Abstract
Genomic selection (GS) is one of the most powerful tools available for maize breeding. Its use of genome-wide marker data to estimate breeding values translates to increased genetic gains with fewer breeding cycles. In this review, we cover the history of GS and highlight particular milestones during its adaptation to maize breeding. We discuss how GS can be applied to developing superior maize inbreds and hybrids. Additionally, we characterize refinements in GS models that could enable the encapsulation of non-additive genetic effects, genotype by environment interactions, and multiple levels of the biological hierarchy, all of which could ultimately result in more accurate predictions of breeding values. Finally, we suggest the stages in a maize breeding program where it would be beneficial to apply GS. Given the current sophistication of high-throughput phenotypic, genotypic, and other -omic level data currently available to the maize community, now is the time to explore the implications of their incorporation into GS models and thus ensure that genetic gains are being achieved as quickly and efficiently as possible.
Keywords: Genomic selection, Maize, Hybrid prediction, Omics, Multi-kernel, GBLUP
Introduction
One of the most beneficial outcomes of plant breeding and crop management improvements is the continuous increase of maize (Zea mays L.) yields since the 1930s (Duvick 2005). Besides being a staple food in multiple cultures, maize is used as animal feed, food additive, biofuel, fiber, and raw materials (Ranum et al. 2014). Continued yield gains in maize are paramount for global food security and might only be possible with further advancements in plant breeding. The amelioration of genomic selection (GS), already a mainstay tool in modern crop breeding (Heffner et al. 2009), is one arena that is critical for achieving this goal. Maize is among the numerous crop species to have benefited from both GS and high-throughput phenotypic collection techniques (Beyene et al. 2015; Vivek et al. 2017; Zhao et al. 2012). Since the adoption of GS, techniques for collecting mass phenotypes and gene regulatory information have improved substantially (Mejia-Guerra et al. 2012; Tardieu et al. 2017). Adapting GS approaches to take advantage of new sources of information may prove integral for meeting increasing global demand for maize products.
Both direct and indirect selections are fundamental concepts of human and plant coevolution and are widely considered to be the foundation for plant domestication (Ross-Ibarra et al. 2007). Teosinte (Zea mays subsp. parviglumis), the hypothesized wild ancestor to maize, overcame several pre-domestication traits around 10,000 years ago to become maize (Doebley 2004). For example, ancient humans choosing to save seed late in the growing season for replanting rather than consumption is theorized to be responsible for non-shattering genotypes (Lin et al. 2012). This type of human selection is further supported by evidence of reduced allelic variation of several genes and major loci in maize compared to teosinte (Chen et al. 2020). Through this coevolution and a high degree of local adaptation (Buckler et al. 2009), maize has become one of the most important crop species economically. Moreover, maize has been a focus of plant breeding efforts partly due to its separate flowering structures that facilitate easy control of cross- and self-pollinations (Brown et al. 2011). Self-pollination produces inbred lines that can be considered genotypic replicates, convenient for collecting replicated data. Simple cross-pollination techniques allow for the production of hybrids, which is the predominant method for growing maize commercially due to its high degree of heterosis (Hallauer et al. 1988; Hayes 1912).
The rediscovery of Mendel’s laws in the early 1900s and the subsequent quantitative genetic ramifications elucidated by R.A. Fisher, Sewall Wright, and J.B.S. Haldane have helped make quantitative techniques common practice in modern plant and animal breeding (Hallauer 2007). For example, Mendel’s laws of inheritance can quantify identity by descent between individuals and therefore define relatedness for pedigree selection (Genetic Alliance 2010). Pedigrees are largely responsible for the development of heterotic groups in US germplasm, capturing high levels of heterosis (Gerdes and Tracy 1993) and the estimation of general and specific combining ability (Box 1; Henderson 1952). For a comprehensive overview on the history of selection methodologies in commercial breeding, see Duvick et al. (2004).
Box 1.
General and specific combining ability
|
Arguably, the most common approach for subdividing the genetic contributions to trait variation is through additive (i.e., the contribution of an allele at a locus), dominance (interaction between alleles at the same locus), and epistatic (interaction between alleles at different loci) effects. An alternate method for quantifying these subdivisions is through general combining ability (GCA) and specific combining ability (SCA). To understand GCA and SCA, consider a quantitative trait measured in a maize F1 hybrid, where the mother and father are from separate heterotic pools. The genetic contribution of each parent of the F1 hybrid is the GCA. Thus, the F1 hybrid will have two GCAs: one from the mother and one from the father. One common approach for accounting for GCAs in a GS model is to include the mother and father as random effects in the model and then to use an additive genetic relatedness matrix from the respective heterotic pools to model the variance-covariance. The SCA is then the remaining genetic signal of the trait that is not explained by the GCAs of the two parents and can be accounted for in a GS model by including another random effect in the model, where a dominance relationship matrix is used to model the variance-covariance. Without loss of generality, the GCA can be thought of as analogous to additive contributions, while the SCA can be thought of as analogous to dominance contributions. Please see Kadam et al. (2016), Technow et al. (2014), and Sprague and Tatum (1942) for examples of the use of GCA and SCA in maize hybrid breeding. |
Maize breeders have relied on the same general breeding strategies for the past century, crossing lines, and evaluating offspring for increased yield while inadvertently moving a population towards an ideotype (Donald 1968). However, modern maize breeding now has the advantage of molecular marker technology (Bernardo 2008) to predict the outcome of these crosses and perform marker-assisted selection (MAS) (Xu and Crouch 2008). Based on the principals of Mendelian genetics, one can assume that at least some markers from a high-density genotyping platform are in linkage disequilibrium (LD) with ungenotyped causal mutation(s) and therefore explain a statistically significant proportion of trait variation. Through the use of MAS on such markers, it is possible for breeders to advance and cross elite germplasm while ensuring additional favorable characteristics are maintained, such as disease resistance (Poland and Rutkoski 2016).
The technique of MAS has even been successful at the introgression of yield-related quantitative trait loci (QTL; Bouchez et al. 2002), yet this approach has several noteworthy drawbacks. Breeding over the past several decades has reduced allelic variation and fixed large additive effect QTL, making it difficult for markers tagging the remaining QTLs to pass stringent statistical thresholds of significance (Reif et al. 2005). Other issues with MAS include possible weak LD between markers and causal mutations, resulting in a loss of predictive ability in later generations (Zhang and Smith 1992), and the lower relative efficiency of MAS over phenotypic selection for low heritable traits (Hospital et al. 1997). Fisher 1958a, 1958b proposed a theory that quantitative traits are the results of many small-effect causal mutations distributed across the genome. Many agronomically important traits appear to adhere to this complex genetic architecture (Buckler et al. 2009; Peiffer et al. 2014; Prado et al. 2014). Thus, GS models (described in, e.g., Meuwissen et al. 2001) that use dense genome-wide markers to predict genomic estimated breeding values (GEBVs) of such traits have been widely explored in maize (Crossa et al. 2013). Similar in practice to the use of pedigree data, GS can be used to obtain genomic best linear unbiased predictions (GBLUP) of progeny for either inbred development or hybrid production in maize (Bernardo 1994). The accurate prediction of GEBVs translates to a reduction in the number of potential crosses and environmental replicates needed to observe genetic gains (Heffner et al. 2010). Therefore, GS models will continue to be invaluable tools in breeding programs to ensure that the societal demands of maize are met. In this review, we (i) give an overview on the use GS in maize and how models utilize non-genetic information sources, (ii) argue for customizing GS models to reflect a given trait’s genetic architecture and to include -omic information sources, and finally (iii) discuss the factors of when and how GS is applied in maize hybrid breeding.
Overview of genomic selection
The use of a combination of pedigree data and 220 restriction enzyme markers to construct a variance-covariance relationship matrix was demonstrated in Bernardo (1994). The implementation of a marker-based variance-covariance relationship matrix (Habier et al. 2007) to make predictions was popularized under the term GBLUP, with the approach from VanRaden (2008) being the most widely used approach for estimating a genetic relationship matrix. Advances in genome sequencing and decreasing costs per sample have resulted in more individuals that can be genotyped with denser marker sets, including millions of single nucleotide polymorphisms (SNPs) (Bukowski et al. 2018; Heather and Chain 2016). Using such dense marker maps, Meuwissen et al. (2001) expanded GS and showed that selection based on marker haplotype effects could increase genetic gains in plant and animal breeding. Genotyping-by-sequencing technology and SNP arrays make it possible to efficiently genotype thousands of lines for several hundred thousand SNPs, ultimately facilitating the implementation of GS in large breeding populations (Poland and Rife 2012; Rasheed et al. 2017). In general, the availability of dense marker data translates to highly accurate GEBVs, which for some maize traits have been shown to produce annual gains that are three times greater than MAS (Heffner et al. 2010).
