Abstract
The root-lesion nematode Pratylenchus thornei Sher & Allen, 1953 is a damaging parasite of many crop plants, including the grain legume chickpea (Cicer arietinum L.). Within cultivated chickpea, there are no known sources of strong resistance to P. thornei, but some cultivars have partial resistance. In the research reported here, the genetic basis for differences in P. thornei resistance was analysed using a population derived by accelerated single seed descent from a cross between a partially resistant cultivar, PBA HatTrick, and a very susceptible cultivar, Kyabra. A genetic linkage map was constructed from genotyping-by-sequencing data. Two quantitative trait loci were mapped, one on the Ca4 chromosome and one on the Ca7 chromosome. The Ca7 locus had a greater and more consistent effect than the Ca4 locus. Marker assays designed for single nucleotide polymorphisms on Ca7 were applied to a panel of chickpea accessions. Some of these markers should be useful for marker-assisted selection in chickpea breeding. Haplotype analysis confirmed the Iranian landrace ICC14903 to be the source of the resistance allele in PBA HatTrick and indicated that other Australian cultivars inherited the same allele from other Iranian landraces. A candidate region was defined on the Ca7 pseudomolecule. Within that region, 69 genes have been predicted with high confidence. Among these, two have annotations related to biotic stress response. Three others have previously been reported to be expressed in roots of PBA HatTrick and Kyabra, including one that is more highly expressed in PBA HatTrick than in Kyabra.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01271-8.
Keywords: Root-lesion nematode, Pratylenchus thornei, Chickpea, Cicer arietinum, Quantitative trait locus, Molecular markers
Introduction
The root-lesion nematode Pratylenchus thornei Sher & Allen, 1953 is a soil-borne parasite of many plant species, including chickpea (Cicer arietinum L.) and other legumes, as well as cereal crops. As a migratory endoparasite, it migrates through and feeds upon root tissue, causing extensive tissue damage and reducing the stress tolerance of the host plant (Castillo et al. 1995). In Australia, and especially in northeastern Australia, P. thornei is one of the most damaging pathogens of both chickpea (Murray and Brennan 2012) and wheat (Triticum aestivum L.) (Murray and Brennan 2009). As these two crops are often grown in rotation with each other, sustainable control of P. thornei would benefit from the development and adoption of both chickpea and wheat cultivars with improved resistance (the ability to reduce nematode reproduction). For wheat, there have been numerous reports on sources of partial resistance to P. thornei (Thompson and Haak 1997; Thompson 2008; Sheedy and Thompson 2009; Thompson et al. 2009; Sheedy et al. 2012) and some reports on the genetic control of this resistance (Zwart et al. 2004, 2005; Thompson et al. 2012; Linsell et al. 2014; Rahman et al. 2019). For chickpea, some cultivars have been reported to exhibit partial resistance (Tiwari et al. 1992; Thompson et al. 2011; Rodda et al. 2016; Reen et al. 2019). Stronger resistance has been reported in Cicer species from the tertiary gene pool of chickpea (Di Vito et al. 1995) and in C. reticulatum Ladiz. and C. echinospermum P.H. Davis, which are cross-compatible with C. arietinum (Thompson et al. 2011; Rodda et al. 2016; Reen et al. 2019; Zwart et al. 2019; Ayaz et al. 2021). There have been no reports on the genetic control of differences in P. thornei resistance in cultivated chickpea or its wild relatives.
Here, we report on genetic mapping for P. thornei resistance in an experimental population derived from a cross between a partially resistant chickpea cultivar (PBA HatTrick; pedigree Jimbour/ICC14903) and a very susceptible chickpea cultivar (Kyabra; pedigree Amethyst//946-31/Barwon/3/Lasseter/940-26//946-31/Norwin/4/8507-28H//Amethyst/T1069///8507-28H/946-31). Among 13 chickpea cultivars evaluated for P. thornei resistance by Rodda et al. (2016), PBA HatTrick was the most resistant and Kyabra was among the most susceptible. One of PBA HatTrick’s parents, the Iranian landrace ICC14903, is known to be more resistant than the other parent, Jimbour (Thompson et al. 2011; Rodda et al. 2016), indicating that the resistance of PBA HatTrick would have been inherited from ICC14903.
Methods
Population development
Recombinant inbred line (RIL) development was undertaken within plant growth facilities at the University of Western Australia (31.9800° S, 115.8190° E). Population development was undertaken in three stages:
Parental line purification: Seeds of the desi-type chickpea cultivars PBA HatTrick and Kyabra were obtained from the Australian Grains Genebank (Horsham, Vic, Australia). Plants were cycled through four selfing generations using the accelerated single seed descent (aSSD) conditions described by Atieno et al. (2021), with a 20 h photoperiod and light emitting diode-based arrays (red:far-red ratio 2.89; light intensity c. 325 μmol m−2 s−1 at canopy).
Crossing and F2 seed production: Five plants of each purified parental line were grown under aSSD conditions. At flowering, they were cross-pollinated following emasculation as per Kalve and Tadege (2017) using PBA HatTrick as the maternal parent and Kyabra as the paternal parent. Mature F1 seeds were harvested and sown in 4 L pots. F1 seedlings were grown at 22 °C day/18 °C night +− 3 °C under natural day length of approximately 14 h. Hybridity was confirmed using molecular markers. One confirmed hybrid plant was cloned eight times as per Danehloueipour et al. (2006). The resulting clones were allowed to self-pollinate and fully mature before F2 seeds were harvested.
RIL development: In the F2, F3, F4, and F5 generations, plants were grown under aSSD conditions. Pods containing immature seeds were removed at physiological maturity (about 18 d after flowering). The pods from each plant were put into a single 135 80 mm envelope (Tudor Press Seal Plain, Officeworks, Australia). Envelopes were placed in a plastic tray containing a bed (about 3 cm deep) of orange indicator silica gel beads (Silica Gel Australia, Gordon, NSW, Australia) and dried for 5 to 7 days in a controlled temperature cabinet set at 25 °C with relative humidity between 30 and 40%. At about 9% moisture, based on equilibrium relative humidity measured with a water activity meter (Rotronic AG, Bassersdorf Switzerland) and converted to seed moisture based on a moisture sorption isotherm for chickpea (Menkov 2000), seeds were sown to grow the next generation. The F6 seeds were left to fully mature on the F5 plants. One F6 plant of each of 218 recombinant inbred lines (RILs), each tracing back to a different F2 plant, was grown in a shade house in Urrbrae, South Australia (34.9670° S, 138.6360° E) providing F6:7 seed for each RIL.
DNA extraction and genotyping-by-sequencing
Leaf tissue was sampled from each of 213 F6 plants and from each of four plants of each of the parental cultivars. DNA was isolated using a phenol chloroform method (Rogowsky et al. 1991), with the modifications described by Pallotta et al. (2000). Aliquots of DNA were sent to Diversity Arrays Technology Pty. Ltd. (Bruce, ACT, Australia) for genotyping-by-sequencing (GBS) analysis using its chickpea DArTseq 1.0 platform. As described by Aznar-Fernández et al. (2020), DArTseq technology involves processing of DNA samples in digestion/ligation reactions, using adaptors that include barcode sequences and flowcell attachment sequences for Illumina sequencing. Amplification products are bulked across samples, subjected to bridge PCR, and sequenced using an Illumina HiSeq (Illumina, USA). The resulting data were analysed by Diversity Arrays Technology Pty. Ltd. using its proprietary software pipeline. In that pipeline, poor-quality sequences are filtered out, with stringent selection in the barcode region. The remaining sequences are assigned to samples based on barcodes, identical sequences are collapsed into fastqcall files, and genotypes are called for SNP and presence/absence (“SilicoDArT”) markers.
