sensibility of VHHs.
4. Conclusions
The development of a simple, portable, and reliable platform for rapid and sensitive SARS-CoV-2 detection has been highly challenging. To address this demand, biosensors were developed for SARS-CoV-2 spike protein RBD detection in saliva samples using PPy-NTs and Ni(OH)2 modified SPCE electrodes. The use of Ni(OH)2 is important to correctly orient an antibody immobilization on a biosensor surface so that it can avoid loss of antigen binding activity and increase the sensitivity.
Two conventional IgG antibodies (mAb10540 and 105802) and antibody fragment VHH Sb#15-His6 were used as bioreceptors. The VHH Sb#15His6 was inexpensively produced with a high yield and stable structure, which is important for scaling up the production and use in rapid test platforms. It is antigen-specific and interacts with SARS-CoV-2 spike protein RBD by affinity in nM range. Among the developed biosensors, VHH Sb#15-His6 showed more sensitivity and specificity in the detection of spike protein RBD Wuhan strain and Omicron and Delta variants, than conventional antibodies. The VHH biosensor also showed a lower LOQ than commercial antibodies, being possible to quantify 0.01 pg/mL of SARS-CoV-2 spike protein RBD in PBS.
The biosensors were validated by SARS-CoV-2 spike protein RBD detection in pre-characterized saliva samples, and it was possible to demonstrate that Sb#15-His6 biosensor was the only one that met WHO requirements, presenting 93% sensitivity and 100% specificity. The biosensor needs a low volume of saliva sample to perform SARS-CoV-2 detection and it can provide results within 15 min without further sample preparations. The obtained results demonstrate that the biosensor developed with VHH Sb#15-His6 has potential to be used as rapid test to diagnose COVID-19. This rapid and non-invasive evaluation of SARS-CoV-2 enables COVID-19 screening in the early stage of the disease due to the low limit of quantification, which can be very helpful to alert cases requiring immediate medical attention. In addition, the proposed platform can be further used with different antigen/antibody systems, showing promising potential to diagnose several other diseases.
Funding sources
This work was partially funded by CAPES (AUXPE 88881.504691/2020-01, 001), including a post-doctoral fellowship assigned to DSBR, a PhD scholarship to APMSB, and a master's scholarship to VZR. NITZ and BGG are supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) research career fellowships (303038/2019-5; 408635/2018-5; 304167/2019-3 and 304788/2018-0) and CNPq/MCTI/FNDCT Nº 18/2021 (408589/2021-3). INCT in Bioanalytics (FAPESP grant no. 2014/50867-3 and CNPq grant no. 465389/2014-7); Fundação Araucária/FIOCRUZ (CP December 2020 – Programa de Pesquisa, Desenvolvimento Tecnológico e Inovação FIOCRUZ); Universidade Federal do Paraná (PROIND-2020).
Credit statement
Alecsandra Santos: Conceptualization, Methodology, Validation, Formal Analysis, Investigation, Writing – Original Draft, Visualization. Ana Paula Macedo de Souza Brandão: Conceptualization, Methodology, Validation, Formal Analysis, Investigation, Writing – Original Draft, Visualization. Bruna M. Hryniewicz: Conceptualization, Methodology, Validation, Formal Analysis, Investigation, Writing – Original Draft, Visualization. Hellen Abreu: Conceptualization, Methodology, Validation, Writing – Original Draft. Larissa Bach-Toledo: Conceptualization, Methodology, Investigation, Writing – Original Draft. Stella Schuster da Silva: Conceptualization, Methodology, Validation Investigation, Writing– Original Draft. Andrei E. Deller: Validation, Investigation, Visualization. Vanessa Zulkievicz Rogerio: Methodology, Validation, Investigation. Dayane Sereno Baêta Rodrigues: Methodology, Validation, Investigation. Priscila Mazzochi Hiraiwa: Methodology, Validation, Investigation. Beatriz Gomes Guimarães: Methodology, Validation, Investigation. Luís F. Marchesi: Software, Formal Analysis, Writing – Review & Editing. Supervision. Jaqueline Carvalho de Oliveira: Methodology, Resources, Supervision, Project Administration, Funding Acquisition. Daniela Fiori Gradia: Methodology, Resources, Supervision, Project Administration, Funding Acquisition. Frederico L.F. Soares: Software, Formal analysis, Writing – Original Draft. Nilson I. T. Zanchin: Methodology, Writing – Review & Editing, Resources, Supervision, Project Administration, Funding Acquisition. Carolina Camargo de Oliveira: Methodology, Writing – Review & Editing, Resources, Supervision, Project Administration, Funding Acquisition Marcio Vidotti: Methodology, Writing – Review & Editing, Resources, Supervision, Project Administration, Funding Acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors acknowledge the FIOCRUZ program of technical platforms for access to the facilities for Integrated Structural Biology (RPT-15A) and Cytometry (RPT-08L). CTI Renato Archer for microscopic facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtchem.2023.101597.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
No data was used for the research described in the article.
