Abstract
Soybean is a major oil crop in the world, and fatty acids are the predominant components for oil bio-synthesis and catabolism metabolisms and also are the most important energy resources for organisms. In view of this, two recombinant inbred line (RIL) populations (ZL-RIL and ZQ-RIL) and one natural population were evaluated for five individual seed fatty acid contents (palmitic acid, stearic acid, oleic acid, linoleic acid, and linolenic acid) under four different environments, simultaneously. In total, sixteen additive QTL clusters were identified in ZL-RIL population, and fifteen were stably expressed across multiple environments or had pleiotropic effects on various fatty acid contents. Furthermore, five and five of these 16 QTL clusters were verified in ZQ-RIL population and natural population, respectively. Among these consistent and stable QTL clusters, one QTL cluster controlling fatty acid on chromosome 5 with pleiotropic effect was identified under all of the environments in ZL-RIL and ZQ-RIL populations and also was validated in the natural population. Meanwhile, another stable QTL cluster was detected on chromosome 9 with pleiotropic effect under multiple environments in ZL-RIL population and was further verified by the natural population. More importantly, some causal genes, such as the genes on chromosome 9, involving in the fatty acid catabolism process were found in these stable QTL clusters, and some of them, such as Gm09G042000, Gm09G041500, and Gm09G047200 on chromosome 9, showed different expressions in ZL-RIL parents (Zheng92116 and Liaodou14) based on the transcriptome sequencing analysis at different seed developmental stages. Thus, the study results provided insights into the genetic basis and molecular markers for regulating seed fatty acid contents in soybean breeding program.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01227-y.
Keywords: Soybean, Fatty acid, Multiple environments, Multiple genetic backgrounds, Pleiotropism QTLs, Causal genes
Introduction
Soybean (Glycine max, 2n = 40) has been cultivated for more than 5000 years, and its genome size is about 1.0 Gb with ~ 50 thousands of protein coding genes (Meng et al. 2016; Shen et al. 2018). Soybean is also the most important oil crop in the world, and more than 60% of the food oil comes from soybean seed (Zhang et al. 2018; Torkamaneh et al. 2020). Fatty acids (FA) are the predominant components of the seed oil and are also the most important energy resources for the organisms (Fan et al. 2015). There are five major types of individual fatty acid in soybean seed, which namely palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3) (Hyten et al. 2004; Spencer et al. 2004; Zhang et al. 2018). For these five individual fatty acids, there exist many differences, no matter the relative contents in seed, the qualities in biochemical field, or the utilization in the food industry and medical field. For instance, linoleic acid and linolenic acid are attributed to the essential fatty acids and are irreplaceable for humans with great health functions (Shibata et al. 2008; Akond et al. 2014). Meanwhile, oleic acid is also very important for human due to its vital function to soften blood vessels and prevent lots of human diseases caused by vascular sclerosis (Fan et al. 2015; Amin et al. 2019). In addition, the fatty acids also play very important roles in the manufacture industry to produce many necessary productions for human daily life and also can function as intra-cellular or extra-cellular signals for plants to defend many diseases (Kachroo et al. 2008; Shibata et al. 2008; Lim et al. 2017).
Although the vital applications of fatty acids have been found, the molecular and physiological mechanisms controlling fatty acids are still largely unknown, and the genetic improvements of soybean varieties with desirable components/contents are still relative slow and lagging behind (Zhang et al. 2018). The reasons for these might mainly due to the fatty acids are controlled by multiple minor-effect genes and attributed to the quantitative characteristics. Goettel et al. (2014) had predicted 1090 genes potentially participated in the soybean acyl lipid synthesis, catabolism, and signaling pathway, and 769 genes were found to express in seed; while 70 genes might associate with the synthesis of fatty acids in plastids, 104 associate with the degradation of lipids and fatty acids. Thus, the mapping of quantitative trait loci (QTL) and mining of related genes become the irreplaceable strategy for illustrating the genetic mechanisms and developing the varieties with desirable fatty acids in soybean (Panthee et al. 2006).
At present, there are some studies focused on the mapping of genetic loci for fatty acid contents in soybean. Bachlava et al. (2009) had identified one QTL controlling oleic acid content on chromosome 13 across multiple environments and genetic backgrounds. Li et al. (2011) detected 27 QTLs for seed fatty acid and oil contents on 12 chromosomes (Gm02, Gm03, Gm04, Gm08, Gm09, Gm10, Gm11, Gm14, Gm15, Gm16, Gm18, and Gm20) via QTLNetwork2.0 software. Wang et al. (2012) had found eight QTLs linked with palmitic acid content on six chromosomes (Gm05, Gm06, Gm09, Gm13, Gm15, and Gm18), five QTLs linked with stearic acid content on four chromosomes (Gm03, Gm05, Gm13, and Gm18), and nine QTLs linked with saturated fatty acid on seven chromosomes (Gm03, Gm05, Gm06, Gm09, Gm13, Gm15, and Gm18). Xie et al. (2012) also found six QTLs responsible for seed linolenic acid content on Gm02, Gm14, and Gm18; four QTLs for linoleic acid on Gm14, Gm16, Gm17, and Gm18; four QTLs for oleic acid and four QTLs for palmitic acid on Gm09, Gm16, Gm17, and Gm18; and one QTL for stearic acid content on Gm02.
Meanwhile, Akond et al. (2014) had detected 16 QTLs responsible for the fatty acid contents on chromosomes 3, 13, 14, 15, 16, and 18. Fan et al. (2015) had identified 35 additive QTLs underlying fatty acid contents, and 17 of them were detected on ten chromosomes (Gm03, Gm06, Gm07, Gm08, Gm09, Gm10, Gm12, Gm13, Gm18, and Gm20) under multiple environments or associated with multiple components via the linkage analysis of RIL population with 161 SSR markers. Zhang et al. (2018) identified 18 QTLs associated with fatty acids on chromosomes 5, 8, 14, and 15 via the genome wide association study (GWAS) of 313 soybean germplasms. Fang et al. (2017) identified 27 genome regions associating with ten fatty acid related traits on chromosomes 2, 3, 4, 5, 6, 7, 10, 11, 13, and 14 via the GWAS of 809 soybean germplasms. Van and Mchale (2017) had summarized 113 QTLs of seed fatty acid contents, and no meta-QTL was obtained due to the limited number and minimal overlap of these reported QTLs. In addition, to discover the genetic loci in wild soybean, Ha et al. (2014) identified nine QTLs controlling alpha-linolenic acid content on chromosomes 3, 5, 6, 11, 12, 14, 15, and 17 in the RIL population derived from the cross of wild soybean germplasm “PI483463” and cultivar variety “Hutcheson.” Leamy et al. (2017) investigated 570 wild soybean germplasms for the fatty acid contents, and 24 associated SNPs were found on chromosomes 7 and 14.
In summary, there were some studies reported QTLs of seed fatty acid contents at present, and the QTLs were located on nearly all of the soybean chromosomes (eighteen chromosomes except for the chromosomes 1 and 19), especially for the chromosomes 3, 5, 13, 14, 15, and 18 reported more times. However, there were very few loci and markers applied in soybean quality improvement due to the un-stability across different environments and genetic backgrounds. Meanwhile, the genetic and molecular mechanisms of soybean fatty acids are still largely unknown to present. Thus, it is necessary to identify the consistent and reliable QTLs across multiple environments and genetic backgrounds. In view of this, the objectives of this study are to (1) identify and validate consistent QTLs controlling soybean seed fatty acid contents across multiple populations and environments; (2) search and validate the causal genes responsible for the seed fatty acids in these consistent QTLs.
Materials and methods
Soybean materials
Two soybean RIL populations named ZL-RIL (F6:7, 345 lines) and ZQ-RIL (F6:7, 289 lines) were used in this study. ZL-RIL was developed from the cross of “Zheng92116” and “Liaodou14”; ZQ-RIL was developed from the cross of “Zheng92116” and “Qihuang30.” There were lots of differences between “Liaodou14” and “Qihuang30,” such as the parents and their origins, variety type, plant height, fuzz color, seed hilum color, and protein content. The seed fatty acid contents of three parents of RIL populations were listed in Table S1. Meanwhile, one natural soybean population consisted of 342 germplasms (69 landraces and 273 cultivars) was used to further verify the QTLs identified in the RIL populations.
Methods
Planting of soybean populations and analyzing of seed fatty acid contents
ZL-RIL and ZQ-RIL populations were planted under four different environments, simultaneously, which designated as E1 ~ E4, respectively. And E1 represented Qingxian-2015 (Cangzhou City, Hebei Province, N37°29′, E115°42′, China), E2 represented Qingyuan-2015 (Baoding City, Hebei Province, N38°10′, E113°40′, China), E3 represented Qingyuan-2016, and E4 represented Hebei Agricultural University Breeding Center-2016 (Baoding City, Hebei Province). For these four experiments, randomized complete block design was used with three replications, and the field planting methods and managements were same as our previous study (Li et al. 2018). In addition, the 342 germplasms of natural population were planted in Hebei Agricultural University Breeding Center in 2017 with two replications.
For fatty acid contents analyzing, the seeds of RIL populations (ZL-RIL and ZQ-RIL) and natural population (342 germplasms) were analyzed by using the DA7200 near-infrared (NIR) analyzer (Perten Instruments, Sweden). The five fatty acids, palmitic acid (PA), stearic acid (SA), oleic acid (OA), linoleic acid (LA), and linolenic acid (LNA), were analyzed. The saturated fatty acid (SFA) content was calculated by the sum of palmitic acid and stearic acid, while the unsaturated fatty acid (USFA) was calculated by the sum of oleic acid, linoleic acid, and linolenic acid.