Basic construction of a GS model
The basic concept behind GS is to use phenotype and genotype information in a training set to obtain GEBVs for non-phenotyped individuals in an independent set of breeding material. For an overview on how GS guides decisions in breeding programs, please see Jannink (2010). Here we present a typical configuration of a GS model that has the trait of interest as the response variable and all genome-wide markers as the explanatory variables. When the number of markers (p) is greater than the number of individuals (n), there are not enough degrees of freedom to obtain unique estimates of all marker effects (Gianola 2013). To overcome this large p, small n (i.e., p>>n) issue, several Bayesian and penalized approaches have been implemented in practice. De los Campos et al. (2013) provides a comprehensive overview of these approaches. An alternate configuration of a GS model is to include the individuals as random effects, with their variance-covariance being set as proportional to an additive genetic relationship matrix calculated from the genome-wide markers (VanRaden 2008). Commonly referred to as GBLUP, this model is similar to a pedigree-based best linear unbiased prediction (BLUP) model (Daetwyler et al. 2010). It is widely established (e.g., in Habier et al. 2007) that the configuration of including p markers in the GS model as explanatory variables and the GBLUP configuration are equivalent, but exact equivalence between these two configurations will respectively depend on the approaches used to account for the p>>n issue and to calculate the genetic relatedness matrix.
Since the introduction of GS, several studies have demonstrated its broad utility in maize by using different models to accommodate a wide variety of various breeding aspects (Bandeira e Sousa et al. 2017; J. Crossa et al. 2014; J. Crossa et al. 2013; Pinho Morais et al. 2020). Nonetheless, GS studies typically fail to obtain high prediction accuracies whenever training and validation sets are distantly related (Pinho Morais et al. 2020) and when non-genetic factors are a large source of trait variation (Hickey et al. 2017). Examination across several biparental maize populations indicated a relationship between genetic variance, i.e., heritability and prediction accuracy (Zhang et al. 2017).
The selection of an ideal training set for fitting a GS model is essential for accurate GEBVs in the breeding material (Olatoye et al. 2020; Schopp et al. 2017; Windhausen et al. 2012). Best practices on how training data are developed and used are still being discussed in the current literature (Dias et al. 2020). There are many factors to consider in optimizing training sets, such as population structure between lines (Isidro et al. 2015) and mating design. Findings from Fristche-Neto et al. (2018) suggest that optimal prediction accuracies can be achieved by maximizing the number of crosses per parent in the training set. They also challenged the use of a testcross mating design, demonstrating that practical considerations such as the development of a training set have strong influence on prediction accuracy. Expanding on this, it is highly recommended that a set of standard checks are incorporated into each field and growing season; these checks are likely to reduce the influence of the unbalanced nature of breeding programs on the training set (Gorjanc et al. 2018; Jarquin et al. 2020; Piepho et al. 2006). These situations underscore that GS and its implementation require continued testing and modification to continue leading the charge for increased genetic gain in crop breeding programs.
Expanding upon the basic GS model and factors influencing prediction accuracy
The need to account for non-genetic sources of trait variation and non-additive modes of gene action in complex traits makes the choice of a best-suited GS model less straightforward. For instance, the task of incorporating environmental fluctuations has resulted in the investigation of GS models that directly incorporate environmental information into the model (Burgueño et al. 2012; Jarquín et al. 2014; Lopez-Cruz et al. 2015). The typical form of such GS models is to include an environmental main effect, as well as a two-way interaction term between the genetic and environmental (GxE) effects (please see, e.g., Heslot et al. 2014, for an in-depth description of these models). When weather and other environmental data are available, it is common to use this information to model both the covariance between environments and the GxE effects (Jarquín et al. 2014). Inclusion of such environmental information into GS models in this manner has been shown to accurately predict GEBVs in diverse sets of environments (Basnet et al. 2019; Jarquín et al. 2014; Saint Pierre et al. 2016).
One could also expand the basic GS model to include dominance and epistatic effects, both of which are particularly important for hybrid breeding (Jiang and Reif 2015; Technow et al. 2012; Vitezica et al. 2013). Prediction using additive variation is essential for obtaining breeding values (Liu et al. 2018), while predicting dominance and epistatic effects could have potentially important commercial benefits in the form of heterosis and hybrid vigor (Chen 2010; McMullen et al. 2009; Sprague 1983). Moreover, it has been shown that modeling multiple types of genetic and non-genetic sources of variation is important because both heritability estimates and prediction accuracy could be maximized (Brachi et al. 2011; Voss-Fels et al. 2019). Please see Rogers et al. (2021) for a maize GS study that included GxE effects and non-additive genetic effects in the GS model.
“Diverse” genomic selection models
Fitting models to match trait genetic architecture
In general, GS has potential to increase genetic gain under reduced resources, but there is nevertheless no evidence for a single best GS model for all scenarios. A factor that influences the prediction accuracy of GS models to various extents is the choice of penalty, priors, and/or which markers to include as fixed-effect covariates (Heslot et al. 2012b; Zhang et al. 2019). Depending on the genetic architecture of the studied trait, some penalties or priors are theoretically better suited than others. For example, random regression-BLUP (RR-BLUP) (Meuwissen et al. 2001; Whittaker et al. 2000) should be ideal for predicting breeding values of complex traits because each of the marker effects are drawn from the same normal distribution, and the penalty is equally applied to their predictions. Alternatively, if the genetic architecture of the studied trait is oligogenic, then other penalties such as the LASSO (Tibshirani 1996) that perform model selection or Bayesian approaches (reviewed in Gianola 2013) that assign mixture prior densities to marker effects should hypothetically yield the highest prediction accuracies. However, in general these various types of penalties and priors tend to yield approximately the same prediction accuracies (De los Campos et al. 2013; Heslot et al. 2012a).
Several studies have demonstrated the utility of adjusting penalties and/or kernels (see Box 2; Cuevas et al. 2016, 2017) in the GS model so that the impact of markers in the vicinity of a priori candidate genes differs from those of the remaining genomic markers (Edwards et al. 2015; Fang et al. 2017; MacLeod et al. 2016; Speed and Balding 2014; Turner-Hissong et al. 2020). For example, Turner-Hissong et al. (2020) used such an approach to explore the influence of genes from various free amino acid (FAA) -related pathways on genomic prediction of FAA-derived traits in Arabidopsis thaliana seeds. A GS model that created a separate kernel for markers in the vicinity of these candidate genes yielded higher prediction accuracies than a standard GBLUP model. In addition to suggesting that such a multi-kernel BLUP model is useful for genomic prediction, this study also demonstrated how GS models that incorporate a priori information into their kernels could be useful for making inferences on the genetic architecture of the studied trait.
Box 2.
Kernels
|
One of the most commonly used but potentially most misunderstood terms in GS is the kernel. To introduce this term in context, consider a GBLUP-style model that includes at least one random effect to account for the individuals. These random effects are assumed to follow a multivariate normal distribution with population mean vector 0 and population variance-covariance matrix Σ. One common practice is to set Σ to be proportional to an additive genetic relatedness matrix, for example, the matrix described in VanRaden (2008). Colloquially, the methodology used to calculate each element of this relatedness matrix is called the kernel. Different kernels, or methodologies used to estimate elements of a relatedness matrix, are used in practice. For instance, suppose one was interested in accounting for additive, dominance, and additive × additive epistasis in a GS model (the so-called ADE model from Covarrubias-Pazaran (2016)). Then one could fit a GBLUP-style model, where there are three individual random effects. To account for additive and dominance genetic effects, relatedness matrices could be calculated respectively using additive and dominance kernels (e.g., described in Su et al. (2012)), and the resulting relatedness matrices could be used to model the variance-covariance of two of the random effects. To account for additive × additive epistasis, a similar relatedness matrix could be created where the kernel used is the Hadamard product (i.e., element-wise multiplication) between each element of two additive relatedness matrices. Other kernels, for example, the reproducing kernel Hilbert space (described in Neves et al. (2012) and Pérez, and de los Campos, G. (2014)) or the support vector machine regression (described in Howard et al. (2014)), are also commonly used to account for non-additive genetic effects in GS models. Approaches for calculating kernels are critical for future GS research because they can be used to account for additional sources of trait variability in a model. For example, suppose one wanted to account for small RNA (sRNA) in a GS model (as done in Schrag et al. (2018)). In this instance, they could include an additional random effect for the individuals in the model and then incorporate sRNA information into kernels that ultimately create a relatedness matrix. In summary, the increasing amount of available data has a potential to increase the effectiveness of genomic prediction, and these data can be easily incorporated into GS models through kernels. |
The last modification to the GS model attempts to account for traits that are controlled by both large- and small-effect loci. In this approach, markers tagging large-effect genes are included as fixed-effect covariates in a standard GS model. Because the fixed-effect estimates of such markers are not subject to the penalties applied to the prediction of the remaining random marker effects, a GS model augmented with these fixed-effect covariates could potentially increase prediction accuracy whenever large-effect genes account for at least 10% of the total genetic variability (Bernardo 2014). Nonetheless, studies using GS models that included fixed-effect covariates tagging quantitative trait nucleotides (QTNs) have not consistently reported increased prediction accuracy over the standard GS models without any covariates (Arruda et al. 2016; Rice and Lipka 2019; Spindel et al. 2016; Zhang et al. 2014). In particular, the simulation study conducted by Rice and Lipka (2019) demonstrated that the use of fixed-effect covariates tagging large-effect genes in a GS model could potentially be more likely to decrease instead of increase prediction accuracy and that a preliminary cross-validation study into the usefulness of such an augmented GS model is warranted prior to its implementation into a breeding program.
