Abstract
Meta-QTL analysis for thermotolerance in wheat was conducted to identify robust meta-QTLs (MQTLs). In this study, 441 QTLs related to 31 heat-responsive traits were projected on the consensus map with 50,310 markers. This exercise resulted in the identification of 85 MQTLs with confidence interval (CI) ranging from 0.11 to 34.9 cM with an average of 5.6 cM. This amounted to a 2.96-fold reduction relative to the mean CI (16.5 cM) of the QTLs used. Seventy-seven (77) of these MQTLs were also compared and verified with the results of recent genome-wide association studies (GWAS). The 85 MQTLs included seven MQTLs that are particularly useful for breeding purposes (we called them breeders’ MQTLs). Seven ortho-MQTLs between wheat and rice genomes were also identified using synteny and collinearity. The MQTLs were used for the identification of 1,704 candidate genes (CGs). In silico expression analysis of these CGs permitted identification of 182 differentially expressed genes (DEGs), which included 36 high confidence CGs with known functions previously reported to be important for thermotolerance. These high confidence CGs encoded proteins belonging to the following families: protein kinase, WD40 repeat, glycosyltransferase, ribosomal protein, SNARE associated Golgi protein, GDSL lipase/esterase, SANT/Myb domain, K homology domain, etc. Thus, the present study resulted in the identification of MQTLs (including breeders’ MQTLs), ortho-MQTLs, and underlying CGs, which could prove useful not only for molecular breeding for the development of thermotolerant wheat cultivars but also for future studies focused on understanding the molecular basis of thermotolerance.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01264-7.
Keywords: Wheat, Heat stress tolerance, Meta-QTLs, Ortho-MQTLs, GWAS, Candidate genes
Introduction
Wheat (Triticum aestivum L.) is grown in the tropical and sub-tropical regions of the world, and is plagued with various abiotic stresses (Gupta et al. 2020). Heat, drought, floods, salinity, cold, and toxic chemicals are all prominent abiotic stresses, which adversely affect crop productivity (Lesk et al. 2016). At the global level, heat and drought are recognized as the two most widely occurring abiotic stresses reducing wheat yield significantly (Gupta et al. 2012, 2020). According to a global climate change model, the average ambient temperature is expected to rise by 6 °C by the end of the twenty-first century (De Costa 2011). In India, the temperature during the wheat growing season increased at a rate of 0.04 °C per decade during the twentieth century (1901–2000) (Chauhan et al. 2014). A recent estimate suggested that wheat yield can sometimes be reduced up to 60% in the field due to extreme heat stress (Zampieri et al. 2017). According to several studies on the effect of rising temperature on wheat grain yield, it is now known that every 1 °C rise in average temperature reduces global wheat yield by 6% (Asseng et al. 2011). This rise of 1 °C during the grain filling period can cause a yield loss of 0.25 tons ha−1 (~ 8%, assuming a global average yield of 3,200 kg ha−1) in winter wheat (Kristensen et al. 2011). In another study, a rise of 2 °C was estimated to reduce global wheat yields by 11% (Zaveri and Lobell 2019). Also, based on simulation studies involving agronomic trials conducted during 1985 to 2011, a 1 °C rise in mean temperature during reproductive stages was shown to cause 21% reduction in wheat yield (Barkley et al. 2014).
Studies have also been conducted on the effect of heat stress on physiological traits associated with yield losses. Based on these studies, reduction in wheat yield due to heat stress has been shown to be associated with the following traits: poor seed germination, inactivation of photosynthetic enzymes, reduced photosynthetic capability, slower nutrient transport, lower chlorophyll content, premature leaf senescence, poor pollen viability/germination, shorter grain filling period, fewer and lighter grains and poor starch/protein content resulting into poor bread-making quality (Akter and Rafiqul Islam 2017; Bheemanahalli et al. 2019; Jacott and Boden 2020; Jagdish 2020; Djanaguiraman et al. 2020; Poudel and Poudel 2020).
In view of the above, a breeding strategy that enables plants to become resilient to high-temperature stress appears to be an effective and sustainable solution. Therefore, the development of heat-tolerant, high-yielding wheat varieties, and/or pre-breeding materials for future breeding programs aimed at breeding for heat tolerance is crucial for food and nutritional security. Therefore, a complete understanding of the genetics of thermotolerance, which depends on a number of interacting QTLs/genes, should certainly help in the development of heat-tolerant cultivars.
Hundreds of QTLs are now known in wheat {including both tetraploid (4x) and hexaploid (6x)}, which control various agronomic and quality traits under heat stress (WheatQTLdb by Singh et al. 2021; http://www.wheatqtldb.net/). However, limited efforts have been made for the discovery of meta-QTLs (MQTLs) for heat tolerance in wheat (Acuña-Galindo et al. 2015; Liu et al. 2020). BioMercator (Arcade et al. 2004; Sosnowski et al. 2012) and MetaQTL (Veyrieras et al. 2007) are the two most versatile software packages available for meta-QTL analysis. Algorithms furnished in BioMercator v2.0 estimate the most appropriate MQTLs using the available information. Meta-QTL analysis may be followed by the selection of promising MQTLs (breeders’ QTLs) for marker-assisted selection (MAS) (Goffinet and Gerber 2000; Yu et al. 2014; Maccaferri et al. 2015) and may also be used for identification of candidate genes (CGs) for future studies.
In the recent past, MQTL analysis in wheat has been carried out successfully to detect robust and stable genomic regions for a variety of traits, including the following: (i) grain yield and associated traits (Gegas et al. 2010; Bilgrami et al. 2020; Saini et al. 2021; Yang et al. 2021), (ii) root morphology traits (Ramandi et al. 2017; Soriano and Alvaro 2019), (iii) potassium use efficiency (Safdar et al. 2020), (iv) nutritional and quality traits (Quraishi et al. 2011, 2017; Shariatipour et al. 2021), (v) fusarium head blight resistance (Liu et al. 2009; Mao et al. 2010; Cai et al. 2019; Venske et al. 2019; Zheng et al. 2020), (vi) leaf rust resistance (Soriano and Royo 2015; Aduragbemi and Soriano 2021), (vii) yellow rust resistance (Jan et al. 2021), (viii) pre-harvest sprouting tolerance (Tyagi and Gupta 2012; Tai et al. 2021), and (ix) drought tolerance (Kumar et al. 2020).
Two studies, which have already been conducted on MQTL analysis for tolerance against heat stress in wheat (Acuña-Galindo et al. 2015; Liu et al. 2020), gave only limited information. In one study, two MQTLs and in the other study, 36 MQTLs for heat tolerance alone were reported (Acuña-Galindo et al. 2015; Liu et al. 2020). Acuña-Galindo et al. (2015) reported 43 MQTLs, which provided tolerance against both heat and drought stresses. The recent study by Liu et al. (2020) reported 59 MQTLs using QTLs associated with grain yield and its major components only, thus leaving out several other important traits such as stay green habit, chlorophyll content, and chlorophyll fluorescence. Since a large number of studies involving both interval mapping and GWAS for heat stress tolerance have been reported in wheat during the past decade (see WheatQTLdb by Singh et al. 2021), further MQTL analysis for heat-responsive traits is warranted.
The present study on MQTL and ortho-MQTL analysis was undertaken using the information on available QTLs for a number of traits related to heat tolerance (or thermotolerance) in wheat. MQTLs in rice along with information on synteny and collinearity between wheat and rice genomes (Ahn et al. 1993; Kumar et al. 2009) were also utilized for identification of ortho-MQTLs. The MQTLs identified in the present study were also verified using available information on GWAS and known genes for thermotolerance in wheat. Important genes including high confidence CGs relevant for thermotolerance in wheat were also identified using the information on MQTLs and the reference sequence of the hexaploid wheat genome (Zhu et al. 2021).
Materials and methods
Bibliographic search and collection of data on QTLs
A comprehensive search using Google Scholar (https://scholar.google.com/) and PubMed (https://pubmed.ncbi.nlm.nih.gov/) was made for reports on QTLs associated with thermotolerance in wheat {including both 4 × and 6x (spring and winter) wheats}; this information was supplemented by the information available in WheatQTLdb (see Singh et al. 2021; http://www.wheatqtldb.net/). The details of parental crosses, populations type/size, traits and markers associated with QTL are given in Online Resource 1. The following information for each QTL was collected: (i) traits/parameters relevant to thermotolerance, (ii) most closely associated markers, (iii) chromosomal location of associated markers and QTLs, (iv) logarithm of the odds (LOD) value for each QTL, and (v) percent phenotypic variation explained (PVE%) or R2 value for individual QTLs (Online Resource 2). All QTLs were treated as single QTLs, even if they were discovered for more than one trait or detected in more than one environment. Some studies reported QTLs separately under normal (control) and heat-stress conditions; however, we considered only QTLs that were identified under heat stress conditions.
Construction of the consensus map
A saturated consensus genetic map (described as Wheat_Consensus_map_2021) was generated using the following integrated genetic maps: (i) the “Wheat Consensus SSR 2004” with 1258 marker loci (Somers et al. 2004); (ii) an integrated map for durum wheat with 3618 markers (Marone et al. 2013); (iii) the SNP-based consensus map of tetraploid wheat with 30,144 markers (Maccaferri et al. 2015); (iv) SNP-based genetic map constructed with 46,977 loci from the Illumina iSelect 90K SNP Array (Wang et al. 2014); and (v) individual genetic maps for different mapping populations reported in previous studies on QTL analysis (Online Resources 1 and 2). The number of common and unique markers among the four individual maps was calculated and illustrated using custom Venn diagrams webtool (https://bioinformatics.psb.ugent.be/webtools/Venn/). The R package LPMerge (Endelman and Plomion, 2014) was used for the construction of the Wheat_Consensus_map_2021. This map was used as a reference map for QTL projection and identification of MQTLs.
Projection of QTLs on the consensus map
The projection of QTLs onto the Wheat_Consensus_map_2021 was performed by BioMercator v4.2 (https://urgi.versailles.inra.fr/Tools/BioMercator-V4; Arcade et al. 2004) using the QTLProj command. QTLs were projected on the consensus map following the homothetic approach proposed by Chardon et al. (2004). When confidence interval (CI) was not reported in the original studies, the genetic distance between flanking markers was taken as the CI. When the QTL peak position was available, CI (95%) was calculated for each locus using different population-specific equations (Darvasi and Soller 1997; Guo et al. 2006; Liu et al. 2009), which included the following: (i) for backcross and F2 populations: CI = 530/NR2; (ii) for doubled haploid (DH) lines: CI = 287/NR2, and (iii) for recombinant inbred lines (RILs), CI was calculated as 163/NR2, where N is the population size and R2 is the PVE% by the QTL.
Meta-analysis of the QTLs
QTL meta-analysis was performed for each wheat chromosome, separately, using algorithms implemented in BioMercator v4.2 (Goffinet and Gerber 2000; Veyrieras et al. 2007). Depending on the number of projected QTLs per chromosome, the following two distinct approaches were applied: (i) when the number of QTLs per chromosome was 10 or less, the methods proposed by Goffinet and Gerber (2000) was applied, and (ii) when the number of QTLs per chromosome was > 10, the method proposed by Veyrieras et al. (2007) was utilized. In the first approach, the model with the lowest Akaike Information Criterion (AIC) value was used to select the best QTL model for determining the number of MQTLs on each consensus chromosome; this approach was followed in several earlier meta-QTL studies (Lu et al. 2018; Venske et al. 2019; Liu et al. 2020). In the second approach, the following five parameters were used to ascertain the correct number of MQTLs (Veyrieras et al. 2007): (i) Akaike Information Criterion (AIC), (ii) AIC correction (AICc), (iii) AIC3 candidate model, (iv) Bayesian Information Criterion (BIC), and (v) Approximate Weight of Evidence Criterion (AWE). The model with the lowest value in at least three of the five parameters was chosen as the best fit (Chardon et al. 2004). In our meta-analysis, the QTLs identified from the best-fit model were considered as the MQTL. The number of MQTLs on each wheat chromosome was determined based on the genetic positions and 95% CI. Detailed information on statistical procedures and algorithms implemented in the BioMercator are available elsewhere (Arcade et al. 2004; Veyrieras et al. 2007; Sosnowski et al. 2012).
The physical positions for individual MQTLs were obtained using the nucleotide sequences of the markers flanking the MQTLs; these nucleotide sequences were obtained from published literature or retrieved from GrainGenes (https://wheat.pw.usda.gov/GG3/) and/or CerealDB (https://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/index NEW.php) databases. The marker sequences were used for BLAST against the hexaploid wheat reference genome (IWGSC RefSeq v1.0). In some cases, the physical positions of SNP markers were directly obtained from the JBrowse wheat genome browser (https://wheat-urgi.versailles.inra.fr/Tools/Jbrowse). The genomic region between the flanking markers was treated as the physical position of the corresponding MQTL.