Linkage map construction
Genotypic data from GBS were processed using the R Statistical Computing Environment (R Core Team 2020). Markers for which parental genotypes were unknown or uncertain (because they had not been called for the parental DNA samples), markers with greater than 25% missing values, and markers exhibiting significant segregation distortion (greater than a Bonferroni corrected p-value for a genome-wide alpha level of 0.05) were temporarily set aside. Using the genotypic data for the remaining markers, an initial linkage map was constructed using the MSTMap (Wu et al. 2008) functionality of the R package ASMap (Taylor and Butler 2017). Markers for which the parental phase was unknown were then added to linkage groups using the pushCross function of ASMap. For markers that remained unlinked, the allelic phase was switched and a second “push” was conducted. Any marker that still remained unlinked was excluded from further consideration. Finally, markers that had been set aside due to missing values and/or segregation distortion were assessed for suitability and additional markers were “pushed” into the linkage map. Linkage maps were assigned to pseudomolecules Ca1 through Ca7 based on the results of a BLASTn analysis (Altschul et al. 1990) of GBS sequence tags against version 2.6.3 (Edwards 2016) of a genome assembly for kabuli-type chickpea (Varshney et al. 2013). A linkage map was drawn using the software MapChart (Voorrips 2002).
To prepare the genotypic data for use in QTL analysis, each set of co-locating markers was condensed to form a representative consensus marker with a unique map position. Marker genotypes were numerically encoded (PBA HatTrick homozygotes as + 1; heterozygotes as 0; Kyabra homozygotes as − 1). Missing genotype calls were numerically imputed using the flanking marker rules of Martínez and Curnow (1992).
Evaluation of resistance to Pratylenchus thornei
Resistance to P. thornei was evaluated in each of two years in a glasshouse in Toowoomba, Queensland (27.5598° S, 151.9507° E). Each experiment consisted of three randomised complete blocks. In the first experiment, conducted in 2018, each of 205 RILs was represented by three F6 siblings of the genotyped F6 plant. In the second experiment, conducted in 2019, each of that 213 RILs that had been included in the genotyping was represented by three F7 selfed progeny of the genotyped F6 plant. Several additional entries were included in each experiment: five RILs that had not been genotyped, the parental lines (PBA HatTrick and Kyabra), a P. thornei-resistant accession of C. reticulatum (ILWC123), three moderately resistant wheat lines (CPI133872, GS50a and QT8447)), two susceptible wheat cultivars (Petrie and Strzelecki), three wheat cultivars of intermediate reaction (Gauntlet, Sunguard and Sunzell), and an inoculated/unsown control treatment. The 2018 (F6) experiment also included two chickpea cultivars of intermediate reaction: Howzat and Sonali. The pots (70 mm square base 150 mm high), which were designed for bottom watering, were partially filled with 264 g of black vertisolic soil, providing a base layer of 80% of the final total soil weight (330 g oven-dry soil). The soil had previously been sieved to remove aggregates larger than 1 cm, pasteurised for 40 min using aerated steam at 85 °C, and fertilised to supply NO3-N. P, K, S, and Zn at 200, 25, 88, 36, and 5 mg/kg of soil, respectively. Two seeds were placed on top of that base.
Inoculum prepared using P. thornei from a culture that originated from ten specimens collected near Jondaryan, Queensland (Thompson 2008) was applied by pipetting 10 mL of a suspension containing 3300 P. thornei (equivalent to 10,000 nematodes per kg of oven-dry soil) around the seeds. The remaining soil was placed over the seeds as a cap. After emergence, seedlings were thinned to one seedling per pot. Both experiments were conducted on benches fitted with a self-regulating continuous bottom-watering system set to a water tension of 2 cm. Air and soil temperatures were maintained in the range of 20 to 25 °C using reverse-cycle air conditioning and evaporative coolers.
At 16 weeks after sowing and inoculation, the roots and soil were removed from the tubes. The soil and roots from the lower half of the pot were removed and stored at 3 °C in sealed plastic bags for later processing. The plants and upper soil and roots were re-potted and returned to the glasshouse so that the plants could grow to maturity to produce seed. The soil and roots of the lower half of each sample were then manually cut into segments approximately 1 cm long and homogenised. A 100 g subsample of the soil-root mixture from each tube was dried at 105 °C for 48 h to determine gravimetric moisture content. Nematodes were extracted from another 150 g subsample at 22 °C for 48 h using the Whitehead tray method (Whitehead and Hemming 1965) and collected on a 20-μm sieve. Samples were stored in 30 mL vials at 3 °C. Nematodes were counted using a 1 mL Peters nematode counting chamber slide (Chalex Corporation, Portland, OR, USA) under a compound microscope (40 ).
Statistical analysis of nematode counts
Nematode counts from each experiment were initially subjected to an analysis of variance containing a factor for distinguishing among entries (RILs, parents, and control cultivars). The profile likelihood methods outlined by Venables and Ripley (2002) were used to determine optimal values for in the transformed response , where Pt is the number of P. thornei nematodes per kg (oven-dry equivalent) of soil and roots.
Transformed trait values were then analysed using a linear mixed model (LMM) that partitioned and accounted for genetic and non-genetic sources of variation (Gilmour et al. 1997). The LMM included fixed-effect indicator terms to allow estimation of mean effects for the RIL population, each parent and each control cultivar and a random-effect term to account for non-genetic variation from experimental design blocks. To account for potential spatial variation across the experimental layout, the variance of the residuals of the LMM were assumed to contain a separable AR1 by AR1 (AR1 = autoregressive structure of order 1) correlation structure in row and column directions. To estimate the genetic variance among the RIL progeny, the LMM also contained a random-effect term to distinguish among the RILs. From the fitted LMM, empirical best linear unbiased estimators (eBLUEs) of the parental lines were extracted and used as total predictions for the parents. For each RIL, a total prediction was calculated from the overall eBLUE for the population and the empirical predictor (eBLUP) for the RIL.
All LMM analyses of phenotypic data were conducted in the R statistical computing environment (R Core Team 2020) with the ASReml-R package (Butler et al. 2017), which uses the REML algorithm of Patterson and Thompson (1971) to estimate model parameters. Additional model diagnosis was conducted using the ASExtras R package (http://www.mmade.org).
Whole-genome QTL analysis
QTL analysis was conducted following the whole-genome average interval mapping (WGAIM) approach of Verbyla et al. (2007) and Verbyla et al. (2012), except that analysis was based on individual marker loci rather than the midpoints of intervals between marker loci. The LMM was extended to partition total genetic effects into additive and residual genetic random components, and then the complete set of markers was incorporated in the variance of the additive effects as a known additive genomic relationship matrix scaled by an unknown additive variance parameter. The inclusion of the additive term was then tested using a simple likelihood ratio test and if found to be significant at an alpha level of 0.05, an outlier detection method was used to select the marker that was most strongly associated with the trait. That marker was then moved to the fixed component of the LMM as a numerical covariate and any markers within 20 cM of the selected marker were removed from further analysis. This forward selection process was repeated until the term containing the remaining set of markers was non-significant. All QTL analyses and summaries were performed using the wgaim R package (Taylor and Verbyla 2011).