If GS models are to be diversified, genome-wide association, QTL mapping, and other exploratory genomic studies that elicit the underlying genetic architecture of traits and identify associated markers and genes (Gage et al. 2020; Glowinski and Flint-Garcia 2018; Lipka et al. 2015) will be essential. In theory, if all marker or haplotype effects were accurate in size and gene action relative to a given environment, one could predict a traits’ breeding value with maximum accuracy, minimum bias, and avoiding any missing heritability (Box 3).
Box 3.
Missing heritability
|
In general, GWAS has not been as successful as originally anticipated. Issues pertaining to multiple testing corrections on a genome-wide scale, accounting for spurious associations due to population structure and familial relatedness, and simplicity of the statistical model being used have resulted in GWAS being able to only declare moderate- to large-effect markers to have statistically significant associations with the tested trait. The percentage of trait variability explained (PVE) by these peak-associated markers is typically lower than the estimated heritability of the trait. This discrepancy in the amount of PVE of GWAS markers and the estimated heritability is known as the missing heritability. Described and debated over since at least 2009 (e.g., in Myles et al. (2009)), missing heritability is thought to arise from multiple factors, including inadequate sample sizes to overcome conservative multiple testing corrections, inadequate marker densities, and over-simplistic statistical models. The former two issues are being addressed by decreasing genotyping and phenotyping costs, which makes it possible to obtain denser marker sets for more individuals. However, the commonplace usage of statistical models such as the unified mixed linear model (MLM; Yu et al. 2006) that only test for the additive effect of one marker at a time translates to an inability to directly quantify dominance, epistatic, and genotype × environment effects. Furthermore, the univariate implementation of the unified MLM to test for one trait at a time makes it impossible to quantify pleiotropic contributions. Thus, one important approach for potentially reducing the magnitude of the missing heritability is to focus on the development and implementation of statistical models that account for a wider variety of non-additive contributions to quantitative trait variation. |
Expanding -omics-based prediction
Modern breeding programs are now able to take advantage of emerging data from high-throughput phenotyping technologies. Breeding can now focus on several physiological traits over multiple time points in a given field season and incorporate them into combined genomic and crop growth models (Araus et al. 2018; Washburn et al. 2020; Zhao et al. 2019). Multivariate derivatives of available models (Jia and Jannink 2012; Velazco et al. 2019) can consider multiple “proxy” traits and take advantage of these large-scale phenotyping platforms (Joo et al. 2016). The “diverse” approaches suggested here are currently available to any breeding program with phenotyping and genotyping capabilities. They only require a priori knowledge about the genetic architecture of the trait(s) and a wilingess to creatively include this knowledge in prediction.
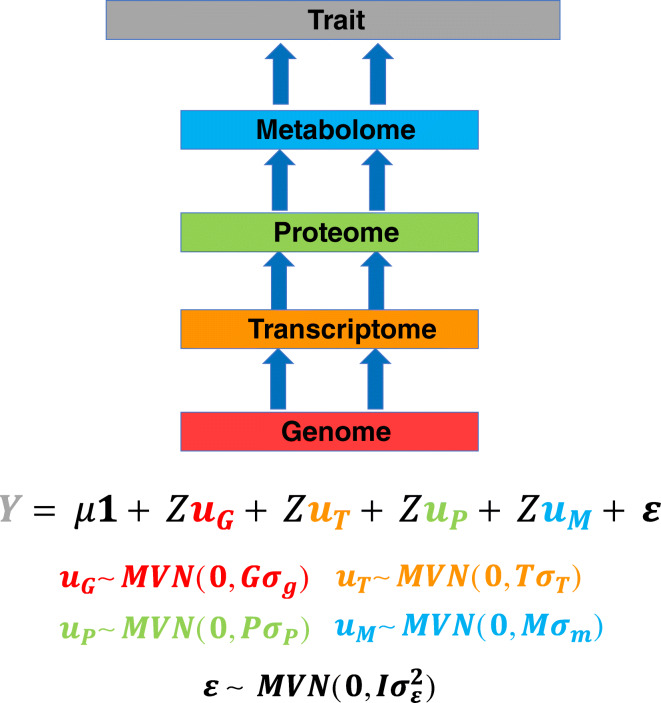
Information from DNA variants alone cannot describe the full contribution of the biological hierarchy (e.g., transcriptomic and proteomic levels) to trait variability. The advent of high-throughput molecular technologies (Fernie and Schauer 2009; Wang et al. 2009; Widłak 2013) for collecting multiple types of genomic regulatory information, referred to as -omics, facilitate the quantification of more components of gene regulation and therefore trait variability. For example, gene expression levels and protein abundance capture heritable variation (Guo et al. 2016; Li et al. 2019) and could be included in GS models as covariates, substitutes for genome-wide markers (as shown in Schrag et al. (2018)), or both. Such -omics data are routinely used in crop improvement studies to identify markers associated with metabolomic (Wen et al. 2014) and gene expression (Kremling et al. 2019; Nica and Dermitzakis 2013). We argue that now is the time to build on the findings of these previous studies and start incorporating -omics information into GS models in a more widespread manner (see Fig. 1 for a generalized schematic of how multiple layers of data can be used to predict a response). We present several reasons for this argument below.
Fig. 1.
Biological hierarchy in genomic selection. Schematic of how various levels of the biological hierarchy of traits could be incorporated into GS models. The availability of the genomic (red), transcriptomic (orange), proteomic (green), and metabolomic (blue) data in maize makes it possible to incorporate multiple levels of the biological hierarchy of an agronomic trait directly into genomic selection (GS) models. Each of the different levels of the biological hierarchy can be used to calculate the correspondingly colored relationship matrices G, T, P, and M. Model terms and abbreviations: Y, observed vector trait values for n individuals; μ, grand mean; 1, n-dimensional vector of 1’s; u, n-dimensional random vector of polygenic effects; Z, incidence matrix relating u to Y; ε, n-dimensional random vector for error terms; MVN, multivariate normal
First, epistatic effect estimates quantified from genomic data are typically negligible in magnitude relative to additive effects (Hill et al. 2008; Sackton and Hartl 2016). Nevertheless, evidence of physiological epistasis has appeared in many previous studies (Carlborg et al. 2004; Doebley et al. 1995; Huang et al. 2012). Because it is argued that -omics data better encapsulates physiological epistasis (Sackton and Hartl 2016), the inclusion of -omics data in GS models could improve their predictive abilities whenever physiological epistasis is an important source of trait variability. The second argument addresses epigenetics modifications that regulate transcription levels through altering accessibility to a given genomic region and explain a large portion of phenotypic variation in plants (Hu et al. 2015; Johannes et al. 2009). Although epigenetic states are reversible (i.e., they can revert states during or between generations; Becker et al. 2011), it has been previously argued that the use of epigenomic data for trait prediction could be useful for human (Vazquez et al. 2014) and animal (González-Recio 2012) studies. Finally, DNA variants are constant, but -omic levels are subject to biotic and abiotic stimuli (Atkinson and Urwin 2012; Cramer et al. 2011). A recent study explored the link between local adaption and variation in gene expression in maize (Blanc et al. 2021). Thus, use of the plethora of data available from the -omics landscape as a prediction tool could help better understand and compare different genotypes and their environmental contingent trait expression (Li et al. 2018; Proulx et al. 2007; Sadras and Lawson 2011; Vu et al. 2015; Zhou et al. 2012). Certainly, with significant phenotypic variation explained by non-genomic sources and their interaction with the environment, our arguments and suggested uses for -omics-based prediction in maize hybrid breeding are far from exhaustive.