References
- 1.McFee R.B. SARS 2 human coronavirus (COVID -19, SARS CoV2) Disease-a-Month. 2020;66 doi: 10.1016/j.disamonth.2020.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruell H., Vanshylla K., Korenkov M., Kurth F., Kreer C., Klein F. SARS-CoV-2 Omicron sublineages exhibit distinct antibody escape patterns short article SARS-CoV-2 Omicron sublineages exhibit distinct antibody escape patterns. Cell Host Microbe. 2022;30:1–11. doi: 10.1016/j.chom.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Marschalek R., Herrmann E., Helfritz F.A., Wolf T., Goetsch U., Ciesek S. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. EBioMedicine. 2022;82 doi: 10.1016/j.ebiom.2022.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao L., Zhu K.-L., Jiang X.-L., Wang X.-J., Zhan B.-D., Gao H.-X., Geng X.-Y., Duan L.-J., Dai E.-H., Ma M.-J. Omicron subvariants escape antibodies elicited by vaccination and BA.2.2 infection. Lancet Infect. Dis. 2022;22:1116–1117. doi: 10.1016/s1473-3099(22)00410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu F., Shang X., Chen M., Zhang C. Joint detection of serum IgM/IgG antibody is an important key to clinical diagnosis of SARS-CoV-2 infection. Can. J. Infect Dis. Med. Microbiol. 2020:2020. doi: 10.1155/2020/1020843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orooji Y., Sohrabi H., Hemmat N., Oroojalian F., Baradaran B., Mokhtarzadeh A., Mohaghegh M., Karimi-Maleh H. An overview on SARS-CoV-2 (COVID-19) and other human coronaviruses and their detection capability via amplification assay, chemical sensing, biosensing, immunosensing, and clinical assays. Nano-Micro Lett. 2021;13:1–30. doi: 10.1007/s40820-020-00533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toptan T., Eckermann L., Pfeiffer A.E., Hoehl S., Ciesek S., Drosten C., Corman V.M. Evaluation of a SARS-CoV-2 rapid antigen test: potential to help reduce community spread? J. Clin. Virol. 2021;135 doi: 10.1016/J.JCV.2020.104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazaca L.C., dos Santos P.L., de Oliveira P.R., Rocha D.P., Stefano J.S., Kalinke C., Abarza Muñoz R.A., Bonacin J.A., Janegitz B.C., Carrilho E. Biosensing strategies for the electrochemical detection of viruses and viral diseases – a review. Anal. Chim. Acta. 2021;1159 doi: 10.1016/J.ACA.2021.338384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peeling R.W., Olliaro P.L., Boeras D.I., Fongwen N. Scaling up COVID-19 rapid antigen tests: promises and challenges. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00048-7. e290–e295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C.C., Chiang Y.H., Chiang H.Y. A label-free electrochemical impedimetric immunosensor with biotinylated-antibody for SARS-CoV-2 Nucleoprotein detection in saliva. Biosensors. 2022;12 doi: 10.3390/bios12050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Souf S. Review article recent advances in diagnostic testing for viral infections. Biosci. Horiz. 2016;9:1–11. [Google Scholar]