Development and application of marker assays for SNPs in QTL regions
For 15 SNPs that were found to be linked with a QTL affecting resistance to P. thornei, primers for Kompetitive Allele Specific PCR™ (KASP™) marker assays were designed using Kraken™ software (LGC Genomics Limited, Hoddlesdon, UK). These assays were applied to single-plant DNA samples from PBA HatTrick, Kyabra, and 66 other chickpea accessions using an automated SNPLine™ system (LGC Genomics Limited, Hoddlesdon, UK) according to the manufacturer’s instructions.
Results
The aSSD process started with the sowing of 250 F2 seeds. In each generation, the plants flowered between 25 and 28 days after sowing. In the F2 through F4 generations, the number of growing degree days from sowing to harvest ranged from 917 to 981 (taking Tb to be 0 °C, as per Lake et al. (2016)). Between 3 and 10 mature F6 seeds were harvested from each of 218 F5 plants (87.2% of 250). Of these lines, 213 were used for genotyping, 205 were included in the F6 phenotyping experiment, and all 218 were included in the F7 phenotyping experiment.
In both phenotyping experiments, nematode counts were lower for PBA HatTrick (eBLUE = 9.56 ± 0.58 in the F6 experiment and 11.58 ± 0.57 in the F7 experiment) than for Kyabra (eBLUE = 10.43 ± 0.58 in the F6 experiment and 12.71 ± 0.57 in the F7 experiment) and varied among the PBA HatTrick Kyabra RILs (Fig. 1). Among the RILs, there was a significant positive phenotypic correlation between the two experiments (r = 0.35, p < 0.00001) (Fig. 2).
Fig. 1.
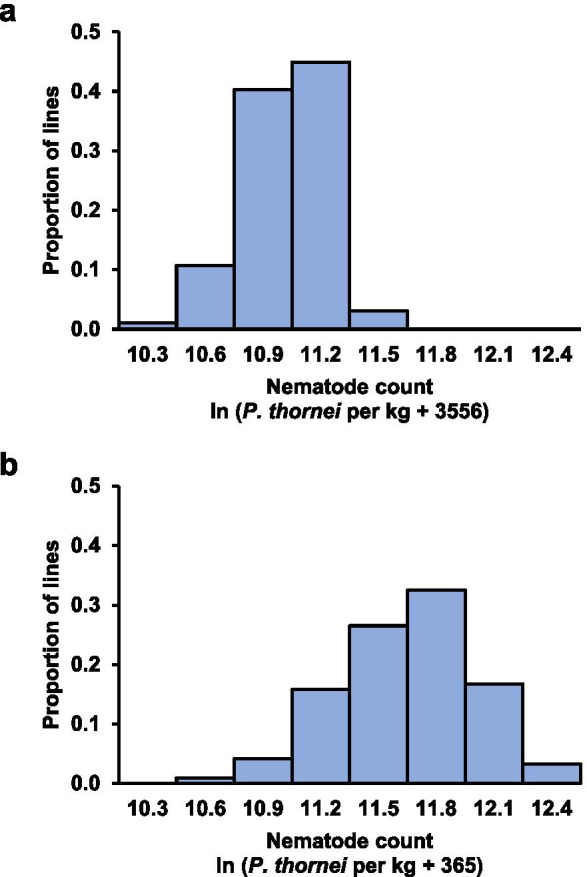
Phenotypic distributions for predictions of log-transformed numbers of Pratylenchus thornei per kg of soil and roots for a population of PBA HatTrick Kyabra recombinant inbred lines (RILs) evaluated in (a) the F6 generation in 2018 and (b) the F7 generation in 2019
Fig. 2.
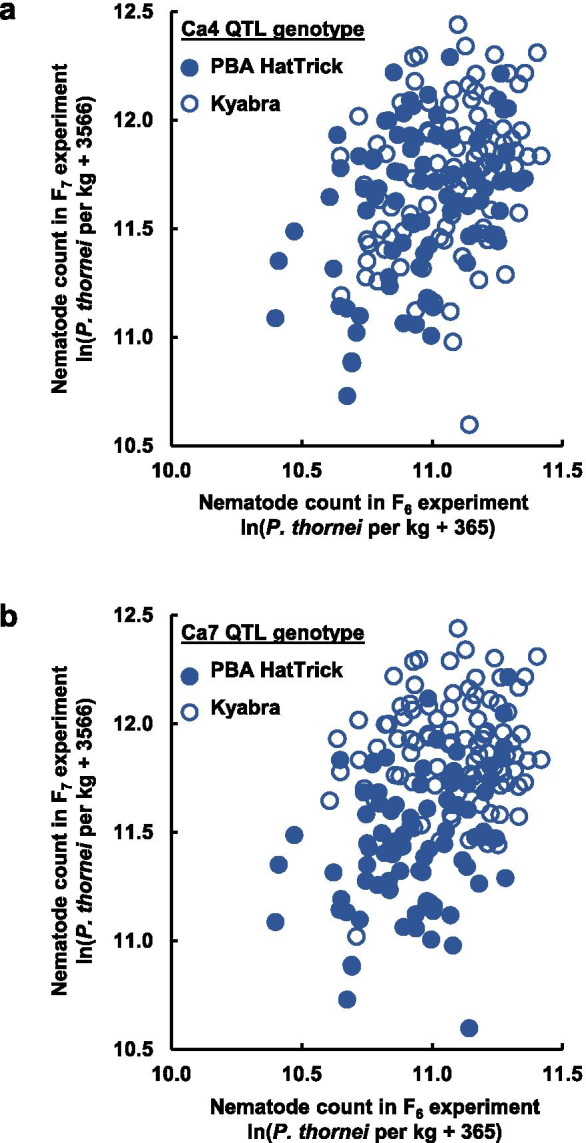
Log-transformed numbers of Pratylenchus thornei per kg of soil and roots, for 181 PBA HatTrick Kyabra recombinant lines evaluated in the F6 generation in 2018 and the F7 generation in 2019. The lines shown are those that are homozygous in QTL regions on the Ca4 and Ca7 chromosomes and that were evaluated in both experiments. Dark and white symbols represent lines with the PBA HatTrick and Kyabra genotypes, respectively at (a) the Ca4 QTL and (b) the Ca7 QTL
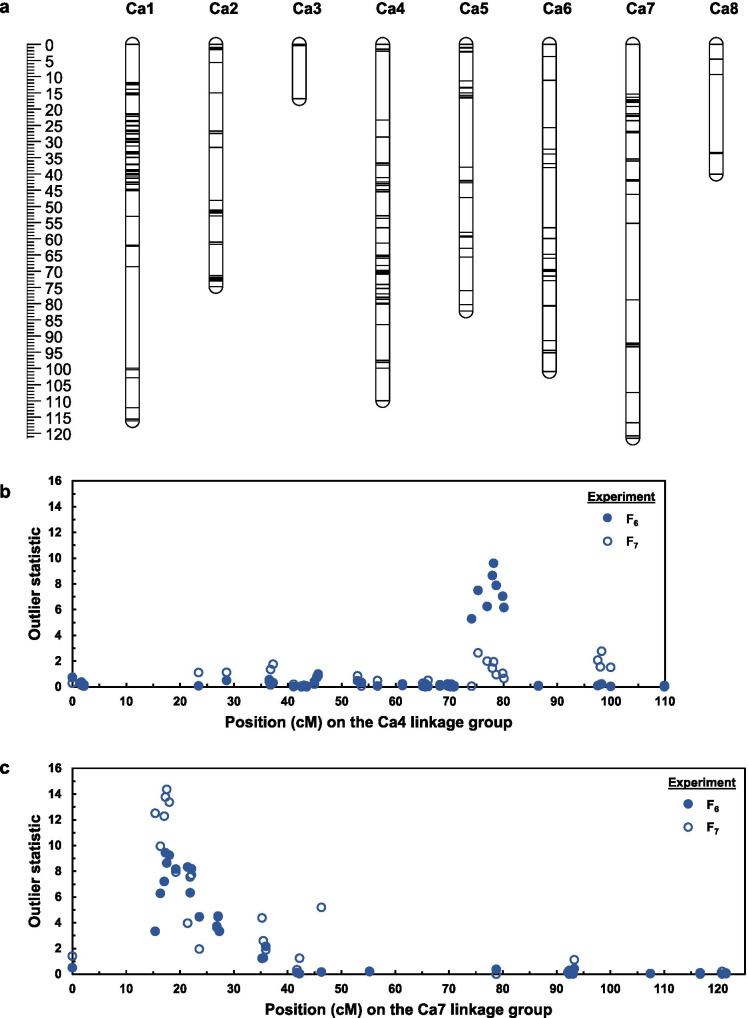
From among 806 tag pairs discovered by DArTseq GBS, 610 were genetically mapped (Online Resource 1) at 211 unique positions (Fig. 3a). Information from physical anchoring of sequence tags to the version 2.6.3 genome assembly for kabuli-type chickpea was sufficient to assign linkage groups to pseudomolecules. Each of the eight chickpea chromosomes was represented by one linkage group. These groups ranged in length from 16.8 to 121.7 cM. The longest intervals between markers on the same linkage group were on the chromosomes corresponding to pseudomolecules Ca1 (31.3 cM), Ca4 (21.3 cM), Ca5 (21.3 cM), Ca7 (23.6 cM), and Ca8 (24.2 cM). Information from physical anchoring of sequence tags was used as basis for orienting linkage groups, but some of the orientations shown in Fig. 3a should be considered tentative, given discrepancies between physical and genetic marker order (Online Resource 1).