Statistical analysis of seed fatty acid contents
The analysis of variance (ANOVA) of seed fatty acid contents was conducted via the SPSS V21.0 software, and the descriptive statistics (mean, standard deviation, coefficient of variation, maximum and minimum value, coefficients of skewness and kurtosis) of fatty acid contents was calculated via the Microsoft Excel 2007 software. The correlation coefficients of fatty acid contents were calculated via SPSS V25.0 software, and the hot-plot of correlation coefficient was obtained via the ggplot2 and corrplot software package of the R statistical program (https://www.r-project.org/).
QTL mapping of the soybean RIL populations
Based on the genetic linkage maps of ZL-RIL and ZQ-RIL populations constructed in our previous study (Li et al. 2018), the QTL mapping works of seed fatty acid contents of these two populations were conducted via the inclusive composite interval mapping method of ICIMapping v4.2 software in the present study. The threshold of LOD scores was determined by using 1000 permutations as our previous study (Li et al. 2018). Meanwhile, for QTL mapping result analysis, the QTLs conferring individual fatty acid content within 5.0 cM in soybean genetic linkage map or physical distances within 2.0 Mb in soybean genome sequence were considered as the same locus according to the reference (Meng et al. 2016) and our previous published studies (Li et al., 2018; 2019). And if the QTLs conferring different fatty acid contents located in the same locus, then designated as QTL cluster with pleiotropic effects. In addition, QTL was named according to the Soybean Genetics Committee recommendation.
Association analysis of fatty acid contents of natural population
To further verify the linkage QTLs conferring seed fatty acid contents obtained from the two RIL populations under multiple environments, one natural population was estimated for the fatty acid contents in Hebei Agricultural University Breeding Center in 2017. Based on the fatty acid contents and SNP genotypes of the natural population, the primary association analysis was conducted by using the MLM model (mixed linear model) via the GAPIT software package of R statistical program (https://www.r-project.org/).
Analysis of candidate genes via the transcriptome sequencing data
To discover the candidate genes in stable QTL intervals associated with seed fatty acid contents, the expression patterns of candidate genes were analyzed by using the transcriptome sequencing data of ZL-RIL parents (Zheng92116 and Liaodou14). In our previous study (Li et al. 2019), the transcriptome sequencing of Zheng92116 and Liaodou14 was conducted with two biological replications at five different soybean seed developmental stages (T1 ~ T5, represented 24th, 31st, 38th, 45th, and 52th days after flowering, respectively). Based on these transcriptome sequencing data, the gene expressions were analyzed. In addition, the physical positions of QTLs and candidate genes were noted as Williams82 reference genome sequence Gmax2.0.
Results
Relative wide genetic variations and significant correlation coefficients of seed fatty acid contents in ZL-RIL population
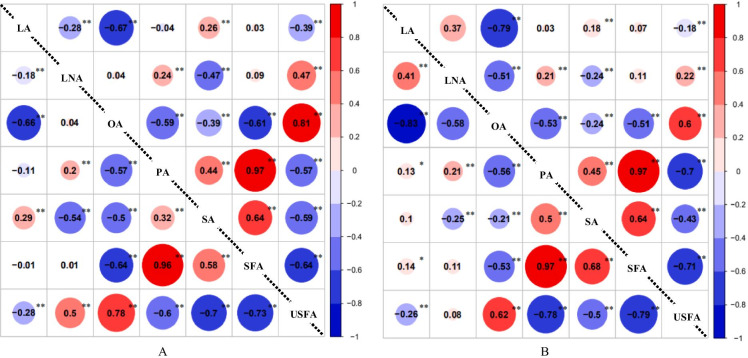
ANOVA results showed that there existed significant differences of seed fatty acid contents among different soybean RIL lines (Table S2). Descriptive statistical results of fatty acid contents under different environments showed that the relative wide genetic variations existed in ZL-RIL population, especially for the linolenic acid and oleic acid contents with the CV% values ranged from 17.9 to 25.9% and 11.4 to 13.3%, respectively (Table S2). The linoleic acid contents ranged from 52.6 to 61.6% with average 57.9% in soybean RIL lines, and the linolenic acid ranged from 1.8 to 9.3% with average 5.4%. The oleic acid contents ranged from 11.7 to 34.1% with average 20.5%, palmitic acid ranged from 6.6 to 12.7% with average 10.8%, and stearic acid ranged from 2.6 to 4.3% with average 3.8% (Table S2). The skewness and kurtosis absolute values indicated that these fatty acid contents displayed the normal distributions and attributed to the quantitative characteristics (Table S2, FigureS1). The correlation analysis found that there existed significant positive and negative correlations between these fatty acids (Fig. 1), such as the extremely significant negative correlations between oleic acid and the other fatty acid components (palmitic acid, stearic acid, linoleic acid, etc.).
Fig. 1.
The correlation coefficients of seed fatty acid contents in ZL-RIL population under different environments. LA, linoleic acid; LNA, linolenic acid; OA, oleic acid; PA, palmitic acid; SA, stearic acid; SFA, saturated fatty acid; USFA, unsaturated fatty acid. *Significant at 0.05 level; **Significant at 0.01 level. Lower triangular in A: correlation coefficients under E1 environment (Qingxian-2015); upper triangular in A: correlation coefficients under E2 environment (Qingyuan-2015); lower triangular in B: correlation coefficients under E3 environment (Qingyuan-2016); upper triangular in B: correlation coefficients under E4 environment (HAU Breeding Center-2016)
Additive pleiotropism QTL clusters of seed fatty acid contents across multiple environments in ZL-RIL population
Based on the fatty acid contents under four environments (E1 ~ E4), sixteen additive QTL clusters were identified in ZL-RIL population, and fifteen of them were detected across multiple environments or underlying multiple fatty acid contents with pleiotropic effects (Table 1). The QTL clusters were located on eleven chromosomes (Gm03, Gm04, Gm05, Gm06, Gm08, Gm09, Gm10, Gm12, Gm13, Gm16, and Gm17) with phenotypic variation explanations (PVEs) ranged from 2.1 to 10.2%, which indicated that there existed environmental stable QTLs with pleiotropic effects controlling soybean seed fatty acid contents in ZL-RIL population.
Table 1.
Additive QTL clusters of soybean seed fatty acid contents in ZL-RIL population
| Clustera | QTLb | Trait | Chr.c | Left marker | Right marker | Left marker positiond | Right marker positione | Pos.f | LODg | PVEh (%) |
Add.i | Left CIj | Right CI | Env.k |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1*# | qLNA-A1-1 | Linolenic acid | 5 | Satt211 | ss715591655 | 40,414,305 | 41,712,419 | 25.6 | 6.4 | 7.7 | − 0.25 | 19.7 | 31.4 | E3 |
| ss715591655 | ss715591780 | 41,712,419 | 41,007,475 | 28.4 | 2.9 | 3.9 | − 0.22 | 19.7 | 34.0 | E1 | ||||
| 28.4 | 3.6 | 3.6 | − 0.17 | 22.4 | 31.8 | E4 | ||||||||
| qLA-A1-1 | Linoleic acid | 5 | Satt211 | ss715591655 | 40,414,305 | 41,712,419 | 26.0 | 8.0 | 8.7 | − 0.37 | 21.7 | 31.0 | E3 | |
| ss715591655 | ss715591780 | 41,712,419 | 41,007,475 | 28.4 | 6.7 | 7.3 | − 0.29 | 24.9 | 31.2 | E4 | ||||
| 30.0 | 8.2 | 10.2 | − 0.36 | 28.5 | 33.4 | E1 | ||||||||
| Satt511 | Sat_217 | – | 41,679,029 | 6.5 | 3.4 | 4.3 | − 0.27 | 0.6 | 13.2 | E2 | ||||
| qOA-A1-1 | Oleic acid | 5 | ss715591655 | ss715591780 | 41,712,419 | 41,007,475 | 28.4 | 7.0 | 6.7 | 0.58 | 23.6 | 32.8 | E3 | |
| 29.0 | 3.5 | 4.0 | 0.44 | 23.4 | 34.0 | E1 | ||||||||
| 2# | qPA-K-1 | Palmitic acid | 9 | ss715603594 | ss715603616 | 3,215,569 | 3,271,593 | 38.8 | 4.8 | 5.1 | − 0.11 | 34.9 | 41.3 | E3 |
| ss715603616 | ss715604428 | 3,271,593 | 4,137,128 | 41.9 | 3.9 | 4.8 | − 0.16 | 38.8 | 54.7 | E2 | ||||
| 45.5 | 3.0 | 3.6 | − 0.09 | 38.8 | 55.2 | E4 | ||||||||
| qOA-K-1 | Oleic acid | 9 | ss715603594 | ss715603616 | 3,215,569 | 3,271,593 | 38.8 | 3.6 | 3.4 | 0.41 | 36.0 | 41.3 | E3 | |