The incorporation of -omics data into GS models has already been shown to be adequate for predicting GEBVs in maize. For example, data from 130 metabolites had equivalent accuracy to SNP data for predicting GCA for seven biomass and bioenergy-related traits (Riedelsheimer et al. 2012). Guo et al. (2016) used a diverse population and an extension to the GBLUP model that incorporated transcriptomic and metabolomic data. The authors showed that the incorporation of transcriptomic and metabolomic effects into the GS models explained more heritable variation for nine of eleven traits compared to any single predictor. They also observed an increase in predictive ability over the standard GBLUP model for several traits when using either transcriptomic data alone or in conjunction with genomic data. Westhues et al. (2017) showed that transcriptomic and metabolomic data could differentiate heterotic groups and improve GCA and SCA estimates for yield and grain quality traits, potentially increasing the likelihood of making optimal hybrid combinations. Expanding on this result, Schrag et al. (2018) demonstrated improved prediction for hybrids with non-phenotyped parents (referred to as T0 hybrids) using only transcriptomic information. The results from these three studies suggest that -omics-based prediction is a viable approach for maize population development and hybrid prediction.
There are challenges to implementing multi-omics in genomic prediction as it requires collecting information on a large number of individuals. Additionally, costs associated with high-throughput collection, data storage, and training technical ability to process data (Argueso et al. 2019; Conesa and Beck 2019) are needed to be taken into consideration. Due to such limitations, implementation of multi-omics data directly into genomic prediction may have more immediate application for high-value breeding targets.
When and how genomic selection is applied matters
The specific stage(s) of a breeding pipeline when GS is applied, as well as the intended outcome of its application (i.e., how), are as important as choosing the best suited model (Hickey et al. 2017). Because of doubled haploid technology in particular, the generation of highly inbred material is no longer a prominent time prohibiting factor in breeding (Chaikam et al. 2019). Instead, the development and evaluation of crosses that advance genetic gain for a target trait becomes the formidable bottleneck (Bernardo 2009). Breeders can overcome this by utilizing GS at multiple points in their program. For example, in the absence of disease pressure, it would be impossible to select for disease resistance based on phenotypic values. Instead, selecting inbred lines during population development based on GEBVs would ensure favorable disease resistance, while reducing the labor required to inoculate and phenotype over multiple growing seasons. Another example of the use of GS early in a breeding pipeline is for the identification of lines with high GCA and SCA, as demonstrated by Kadam et al. (2016). Here, the authors achieved high prediction accuracy for agronomically important traits such as staygreen, height, and grain yield of single crosses. Another important use of GS early in a breeding program would be to predict GEBVs for combining elite and diverse germplasm when introgressing exotic haplotype segments (Bernardo 2009, 2016; Gorjanc et al. 2016; Ru and Bernardo 2020). The germplasm enhancement of maize (GEM) project is an important example of the potential impact of incorporating diverse germplasm with elite breeding lines (Pollak 2003), and applying GS could intensify such a program.
Conclusion: When does genomic selection work?
Criteria for the ideal GS model are far from being rigorously tested for all environments, breeding schemes, and traits; thus no “one size fits all” GS model exists. Instead of focusing on the best penalty or prior to applying to a GS model that accounts for only the additive effects of genomic markers, research into model development should focus on making it more practical to include additional sources of variation such as GxE and -omics data (Burgueño et al. 2012; Schrag et al. 2018). Moreover, additional challenges not discussed in this review exist for programs applying GS. For example, as new phenotypic information becomes available and allele frequencies shift across generations, models require updating (Jannink 2010). Even the choice of molecular markers can have an impact on accuracy (Chang et al. 2018; Habier et al. 2009; Kriaridou et al. 2020).
To end this review, we readdress the question from Lian et al. (2014) that seeks to clarify under which circumstances GS outperforms conventional phenotypic selection. In their study, 969 biparental families were predicted for seven traits, including yield, using the RR-BLUP model (Meuwissen et al. 2001; Whittaker et al. 2000). When they applied the same model to the same traits for different families, they obtained vastly different prediction accuracies. This suggests that further research is needed on topics such as the influence of training set composition, as well as other factors, on the performance and effectiveness of GS. Although it is a potentially daunting task to systematically study the contributions of these factors in a series of field trails potentially across multiple years, simulation software is making it possible to initiate such an undertaking (Bernardo 2017; Faux et al. 2016; Fernandes and Lipka 2020). Therefore, we argue that the challenge of which model to apply to which breeding scheme may never be fully addressed by human users. Perhaps with machine learning, where certain model assumptions are not necessary, we can explore more fully the space of GS model optimization (for machine learning reviews, see González-Camacho et al. (2018), Ogutu et al. (2011), and Washburn et al. (2020)). In reality, GS as a tool will continue to be flexibly applied according to the breeder and likely not replace phenotyping and the breeder’s expertise; instead GS should work in tandem with these other factors to accelerate genetic gain.
Acknowledgements
Special thanks to Dr. Samuel Fernandes and Sarah Widener for their helpful comments on the manuscript.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Funding
This project is funded by the National Science Foundation Plant Genome Research Project, award number 1733606.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
This article is part of the Topical Collection on Maize Genetics, Genomics and Sustainable Improvement
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alliance G (2010) Genetics 101. Understanding genetics: a district of Colombia guide for patients and health professionals:22–32 https://www.resourcerepository.org/documents/1869/understandinggenetics:adistrictofcolumbiaguideforpatientsandhealthprofessionals/%0A, https://www.ncbi.nlm.nih.gov/books/NBK132149/pdf/Bookshelf_NBK132149.pdf
- Araus JL, Kefauver SC, Zaman-Allah M, Olsen MS, Cairns JE. Translating high-throughput phenotyping into genetic gain. Trends in Plant Science. 2018;23(5):451–466. doi: 10.1016/j.tplants.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso CT, Assmann SM, Birnbaum KD, Chen S, Dinneny JR, Doherty CJ, Eveland AL, Friesner J, Greenlee VR, Law JA, Marshall-Colón A, Mason GA, O’Lexy R, Peck SC, Schmitz RJ, Song L, Stern D, Varagona MJ, Walley JW, Williams CM. Directions for research and training in plant omics: big questions and big data. Plant Direct. 2019;3(4):e00133. doi: 10.1002/pld3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda MP, Lipka AE, Brown PJ, Krill AM, Thurber C, Brown-Guedira G, Dong Y, Foresman BJ, Kolb FL. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.) Molecular Breeding. 2016;36(7):1–11. doi: 10.1007/s11032-016-0508-5. [DOI] [Google Scholar]
- Atkinson NJ, Urwin PE. The interaction of plant biotic and abiotic stresses: from genes to the field. Journal of Experimental Botany. 2012;63(10):3523–3543. doi: 10.1093/jxb/ers100. [DOI] [PubMed] [Google Scholar]
- Bandeira e Sousa M, Cuevas J, de Oliveira Couto EG, Pérez-Rodríguez P, Jarquín D, Fritsche-Neto R, Burgueño J, Crossa J. Genomic-enabled prediction in maize using kernel models with genotype × environment interaction. G3. 2017;7(6):1995–2014. doi: 10.1534/g3.117.042341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnet BR, Crossa J, Dreisigacker S, Pérez-Rodríguez P, Manes Y, Singh RP, Rosyara UR, Camarillo-Castillo F, Murua M. Hybrid wheat prediction using genomic, pedigree, and environmental covariables interaction models. The Plant Genome. 2019;12(1):180051. doi: 10.3835/plantgenome2018.07.0051. [DOI] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480(7376):245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- Bernardo R. Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Science. 1994;34(1):20–25. doi: 10.2135/cropsci1994.0011183X003400010003x. [DOI] [Google Scholar]