Fig. 3.
Linkage map (a) for a PBA HatTrick Kyabra chickpea population and outlier statistics for the association of marker positions on the Ca4 (b) and Ca7 (c) chromosomes with log-transformed numbers of Pratylenchus thornei per kg of soil and roots, as evaluated in the F6 generation in 2018 (dark symbols) and the F7 generation in 2019 (white symbols). In panel a, the horizontal lines on the linkage groups represent positions at which genotyping-by-sequencing tag pairs were mapped and the scale bar on the left shows genetic distances in cM
Two P. thornei resistance QTLs were mapped (Table 1), one on the Ca4 chromosome (detected only in the F6 experiment; Fig. 3b) and one on the Ca7 chromosome (detected in both experiments; Fig. 3c) (Table 1). At each of these loci, the favourable allele (reducing the nematode count) was from PBA HatTrick. For the Ca4 QTL, differences (if any) in nematode numbers between the PBA HatTrick and Kyabra genotypic classes are not obvious (Fig. 2a). For the Ca7 QTL, lines with the PBA HatTrick genotype clearly tend to have fewer nematodes than lines with the Kyabra genotype (Fig. 2b).
Table 1.
QTLs detected for numbers of Pratylenchus thornei in a population of PBA HatTrick Kyabra recombinant inbred lines of chickpea
| Experiment | Chromosome | Marker | Position (cM) | LOD | p | % of genetic variance explained |
|---|---|---|---|---|---|---|
| F6 plants evaluated in 2018 | Ca4 | 10266564|F|0-33:T>G | 78.20 | 2.55 | 0.0003 | 18 |
| Ca7 | 29631748|F|0-45:A>G | 17.32 | 5.96 | < 0.0001 | 40 | |
| F7 plants evaluated in 2019 | Ca7 | 5826018|F|0-44:G>A4 | 17.55 | 22.56 | < 0.0001 | 51 |
At each QTL, alleles from PBA HatTrick were associated with a reduction in the nematode count
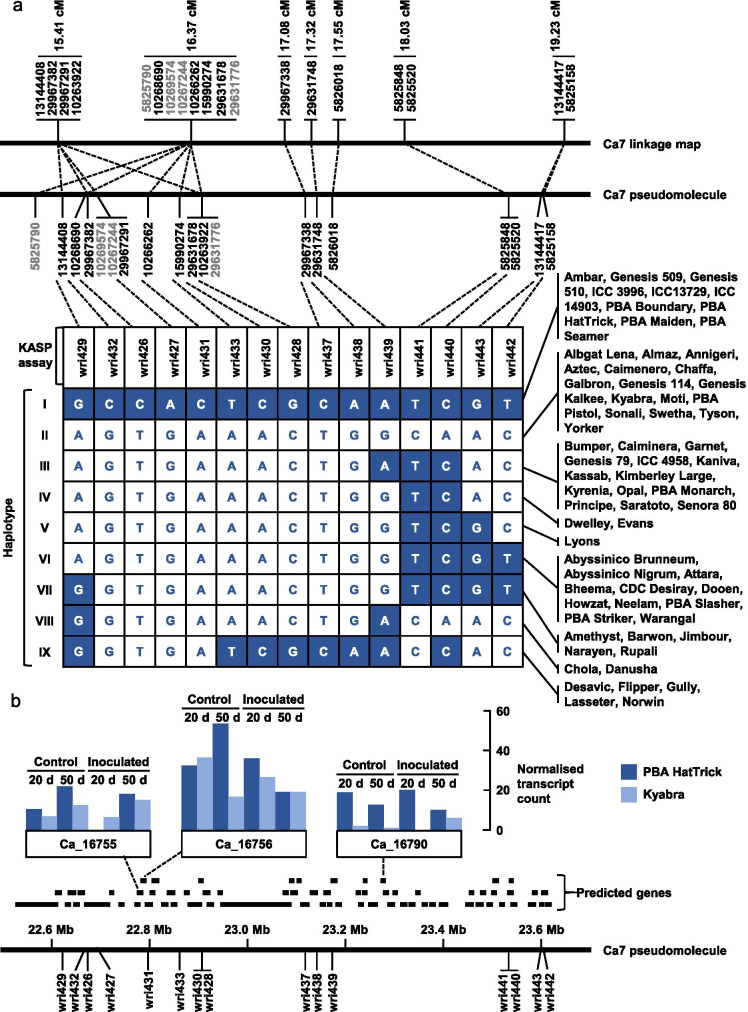
The Ca7 QTL was selected for further investigation. When the 94 GBS tag sequences that were genetically mapped on the same linkage group as this QTL were BLASTed against the version 2.6.3 genome assembly for kabuli-type chickpea, 84 had their best hit on the Ca7 pseudomolecule, nine had their best hit on other pseudomolecules (two on Ca2 and one on each of Ca1, Ca3, Ca4, and Ca6), and three had no hit (Online Resource 1). Within the 56.25 Mb Ca7 pseudomolecule, there are nine regions longer than 3 Mb (ranging from 3.03 to 5.70 Mb) in which no tags were anchored, but there are other regions, including the QTL region, in which many tags were anchored. In the QTL region, all 19 tags that were genetically mapped between 15.41 and 19.23 cM were anchored to physical positions between 22.57 and 23.60 Mb (Online Resource 1). From 16.37 to 19.23 cM, the genetic order was entirely consistent with the physical order (Fig. 4, Online Resource 1).