| ss715603616 | ss715604428 | 3,271,593 | 4,137,128 | 46.3 | 2.8 | 3.7 | 0.56 | 41.4 | 55.2 | E2 | ||||
| qSFA-K-1 | Saturated fatty acid | 9 | ss715603594 | ss715603616 | 3,215,569 | 3,271,593 | 38.8 | 3.7 | 3.7 | − 0.12 | 33.7 | 41.3 | E3 | |
| ss715603616 | ss715604428 | 3,271,593 | 4,137,128 | 43.5 | 3.5 | 4.2 | − 0.18 | 38.8 | 55.2 | E2 | ||||
| ss715605175 | ss715605238 | 5,690,163 | 6,079,042 | 68.8 | 2.8 | 3.1 | − 0.12 | 66.5 | 74.6 | E1 | ||||
| Palmitic acid | 9 | ss715605175 | ss715605238 | 5,690,163 | 6,079,042 | 68.8 | 2.8 | 3.1 | − 0.10 | 66.5 | 73.9 | E1 | ||
| 3# | qOA-O | Oleic acid | 10 | ss715607504 | ss715607483 | 45,629,506 | 45,322,752 | 35.2 | 3.3 | 3.8 | 0.43 | 33.8 | 36.8 | E1 |
| ss715607450 | ss715607435 | 45,023,591 | 44,920,131 | 39.1 | 4.0 | 3.7 | 0.43 | 37.8 | 39.9 | E3 | ||||
| qLNA-O | Linolenic acid | 10 | ss715607504 | ss715607483 | 45,629,506 | 45,322,752 | 35.5 | 6.2 | 6.4 | − 0.23 | 33.8 | 36.8 | E4 | |
| ss715607450 | ss715607435 | 45,023,591 | 44,920,131 | 39.0 | 6.1 | 6.1 | − 0.23 | 37.8 | 39.9 | E3 | ||||
| qLA-O | Linoleic acid | 10 | ss715607504 | ss715607483 | 45,629,506 | 45,322,752 | 36.7 | 2.5 | 2.1 | − 0.18 | 34.3 | 37.7 | E3 | |
| ss715607772 | ss715607250 | 48,147,731 | 43,131,019 | 59.6 | 4.0 | 5.4 | − 0.25 | 58.5 | 60.4 | E4 | ||||
| qSA-O | Stearic acid | 10 | ss715607504 | ss715607483 | 45,629,506 | 45,322,752 | 36.7 | 2.6 | 3.1 | − 0.04 | 34.3 | 37.7 | E1 | |
| 4* | qPA-C1-1 | Palmitic acid | 4 | ss715588385 | ss715588105 | 4,421,661 | 4,206,857 | 96.6 | 4.1 | 4.2 | − 0.09 | 95.3 | 97.9 | E4 |
| ss715589072 | Sat_235 | 5,555,278 | – | 77.4 | 3.1 | 3.3 | 0.09 | 77.1 | 79.2 | E3 | ||||
| qSFA-C1-1 | Saturated fatty acid | 4 | ss715588385 | ss715588105 | 4,421,661 | 4,206,857 | 96.6 | 3.3 | 3.8 | − 0.10 | 95.3 | 97.9 | E4 | |
| ss715589072 | Sat_235 | 5,555,278 | – | 77.4 | 2.9 | 2.9 | 0.10 | 77.1 | 79.0 | E3 | ||||
| 5 | qOA-K-2 | Oleic acid | 9 | ss715603840 | ss715603826 | 39,520,644 | 39,418,052 | 122.2 | 4.9 | 4.6 | 0.48 | 122.0 | 122.3 | E3 |
| qLA-K-2 | Linoleic acid | 9 | ss715603840 | ss715603826 | 39,520,644 | 39,418,052 | 122.2 | 3.8 | 3.3 | − 0.23 | 122.1 | 126.9 | E3 | |
| qUSFA-K-2 | Unsaturated fatty acid | 9 | ss715603840 | ss715603826 | 39,520,644 | 39,418,052 | 122.2 | 3.1 | 3.7 | 0.23 | 122.0 | 122.3 | E3 | |
| 6 | qSA-F | Stearic acid | 13 | ss715617090 | Sat_133 | 21,147,361 | 24,949,516 | 96.8 | 2.9 | 4.1 | − 0.04 | 87.1 | 108.4 | E3 |
| qSFA-F | Saturated fatty acid | 13 | ss715617090 | Sat_133 | 21,147,361 | 24,949,516 | 100.1 | 3.6 | 5.5 | − 0.14 | 91.6 | 111.7 | E3 | |
| qPA-F | Palmitic acid | 13 | ss715617090 | Sat_133 | 21,147,361 | 24,949,516 | 101.0 | 2.9 | 4.3 | − 0.10 | 91.3 | 100.9 | E3 | |
| 7# | qLA-C1-2 | Linoleic acid | 4 | ss715589674 | ss715589686 | 9,532,122 | 9,580,008 | 23.1 | 2.8 | 3.3 | − 0.24 | 22.0 | 24.7 | E2 |
| qSFA-C1-2 | Saturated fatty acid | 4 | ss715589674 | ss715589686 | 9,532,122 | 9,580,008 | 23.5 | 3.6 | 4.0 | 0.10 | 23.1 | 25.5 | E4 | |
| qPA-C1-2 | Palmitic acid | 4 | ss715589674 | ss715589686 | 9,532,122 | 9,580,008 | 23.6 | 4.4 | 4.3 | 0.09 | 23.1 | 25.5 | E4 | |
| 8 | qLA-C2 | Linoleic acid | 6 | ss715593873 | ss715593858 | 21,143,967 | 20,686,842 | 93.1 | 9.3 | 8.3 | 0.36 | 93.0 | 93.2 | E3 |
| qLNA-C2 | Linolenic acid | 6 | ss715593858 | ss715593820 | 20,686,842 | 19,486,921 | 93.4 | 4.4 | 4.4 | 0.19 | 93.3 | 94.1 | E3 | |
| qOA-C2 | Oleic acid | 6 | ss715593858 | ss715593820 | 20,686,842 | 19,486,921 | 93.6 | 6.4 | 6.2 | − 0.56 | 93.0 | 94.1 | E3 | |
| Linoleic acid | 6 | ss715593801 | ss715593768 | 18,788,512 | 17,672,411 | 98.3 | 3.8 | 4.3 | 0.24 | 96.8 | 100.3 | E1 | ||
| 9# | qOA-A2 | Oleic acid | 8 | ss715599406 | ss715599449 | 12,393,450 | 13,275,688 | 100.1 | 2.6 | 2.5 | 0.36 | 97.4 | 104.0 | E3 |
| qLNA-A2 | Linolenic acid | 8 | ss715599492 | ss715599497 | 13,890,148 | 13,934,839 | 107.4 | 3.5 | 3.4 | − 0.17 | 106.9 | 108.1 | E3 | |
| ss715599492 | ss715599497 | 13,890,148 | 13,934,839 | 107.4 | 4.8 | 4.9 | − 0.20 | 106.9 | 108.1 | E4 | ||||
| 10 | qLNA-C1-3 | Linolenic acid | 4 | ss715588097 | ss715588036 | 44,684,894 | 43,971,465 | 17.5 | 2.6 | 2.7 | 0.15 | 15.9 | 18.7 | E4 |
| qPA-C1-3 | Palmitic acid | 4 | ss715588097 | ss715588036 | 44,684,894 | 43,971,465 | 17.8 | 4.7 | 5.2 | 0.13 | 16.1 | 17.9 | E1 | |
| qSFA-C1-3 | Saturated fatty acid | 4 | ss715588054 | ss715587943 | 44,142,247 | 42,905,456 | 19.8 | 4.0 | 4.7 | 0.15 | 18.9 | 20.8 | E1 | |
| 11* | qOA-J | Oleic acid | 16 | ss715624870 | ss715624875 | 36,583,392 | 36,635,344 | 156.4 | 3.0 | 3.3 | 0.40 | 156.0 | 156.8 | E1 |
| qPA-J | Palmitic acid | 16 | ss715624993 | Sat_395 | 37,518,951 | 37,577,362 | 163.8 | 4.3 | 4.7 | − 0.13 | 161.1 | 175.4 | E1 | |
| qSFA-J | Saturated fatty acid | 16 | ss715624993 | Sat_395 | 37,518,951 | 37,577,362 | 165.5 | 3.3 | 4.1 | − 0.14 | 160.9 | 178.7 | E1 | |
| 12 | qLNA-H | Linolenic acid | 12 | ss715611672 | ss715611537 | 1,766,819 | 1,464,938 | 124.6 | 3.7 | 3.6 | − 0.17 | 123.8 | 125.8 | E3 |
| qLA-H | Linoleic acid | 12 | ss715611564 | ss715611525 | 1,536,763 | 1,438,251 | 126.2 | 4.1 | 3.5 | − 0.24 | 125.9 | 127.1 | E3 | |
| qOA-H | Oleic acid | 12 | ss715611435 | ss715611407 | 1,234,005 | 1,166,537 | 129.3 | 3.6 | 3.3 | 0.41 | 127.1 | 129.3 | E3 | |
| 13* | qPA-D2 | Palmitic acid | 17 | ss715627538 | ss715627484 | 3,644,762 | 3,611,437 | 194.2 | 3.3 | 3.5 | − 0.09 | 193.2 | 194.4 | E3 |
| qSFA-D2 | Saturated fatty acid | 17 | ss715627538 | ss715627484 | 3,644,762 | 3,611,437 | 194.2 | 2.8 | 2.7 | − 0.10 | 193.2 | 194.4 | E3 | |
| 14 | qOA-N | Oleic acid | 3 | ss715585348 | ss715585205 | 3,582,559 | 3,362,229 | 162.1 | 3.7 | 4.1 | 0.45 | 157.5 | 162.8 | E1 |
| qLA-N | Linoleic acid | 3 | ss715585205 | ss715585138 | 3,362,229 | 3,192,661 | 163.3 | 2.8 | 3.1 | − 0.20 | 162.8 | 165.3 | E1 | |
| 15 | qSFA-A1-2 | Saturated fatty acid | 5 | ss715591194 | ss715591168 | 35,722,019 | 35,533,047 | 83.8 | 2.9 | 3.2 | − 0.09 | 81.1 | 84.6 | E4 |
| qOA-A1-2 | Oleic acid | 5 | ss715591168 | ss715591141 | 35,533,047 | 35,309,800 | 84.1 | 4.1 | 5.4 | 0.46 | 82.3 | 84.6 | E4 | |
| 16* | qPA-A2 | Palmitic acid | 8 | ss715599230 | ss715599242 | 10,700,780 | 10,963,012 | 85.0 | 3.3 | 3.3 | − 0.08 | 84.1 | 85.0 | E4 |
Note: a* indicated the QTL clusters further verified by ZQ-RIL population in this study. # indicated the QTL clusters further verified by the natural population in this study
bThe nomenclature of QTL included: QTL, trait, and linkage group
cChromosome
dPhysical position of left marker in soybean genome (Gmax2.0)
ePhysical position of right marker in soybean genome (Gmax2.0)
fPosition
gLikelihood odds ratio
hProportion of phenotypic variation explained by QTL
iAdditive effect
jConfidence interval
kEnvironment: E1, Qingxian-2015; E2, Qingyuan-2015; E3, Qingyuan-2016; E4, HAU Breeding Center-2016
Among these QTL clusters, the first pleiotropism QTL cluster was detected to control linolenic acid, linoleic acid, and oleic acid contents under multiple environments, with PVEs ranged from 3.6 to 10.2% on chromosome 5 (Table 1, Fig. 2). Further analysis found that this QTL cluster associated with linolenic acid content under three environments (E1, E3, and E4), associated with linoleic acid under all of the environments, and also associated with oleic acid under two environments (E1 and E3). The favorable alleles of this QTL cluster for linolenic acid and linoleic acid contents were derived from the high fatty acid content ZL-RIL parent “Liaodou14.”