- 13.Mehmandoust M., Gumus Z.P., Soylak M., Erk N. Electrochemical immunosensor for rapid and highly sensitive detection of SARS-CoV-2 antigen in the nasal sample. Talanta. 2022;240 doi: 10.1016/j.talanta.2022.123211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avelino K.Y.P.S., dos Santos G.S., Frías I.A.M., Silva-Junior A.G., Pereira M.C., Pitta M.G.R., de Araújo B.C., Errachid A., Oliveira M.D.L., Andrade C.A.S. Nanostructured sensor platform based on organic polymer conjugated to metallic nanoparticle for the impedimetric detection of SARS-CoV-2 at various stages of viral infection. J. Pharm. Biomed. Anal. 2021;206 doi: 10.1016/j.jpba.2021.114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahmati Z., Roushani M., Hosseini H., Choobin H. An electrochemical immunosensor using SARS-CoV-2 spike protein-nickel hydroxide nanoparticles bio-conjugate modified SPCE for ultrasensitive detection of SARS-CoV-2 antibodies. Microchem. J. 2021;170 doi: 10.1016/j.microc.2021.106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji D., Guo M., Wu Y., Liu W., Luo S., Wang X., Kang H., Chen Y., Dai C., Kong D., Ma H., Liu Y., Wei D. Electrochemical detection of a few copies of unamplified SARS-CoV-2 nucleic acids by a self-actuated molecular system. J. Am. Chem. Soc. 2022;144:13526–13537. doi: 10.1021/jacs.2c02884. [DOI] [PubMed] [Google Scholar]
- 18.Hryniewicz B.M., Volpe J., Bach-Toledo L., Kurpel K.C., Deller A.E., Soares A.L., Nardin J.M., Marchesi L.F., Simas F.F., Oliveira C.C., Huergo L., Souto D.E.P., Vidotti M. Development of polypyrrole (nano)structures decorated with gold nanoparticles towards immunosensing for COVID-19 serological diagnosis. Mater. Today Chem. 2022;24 doi: 10.1016/j.mtchem.2022.100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogomolova A., Komarova E., Reber K., Gerasimov T., Yavuz O., Bhatt S., Aldissi M. Challenges of electrochemical impedance spectroscopy in protein biosensing. Anal. Chem. 2009;81:3944–3949. doi: 10.1021/ac9002358. [DOI] [PubMed] [Google Scholar]
- 20.Zeng X., Shen Z., Mernaugh R. Recombinant antibodies and their use in biosensors. Anal. Bioanal. Chem. 2012;402:3027–3038. doi: 10.1007/s00216-011-5569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahadir E.B., Sezgintürk M.K. Label-free, ITO-based immunosensor for the detection of a cancer biomarker: receptor for activated C kinase 1. Analyst. 2016;141:5618–5626. doi: 10.1039/c6an00694a. [DOI] [PubMed] [Google Scholar]
- 22.Aydın M., Aydın E.B., Sezgintürk M.K. A highly selective electrochemical immunosensor based on conductive carbon black and star PGMA polymer composite material for IL-8 biomarker detection in human serum and saliva. Biosens. Bioelectron. 2018;117:720–728. doi: 10.1016/j.bios.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Arshak K., Velusamy V., Korostynska O., Oliwa-Stasiak K., Adley C. Conducting polymers and their applications to biosensors: emphasizing on foodborne pathogen detection. IEEE Sens J. 2009;9:1942–1951. doi: 10.1109/JSEN.2009.2032052. [DOI] [Google Scholar]
- 24.Guimard N.K., Gomez N., Schmidt C.E. Conducting polymers in biomedical engineering. Prog. Polym. Sci. 2007;32:876–921. doi: 10.1016/j.progpolymsci.2007.05.012. [DOI] [Google Scholar]
- 25.Ramanavičius A., Ramanavičiene A., Malinauskas A. Electrochemical sensors based on conducting polymer-polypyrrole. Electrochim. Acta. 2006;51:6025–6037. doi: 10.1016/j.electacta.2005.11.052. [DOI] [Google Scholar]