- Bernardo R. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Science. 2008;48(5):1649–1664. doi: 10.2135/cropsci2008.03.0131. [DOI] [Google Scholar]
- Bernardo R. Genomewide selection for rapid introgression of exotic germplasm in maize. Crop Science. 2009;49(2):419–425. doi: 10.2135/cropsci2008.08.0452. [DOI] [Google Scholar]
- Bernardo R. Genomewide predictions for backcrossing a quantitative trait from an exotic to an adapted line. Crop Science. 2016;56(3):1067–1075. doi: 10.2135/cropsci2015.09.0586. [DOI] [Google Scholar]
- Bernardo R. BreedingGames Software. Crop Science. 2017;57(5):2313–2313. doi: 10.2135/cropsci2017.07.0419le. [DOI] [Google Scholar]
- Beyene Y, Semagn K, Mugo S, Tarekegne A, Babu R, Meisel B, Sehabiague P, Makumbi D, Magorokosho C, Oikeh S, Gakunga J, Vargas M, Olsen M, Prasanna BM, Banziger M, Crossa J. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Science. 2015;55(1):154–163. doi: 10.2135/cropsci2014.07.0460. [DOI] [Google Scholar]
- Blanc J, Kremling KAG, Buckler E, Josephs EB (2021) Local adaptation contributes to gene expression divergence in maize. G3 Genes|Genomes|Genetics:2021. 10.1093/g3journal/jkab004 [DOI] [PMC free article] [PubMed]
- Bouchez A, Hospital F, Causse M, Gallais A, Charcosset A. Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics. 2002;162(4):1945–1959. doi: 10.1093/genetics/162.4.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Morris GP, Borevitz JO. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biology. 2011;12(10):232. doi: 10.1186/gb-2011-12-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, Holland JB, Flint-Garcia S, McMullen MD, Buckler ES, Rocheford TR. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genetics. 2011;7(11):e1002383. doi: 10.1371/journal.pgen.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM, Harjes C, Guill K, Kroon DE, Larsson S. The genetic architecture of maize flowering time. Science. 2009;325(2009):714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- Bukowski R, Guo X, Lu Y, Zou C, He B, Rong Z, Wang B, Xu D, Yang B, Xie C, Fan L, Gao S, Xu X, Zhang G, Li Y, Jiao Y, Doebley JF, Ross-Ibarra J, Lorant A, Buffalo V, Romay MC, Buckler ES, Ware D, Lai J, Sun Q, Xu Y. Construction of the third-generation Zea mays haplotype map. GigaScience. 2018;7(4):1–12. doi: 10.1093/gigascience/gix134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgueño J, De los Campos G, Weigel K, Crossa J. Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Science. 2012;52(2):707–719. doi: 10.2135/cropsci2011.06.0299. [DOI] [Google Scholar]
- Carlborg R, HOCKING PM, BURT DW, HALEY CS. Simultaneous mapping of epistatic QTL in chickens reveals clusters of QTL pairs with similar genetic effects on growth. Genetical Research. 2004;83(3):197–209. doi: 10.1017/S0016672304006779. [DOI] [PubMed] [Google Scholar]
- Chaikam V, Molenaar W, Melchinger AE, Boddupalli PM. Doubled haploid technology for line development in maize: technical advances and prospects. Theoretical and Applied Genetics. 2019;132(12):3227–3243. doi: 10.1007/s00122-019-03433-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L-Y, Toghiani S, Ling A, Aggrey SE, Rekaya R. High density marker panels, SNPs prioritizing and accuracy of genomic selection. BMC Genetics. 2018;19(1):4. doi: 10.1186/s12863-017-0595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends in Plant Science. 2010;15(2):57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Samayoa LF, Yang CJ, Bradbury PJ, Olukolu BA, Neumeyer MA, Romay MC, Sun Q, Lorant A, Buckler ES, Ross-Ibarra J, Holland JB, Doebley JF. The genetic architecture of the maize progenitor, teosinte, and how it was altered during maize domestication. PLOS Genetics. 2020;16(5):1–21. doi: 10.1371/journal.pgen.1008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Beck S. Making multi-omics data accessible to researchers. Scientific Data. 2019;6(1):251. doi: 10.1038/s41597-019-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias-Pazaran G. Genome-assisted prediction of quantitative traits using the R package sommer. PLOS ONE. 2016;11(6):e0156744. doi: 10.1371/journal.pone.0156744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biology. 2011;11(1):163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Beyene Y, Kassa S, Pérez P, Hickey JM, Chen C, De los Campos G, Burgueño J, Windhausen VS, Buckler E, Jannink J-L, Lopez Cruz MA, Babu R. Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3: Genes|Genomes|Genetics. 2013;3(11):1903–1926. doi: 10.1534/g3.113.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Pérez P, Hickey J, Burgueño J, Ornella L, Cerón-Rojas J, Zhang X, Dreisigacker S, Babu R, Li Y, Bonnett D, Mathews K. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity. 2014;112(1):48–60. doi: 10.1038/hdy.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas J, Crossa J, Soberanis V, Pérez-Elizalde S, Pérez-Rodríguez P, De los Campos G, Montesinos-López OA, Burgueño J. Genomic prediction of genotype × environment interaction kernel regression models. The Plant Genome. 2016;9(3):1–20. doi: 10.3835/plantgenome2016.03.0024. [DOI] [PubMed] [Google Scholar]
- Cuevas J, Crossa J, Montesinos-López OA, Burgueño J, Pérez-Rodríguez P, De los Campos G. Bayesian genomic prediction with genotype × environment interaction kernel models. G3: Genes|Genomes|Genetics. 2017;7(1):41–53. doi: 10.1534/g3.116.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA. The impact of genetic architecture on genome-wide evaluation methods. Genetics. 2010;185(3):1021–1031. doi: 10.1534/genetics.110.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De los Campos G, Hickey JM, Pong-Wong R, Daetwyler HD, Calus MPL. Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics. 2013;193(2):327–345. doi: 10.1534/genetics.112.143313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias KOG, Piepho HP, Guimarães LJM, Guimarães PEO, Parentoni SN, Pinto MO, Noda RW, Magalhães JV, Guimarães CT, Garcia AAF, Pastina MM. Novel strategies for genomic prediction of untested single-cross maize hybrids using unbalanced historical data. Theoretical and Applied Genetics. 2020;133(2):443–455. doi: 10.1007/s00122-019-03475-1. [DOI] [PubMed] [Google Scholar]
- Doebley J. The genetics of maize evolution. Annual Review of Genetics. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C. Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141(1):333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald CM. The breeding of crop ideotypes. Euphytica. 1968;17(3):385–403. doi: 10.1007/BF00056241. [DOI] [Google Scholar]
- Duvick DN. The contribution of breeding to yield advances in maize (Zea mays L.) Advances in Agronomy. 2005;86:83–145. doi: 10.1016/S0065-2113(05)86002-X. [DOI] [Google Scholar]
- Duvick DN, Smith JSC, Cooper M (2004) Long-term selection on a commercial hybrid maize breeding program. In Plant breeding reviews 24
- Edwards SM, Thomsen B, Madsen P, Sørensen P. Partitioning of genomic variance reveals biological pathways associated with udder health and milk production traits in dairy cattle. Genetics Selection Evolution. 2015;47(1):60. doi: 10.1186/s12711-015-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Sahana G, Ma P, Su G, Yu Y, Zhang S, Lund MS, Sørensen P. Exploring the genetic architecture and improving genomic prediction accuracy for mastitis and milk production traits in dairy cattle by mapping variants to hepatic transcriptomic regions responsive to intra-mammary infection. Genetics Selection Evolution. 2017;49(1):1–18. doi: 10.1186/s12711-017-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux A, Gorjanc G, Gaynor RC, Battagin M, Edwards SM, Wilson DL, Hearne SJ, Gonen S, Hickey JM. AlphaSim: software for breeding program simulation. The Plant Genome. 2016;9(3):1–14. doi: 10.3835/plantgenome2016.02.0013. [DOI] [PubMed] [Google Scholar]
- Fernandes SB, Lipka AE. simplePHENOTYPES: simulation of pleiotropic, linked and epistatic phenotypes. BMC Bioinformatics. 2020;21(1):491. doi: 10.1186/s12859-020-03804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Schauer N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends in Genetics. 2009;25(1):39–48. doi: 10.1016/j.tig.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. 2. New York: Dover Publication; 1958. [Google Scholar]
- Fisher RA (1958b) The genetical theory of natural selection, 2nd edn. Dover Publication. 10.5962/bhl.title.27468