Comparison of MQTLs with known GWAS-MTAs and genes
MTAs reported in five earlier genome-wide association studies (GWAS; published during 2019–2020) on heat stress-responsive traits were available; these were utilized for a comparison with MQTLs detected in the present study. The details of the size of association mapping panel, traits-parameters, SNP markers used, and MTAs identified in these studies are given in Online Resource 3. The physical positions of the MTAs obtained from different databases (such as CerealDB and JBrowse wheat genome browser) were compared with the physical coordinates of the MQTL regions on the same chromosome; an individual MTA occurring within a specific MQTL region was considered co-located. In this manner, MQTLs detected in this study were verified by the MTAs reported in at least one of the five GWAS reports.
In addition to the above, the previously known heat stress-responsive gene sequences were subjected to BLAST analysis against the IWGSC RefSeq v1.0 and the physical coordinates of known genes were extracted, and compared with the physical positions of the MQTL regions. The MQTLs matching with sequences of heat-responsive genes were treated as coincident.
Identification of genes and their expression analysis
The genes underlying individual MQTL regions were identified in the 1-Mb interval on either side (upstream and downstream) of the peak position of the MQTL (total 2 Mb region; Jan et al. 2021); for this purpose, the BioMart tool of EnsemblPlants (https://plants.ensembl.org/biomart/martview) was used. In silico expression analysis of genes was carried out using transcriptome data available at wheat expression browser (expression and visualization platform) powered by expVIP (http://www.wheat-expression.com; Ramírez-González et al. 2018). For this purpose, the relevant expression dataset including expression data related to heat stress was utilized (Liu et al. 2015). This RNA-seq dataset consisted of differential expression of genes in a heat and drought tolerant wheat cv. TAM107, grown under heat-stressed condition, with leaf samples collected separately at 1 h and 6 h after heat stress treatment (40 °C); seedlings grown in normal growth condition (22 °C, well-watered) were taken as control (Liu et al. 2015). The data on expression was available as log2 transformed TPM (transcripts per million) values. Only genes showing fold change (FC ≥ 2 or FC ≤ − 2 TPM values) relative to control were considered as differentially expressed. Gene ontology (GO) analysis was conducted using the BioMart tool and the heat maps of the expression patterns were generated using the Morpheus program (https://software.broadinstitute.org/morpheus/).
Identification of ortho-MQTLs
Seven wheat MQTLs were used for the identification of ortho- or syntenic MQTLs in rice. Following steps were involved: (i) genes detected in the regions of the above seven MQTLs were subjected to BLAST analysis against rice genome database at EnsemblPlants to identify rice orthologues. (ii) The rice orthologues were extracted with their physical positions from the database. (iii) Physical positions of genes underlying wheat MQTLs were compared with the orthologous genes underlying rice MQTLs for heat tolerance (Raza et al. 2020), and (iv) MQTLs harboring similar genes located on known syntenic genomic positions of wheat and rice were considered as ortho-MQTLs.
Results and discussion
The QTL data used for conducting meta-QTL (MQTL) analysis in the present study were largely similar to those used in two previous studies (Acuña-Galindo et al. 2015; Liu et al. 2020). This also facilitated a comparison across three studies and interpretation of results. A detailed comparison with respect to the data used and the results obtained are presented in Online Resource 4. For identification of MQTLs involved in thermotolerance, integration of GWAS (for MQTL verification), heat-responsive genes, genes underlying MQTLs and their expression, and homology comparison with rice (ortho-MQTL) offer an interesting contrast between the present study and two earlier studies. The chromosomes in the consensus map were more densely populated with markers and the QTLs used in the present study. In our analysis, Wheat_Consensus_map_2021 was prepared with 50,310 markers and 441 QTLs were projected for 154 heat-responsive traits belonging to 31 categories (Online Resource 5), which was higher relative to those in previous two studies (Acuña-Galindo et al. 2015; Liu et al. 2020). Further, out of 85 MQTLs detected in our study, the genetic position of 18 MQTLs matched with the MQTLs reported in above two studies (Acuña-Galindo et al. 2015; Liu et al. 2020), whereas the remaining 67 MQTLs were perhaps novel which were not reported earlier. Interestingly, two of the 18 MQTLs were exclusively associated with thermotolerance, 14 MQTLs were associated either only with drought or heat/drought tolerance. However, the remaining two MQTLs were neither associated with heat tolerance nor with drought tolerance. Overall, the results suggest that ~ 20% (16/85) of MQTLs involved in thermotolerance as well as drought were common among the three studies including our own study and therefore provide a genetic basis for future studies on abiotic stress tolerance in wheat. Moreover, above 16 MQTL regions associated with heat and drought tolerance-responsive traits can be targeted for future studies involving fine mapping, cloning, functional analysis, and development of markers for use in marker-assisted selection.
QTLs associated with thermotolerance
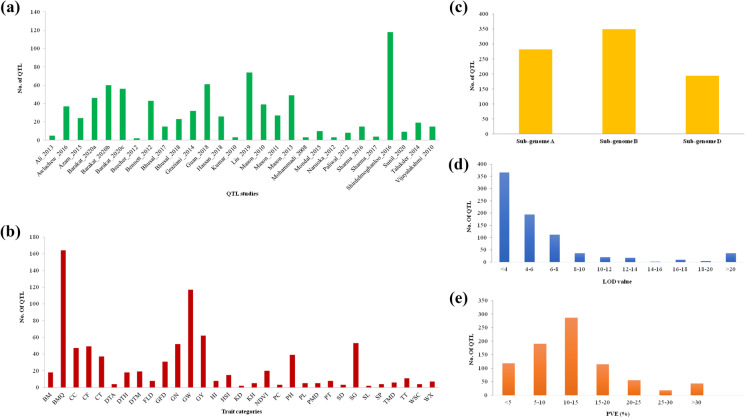
Information on QTLs from 31 interval mapping reports (published during 2008–2020) for heat-responsive traits was collected (Online Resource 1). Only 28 of these studies (25 for hexaploid and three for tetraploid wheats) had complete information (marker positions and PVE values) needed for conducting meta-QTL analysis; 826 individual QTLs for 154 relevant traits were available from these 28 studies (Online Resources 1 and 2). The populations involved in these 28 studies included 13 recombinant inbred line (RIL) populations, 6 doubled haploid (DH) populations, 5 F2/backcross populations, and one family-based population (rarely same population was used in more than one study). The population size in different interval mapping studies varied from 64 (Mason et al. 2010) to 420 (Sharma et al. 2017) and the number of QTLs reported in individual studies ranged from 2 (Beecher et al. 2012) to 118 QTLs (Shirdelmoghanloo et al. 2016) (Fig. 1a).
Fig. 1.
Characteristic features of the collected QTLs and their distribution at different levels. (a) Number of QTLs in different studies. (b) Number of QTLs across trait categories. (c) Number of QTLs in wheat sub-genomes A, B, and D. (d) Frequencies of QTLs with different levels of LOD values. (e) Frequencies of QTLs with different levels of PVE (%). Traits abbreviations include GY grain yield, GW grain weight, GN grain number, DTH days to heading, DTA days to anthesis, DTM days to maturity, GFD grain filling duration, HI harvest index, BM biomass, PT productive tiller, KD kernel diameter, SL spike length, SD spike density, SP spikes per plant, PH plant height, BMQ bread-making quality traits, SG stay green, CF chlorophyll fluorescence, CC chlorophyll content, CT canopy temperature, NDVI normalized difference vegetation index, HSI heat susceptibility index, TT thermotolerance, FLD flag leaf dimension, WX waxiness, TMD thylakoid membrane damage, KH kernel hardness, PL length of the ear peduncle, PMD plasma membrane damage, WSC water soluble carbohydrates, and PC proline content
The number of QTLs on individual chromosomes ranged from a minimum of 13 QTLs on chromosome 1D to a maximum of 90 on chromosome 3B (Online Resource 2). Distribution of QTLs on individual homoeologous groups also differed, with homoeologous group 2 having a maximum of 197 QTLs, and group 6 having a minimum of 66 QTLs. This distribution of QTLs on the seven homoeologous groups may be attributed to higher gene density of stress-related expressed sequence tags (ESTs) on group 2 chromosomes and the lowest gene density on group 6 chromosomes as reported in an earlier study in wheat (Ramalingam et al. 2006). The number of QTLs for different traits ranged from 2 (spike length; SL) to 164 (bread-making quality; BMQ), which comprise 25 individual traits-parameters, with an average of ~ 26 QTLs per category of traits (Fig. 1b). The distribution of QTLs in three sub-genomes also differed with sub-genome B carrying the maximum of 350 QTLs, followed by sub-genomes A with 282 and sub-genome D with 194 QTLs (Fig. 1c). This distribution of QTLs on homoeologous groups and sub-genomes is also in agreement with previous MQTL studies in wheat for thermotolerance (Acuña-Galindo et al. 2015; Liu et al. 2020) and drought tolerance (Kumar et al. 2020). The 826 QTLs were further categorized based on LOD scores and PVE%. The LOD score of the individual QTLs ranged from 2 to 60 with an average of 6.22 (Fig. 1d). Nearly half (46.7%) of all QTLs had their LOD scores ranging from 2 to 4. The PVE% for individual QTLs ranged from 1 to 60.1% with a mean PVE of 12%. Approximately 75% (619/826) QTLs had up to 15% PVE, whereas about 5% (41/826) QTLs exhibited > 30% PVE (Fig. 1e), suggesting the availability of both major and minor QTLs for thermotolerance.
Consensus map of wheat (Wheat_Consensus_map_2021)
The marker types on the Wheat_Consensus_map_2021 included RFLPs, AFLPs, SSRs, DArTs, SNPs, and some well-known gene loci, such as Vrn, Ppd, Rht, and Glu. The map contained 50,310 marker loci with a total genetic length of 12,280.7 cM, giving a density of 4.1 marker loci/cM (Fig. S1 and S2, Online Resource 6). The length of the linkage maps for the individual chromosomes ranged from 280.1 cM (4D) to 894.1 cM (4A), and the number of markers in individual linkage groups ranged from 392 on 4D to 4,121 on chromosome 1B. The marker density for individual chromosomes ranged from 1.4 markers/cM for 4D to 11.1 markers/cM for 2A. Sub-genome A covered 4,102.7 cM with 19,518 markers (4.76 markers/cM), sub-genome B covered 4,579.9 cM with 23,058 markers (5.03 markers/cM), and sub-genome D spanned 3,598.1 cM size with 7,734 markers (2.15 markers/cM) (Fig. S2; Supplementary Material 2). This consensus map is relatively more saturated (dense) than the consensus maps used in earlier two MQTLs studies on thermotolerance (Acuña-Galindo et al. 2015; Liu et al. 2020) (Online Resource 4). This resulted in the identification of many more MQTLs in the present study.
QTL projection and MQTL analysis
The distribution of the number of available QTL, the QTLs utilized for projection, and the MQTLs identified during the present study among different wheats (durum wheats, spring 6 × wheats, and winter 6 × wheats) are summarized in Table 1. From the 826 individual QTLs, 441 (53.4%) QTLs for different heat-responsive traits were successfully projected onto the above Consensus map. These 441 QTLs included 27 QTLs derived from tetraploid wheat (durum) and 414 from hexaploid wheat (involving 244 from spring wheats, 65 from winter wheats, 105 from spring × winter crosses). The remaining 385 (46.6%) QTLs could not be projected due to the following reasons: (i) use of marker types that were not used in consensus map, (ii) lack of commonly associated markers between consensus and initial maps, and (iii) very long CI.
Table 1.
Number of QTLs available from tetraploid (4x) and hexaploid (6x) wheats
| Type of wheat | QTL collected | QTL projected on map* | QTL involved in MQTLs |
|---|---|---|---|
| Tetraploid wheat (4x) | 74 | 27 | 24 |
| Spring wheat (6x) | 391 | 244 | 234 |
| Winter wheat (6x) | 236 | 65 | 54 |
| Spring × winter wheat (6x) | 125 | 105 | 99 |
| Total | 826 | 441 | 411 |
*Wheat_Consensus_map_2021
As a result of meta-analysis, as many as 85 MQTL regions were identified from 411 QTL with at least one MQTL on each of the 21 wheat chromosomes (Table 2, Online Resource 7). The remaining 30 QTLs stayed singletons and were therefore not used for further analysis. Identification of 85 MQTLs from 411 QTLs amounted to a 4.83-fold (411/85) reduction in the number of genomic regions or QTLs (MQTLs) associated with thermotolerance in wheat. Earlier, while conducting a combined MQTL analysis for heat and drought tolerance in wheat, reductions of 8.09-fold and 4.43-fold were reported in two earlier studies (Acuña-Galindo et al. 2015; Liu et al. 2020). A relatively large reduction in the number of QTLs reported by Acuña-Galindo et al. (2015) may be attributed to inclusion of QTLs for both heat and drought tolerance.