- 26.Sargent A., Sadik O.A. Monitoring antibody-antigen reactions at conducting polymer-based immunosensors using impedance spectroscopy. Electrochim. Acta. 1999;44:4667–4675. doi: 10.1016/S0013-4686(99)00265-0. [DOI] [Google Scholar]
- 27.Magar H.S., Hassan R.Y.A., Mulchandani A. Electrochemical impedance spectroscopy (EIS): Principles, construction, and biosensing applications. Sensors. 2021;21:6578. doi: 10.3390/S21196578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain R., Jadon N., Pawaiya A. Polypyrrole based next generation electrochemical sensors and biosensors: a review. Trends Anal. Chem. 2017;97:363–373. doi: 10.1016/j.trac.2017.10.009. [DOI] [Google Scholar]
- 29.Hahm J.I. Biomedical detection via macro- and nano-sensors fabricated with metallic and semiconducting oxides. J. Biomed. Nanotechnol. 2013;9:1–25. doi: 10.1166/JBN.2013.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ley C., Holtmann D., Mangold K.M., Schrader J. Immobilization of histidine-tagged proteins on electrodes. Colloids Surf. B Biointerfaces. 2011;88:539–551. doi: 10.1016/j.colsurfb.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 31.Madoz-Gúrpide J., Abad J.M., Fernández-Recio J., Vélez M., Vázquez L., Gómez-Moreno C., Fernández V.M. Modulation of electroenzymatic NADPH oxidation through oriented immobilization of ferredoxin:NADP+ reductase onto modified gold electrodes. J. Am. Chem. Soc. 2000;122:9808–9817. doi: 10.1021/ja001365m. [DOI] [Google Scholar]
- 32.Chebil S., Miodek A., Ambike V., Sauriat-Dorizon H., Policar C., Korri-Youssoufi H. Polypyrrole functionalized with new copper complex as platform for His-tag antibody immobilization and direct antigen detection. Sens. Actuators B Chem. 2013;185:762–770. doi: 10.1016/j.snb.2013.05.024. [DOI] [Google Scholar]
- 33.Graham B., Spiccia L., Hearn M.T.W. Comparison of the binding behavior of several histidine-containing proteins with immobilized copper(II) complexes of 1,4,7-triazacyclononane and 1,4-bis(1,4,7-triazacyclononan-1-yl)butane. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2011;879:844–852. doi: 10.1016/j.jchromb.2011.02.029. [DOI] [PubMed] [Google Scholar]
- 34.Noorbakhsh A., Salimi A. Amperometric detection of hydrogen peroxide at nano-nickel oxide/thionine and celestine blue nanocomposite-modified glassy carbon electrodes. Electrochim. Acta. 2009;54:6312–6321. doi: 10.1016/j.electacta.2009.05.078. [DOI] [Google Scholar]
- 35.Ganesana M., Istarnboulie G., Marty J.L., Noguer T., Andreescu S. Site-specific immobilization of a (His)6-tagged acetylcholinesterase on nickel nanoparticles for highly sensitive toxicity biosensors. Biosens. Bioelectron. 2011;30:43–48. doi: 10.1016/j.bios.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder H.W., Cavacini L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010;125 doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood E.J. In: Biochemistry and Molecular Biology Education. fifth ed. Abbas A.K., Lichtman A.H., editors. vol. 32. 2004. Cellular and molecular immunology; pp. 65–66. [DOI] [Google Scholar]