Fig. 4.
a Genetic positions of 19 genotyping-by-sequencing markers that are within 2 cM of the estimated position of a QTL for resistance of chickpea to Pratylenchus thornei, physical positions at which those markers were anchored on the Ca7 pseudomolecule of version 2.6.3 of a genome assembly for kabuli-type chickpea; and haplotypes observed among 68 chickpea accessions genotyped with KASP assays for 15 of those markers. In the haplotypes, nucleotides shown in white text on a dark background are the same as those carried by the partially resistant parent PBA HatTrick and nucleotides shown in dark text on a white background are the same as those carried by the susceptible parent Kyabra. b Physical positions of 69 genes predicted with high confidence within a 1.03 Mb region of the Ca7 pseudomolecule, with transcript abundance data (from Channale et al. 2021) for three of those genes
KASP assays were designed for 15 SNPs that mapped between 15.41 and 19.23 cM on the Ca7 linkage group (Fig. 4, Online Resource 2). When these assays were applied to 68 chickpea accessions, ten accessions exhibited the PBA HatTrick haplotype, 16 accessions exhibited the Kyabra haplotype, and seven other haplotypes were observed (Fig. 4).
Within the 1.03 Mb region from 22.57 to 23.60 Mb on the Ca7 pseudomolecule, 69 genes have been predicted with high confidence (Online Resource 3). Among these, five (Ca_16754, Ca_16755, Ca_16756, Ca_16784, and Ca_16790) have previously been reported (Channale et al. 2021) to be expressed in roots of PBA HatTrick and Kyabra, with and/or without inoculation with P. thornei (Online resource 4). Transcript abundance results (from Channale et al. 2021) for the three most highly expressed genes (Ca_16755, Ca_16756, and Ca_16790) are shown in Fig. 4b. For Ca_16790, transcripts were consistently more abundant in root tissue from PBA HatTrick than in root tissue from Kyabra.
Discussion
Prior to this research, resistance to P. thornei had been identified within wild relatives of chickpea, including C. arietinum ssp. reticulatum and C. echinospermum, which are cross-compatible with cultivated chickpea. While effort has been made to introgress this resistance into chickpea, this is a lengthy process and may not result in lines that combine adequate resistance with good agronomic performance. To complement this effort, we sought to exploit intraspecific variation within C. arietinum. As there were no reports of any sources of strong resistance in cultivated chickpea, we chose to investigate the genetic basis of the difference between a partially resistant cultivar (PBA HatTrick) and a very susceptible cultivar (Kyabra). To accelerate the development of PBA HatTrick Kyabra RILs for use in genetic mapping, we employed aSSD methods similar to those reported by Croser et al. (2016) and Ribalta et al. (2017) for narrow-leaf lupin (Lupinus angustifolius L.) and pea (Pisum sativum L.) and as applied to chickpea by Atieno et al. (2021). After confirmation of hybridity, F1 plants were vegetatively propagated so that a large number of F2 seeds could be obtained to initiate aSSD. The entire process, from sowing of the parents for crossing through to harvest of F6 seed was completed within just 18 months (compared to 3 to 4 years using conventional glasshouse-based single seed descent). From just 250 F2 seeds sown, 218 F6:7 lines were developed, representing an average attrition rate of just 2.6% per generation.
With GBS, hundreds of SNPs were discovered and genetically mapped. While some regions of the genome had many polymorphic SNPs, others had none, presumably due to PBA HatTrick (pedigree Jimbour/ICC14903, which can be expanded to Amethyst/Barwon//ICC14903) and Kyabra (pedigree Amethyst//946-31/Barwon///Lasseter/940-26//946-31/Norwin////8507-28H//Amethyst/T1069///8507-28H/946-31) having inherited some identical chromosome regions from their common ancestors Amethyst and Barwon. Anchoring of the markers to a genome assembly revealed numerous discrepancies between genetic and physical marker orders, and for some chromosomes, the correct orientation of the linkage group(s) was not clear. This will need to be taken into account if the PBA HatTrick Kyabra population and linkage map are used for other traits. Discrepancies in marker order can be due to errors in the linkage map, errors in the genome assembly or true biological differences in materials. In this case, it seems unlikely that the differences are due to differences between desi-type chickpea (PBA HatTrick, Kyabra, and their progeny) and kabuli-type chickpea (CDC Frontier, the cultivar used for the genome assembly), because we obtained similar results (not shown) with a desi-type genome assembly. Fortunately, there were no discrepancies in marker order within the QTL regions mapped in this research.
As expected, PBA HatTrick exhibited more resistance than Kyabra in both inoculated experiments. The distributions of the phenotypic values observed in the two experiments overlapped with each other, but nematode numbers were considerably higher in the F7 experiment than in the F6 experiment. Such differences between experiments are not unusual for this trait. They may be partly due to seasonal variations in environmental conditions, as the experiments were conducted in a regular glasshouse, not a controlled environment facility. Seed source effects could also be involved in this case, with the F6 seeds having come from F5 plants grown under conditions used for aSSD and the F7 seeds having come from F6 plants grown under more favourable conditions. Despite the differences between experiments, nematode counts varied widely among the RILs in both experiments and were significantly correlated between experiments, providing a basis for QTL mapping.
Of two QTLs that were mapped here, the Ca7 QTL was detected in both experiments, at almost exactly the same position, explaining 40% of genetic variation in one experiment and 51% of genetic variation in the other experiment. That QTL was chosen for further investigation. The Ca4 QTL, which explained less variation in the first experiment and was not detected in the second experiment, was not pursued.
The development and application of KASP assays for 15 SNPs in the Ca7 QTL region made it possible to investigate haplotypes across a panel of chickpea accessions. Given that Jimbour is known to be more susceptible to P. thornei than ICC14903 (Thompson et al. 2011; Rodda et al. 2016), it was expected that PBA HatTrick (Jimbour/ICC14903) would have inherited its partial resistance from its other parent, the Iranian landrace ICC14903. Consistent with this, ICC14903 exhibited the same haplotype as PBA HatTrick. That haplotype was also observed in two other Iranian landraces (ICC3996 and ICC13729) and in six other Australian chickpea cultivars (Ambar, Genesis 509, Genesis 510, PBA Boundary, PBA Maiden and PBA Seamer), all of which have partial resistance (Thompson et al. 2011; Rodda et al. 2016). PBA Seamer (pedigree 9081-3024/PBA HatTrick) would have inherited the resistance-associated haplotype from ICC14903 via PBA HatTrick. Ambar (pedigree ICCV92501/ICC13729//WACPE2021/ICCV96808) could have inherited the same haplotype from ICC13729, while Genesis 509 (pedigree ICC3996/ILC/5928), Genesis 510 (pedigree ICC3996/ILC5928), PBA Boundary (pedigree Jimbour/ICC3996), and PBA Maiden (pedigree Howzat//940-105/ICC3996) would all have inherited it from ICC3996.