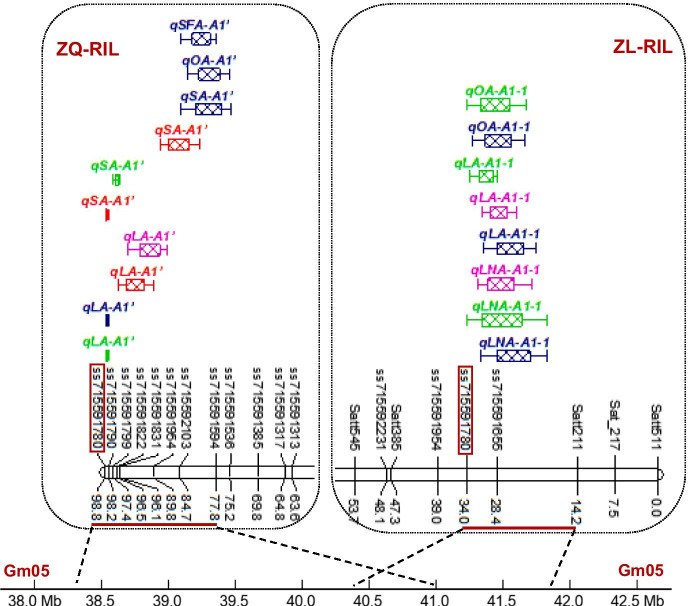
Fig. 2.
The pleiotropism QTL cluster controlling fatty acid contents on chromosome 5 across multiple environments and genetic backgrounds. The common SNP marker ss715591780 across two populations was highlighted with red rectangle; different colors of QTLs indicating different environments, green: E1 environment; red: E2 environment; blue: E3 environment; pink: E4 environment
The second pleiotropism QTL cluster was detected to associate with palmitic acid, oleic acid, and saturated fatty acid contents across multiple environments with PVEs ranged from 3.1 to 5.1% (Table 1, Fig. 3). This QTL cluster was identified on chromosome 9, linked with palmitic acid content under all of the four environments, linked with oleic acid content under two environments (E2 and E3), and linked with saturated fatty acid content under three environments (E1, E2, and E3). The favorable alleles of this QTL cluster for palmitic acid and saturated fatty acid contents were also derived from “Liaodou14.”
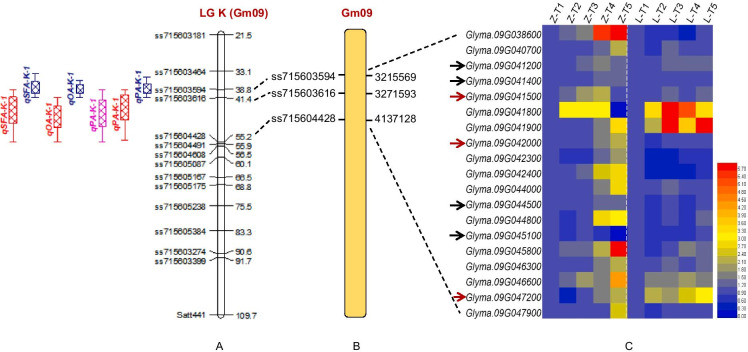
Fig. 3.
The consistent and stable QTL cluster and causal genes for seed fatty acid contents on chromosome 9 in soybean. A Consistent QTL cluster on chromosome 9 in ZL-RIL population, different colors indicated QTLs detected under different environments (red: E2 environment; blue: E3 environment; pink: E4 environment), the SNP markers and their genetic positions were showed on the chromosome 9. B Physical position of QTL cluster on chromosome 9, the linkage SNP markers and their physical positions were showed on the chromosome 9, the physical positions were obtained via the Williams82 reference genome (Gmax2.0). C Expressions of causal genes in the QTL cluster interval on chromosome 9, data from transcriptome sequencing of ZL-RIL parents Zheng92116 and Liaodou14 (all the arrows indicated causal genes participating in the catabolism process of soybean fatty acids, and the red arrows indicated the causal genes with differential expressions between Zheng92116 and Liaodou14; Z-T1 ~ Z-T5 represented the gene expressions at five seed developmental stages in Zheng92116; L-T1 ~ L-T5 represented the gene expressions at five seed developmental stages in Liaodou14)
The third pleiotropism QTL cluster was detected to control four types of fatty acid contents, oleic acid, linolenic acid, linoleic acid, and stearic acid, under multiple environments with PVEs ranged from 2.1 to 6.4% (Table 1). The QTL cluster was identified on chromosome 10 and associated with oleic acid, linolenic acid, and linoleic acid contents under two environments, respectively. And also, the favorable alleles of this QTL cluster for linolenic acid, linoleic acid, and stearic acid contents were derived from “Liaodou14.” Thus, it could be seen that the RIL parent “Liaodou14” contributed many favorable alleles for linolenic acid, linoleic acid, palmitic acid, and stearic acid contents due to its high fatty acid contents in seed.
Apart from these pleiotropism QTL clusters across multiple environments above, there were other twelve QTL clusters controlling different fatty acids under multiple environments or conferring multiple fatty acids identified in ZL-RIL population (Table 1). The favorable alleles of these QTLs for linolenic acid, linoleic acid, palmitic acid, and stearic acid contents mainly derived from “Liaodou14,” while the favorable alleles for oleic acid content mainly derived from the another RIL parent “Zheng92116,” which indicated that the recombination of these two RIL parents could produce the favorite lines with higher five individual fatty acid contents.
Verification of additive QTL clusters conferring seed fatty acid contents under multiple environments via ZQ-RIL population
To further verify the QTL clusters obtained in ZL-RIL population, the fatty acid contents of ZQ-RIL population were assessed under the same four environments (E1 ~ E4) via the same methods (Table S3, FigureS2). ANOVA results showed that there existed significant differences of fatty acid contents among different RIL lines (TableS3). Based on these, the QTL mapping was conducted. In total, eight additive QTL clusters with PVEs 3.1 ~ 9.4% were identified on seven chromosomes (Gm04, Gm05, Gm06, Gm08, Gm15, Gm16, and Gm17) in ZQ-RIL population, and five of them were consistent with the QTL clusters of ZL-RIL population (Table 2). These consistent QTL clusters offered the stable QTLs for marker assisted selection and candidate gene mining for seed fatty acid contents in soybean molecular breeding program.
Table 2.