- 38.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hammers C., Songa E.B., Bendahman N., Hammers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 39.Sandomenico A., Sivaccumar J.P., Ruvo M. Evolution of Escherichia coli expression system in producing antibody recombinant fragments. Int. J. Mol. Sci. 2020;21:1–39. doi: 10.3390/ijms21176324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dumoulin M., Conrath K., Van Meirhaeghe A., Meersman F., Heremans K., Frenken L.G.J., Muyldermans S., Wyns L., Matagne A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2009;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hussack G., Hirama T., Ding W., MacKenzie R., Tanha J. Engineered single-domain antibodies with high protease resistance and thermal stability. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 43.Yang E., Liu Q., Huang G., Liu J., Wei W. Engineering nanobodies for next-generation molecular imaging. Drug Discov. Today. 2022;27:1622–1638. doi: 10.1016/j.drudis.2022.03.013. [DOI] [PubMed] [Google Scholar]
- 44.Genelhoud G., Adamoski D., Spalanzani R.N., Bochnia-Bueno L., de Oliveira J.C., Gradia D.F., Bonatto A.C., Wassem R., Raboni S.M., Nogueira M.B., de Araujo-Souza P.S. Comparison of SARS-CoV-2 molecular detection in nasopharyngeal swab, saliva, and gargle samples. Diagn. Microbiol. Infect. Dis. 2022;103 doi: 10.1016/j.diagmicrobio.2022.115678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prodromidis M.I. Impedimetric immunosensors-A review. Electrochim. Acta. 2010;55:4227–4233. doi: 10.1016/j.electacta.2009.01.081. [DOI] [Google Scholar]
- 46.Kaya S.I., Karadurmus L., Ozcelikay G., Bakirhan N.K., Ozkan S.A. INC; 2020. Electrochemical Virus Detections with Nanobiosensors. [DOI] [Google Scholar]
- 47.Walter J.D., Hutter C.A., Garaeva A.A., Scherer M., Zimmermann I., Wyss M., Rheinberger J., Ruedin Y., Earp J.C., Egloff P., Hürlimann L.M., Gonda I., Meier G., Remm S., Thavarasah S., van Geest G., Bruggman R., Zimmer G., Slotboom D.J., Paulino C., Seeger M.A. Biparatopic sybody constructs neutralize SARS-CoV-2 variants of 1 concern and mitigate emergence of drug resistance. EMBO Rep. 2022;23 doi: 10.1101/2020.11.10.376822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roof W.D., Horne S.M., Young K.D., Young R. slyD, a host gene required for phi X174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. FEMS Microbiol. Rev. 1995;17:213–218. doi: 10.1111/J.1574-6976.1995.TB00204.X. [DOI] [PubMed] [Google Scholar]
- 50.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 51.Hawley-Nelson P., Ciccarone V., Moore M.L. Transfection of cultured eukaryotic cells using cationic lipid reagents. Curr. Protoc. Mol. Biol. 2008;81:9.4.1–9.4.17. doi: 10.1002/0471142727.MB0904S81. [DOI] [PubMed] [Google Scholar]
- 52.Hryniewicz B.M., Lima R.V., Wolfart F., Vidotti M. Influence of the pH on the electrochemical synthesis of polypyrrole nanotubes and the supercapacitive performance evaluation. Electrochim. Acta. 2019;293:447–457. doi: 10.1016/j.electacta.2018.09.200. [DOI] [Google Scholar]