- Fristche-Neto R, Akdemir D, Jannink J-L. Accuracy of genomic selection to predict maize single-crosses obtained through different mating designs. Theoretical and Applied Genetics. 2018;131(5):1153–1162. doi: 10.1007/s00122-018-3068-8. [DOI] [PubMed] [Google Scholar]
- Gage JL, Monier B, Giri A, Buckler ES. Ten years of the maize nested association mapping population: impact, limitations, and future directions. The Plant Cell. 2020;32(7):2083–2093. doi: 10.1105/tpc.19.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JT, Tracy WF. Pedigree diversity within the Lancaster Surecrop heterotic group of maize. Crop Science. 1993;33(2):334. doi: 10.2135/cropsci1993.0011183X003300020025x. [DOI] [Google Scholar]
- Gianola D. Priors in whole-genome regression: the Bayesian alphabet returns. Genetics. 2013;194(3):573–596. doi: 10.1534/genetics.113.151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowinski A, Flint-Garcia S (2018) Germplasm resources for mapping quantitative traits in maize:143–159. 10.1007/978-3-319-97427-9_10
- González-Camacho JM, Ornella L, Pérez-Rodríguez P, Gianola D, Dreisigacker S, Crossa J. Applications of machine learning methods to genomic selection in breeding wheat for rust resistance. The Plant Genome. 2018;11(2):170104. doi: 10.3835/plantgenome2017.11.0104. [DOI] [PubMed] [Google Scholar]
- González-Recio O. Epigenetics: a new challenge in the post-genomic era of livestock. Frontiers in Genetics. 2012;2(JAN):2010–2013. doi: 10.3389/fgene.2011.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanc G, Jenko J, Hearne SJ, Hickey JM. Initiating maize pre-breeding programs using genomic selection to harness polygenic variation from landrace populations. BMC Genomics. 2016;17(1):30. doi: 10.1186/s12864-015-2345-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanc G, Gaynor RC, Hickey JM. Optimal cross selection for long-term genetic gain in two-part programs with rapid recurrent genomic selection. Theoretical and Applied Genetics. 2018;131(9):1953–1966. doi: 10.1007/s00122-018-3125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Magwire MM, Basten CJ, Xu Z, Wang D. Evaluation of the utility of gene expression and metabolic information for genomic prediction in maize. Theoretical and Applied Genetics. 2016;129(12):2413–2427. doi: 10.1007/s00122-016-2780-5. [DOI] [PubMed] [Google Scholar]
- Habier D, Fernando RL, Dekkers JCM. The impact of genetic relationship information on genome-assisted breeding values. Genetics. 2007;177(4):2389–2397. doi: 10.1534/genetics.107.081190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D, Fernando RL, Dekkers JCM. Genomic selection using low-density marker panels. Genetics. 2009;182(1):343–353. doi: 10.1534/genetics.108.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallauer AR. History, contribution, and future of quantitative genetics in plant breeding: lessons from maize. Crop Science. 2007;47(SUPPL. DEC.):S-4–S-19. doi: 10.2135/cropsci2007.04.0002IPBS. [DOI] [Google Scholar]
- Hallauer AR, Russell WA, Lamkey KR (1988) Corn breeding. In Agronomy Publications:463–564. 10.2134/agronmonogr18.3ed.c8
- Hayes HK. Methods of corn breeding. Journal of Heredity. 1912;3(2):99–108. doi: 10.1093/oxfordjournals.jhered.a105896. [DOI] [Google Scholar]
- Heather JM, Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107(1):1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner EL, Sorrells ME, Jannink JL. Genomic selection for crop improvement. Crop Science. 2009;49(1):1–12. doi: 10.2135/cropsci2008.08.0512. [DOI] [Google Scholar]
- Heffner EL, Lorenz AJ, Jannink J-L, Sorrells ME. Plant breeding with genomic selection: gain per unit time and cost. Crop Science. 2010;50(5):1681–1690. doi: 10.2135/cropsci2009.11.0662. [DOI] [Google Scholar]
- Henderson CR (1952) In: Gowen JW (ed) Specific and general combining ability, Iowa State College Press.
- Heslot N, Yang H-P, Sorrells ME, Jannink J-L. Genomic selection in plant breeding: a comparison of models. Crop Science. 2012;52(1):146. doi: 10.2135/cropsci2011.09.0297. [DOI] [Google Scholar]
- Heslot N, Yang H-P, Sorrells ME, Jannink J-L. Genomic selection in plant breeding: a comparison of models. Crop Science. 2012;52(1):146–160. doi: 10.2135/cropsci2011.06.0297. [DOI] [Google Scholar]
- Heslot N, Akdemir D, Sorrells ME, Jannink J-L. Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theoretical and Applied Genetics. 2014;127(2):463–480. doi: 10.1007/s00122-013-2231-5. [DOI] [PubMed] [Google Scholar]
- Hickey JM, Chiurugwi T, Mackay I, Powell W. Genomic prediction unifies animal and plant breeding programs to form platforms for biological discovery. Nature Genetics. 2017;49(9):1297–1303. doi: 10.1038/ng.3920. [DOI] [PubMed] [Google Scholar]
- Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genetics. 2008;4(2):e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F, Moreau L, Lacoudre F, Charcosset A, Gallais A. More on the efficiency of marker-assisted selection. Theoretical and Applied Genetics. 1997;95(8):1181–1189. doi: 10.1007/s001220050679. [DOI] [Google Scholar]
- Howard R, Carriquiry AL, Beavis WD. Parametric and nonparametric statistical methods for genomic selection of traits with additive and epistatic genetic architectures. G3: Genes|Genomes|Genetics. 2014;4(6):1027–1046. doi: 10.1534/g3.114.010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Morota G, Rosa GJM, Gianola D. Prediction of plant height in Arabidopsis thaliana using DNA methylation data. Genetics. 2015;201(2):779–793. doi: 10.1534/genetics.115.177204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Richards S, Carbone MA, Zhu D, Anholt RRH, Ayroles JF, Duncan L, Jordan KW, Lawrence F, Magwire MM, Warner CB, Blankenburg K, Han Y, Javaid M, Jayaseelan J, Jhangiani SN, Muzny D, Ongeri F, Perales L, Wu YQ, Zhang Y, Zou X, Stone EA, Gibbs RA, Mackay TFC. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proceedings of the National Academy of Sciences. 2012;109(39):15553–15559. doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidro J, Jannink J-L, Akdemir D, Poland J, Heslot N, Sorrells ME. Training set optimization under population structure in genomic selection. Theoretical and Applied Genetics. 2015;128(1):145–158. doi: 10.1007/s00122-014-2418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannink J-L. Dynamics of long-term genomic selection. Genetics Selection Evolution. 2010;42(1):35. doi: 10.1186/1297-9686-42-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarquín D, Crossa J, Lacaze X, Du Cheyron P, Daucourt J, Lorgeou J, Piraux F, Guerreiro L, Pérez P, Calus M, Burgueño J, De los Campos G. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theoretical and Applied Genetics. 2014;127(3):595–607. doi: 10.1007/s00122-013-2243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarquin D, Howard R, Crossa J, Beyene Y, Gowda M, Martini JWR, Covarrubias Pazaran G, Burgueño J, Pacheco A, Grondona M, Wimmer V, Prasanna BM. Genomic prediction enhanced sparse testing for multi-environment trials. G3: Genes|Genomes|Genetics. 2020;10(8):2725–2739. doi: 10.1534/g3.120.401349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Jannink JL. Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics. 2012;192(4):1513–1522. doi: 10.1534/genetics.112.144246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Reif JC. Modeling epistasis in genomic selection. Genetics. 2015;201(2):759–768. doi: 10.1534/genetics.115.177907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, Bulski A, Albuisson J, Heredia F, Audigier P, Bouchez D, Dillmann C, Guerche P, Hospital F, Colot V. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genetics. 2009;5(6):e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JWJ, Kang EY, Org E, Furlotte N, Parks B, Hormozdiari F, Lusis AJ, Eskin E. Efficient and accurate multiple-phenotype regression method for high dimensional data considering population structure. Genetics. 2016;204(4):1379–1390. doi: 10.1534/genetics.116.189712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam DC, Potts SM, Bohn MO, Lipka AE, Lorenz AJ. Genomic prediction of single crosses in the early stages of a maize hybrid breeding pipeline. G3: Genes|Genomes|Genetics. 2016;6(11):3443–3453. doi: 10.1534/g3.116.031286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremling KAG, Diepenbrock CH, Gore MA, Buckler ES, Bandillo NB. Transcriptome-wide association supplements genome-wide association in Zea mays. G3: Genes|Genomes|Genetics. 2019;9(9):3023–3033. doi: 10.1534/g3.119.400549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaridou C, Tsairidou S, Houston RD, Robledo D. Genomic prediction using low density marker panels in aquaculture: performance across species, traits, and genotyping platforms. Frontiers in Genetics. 2020;11(February):1–8. doi: 10.3389/fgene.2020.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Guo T, Mu Q, Li X, Yu J. Genomic and environmental determinants and their interplay underlying phenotypic plasticity. Proceedings of the National Academy of Sciences. 2018;115(26):6679–6684. doi: 10.1073/pnas.1718326115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gao N, Martini JWR, Simianer H. Integrating gene expression data into genomic prediction. Frontiers in Genetics. 2019;10(FEB):1–11. doi: 10.3389/fgene.2019.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian L, Jacobson A, Zhong S, Bernardo R. Genomewide prediction accuracy within 969 maize biparental populations. Crop Science. 2014;54(4):1514–1522. doi: 10.2135/cropsci2013.12.0856. [DOI] [Google Scholar]