Table 2.
Details of MQTLs detected for thermotolerance and associated traits in wheat
| MQTL (CI range) | bSource of QTLs | Flanking marker | LOD value | PVE (%) | cTrait (no. of QTL) |
|---|---|---|---|---|---|
| MQTL1A.1 (33.4–59.2) | Spring wheat (2) | P3615.2/BobWhite_c863_1064 | 3.20 | 10.00 | BMQ (2) |
| MQTL1A.2 (82.9–91.6) | Spring wheat (2) | RAC875_c25125_210/gwm905 | 5.40 | 14.87 | SG (1), GW (1) |
| MQTL1A.3 (99.7–101.8) | Tetraploid wheat (1), winter wheat (2) | gdm33/gwm33a | 2.67 | 6.10 | CF (2), GW (1) |
| MQTL1A.4 (105.6–107.7) | Spring wheat (4), winter wheat (1), winter × spring (2) | gwm33a/gwm1104 | 4.44 | 13.09 | GN (1), GW (1), CF (1), GY (1), BMQ (3) |
| MQTL1A.5 (136.8–139.2) | Spring wheat (1), winter wheat (1) | Tdurum_contig11756_264/gwm691 | 5.04 | 12.00 | GW (1), BMQ (1) |
| MQTL1B.1 (33.2–42.1) | Spring wheat (2), winter wheat (1) | RFL_Contig293_498/Excalibur_rep_c15165_367 | 3.69 | 22.60 | HSI (1), BMQ (1), CF (1), |
| MQTL1B.2 (100.2–100.6) | Tetraploid wheat (1), spring wheat (3), winter wheat (1), winter × spring (1) | wpt-0944/cwm90 | 3.50 | 15.42 | GY (1), CC (2), GY (1), GFD (1), BMQ (1) |
| MQTL1B.3 (105.4–111.7) | Spring × winter (1), spring wheat (1) | wpt-0944/D_contig37179_75 | 3.54 | 9.40 | FLD (1), GFD (1) |
| MQTL1B.4 (120.8–120.9) | Spring wheat (4), Ae. speltoides × Triticum durum (3), winter wheat (1), spring × winter (1) | wsnp_Ex_c1137_2183723/wsnp_Ku_c7822_13408189 | 3.91 | 12.17 | BMQ (3), GW (1), TT (1), PT (1), SG (1), HSI (1) |
| MQTL1D.1 (117.4–134.6) | Spring wheat (2) | 1228670 s/BS00000721_51 | 4.15 | 13.50 | BMQ (2) |
| MQTL1D.2 (213.9–216.9) | Spring × winter (1), winter wheat (3) | Excalibur_c16851_835/Kukri_c8533_1398 | 3.02 | 12.84 | GFD (1), PMD (1), CC (1), TMD (1) |
| MQTL1D.3 (224.5–248.0) | Spring × winter (1), spring wheat (1) | Kukri_cA592_935/Ku_c17846_363 | 3.08 | 7.90 | FLD (1), GY (1) |
| MQTL2A.1 (33.7–39.7) | Spring wheat (1), spring × winter (3) | wmc382b/barc124 | 5.00 | 17.13 | GFD (2) (1), GN (1), BMQ (1) |
| MQTL2A.2 (56.0–65.3) | Tetraploid wheat (1), winter wheat (1) | AX-111714220/Ra_c69660_1577 | 5.02 | 5.00 | DTM (1), PH (1), NDVI (1) |
| MQTL2A.3 (87.8–89.8) | Spring wheat (5), winter wheat (1), spring × winter (1) | Ra_c31463_148/RAC875_c54668_102 | 8.35 | 16.97 | BMQ (2), GW (1), WX (1), GN (1) |
| aMQTL2A.4 (95.7–96.4) | Spring wheat (6), winter wheat (1) | Excalibur_c38761_288/BS00104558_51 | 5.62 | 10.32 | BMQ (4), GW(2), PH (1) |
| MQTL2B.1 (73.8–75.2) | Spring wheat (7) | 979087S/BS00075855_51 | 6.17 | 7.78 | DTM (1), NDVI (1), BMQ (2), GN (2), CT (1) |
| MQTL2B.2 (119.6–125.9) | Ae. speltoides × Triticum durum (4), tetraploid wheat (1), spring wheat (7), winter wheat (2), spring × winter (4) | BobWhite_c19433_185/1243120D | 7.59 | 16.66 | GY (1), SG (2), GW (3), GN (2), HSI (1), CF (1), FLD (1), CC (1), CT (1), BMQ (2), GFD (2), GY (1) |
| MQTL2B.3 (140.7–145.9) | Tetraploid wheat (1), spring wheat (9), winter wheat (2) | 1125044D/wmc764 | 4.10 | 10.54 | BMQ (6), NDVI (1), PMD (1), GY (1), GW (1), CF (1), TT (1) |
| MQTL2B.4 (158.1–163.7) | Spring wheat (3) | wPt-2106/wPt-4195 | 3.39 | 9.46 | CC (1), GY (1) |
| MQTL2B.5 (332.8–352.6) | Spring wheat (2) | RAC875-c37639_94/gwm935 | 3.25 | 18.99 | GW (1), GFD (1) |
| MQTL2D.1 (112.4–115.4) | Spring wheat (4), spring × winter wheat (6) | GENE-0644_42/RAC875_C4314_993 | 5.28 | 12.55 | GW (2), DTH (1), CT (3), GY (1), DTM (1), BMQ (1), PH (1) |
| MQTL2D.2 (124.3–128.7) | Spring wheat (9), spring × winter wheat (13) | Kukri_c11566_386/gdm87a | 6.88 | 15.73 | BMQ (4), NDVI (1), GY (4), CT (1), KD (1), DTM (1), KH (1), GW (3), DTA (1), DTH (1), HI (1), FLD (2), HSI (1), NDVI (1) |
| MQTL2D.3 (153.0–154.8) | Spring wheat (8) | D_GA8KES401CIV9Z_240/BS00022276_51 | 4.89 | 11.38 | BMQ (5), GY (3) |
| MQTL3A.1 (82.6–117.5) | Spring wheat (2) | 2278441D/BobWhite_c17386_221 | 3.85 | 11.50 | BMQ (2) |
| MQTL3A.2 (145.8–148.3) | Spring wheat (2) | Excalibur_c21521_1318/Kukri_c35153_956 | 3.36 | 5.22 | GY (2) |
| MQTL3A.3 (166.1–167.3) | Tetraploid wheat (1), spring wheat (2), winter wheat (1) | Kukri_c35153_956/BobWhite_c15763_205 | 11.97 | 12.11 | PL (1), SG (1), GY (1), GN (1) |
| MQTL3A.4 (189.8–190.1) | Ae. speltoides × Triticum durum (3) | BobWhite_c15763_205/barc54 | 4.77 | 19.70 | TT (1), HSI (1), SG (1) |
| MQTL3B.1 (57.3–60.0) | Spring wheat (8), spring × winter wheat (2) | wsnp_Ex_c1097_2105209(R)/Tdurum_contig44013_164 | 4.27 | 14.32 | CT (1), BMQ (6), FLD (1), GN (1), SG (1), |
| aMQTL3B.2 (84.9–86.3) | Ae. speltoides × Triticum durum (3), spring wheat (5), spring × winter wheat (6) | wPt-7472/wPt-10552 | 4.84 | 11.48 | BMQ (2), GY(4) (1), CC (1), SG (1), CT (1), GW (1), BM (1), HSI (1), SD (1), CT (1) |
| MQTL3B.3 (106.9–110.5) | Spring wheat (5), spring × winter (2) | Kukri_c639_2168/BS00110416_51 | 5.83 | 15.48 | CT (1), GY (4), TT (1), GW (1) |
| MQTL3D.1 (67.7–74.3) | Spring wheat (7), spring × winter (1) | gwm383/RAC875_c35399_497 | 5.03 | 9.20 | GY (3) (1), BMQ (4) (1), HSI (1) |
| MQTL3D.2 (140.2–145.0) | Spring wheat (2) | wmc529/D_GBF1XID02GHPCU_167 | 4.84 | 6.29 | NDVI (1), BMQ (1) |
| MQTL3D.3 (176.4–177.3) | Spring wheat (3) | Kukri_rep_c96809_352/gwm114a | 5.89 | 7.33 | GN (1), NDVI (1), GY (1) |
| MQTL4A.1 (631.6–634.7) | Winter wheat (2) | BS00018120_51/tplb0043j11_1190 | 8.42 | 2.50 | GW (1), SP (1) |
| MQTL4A.2 (648.4–649.1) | Spring wheat (1), winter wheat (1), spring × winter (1) | tplb0043j11_1190/BS00069271_51 | 6.31 | 8.49 | WSC (1), GN (1), GW |
| MQTL4A.3 (676.5–681.9) | Spring wheat (5), spring × winter (2) | cfa2256/mgb7 | 4.21 | 11.13 | BMQ (4), GW(3) |
| MQTL4B.1 (70.7–76.7) | Spring wheat (2), winter wheat (1) | wsnp_Ex_rep_c73742_71687509(R)/wsnp_Ex_c30695_39579408(R) | 4.38 | 9.33 | GW (1), BMQ (1) |
| MQTL4B.2 (100.1–106.1) | Spring wheat (1), winter wheat (1) | Tdurum_contig60051_169/wsnp_Ex_c18433_27269748 | 7.60 | 19.70 | BMQ (1), DTM (1), GFD (1), GW (1), BM (1), PH (1), HI (1) |
| MQTL4B.3 (127.5–130.3) | Spring × winter (2) | Kukri_c27037_112/barc340a | 4.19 | 8.15 | KH (1), PH (1) |
| MQTL4B.4 (134.7–136.3) | Spring wheat (2), winter wheat (4), spring × winter (1) | Kukri_c27037_112/barc340a | 14.77 | 7.44 | DTH (1), SP (1), PH (1), GW (2), TT (1), GN (1) |
| MQTL4B.5 (146.1–149.7) | Tetraploid wheat (1), spring wheat (2) | Kukri_c27037_112/barc340a | 4.06 | 5.53 | GY (2), GW (1) |
| MQTL4B.6 (157.1–159.5) | Spring wheat (1), winter wheat (1) | wPt-11509/wPt-4537 | 5.16 | 8.84 | DTM (2), GFD (1), GW (1), GN (1), BM (1), SG (1), PH (1) |
| MQTL4D.1 (0–7.3) | Spring wheat (2) | IACX11246/CAP8_c2722_258 | 5.10 | 10.00 | BMQ (2) |
| MQTL4D.2 (30.1–39.0) | Spring wheat (2) | GENE-2463_463/Kukri_c29521_123 | 3.15 | 10.00 | BMQ (2) |
| MQTL4D.3 (58.9–62.8) | Spring wheat (3) | Ex_c16174_685/D_contig26950_309 | 3.47 | 7.52 | PH (1), GY (2) |
| MQTL4D.4 (72.6–75.0) | Winter wheat (2) | barc334/wmc89 | 29.76 | 4.50 | SP (1), PH (1) |
| MQTL5A.1 (347.7–350.8) | Spring wheat (1), spring × winter (3) | gwm865/gwm330 | 4.49 | 9.17 | GW (1), CT (2), GFD (1) |
| MQTL5A.2 (358.1–360.4) | Tetraploid wheat (1), spring wheat (5), winter wheat (2), spring × winter (4) | utv711/wmc189_1 | 4.13 | 15.97 | GW (3), GFD (2), WX (1), BMQ (1), KD (1), PC (1), GN (2), PH (1) |
| MQTL5A.3 (380.0–380.4) | Spring wheat (3), winter wheat (1), spring × winter (6) | 1146968S/2255033D | 4.55 | 9.58 | GY (1), CT (3), GW (3), WSC (1), PH (2) |
| MQTL5B.1 (105.8–112.5) | Spring wheat (1), spring × winter (2) | 1032121S/Ra_c28545_196 | 5.87 | 15.00 | FLD (1), DTA (1), GW (1) |
| MQTL5B.2 (200.3–202.2) | Ae. speltoides × Triticum durum, spring wheat (1) | 1026809D/BooWhite_c1973_383 | 5.69 | 10.19 | TT (1), DTM (1) |
| aMQTL5B.3 (216.9–217.5) | Spring wheat (7), spring × winter (1) | 1026809D/BooWhite_c1973_383 | 4.63 | 11.70 | GY (1), GW (1), PC (1), BMQ (5) |
| MQTL5B.4 (234.7–241.1) | Spring wheat (2), spring × winter (6) | BooWhite_c1973_383/BooWhite_c20282_164 | 4.37 | 13.15 | HSI (1), BMQ (1), SD (1), KH (1), DTM (1), DTH (1), HI (1), GN (1) |
| MQTL5B.5 (289.8–304.5) | Spring wheat (2) | wsnp_Ex_c12127_19394952(R)/Excalibur_c80450_75 | 6.00 | 13.50 | BMQ (2) |