Additive QTL clusters of soybean seed fatty acid contents in ZQ-RIL verification population
| Clustera | QTLb | Trait | Chr.c | Left marker | Right marker | Left marker Pos.d | Right marker Pos.e | Pos.f | LODg | PVEh (%) |
Add.i | Left CIj | Right CI | Env.k |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1* | qLA-A1’ | Linoleic acid | 5 | ss715591790 | ss715591780 | 40,974,254 | 41,007,475 | 98.7 | 5.7 | 8.6 | − 0.32 | 98.2 | 98.7 | E1 |
| 98.7 | 5.7 | 6.9 | − 0.22 | 98.2 | 98.7 | E3 | ||||||||
| ss715591954 | ss715591831 | 39,886,822 | 40,684,517 | 93.2 | 4.8 | 6.8 | − 0.24 | 89.8 | 96.4 | E2 | ||||
| 89.9 | 5.5 | 7.3 | − 0.22 | 87.0 | 94.5 | E4 | ||||||||
| qSA-A1’ | Stearic acid | 5 | ss715591790 | ss715591780 | 40,974,254 | 41,007,475 | 98.7 | 13.1 | 9.4 | − 0.08 | 98.2 | 98.7 | E2 | |
| ss715591822 | ss715591799 | 40,772,548 | 40,916,521 | 96.5 | 2.9 | 4.2 | − 0.05 | 96.1 | 97.3 | E1 | ||||
| ss715592103 | ss715591954 | 39,151,875 | 39,886,822 | 85.3 | 5.4 | 3.8 | 0.05 | 80.9 | 88.5 | E2 | ||||
| ss715591594 | ss715592103 | 38,280,990 | 39,151,875 | 78.6 | 4.0 | 4.8 | − 0.04 | 75.1 | 84.6 | E3 | ||||
| qOA-A1’ | Oleic acid | 5 | ss715591594 | ss715592103 | 38,280,990 | 39,151,875 | 78.9 | 4.6 | 5.6 | 0.44 | 75.4 | 83.3 | E3 | |
| qSFA-A1’ | Saturated fatty acid | 5 | ss715591594 | ss715592103 | 38,280,990 | 39,151,875 | 80.1 | 3.5 | 5.1 | − 0.13 | 77.8 | 84.6 | E3 | |
| 2* | qPA-C1’ | Palmitic acid | 4 | ss715587949 | ss715587994 | 4,045,439 | 4,104,193 | 22.7 | 3.5 | 3.8 | − 0.11 | 21.8 | 24.8 | E1 |
| ss715587994 | ss715588105 | 4,104,193 | 4,206,857 | 24.7 | 5.6 | 7.6 | − 0.13 | 22.8 | 27.2 | E3 | ||||
| qSFA-C1’ | Saturated fatty acid | 4 | ss715587994 | ss715588105 | 4,104,193 | 4,206,857 | 24.7 | 5.0 | 6.5 | − 0.14 | 22.8 | 27.2 | E3 | |
| qOA-C1’ | Oleic acid | 4 | ss715588105 | ss715588385 | 4,206,857 | 4,421,661 | 25.6 | 4.3 | 4.9 | 0.41 | 22.8 | 27.2 | E3 | |
| ss715587789 | ss715587843 | 3,590,478 | 3,739,404 | 18.7 | 3.0 | 4.7 | 0.61 | 18.2 | 19.4 | E1 | ||||
| qUSFA-C1’ | Unsaturated fatty acid | 4 | ss715589110 | ss715589231 | 6,078,937 | 7,191,821 | 47.1 | 3.0 | 3.9 | 0.24 | 44.2 | 55.7 | E3 | |
| 3 | qLA-A2-1’ | Linoleic acid | 8 | ss715602131 | ss715602331 | 43,896,715 | 45,913,059 | 261.3 | 2.7 | 5.2 | 0.21 | 244.8 | 269.4 | E2 |
| 262.2 | 4.3 | 7.9 | 0.23 | 252.4 | 269.4 | E3 | ||||||||
| 269.4 | 6.4 | 8.5 | 0.24 | 261.2 | 269.4 | E4 | ||||||||
| qOA-A2-1’ | Oleic acid | 8 | ss715602131 | ss715602331 | 43,896,715 | 45,913,059 | 262.6 | 3.0 | 5.0 | − 0.42 | 249.9 | 269.4 | E3 | |
| 266.9 | 4.3 | 6.2 | − 0.51 | 257.3 | 269.4 | E4 | ||||||||
| 4 | qSFA-E’ | Saturated fatty acid | 15 | ss715622827 | ss715622951 | 5,222,329 | 6,841,404 | 69.1 | 4.0 | 6.7 | − 0.14 | 58.2 | 80.8 | E3 |
| qPA-E’ | Palmitic acid | 15 | ss715622827 | ss715622951 | 5,222,329 | 6,841,404 | 72.7 | 4.2 | 7.3 | − 0.13 | 60.3 | 72.6 | E3 | |
| qUSFA-E’ | Unsaturated fatty acid | 15 | ss715622827 | ss715622951 | 5,222,329 | 6,841,404 | 77.1 | 4.8 | 8.0 | 0.34 | 66.2 | 77.0 | E3 | |
| 5 | qOA-C2’ | Oleic acid | 6 | ss715594793 | ss715594746 | 47,869,389 | 47,722,575 | 46.3 | 3.1 | 3.6 | − 0.39 | 39.7 | 47.0 | E4 |
| 47.0 | 3.8 | 4.3 | − 0.39 | 45.5 | 48.3 | E3 | ||||||||
| 6* | qPA-A2-2’ | Palmitic acid | 8 | ss715599171 | ss715599202 | 10,243,564 | 10,482,820 | 94.5 | 4.4 | 4.9 | − 0.12 | 93.5 | 95.7 | E1 |
| qSFA-A2-2’ | Saturated fatty acid | 8 | ss715599171 | ss715599202 | 10,243,564 | 10,482,820 | 95.7 | 3.7 | 4.0 | − 0.14 | 94.4 | 96.4 | E1 | |
| 7* | qPA-J’ | Palmitic acid | 16 | ss715624939 | ss715624938 | 37,125,034 | 37,086,672 | 5.5 | 4.8 | 5.3 | − 0.13 | 5.2 | 5.6 | E1 |
| 8* | qPA-D2’ | Palmitic acid | 17 | ss715626740 | ss715626718 | 3,244,333 | 3,164,428 | 133.1 | 3.4 | 3.8 | − 0.11 | 132.7 | 133.8 | E1 |
Note: a* indicated the QTL clusters co-located with ZL-RIL population in this study
bThe nomenclature of QTL included: QTL, trait and linkage group
cChromosome
dPhysical position of left marker in soybean genome (Gmax2.0)
ePhysical position of right marker in soybean genome (Gmax2.0)
fPosition
gLikelihood odds ratio
hProportion of phenotypic variation explained by QTL
iAdditive effect
jConfidence interval
kEnvironment: E1, Qingxian-2015; E2, Qingyuan-2015; E3, Qingyuan-2016; E4, HAU Breeding Center-2016
Among these QTL clusters in ZQ-RIL population, the first consistent pleiotropism QTL cluster was identified to associate with linoleic acid, stearic acid, oleic acid, and saturated fatty acid on chromosome 5 across multiple environments (Table 2, Fig. 2). In details, this QTL cluster was detected to associate with linoleic acid under all of the environments with PVEs 6.8 ~ 8.6% and link with stearic acid content under three different environments (E1, E2, and E3) with PVEs 3.8 ~ 9.4% (Table 2), and also responsible for the oleic acid and saturated fatty acid contents under E3 environment. More importantly, this consistent QTL cluster in ZQ-RIL population shared one common SNP marker ss715591780 with the QTL cluster in ZL-RIL population (Tables 1 and 2, Fig. 2), which indicated that it was a stable QTL cluster controlling multiple fatty acid contents on chromosome 5 across different soybean genetic backgrounds and multiple environments.
The second consistent pleiotropism QTL cluster was identified to associate with palmitic acid, oleic acid, saturated fatty acid, and unsaturated fatty acid on chromosome 4 under multiple environments in ZQ-RIL population (Table 2). In details, this QTL cluster was detected to link with palmitic acid and oleic acid across E1 and E3 environments with PVEs 3.8 ~ 7.6% and 4.7 ~ 4.9%, respectively. It was also detected to link with saturated fatty acid and unsaturated fatty acid under E3 environment (Table 2). And more importantly, this consistent QTL cluster in ZQ-RIL shared two common SNP markers, ss715588105 and ss715588385, with the QTL cluster in ZL-RIL population (Tables 1 and 2), indicated that it was another stable QTL cluster controlling multiple fatty acid contents on chromosome 4 across multiple genetic backgrounds and environments.
Apart from these two consistent pleiotropism QTL clusters across ZL-RIL and ZQ-RIL populations, there were other three consistent QTL clusters on chromosomes 8, 16, and 17 (Tables 1 and 2). Furthermore, there were three specific QTL clusters detected in ZQ-RIL under multiple environments (Table 2). The pleiotropism QTL cluster, qLA-A2-1’ and qOA-A2-1’ (ss715602131 ~ ss715602331), was associated with linoleic acid under three environments (E2, E3, and E4) with PVEs 5.2 ~ 8.5% and also associated with oleic acid under two environments (E3 and E4) with PVEs 5.0 ~ 6.2% (Table 2). More importantly, the favorable allele of qLA-A2-1’ for linoleic acid was derived from the RIL parent “Zheng92116,” which was different from the other QTLs in ZQ-RIL population. In summary, these consistent and stable QTL clusters in ZL-RIL and ZQ-RIL populations could be detected across multiple environments and genetic backgrounds and could be applied in the breeding program for seed fatty acid genetic improvement.
Verification of additive QTLs for fatty acid contents via the natural population
To further demonstrate the QTLs identified in ZL-RIL population, the fatty acid contents of natural population (342 germplasms) were assessed, and the coefficient of variations ranged from 1.8 to 18.8% with the relative wide variations on linolenic acid and oleic acid contents (Table S4). The skewness and kurtosis absolute values showed that these fatty acid contents displayed the normal distributions and attributed to the quantitative characteristics (TableS4 , FigureS3). Based on these, the primary GWAS analysis was conducted. The results showed that there were six associated SNPs identified to consistent with five QTL clusters in ZL-RIL population, which located on chromosomes 4, 5, 8, 9, and 10 (Table S5), and three of them were justly located in the QTL intervals of ZL-RIL population (Table 1).
