Abstract
Founder wheat lines have played key role in Chinese wheat improvement. Wheat-Dasypyrum villosum translocation T6VS·6AL has been widely used in wheat breeding in recent years due to its high level of powdery mildew resistance and other beneficial genes. Reference oligo-nucleotide multiplex probe (ONMP)–FISH karyotypes of six T6VS·6AL donor lines were developed and used for characterizing 32 derivative cultivars and lines. T6VS·6AL was present in 27 cultivar/lines with 20 from southern China. Next, ONMP–FISH was used to study chromosome constitution of randomly collected wheat cultivars and advanced breeding lines from southern and northern regions of China: 123 lines from the regional test plots of southern China and 110 from northern China. In southern China, T6VS·6AL (35.8%) was the most predominant variation, while T1RS·1BL (27.3%) was the most predominant in northern China. The pericentric inversion perInv 6B derived from its founder wheat Funo and Abbondaza was the second most predominant chromosome variant in both regions. Other chromosome variants were present in very low frequencies. Additionally, 167 polymorphic chromosome types were identified. Based on these variations, 271 cultivars and lines were clustered into three groups, including southern, northern, and mixed groups that contained wheat from both regions. Different dominant chromosome variations were seen, indicating chromosome differentiation in the three groups of wheat. The clearly identified wheat lines with T6VS·6AL in different backgrounds and oligonucleotide probe set will facilitate their utilization in wheat breeding and in identifying other beneficial traits that may be linked to this translocation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01206-3.
Keywords: Founder wheat, T6VS·6AL, Oligonucleotide FISH, Chromosome fingerprinting, Powdery mildew resistance
Introduction
Wheat (Triticum aestivum L.) is a major crop worldwide (Fu et al. 2018; IWGSC 2018). China became the largest wheat producer in the 1990s, and more than 2000 wheat cultivars were released during 1949–2000 (Zhuang 2003). Many researches found that 19 wheat founders: “Mazhamai,” “Yanda 1817,” “Jangdongmen,” “Chenduguangtou,” “Youzimai,” “Bima 4,” “Beijing 8,” “Xinong 6028,” “Wuyimai,” “Nanda 2419,” “Funo,” “Abbondaza,” “Orofen,” “Zaoyangmai,” “Lovrin 10,” “Mexipa 66,” “Fan 6,” “Aimengniu,” and “Xiaoyan 6”, have palyed key roles in developing these cultivars (Zhuang 2003; Xu and Li 2012; Hao et al. 2017; Li et al. 2019a). Molecular genetic studies of founder wheats identified genomic regions and markers related to wheat productivity traits and markers for marker assisted selection (MAS) breeding were developed (Ge et al. 2009; Han et al. 2009; Li et al. 2009; Xiao et al. 2011; Liu et al. 2012; Yu et al. 2012; Xiang et al. 2013; Hao et al. 2017).
A cytogenetic survey of 373 widely grown Chinese cultivars/breeding lines identified 5 of 14 structural chromosome rearrangements in 148 accessions tracing to founder varieties (Huang et al. 2018). Xu and Li (2012) reported that chromosome rearrangement associated elite traits and a high level of combining ability of founder lines facilitated their use in wheat breeding. For example, reciprocal chromosome translocations T4AS∙4AL-1DS/1DL∙1DS-4AL in Mazhamai, the wheat-rye translocation T1RS∙1BL in Lovrin series, and a pericentric inversion (perInv) 6B in Funo, Abbondaza, and Fan 6 (Huang et al. 2018), may reduce recombination in the translocated chromosome regions and thus transmit beneficial genes in tightly linked blocks in founder cultivars.
In recent years, several newly developed lines or cultivars such as “Zhou 8425B,” “Lumai 14,” and wheat-Dasypyrum villosum translocation line T6VS·6AL (the short arm 6AS of wheat was replaced by arm 6 VS of D. villosum) were extensively used in Chinese breeding programs for new cultivar development (He et al. 2011). Lines Zhou 8425B and Lumai 14 also carried wheat-rye T1RS∙1BL translocation (Hao et al. 2017; Huang et al. 2018) that was already widely present in released cultivars. However, the translocation T6VS·6AL was a relatively new alien chromosome translocation containing gene Pm21 conferring excellent resistance to powdery mildew (Chen et al. 1995). A set of six lines carrying wheat–D. villosum T6VS·6AL translocation (92R series) in different genetic backgrounds were released by the Cytogenetics Institute of Nanjing Agricultural University (CINAU), and were widely used in many Chinese wheat breeding programs (Li et al. 2006; Cao et al. 2011; Du et al. 2019). However, only a few of the cultivars or lines involving 92R series in their pedigrees have been characterized by cytogenetic techniques for their chromosome constitution (Li et al. 2006; Wang et al. 2017a; Huang et al. 2018; Du et al. 2019).
Previously, Liu et al. (1999a) found that powdery mildew resistance governed by T6VS·6AL was inherited as a single dominant gene mendelian trait. Zhao et al. (2019) also reported that T6VS·6AL can be transmitted across generations more frequently than T6VS·6DL, the translocation involving chromosomes 6D and 6V. Small segmental translocations were induced and lead to fine mapping and isolation of the powdery mildew resistance gene Pm21 and its key members (Cao et al. 2011; Chen et al. 2013; Xing et al. 2018). Also, markers specific to a segment of 6VS in T6VS·6AL were used in MAS to improve powdery mildew resistance (Bie et al. 2015; Cao et al. 2009; Chen and Chen 2010; He et al. 2017; He et al. 2016; He et al. 2013; Li et al. 2005; Li et al. 2006; Liu et al. 1999b; Qi et al. 1998; Wang et al. 2007; Zhu et al. 2018). Other studies also indicated that T6VS·6AL translocation may carry genes controlling kernel weight, stem diameter, spike length, grain number per spike, top spikelet fertility, plant height, and tillering (fewer tillers/spikes per plant) (Li et al. 2006; Ma 2007; Zhao et al. 2019). Wang et al. (2009) reported that T6VS·6AL may carry genes for improving water absorption, maximum resistance to extension, resistance to extension at 50 mm, and stability time of the dough but negatively affect test weight, falling number, peak viscosity, and softening degree, probably due to different gliadin genes on T6VS·6AL (Xu et al. 2011). Thus, a survey of T6VS·6AL in the recently released cultivars or elite breeding lines will provide additional information on many of these and other traits, in addition to powdery mildew resistance.
Fluorescence in situ hybridization (FISH) using oligo-nucleotide multiplex probe (ONMP) provides a new powerful tool for tracking specific chromosome segments and dissecting chromosome constitution (Du et al. 2017; Huang et al. 2018; Wang et al. 2017a). FISH with ONMP can produce a unique signal pattern in each chromosome and thus can detect variations in both target region and background simultaneously, which provides the whole image of the chromosomal composition in a genetic stock. For example, in a wheat line “NAU 1258,” both the translocation T6VS·6AL and a reciprocal translocation RT5AL·3BL/5AS·3BS were revealed through FISH with ONMP (Wang et al. 2017a).
In this study, we used ONMP FISH, genomic in situ hybridization (GISH), and molecular markers to analyze six T6VS·6AL donor lines and their 32 derivative cultivars and lines, and 233 newly developed wheat cultivars/breeding lines from different regions of China. The objectives of the study were: (1) detect the chromosome variations among those materials; (2) identify predominant chromosome variations in those new cultivars/lines from southern and northern China; and (3) determine breeding potential of the chromosome variations and develop high resolution karyotypes for accurate variation identification.
Materials and methods
Plant materials
Three wheat-D. villosum alien chromosome lines, DA6V (disomic addition of chromosome 6V), DS6V(6A) (disomic substitution of chromosome 6A with 6V) and del6VL (carried deletion on the arm 6VL), along with six T6VS·6AL translocation donor lines (92R89, 92R137, 92R139, 92R141, 92R149, and 92R178, designated as 92R series hereafter) with high level of resistance to powdery mildew and yellow rust developed by Cytogenetics Institute of Nanjing Agricultural University (CINAU) (Chen et al. 1995) and the landrace Chinese Spring (CS) were used for developing reference karyotypes of chromosomes 6V and T6VS·6AL by ONMP FISH. The six 92R series lines showed different phenotypes although they carried the same translocation. Cytogenetic survey using the ONMP FISH technique was conducted on 265 wheat cultivars/breeding lines, including 29 cultivars and three elite breeding lines that have 92R series in their pedigree, a set of 123 advanced breeding lines from southern China (including 24 from regional yield trials of winter wheat zone in middle and lower Yangtze River valley (MLYRV), 29 from regional yield trials of winter wheat zone in southern Huai River valley (SHRV) of Jiangsu Province, and 32 and 38 from preliminary yield trials in MLYRV of China and SHRV of Jiangsu Province in 2018–2019, respectively) and 110 cultivars/lines from northern China (including 44 lines from regional yield trials of northern Yellow-Huai River Valleys of China in 2018–2019), and 53 modern cultivars and 13 breeding lines randomly collected from provinces of Shandong, Shanxi, Hebei, and Henan in northern China. The breeding lines used in this research are all entered in regional yield trials in southern or northern wheat grown regions and thus represent the future new cultivars in China. The cultivars used here were all randomly picked and were newly released (Table S1); therefore, materials used in this research covered most of the typical gene sources in current Chinese wheat breeding.