Assessment of resistance to root-lesion nematodes requires either substantial labour for nematode counting (as done here and by Reen et al. (2019)) or DNA-based quantification of nematodes in soil and roots (as done by Rodda et al. (2016)). Both methods are too costly for large-scale application in breeding programmes. Accordingly, this trait is a good candidate for marker-assisted selection. The best marker(s) for this purpose would be closely linked with the QTL position and would distinguish the favourable haplotype from other haplotypes in chickpea breeding germplasm. All of the markers shown in Fig. 4 are very closely linked with the estimated QTL position. Among these markers, the one that is most closely linked with the QTL position, wri438 (at 17.3 cM and 23.1 Mb), distinguishes the PBA HatTrick haplotype from all other haplotypes except haplotype IX, as do the nearby markers wri437 (at 23.1 Mb) and wri428, wri430, and wri530 (at 22.9 Mb). The markers wri431 (at 22.8 Mb) and wri427, wri426, and wri432 (at 22.7 Mb) all distinguish the PBA HatTrick haplotype from all other observed haplotypes. Given that Desavic, one of the haplotype IX cultivars, is known to be quite susceptible (Thompson et al. 2011), it seems unlikely that haplotype IX confers resistance. On this basis, the four markers at 22.7–22.8 Mb (wri426, wri427, wri431, and wri432) can be expected to be more diagnostic of resistance across a wide range of germplasm than the five markers at 22.9–23.1 Mb.
Based on anchoring of marker sequences to the Ca7 pseudomolecule, a candidate region of 1.03 Mb was selected and a list of 69 high-confidence predicted genes was compiled (Online Resource 3). Given that no genes for resistance against migratory endoparasitic nematodes have been isolated from any host plant and that little is known about resistance mechanisms (Mathew and Opperman 2020), none of these genes can be excluded solely based on annotation. Nevertheless, two of these predicted genes (Ca_16763 at 22.9 Mb and Ca_16772 at 23.1 Mb) may be stronger candidates than others, in that they are similar to the Medicago truncatula genes Medtr8g020290.1 and Medtr8g020300.1, respectively. These genes belong to the Toll interleukin-1 receptor-nucleotide binding site-leucine rich repeat (TIR-NBS-LRR) family. TIR-NBS-LRR genes have been implicated in responses to biotic stress responses in many plants, including chickpea (Sagi et al. 2017). The potato (Solanum tuberosum L.) Gro1-4 gene, which confers resistance to the golden nematode Globodera rostochiensis Wollenwebber, 1923 (a sedentary endoparasite), is a TIR-NBS-LRR gene (Paal et al. 2004).
Five predicted genes in the candidate region have previously been reported to be expressed in roots of PBA HatTrick and Kyabra (Channale et al. 2021). For two of these genes (Ca_16754 and Ca_16784), Channale et al. (2021) detected transcripts only at low levels and only in mock-inoculated control plants of PBA HatTrick. For the other three genes (Ca_16755, Ca_16756, and Ca_16790), transcripts were detected at high levels in both PBA HatTrick and Kyabra and in both P. thornei-inoculated and mock-inoculated control plants. Two of these genes are similar to genes encoding transcription factors in M. truncatula: an alfin-like transcription factor (Medtr8g019520.2, similar to Ca_16755) and a CCAAT-binding transcription factor (Medtr8g019540.1, similar to Ca_16756). Both of these genes are between two of the most diagnostic markers: 29967291|F|0-44:G>A (wri427; 16.3 cM ; 22.7 Mb) and 10266262|F|0-30:C>A (wri431; 16.37 cM; 22.8 Mb). The third highly expressed gene (Ca_16790) has no known homolog in either M. truncatula or Arabidopsis thaliana, but is of particular interest because its transcript abundance was consistently higher in the roots of PBA HatTrick than in the roots of Kyabra (Fig. 4b). The position of this gene on the Ca7 pseudomolecule (23.3 Mb) is just distal to the marker that was the most significant in the F7 experiment: marker 5826018|F|0-44:G>A (wri439; 17.55 cM; 23.2 Mb). In future research aimed at identifying the resistance gene, Ca_16790 could be given priority for functional analysis, and Ca_16755, Ca_16756, Ca_16763, and Ca16772 may also be worth investigating. However, based on current evidence, it would be premature to definitively exclude any of the other 64 predicted genes in the candidate region. Higher resolution genetic mapping could be pursued to narrow the candidate region. Within the PBA HatTrick Kyabra population, there are a number of lines for which the genotyped F6 plant was heterozygous in the Ca7 QTL region (Online Resource 1). These materials could be useful for generating recombinant progeny for fine mapping.
Given that the favourable allele at the QTL discovered here is present in PBA HatTrick and some other recent cultivars (Ambar, PBA Boundary, PBA Maiden, and PBA Seamer), there is an immediate opportunity to apply marker-assisted selection in chickpea breeding, providing some protection to chickpea and to wheat and other crops that are grown in rotation with chickpea. More complete protection would require discovery of additional resistance loci within cultivated chickpea and/or introgression of resistance alleles from wild relatives. Potential sources that have already been identified in C. reticulatum and C. echinospermum (Thompson et al. 2011; Rodda et al. 2016; Reen et al. 2019) could be used as parents for further genetic mapping and to develop molecular markers for the loci responsible for their resistance.
Supplementary Information
Below is the link to the electronic supplementary material.
Genotypic data from genotyping-by-sequencing, and phenotypic data for resistance against Pratylenchus thornei for a PBA HatTrick × Kyabra chickpea mapping population, with marker positions on a genetic linkage map and on the pseudomolecules of version 2.6.3 of a genome assembly for kabuli-type chickpea (XLSX 573 KB)
rimer sequences for 15 KASP assays designed for single nucleotide polymorphisms in a QTL region on the Ca7 chromosome of chickpea (XLSX 16 KB)
Genes predicted with high confidence within a candidate region on the Ca7 pseudomolecule of kabuli chickpea (XLSX 67 KB)
Normalised transcript counts and PBA HatTrick:Kyabra fold differences for five predicted genes that had previously been reported to be expressed in root tissue of PBA HatTrick and/or Kyabra chickpea (XLSX 15 KB)
Acknowledgements
This work was supported by the Grains Research and Development Corporation (project UA00157). The authors thank Jing Lin, Federico Ribalta, Hannah Rostad, Karri Thomas, Michelle Thompson, Mei Wang and Xiaoyun Wang for assistance with the experimental work, and Kristy Hobson and Yongle Li for helpful advice.
Author contribution
Conceptualisation: Diane Mather, Kelvin Khoo, Tim Sutton, John Thompson, and Jason Sheedy; Materials development: Janine Croser, Julie Hayes, and Kelvin Khoo; Assay design and application: Kelvin Khoo; Data collection: Kelvin Khoo and Jason Sheedy; Experimental design and statistical analysis: Julian Taylor; Writing (initial drafts): Diane Mather and Kelvin Khoo; Writing (review and editing): all authors. Funding acquisition and project supervision: Diane Mather and Tim Sutton.
Funding
This work was supported by the Grains Research and Development Corporation (project UA00157).
Availability of data and materials
The data generated during this research are included in this published article and its supplementary information files. Seed of mapping lines can be made available upon reasonable request to the corresponding authors, subject to approval by the Grains Research and Development Corporation.