| MQTL5D.1 (27.9–28.9) | Spring wheat (3) | BS00034658_51/D_GCE8AKX02GJQ22_146 | 7.73 | 13.81 | DTH (1), GW (1), BMQ (1) |
| MQTL5D.2 (76.0–102.0) | Spring wheat (2) | Ra_c21355_446/RAC875_rep_c113555_122 | 6.20 | 10.50 | BMQ (2) |
| MQTL5D.3 (110.3–118.9) | Spring wheat (2) | RAC875_rep_c113555_122/wsnp_Ex_c30078_39930134 | 6.69 | 10.72 | NDVI (1), CT (1) |
| MQTL5D.4 (135.9–142.7) | Spring wheat (1), spring × winter (1) | wsnp_Ex_rep_c69631_68583363/D_GDEEGVY01D91EB_346 | 6.78 | 10.90 | HSI (1), NDVI (1) |
| MQTL5D.5 (331.1–337.2) | Spring × winter (3) | GENE-4974_839/BobWhite_c9761_225 | 4.76 | 7.23 | GY (1), KH (1), CT (1) |
| MQTL6A.1 (444.5–447.0) | Spring wheat (1), winter wheat (2) | wsnp_Ra_rep_c79566_75592262(R)/gdm36b | 3.11 | 7.33 | BMQ (1), GW (2) |
| MQTL6A.2 (470.9–472.2) | Spring wheat (2), winter wheat (4) | RZ876/GTG.ACGC-108 | 5.09 | 22.45 | CF (1), SG (2), CC (1), TMD (1), BMQ (1) |
| MQTL6A.3 (537.7–539.6) | Spring wheat (2) | wsnp_Ku_c1318_2624758(R)/TGCGCG349 | 7.40 | 10.49 | PH (1), BMQ (1) |
| MQTL6B.1 (145.4–147.4) | Spring wheat (2), winter wheat (2) | barc211/BobWhite_c11652_421 | 2.85 | 9.71 | CF (1), BMQ (1), SG (2) |
| MQTL6B.2 (159.0–162.8) | Spring wheat (2) | BobWhite_c32377_278/wPt-689 | 13.41 | 20.28 | BMQ (1), GN (1) |
| MQTL6B.3 (180.3–192.4) | Spring wheat (3) | RAC875_c16962_288/wmc419 | 3.26 | 9.33 | GY (2), BMQ (1) |
| MQTL6B.4 (195.7–199.7) | Spring wheat (2) | wmc486/wsnp_Ex_c6057_10611952(R) | 4.45 | 13.00 | PT (1), GY (1) |
| MQTL6B.5 (692.8–704.8) | Spring wheat (3) | wsnp_JD_c1119_1642176/IACX3288 | 5.03 | 11.67 | BMQ (1) |
| aMQTL6D.1 (53.7–54.6) | Spring wheat (3), spring × winter (4) | CAP11_rep_c6864_291/Ra_c23837_298 | 12.28 | 26.45 | GW (2), GFD (1), CT (3), GY (1) |
| aMQTL7A.1 (94.5–96.2) | Spring wheat (4), winter wheat (1), Ae. speltoides × Triticum durum | wPt-6460/gwm1187 | 4.24 | 14.74 | SG (2), HSI (1), BMQ (3), GY (1) |
| MQTL7A.2 (97.8–98.3) | Spring wheat (3) | gwm1187/EXCALIBUR_C1625_1215 | 3.01 | 5.00 | PC (1), GY (2) |
| MQTL7A.3 (104.1–108.4) | Spring wheat (2), spring × winter (1) | gwm1187/EXCALIBUR_C1625_1215 | 4.57 | 8.33 | FLD (1), GY (2) |
| MQTL7A.4 (110.6–111.9) | Spring wheat (1), winter wheat (2) | gwm1187/EXCALIBUR_C1625_1215 | 3.95 | 4.89 | TT (1), GN (1), GW (1) |
| MQTL7A.5 (115.3–116.3) | Winter wheat (1), spring × winter (2) | EXCALIBUR_C1625_1215/wPt-2 | 4.18 | 17.37 | PH (1), GW (1), DTM (1) |
| MQTL7A.6 (122.1–124.5) | Winter wheat (5), spring × winter (2) | wPt-5153/wPt-5533 | 5.17 | 19.18 | DTH (1), CF (2), PMD (1), CC (1), TMD (1), GN (1) |
| MQTL7A.7 (152.3–154.8) | Spring wheat (1), winter wheat (1), spring × winter (4) | RAC875_c9506_343/Kukri_c38390_218 | 5.25 | 12.75 | HSI (1), CT (1), GFD (1), GW (1), DTH (1), BMQ (1) |
| aMQTL7B.1 (79.9–80.6) | Spring wheat (6), winter wheat (1), spring × winter (1) | psr129/Ex_c24068_652 | 8.56 | 11.97 | GY (3), HI (1), DTM (2), BM (1), GFD (1), GW (1), NDVI (1), DTH (1), GN (1) |
| MQTL7B.2 (101.4–104.1) | Spring wheat (2) | wmc546.1/1102954|F|0–30:C > T-30:C > T | 3.00 | 4.10 | TT (1), CF (1) |
| aMQTL7B.3 (120.8–122.4) | Spring wheat (11), winter wheat (1), Ae. speltoides × Triticum durum (1) | 1220186D/GENE-4507_56 | 4.18 | 13.89 | BMQ (6), GW (1), PH (1), GY (2) (1), CC (1), HSI (1), CT (1) |
| MQTL7D.1 (44.5–71.2) | Spring wheat (2) | BobWhite_c28137_293/gwm332B | 5.15 | 10.00 | GY (1), BMQ (1) |
| MQTL7D.2 (232.2–245.7) | Spring wheat (1), spring × winter (1) | RAC875_rep_c105462_468/gwm535_1 | 3.36 | 9.85 | HI (1), BMQ (1) |
| MQTL7D.3 (286.4–287.6) | Spring wheat (5) | gwm3100/Tdurum_contig74815_800 | 3.20 | 11.43 | WSC (1), SG (1), BMQ (1), GW (1), DTM (1) |
| MQTL7D.4 (312.8–318.2) | Winter wheat (1), spring × winter (7) | wmc473_2/wmc94 | 4.79 | 8.13 | SD (1), CF (1), DTH (1), GY (1), KH (1), GW (1), GN (1), DTM (1) |
| MQTL7D.5 (354.1–354.8) | Spring wheat (3) | RAC875_c39217_314/gwm1123 | 3.85 | 5.95 | DTH (1), PH (1), WX (1) |
| MQTL7D.6 (377.1–383.4) | Spring wheat (2) | gwm350/Kukri_c37802_1215 | 7.04 | 11.55 | DTM (1), NDVI (1) |
aBreeder’s QTL, bvalues given in the parenthesis represent number of QTLs derived from different wheat types, CI confidence interval, PVE (%) percent phenotypic variation explained by QTL, ctrait abbreviations: GY grain yield, GW grain weight, GN grain number, DTH days to heading, DTA days to anthesis, DTM days to maturity, GFD grain filling duration, HI harvest index, BM biomass, PT productive tiller, KD kernel diameter, SL spike length, SD spike density, SP spikes per plant, PH plant height, BMQ bread making quality traits, SG stay green, CF chlorophyll fluorescence, CC chlorophyll content, CT canopy temperature, NDVI normalized difference vegetation index, HSI heat susceptibility index, TT thermotolerance, FLD flag leaf dimension, WX waxiness, TMD thylakoid membrane damage, KH kernel hardness, PL length of the ear peduncle, PMD plasma membrane damage, WSC water soluble carbohydrates, and PC proline content
In addition to reduction in the number of QTLs, MQTL analysis also leads to reduction in the size of CI by accumulating QTLs information from different populations of diverse genetic origins. In the present study, the CI of the MQTLs ranged from 0.11 to 34.9 cM with a mean of 5.6 cM. The average length of CI of MQTLs was reduced 2.95-fold relative to the CIs in QTLs, which is quite similar to those reported in previous meta-QTL studies in wheat (Acuña-Galindo et al. 2015; Liu et al. 2020; Yang et al. 2021). There were significant differences in average CIs of MQTLs that mapped on different chromosomes; CI of MQTLs on 6D was reduced 14.6 times and that on 5A was reduced 8.9 times, followed by 8.8 times for 7A and 8.1 times for 7B (Fig. S3). Twenty-seven (27; 31.76%) MQTLs had CI < 2 cM. The physical lengths for individual 85 MQTLs also differed and ranged from 0.058 Mb for MQTL7D.5 to 735.48 Mb for MQTL2B.3, with an average of 198.83 Mb (26 of these MQTLs each covered a physical distance of < 20 Mb) (Online Resource 7). Our results in the present study support the finding of an earlier study by Yang et al. (2021) showing that the CI was proportional to the number of QTLs on which the MQTL was based. These results and those of the present study suggest that an increase in the number of QTLs used for meta-QTL analysis could significantly reduce the size of the CI of MQTL relative to those in the initial QTLs.
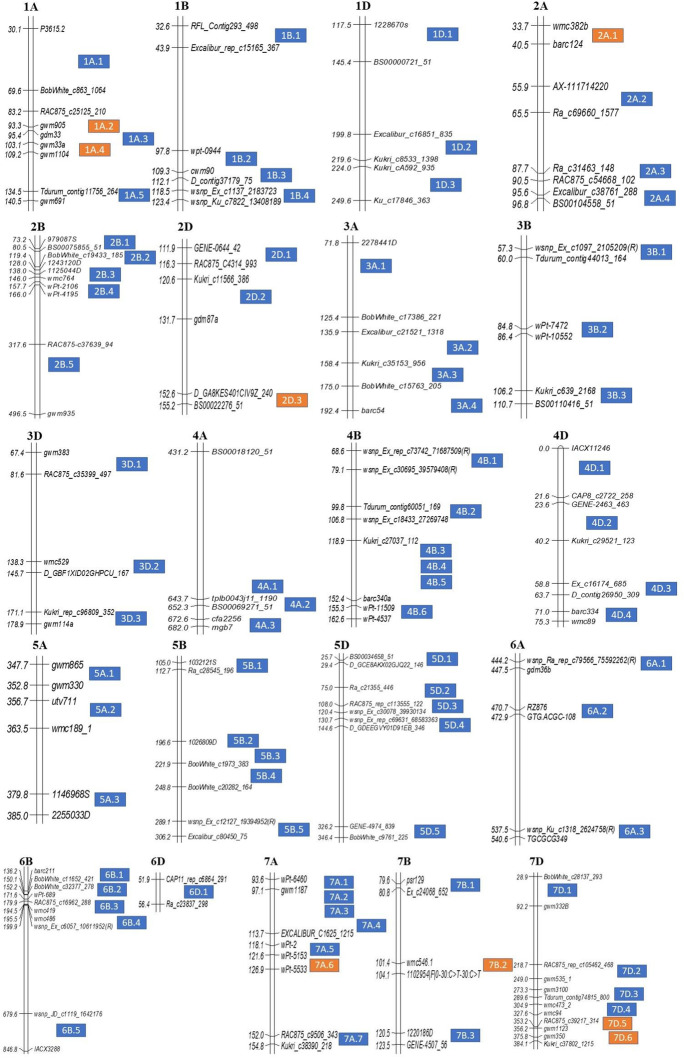
As expected, a solitary MQTL based exclusively on tetraploid (durum) wheat and those based on both tetraploid and hexaploid wheats belong to only sub-genomes A and B (none belonging to sub-genome D), while those belonging exclusively to hexaploid wheat (spring and/or winter wheats) carried MQTLs belonging to all the three sub-genomes. It is interesting to note that out of 85 MQTLs identified in the present study, 25 MQTLs contained original QTLs exclusively from spring wheat and two MQTLs were such which contained original QTLs belonging exclusively to winter wheat; a solitary MQTL was however based exclusively on the original QTL from durum wheat (Table 2). Thus, our findings clearly dissected specific genomic regions that contribute to heat tolerance in spring and winter wheats, separately. The distribution of MQTLs on 21 individual wheat chromosomes ranged from 1 on 6D to 7 on 7A (Fig. 2, Online Resource 7) and this number did not depend on the number of QTLs on individual chromosomes utilized for MQTL analysis. For instance, chromosomes 2A, 2D, and 3B each had many more QTLs but relatively fewer MQTLs. Several MQTLs had a clustered distribution in the genome, with some having overlapping regions. One of these clusters, consisting of 4 MQTLs, was identified on chromosome 1A physically located between 2.54 and 12.09 Mb (1A.1-1A.4). All MQTLs (except 4B.2) on chromosome 4B were physically clustered in 5.19-255.38 Mb region. Similarly, five MQTLs (5B.1–5B.5) were found in the 64.74–711.63-Mb genomic region of 5B. Clustered MQTLs were also available on 5D (245.29–498.4 Mb), 6B (16.82–119.97 Mb), and 7D (4.19–44.56 Mb) (Online Resource 7). Such a clustered pattern of distributions of MQTLs in wheat genome was also reported in three recent studies (Jan et al. 2021; Saini et al. 2021; Yang et al. 2021).