Methods
Cytological analysis
Chromosome preparations followed procedures described in Doležel et al. (1992), Kato (1999), and Huang et al. (2018). In ONMP FISH, eight single-strand oligonucleotides with TAMRA (6-carboxytetramethylrhodamine) modified probes pAs1–1, pAs1–3, pAs1–4, pAs1–6, AFA-3, and AFA-4, and FAM (6-carboxyfluorescein) modified probes (GAA)10 and pSc119.2–1 were used to form multiplex probes (Du et al. 2017; Wang et al. 2017a; Huang et al. 2018). All oligonucleotides were synthesized and modified at General Biosystems Inc. (Chuzhou, China). GISH using total genomic DNA of D. villosum and Secale cereale as probes were performed as those described in Zhang et al. (2012) and Wu et al. (2018a). After hybridization with probes, chromosomes were visualized under an Olympus BX60 or BX53 microscope (Olympus Inc., Japan), and images were captured through a SPOT CCD (SPOT Cooled Color Digital, Olympus DP72 or DP80) camera (Olympus Inc., Japan). Image analysis was conducted using the computer program Photoshop (Adobe Inc., USA).
Molecular marker analysis
Markers XCINAU 272 and XCINAU 275 that are specific for chromosome 6V of D. villosum were used to select plants carrying 6V chromatin (Chen et al. 2013). Primers for producing the two markers were synthesized by General Biosystems Inc. (Chuzhou, China), and PCR was conducted in a 10-μL volume containing 2 μL (ca. 25 ng μL-1) of template DNA, 0.2 μL (10 μm) of each primer, 5 μL of Taq MIX (TsingKe Biological Technology, Beijing, China) and 2.6 μL of ddH2O. Amplifications were conducted for 3 min at 94 °C, followed by 34 cycles of 30 s each at 94 °C, 50 s at 50–65 °C according to specific primers, 1 min 10 s at 72 °C, and a final step of 10 min at 72 °C. PCR products were separated in 8% polyacrylamide gels and visualized with an automatic gel imager JS-680B (Peiqing Science and Technology, Shanghai, China) after silver staining.
Data analysis
For karyotyping and chromosome diversity analysis, 3~10 plants of each accession were used, 9~38 cells were analyzed to obtain 18~76 observations of each chromosome. Most karyotypes were developed from a single cell except for a few that were from two or three cells due to chromosome overlapping or incomplete cells. If the accessions displayed heterogeneous chromosomes in different plants, different karyotypes were developed. For chromosome diversity analysis, a numerical scale was used, with the number “1” and “0” as presence and absence of the structural rearrangements and/or block types, respectively, and “0.5” as heterozygous genotype. All polymorphic types were named based on the system developed in Huang et al. (2018). Chromosome polymorphic information content (CPIC) was calculated using formula (Thiel et al. 2003), where k is the total number of structural rearrangements and polymorphic block types for a chromosome detected, and Pi is the frequency of the ith block type among all the same class accessions. For each chromosome, structural variation denoted either a translocation or an inversion; a block means a unique signal pattern of ONMP FISH (Huang et al. 2018). The Stack Column figures were developed using computer package Origin Pro 2018 (OriginLab, Northampton, MA, USA). Principal component analysis (PCA) was performed using prcomp function of R package ggplot2 (Price et al. 2006).
Results
Reference karyotype and detection of T6VS·6AL in derived commercial cultivars
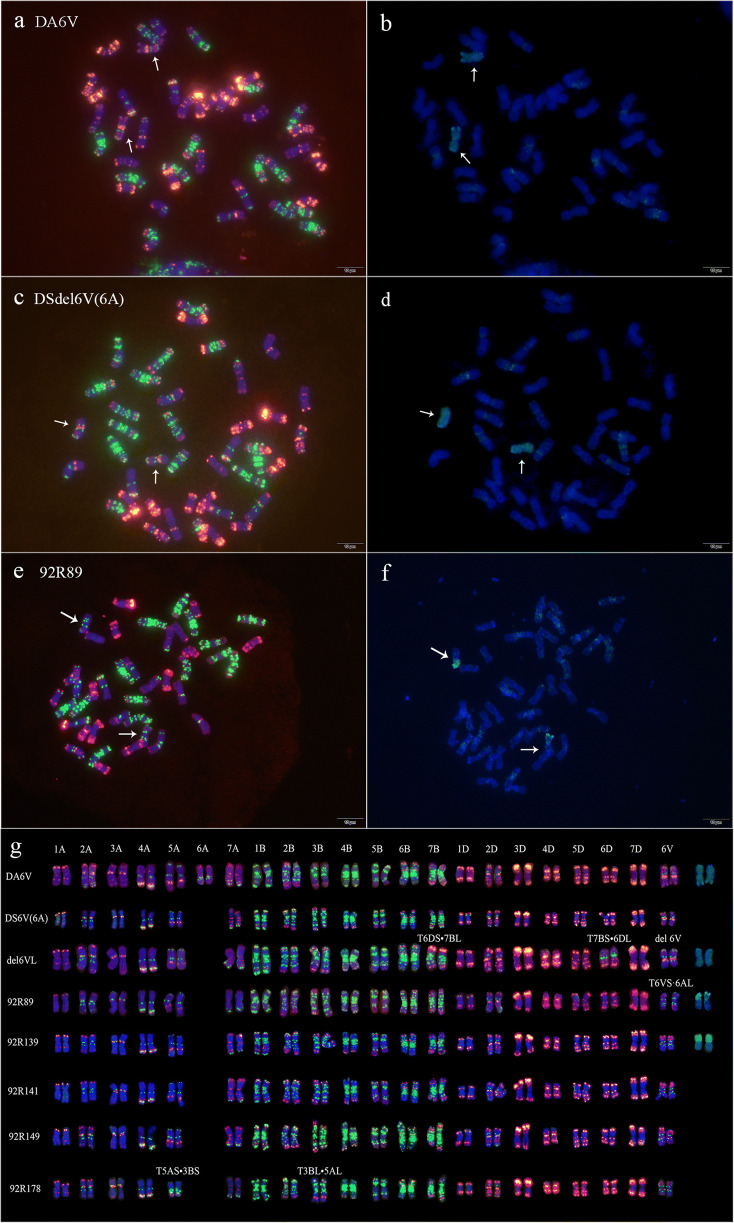
From six 92R series lines and three wheat alien chromosome lines DA6V, DS6V(6A), and del6VL, ONMP FISH detected the unique signal patterns on chromosome 6V and T6VS·6AL (Fig. 1; Fig. S1), which were used to develop the reference karyotypes for tracking the transmission of the translocation T6VS·6AL or 6V segment across generations. Of the six 92R series lines, five (92R89, 92R137, 92R139, 92R141, and 92R149) carried only the translocation T6VS·6AL, whereas line 92R178 also carried, in addition to T6VS·6AL, reciprocal translocation T5AS•3BS/T5AL•3BL. In line del6VL, where del6VL substituted for chromosome 6A of wheat, a reciprocal translocation T6DS·7BL/T7BS·6DL was also detected (Fig. 1).
Fig. 1.
Chromosomes of wheat-Dasypyrum villosum alien chromosome lines DA6V, del6VL, and T6VS·6AL after ONMP FISH/GISH and their karyotypes. Blue color indicates chromosomes counterstained with DAPI. Green shows pSc119.2–1 and (GAA)10 modified with FAM. Red shows oligonucleotides pAs1–1, pAs1–3, pAs1–4, pAs1–6, AFA-3, and AFA-4 modified with TAMRA. The VV genome was labeled with Fluorescein-12-dUTP (green) in GISH. a & b DA6V after ONMP FISH and sequential GISH; c & d del6VL after ONMP FISH and sequential GISH; e & f T6VS·6AL(92R89) after ONMP FISH and sequential GISH; g kayotypes of DA6V, del6VL, 92R89 and other lines of 92R series after ONMP FISH and sequential GISH. Arrows indicate different chromosome 6V
Since the release of six 92R series lines in 1992, more than 30 commercial cultivars have been bred from them and released for wheat production in China (Cao et al. 2011; Du et al. 2019). Several elite breeding lines involving T6VS·6AL have also been developed and used as new parents for wheat breeding. In this study, 29 cultivars and three lines with 92R series in their pedigree were identified, 27 (84.4%) carried T6VS·6AL. Among them, 11 also had other chromosome variations: five had reciprocal translocation T5AS•3BS/T5AL•3BL, three had perInv 6B, two carried translocation T1RS•1BL and one simultaneously carried reciprocal translocation T5AS•3BS/T5AL•3BL and perInv 6B. The remaining five cultivars had no T6VS·6AL, but one had T1RS•1BL, and four had normal karyotype (Fig. S2; Table S2). According to the pedigree (Fig. 2e), 92R137 and 92R178 served as the new founder wheats, T6VS·6AL were present in 27 of the 32 cultivars/breeding lines, of which 20 were from southern China and seven from northern China. It is necessary to compare the frequency of T6VS·6AL in the newly developed lines or cultivars in southern and northern China.