Among these consistent SNPs, the first SNP ss715591621 was identified to associate with oleic acid content on chromosome 5 (Table S5) and was adjacent to the pleiotropism QTL cluster (qLNA-A1-1, qLA-A1-1, and qOA-A1-1 on chromosome 5, Table 1) in ZL-RIL population, and the physical distance to this QTL cluster marker ss715591655 was 407 kb (Table 1 and Table S5). The second consistent associated SNP ss715603674 was found to responsible for the saturated fatty acid content on chromosome 9 and was justly located in the QTL cluster interval (qPA-K-1, qOA-K-1, and qSFA-K-1) of ZL-RIL population (Table 1 and Table S5).
The third consistent associated SNP ss715607502 was also found to responsible for the saturated fatty acid content on chromosome 10 and was justly located in the QTL cluster interval (qOA-O, qLNA-O, qLA-O, and qSA-O) of ZL-RIL population (Table 1 and Table S5). Meanwhile, SNP markers ss715589728 and ss715589724 were identified to associate with palmitic acid content on chromosome 4, and the distances of these two markers to the adjacent QTL cluster (qLA-C1-2, qSFA-C1-2, and qPA-C1-2) were 207 kb and 241 kb, respectively (Table 1 and Table S5). In addition, the SNP marker ss715599409 was detected to associate with stearic acid content on chromosome 8 and was located in the QTL cluster interval (qOA-A2 and qLNA-A2) of ZL-RIL population (Table 1 and Table S5).
Causal genes in the consistent pleiotropism QTL clusters involved in the synthesis and catabolism metabolism of seed fatty acids in soybean
To further discover the causal genes involved in the synthesis and catabolism metabolism of seed fatty acids in the consistent and stable pleiotropism QTL clusters under multiple environments identified in this study, the expressions of candidate genes in the QTL intervals reference to Williams82 genome (Gmax2.0) were analyzed based on the transcriptome sequencing data of soybean seeds at different developmental stages of the ZL-RIL population parents (Zheng92116 and Liaodou14). As expected, some causal genes were found with different expression patterns between Zheng92116 and Liaodou14 in these QTL clusters participating in the synthesis or catabolism metabolism process of fatty acids in soybean (Table S6).
Among these genes, there were several genes (Glyma.09G041200, Glyma.09G041400, Glyma.09G042000, etc.) identified in the QTL cluster interval (qPA-K-1, qOA-K-1, and qSFA-K-1) on chromosome 9 involving in the seed fatty acid catabolism process (Table S6). In details, Glyma.09G041200 and Glyma.09G041400 were predicted to encode the diacylglycerol lipase and phospholipase, respectively, which were the important enzymes to catalyze the catabolism of triglyceride (seed oil) to generate fatty acid in soybean. Meanwhile, Glyma.09G042000 was predicted to encode the acyl-CoA dehydrogenase, which was the key enzyme for fatty acid catabolism to generate acetyl-CoA via the β-oxidation pathway in soybean. And more importantly, the causal gene Glyma.09G042000 displayed relative higher expression levels in the Zheng92116 at the T4 and T5 stages (Fig. 3), which indicated that there might exist a relative higher catabolism process of fatty acid at these two stages in Zheng92116, and resulted in the relative lower fatty acid contents of this variety. Conversely, the relative lower expressions of Glyma.09G042000 in Liaodou14 led to the relative higher fatty acid contents in seed (Fig. 3).
Apart from the causal genes on chromosome 9, there were several genes identified in the QTL cluster (qOA-O, qLNA-O, qLA-O, and qSA-O) on chromosome 10 (Table S6). In details, Glyma.10G222400 and Glyma.10G222500 were predicted to encode the peroxidase, and peroxidase was the important enzyme to catabolism the fatty acid in peroxisome in plant seed. And more importantly, the Glyma.10G222400 showed relative higher expression levels in the Zheng92116 at the T4 and T5 stages, which indicated the relative higher catabolism process of fatty acid at these stages in Zheng92116, and resulted in the relative lower fatty acid contents in this variety, while the relative lower expressions of Glyma.10G222400 in Liaodou14 led to the relative higher fatty acid contents. In summary, the causal genes in the consistent and stable QTLs not only further demonstrated the mapping results of this study, but also offered the important genes to be further studied in the fatty acid genetic improvement in soybean molecular breeding program.
4. Discussion
The consistent and stable pleiotropism QTL cluster responsible for soybean seed fatty acid and oil contents on chromosome 5 should be more in-depth researched
Soybean is the most planted oil crop in the world, and nearly more than 60% of the editable oil is offered by soybean seed (Li et al. 2011; Zhang et al. 2020). Meanwhile, fatty acids are the most predominant components for seed oil process of synthesis and catabolism and are usually regarded as the most important energy resources for the bio-system (Li-Beisson et al. 2013). Thus, understanding the genetic mechanisms and illustrating the molecular mechanisms of seed fatty acid metabolism process is necessary for the seed oil genetic improvement in soybean breeding program. In view of this, two soybean RIL populations and one natural population were analyzed with the five seed fatty acid components, simultaneously. Based on these, the linkage and association analysis were conducted, and several confirmed QTL clusters controlling multiple seed fatty acid contents were identified across multiple environments and genetic backgrounds in present study.
Among these confirmed QTL clusters, one stable pleiotropism QTL cluster controlling multiple seed fatty acid contents was identified on chromosome 5 across multiple environments and genetic backgrounds. In details, this QTL cluster was detected on chromosome 5 in the ZL-RIL and ZQ-RIL populations under all of the four environments, with the sharing common SNP marker ss715591780 (Tables 1 and 2; Fig. 2); furthermore, it was verified in the natural population (Table S5). More importantly, there was one stable QTL qOIL-A1 controlling soybean seed oil content under multiple environments identified in our previous study based on the same RIL populations with the same sharing marker ss715591780 (Li et al. 2019). Thus, these results indicated that this QTL cluster on chromosome 5 was very important for seed fatty acid and oil contents in soybean and worth more in-depth studies to illustrate the formation mechanisms of these important quality characteristics in soybean.
Apart from our studies for fatty acid and oil contents on chromosome 5, there were some other studies that had also reported QTLs associated with these characteristics in the similar regions (Fig. 4) (Brummer et al. 1997; Orf et al. 1999; Specht et al. 2001; Bachlava et al. 2009; Liang et al. 2009; Wang et al. 2012; Pathan et al. 2013; Vaughn et al. 2014; Wang et al. 2014; Cao et al. 2017; Zhang et al. 2018; Lee et al. 2019; Yan et al. 2019). Bachlava et al. (2009) identified QTL (Satt225 ~ Sat_217) for linoleic acid with the physical position 40.26 ~ 41.68 Mb (Gmax2.0), which was very near to the qLA-A-1 (40.41 ~ 41.72 Mb) and qLA-A’ (39.88 ~ 41.01 Mb) in ZL-RIL and ZQ-RIL in our study (Tables 1 and 2, Fig. 4). Wang et al. (2012) detected two QTLs qSTE-A1-1 (BARC-041257–07,953 ~ BARC-050849–09,931) and qSTE-A1-2 (Sat_374 ~ BARC-029787–06,340) for stearic acid with the physical positions 40.64 ~ 41.27 Mb and 40.43 ~ 41.92 Mb (Gmax2.0), respectively, which was also very near to the qLA-A-1 and qLA-A’ in ZL-RIL and ZQ-RIL in our study (Tables 1 and 2, Fig. 4). Cao et al. (2017) had located the QTL for seed oil content in the region of 38.13 ~ 38.57 Mb (Gmax1.0, equal to 41.78 ~ 42.20 Mb in Gmax2.0) on chromosome 5, and 13 candidate genes (Glyma05g33510, Glyma05g33540, Glyma05g33620, Glyma05g33630, Glyma05g33770, Glyma05g33820, Glyma05g33850, Glyma05g33930, Glyma05g33990, Glyma05g34020, Glyma05g34030, Glyma05g34070, and Glyma05g34090) displayed relative higher expressions in soybean seed.
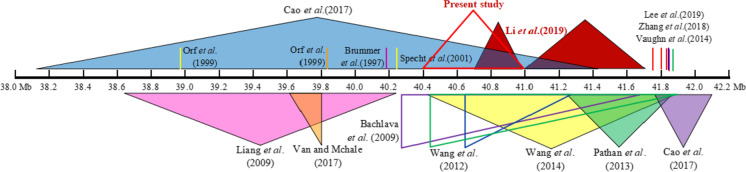
Fig. 4.
The consistent QTL mapping results for seed fatty acid and oil contents on chromosome 5 (triangle indicated the QTL intervals; vertical line indicated the associated SNPs; the common QTL interval for fatty acids of ZL-RIL and ZQ-RIL in the present study was highlighted with red hollow triangle; the QTL interval for oil content of ZL-RIL and ZQ-RIL in our previous study was highlighted with red solid triangle)
Meanwhile, Zhang et al. (2018) had identified the SNP marker ss715591638 on chromosome 5 (physical position 41,883,826, Gmax2.0) associating with oil content in soybean seed (Fig. 4), which was nearing (physical distance ~ 171 kb) to the fatty acid and oil contents linkage marker ss715591655 (physical position 41,712,419, Gmax2.0) in our studies (Table 1). Furthermore, Lee et al. (2019) identified five adjacent SNP markers on chromosome 5 (ss715591649, ss715591647, ss715591644, ss715591642, and ss715591641) associating with seed oil content with the physical position 41.78 ~ 41.86 Mb (Fig. 4), and the marker ss715591649 only had ~ 68 kb physical distance to the linkage marker ss715591655 in our study. More importantly, some candidate genes, including the lipase related gene Glyma.05g232700, were found in this QTL region with different expressions between the ZL-RIL and ZQ-RIL parents in our previous study (Li et al. 2019).