Fig. 2.
Distribution of meta-QTLs (MQTLs) on all the 21 wheat chromosomes. Only the flanking markers are shown for better visualization. MQTLs in blue boxes (right side) were co-located with marker-trait associations (MTAs) identified using genome-wide association studies
The number of individual QTLs covered within an individual MQTL ranged from 2 to 22 (Table 2) and a large proportion of MQTLs (65%, 55/85) was each based on > 3 QTLs derived from different mapping populations. The following six MQTLs comprised ≥ 12 QTLs: MQTL2B.2 (18), MQTL2B.3 (12), MQTL2D.2 (22), MQTL3B.2 (14), MQTL5A.2 (12), and MQTL7B.3 (13). This is in sharp contrast to earlier studies, where a single MQTL associated with thermotolerance was based on no more than 10 QTLs (Acuña-Galindo et al. 2015; Liu et al. 2020). The MQTLs that harbored original QTLs derived from more than one mapping population and consistently appeared in several environments may be the most reliable, robust, and stable for wheat breeding.
The number of traits controlled by an individual MQTL ranged from a solitary trait controlled by each of many MQTLs to as many as 14 traits controlled by a solitary MQTL (e.g., MQTL2D.1); 76 of the 85 MQTLs were multi-trait MQTLs (Table 2), each controlling more than one trait indicating either the occurrence of pleiotropic genes or tight linkage among genes for different traits. It could also be due to the use of related traits as also shown in the case of MQTL analysis in wheat for grain yield and related traits (Saini et al. 2021) and root-related traits in wheat (Soriano and Alvaro, 2019).
The LOD score of individual MQTLs ranged from 2.7 to 29.8 with an average of 5.5. Similarly, the PVE ranged from 2.5 to 26.5% with a mean of 11.5%. The MQTLs were subjected to further selection for identification of some promising MQTLs that we liked to call “breeders’ MQTLs,” each fulfilling the following conditions: (i) CI < 2 cM, (ii) average PVE > 10%, (iii) average LOD > 4, and (iv) involvement of at least 7 QTLs within the MQTL. This exercise resulted in the selection of seven promising MQTLs (2A.4, 3B.2, 5B.3, 6D.1, 7A.1, 7B.1, and 7B.3) that contained original QTLs belonging to both spring and winter wheats. These seven promising MQTL regions overlapped genomic regions carrying MTAs reported in earlier GWA studies (see later). Although three more MQTLs (viz., 1B.1, 6A.2, and 6B.2) each had high PVE (> 20%), these were not included in the list of breeders’ MQTLs, due to their association with large CI or due to the fact that these were each based on fewer QTLs. The so-called breeders’ MQTLs should be subjected to further scrutiny (including validation) for their utility in MAS and may also be used as a resource for fine mapping and cloning.
Verification of MQTLs using MTAs from GWAS
The physical coordinates of MQTLs were also compared with physical positions of 1,034 MTAs for the traits associated with thermotolerance reported earlier in GWA studies; this resulted in the identification of MQTLs matching with as many as 704 MTAs (involving 139 MTAs, each associated with multiple heat-responsive traits within an individual or across several GWA studies) (Fig. 2, Online Resource 8). As many as 63 MQTLs were verified with MTAs obtained from GWAS involving spring wheats and 67 MQTLs were identified for winter wheat populations. However, 53 MQTLs were verified using GWAS involving both spring and winter wheats. The MQTL6B.5 matched with 110 MTAs and MQTL6B.1 matched with 103 MTAs. There were other MQTLs (e.g., 2B.2, 2B.3, and 5A.3) each involving 10 or more QTLs and matched with more than 60 MTAs (Online Resource 8). Overall, ~ 90% of the total MQTLs matched GWAS results. These findings are in sharp contrast to recent studies by Aduragbemi and Soriano (2021) and Yang et al. (2021), where only 38.66% and 62.68% MQTLs, respectively, overlapped MTAs obtained from GWA studies. This might be due to the availability of a comparatively large number of MTAs for comparison in the present study. The MQTLs, which matched GWAS-based MTAs, provide a basis for accurately mining of high confidence candidate genes associated with thermotolerance in wheat.
MQTLs co-located with known genes for thermotolerance
A comparison of MQTLs with known genes for thermotolerance in wheat may also facilitate the efforts being made towards molecular dissection of genomic regions involved in thermotolerance. Therefore, association of MQTLs with known thermotolerance genes was also examined. Nine such MQTLs were associated with seven genes known for thermotolerance in wheat (Table 3). These known genes associated with thermotolerance included three different classes: (i) Following four HSP/HSF genes were found in the following seven MQTL regions: TaHSP17 in MQTL3D.1, TaHSP101 in MQTL3D.1, 3D.3, TaHSP90 in MQTL7A.2, 7A.3, 7A.4, 7A.5, and TaHsfA6f in MQTL5D.4. The HSPs are a large family of chaperones that play a major role in providing thermotolerance (Kumar et al. 2012; Campbell et al. 2001; Vishwakarma et al. 2018; Xue et al. 2015). (ii) The gene TaGF14-JM22 (encoding 14–3-3 protein-like) corresponding to MQTL4A.3 is known to be involved in starch biosynthesis. This gene has a negative association with thermotolerance so that its reduced expression enhances thermotolerance (Guo et al. 2018). (iii) TaFER-5B and TaPEPKR2 genes, respectively, encoding for ferritin and phosphoenolpyruvate carboxylase kinase-related kinase overlap MQTL5B.1. TaFER-5B is known to provide thermotolerance through scavenging reactive oxygen species (Zang et al. 2017) and TaPEPKR2 has been characterized for thermotolerance and drought tolerance in wheat and Arabidopsis (Zang et al. 2018). These data suggest that the nine MQTLs (Table 3), which correspond to seven known genes for thermotolerance, are promising candidates for the improvement of this trait and could be useful in wheat breeding for climate resilience. Additional efforts are needed to identify functional variants of these genes in the targeted MQTL regions.
Table 3.
Co-location of MQTLs with known genes for thermotolerance in wheat
| Chr | MQTL designation | Co-located known gene (accession ID; physical coordinates, bp) | Encoded protein | Associated traits | Reference |
|---|---|---|---|---|---|
| 3D | MQTL3D.1 | TaHSP17 (JN572711; 68,325,094–68,325,579) | Small heat shock proteins | Confers thermotolerance | Kumar et al. (2012) |
| 3D | MQTL3D.1, 3D.3 | TaHSP101 (AF174433; 379,536,976–379,539,614) | Heat shock protein | Involves in both heat and drought responses | Campbell et al. (2001) |
| 4A | MQTL4A.3 | TaGF14-JM22 (JF957590: 580,906,528–580,908,057) | 14–3-3 protein-like | Involves in starch biosynthesis | Guo et al. (2018) |
| 5B | MQTL5B.1 | TaFER-5B (KX025176; 278,795,539–278,797,502) | Ferritin | Enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging | Zang et al. (2017) |
| 5B | MQTL5B.1 | TaPEPKR2 (GU213488.1; 424,332,735–424,335,643) | Phosphoenolpyruvate carboxylase kinase-related kinase | Enhances heat and dehydration tolerance | Zang et al. (2018) |
| 5D | MQTL5D.4 | TaHsfA6f (KJ685918; 461,911,512–461,912,559) | Heat shock protein | Confers thermotolerance | Xue et al. (2015) |
| 7A | MQTL7A.2-7A.5 | TaHSP90 (MF383197; 217,556,415–217,557,203) | Heat shock protein | Confers tolerance to heat and drought stresses | Vishwakarma et al. (2018) |
Identification of candidate genes, in silico expression, and GO analysis
Mining of genes in genomic regions of all the MQTLs (except MQTL1D.1, 4B.2, 5A.3) allowed identification of 1,704 non-redundant CGs (Online Resource 9), which included the following: (i) 165 CGs including 55 CGs with each of the three MQTLs (MQTL1A.4, 2A.1, 7D.4) and a solitary CG each with two MQTL (MQTL4B.5 and 4D.4). (ii) 260 genes, for which no description for functions was available, and another 78 CGs, which were either uncharacterized or with no known encoded proteins (Online Resource 9). (iii) A large number of CGs, each encoding a variety of proteins products in more than one MQTL region showing redundancy of genes in the wheat genome. These included 102 CGs for protein kinases, 55 CGs for F-box-like domains, 43 CGs for cytochrome P450, 38 CGs for proteins with leucine-rich repeats, 37 CGs for zinc finger RING/FYVE/PHD-type, and 27 CGs for pentatricopeptide repeats (Fig. S4).
Functional relevance of the CGs/gene families listed above in category (iii) with thermotolerance has also been documented previously (Park et al. 2014; Agarwal and Khurana 2018; Chen et al. 2018; Pandian et al. 2020; Kumar et al. 2021; Guérin et al. 2021). The following are some examples: (i) a mitogen-activated protein kinase (MAPK) was recently shown to be associated with oxidative stress tolerance and carbon assimilation in wheat under terminal heat stress (Kumar et al. 2021); (ii) F-box-like proteins are known to play a major role in plant responses to diverse developmental and stress conditions in wheat (Guérin et al. 2021); (iii) cytochrome P450s are the largest enzyme family involved in O2 and/or NADPH-dependent hydroxylation reactions. The expressions of cytochrome P450 genes have been shown to play a prominent role in the crosstalk between abiotic and biotic stress responses (Pandian et al. 2020); (iv) leucine-rich repeats are known to act as receptors of external signals, e.g., abiotic stresses such as heat, salt, and osmotic stresses (Park et al. 2014).
The above CGs were also subjected to an in silico expression analysis under 1 h and 6 h of heat stress using RNA-seq data available in a public database. As many as 182 CGs, belonging to 71 MQTLs, exhibited differential expression; these differentially expressed CGs (DECGs) were categorized into up-regulating (FC ≥ 2) and down-regulating (FC ≤ − 2) DECGs. From these DECGs, 92 were differentially expressed at 1 h of heat stress (16 up-regulated and 76 down-regulated), 116 were differentially expressed at 6 h of heat stress (55 up-regulated and 61 down-regulated), and only 31 were differentially expressed at both conditions (10 up-regulated, 15 down-regulated, 6 up-regulated at one time-point and down-regulated at the other). A representative heat map of some important DECGs is shown in Fig. 3. The genes with uncharacterized proteins need to be subjected to further investigations for determining their roles in thermotolerance.
Fig. 3.
Heat maps representing differential expression of some important genes underlying 33 MQTLs (shown on the left). The scale drawn on the top of heat maps exhibits the level of expression. Proteins which are known to be associated with thermotolerance are marked by an asterisk (*)
The above 182 DECGs (belonging to 71 MQTLs) were also analyzed for gene ontology (GO) enrichment (Online Resource 9). The GO terms associated with biological processes belonged to metabolic (12 CGs) and cellular processes (6 CGs) (Fig. S5). Similarly, GO terms associated with molecular function were for heterocyclic compound binding (25 CGs) and catalytic activity (19 CGs). In relation with cellular components, the CGs were enriched mainly in the cell membrane and its components such as intracellular anatomical structure and cell periphery.
Among the above 1,704 genes, thirty-six (36) high confidence CGs were also detected (Table 4), which either exhibited differential expressions under heat stress or were known to be important for traits related to thermotolerance. The role of some of the important high confidence CGs in providing thermotolerance can be summarized as follows: (i) CGs encoding UDP-glucosyltransferase redirect the metabolic flux from lignin biosynthesis to flavonoid biosynthesis under abiotic stress (heat, drought, and salt stress) and the accumulation of flavonoid glycosides providing protection against abiotic stress leading to improvement in grain size (Dong et al. 2020). (ii) Over-expression of a CG encoding a zinc finger transcription factor conferred improved thermotolerance and grain yield, as demonstrated in Arabidopsis thaliana (Agarwal and Khurana 2018), (iii) CGs encoding soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are also known to be involved in abiotic and biotic stresses, as reported in several plant species (Kwon et al. 2020), (iv) over-expression of MYB genes provided improved thermotolerance and drought tolerance; this has been attributed to enhanced levels of cellular abscisic acid in wheat (Zhao et al. 2017), (v) a KH domain-containing putative RNA-binding protein was reported to be crucial for the regulation of heat stress-responsive CG and thermotolerance in A. thaliana (Guan et al. 2013), and (vi) a CG encoding C2 domain protein was reported to play a key role in the improvement of A. thaliana thermotolerance, mainly through the enhancement of antioxidant enzyme activity, leading to enhanced expression of genes for response to heat stress response (Ye et al. 2020). While these potential CGs are promising targets for future breeding and genetic studies, it is not apparent whether they are the actual regulators behind the MQTLs, since many other genes occur within the CIs of the MQTLs. Further studies are warranted to validate the role of these CGs.