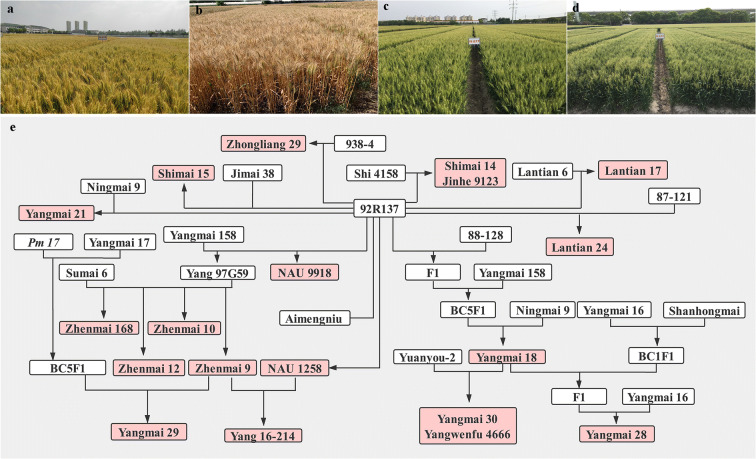
Fig. 2.
Field performance of new cultivars containing T6VS·6AL and pedigree of new cultivars and elite breeding lines involving T6VS·6AL donor line 92R137. a Yangmai 28; b Jinqiang 12; c Yangmai 29; d Yangmai 30; e the pedigree of new cultivars and elite breeding lines involving T6VS·6AL donor line 92R137. Pictures a, c and d courtesy of Zunjie Wang, Yangzhou Academy of Agricultural Sciences, China
Predominant chromosome variations in newly developed lines of southern China
From 123 lines entered in four regional yield trials in southern China, six types of chromosome variation were identified, which included three types of translocations (T6VS·6AL, T1RS·1BL, and T6VS·6DL), one pericentric inversion (perInv 6B), and two types of reciprocal translocation (T1AL·3DL/T3DS·1AS and T2AL·5BL/T5BS·2AS) (Figs. S3–6). According to a cytogenetic survey of those lines, 44 (35.8%) carried T6VS·6AL, 19 (15.4%) carried perInv 6B, seven (5.7%) carried T1RS·1BL, one (0.8%) had T6VS·6DL, one (0.8%) had RT1AL·3DL/3DS·1AS, and one (0.8%) had RT2AL·5BL/5BS·2AS (Figs. 3 and 4, Table 1). Among the 44 lines that carried T6VS·6AL, six were from regional yield trials of MLYRV, nine were from regional yield trials of SHRV, 11 were from preliminary yield trials in MLYRV, and 18 were from preliminary yield trials of SHRV. Interestingly, “Yangmai 17G83 (X215)” contained both T6VS·6AL and RT1AL·3DL/3DS·1AS, “Yang 16-76 (X198)” and “Runyangmai 4155 (X218)” contained both T6VS·6AL and perInv 6B, and “Ningmai 1633 (X242)” contained RT2AL·5BL/5BS·2AS. A high proportion of lines carried T6VS·6AL (35.8%) in breeding programs of southern China indicating that T6VS·6AL translocation lines have become new founder wheats in the southern wheat growing region of China.
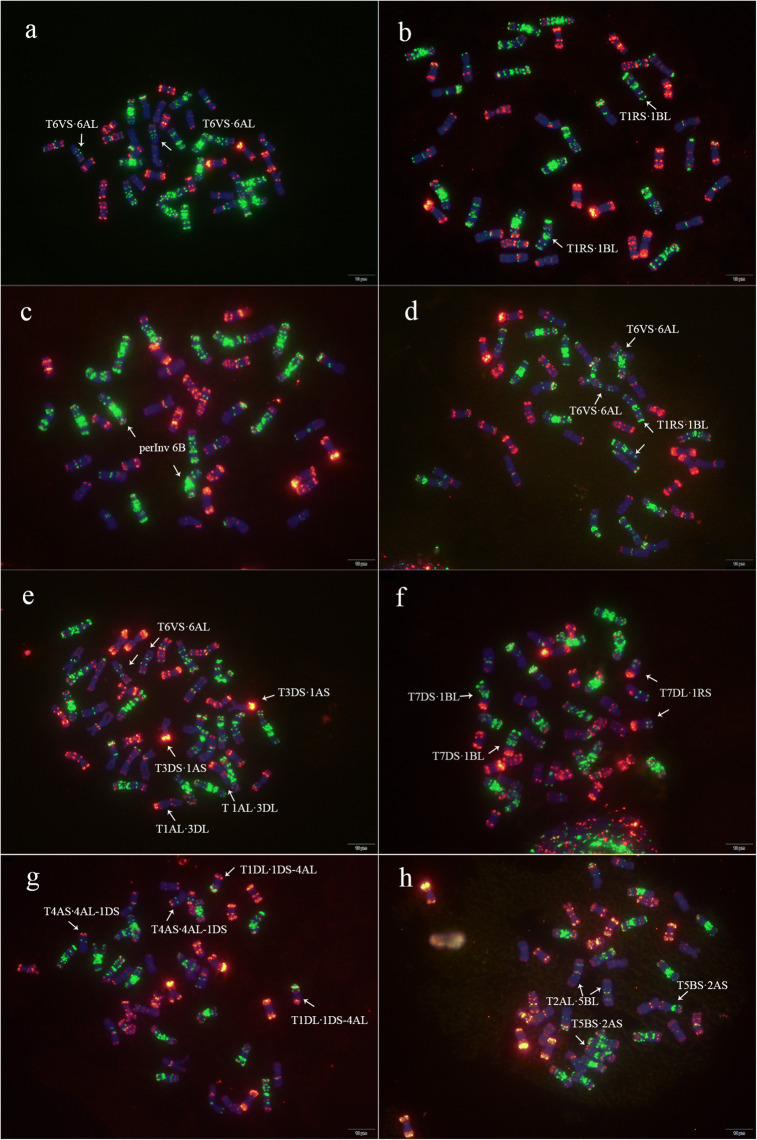
Fig. 3.
Chromosome structural variations of eight cultivars revealed through ONMP FISH. Chromatin indicated by colors was the same as shown Fig. 1. a T6VS∙6AL in X30 (Yangfumai 5145); b T1RS∙1BL in X104 (Shannong 32); c perInv 6B in X185 (Yangmai 15G70); d T6VS∙6AL/T1RS∙1BL in X72 (Jinhe 12,339); e T6VS·6AL/RT1AL·3DL/3DS·1AS in X215 (Yangmai 17G83); f RT7DS·1BL/7DL·1RS in X176 (Luohan 7); g RT4AS·4AL-1DS/1DL·1DS-4AL in X151 (Chang 6878); h RT2AL·5BL/5BS·2AS in X242 (Ningmai 1633)
Fig. 4.
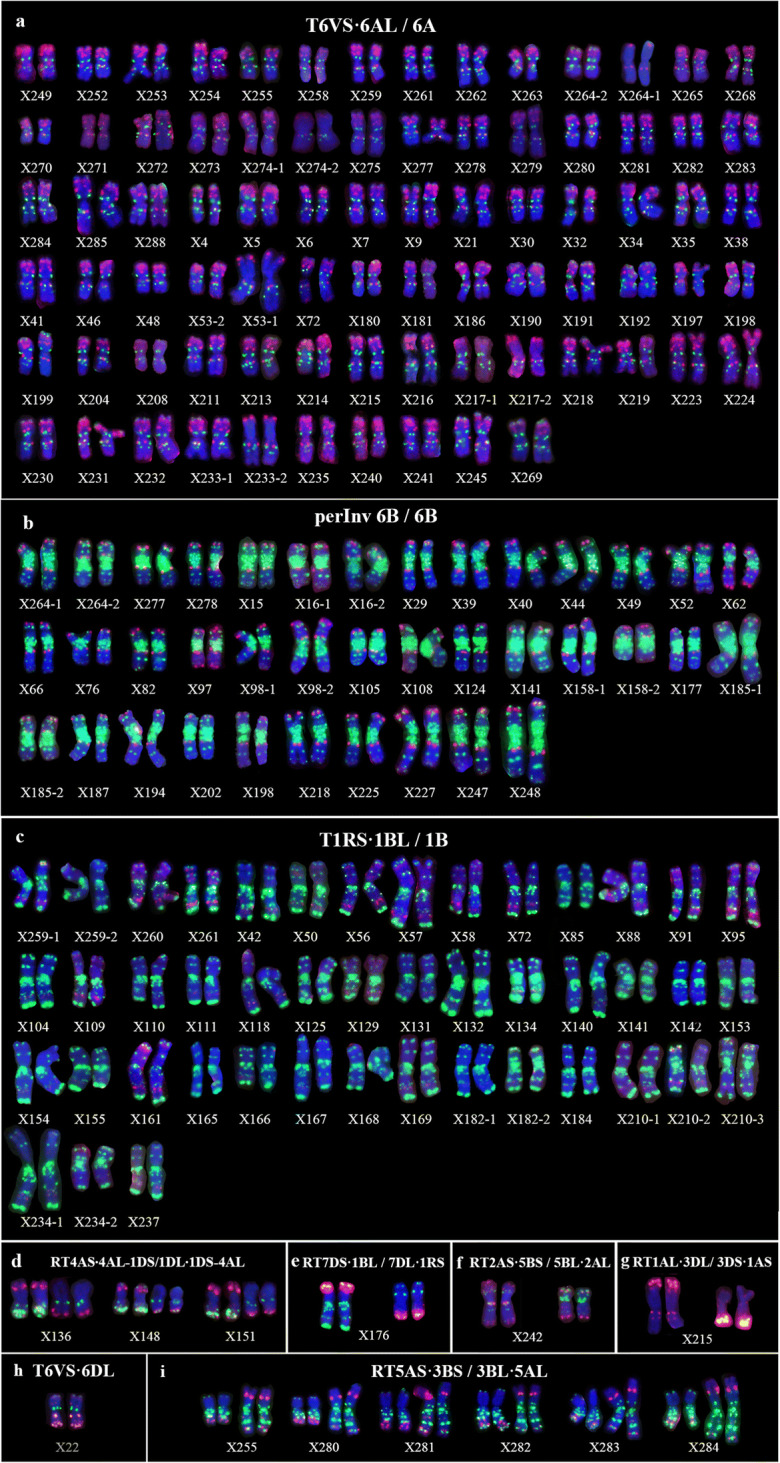
All structural chromosome variations detected in 271 wheat cultivars/lines. Colors are the same as indicated in Fig. 1. Line names were shown in Table S1
Table 1.