- Lin Z, Li X, Shannon LM, Yeh C-T, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, Doebley J, Schnable PS, Tuinstra MR, Tesso TT, White F, Yu J. Parallel domestication of the Shattering1 genes in cereals. Nature Genetics. 2012;44(6):720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Kandianis CB, Hudson ME, Yu J, Drnevich J, Bradbury PJ, Gore MA. From association to prediction: statistical methods for the dissection and selection of complex traits in plants. Current Opinion in Plant Biology. 2015;24:110–118. doi: 10.1016/j.pbi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang H, Wang H, Guo Z, Xu X, Liu J, Wang S, Li W-X, Zou C, Prasanna BM, Olsen MS, Huang C, Xu Y. Factors affecting genomic selection revealed by empirical evidence in maize. The Crop Journal. 2018;6(4):341–352. doi: 10.1016/j.cj.2018.03.005. [DOI] [Google Scholar]
- Lopez-Cruz M, Crossa J, Bonnett D, Dreisigacker S, Poland J, Jannink J-L, Singh RP, Autrique E, De los Campos G. Increased prediction accuracy in wheat breeding trials using a marker × environment interaction genomic selection model. G3: Genes|Genomes|Genetics. 2015;5(4):569–582. doi: 10.1534/g3.114.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod IM, Bowman PJ, Vander Jagt CJ, Haile-Mariam M, Kemper KE, Chamberlain AJ, Schrooten C, Hayes BJ, Goddard ME. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genomics. 2016;17(1):144. doi: 10.1186/s12864-016-2443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Bradbury P, Flint-Garcia S, Browne C, Eller M, Guill K, Lepak N, Peterson B, Romero S, Salvo S, Ware D, Holland JB, Buckler ES, Villeda HS, Bottoms C, Kresovich S, Li H, Acharya C, Brown P, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325(5941):737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- Mejia-Guerra MK, Pomeranz M, Morohashi K, Grotewold E. From plant gene regulatory grids to network dynamics. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012;1819(5):454–465. doi: 10.1016/j.bbagrm.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157(4):1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES. Association mapping: critical considerations shift from genotyping to experimental design. The Plant Cell. 2009;21(8):2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves HHR, Carvalheiro R, Queiroz SA. A comparison of statistical methods for genomic selection in a mice population. BMC Genetics. 2012;13(1):100. doi: 10.1186/1471-2156-13-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Dermitzakis ET. Expression quantitative trait loci: present and future. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1620):20120362. doi: 10.1098/rstb.2012.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogutu JO, Piepho H-P, Schulz-Streeck T. A comparison of random forests, boosting and support vector machines for genomic selection. BMC Proceedings. 2011;5(S3):S11. doi: 10.1186/1753-6561-5-S3-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olatoye MO, Clark LV, Labonte NR, Dong H, Dwiyanti MS, Anzoua KG, Brummer JE, Ghimire BK, Dzyubenko E, Dzyubenko N, Bagmet L, Sabitov A, Chebukin P, Głowacka K, Heo K, Jin X, Nagano H, Peng J, Yu CY, Yoo JH, Zhao H, Long SP, Yamada T, Sacks EJ, Lipka AE. Training population optimization for genomic selection in miscanthus. G3: Genes|Genomes|Genetics. 2020;10(7):2465–2476. doi: 10.1534/g3.120.401402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, Gardner CAC, McMullen MD, Holland JB, Bradbury PJ, Buckler ES. The genetic architecture of maize height. Genetics. 2014;196(4):1337–1356. doi: 10.1534/genetics.113.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez P, De los Campos G. Genome-wide regression and prediction with the BGLR statistical package. Genetics. 2014;198(2):483–495. doi: 10.1534/genetics.114.164442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho HP, Williams ER, Fleck M. A note on the analysis of designed experiments with complex treatment structure. HortScience. 2006;41(2):446–452. doi: 10.21273/HORTSCI.41.2.446. [DOI] [Google Scholar]
- Pinho Morais PP, Akdemir D, Braatz de Andrade LR, Jannink J, Fritsche-Neto R, Borém A, Couto Alves F, Hottis Lyra D, Granato ÍSC. Using public databases for genomic prediction of tropical maize lines. Plant Breeding. 2020;139(4):697–707. doi: 10.1111/pbr.12827. [DOI] [Google Scholar]
- Poland JA, Rife TW. Genotyping-by-sequencing for plant breeding and genetics. The Plant Genome. 2012;5(3):92–102. doi: 10.3835/plantgenome2012.05.0005. [DOI] [Google Scholar]
- Poland J, Rutkoski J. Advances and challenges in genomic selection for disease resistance. Annual Review of Phytopathology. 2016;54(1):79–98. doi: 10.1146/annurev-phyto-080615-100056. [DOI] [PubMed] [Google Scholar]
- Pollak LM (2003) The history and success of the public–private project on germplasm enhancement of maize (GEM). 78:45–87. 10.1016/S0065-2113(02)78002-4
- Prado SA, López CG, Senior ML, Borrás L. The genetic architecture of maize (Zea mays L.) kernel weight determination. G3: Genes, Genomes, Genetics. 2014;4(9):1611–1621. doi: 10.1534/g3.114.013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx SR, Nuzhdin S, Promislow DEL. Direct selection on genetic robustness revealed in the yeast transcriptome. PLoS ONE. 2007;2(9):e911. doi: 10.1371/journal.pone.0000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum P, Peña-Rosas JP, Garcia-Casal MN. Global maize production, utilization, and consumption. Annals of the New York Academy of Sciences. 2014;1312(1):105–112. doi: 10.1111/nyas.12396. [DOI] [PubMed] [Google Scholar]
- Rasheed A, Hao Y, Xia X, Khan A, Xu Y, Varshney RK, He Z. Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Molecular Plant. 2017;10(8):1047–1064. doi: 10.1016/j.molp.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Reif JC, Hamrit S, Heckenberger M, Schipprack W, Maurer HP, Bohn M, Melchinger AE. Trends in genetic diversity among European maize cultivars and their parental components during the past 50 years. Theoretical and Applied Genetics. 2005;111(5):838–845. doi: 10.1007/s00122-005-0004-5. [DOI] [PubMed] [Google Scholar]
- Rice B, Lipka AE. Evaluation of RR-BLUP genomic selection models that incorporate peak genome-wide association study signals in maize and sorghum. The Plant Genome. 2019;12(1):180052. doi: 10.3835/plantgenome2018.07.0052. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nature Genetics. 2012;44(2):217–220. doi: 10.1038/ng.1033. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Dunne JC, Romay C, Bohn M, Buckler ES, Ciampitti IA, Edwards J, Ertl D, Flint-Garcia S, Gore MA, Graham C, Hirsch CN, Hood E, Hooker DC, Knoll J, Lee EC, Lorenz A, Lynch JP, McKay J et al (2021) The importance of dominance and genotype-by-environment interactions on grain yield variation in a large-scale public cooperative maize experiment. G3 Genes|Genomes|Genetics. 10.1093/g3journal/jkaa050 [DOI] [PMC free article] [PubMed]
- Ross-Ibarra J, Morrell PL, Gaut BS. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proceedings of the National Academy of Sciences. 2007;104(Supplement 1):8641–8648. doi: 10.1073/pnas.0700643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru S, Bernardo R. Predicted genetic gains from introgressing chromosome segments from exotic germplasm into an elite soybean cultivar. Theoretical and Applied Genetics. 2020;133(2):605–614. doi: 10.1007/s00122-019-03490-2. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Hartl DL. Genotypic context and epistasis in individuals and populations. Cell. 2016;166(2):279–287. doi: 10.1016/j.cell.2016.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadras VO, Lawson C. Genetic gain in yield and associated changes in phenotype, trait plasticity and competitive ability of South Australian wheat varieties released between 1958 and 2007. Crop and Pasture Science. 2011;62(7):533. doi: 10.1071/CP11060. [DOI] [Google Scholar]