To our knowledge, this is the first time to report the consistent linkage markers on chromosome 5 for seed oil and various seed fatty acid contents across different genetic backgrounds under multiple environments. Thus, this stable pleiotropism QTL cluster responsible for soybean seed fatty acid and oil contents on chromosome 5 offered the reliable QTLs and candidate genes for seed fatty acid and oil contents genetic improvement in soybean molecular breeding program. And the KASP markers of ss715591780 and ss715591790 on chromosome 5 are being developed for applying in the soybean fatty acid content genetic improvement in our research group.
The pleiotropism QTL cluster and causal genes conferring soybean seed fatty acid contents on chromosome 9 is also worth further study
Apart from the stable QTL cluster on chromosome 5, there was another stable QTL cluster conferring soybean seed fatty acid contents identified on chromosome 9 in the present study. This pleiotropism QTL cluster was detected in ZL-RIL population and was verified in the natural population (Table 1 and Table S5). Li et al. (2011) had identified QTLs conferring linolenic acid, palmitic acid, and stearic acid contents with physical positions 37.8 ~ 42.5 Mb on chromosome 9, which was very far from the QTL cluster in our study. Xie et al. (2012) mapped QTL QPAK-1 conferring palmitic acid with physical position ~ 21.8 Mb and QTL QOLK-1 conferring oleic acid with physical positions ~ 11.26 Mb on chromosome 9, which were also far from the QTL cluster in our study. Wang et al. (2012) detected QTLs conferring palmitic acid and saturated fatty acid contents with physical positions 7.8 ~ 12.85 Mb and 42.17 ~ 46.06 Mb on chromosome 9; Fan et al. (2015) identified QTL qFA9-1 conferring linoleic acid and stearic acid contents with physical position 11.22 ~ 22.97 Mb on chromosome 9, which was also far from the QTL cluster in our study. Thus, we deduced that the QTL cluster on chromosome 9 identified in the present study might be a novel genetic locus controlling seed fatty acid contents in soybean.
And more importantly, several causal genes, Gm09G042000 encoding acyl-CoA dehydrogenase, Gm09G041200 encoding diacylglycerol lipase, Gm09G041400 encoding phospholipase, and Gm09G044500 encoding ABC transporter, involving in the metabolism process of fatty acid were found in this QTL cluster interval (Table S6, Fig. 3). And some of them, Gm09G042000, Gm09G041500, and Gm09G047200, displayed different expressions in ZL-RIL parents (Zheng92116 and Liaodou14) based on the transcriptome sequencing analysis (Fig. 3).
Among these causal genes, Glyma.09G041200 and Gm09G042000 were predicted to encode diacylglycerol lipase and acyl-CoA dehydrogenase, respectively. In the catabolism process of seed fatty acid, the seed oil (triacylglycerol) is firstly degraded by the lipase, diacylglycerol lipase (such as Glyma.09G041200 encoding enzyme), and generates fatty acid and glycerol (Li-Beisson et al. 2013). Afterwards, the fatty acid is degraded by the acyl-CoA dehydrogenase (such as Gm09G042000 encoding enzyme) to generate enoyl-CoA, and enoyl-CoA is successively degraded, and lastly generates the acetyl-CoA (also known as fatty acid β-oxidation pathway), then enters the tricarboxylic acid cycle (TCA) to produce the energy for organism. In other words, Glyma.09G041200 and Gm09G042000 on chromosome 9 were predicted to encode the key enzymes in the front steps for fatty acid catabolism in soybean seed. And more importantly, the Glyma.09G042000 displayed relative higher expressions in the RIL parent Zheng92116 (low oil and fatty acid) at the T4 and T5 stages (Fig. 3), which indicated that there might exist relative higher resolve metabolism of fatty acid in Zheng92116, and lastly, resulted in the relative lower oil and fatty acid contents in this variety.
In addition to the genes participated in the bio-synthesis and catabolism process of fatty acid, the genes involved in the lipid metabolism process could also influence the fatty acid contents in soybean seed. In the present study, Gm09G041400 was predicted to encode phospholipase in the common QTL cluster on chromosome 9, and phospholipase is an important enzyme to hydrolyze the phospholipids to generate fatty acid and other lipophilic substances (Li-Beisson et al. 2013). Meanwhile, Gm09G044500 was predicted to encode the ABC (ATP-binding cassette) transporter, which was the important pathway to transport the fatty acid from plastids to the endoplasmic reticulum (ER). The AtABCA9, an ABC transporter reported in Arabidopsis thaliana, could mediate the transport of fatty acid from the synthesized position (plastids) to the ER for further assimilation (Kim et al. 2013). To our knowledge, this is the first report to identify a series of candidate genes participating in the bio-synthesis and catabolism process of seed fatty acid contents on chromosome 9 via the combination results of linkage mapping, association analysis, and transcriptome sequencing in soybean. The results provided insights into the genetic basis of soybean seed fatty acids for genetic improvement.
Thus, the discovery and verification of these consistent, pleiotropism and stable QTL clusters controlling seed fatty acids in present study could be applied in the soybean molecular genetic improvement via the combination of multiple linkage/associated markers in selection of elite breeding lines with desirable fatty acid contents through the conventional hybridization of different germplasms to integrate various controlling QTLs in breeding program.
Conclusion
Overall, sixteen additive QTL clusters were identified in ZL-RIL population, and fifteen of them were detected across multiple environments or underlying multiple fatty acid contents. Five of the QTL clusters were verified in ZQ-RIL population, and five were demonstrated in natural population. One QTL cluster controlling fatty acid contents on chromosome 5 with pleiotropic effect was identified under all of the environments in ZL-RIL and ZQ-RIL populations and also in the natural population. Another stable QTL cluster was detected on chromosome 9 with pleiotropic effect under multiple environments in ZL-RIL population and was further verified by the natural population. Some causal genes, such as Gm09G042000 (encoding acyl-CoA dehydrogenase), Gm09G041200 (encoding diacylglycerol lipase), Gm09G041400 (encoding phospholipase), and Gm09G044500 (encoding ABC transporter) on chromosome 9 involving in the fatty acid catabolism process, were found in these QTL clusters, and some of them, such as Gm09G042000, Gm09G041500, and Gm09G047200, showed different expressions in ZL-RIL parents at different soybean seed developmental stages. Thus, these identified and verified consistent QTL clusters and causal genes could be applied in marker assisted selection breeding and gene cloning for fatty acid contents genetic improvement in soybean.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
Xihuan Li and Caiying Zhang conceived and designed the research. Xihuan Li and Rui Tian conducted the experiments and analyzed the data. Zhenqi Shao, Hua Zhang, Jiahao Chu, Wenlong Li, Youbin Kong, and Hui Du participated in parts of the experiments. Xihuan Li drafted the manuscript, and Caiying Zhang revised the manuscript.
Funding
This research was funded by the Hebei Province Key Research and Development Project (19226356D) and Hebei Province Modern Agricultural Industry Technology System Innovation Team Project (HBCT2019190203).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code availability
Not applicable.