The frequency of chromosome structural variations in 167 newly developed wheat lines, 53 cultivars and 13 elite breeding lines from southern and northern China
| Chromosome variations | Newly developed wheat lines, cultivars, and elite breeding linesa | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| Total cultivars or lines | 24 | 29 | 32 | 38 | 44 | 66 |
| T1RS·1BL | – | 6.9% | 6.3% | 7.9% | 15.9% | 31.8% |
| T6VS·6AL | 25.0% | 31.0% | 31.3% | 42.1% | – | – |
| perInv 6B | 8.3% | 20.7% | 15.6% | 10.5% | 13.6% | 6.1% |
| T1RS·1BL/T6VS·6AL | – | – | – | – | 2.3% | – |
| T6VS·6AL/perInv 6B | – | – | 3.1% | 2.6% | – | – |
| T1RS·1BL/perInv 6B | – | – | – | – | – | 1.5% |
| T6VS·6AL/RT1AL·3DL/3DS·1AS | – | – | – | 2.6% | – | – |
| T6VS·6DL | 4.2% | – | – | – | – | – |
| RT2AL·5BL/5BS·2AS | – | – | – | 2.6% | – | – |
| RT4AS·4AL-1DS/1DL·1DS-4AL | – | – | – | – | – | 4.5% |
| RT7DS·1BL/7DL·1RS | – | – | – | – | – | 1.5% |
| Total frequency of cultivrars or lines with chromosome variations | 37.5% | 58.6% | 56.3% | 68.4% | 31.8% | 45.5% |
a A lines from regional yield trials of the winter wheat zone in the middle and lower Yangtze River valley (MLYRV); B lines from regional yield trials of the winter wheat zone in the southern Huai River valley (SHRV); C lines from preliminary yield trials of the winter wheat zone in MLYRV; D lines from preliminary yield trials of the winter wheat zone in SHRV; E lines from regional yield trials of northern Yellow-Huai River Valleys; F cultivars or elite breeding lines from provinces of Shandong, Shanxi, Hebei, and Henan that all belong to northern wheat grown regions in China
Predominant chromosome variations in northern China
To compare chromosome constitutions of northern Chinese wheat, 44 newly developed lines entered in regional yield trials in northern Yellow-Huai River Valleys, and 53 cultivars and 13 lines that were randomly collected from wheat growing regions in northern China were analyzed. Five types of chromosome variations were detected, including translocations T1RS·1BL and T6VS·6AL, pericentric inversion perInv6B, reciprocal translocations RT4AS·4AL-1DS/1DL·1DS-4AL and RT7DS·1BL/7DL·1RS. T1RS·1BL was found in 30 (27.3%) cultivars/lines, perInv6B was detected in 11 (10.0%) cultivars/lines, and T6VS·6AL was detected in only one line (0.9%) (Figs. 3 and 4, Figs. S7–8, Table 1). The two reciprocal translocations, RT4AS∙4AL-1DS/1DL∙1DS-4AL derived from Mazhamai and RT7DS·1BL/7DL·1RS derived from Aimengniu (Huang et al. 2018), were detected in three (2.7%) and one (0.9%) cultivar, respectively.
“Jinhe 12,339(X72)” was the only cultivar that carried both translocation of T1RS·1BL and T6VS·6AL. “Lunxuan 987 (X141)” carried both T1RS·1BL and perInv6B. Overall, T1RS·1BL was found in 27.3% of 110 newly developed lines or cultivars in northern China, which suggested that T1RS·1BL remains predominant in northern China.
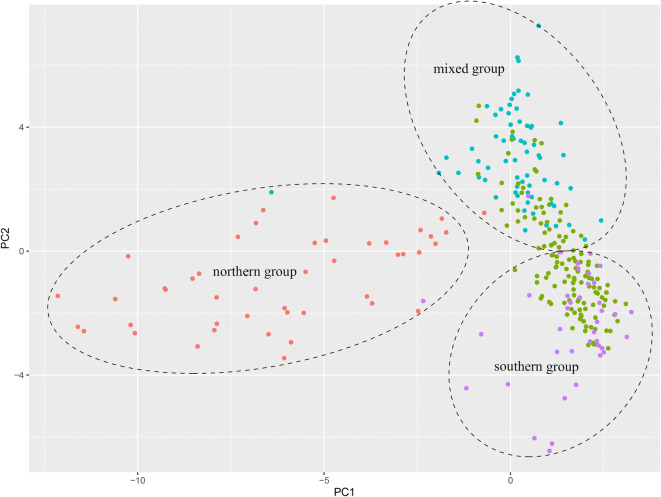
Clustering analysis of southern and northern Chinese wheat
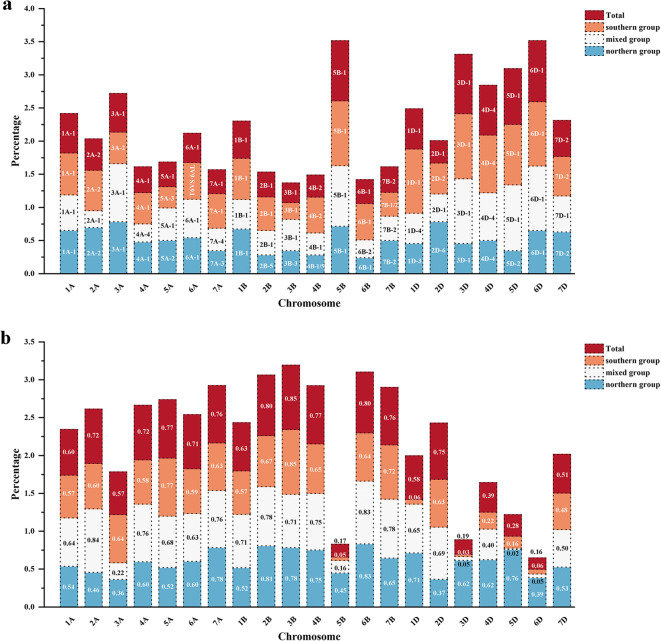
In addition to structural variations, 167 polymorphic chromosome blocks were also identified in a total 271 lines (Fig. S9). Due to heterogeneous chromosome composition in 14 lines, a total of 288 karyotypes were obtained and compared. Based on polymorphic and structural chromosome variations, the analyzed lines were clustered into three types. The northern group included lines mainly from yield trials in northern Yellow-Huai River Valleys. The southern group included lines mainly from southern China. Lastly, the mixed group included lines from both southern and northern regions (Fig. 5; Table S3). Chromosome structural variations and polymorphic blocks and their frequencies varied in the three groups. For example, in genome A, the frequency of T6VS·6AL was highest in the southern group, but the other two groups shared 6A-1 as the highest; for 5A, 5A-3 was the highest in the southern group, but 5A-2 was the highest in the northern group, and 5A-1 was the highest in the mixed group. Similarly, in genome D, 2D-2 was the highest in the southern group, but 2D-6 was the highest in the northern group, and 2D-1 was the highest in the mixed group (Fig. 6a, Table S3).
Fig. 5.
PCA analysis of 271 newly developed Chinese wheat cultivars/lines. Red dots represent lines from northern China (northern group). Green dots represent lines from southern China (southern group). Blue dots represent lines from provinces of Shandong, Shanxi, Hebei, and Henan in northern China and purple dots represent 92R series-derived cultivars or elite breeding lines in southern China, which formed the mixed group
Fig. 6.
The relationship of three groups of wheat. a Types and percentage among the highest proportion of polymorphic blocks on each chromosome in the three groups and total materials. b The mean CPIC of all chromosomes for the three groups and total materials
Chromosome polymorphic information content (CPIC) was calculated for the three groups. The average CPIC of the northern group was the highest, followed by the mixed group, the southern group was the lowest (Table S4). The average CPIC of 21 chromosomes also varied in the three groups, for example, chromosome 3A varied most in the southern group, followed by the northern, and the mixed group; 5B and 3D both varied most in the northern group, followed in the mixed, and southern group (Table S4; Fig. 6b; Fig. S10).