Table 4.
High confidence candidate genes (CGs) underlying MQTLs for thermotolerance
| MQTL designation | Gene ID (expression pattern) | GO term | Functional description |
|---|---|---|---|
| MQTL1A.1 | TraesCS1A02G009320 (↓) | ADP binding | NB-ARC |
| MQTL1A.1 | TraesCS1A02G010400 (↓) | ADP binding | Leucine-rich repeat |
| MQTL1A.3 | TraesCS1A02G016200 (↓) | ADP binding | NB-ARC |
| MQTL2A.1 | TraesCS2A02G005500 (↑) | Integral component of membrane | Late embryogenesis abundant protein |
| MQTL2A.1 | TraesCS2A02G007500 (↓) | Integral component of membrane | Proton-dependent oligopeptide transporter family |
| MQTL2A.2 | TraesCS2A02G085000 (↑) (↓) | Integral component of membrane | WD40 repeat |
| MQTL2A.3 | TraesCS2A02G093700 (↓) | Protein serine/threonine phosphatase activity | PPM-type phosphatase domain |
| MQTL2B.1 | TraesCS2B02G067000 (↓) | Integral component of membrane | Protein kinase domain |
| MQTL2B.5 | TraesCS2B02G539600 (↑) (↓) | Membrane | Glycosyl transferase |
| MQTL3A.4 | TraesCS3A02G454600 (↑) (↓) | DNA binding | Homeobox domain |
| MQTL3A.4 | TraesCS3A02G455000 (↑) | - | Alpha/beta hydrolase fold |
| MQTL3A.4 | TraesCS3A02G456200 (↑) | - | Alpha/beta hydrolase fold-1 |
| MQTL3A.4 | TraesCS3A02G456300 (↓) | - | Alpha/beta hydrolase fold-1 |
| MQTL3D.1 | TraesCS3D02G127000 (↓) | Protein binding | F-box domain |
| MQTL3D.2 | TraesCS3D02G037200 (↓) | Lipid binding | C2 domain |
| MQTL3D.2 | TraesCS3D02G037600 (↓) | Structural constituent of ribosome | Ribosomal protein S10 |
| MQTL3D.2 | TraesCS3D02G037900 (↓) | Regulation of systemic acquired resistance | NPR1/NH1-interacting protein |
| MQTL4A.1 | TraesCS4A02G016900 (↑) (↓) | Oxidoreductase activity | 3-dehydroquinate synthase |
| MQTL4A.1 | TraesCS4A02G017700 (↓) | Nucleus | Zinc finger, Dof-type |
| MQTL4A.1 | TraesCS4A02G019000 (↓) | Structural constituent of ribosome | Ribosomal protein |
| MQTL4B.6 | TraesCS4B02G041800 (↓) | Integral component of membrane | SNARE associated Golgi protein |
| MQTL4D.3 | TraesCS4D02G008500 (↓) | Hydrolase activity, acting on ester bonds | GDSL lipase/esterase |
| MQTL5B.1 | TraesCS5B02G412200 (↑) (↓) | ATP binding | Protein kinase domain |
| MQTL5B.1 | TraesCS5B02G412900 (↓) | Hydrolase activity | Pectinacetylesterase/NOTUM |
| MQTL5D.1 | TraesCS5D02G205200 (↓) | Nucleic acid binding | RNA recognition motif domain |
| MQTL5D.2 | TraesCS5D02G217000 (↓) | rRNA binding | Brix domain |
| MQTL5D.5 | TraesCS5D02G339700 (↑) (↓) | Integral component of membrane | Protein kinase domain |
| MQTL5D.5 | TraesCS5D02G340000 (↑) (↓) | Regulation of protein catabolic process | Proteasome component (PCI) domain |
| MQTL6A.1 | TraesCS6A02G022100 (↓) | Transferase activity, transferring acyl groups other than amino-acyl groups | Transferase |
| MQTL6D.1 | TraesCS6D02G328700 (↑) (↓) | - | Zinc finger, RING-type |
| MQTL7A.1 | TraesCS7A02G287000 (↑) (↓) | RNA binding | K Homology domain, type 2 |
| MQTL7A.5 | TraesCS7A02G193000 (↓) | protein binding | Ankyrin repeat |
| MQTL7A.7 | TraesCS7A02G184700 (↑) (↓) | ATP binding | AAA + ATPase domain |
| MQTL7D.2 | TraesCS7D02G049300 (↓) | Integral component of membrane | Proton-dependent oligopeptide transporter family |
| MQTL7D.4 | TraesCS7D02G011800 (↓) | ATP binding | Protein kinase domain |
| MQTL7D.4 | TraesCS7D02G014900 (↓) | Nucleus | SANT/Myb domain |
GO gene ontology, ↑ up-regulated genes, ↓ down-regulated genes, ↑↓ up-regulated under at one time-point and down-regulated at other
Ortho-MQTLs for wheat and rice
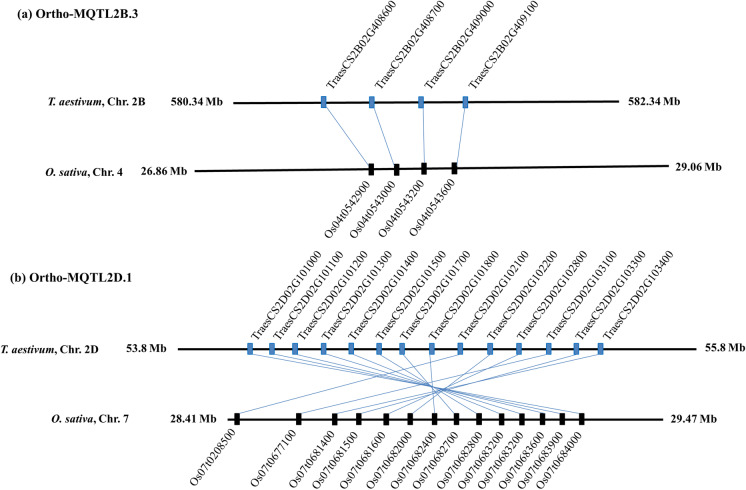
Chromosomal regions carrying MQTLs that are conserved across more than one cereal were also identified. For this purpose, MQTLs for different traits relevant to thermotolerance in the present study in wheat were compared with MQTLs reported in rice (Raza et al. 2020). Ortho-MQTLs could be identified for only seven wheat MQTLs positioned on chromosomes 1B, 2B, 2D, 3B, 7B, and 7D that correspond to rice-MQTLs (rMQTLs) located on rice chromosomes 1, 4, 5, 6, 7, and 8. This confirms the occurrence of wheat MQTLs that are orthologous to rMQTLs. Detailed information related to ortho-MQTLs and underlying genes along with physical positions and functional description is presented in Table 5 and Online Resource 10. Among ortho-MQTLs, two ortho-MQTLs (ortho-MQTL2B.3 and ortho-MQTL2D.1) were considered most promising for revealing gene collinearity within a syntenic region between wheat and rice. For ortho-MQTL2B.3, four underlying CGs on chromosome 2B were found to be syntenous and collinear with similar CGs belonging to rMQTL4.4 on rice chromosome 4 (Fig. 4a and Table 5). The wheat ortho-MQTL2D.1 on chromosome 2D was syntenous with rMQTL7.1 on rice chromosome 7 (Fig. 4b and Table 5). The orthologous genes located at the two most promising syntenic regions (viz., ortho-MQTL2B.3 and 2D.1) were subjected to further analysis. Ortho-MQTL2B.3 contained at least two heat-responsive genes encoding for protein kinase (Sinha et al. 2011) and amino acid/polyamine transporter (Shen et al. 2016). Ortho-MQTL2D.1 included at least 5 heat-responsive genes encoding for the following proteins/products: NAC domain-containing protein (Ren et al. 2021), nucleoside phosphatase (Escobar et al. 2001), PLD-regulated protein1-like (Mishkind et al. 2009), RNA recognition motif domain (Lee et al. 2017), and helicase (Gong et al. 2005).
Table 5.
Identification of ortho-MQTLs between wheat and rice genomes
| Ortho-MQTL designation | Wheat MQTL (physical position, Mb)a | Rice MQTL (chromosome; physical position, Mb)b |
|---|---|---|
| Ortho-MQTL1B.4 | MQTL1B.4 (7.33–9.33) | rMQTL5.3 (5; 24.26–27.34) |
| Ortho-MQTL2B.3 | MQTL2B.3 (580.34–582.34) | rMQTL4.4 (4; 26.86–29.06) |
| Ortho-MQTL2D.1 | MQTL2D.1 (53.8–55.8) | rMQTL7.1 (7; 28.41–29.47) |
| Ortho-MQTL2D.3 | MQTL2D.3 (26.18–28.18) | rMQTL4.3 (4; 21.45–24.16), rMQTL4.4 (4; 26.86–29.06) |
| Ortho-MQTL3B.2 | MQTL3B.2 (9.74–11.74) | rMQTL1.2 (1; 34.95–36.73) |
| Ortho-MQTL7B.3 | MQTL7B.3 (82.78–84.78) | rMQTL6.3 (6:16.75–19.34), rMQTL6.2 (6:3.17–5.09), rMQTL8.2 (8:22.47–25.59) |
| Ortho-MQTL7D.4 | MQTL7D.4 (4.55–6.55) | rMQTL8.1 (8; 3.93–9.78) |
aGenomic position of meta-QTL (MQTL) peak region (total 2 Mb region), bRaza et al. (2020), MB mega base
Fig. 4.
Representing syntenic regions of two ortho-MQTLs between wheat and rice. The chromosome number, physical position, and common genes between the wheat and rice are indicated. More details are given in Online Resource 10
The crucial roles of some of these genes associated with ortho-MQTLs in conferring thermotolerance can be summarized as follows: (i) mitogen-activated protein kinase cascades have been shown to be involved in signalling pathways activated by different abiotic stresses including heat stress in different plant species (Sinha et al. 2011); (ii) polyamine transporter is required for the uptake of extracellular polyamines and is known to play a key role in stabilizing the mRNAs of several important heat stress-responsive genes under heat stress in Arabidopsis (Shen et al. 2016); (iii) CRISPR/Cas9 based genome editing and transgenesis demonstrated a transcriptional regulatory network with two NAC genes (ONAC127 and ONAC129) coordinating multiple pathways to modulate grain development and heat stress responses at reproductive stages in rice (Ren et al. 2021); (iv) a mitochondrial nucleoside diphosphate kinase (mtNDPK) is known to be involved in heat stress response in pea, possibly as a modulator of a protein (86 kilo-dalton protein) synthesized upon exposure to heat (Escobar et al. 2001); (v) phospholipase D and phosphatidylinositol 4,5-bisphosphate are accumulated at the plasma membrane and nucleus, thereby regulate the ion channels and the cytoskeleton under heat stress in Arabidopsis and rice (Mishkind et al. 2009); and (vi) the RNA recognition motif of Arabidopsis splicing factor affects the alternative splicing of pre-mRNAs of genes, including FLOWERING LOCUS M and a heat shock transcription factor involved in the regulation of flowering and adaptation to heat stress (Lee et al. 2017). Among the remaining five ortho-MQTLs, only one ortholog matched between wheat and rice MQTLs (Table 5).
Ortho-MQTLs in wheat in particular and cereals in general have been examined sparingly; at least three such studies are available (Khahani et al. 2020, 2021; Saini et al. 2021). The discovery of ortho-MQTLs in closely related species increases their significance and verifies their stability, as well as the confidence of associated genes (Khahani et al. 2020, 2021). The orthologous genes detected in the present study may be functionally validated and gene-based functional markers can be developed for their use in cereal breeding programs (Quraishi et al. 2009).