Code availability
Not applicable
Declarations
Conflict of interest/Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Atieno J, Colmer TD, Taylor J, Li Y, Quealy J, Kotula L, Nicol D, Nguyen DT, Brien C, Langridge P, Croser J, Hayes JE, Sutton T. Novel salinity tolerance loci in chickpea identified in glasshouse and field environments. Front Plant Sci. 2021;12:629. doi: 10.3389/fpls.2021.667910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz İ, Kasapoğlu Uludamar EB, Behmand T, Elekcioğlu H. Investigation of the development of root lesion nematodes, Pratylenchus spp. (Tylenchida: Pratylenchidae) in three chickpea cultivars. Turk J Entomol. 2021;45:23–31. [Google Scholar]
- Aznar-Fernández T, Barilli E, Cobos MJ, Kilian A, Carling J, Rubiales D. Identification of quantitative trait loci (QTL) controlling resistance to pea weevil (Bruchus pisorum) in a high-density integrated DArTseq SNP-based genetic map of pea. Sci Rep. 2020;10:33. doi: 10.1038/s41598-019-56987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler DG, Cullis BR, Gilmour AR, Gogel BJ. Thompson R (2017) ASReml-R Reference Manual Version 4VSN International Ltd, Hemel Hempstead UK https://asreml.kb.vsni.co.uk/wp-content/uploads/sites/3/ASReml-R-Reference-Manual-4.pdf
- Castillo P, Jiménez-Díaz RM, Gomez-Barcina A, Vovlas N. Parasitism of the root-lesion nematode Pratylenchus thornei on chickpea. Plant Pathol. 1995;44:728–733. doi: 10.1111/j.1365-3059.1995.tb01697.x. [DOI] [Google Scholar]
- Channale S, Kalavikatte D, Thompson JP, Kudapa H, Bajag P, Varshney RK, Zwart RS, Thudi M. Transcriptome analysis reveals key genes associated with root-lesion nematode Pratylenchus thornei resistance in chickpea. Sci Rep. 2021;11:17491. doi: 10.1038/s41598-021-96906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croser JS, Pazos-Navarro M, Bennett R, Tschirren S, Edwards K, Erskine W, Creasy R, Ribalta F. Time to flower of temperate pulses in vivo and generation turnover in vivo-in vitro of narrow-leaf lupin accelerated by low red to far-red ratio and high intensity in the far-red region. Plant Cell Tiss Organ Cult. 2016;12:591–599. doi: 10.1007/s11240-016-1092-4. [DOI] [Google Scholar]
- Danehloueipour N, Yan G, Clarke HJ, Siddique KHM. Successful stem cutting propagation of chickpea, its wild relatives and their interspecific hybrids. Aust J Exp Agric. 2006;46:1349–1354. doi: 10.1071/EA05207. [DOI] [Google Scholar]
- Di Vito M, Zaccheo G, Catalano F. Response of chickpea lines to Meloidogyne artiellia and Pratylenchus thornei. Nematol Medit. 1995;23:81–83. [Google Scholar]
- Edwards D (2016) Improved kabuli reference genome. CyVerse Data Commons.10.7946/P2G596https://datacommons.cyverse.org Accessed 23 November 2020
- Gilmour A, Cullis B, Verbyla A. Accounting for natural and extraneous variation in the analysis of field experiments. J Agric Biol Environ Stat. 1997;2:269–293. doi: 10.2307/1400446. [DOI] [Google Scholar]
- Kalve S, Tadege M. A comprehensive technique for artificial hybridization in chickpea (Cicer arietinum) Plant Methods. 2017;13:52. doi: 10.1186/s13007-017-0202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake L, Chenu K, Sadras VO. Patterns of water stress and temperature for Australian chickpea production. Crop Pasture Sci. 2016;67:204–214. doi: 10.1071/CP15253. [DOI] [Google Scholar]
- Linsell KJ, Rahman MS, Taylor JD, Davey RS, Gogel BJ, Wallwork H, Forrest KL, Hayden MJ, Taylor SP, Oldach KH. QTL for resistance to root-lesion nematode (Pratylenchus thornei) from a synthetic hexaploid wheat source. Theor Appl Genet. 2014;127:1409–1421. doi: 10.1007/s00122-014-2308-9. [DOI] [PubMed] [Google Scholar]
- Martínez O, Curnow RN. Estimating the locations and the sizes of the effects of quantitative trait loci using flanking markers. Theor Appl Genet. 1992;85:480–488. doi: 10.1007/BF00222330. [DOI] [PubMed] [Google Scholar]
- Mathew R, Opperman CH. Current insights into migratory endoparasitism: deciphering the biology, parasitic mechanisms and management strategies of key migratory endoparasitic phytonematodes. Plants. 2020;9:671. doi: 10.3390/plants9060671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkov ND. Moisture sorption isotherms of chickpea seeds at several temperatures. J Food Eng. 2000;45:189–194. doi: 10.1016/S0260-8774(00)00052-2. [DOI] [Google Scholar]
- Murray GM, Brennan JP. Estimating disease losses to the Australian wheat industry. Australas Plant Pathol. 2009;38:558–570. doi: 10.1071/AP09053. [DOI] [Google Scholar]
- Murray GM, Brennan JP. The current and potential costs from diseases of pulse crops in Australia. Kingston, ACT, Australia: Grains Research and Development Corporation; 2012. [Google Scholar]
- Paal J, Henselewski H, Muth J, Meksem K, Menéndez CM, Salamini F, Balvora A, Gebhardt C. Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype R01 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 2004;2:285–297. doi: 10.1111/j.1365-313X.2004.02047.x. [DOI] [PubMed] [Google Scholar]
- Pallotta MA, Graham RD, Langridge P, Sparrow DHB, Barker SJ. RFLP mapping of manganese efficiency in barley. Theor Appl Genet. 2000;101:1100–1108. doi: 10.1007/s001220051585. [DOI] [Google Scholar]
- R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria https://www.R-project.org
- Rahman MS, Linsell KJ, Taylor JD, Hayden MJ, Collins NC, Oldach KH. Fine mapping of root-lesion nematode (Pratylenchus thornei) resistance loci on chromosomes 6D and 2B of wheat. Theor Appl Genet. 2019;133:635–652. doi: 10.1007/s00122-019-03495-x. [DOI] [PubMed] [Google Scholar]
- Reen RA, Mumford MH, Thompson JP. Novel sources of resistance to root-lesion nematode (Pratylenchus thornei) in a new collection of wild Cicer species (C. reticulatum and C. echinospermum) to improve resistance in cultivated chickpea (C. arietinum) Phytopathology. 2019;109:1270–1279. doi: 10.1094/PHYTO-02-19-0047-R. [DOI] [PubMed] [Google Scholar]
- Ribalta FM, Pazos-Navarro M, Nelson K, Edwards K, Bennett RG, Munday C, Ross JJ, Erskine W, Ochatt SJ, Croser JS. Precocious floral initiation and identification of exact timing of embryo physiological maturity facilitate germination of immature seeds to truncate the lifecycle of pea. Plant Growth Regul. 2017;81:3456–3353. doi: 10.1007/s10725-016-0211-x. [DOI] [Google Scholar]