- 53.Vogels C.B.F., Breban M.I., Ott I.M., Alpert T., Petrone M.E., Watkins A.E., Kalinich C.C., Earnest R., Rothman J.E., de Jesus J.G., Claro I.M., Ferreira G.M., Crispim M.A.E., Singh L., Tegally H., Anyaneji U.J., Hodcroft E.B., Mason C.E., Khullar G., Metti J., Dudley J.T., MacKay M.J., Nash M., Wang J., Liu C., Hui P., Murphy S., Neal C., Laszlo E., Landry M.L., Muyombwe A., Downing R., Razeq J., de Oliveira T., Faria N.R., Sabino E.C., Neher R.A., Fauver J.R., Grubaugh N.D., da Silva Sales F.C., Ramundo M.S., Candido D.S., Silva C.A.M., de Pinho M.C., Coletti T. de M., Andrade P. dos S., de Souza L.M., Rocha E.C., Gomes Jardim A.C., Manuli E., Gaburo N., Granato C., Levi J.E., Costa S., de Souza W.M., Salum M.A., Pereira R., de Souza A., Matkin L.E., Nogueria M.L., Levin A.S., Mayaud P., Alexander N., Souza R., Acosta A.L., Prete C., Quick J., Brady O., Messina J., Kraemer M., Gouveia N. da C., Oliva I., de Souza M., Lazari C., Alencar C.S., Thézé J., Buss L., Araujo L., Cunha M.S., Loman N.J., Pybus O.G., Aguiar R.S., Wilkinson E., Msomi N., Iranzadeh A., Fonseca V., Doolabh D., San E.J., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Kosakovsky Pond S.L., Weaver S., Giovanetti M., Alcantara L.C.J., Martin D., Bhiman J.N., Williamson C. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blangero Y., Rabilloud M., Ecochard R., Subtil F. A Bayesian method to estimate the optimal threshold of a marker used to select patients' treatment. Stat. Methods Med. Res. 2020;29:29–43. doi: 10.1177/0962280218821394. [DOI] [PubMed] [Google Scholar]
- 55.Marquetti I., Link J.V., Lemes A.L.G., dos S. Scholz M.B., Valderrama P., Bona E. Partial least square with discriminant analysis and near infrared spectroscopy for evaluation of geographic and genotypic origin of arabica coffee. Comput. Electron. Agric. 2016;121:313–319. doi: 10.1016/j.compag.2015.12.018. [DOI] [Google Scholar]
- 56.Marcelo M.C.A., Soares F.L.F., Ardila J.A., Dias J.C., Pedó R., Kaiser S., Pontes O.F.S., Pulcinelli C.E., Sabin G.P. Fast inline tobacco classification by near-infrared hyperspectral imaging and support vector machine-discriminant analysis. Anal. Methods. 2019;11:1966–1975. doi: 10.1039/c9ay00413k. [DOI] [Google Scholar]
- 57.Thiagarajan G., Semple A., James J.K., Cheung J.K., Shameem M. 2016. A Comparison of Biophysical Characterization Techniques in Predicting Monoclonal Antibody Stability; pp. 1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfart F., Dubal D.P., Vidotti M., Gómez-Romero P. Hybrid core-shell nanostructured electrodes made of polypyrrole nanotubes coated with Ni(OH)2 nanoflakes for high energy-density supercapacitors. RSC Adv. 2016;6:15062–15070. doi: 10.1039/c5ra23671a. [DOI] [Google Scholar]
- 59.Jiang H., Zhao T., Li C., Ma J. Hierarchical self-assembly of ultrathin nickel hydroxide nanoflakes for high-performance supercapacitors. J. Mater. Chem. 2011;21:3818–3823. doi: 10.1039/c0jm03830j. [DOI] [Google Scholar]
- 60.Lorenzen A.L., Rossi T.S., Riegel-Vidotti I.C., Vidotti M. Influence of cationic and anionic micelles in the (sono)chemical synthesis of stable Ni(OH)2 nanoparticles: “In situ” zeta-potential measurements and electrochemical properties. Appl. Surf. Sci. 2018;455:357–366. doi: 10.1016/j.apsusc.2018.05.198. [DOI] [Google Scholar]
- 61.Gao M., Sheng W., Zhuang Z., Fang Q., Gu S., Jiang J., Yan Y. Efficient water oxidation using nanostructured α-nickel-hydroxide as an electrocatalyst. J. Am. Chem. Soc. 2014;136:7077–7084. doi: 10.1021/ja502128j. [DOI] [PubMed] [Google Scholar]
- 62.Skinner N.G., Hall E.A.H. The relevance of an equivalent circuit for polyaniline using immittance spectroscopy. Synth. Met. 1994;63:133–145. doi: 10.1016/0379-6779(94)90261-5. [DOI] [Google Scholar]