- Saint Pierre C, Burgueño J, Crossa J, Fuentes Dávila G, Figueroa López P, Solís Moya E, Ireta Moreno J, Hernández Muela VM, Zamora Villa VM, Vikram P, Mathews K, Sansaloni C, Sehgal D, Jarquin D, Wenzl P, Singh S. Genomic prediction models for grain yield of spring bread wheat in diverse agro-ecological zones. Scientific Reports. 2016;6(1):27312. doi: 10.1038/srep27312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopp P, Müller D, Wientjes YCJ, Melchinger AE. Genomic prediction within and across biparental families: means and variances of prediction accuracy and usefulness of deterministic equations. G3: Genes, Genomes, Genetics. 2017;7(11):3571–3586. doi: 10.1534/g3.117.300076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag TA, Westhues M, Schipprack W, Seifert F, Thiemann A, Scholten S, Melchinger AE. Beyond genomic prediction: combining different types of omics data can improve prediction of hybrid performance in maize. Genetics. 2018;208(4):1373–1385. doi: 10.1534/genetics.117.300374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Balding DJ. MultiBLUP: improved SNP-based prediction for complex traits. Genome Research. 2014;24(9):1550–1557. doi: 10.1101/gr.169375.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindel JE, Begum H, Akdemir D, Collard B, Redoña E, Jannink JL, McCouch S. Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity. 2016;116(4):395–408. doi: 10.1038/hdy.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague GF. Heterosis in maize: theory and practice. In: Frankel R, editor. Heterosis: Reappraisal of Theory and Practice. Berlin Heidelberg: Springer; 1983. pp. 47–70. [Google Scholar]
- Sprague GF, Tatum LA. General vs. specific combining ability in single crosses of corn. Agronomy Journal. 1942;34(10):923–932. doi: 10.2134/agronj1942.00021962003400100008x. [DOI] [Google Scholar]
- Su G, Christensen OF, Ostersen T, Henryon M, Lund MS. Estimating additive and non-additive genetic variances and predicting genetic merits using genome-wide dense single nucleotide polymorphism markers. PLoS ONE. 2012;7(9):e45293. doi: 10.1371/journal.pone.0045293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Cabrera-Bosquet L, Pridmore T, Bennett M. Plant phenomics, from sensors to knowledge. Current Biology. 2017;27(15):R770–R783. doi: 10.1016/j.cub.2017.05.055. [DOI] [PubMed] [Google Scholar]
- Technow F, Riedelsheimer C, Schrag TA, Melchinger AE. Genomic prediction of hybrid performance in maize with models incorporating dominance and population specific marker effects. Theoretical and Applied Genetics. 2012;125(6):1181–1194. doi: 10.1007/s00122-012-1905-8. [DOI] [PubMed] [Google Scholar]
- Technow F, Schrag TA, Schipprack W, Bauer E, Simianer H, Melchinger AE. Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics. 2014;197(4):1343–1355. doi: 10.1534/genetics.114.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statistical Society: Series B (Methodological) 1996;58(1):267–288. doi: 10.1111/j.2517-6161.1996.tb02080.x. [DOI] [Google Scholar]
- Turner-Hissong SD, Bird KA, Lipka AE, King EG, Beissinger TM, Angelovici R. Genomic prediction informed by biological processes expands our understanding of the genetic architecture underlying free amino acid traits in dry arabidopsis seeds. G3: Genes|Genomes|Genetics. 2020;10(11):4227–4239. doi: 10.1534/g3.120.401240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Science. 2008;91(11):4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Vazquez, A., Wiener, H., Shrestha, S., Tiwari, H., & De los Campos, G. (2014). Integration of multi-layer omic data for prediction of disease risk in humans. Proceedings, 10th World Congress of Genetics Applied to Livestock Production, August, 6. 10.13140/2.1.4769.9200
- Velazco JG, Jordan DR, Mace ES, Hunt CH, Malosetti M, van Eeuwijk FA. Genomic prediction of grain yield and drought-adaptation capacity in sorghum is enhanced by multi-trait analysis. Frontiers in Plant Science. 2019;10(July):1–12. doi: 10.3389/fpls.2019.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitezica ZG, Varona L, Legarra A. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics. 2013;195(4):1223–1230. doi: 10.1534/genetics.113.155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivek BS, Krishna GK, Vengadessan V, Babu R, Zaidi PH, Kha LQ, Mandal SS, Grudloyma P, Takalkar S, Krothapalli K, Singh IS, Ocampo ETM, Xingming F, Burgueño J, Azrai M, Singh RP, Crossa J (2017) Use of genomic estimated breeding values results in rapid genetic gains for drought tolerance in maize. The Plant Genome 10(1) plantgenome2016.07.0070. 10.3835/plantgenome2016.07.0070 [DOI] [PubMed]
- Voss-Fels KP, Cooper M, Hayes BJ. Accelerating crop genetic gains with genomic selection. Theoretical and Applied Genetics. 2019;132(3):669–686. doi: 10.1007/s00122-018-3270-8. [DOI] [PubMed] [Google Scholar]
- Vu WT, Chang PL, Moriuchi KS, Friesen ML. Genetic variation of transgenerational plasticity of offspring germination in response to salinity stress and the seed transcriptome of Medicago truncatula Speciation and evolutionary genetics. BMC Evolutionary Biology. 2015;15(1):1–14. doi: 10.1186/s12862-015-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn JD, Burch MB, Franco JAV. Predictive breeding for maize: Making use of molecular phenotypes, machine learning, and physiological crop models. Crop Science. 2020;60(2):622–638. doi: 10.1002/csc2.20052. [DOI] [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, Yan J. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nature Communications. 2014;5(1):3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhues M, Schrag TA, Heuer C, Thaller G, Utz HF, Schipprack W, Thiemann A, Seifert F, Ehret A, Schlereth A, Stitt M, Nikoloski Z, Willmitzer L, Schön CC, Scholten S, Melchinger AE. Omics-based hybrid prediction in maize. Theoretical and Applied Genetics. 2017;130(9):1927–1939. doi: 10.1007/s00122-017-2934-0. [DOI] [PubMed] [Google Scholar]
- Whittaker JC, Thompson R, Denham MC. Marker-assisted selection using ridge regression. Genetics Research. 2000;75(2):249–252. doi: 10.1017/S0016672399004462. [DOI] [PubMed] [Google Scholar]
- Widłak W. High-throughput technologies in molecular biology. In American Psychologist. 2013;65(8):139–153. doi: 10.1007/978-3-642-45361-8_9. [DOI] [Google Scholar]
- Windhausen VS, Atlin GN, Hickey JM, Crossa J, Jannink JL, Sorrells ME, Raman B, Cairns JE, Tarekegne A, Semagn K, Beyene Y, Grudloyma P, Technow F, Riedelsheimer C, Melchinger AE. Effectiveness of genomic prediction of maize hybrid performance in different breeding populations and environments. G3: Genes, Genomes. Genetics. 2012;2(11):1427–1436. doi: 10.1534/g3.112.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Crouch JH. Marker-assisted selection in plant breeding: from publications to practice. Crop Science. 2008;48(2):391–407. doi: 10.2135/cropsci2007.04.0191. [DOI] [Google Scholar]
- Zhang W, Smith C. Computer simulation of marker-assisted selection utilizing linkage disequilibrium. TAG. Theoretical and Applied Genetics. Theoretische Und Angewandte Genetik. 1992;83(6–7):813–820. doi: 10.1007/BF00226702. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ober U, Erbe M, Zhang H, Gao N, He J, Li J, Simianer H. Improving the accuracy of whole genome prediction for complex traits using the results of genome wide association studies. PLoS ONE. 2014;9(3):1–12. doi: 10.1371/journal.pone.0093017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wang H, Beyene Y, Semagn K, Liu Y, Cao S, Cui Z, Ruan Y, Burgueño J, San Vicente F, Olsen M, Prasanna BM, Crossa J, Yu H, Zhang X. Effect of trait heritability, training population size and marker density on genomic prediction accuracy estimation in 22 bi-parental tropical maize populations. Frontiers in Plant Science. 2017;8(November):1–12. doi: 10.3389/fpls.2017.01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yin L, Wang M, Yuan X, Liu X. Factors affecting the accuracy of genomic selection for agricultural economic traits in maize, cattle, and pig populations. Frontiers in Genetics. 2019;10(MAR):1–10. doi: 10.3389/fgene.2019.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gowda M, Liu W, Würschum T, Maurer HP, Longin FH, Ranc N, Reif JC. Accuracy of genomic selection in European maize elite breeding populations. Theoretical and Applied Genetics. 2012;124(4):769–776. doi: 10.1007/s00122-011-1745-y. [DOI] [PubMed] [Google Scholar]
- Zhao C, Zhang Y, Du J, Guo X, Wen W, Gu S, Wang J, Fan J (2019) Crop phenomics: current status and perspectives. Frontiers in Plant Science 10(June). 10.3389/fpls.2019.00714 [DOI] [PMC free article] [PubMed]
- Zhou S, Campbell TG, Stone EA, Mackay TFC, Anholt RRH. Phenotypic plasticity of the drosophila transcriptome. PLoS Genetics. 2012;8(3):e1002593. doi: 10.1371/journal.pgen.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.