- Rodda MS, Hobson KB, Forknall CR, Daniel RP, Fanning JP, Pounsett DD, Simpfendorfer S, Moore KJ, Owen KJ, Sheedy JG, Thompson JP, Hollaway GJ, Slater AT. Highly heritable resistance to root-lesion nematode (Pratylenchus thornei) in Australian chickpea germplasm observed using an optimised glasshouse method and multi-environment trial analysis. Australas Plant Pathol. 2016;45:309–319. doi: 10.1007/s13313-016-0409-4. [DOI] [Google Scholar]
- Rogowsky PM, Guidet FLY, Langridge P, Shepherd KW, Koebner RMD. Isolation and characterization of wheat-rye recombinants involving chromosome arm 1DS of wheat. Theor Appl Genet. 1991;82:537–544. doi: 10.1007/BF00226788. [DOI] [PubMed] [Google Scholar]
- Sagi MS, Deokar AA, Taran B. Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to aschochyta blight infection. Front Plant Sci. 2017;8:838. doi: 10.3389/fpls.2017.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy JG, Thompson JP. Resistance to the root-lesion nematode Pratylenchus thornei of Iranian landrace wheat. Australas Plant Pathol. 2009;38:478–489. doi: 10.1071/AP09030. [DOI] [Google Scholar]
- Sheedy JG, Thompson JP, Kelly A. Diploid and tetraploid progenitors of wheat are valuable sources of resistance to the root-lesion nematode Pratylenchus thornei. Euphytica. 2012;186:377–391. doi: 10.1007/s10681-011-0617-5. [DOI] [Google Scholar]
- Taylor J, Butler D. R Package ASMap: efficient genetic linkage map construction and diagnosis. J Stat Softw. 2017;79(6):1–29. doi: 10.18637/jss.v079.i06. [DOI] [Google Scholar]
- Taylor J, Verbyla A. R Package wgaim: QTL analysis in bi-parental populations using linear mixed models. J Stat Softw. 2011;40(7):1–18. doi: 10.18637/jss.v040.i07. [DOI] [Google Scholar]
- Thompson JP. Resistance to root-lesion nematodes (Pratylenchus thornei and P. neglectus) in synthetic hexaploid wheats and their durum and Aegilops tauschii parents. Aust Agric Res. 2008;59:432–446. doi: 10.1071/AR07222. [DOI] [Google Scholar]
- Thompson JP, Haak MI. Resistance to root-lesion nematode (Pratylenchus thornei) in Aegilops tauschii Coss, the D-genome donor to wheat. Aust J Agric Res. 1997;48:553–559. doi: 10.1071/A96167. [DOI] [Google Scholar]
- Thompson JP, O’Reilly MM, Clewett TG. Resistance to the root-lesion nematode Pratylenchus thornei in wheat landraces and cultivars from the West Asia and North Africa (WANA) region. Crop Pasture Sci. 2009;60:1209–1217. doi: 10.1071/CP09159. [DOI] [Google Scholar]
- Thompson JP, Reen RA, Clewett TG, Sheedy JG, Kelly AM, Gogel BJ, Knights EJ. Hybridisation of Australian chickpea cultivars with wild Cicer spp. increases resistance to root-lesion nematodes (Pratylenchus thornei and P. neglectus) Australas Plant Pathol. 2011;40:601–611. doi: 10.1007/s13313-011-0089-z. [DOI] [Google Scholar]
- Thompson JP, Zwart RS, Butler D. Inheritance of resistance to root-lesion nematodes (Pratylenchus thornei and P. neglectus) in five doubled-haploid populations of wheat. Euphytica. 2012;188:209–219. doi: 10.1007/s10681-012-0689-x. [DOI] [Google Scholar]
- Tiwari SP, Vadhera I, Shukla BN, Bhatt J. Studies on the pathogenicity and relative reactions of chickpea lines to Pratylenchus thornei (Filipjev. 1936) Sher & Allen. 1953. Indian J Mycol Plant Pathol. 1992;22:255–259. [Google Scholar]
- Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Rosen BD, Tar’an B, Millan T, Zhang X, Ramsay LD, Iwata A, Wang Y, Nelson W, Farmer AD, Gaur PM, Soderlund C, Varma Penmetsa V, Xu C, Bharti AK, He W, Winter P, Zhao S, Hane JK, Carrasquilla-Garcia N, Condie JA, Upadhaya HD, Luo M-C, Thudi M, Gowda CLL, Singh NP, Lichtenzveig J, Gali KK, Rubio J, Nadarajan N, Dolezel J, Bansal KC, Xu X, Edwards D, Zhang G, Kahl G, Gil J, Singh KB, Datta SK, Jackson SA, Wang J, Cook DR. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotech. 2013;31:240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S. 4. New York: Springer; 2002. [Google Scholar]
- Verbyla AP, Cullis BR, Thompson R. The analysis of QTL by simultaneous use of the full linkage map. Theor Appl Genet. 2007;116:95–111. doi: 10.1007/s00122-007-0650-x. [DOI] [PubMed] [Google Scholar]
- Verbyla AP, Taylor JD, Verbyla KL. RWGAIM: an efficient high-dimensional whole genome average (QTL) interval mapping approach. Genet Res. 2012;94:291–306. doi: 10.1017/S0016672312000493. [DOI] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Whitehead AG, Hemming JR. A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann Appl Biol. 1965;55:25–38. doi: 10.1111/j.1744-7348.1965.tb07864.x. [DOI] [Google Scholar]
- Wu Y, Bhat PR, Close TJ, Lonardi S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLOS Genet. 2008;4:e1000212. doi: 10.1371/journal.pgen.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart RS, Thompson JP, Godwin ID. Genetic analysis of resistance to root-lesion nematode (Pratylenchus thornei) in wheat. Plant Breed. 2004;123:209–212. doi: 10.1111/j.1439-0523.2004.00986.x. [DOI] [Google Scholar]
- Zwart RS, Thompson JP, Godwin ID. Identification of quantitative trait loci for resistance to two species of root-lesion nematode (Pratylenchus thornei and P. neglectus) in wheat. Aust J Agric Res. 2005;56:345–352. doi: 10.1071/AR04223. [DOI] [Google Scholar]
- Zwart RS, Thudi M, Channale S, Manchikatla PK, Varshney RK, Thompson JP. Resistance to plant-parasitic nematodes in chickpea: current status and future perspectives. Front Plant Sci. 2019;10:966. doi: 10.3389/fpls.2019.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotypic data from genotyping-by-sequencing, and phenotypic data for resistance against Pratylenchus thornei for a PBA HatTrick × Kyabra chickpea mapping population, with marker positions on a genetic linkage map and on the pseudomolecules of version 2.6.3 of a genome assembly for kabuli-type chickpea (XLSX 573 KB)
rimer sequences for 15 KASP assays designed for single nucleotide polymorphisms in a QTL region on the Ca7 chromosome of chickpea (XLSX 16 KB)
Genes predicted with high confidence within a candidate region on the Ca7 pseudomolecule of kabuli chickpea (XLSX 67 KB)
Normalised transcript counts and PBA HatTrick:Kyabra fold differences for five predicted genes that had previously been reported to be expressed in root tissue of PBA HatTrick and/or Kyabra chickpea (XLSX 15 KB)
Data Availability Statement
The data generated during this research are included in this published article and its supplementary information files. Seed of mapping lines can be made available upon reasonable request to the corresponding authors, subject to approval by the Grains Research and Development Corporation.
Not applicable