Declaration
Ethics approval
The experiments were performed in compliance with the current laws of China.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Xihuan Li and Rui Tian contributed equally to this work.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akond M, Liu SM, Boney M, Kantartzi SK, Meksem K, Bellaloui N, Lightfoot DA, Kassem MA. Identification of quantitative trait loci (QTL) underlying protein, oil, and five major fatty acids’ contents in soybean. Am J Plant Sci. 2014;5:158–167. doi: 10.4236/ajps.2014.51021. [DOI] [Google Scholar]
- Amin N, Ahmad N, Wu N, Pu XM, Ma T, DuYY, Bo XX, Wang N, Sharif R, Wang PW (2019) CRISPR-Cas9 mediated targeted disruption of FAD2–2 microsomal omega-6 desaturase in soybean (Glycine max.L). BMC Biotechnol 19:9 [DOI] [PMC free article] [PubMed]
- Bachlava E, Dewey RE, Burton JW, Cardinal AJ. Mapping and comparison of quantitative trait loci for oleic acid seed content in two segregating soybean populations. Crop Sci. 2009;49:433–442. doi: 10.2135/cropsci2008.06.0324. [DOI] [Google Scholar]
- Brummer EC, Graef GL, Orf J, Wilcox JR, Shoemaker RC. Mapping QTL for seed protein and oil content in eight soybean populations. Crop Sci. 1997;37:370–378. doi: 10.2135/cropsci1997.0011183X003700020011x. [DOI] [Google Scholar]
- Cao YC, Li SG, Wang ZL, Chang FG, Kong JJ, Gai JY, Zhao TJ. Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front Plant Sci. 2017;8:1222. doi: 10.3389/fpls.2017.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan SX, Li B, Yu FK, Han FX, Yan SR, Wang LZ, Sun JM. Analysis of additive and epistatic quantitative trait loci underlying fatty acid concentrations in soybean seeds across multiple environments. Euphytica. 2015;206:689–700. doi: 10.1007/s10681-015-1491-3. [DOI] [Google Scholar]
- Fang C, Ma YM, Wu SW, Liu Z, Wang Z, Yang R, Hu GH, Zhou ZK, Yu H, Zhang M, Pan Y, Zhou GA, Ren HX, Du WG, Yan HR, Wang YP, Han DZ, Shen YT, Liu SL, Liu TF, Zhang JX, Qin H, Yuan J, Yuan XH, Kong FJ, Liu BH, Li JY, Zhang ZW, Wang GD, Zhu BG, Tian ZX. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017;18(1):161. doi: 10.1186/s13059-017-1289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goettel W, Xia E, Upchurch R, Wang ML, Chen P, An YQ. Identification and characterization of transcript polymorphisms in soybean lines varying in oil composition and content. BMC Genomics. 2014;15(1):299. doi: 10.1186/1471-2164-15-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha BK, Kim HJ, Velusamy V, Vuong TD, Nguyen HT, Shannon JG, Lee JD. Identification of quantitative trait loci controlling linolenic acid concentration in PI483463 (Glycine soja) Theor Appl Genet. 2014;127(7):1501–1512. doi: 10.1007/s00122-014-2314-y. [DOI] [PubMed] [Google Scholar]
- Hytena DL, Pantalonea VR, Saxtonb AM, Schmidtc ME, Samsa CE. Molecular mapping and identification of soybean fatty acid modifier quantitative trait loci. JAOCS. 2004;81(12):1115–1118. doi: 10.1007/s11746-004-1027-z. [DOI] [Google Scholar]
- Kachroo A, Fu DQ, Havens W, Navarre D, Kachroo P, Ghabrial SA. An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol Plant Microbe Interact. 2008;21(5):564–575. doi: 10.1094/MPMI-21-5-0564. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamaok Y, Ono H, Kim H, Shim D, Maeshima M, Martinoia E, Cahoon EB, Nishida I, Lee Y. AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc Natl Acad Sci USA. 2013;110:773–778. doi: 10.1073/pnas.1214159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy LJ, Zhang HY, Li CB, Chen CY, Song BH. A genome-wide association study of seed composition traits in wild soybean (Glycine soja) BMC Genomics. 2017;18(1):18. doi: 10.1186/s12864-016-3397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Van K, Sung M, Nelson R, LaMantia J, Rouf MLK, Mian MA. Genome-wide association study of seed protein, oil and amino acid contents in soybean from maturity groups I to IV. Theor Appl Genet. 2019;132:1639–1659. doi: 10.1007/s00122-019-03304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HW, Zhao TJ, Wang YF, Yu DY, Chen SY, Zhou RB, Gai JY. Genetic structure composed of additive QTL, epistatic QTL pairs and collective unmapped minor QTL conferring oil content and fatty acid components of soybeans. Euphytica. 2011;182:117–132. doi: 10.1007/s10681-011-0524-9. [DOI] [Google Scholar]
- Li XH, Kamala S, Tian R, Du H, Li WL, Kong YB, Zhang CY. Identification and validation of quantitative trait loci controlling seed isoflavone content across multiple environments and backgrounds in soybean. Mol Breeding. 2018;38(1):8. doi: 10.1007/s11032-017-0768-8. [DOI] [Google Scholar]
- Li XH, Shao ZQ, Tian R, Zhang H, Du H, Kong YB, Li WL, Zhang CY. Mining QTLs and candidate genes for seed protein and oil contents across multiple environments and backgrounds in soybean. Mol Breeding. 2019;39:139. doi: 10.1007/s11032-019-1055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HZ, Wang SF, Yu YL, Lian Y, Wang TF, Wei YL, Gong PT, Liu XY, Fang XJ. QTL mapping of isoflavone, oil and protein content in soybean. Sci Agric Sin. 2009;42(8):2652–2660. [Google Scholar]
- Li-Beisson YH, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, Franke RB, Graham IA, Katayama K, Kelly AA, Larson T, Markham JE, Miquel M, Molina I, Nishida I, Rowland O, Samuels L, Schmid KM, Wada H, Welti R, Xu CC, Zallot R, Ohlrogge J. Acyl-lipid metabolism. The Arabidopsis Book, the American Society of Plant Biologists. 2013 doi: 10.1199/tab.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GH, Singhal R, Kachroo A, Kachroo P. Fatty acid- and lipid-mediated signaling in plant defense. Annu Rev Phytopathol. 2017;55:505–536. doi: 10.1146/annurev-phyto-080516-035406. [DOI] [PubMed] [Google Scholar]
- Meng S, He JB, Zhao TJ, Xing GN, Li Y, Yang SP, Lu JJ, Wang YF, Gai JY. Detecting the QTL-allele system of seed isoflavone content in Chinese soybean landrace population for optimal cross design and gene system exploration. Theor Appl Genet. 2016;129(8):1557–1576. doi: 10.1007/s00122-016-2724-0. [DOI] [PubMed] [Google Scholar]
- Orf JH, Chase K, Jarvik T, Mansur LM, Cregan PB, Adler FR, Lark KG. Genetics of soybean agronomic traits: I. Comparison of three related recombinant inbred populations. Crop Sci. 1999;39:1642–1651. doi: 10.2135/cropsci1999.3961642x. [DOI] [Google Scholar]
- Panthee DR, Pantalone VR, Saxton AM. Modifier QTL for fatty acid composition in soybean oil. Euphytica. 2006;52:67–73. doi: 10.1007/s10681-006-9179-3. [DOI] [Google Scholar]
- Pathan SM, Vuong T, Clark K, Lee JD, Shannon GJ, Roberts CA, Ellersieck MR, Burton JW, Cregan PB, Hyten DL, Nguyen HT, Sleper DA. Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop Sci. 2013;53(3):765–774. doi: 10.2135/cropsci2012.03.0153. [DOI] [Google Scholar]
- Shen YT, Liu J, Geng HY, Zhang JX, Liu YC, Zhang HK, Xing SL, Du JC, Ma SS, Tian ZX. De novo assembly of a Chinese soybean genome. Science China. 2018;61(8):871–884. doi: 10.1007/s11427-018-9360-0. [DOI] [PubMed] [Google Scholar]
- Shibata M, Takayama K, Ujiie A, Yamada T, Abe J, Kitamura K. Genetic relationship between lipid content and linolenic acid concentration in soybean seeds. Breeding Sci. 2008;58(1):361–366. doi: 10.1270/jsbbs.58.361. [DOI] [Google Scholar]
- Specht JE, Chase K, Macrander M, Graef GL, Chung J, Markwell JP, Germann M, Orf JH, Lark KG (2001) Soybean response to water: a QTL analysis of drought tolerance. Crop Sci 41(2):493–509
- Spencer MM, Landau-Ellis D, Meyer EJ, Pantalone VR. Molecular markers associated with linolenic acid content in soybean. JAOCS. 2004;81(6):559–562. doi: 10.1007/s11746-006-0941-4. [DOI] [Google Scholar]
- Torkamaneh D, Laroche J, Valliyodan B, O’Donoughue L, Cober E, Rajcan I, Abdelnoor RV, Sreedasyam A, Schmutz J, Nguyen HT, Belzile F (2020) Soybean (Glycine max) haplotype map (GmHapMap): a universal resource for soybean translational and functional genomics. Plant Biotechnol J 1–11 [DOI] [PMC free article] [PubMed]
- Van K, Mchale LK. Meta-analyses of QTLs associated with protein and oil contents and compositions in soybean seed [Glycine max (L.) Merr.] Int J Mol Sci. 2017;18(6):1180. doi: 10.3390/ijms18061180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn JN, Nelson RL, Song QJ, Cregan PB, Li ZL (2014). The genetic architecture of seed composition in soybean is refined by genome-wide association scans across multiple populations G3 4(11):2283–2294 [DOI] [PMC free article] [PubMed]
- Wang XZ, Jiang GL, Green M, Scott RA, Hyten DL, Cregan PB. Quantitative trait locus analysis of saturated fatty acids in a population of recombinant inbred lines of soybean. Mol Breeding. 2012;30:1163–1179. doi: 10.1007/s11032-012-9704-0. [DOI] [Google Scholar]
- Wang XZ, Jiang GL, Green M, Scott RA, Song QJ, Hyten DL, Cregan PB. Identification and validation of quantitative trait loci for seed yield, oil and protein contents in two recombinant inbred line populations of soybean. Mol Gen Genomics. 2014;289(5):935–949. doi: 10.1007/s00438-014-0865-x. [DOI] [PubMed] [Google Scholar]
- Xie DW, Han YP, Zeng YH, Chang W, Teng WL, Li WB. SSR- and SNP-related QTL underlying linolenic acid and other fatty acid contents in soybean seeds across multiple environments. Mol Breeding. 2012;30:169–179. doi: 10.1007/s11032-011-9607-5. [DOI] [Google Scholar]
- Yan L, Di R, Wu CJ, Liu Q, Wei Y, Hou WH, Zhao QS, Liu BQ, Yang CY, Song QJ, Zhang MC. Haplotype analysis of a major and stable QTL underlying soybean (Glycine max) seed oil content reveals footprint of artificial selection. Mol Breeding. 2019;39:57. doi: 10.1007/s11032-019-0951-1. [DOI] [Google Scholar]
- Zhang JP, Wang XZ, Lu YM, Bhusal SJ, Song QJ, Cregan PB, Yen Y, Brown M, Jiang GL. Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding. Mol Plant. 2018;11(3):460–472. doi: 10.1016/j.molp.2017.12.016. [DOI] [PubMed] [Google Scholar]
- Zhang SS, Hao DR, Zhang SY, Zhang D, Wang H, Du HP, Kan GZ, Yu DY. Genome-wide association mapping for protein, oil and water-soluble protein contents in soybean. Mol Genet Genomics. 2020 doi: 10.1007/s00438-020-01704-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Not applicable.