Conclusions
The present study involved identification of MQTLs using information about the known QTLs for thermotolerance. Improved meta-analysis strategy was used leading to the identification of 85 most stable and robust MQTLs; 77 of these MQTLs were confirmed using available results of recent GWA studies. Seven promising MQTLs (breeders’ MQTLs) were recommended for use in MAB for improvement of thermotolerance in wheat. Ortho-MQTLs for wheat and rice identified in the present study allowed identification of conserved genomic regions associated with heat-responsive traits in rice. As many as 1,704 CGs underlying 82 MQTLs were also identified which included 182 CGs with differential expression under heat stress. In addition, 36 high confidence CGs underlying 24 MQTLs with known functions were identified as being reliable for thermotolerance. These CGs may be validated using one or more of the available approaches (including transgenesis, gene editing, and gene knockout strategies). The results may also be used for studies involving proteomics and metabolomics. On validation, these CGs can be used in breeding programs for improving thermotolerance and may also be used as a resource for designing research programs for understanding the mechanism for thermotolerance in wheat.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The present study was supported by the Biotechnology Industry Research Assistance Council (BIRAC), Government of India, New Delhi and United States Agency for International Development (USAID), USA. HSB was awarded Honorary Scientist position by the Indian National Science Academy (INSA), New Delhi during the course of this study. The Head, Department of Genetics and Plant Breeding, Ch. Charan Singh University, Meerut provided necessary facilities for this study.
Author contribution
SK, HSB, and PKG planned this study. SoK, VPS, DKS, HS, and SGM performed the literature search, retrieved data, developed consensus map, conducted meta-analysis, and wrote the first draft of the manuscript. SK, HSB, and PKG critically revised and edited the manuscript. All authors have read and agree to the final version of the manuscript.
Funding
This research was funded by the “Biotechnology Industry Research Assistance Council (BIRAC), Government of India, New Delhi” and “United States Agency for International Development (USAID), USA” (Grant No. BIRAC/TG/USAID/08/2014).
Availability of data and material
Relevant data are included in this paper and its associated Supplementary Information (SI).
Code availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Acuña-Galindo MA, Mason RE, Subramanian NK, Hays DB. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 2015;55:477–492. doi: 10.2135/cropsci2013.11.0793. [DOI] [Google Scholar]
- Aduragbemi A, Soriano JM (2021) Unravelling consensus genomic regions conferring leaf rust resistance in wheat via meta-QTL analysis. bioRxiv. 10.1101/2021.05.11.443557 [DOI] [PubMed]
- Agarwal P, Khurana P. Characterization of a novel zinc finger transcription factor (TaZnF) from wheat conferring thermotolerance in Arabidopsis. Cell Stress Chaperones. 2018;23:253–267. doi: 10.1007/s12192-017-0838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Anderson JA, Sorrells ME, Tanksley SD. Homoeologous relationships of rice wheat and maize chromosomes. Mol Gen Genet. 1993;241:483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- Akter N, Rafiqul Islam M. Heat stress effects and management in wheat. A Review Agron Sustain Dev. 2017;37:37. doi: 10.1007/s13593-017-0443-9. [DOI] [Google Scholar]
- Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J. BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics. 2004;20:2324–2326. doi: 10.1093/bioinformatics/bth230. [DOI] [PubMed] [Google Scholar]
- Asseng S, Foster IAN, Turner NC. The impact of temperature variability on wheat yields. Glob Change Biol. 2011;17:997–1012. doi: 10.1111/j.1365-2486.2010.02262.x. [DOI] [Google Scholar]
- Barkley A, Tack J, Nalley LL, Bergtold J, Bowden R, Fritz A. Weather disease and wheat breeding effects on Kansas wheat varietal yields 1985 to 2011. Agron J. 2014;106:227–235. doi: 10.2134/agronj2013.0388. [DOI] [Google Scholar]
- Beecher FW, Mason E, Mondal S, Awika J, Hays D, Ibrahim A. Identification of quantitative trait loci (QTLs) associated with maintenance of wheat (Triticum aestivum L.) quality characteristics under heat stress conditions. Euphytica. 2012;188:361–368. doi: 10.1007/s10681-012-0687-z. [DOI] [Google Scholar]
- Bheemanahalli R, Sunoj VJ, Saripalli G, Prasad PV, Balyan HS, Gupta PK, Grant N, Gill KS, Krishna Jagdish SV. Quantifying the impact of heat stress on pollen germination seed set and grain filling in spring wheat. Crop Sci. 2019;59:684–696. doi: 10.2135/cropsci2018.05.0292. [DOI] [Google Scholar]
- Bilgrami SS, Ramandi HD, Shariati V, Razavi K, Tavakol E, Fakheri BA, Nezhad NM, Ghaderian M. Detection of genomic regions associated with tiller number in iranian bread wheat under different water regimes using genome-wide association study. Sci Rep. 2020;10:1–17. doi: 10.1038/s41598-020-69442-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Wang S, Su Z, Li T, Zhang X, Bai G. Meta-analysis of QTL for fusarium head blight resistance in Chinese wheat landraces. Crop J. 2019;7:784–798. doi: 10.1016/j.cj.2019.05.003. [DOI] [Google Scholar]
- Campbell JL, Natalya Y, Hong-gang ZK, Jorge NS, T-HD H, Henry TN, Cloning of new members of heat shock protein HSP101 gene family in wheat (Triticum aestivum (L.) Moench) inducible by heat dehydration and ABA. Biochim Biophys Acta. 2001;15172:270–277. doi: 10.1016/S0167-4781(00)00292-X. [DOI] [PubMed] [Google Scholar]
- Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics. 2004;168:2169–2185. doi: 10.1534/genetics.104.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BS, Prabhjyot K, Mahajan G, Randhawa RK, Singh H, Kang MS. Global warming and its possible impact on agriculture in India. Adv Agron. 2014;123:65–121. doi: 10.1016/B978-0-12-420225-2.00002-9. [DOI] [Google Scholar]
- Chen G, Zou Y, Hu J, Ding Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics. 2018;19:1–14. doi: 10.1186/s12864-018-5088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav Genet. 1997;27:125–132. doi: 10.1023/A:1025685324830. [DOI] [PubMed] [Google Scholar]
- De Costa WAJM. A review of the possible impacts of climate change on forests in the humid tropics. J Natl Sci Found Sri. 2011;39:281–302. [Google Scholar]
- Djanaguiraman M, Narayanan S, Erdayani E. Effects of high temperature stress during anthesis and grain filling periods on photosynthesis lipids and grain yield in wheat. BMC Plant Biol. 2020;20:268. doi: 10.1186/s12870-020-02479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong NQ, Sun Y, Guo T, Shi CL, Zhang YM, Kan Y, Xiang YH, Zhang H, Yang YB, Li YC, Zhao HY. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat Commun. 2020;11:1–16. doi: 10.1038/s41467-020-16403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman JB, Plomion C. LPmerge: an R package for merging genetic maps by linear programming. Bioinformatics. 2014;30:1623–1624. doi: 10.1093/bioinformatics/btu091. [DOI] [PubMed] [Google Scholar]
- Escobar GML, Marttila S, Håkansson G, Forsberg J, Knorpp C. Heat stress response in pea involves interaction of mitochondrial nucleoside diphosphate kinase with a novel 86-kilodalton protein. Plant Physiol. 2001;126:69–77. doi: 10.1104/pp.126.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegas VC, Nazari A, Griffiths S, Simmonds J, Fish L, Orford S, Sayers L, Doonan JH, Snape JW. A genetic framework for grain size and shape variation in wheat. Plant Cell. 2010;22:1046–1056. doi: 10.1105/tpc.110.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet B, Gerber S. Quantitative trait loci: a meta-analysis. Genetics. 2000;155:463–473. doi: 10.1093/genetics/155.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17:256–267. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Wen C, Zeng H, Zhu J. A KH domain-containing putative RNA-binding protein is critical for heat stress-responsive gene regulation and thermo-tolerance in Arabidopsis. Mol Plant. 2013;6:386–395. doi: 10.1093/mp/sss119. [DOI] [PubMed] [Google Scholar]
- Guérin C, Mouzeyar S, Roche J. The landscape of the genomic distribution and the expression of the F-Box genes unveil genome plasticity in hexaploid wheat during grain development and in response to heat and drought stress. Int J Mol Sci. 2021;22:3111. doi: 10.3390/ijms22063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Sleper DA, Lu P, Shannon JG, Nguyen HT, Arelli PR. QTLs associated with resistance to soybean cyst nematode in soybean meta-analysis of QTL locations. Crop Sci. 2006;46:202–202. doi: 10.2135/cropsci2005.04-0036. [DOI] [Google Scholar]
- Guo J, Dai S, Li H, Liu A, Liu C, Cheng D, Cao X, Chu X, Zhai S, Liu J, Zhao Z. Identification and expression analysis of wheat TaGF14 genes. Front Genet. 2018;9:12. doi: 10.3389/fgene.2018.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK, Balyan HS, Kulwal P, Gahlaut V. Phenotyping genetic dissection and breeding for tolerance to drought and heat in common wheat: present status and future prospects. Plant Breed Rev. 2012;36:82–168. [Google Scholar]
- Gupta PK, Balyan HS, Sharma S, Kumar R. Genetics of yield, abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.) Theor Appl Genet. 2020;133:1569–1602. doi: 10.1007/s00122-020-03583-3. [DOI] [PubMed] [Google Scholar]
- Jacott CN, Boden SA. Feeling the heat: developmental and molecular responses of wheat and barley to high ambient temperatures. J Exp Bot. 2020;71:5740–5751. doi: 10.1093/jxb/eraa326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagdish SVK. Heat stress during flowering in cereals–effects and adaptation strategies. New Phytol. 2020;226:1567–1572. doi: 10.1111/nph.16429. [DOI] [PubMed] [Google Scholar]
- Jan I, Saripalli G, Kumar K, Kumar A, Singh R, Batra R, Sharma PK, Balyan HS, Gupta PK (2021) Meta-QTL analysis for stripe rust resistance in wheat. Res Square. 10.21203/rs.3.rs-380807/v1 [DOI] [PMC free article] [PubMed]
- Khahani B, Tavakol E, Shariati V, Fornara F. Genome wide screening and comparative genome analysis for meta-QTLs ortho-MQTLs and candidate genes controlling yield and yield-related traits in rice. BMC Genomics. 2020;21:1–24. doi: 10.1186/s12864-020-6702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khahani B, Tavakol E, Shariati V, Rossini L. Meta-QTL and ortho-MQTL analyses identified genomic regions controlling rice yield yield-related traits and root architecture under water deficit conditions. Sci Rep. 2021;11:1–18. doi: 10.1038/s41598-021-86259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen K, Schelde K, Olesen JE. Winter wheat yield response to climate variability in Denmark. J Agric Sci. 2011;149:33–47. doi: 10.1017/S0021859610000675. [DOI] [Google Scholar]
- Kumar A, Saripalli G, Jan I, Kumar K, Sharma PK, Balyan HS, Gupta PK. Meta-QTL analysis and identification of candidate genes for drought tolerance in bread wheat (Triticum aestivum L.) Physiol Mol Biol Plants. 2020;26:1713–1725. doi: 10.1007/s12298-020-00847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RR, Dubey K, Arora K, Dalal M, Rai GK, Mishra D, Chaturvedi KK, Rai A, Kumar SN, Singh B, Chinnusamy V. Characterizing the putative mitogen-activated protein kinase (MAPK) and their protective role in oxidative stress tolerance and carbon assimilation in wheat under terminal heat stress. Biotechnol Rep. 2021;29:00597. doi: 10.1016/j.btre.2021.e00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RR, Singh GP, Sharma SK, Singh K, Goswami S, Rai RD. Molecular cloning of HSP17 gene (sHSP) and their differential expression under exogenous putrescine and heat shock in wheat (Triticum aestivum) Afr J Biotechnol. 2012;11:16800–16808. [Google Scholar]
- Kumar S, Mohan A, Balyan HS, Gupta PK. Orthology between genomes of brachypodium wheat and rice. BMC Res Notes. 2009;2:1–9. doi: 10.1186/1756-0500-2-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Lee JH, Yun HS. SNAREs in plant biotic and abiotic stress responses. Mol Cells. 2020;43:501. doi: 10.14348/molcells.2020.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Jang YH, Kim SK, Park HY, Thu MP, Lee JH, Kim JK. RRM domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Rep. 2017;36:1083–1095. doi: 10.1007/s00299-017-2140-1. [DOI] [PubMed] [Google Scholar]