- 63.Bisquert J. Theory of the impedance of electron diffusion and recombination in a thin layer. J. Phys. Chem. B. 2002;106:325–333. [Google Scholar]
- 64.Wolfart F., Hryniewicz B.M.B.M., Marchesi L.F.L.F., Orth E.S.E.S., Dubal D.P.D.P., Gómez-Romero P., Vidotti M. Direct electrodeposition of imidazole modified poly(pyrrole) copolymers: synthesis, characterization and supercapacitive properties. Electrochim. Acta. 2017;243:260–269. doi: 10.1016/j.electacta.2017.05.082. [DOI] [Google Scholar]
- 65.Soares A.L., Zamora M.L., Marchesi L.F., Vidotti M. Adsorption of catechol onto PEDOT films doped with gold nanoparticles: electrochemical and spectroscopic studies. Electrochim. Acta. 2019;322 doi: 10.1016/j.electacta.2019.134773. [DOI] [Google Scholar]
- 66.Gazotti W.A., Matencio T., de Paoli M.A. Electrochemical impedance spectroscopy studies for chemically prepared poly(o-methoxyaniline) doped with functionalized acids. Electrochim. Acta. 1997;43:457–464. doi: 10.1016/S0013-4686(97)00120-5. [DOI] [Google Scholar]
- 67.Garcia-Belmonte G., Bisquert J., Pereira E.C., Fabregat-Santiago F. Switching behaviour in lightly doped polymeric porous film electrodes. Improving distributed impedance models for mixed conduction conditions. J. Electroanal. Chem. 2001;508:48–58. doi: 10.1016/S0022-0728(01)00504-6. [DOI] [Google Scholar]
- 68.Pesqueira C., Hryniewicz B.M., Bach-Toledo L., Tenório L.N., Marchesi L.F., Mazon T., Vidotti M. Interfacial characterization of polypyrrole/AuNP composites towards electrocatalysis of Ascorbic acid oxidation. Molecules. 2022;27:5776. doi: 10.3390/molecules27185776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Levie R. On porous electrodes in electrolyte solutions: I. Capacitance effects. Electrochim. Acta. 1963;8:751–780. doi: 10.1016/0013-4686(63)80042-0. [DOI] [Google Scholar]
- 70.Barsoukov Evgenij, Ross Macdonald J. John Wiley & Sons, Inc.; 2018. Impedance Spectroscopy: Theory, Experiment, and Applications. [DOI] [Google Scholar]
- 71.Garcia-Belmonte G., Bisquert J. Impedance analysis of galvanostatically synthesized polypyrrole films. Correlation of ionic diffusion and capacitance parameters with the electrode morphology. Electrochim. Acta. 2002;47:4263–4272. doi: 10.1016/S0013-4686(02)00510-8. [DOI] [Google Scholar]
- 72.Barral G., Maximovitch S., Njanjo-Eyoke F. 1996. Study of Electrochemically Formed Ni(OH), Layers by EIS. [Google Scholar]
- 73.Zhang X., Zhang J., Liu Z., Robinson C. Inorganic/organic mesostructure directed synthesis of wire/ribbon-like polypyrrole nanostructures. Chem. Commun. 2004:1852–1853. doi: 10.1039/b405255b. [DOI] [PubMed] [Google Scholar]
- 74.Yang X., Zhu Z., Dai T., Lu Y. Facile fabrication of functional polypyrrole nanotubes via a reactive self-degraded template. Macromol. Rapid Commun. 2005;26:1736–1740. doi: 10.1002/marc.200500514. [DOI] [Google Scholar]
- 75.Xu L., Ding Y.S., Chen C.H., Zhao L., Rimkus C., Joesten R., Suib S.L. 3D flowerlike α-nickel hydroxide with enhanced electrochemical activity synthesized by microwave-assisted hydrothermal method. Chem. Mater. 2008;20:308–316. doi: 10.1021/cm702207w. [DOI] [Google Scholar]