Discussion
At present, Fusarium head blight (scab) and powdery mildew are two major diseases in both southern and northern wheat grown areas of China. Previously, T1RS·1BL originating from the Lovrin series was predominant in Chinese wheat cultivars, mainly due to its resistance to multiple diseases such as powdery mildew and other beneficial genes (Zhuang 2003). Our results indicated this translocation continues to play an important role in northern China but with a reduced frequency since its major resistance gene Pm8 was defeated by new virulent isolates. Thus, introducing using new T1RS·1BL seems necessary and possible for further mining and utilizing the beneficial genes in rye chromosome arm 1RS because rapid chromosome evolution has been inferred in the open pollinated rye (Guo et al. 2019). However, in southern China, T6VS·6AL is the predominant chromosome variation and 92R series as the donor lines of this translocation have become the new founder wheat in this region. Among 27 commercial cultivars/lines with T6VS·6AL, only seven were developed by breeding programs in northern China, all others were bred in southern China. A cytogenetic survey of 123 newly developed lines from yield trials in southern China found 35.8% of them carried T6VS·6AL, which was much higher than lines developed in northern China. Why was T6VS·6AL more used in southern Chinese wheat? Probably, mainly because T6VS·6AL donor lines were developed in a genetic background of well-known southern wheat cultivar “Yangmai 5.” Moreover, 92R series lines are spring type with resistance to powdery mildew, scab and pre-harvest sprouting, and have red kernels, relatively taller plant height, larger spikes and fewer tillers, which are more adaptive to the climate of southern China. Particularly, the release of the second and third generation derivative cultivars of 92R series such as Zhenmai 9 and Yangmai 18 facilitated the application of T6VS·6AL to many more lines of southern China. In comparison to the initial donor lines, the new cultivars had much improved yield traits and were widely used as parents. However, in northern China, the different climate needs quite different genetic backgrounds for T6VS·6AL; the release of elite T6VS·6AL cultivars and lines with better winter hardiness, dwarf habit, more tillers, and better quality will facilitate its application in this region.
To date, more than 100 powdery mildew resistance genes of wheat were identified (McIntosh et al. 2017), only a few genes originated from wheat relatives have been found in founder wheat and widely used in breeding. The typical example is gene Pm8 from Secale cereale was transmitted into many wheat cultivars via “Lovrin” series in China (Xu et al. 2010). This study presented another example of gene Pm21 from D. villosum introgressed into many southern wheat lines through 92R series. Unfortunately, Pm8 was defeated by new races of Blumeria graminis f. sp. tritici (syn. Erysiphe graminis) that causes powdery mildew, but gene Pm21 still shows an excellent level of resistance to all isolates so far. Therefore, lines of 92R series and their derivatives will play important roles in developing new cultivars particularly adapted to southern China. It has been reported that T6VS·6AL may reduce number of wheat tillers but increase plant height and thousand kernel weight (Li et al. 2006; Ma 2007; Zhao et al. 2019). Additionally, other studies indicated that T6VS·6AL probably involved resistance to wheat streak mosaic virus and FHB (Li et al. 2002; Oliver et al. 2005). A stripe rust resistance gene Yr26 was also mapped on chromosome 1B of the donor lines of T6VS·6AL (Ma et al. 2001; Wang et al. 2008; Wu et al. 2018b). Further analysis and discovery of all other beneficial genes or gene clusters of T6VS·6AL will promote wide utilization of T6VS·6AL in wheat breeding programs of China and other regions of the world. At present, T6VS·6AL has been distributed to the nations and international organizations such as the USA, Germany, Poland, Czech Republic, and CIMMYT, where this translocation all shows good resistance to powdery mildew, indicating its potential in future wheat breeding. However, due to its genetic background of spring type, this translocation should be introgressed to the local elite cultivar via backcrossing, similar to in southern China, with the development of second or third generations of T6VS·6AL with elite traits, this translocation will become more used in wheat breeding.
Genetic differentiation between wheats in southern and northern China has been analyzed through molecular markers (Chen et al. 2019; Cheng et al. 2019; Li et al. 2018a; Rasheed et al. 2019; Ren et al. 2018; Zhou et al. 2018). In this study, cluster analysis grouped all wheat lines in this research into southern, northern, and mixed groups. Among three genomes, the D-genome showed the lowest CPIC, while the A- and B-genomes showed similar CPIC. However, among the three groups of wheat categorized through PCA analysis, chromosome variations in the northern group were the highest in the D-genome mainly due to the selection of lines specifically adapted to the geography and climate conditions in different regions of northern China. Tolerance to salinity, drought and cold temperature are the main concerns to improve wheat production in northern China, and the D-genome chromosomes mostly contain genes governing tolerance to abiotic stresses (Feldman and Levy 2012; Gorafi et al. 2018; Kishii et al. 2019). The A-genome chromosomes contain more genes controlling traits of plant height, maturity, and yield components (Ling et al. 2018; Talini et al. 2020). Lines from northern China generally have similar phenotypes for plant height, spike type, number of tillers, and other yield components due to intensive selection on grain yield in this region but application of a limited number of founder wheats may be the main reason leading to relatively low genetic diversity in A-genome chromosomes of northern lines. This result indicates that introgression of new A- and D-genome chromosome variations will help improve the northern and southern Chinese wheat. The application of synthetic wheat lines with diversified D-genome or A-genome chromosomes will bring in more diversity for efficient wheat improvement (Kishii et al. 2019).
In addition to T6VS·6AL and T1RS·1BL as predominant variations, dominant chromosome polymorphic types of 21 chromosomes were also identified in the analyzed wheat lines. Chromosome polymorphisms mainly occur in heterochromatin regions because of uneven recombination or duplication of tandem repeats, but their biological significance is unclear. However, considering T6VS·6AL and T1RS·1BL both involved beneficial genes of disease resistance and yield (Wang et al. 2009; Xiao et al. 2006), the polymorphic chromosome blocks widely observed in the current study probably also involve some selection advantages. Chromosome differentiation in the three groups of wheat mainly occurred in homoeologous group 2, 5, and 7 chromosomes, indicating that such polymorphic blocks probably involve important growth habit-, disease resistance- or yield-related genes. As previously indicated, chromosomes 2B, 2D, and 6B carried favorite alleles affecting grain size (Hu et al. 2016; Li et al. 2019b; Liu et al. 2020), chromosomes 5A, 2B, and 2D have genes controlling the heading date (Beales et al. 2007; Li et al. 2018a, b; Yan et al. 2003), chromosomes 2B, 2D, 4B, 5A, and 7A have genes for plant height (Mo et al. 2018; Herter et al. 2018; Yu et al. 2020), and chromosomes 7A, 4B, and 7B carry genes conferring disease resistance (Wang et al. 2017b; Zhang et al. 2017; Zou et al. 2018). Additionally, QTLs affecting the above-mentioned traits were also identified (Aoun et al. 2016; Cheng et al. 2019; Li et al. 2015; Edae et al. 2014; Ma et al. 2019; Pang et al. 2020). Genome wide association mapping using the present lines as a natural population may be helpful for validating the inferred association between adaptive genes and dominant chromosome blocks.
Conclusion
Based on ONMP FISH, GISH, and molecular marker analysis, the reference karyotypes of wheat-D. villosum T6VS·6AL donor lines were developed for tracing this translocation in wheat breeding programs. The wheat-D. villosum translocation T6VS·6AL lines have become new founder wheat material in southern China, but wheat-S. cereale translocation T1RS·1BL lines remain predominant in northern China. Additionally, 167 chromosome polymorphic types were identified in the analyzed wheat lines, and dominant chromosome polymorphic blocks for the southern, northern, and the mixed group wheats were identified. These clearly identified wheat lines with T6VS·6AL and oligonucleotide probes provide new tools for further using, tracing, and identifying the beneficial genes associated with this translocation for breeding and genetic research.
Supplementary Information
Wheat accessions used in this study. (XLSX 21.4 kb)
92R series and their derived cultivars (lines) used in production and breeding. (DOCX 19 kb)
Number of wheat cultivars/lines with different polymorphic chromosome blocks in the three groups of wheat. (XLSX 42 kb)
The CPIC data of total wheat and the three groups. (XLSX 14 kb)
PCR analysis of 6 VS specific markers XCINAU 272 (a) and XCINAU 275 (b) analysis in DA6V, del6VL, and 21 wheat accessions M: DL2000; 1: Chinese Spring; 2: Dasypyrum villosum; 3: Durum wheat-D. villosum amphiploid; 4: DA6V; 5: DSdel6VL(6A); 6–7: X249-X250; 8–10: X252-X254; 11–16: X258-X263; 17: X265; 18–20: X268-X270; 21: X275; 22–24:X277-X279; 25–26: X286-X287; Arrows indicate specific loci of 6VS chromosome. (PNG 2738 kb)
Karyotypes of 32 92R series derivatives after ONMP FISH. (PNG 1760 kb)
Karyotypes of 24 newly developed wheat lines from regional yield trials of the winter wheat zone in MLYRV after ONMP FISH. (PNG 1753 kb)
Karyotypes of 29 newly developed wheat lines from regional yield trials of the winter wheat zone in SHRV after ONMP FISH. (PNG 3219 kb)
Karyotypes of 32 newly developed wheat lines from preliminary yield trials of the winter wheat zone in MLYRV after ONMP FISH. (PNG 2050 kb)
Karyotypes of 38 newly developed wheat lines from preliminary yield trials of the winter wheat zone in SHRV after ONMP FISH. (PNG 2485 kb)
Karyotypes of 44 newly developed wheat lines from regional yield trials of the winter wheat zone in northern Yellow-Huai River Valleys after ONMP FISH. (PNG 2382 kb)
Karyotypes of 66 cultivars/lines from Shandong, Shanxi, Hebei, and Henan Provinces in northern China after ONMP FISH. (PNG 5243 kb)
Polymorphic chromosome blocks and their frequencies occurred in 271 wheat cutivars/lines. The chromosome type numbers in yellow were newly identified in this study, the number in white was based on Huang et al. (2018). (PNG 763 kb)
The mean CPIC of A-genome, B-genome, D-genome, and total chromosomes for three group wheat cultivars/lines. (PNG 173 kb)
Acknowledgements
We thank Prof. B. S. Gill, Department of Plant Pathology, Kansas State University, USA, for kind review and editing of the English, and Mr. Yang Yang, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, China for kind help in data analysis.