- Lesk C, Rowhani P, Ramankutty N. Influence of extreme weather disasters on global crop production. Nature. 2016;529:84–87. doi: 10.1038/nature16467. [DOI] [PubMed] [Google Scholar]
- Liu H, Mullan D, Zhang C, Zhao S, Li X, Zhang A, Lu Z, Wang Y, Yan G. Major genomic regions responsible for wheat yield and its components as revealed by meta-QTL and genotype–phenotype association analyses. Planta. 2020;252:1–22. doi: 10.1007/s00425-020-03466-3. [DOI] [PubMed] [Google Scholar]
- Liu S, Hall MD, Griffey CA, Mckendry AL. Meta-analysis of QTL associated with fusarium head blight resistance in wheat. Crop Sci. 2009;49:1955–1968. doi: 10.2135/cropsci2009.03.0115. [DOI] [Google Scholar]
- Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, Sun Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.) BMC Plant Biol. 2015;15:1–20. doi: 10.1186/s12870-015-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Liu H, Hong Y, Li H, Liu H, Li X, Wen S, Zhou G, Li S, Chen X, Liang X. Consensus map integration and QTL meta-analysis narrowed a locus for yield traits to 0.7 cM and refined a region for late leaf spot resistance traits to 0.38 cM on linkage group A05 in peanut (Arachis hypogaea L.) BMC Genomics. 2018;19:887. doi: 10.1186/s12864-018-5288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, Ricci A, Salvi S, Milner SG, Noli E, Martelli PL, Casadio R, Akhunov E, Scalabrin S, Vendramin V, Ammar K. A high-density SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol J. 2015;13:648–663. doi: 10.1111/pbi.12288. [DOI] [PubMed] [Google Scholar]
- Mao SL, Wei YM, Cao W, Lan XJ, Yu M, Chen ZM, Chen GY, Zheng YL. Confirmation of the relationship between plant height and Fusarium head blight resistance in wheat (Triticum aestivum L.) by QTL meta-analysis. Euphytica. 2010;174:343–356. doi: 10.1007/s10681-010-0128-9. [DOI] [Google Scholar]
- Marone D, Russo MA, Laidò G, Vita PD, Papa R, Blanco A, Gadaleta A, Rubiales D, Mastrangelo AM. Genetic basis of qualitative and quantitative resistance to powdery mildew in wheat: from consensus regions to candidate genes. BMC Genomics. 2013;14:562. doi: 10.1186/1471-2164-14-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RE, Mondal S, Beecher FW, Pacheco A, Jampala B, Ibrahim AM, Hays DB. QTL associated with heat susceptibility index in wheat (Triticum aestivum L.) under short-term reproductive stage heat stress. Euphytica. 2010;174:423–436. doi: 10.1007/s10681-010-0151-x. [DOI] [Google Scholar]
- Mishkind M, Vermeer JE, Darwish E, Munnik T. Heat stress activates phospholipase D and triggers PIP2 accumulation at the plasma membrane and nucleus. Plant J. 2009;60:10–21. doi: 10.1111/j.1365-313X.2009.03933.x. [DOI] [PubMed] [Google Scholar]
- Pandian BA, Sathishraj R, Djanaguiraman M, Prasad PV, Jugulam M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants. 2020;9:454. doi: 10.3390/antiox9050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Moon JC, Park YC, Kim JH, Kim DS, Jang CS. Molecular dissection of the response of a rice leucine-rich repeat receptor-like kinase (LRR-RLK) gene to abiotic stresses. J Plant Physiol. 2014;171:1645–1653. doi: 10.1016/j.jplph.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Poudel PB, Poudel MR. Heat stress effects and tolerance in wheat: a review. J Biol Today's World. 2020;9:217. [Google Scholar]
- Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, Charmet G. Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genomics. 2009;9:473–484. doi: 10.1007/s10142-009-0129-8. [DOI] [PubMed] [Google Scholar]
- Quraishi UM, Murat F, Abrouk M, Pont C, Confolent C, Oury FX, Ward J, Boros D, Gebruers K, Delcour JA, Courtin CM. Combined meta-genomics analyses unravel candidate genes for the grain dietary fiber content in bread wheat (Triticum aestivum L.) Funct Integr Genomics. 2011;11:71–83. doi: 10.1007/s10142-010-0183-2. [DOI] [PubMed] [Google Scholar]
- Quraishi UM, Pont C, Ain QU, Flores R, Burlot L, Alaux M, Quesneville H, Salse J. Combined genomic and genetic data integration of major agronomical traits in bread wheat (Triticum aestivum L.) Front Plant Sci. 2017;8:1843. doi: 10.3389/fpls.2017.01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam J, Pathan MS, Feril O, Miftahudin M, Ross K, Ma XF, Mahmoud AA, Layton J, Rodriguez MA, Chikmawati T, Valliyoda B. Structural and functional analyses of the wheat genomes based on expressed sequence tags (ESTs) related to abiotic stresses. Genomics. 2006;49:1324–1340. doi: 10.1139/g06-094. [DOI] [PubMed] [Google Scholar]
- Ramandi DH, Shariati JV, Tavakol E, Zarini NH, Bilgrami SS, Razavi K. Detection of consensus genomic regions associated with root architecture of bread wheat on groups 2 and 3 chromosomes using QTL meta-analysis. Aust J Crop Sci. 2017;11:777–785. doi: 10.21475/ajcs.17.11.07.pne312. [DOI] [Google Scholar]
- Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, Van Ex F, Pasha A, Khedikar Y, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361:6403. doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- Raza Q, Riaz A, Bashir K, Sabar M. Reproductive tissues-specific meta-QTLs and candidate genes for development of heat-tolerant rice cultivars. Plant Mol Biol. 2020;104:97–112. doi: 10.1007/s11103-020-01027-6. [DOI] [PubMed] [Google Scholar]
- Ren Y, Huang Z, Jiang H, Wang Z, Wu F, Xiong Y, Yao J. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J Exp Bot. 2021;72:2947–2964. doi: 10.1093/jxb/erab027. [DOI] [PubMed] [Google Scholar]
- Safdar LB, Andleeb T, Latif S, Umer MJ, Tang M, Li X, Liu S, Quraishi UM. Genome-wide association study and QTL meta-analysis identified novel genomic loci controlling potassium use efficiency and agronomic traits in bread wheat. Front Plant Sci. 2020;11:70. doi: 10.3389/fpls.2020.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini DK, Srivastava P, Pal N, Gupta PK (2021) Meta-QTLs Ortho-Meta-QTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.). Res Square. 10.21203/rs.3.rs-430452/v1 [DOI] [PubMed]
- Shariatipour N, Heidari B, Richards CM. Meta-analysis of QTLome for grain zinc and iron contents in wheat (Triticum aestivum L.) Euphytica. 2021;217:1–14. doi: 10.1007/s10681-021-02818-8. [DOI] [Google Scholar]
- Sharma DK, Torp AM, Rosenqvist E, Ottosen CO, Andersen SB. QTLs and potential candidate genes for heat stress tolerance identified from the mapping populations specifically segregating for Fv/Fm in wheat. Front Plant Sci. 2017;8:1668. doi: 10.3389/fpls.2017.01668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Ruan Q, Chai H, Yuan Y, Yang W, Chen J, Xin Z, Shi H. The Arabidopsis polyamine transporter LHR 1/PUT 3 modulates heat responsive gene expression by enhancing mRNA stability. Plant J. 2016;88:1006–1021. doi: 10.1111/tpj.13310. [DOI] [PubMed] [Google Scholar]
- Shirdelmoghanloo H, Taylor JD, Lohraseb I, Rabie H, Brien C, Timmins A, Martin P, Mather DE, Emebiri L, Collins NC. A QTL on the short arm of wheat (Triticum aestivum L.) chromosome 3B affects the stability of grain weight in plants exposed to a brief heat shock early in grain filling. BMC Plant Biol. 2016;16:1–15. doi: 10.1186/s12870-016-0784-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Batra R, Sharma S, Saripalli G, Gautam T, Singh R, Pal S, Malik P, Kumar M, Jan I, Singh S, et al. WheatQTLdb: a QTL database for wheat. Mol Genet Genomics. 2021;2965:1051–1056. doi: 10.1007/s00438-021-01796-9. [DOI] [PubMed] [Google Scholar]
- Sinha AK, Jaggi M, Raghuram B, Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav. 2011;6:196–203. doi: 10.4161/psb.6.2.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L) Theor Appl Genet. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Soriano JM, Alvaro F. Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci Rep. 2019;9:1–14. doi: 10.1038/s41598-019-47038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano JM, Royo C. Dissecting the genetic architecture of leaf rust resistance in wheat by QTL meta-analysis. Phytopathology. 2015;105:1585–1593. doi: 10.1094/PHYTO-05-15-0130-R. [DOI] [PubMed] [Google Scholar]
- Sosnowski O, Charcosset A, Joets J. BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics. 2012;28:2082–2083. doi: 10.1093/bioinformatics/bts313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai L, Wang HJ, Xu XJ, Sun WH, Ju L, Liu WT, Chen KM. Cereal pre-harvest sprouting: a global agricultural disaster regulated by complex genetic and biochemical mechanisms. J Exp Bot. 2021;72:2857–2876. doi: 10.1093/jxb/erab024. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Gupta PK. Meta-analysis of QTLs involved in pre-harvest sprouting tolerance and dormancy in bread wheat. Triticeae Genomics Genet. 2012;3:1. [Google Scholar]
- Venske E, Santos RSD, Farias DDR, Rother V, Maia LCD, Pegoraro C, Oliveira ACD. Meta-analysis of the QTLome of fusarium head blight resistance in bread wheat: Refining the current puzzle. Front Plant Sci. 2019;10:727. doi: 10.3389/fpls.2019.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Goffinet B, Charcosset A. MetaQTL: a package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinformatics. 2007;8:1–16. doi: 10.1186/1471-2105-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma H, Junaid A, Manjhi J, Singh GP, Gaikwad K, Padaria JC. Heat stress transcripts differential expression and profiling of heat stress tolerant gene TaHsp90 in Indian wheat (Triticum aestivum L.) cv C306. PloS one. 2018;13:0198293. doi: 10.1371/journal.pone.0198293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM. Characterization of polyploid wheat genomic diversity using a high density 90 000 single nucleotide polymorphism array. Plant Biotechnol J. 2014;12:787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP, Drenth J, Mcintyre CL. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J Exp Bot. 2015;66:1025–1039. doi: 10.1093/jxb/eru462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Amo A, Wei D, Chai Y, Zheng J, Qiao P, Cui C, Lu S, Chen L, Hu YG (2021) Large-scale integration of meta-QTL and genome-wide association study discovers the genomic regions and candidate genes for yield and yield-related traits in bread wheat. Theor Appl Genet 1–27 [DOI] [PubMed]
- Ye Q, Yu J, Zhang Z, Hou L, Liu X (2020) VvBAP1 a grape C2 domain protein plays a positive regulatory role under heat stress. Front Plant Sci 11 [DOI] [PMC free article] [PubMed]
- Yu Q, Li L, Luo Q, Eamus D, Xu S, Chen C. Year patterns of climate impact on wheat yields. Int J Climatol. 2014;34:518–528. doi: 10.1002/joc.3704. [DOI] [Google Scholar]
- Zampieri M, Ceglar A, Dentener F, Toreti A. Wheat yield loss attributable to heat waves drought and water excess at the global national and subnational scales. Environ Res Lett. 2017;12:6. doi: 10.1088/1748-9326/aa723b. [DOI] [Google Scholar]
- Zang X, Geng X, He K, Wang F, Tian X, Xin M, Yao Y, Hu Z, Ni Z, Sun Q, Peng H. Overexpression of the wheat (Triticum aestivum L.) TaPEPKR2 gene enhances heat and dehydration tolerance in both wheat and Arabidopsis. Front Plant Sci. 2018;9:1710. doi: 10.3389/fpls.2018.01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X, Geng X, Wang F, Liu Z, Zhang L, Zhao Y, Tian X, Ni Z, Yao Y, Xin M, Hu Z, Sun Q, Peng H. Overexpression of wheat ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging. BMC Plant Biol. 2017;17:1–13. doi: 10.1186/s12870-016-0958-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaveri E, Lobell DB. The role of irrigation in changing wheat yields and heat sensitivity in India. Nat Commun. 2019;10:1–7. doi: 10.1038/s41467-019-12183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tian X, Wang F, Zhang L, Xin M, Hu Z, Yao Y, Ni Z, Sun Q, Peng H. Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 2017;17:1–14. doi: 10.1186/s12870-017-1158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Hua C, Li L, Sun Z, Yuan M, Bai G, Humphreys G, Li T. Integration of meta-QTL discovery with omics: towards a molecular breeding platform for improving wheat resistance to Fusarium head blight. Crop J. 2020;9:739–749. doi: 10.1016/j.cj.2020.10.006. [DOI] [Google Scholar]
- Zhu T, Wang L, Rimbert H, Rodriguez JC, Deal KR, Oliveira RD, Choulet F, Keeble Gagnère GK, Tibbits J, Rogers J, Eversole K, Appels R, Gu YQ, Mascher M, Dvorak J, Luo MC. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021;107:303–314. doi: 10.1111/tpj.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relevant data are included in this paper and its associated Supplementary Information (SI).
Not applicable.