- 76.Ramesh T.N., Kamath P.V. Synthesis of nickel hydroxide: effect of precipitation conditions on phase selectivity and structural disorder. J. Power Sources. 2006;156:655–661. doi: 10.1016/j.jpowsour.2005.05.050. [DOI] [Google Scholar]
- 77.Barbucci R., Macnani A., Roncolini C., Silvestri S. Antigen-antibody recognition by Fourier transform IR spectroscopy/attenuated total reflection studies: biotin-avidin complex as an example. Biopolymers. 1991;31:827–834. doi: 10.1002/bip.360310703. [DOI] [PubMed] [Google Scholar]
- 78.Ahmad F., Zhou Y., Ling Z., Xiang Q., Zhou X. Systematic elucidation of interactive unfolding and corona formation of bovine serum albumin with cobalt ferrite nanoparticles. RSC Adv. 2016;6:35719–35730. doi: 10.1039/c6ra02850k. [DOI] [Google Scholar]
- 79.Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007;1767:1073–1101. doi: 10.1016/J.BBABIO.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 80.Ehsan M.A., Khan S.A., Rehman A. Screen-printed graphene/carbon electrodes on paper substrates as impedance sensors for detection of coronavirus in nasopharyngeal fluid samples. Diagnostics. 2021;11 doi: 10.3390/diagnostics11061030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rahmati Z., Roushani M., Hosseini H., Choobin H. Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Microchim. Acta. 2021;188 doi: 10.1007/s00604-021-04762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aydın E.B., Aydın M., Sezgintürk M.K. Highly selective and sensitive sandwich immunosensor platform modified with MUA-capped GNPs for detection of spike receptor binding domain protein: a precious marker of COVID 19 infection. Sens. Actuators B Chem. 2021;345 doi: 10.1016/j.snb.2021.130355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Torres M.D.T., de Araujo W.R., de Lima L.F., Ferreira A.L., de la Fuente-Nunez C. Low-cost biosensor for rapid detection of SARS-CoV-2 at the point of care. Matter. 2021;4:2403–2416. doi: 10.1016/j.matt.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salahandish R., Haghayegh F., Ayala-Charca G., Hyun J.E., Khalghollah M., Zare A., Far B., Berenger B.M., Niu Y.D., Ghafar-Zadeh E., Sanati-Nezhad A. Bi-ECDAQ: An electrochemical dual-immuno-biosensor accompanied by a customized bi-potentiostat for clinical detection of SARS-CoV-2 nucleocapsid proteins. Biosens. Bioelectron. 2022;203 doi: 10.1016/j.bios.2022.114018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zaccariotto G.C., Silva M.K.L., Rocha G.S., Cesarino I. A novel method for the detection of sars-cov-2 based on graphene-impedimetric immunosensor. Materials. 2021;14 doi: 10.3390/ma14154230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karaman C., Yola B.B., Karaman O., Atar N., Polat İ., Yola M.L. Sensitive sandwich-type electrochemical SARS-CoV-2 nucleocapsid protein immunosensor. Microchim. Acta. 2021;188 doi: 10.1007/s00604-021-05092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rahmati Z., Roushani M., Hosseini H. Amorphous Ni(OH)2 nano-boxes as a high performance substrate for aptasensor application. Measurement. 2022;189 doi: 10.1016/j.measurement.2021.110649. [DOI] [Google Scholar]
- 88.World Health Organization . Interim Guidance; 2021. Antigen-detection in the Diagnosis of SARS-CoV-2 Infection. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.