Author contribution
Project design: ZQ, PC, XY, and TB; Experimental work: NW, YL, DP, HW, XL, JF, JG, JZ, JG, AL, and BZ; Data analysis: NW, YL, DP, HW, JZ, JG, AL, ZQ, XY, and TB; Writing manuscript: NW, YL, DP, HW, ZQ, XY, and TB. All authors reviewed and approved the manuscript.
Funding
This project was supported by Postgraduate Research & Practice Innovation Programs of Jiangsu Province KYCX18_0652 and SJCX19_0117, Jiangsu Key Research Program for Modern Agriculture (BE2018350), and Jiangsu Agricultural Science and Technology Innovation fund (CX(19)1001). Bioinformatics analyses were supported by the Bioinformatics Center of Nanjing Agricultural University, China.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nan Wu, Yanhong Lei, Dan Pei and Hao Wu contributed equally to this work.
Contributor Information
Zengjun Qi, Email: zjqi@njau.edu.cn.
Xueming Yang, Email: xmyang@jaas.ac.cn.
Tongde Bie, Email: bietd@126.com.
References
- Aoun M, Breiland M, Kathryn Turner M, Loladze A, Chao S, Xu SS, Ammar K, Anderson JA, Kolmer JA, Acevedo M (2016) Genome-wide association mapping of leaf rust response in a durum wheat worldwide germplasm collection. Plant Genome 9. 10.3835/plantgenome2016.01.0008 [DOI] [PubMed]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733. 10.1007/s00122-007-0603-4 [DOI] [PubMed]
- Bie T, Zhao R, Jiang Z, Gao D, Zhang B, He H. Efficient marker-assisted screening of structural changes involving Haynaldia villosa chromosome 6V using a double-distal-marker strategy. Mol Breeding. 2015;35:34. doi: 10.1007/s11032-015-0211-y. [DOI] [Google Scholar]
- Cao A, Xing L, Wang X, Yang X, Wang W, Sun Y, Qian C, Ni J, Chen Y, Liu D, Wang X, Chen P (2011) Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc Natl Acad Sci U S A 108:7727–7732. 10.1073/pnas.1016981108 [DOI] [PMC free article] [PubMed]
- Cao Y, Cao A, Wang X, Chen P (2009) Screening and application of EST-based PCR markers specific to individual chromosomes of Haynaldia villosa. Acta Agron Sin 35:1–10. 10.3724/sp.J.1006.2009.00001. [In Chinese with English abstract.]
- Chen J, Zhang F, Zhao C, Lv G, Sun C, Pan Y, Guo X, Chen F (2019) Genome-wide association study of six quality traits reveals the association of the TaRPP13L1 gene with flour colour in Chinese bread wheat. Plant Biotechnol J 17:2106–2122. 10.1111/pbi.13126 [DOI] [PMC free article] [PubMed]
- Chen P, Qi L, Zhou B, Zhang Z, Liu D (1995) Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128. 10.1007/bf00223930 [DOI] [PubMed]
- Chen P, You C, Hu Y, Chen S, Zhou B, Cao A, Wang X. Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol Breeding. 2013;31:477–484. doi: 10.1007/s11032-012-9804-x. [DOI] [Google Scholar]
- Chen S, Chen P (2010) Development of the specific EST markers for the 6V chromosome short arm of Haynaldia Villosa and location of their deletion. J Triticeae Crops 30:789–795. 10.1080/00949651003724790
- Cheng H, Liu J, Wen J, Nie X, Xu L, Chen N, Li Z, Wang Q, Zheng Z, Li M, Cui L, Liu Z, Bian J, Wang Z, Xu S, Yang Q, Appels R, Han D, Song W, Sun Q, Jiang Y. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 2019;20:136. doi: 10.1186/s13059-019-1744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Li S, He Y, Tang Z, Ren Y (2019) Structural variation of 6VS/6AL translocation chromosomes in Mianmai 37 and its derivativs. J Triticeae Crops 39:659–665. 10.4606/j.issn.1009-1041.2019.06.05
- Du P, Zhuang L, Wang Y, Yuan L, Wang Q, Wang D, Dawadondup TL, Shen J, Xu H, Zhao H, Chu C, Qi Z. Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome. 2017;60:93–103. doi: 10.1139/gen-2016-0095. [DOI] [PubMed] [Google Scholar]
- Edae EA, Byrne PF, Haley SD, Lopes MS, Reynolds MP (2014) Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet 127:791–807. 10.1007/s00122-013-2257-8 [DOI] [PubMed]
- Feldman M, Levy AA (2012) Genome evolution due to allopolyploidization in wheat. Genetics 192:763–774. 10.1534/genetics.112.146316 [DOI] [PMC free article] [PubMed]
- Fu X, Liu Q, Li Z, Zhang A, Ling H, Tong Y, Liu Z (2018) Research achievement and prospect development on wheat genome. Bulletin of Chinese Academy of Sciences 33:909–914. 10.16418/j.issn.1000-3045.2018.09.003. [In Chinese with English abstract]
- Ge H, Wang L, You G, Hao C, Dong Y, Zhang X (2009) Fundamental roles of cornerstone breeding lines in wheat reflected by SSR random scanning. Sci Agric Sin 42:1503–1511. 10.3864/j.issn.0578-1752.2009.05.001. [In Chinese with English abstract]
- Gorafi Y, Kim J, Elbashir A, Tsujimoto H (2018) A population of wheat multiple synthetic derivatives: an effective platform to explore, harness and utilize genetic diversity of Aegilops tauschii for wheat improvement. Theor Appl Genet 131:1615–1626. 10.1007/s00122-018-3102-x [DOI] [PMC free article] [PubMed]
- Guo J, Lei Y, Zhang H, Song D, Liu X, Cao Z, Chu C, Zhuang L, Qi Z (2019) Frequent variations in tandem repeats pSc200 and pSc119.2 cause rapid chromosome evolution of open-pollinated rye. Mol Breeding 39. 10.3724/SP.J.1006.2009.01395
- Han J, Zhang L, Li J, Shi L, Xie C, You M, Yang Z, Liu G, Sun Q, Liu Z (2009) Molecular dissection of core parental cross “Triumph/Yanda1817” and its derivatives in wheat breeding program. Acta Agron Sin 35:1395–1404. 10.3724/SP.J.1006.2009.01395. [In Chinese with English abstract]
- Hao C, Wang Y, Chao S, Li T, Liu H, Wang L, Zhang X. The iSelect 9 K SNP analysis revealed polyploidization induced revolutionary changes and intense human selection causing strong haplotype blocks in wheat. Sci Rep. 2017;7:41247. doi: 10.1038/srep41247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Ji Y, Zhu S, Li B, Zhao R, Jiang Z, Bie T. Genetic, physical and comparative mapping of the powdery mildew resistance gene Pm21 originating from Dasypyrum villosum. Frontiers Plant Sci. 2017;8:1914. doi: 10.3389/fpls.2017.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Zhu S, Jiang Z, Ji Y, Wang F, Zhao R, Bie T (2016) Comparative mapping of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor Appl Genet 129:819–829. 10.1007/s00122-016-2668-4 [DOI] [PubMed]
- He H, Zhu S, Sun W, Gao D, Bie T, Igartua E. Efficient development of Haynaldia villosa chromosome 6VS-specific DNA markers using a CISP-IS strategy. Plant Breed. 2013;132:290–294. doi: 10.1111/pbr.12035. [DOI] [Google Scholar]
- He Z, Xia X, Chen X, Zhuang Q (2011) Progress of wheat breeding in China and the future perspective. Acta Agron Sin 37:202–215. 10.3724/sp.J.1006.2011.00202. [In Chinese with English abstract]
- Herter CP, Ebmeyer E, Kollers S, Korzun V, Leiser WL, Wurschum T, Miedaner T (2018) Rht24 reduces height in the winter wheat population ‘Solitar x Bussard’ without adverse effects on Fusarium head blight infection. Theor Appl Genet 131:1263–1272. 10.1007/s00122-018-3076-8 [DOI] [PubMed]
- Hu M, Zhang H, Liu K, Cao J, Wang S, Jiang H, Wu Z, Lu J, Zhu X, Xia X, Sun G, Ma C, Chang C (2016) Cloning and characterization of TaTGW-7A gene associated with grain weight in wheat via SLAF-seq-BSA. Front Plant Sci 7:1902. 10.3389/fpls.2016.01902 [DOI] [PMC free article] [PubMed]
- Huang X, Zhu M, Zhuang L, Zhang S, Wang J, Chen X, Wang D, Chen J, Bao Y, Guo J, Zhang J, Feng Y, Chu C, Du P, Qi Z, Wang H, Chen P (2018) Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor Appl Genet 131:1967–1986. 10.1007/s00122-018-3126-2 [DOI] [PubMed]
- IWGSC Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:1–13. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- Kishii M, Huerta J, Tsujimoto H, Matsuoka Y (2019) Stripe rust resistance in wild wheat Aegilops tauschii Coss.: genetic structure and inheritance in synthetic allohexaploid Triticum wheat lines. Genet Resour Crop Evol 66:909–920. 10.1007/s10722-019-00758-w
- Li F, Wen W, He Z, Liu J, Jin H, Cao S, Geng H, Yan J, Zhang P, Wan Y, Xia X (2018b) Genome-wide linkage mapping of yield-related traits in three chinese bread wheat populations using high-density SNP markers. Theor Appl Genet 131:1–22. 10.1007/s00122-018-3122-6 [DOI] [PubMed]
- Li G, Chen P, Zhang S, Wang X, He Z, Zhang Y, Zhao H, Huang H, Zhou X. Effects of the T6VS·6AL translocation on agronomic traits and dough properties of wheat. Euphytica. 2006;155:305–313. doi: 10.1007/s10681-006-9332-z. [DOI] [Google Scholar]
- Li H, Chen Q, Jia X, Li H, Graf R, Laroche A, Kuzyk A (2002) Different reactions to the wheat curl mite and wheat streak mosaic virus in various wheat-Haynaldia villosa 6V and 6VS lines. Plant Dis 86:423–428. 10.1094/PDIS.2002.86.4.423 [DOI] [PubMed]
- Li H, Chen X, Shi A, Kong F, Leath S, Murphy J, Jia X (2005) Characterization of RAPD markers and RFLP marker linked to powdery mildew resistant gene derived from different H. villosa. Sci Agric Sin 38:439–445. [In Chinese with English abstract]
- Li T, Liu H, Mai C, Yu G, Li H, Meng L, Jian D, Yang L, Zhou Y, Zhang H, Li H. Variation in allelic frequencies at loci associated with kernel weight and their effects on kernel weight-related traits in winter wheat. Crop J. 2019;7:30–37. doi: 10.1016/j.cj.2018.08.002. [DOI] [Google Scholar]
- Li X, Wang T, Li Y (2019a) Formation, research and utilization of founder parents in major crops. J Plant Genetic Resour 20:1093–1102. 10.13430/j.cnki.jpgr.20190505003. [In Chinese with English abstract]
- Li X, Xia X, Xiao Y, He Z, Wang D, Trethowan R, Wang H, Chen X. QTL mapping for plant height and yield components in common wheat under water-limited and full irrigation environments. Crop Pasture Sci. 2015;66:660–670. doi: 10.1071/CP14236. [DOI] [Google Scholar]
- Li X, Xu X, Liu W, Li X, Li L (2009) Genetic diversity of the founder parent Orofen and its progenies revealed by SSR markers. Sci Agric Sin 42:3397–3404. 10.3864/j.issn.0578-1752.2009.10.003. [In Chinese with English abstract]
- Li Z, Yang J, Li R, Wang X, Zhang X, Zhang L (2018a) Composition and distribution of major vernalization genes in wheat cultivars from main production areas in China. J Northwest A&F Univ 46:35–40. 10.13207/j.cnki.jnwafu.2018.09.005. [In Chinese with English abstract]
- Liu H, Li H, Hao C, Wang K, Wang Y, Qin L, An D, Li T, Zhang X (2020) TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol J 18:1330–1342. 10.1111/pbi.13298 [DOI] [PMC free article] [PubMed]
- Liu J, Tao W, Liu D, Chen P, Li W (1999a) Transmission of 6VS chromosome in wheat-Haynaldia villosa translocation lines and genetic stability of Pm21 carried by 6VS. Acta Bot Sin 41:1058–1060. 10.1016/S0168-9452(99)00123-5. [In Chinese with English abstract]
- Liu X, Si Q, Li Q, Wang C, Wang Y, Zhang H, Ji W (2012) SSR analysis of genetic diversity and temporal trends of the core wheat (Triticum aestivum L.) parent Funo and its derivative varieties (lines). J Agric Biotechnol 20:983–995. 10.3969/j.issn.1674-7968.2012.09.002. [In Chinese with English abstract]
- Liu Z, Sun Q, Ni Z, Yang T. Development of SCAR markers linked to the Pm21 gene conferring resistance to powdery mildew in common wheat. Plant Breed. 1999;118:215–219. doi: 10.1046/j.1439-0523.1999.118003215.x. [DOI] [Google Scholar]
- Ling H, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, Yu Y, Du H, Qi M, Li Y, Lu H, Yu H, Cui Y, Wang N, Chen C, Wu H, Zhao Y, Zhang J, Li Y, Zhou W, Zhang B, Hu W, van Eijk MJ, Tang J, Witsenboer HMA, Zhao S, Li Z, Zhang A, Wang D, Liang C (2018) Genome sequence of the progenitor of wheat a subgenome Triticum urartu. Nature 557:424–428. 10.1038/s41586-018-0108-0 [DOI] [PMC free article] [PubMed]
- Ma J, Zhou R, Dong Y, Wang L, Wang X, Jia J. Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica. 2001;120:219–226. doi: 10.1023/A:1017510331721. [DOI] [Google Scholar]
- Ma J, Qin N, Cai B, Chen G, Ding P, Zhang H, Yang C, Huang L, Mu Y, Tang H, Liu Y, Wang J, Qi P, Jiang Q, Zheng Y, Liu C, Lan X, Wei Y (2019) Identification and validation of a novel major QTL for all-stage stripe rustresistance on 1BL in the winter wheat line 20828. Theor Appl Genet 132:1363–1137. 10.1007/s00122-019-03283-7 [DOI] [PubMed]
- Ma Q (2007) Genetic analysis of new wheat recombinant inbred lines population derived from wheat-Haynaldia villosa 6VS·6AL translocation and Huixianhong and some agronomic traits. Master's thesis, Nanjing Agricultural University, Nanjing, China. [In Chinese with English abstract]
- McIntosh R, Dubcovsky J, Rogers W, Morris C, Xia X (2017) Catalogue of gene symbols for wheat: 2017 supplement. In: KOMUGI wheat genetic resource database. https://wheat.pw.usda.gov/GG3/Wheat_Gene_Catalog_Documents
- Mo Y, Vanzetti L, Hale I, Spagnolo E, Guidobaldi F, Al-Oboudi J, Odle N, Pearce S, Helguera M, Dubcovsky J (2018) Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor Appl Genet 131:2021–2035. 10.1007/s00122-018-3130-6 [DOI] [PubMed]
- Oliver R, Cai X, Xu S, Chen X, Stack R. Wheat alien species derivatives: a novel source of resistance to Fusarium head blight in wheat. Crop Sci. 2005;45:1353–1360. doi: 10.2135/cropsci2004.0503. [DOI] [Google Scholar]
- Pang Y, Liu C, Wang D, St. Amand P, Bernardo A, Li W, He F, Li L, Wang L, Yuan X, Dong L, Su Y, Zhang H, Zhao M, Liang Y, Jia H, Shen X, Lu Y, Jiang H, Wu Y, Li A, Wang H, Kong L, Bai G, Liu S (2020) High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat. Mol Plant 13(9):1311-1327. 10.1016/j.molp.2020.07.008 [DOI] [PubMed]
- Price A, Patterson N, Plenge R, Weinblatt M, Shadick N, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Qi L, Wang S, Chen P, Liu D, Gill B (1998) Identification and physical mapping of three Haynaldia villosa chromosome-6V deletion lines. Theor Appl Genet 97:1042–1046. 10.1007/s001220050989
- Rasheed A, Jin H, Xiao Y, Zhang Y, Hao Y, Zhang Y, Hickey L, Morgounov A, Xia X, He Z. Allelic effects and variations for key bread-making quality genes in bread wheat using high-throughput molecular markers. J Cereal Sci. 2019;85:305–309. doi: 10.1016/j.jcs.2018.12.004. [DOI] [Google Scholar]
- Ren Z, Li Z, Shi L, Wang X, Zhu L, Li X, Liu D. Molecular identification of wheat leaf rust resistance genes in sixty Chinese wheat cultivars. Czech J Genet Plant Breeding. 2018;54:1–8. doi: 10.17221/6/2016-cjgpb. [DOI] [Google Scholar]
- Talini RF, Brandolini A, Miculan M, Brunazzi A, Vaccino P, Pe ME, Dell’Acqua M. Genome-wide association study of agronomic and quality traits in a world collection of the wild wheat relative Triticum urartu. Plant J. 2020;102:555–568. doi: 10.1111/tpj.14650. [DOI] [PubMed] [Google Scholar]
- Wang C, Bie T, Chen Q, Cao A, Chen P (2007) Development and application of molecular markers specific to chromosome 6VS of Haynaldia villosa. Acta Agron Sin 33:1595–1600. [In Chinese with English abstract]
- Wang C, Zhang Y, Han D, Kang Z, Li G, Cao A, Chen P (2008) SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica 159:359–366. 10.1007/s10681-007-9524-1
- Wang C, Ma Q, Qi Z, Zhuang L, Feng J, Jiang D, Hu L, Qi X, Niu J, Feng Y, Chen P (2009) Effects of wheat—Haynaldia Villosa T6VS·6AL translocation on grain and flour quality of wheat. Journal of Triticeae Crops 29:787–792. [In Chinese with English abstract]
- Wang D, Du P, Pei Z, Zhuang L, Qi Z (2017a) Development and application of high resolution karyotypes of wheat “Chinese spring” aneuploids. Acta Agron Sin 43(11):1575-1587 10.3724/sp.J.1006.2017.01575. [In Chinese with English abstract]
- Wang Y, Xie J, Zhang H, Guo B, Ning S, Chen Y, Lu P, Wu Q, Li M, Zhang D, Guo G, Zhang Y, Liu D, Zou S, Tang J, Zhao H, Wang X, Li J, Yang W, Cao T, Yin G, Liu Z (2017b) Mapping stripe rust resistance gene YrZH22 in Chinese wheat cultivar Zhoumai 22 by bulked segregant RNA-Seq (BSR-Seq) and comparative genomics analyses. Theor Appl Genet 130:2191–2201. 10.1007/s00122-017-2950-0 [DOI] [PubMed]
- Wu J, Zeng Q, Wang Q, Liu S, Yu S, Mu J, Huang S, Sela H, Distelfeld A, Huang L, Han D, Kang Z (2018b) SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor Appl Genet 131:1481–1496. 10.1007/s00122-018-3092-8 [DOI] [PubMed]
- Wu N, Li M, Sun H, Cao Z, Liu P, Ding T, Xu H, Chu C, Zhuang L, Qi Z (2018a) RNA-seq facilitates development of chromosome-specific markers and transfer of rye chromatin to wheat. Mol Breeding 38(1):6. 10.1007/s11032-017-0762-1
- Xiang J, Yang X, Li X, Liu W, Gao A, Li L, Ma X (2013) The analysis of HMW-GS evolution in mentana and its derivatives. J Plant Genetic Resour 14:1053–1058. [In Chinese with English abstract]
- Xiao Y, Yin G, Li H, Xia X, Yan J, Zheng T, Ji W, He Z (2011) Genetic diversity and genome-wide association analysis of stripe rust resistance among the core wheat parent Zhou 8425B and its derivatives. Sci Agric Sin 44:3919–3929. 10.3864/j.issn.0578-1752.2011.19.001. [In Chinese with English abstract]
- Xiao Y, Yan J, He Z, Zhang Y, Zhang X, Liu L, Li T, Qu Y, Xia X. Effect of 1BL / 1RS translocation on yield traits and powdery mildew resistance in common wheat and QTL analysis. Crop J. 2006;32:1636–1641. doi: 10.3321/j.issn:0496-3490.2006.11.007. [DOI] [Google Scholar]
- Xing L, Hu P, Liu J, Witek K, Zhou S, Xu J, Zhou W, Gao L, Huang Z, Zhang R, Wang X, Chen P, Wang H, Jones JDG, Karafiatova M, Vrana J, Bartos J, Dolezel J, Tian Y, Wu Y, Cao A. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol Plant. 2018;11:874–878. doi: 10.1016/j.molp.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Xu T, Cai J, Wang B, Qi Z, Dai T, Cao W, Jiang D. Relationship between T6VS·6AL chromosome translocation and accumulations of high molecular weight glutenin subunits and glutenin macropolymer in wheat grain. Acta Agron Sin. 2011;37:2059–−2065. doi: 10.3724/SP.J.1006.2011.02059. [DOI] [Google Scholar]
- Xu X, Li X. Research progress of founder parents in wheat. J Henan Agric Sci. 2012;41:5–8. doi: 10.15933/j.cnki.1004-3268.2012.02.016. [DOI] [Google Scholar]
- Xu X, Li X, Li X, Yang X, Liu W, Gao A, Li L (2010) Inheritance of 1BL/1RS of founder parent Lovrin 10 in its progeny. J Triticeae Crops 30:221–226. 10.1080/00949651003724790
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A. 2003;100(10):6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xiao J, Tian J. Genetic dissection of milestone parent Aimengniu and its derivatives. Sci Agric Sin. 2012;45:199–207. doi: 10.3864/j.issn.0578-1752.2012.02.001. [DOI] [Google Scholar]
- Yu M, Liu Z, Yang B, Chen H, Zhang H, Hou D (2020) The contribution of photosynthesis traits and plant height components to plant height in wheat at the individual quantitative trait locus level. Sci Rep 10(1):12261. 10.1038/s41598-020-69138-0 [DOI] [PMC free article] [PubMed]
- Zhang H, Xie J, Cheng Y, Liu X, Wang Y, Yan S, Yang Z, Zhao H, Wang X, Jia L, Cao T, Liu Z. Mapping stripe rust resistance gene YrZM103 in wheat cultivar Zhengmai 103 by BSR-seq. Acta Agron Sin. 2017;43:1643–1649. doi: 10.3724/SP.J.1006.2017.01643. [DOI] [Google Scholar]
- Zhang R, Wang X, Chen P (2012) Molecular and cytogenetic characterization of a small alien-segment translocation line carrying the softness genes of Haynaldia villosa. Genome 55:639–646. 10.1139/g2012-051 [DOI] [PubMed]
- Zhao R, Jiang Z, Chen T, Wang L, Ji Y, Hu Z, He H, Bie T. Comparative analysis of genetic effects of wheat-Dasypyrum villosum translocations T6V#2S·6AL and T6V#4S·6DL. Plant Breed. 2019;138:503–512. doi: 10.1111/pbr.12711. [DOI] [Google Scholar]
- Zhou Y, Chen Z, Cheng M, Chen J, Zhu T, Wang R, Liu Y, Qi P, Chen G, Jiang Q, Wei Y, Luo MC, Nevo E, Allaby RG, Liu D, Wang J, Dvorak J, Zheng Y. Uncovering the dispersion history, adaptive evolution and selection of wheat in China. Plant Biotechnol J. 2018;16:280–291. doi: 10.1111/pbi.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Ji Y, Ji J, Bie T, Gao A, He H (2018) Fine physical bin mapping of the powdery mildew resistance gene Pm21 based on chromosomal structural variations in wheat. Int J Mol Sci 19:643. 10.3390/ijms19020643 [DOI] [PMC free article] [PubMed]
- Zhuang Q. Chinese wheat improvement and pedigree analysis. Beijing: China Agric Press; 2003. [Google Scholar]
- Zou S, Wang H, Li Y, Kong Z, Tang D (2018) The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytologist 218:298–309. 10.1111/nph.14964 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wheat accessions used in this study. (XLSX 21.4 kb)
92R series and their derived cultivars (lines) used in production and breeding. (DOCX 19 kb)
Number of wheat cultivars/lines with different polymorphic chromosome blocks in the three groups of wheat. (XLSX 42 kb)
The CPIC data of total wheat and the three groups. (XLSX 14 kb)
PCR analysis of 6 VS specific markers XCINAU 272 (a) and XCINAU 275 (b) analysis in DA6V, del6VL, and 21 wheat accessions M: DL2000; 1: Chinese Spring; 2: Dasypyrum villosum; 3: Durum wheat-D. villosum amphiploid; 4: DA6V; 5: DSdel6VL(6A); 6–7: X249-X250; 8–10: X252-X254; 11–16: X258-X263; 17: X265; 18–20: X268-X270; 21: X275; 22–24:X277-X279; 25–26: X286-X287; Arrows indicate specific loci of 6VS chromosome. (PNG 2738 kb)
Karyotypes of 32 92R series derivatives after ONMP FISH. (PNG 1760 kb)
Karyotypes of 24 newly developed wheat lines from regional yield trials of the winter wheat zone in MLYRV after ONMP FISH. (PNG 1753 kb)
Karyotypes of 29 newly developed wheat lines from regional yield trials of the winter wheat zone in SHRV after ONMP FISH. (PNG 3219 kb)
Karyotypes of 32 newly developed wheat lines from preliminary yield trials of the winter wheat zone in MLYRV after ONMP FISH. (PNG 2050 kb)
Karyotypes of 38 newly developed wheat lines from preliminary yield trials of the winter wheat zone in SHRV after ONMP FISH. (PNG 2485 kb)
Karyotypes of 44 newly developed wheat lines from regional yield trials of the winter wheat zone in northern Yellow-Huai River Valleys after ONMP FISH. (PNG 2382 kb)
Karyotypes of 66 cultivars/lines from Shandong, Shanxi, Hebei, and Henan Provinces in northern China after ONMP FISH. (PNG 5243 kb)
Polymorphic chromosome blocks and their frequencies occurred in 271 wheat cutivars/lines. The chromosome type numbers in yellow were newly identified in this study, the number in white was based on Huang et al. (2018). (PNG 763 kb)
The mean CPIC of A-genome, B-genome, D-genome, and total chromosomes for three group wheat cultivars/lines. (PNG 173 kb)