Abstract
Edwardsiella tarda is one of the most significant fish pathogens, causes edwardsiellosis in a variety of freshwater fish species, and its antibiotic resistance against multiple drugs has made it a health risk worldwide. In this study, we aimed to investigate the antibiotic resistance (ABR) genes of E. tarda and establish its antibiotic susceptibility. Thus, 540 fish (299 Oreochromis niloticus, 138 O.mossambicus, and 103 O. aureus) were collected randomly from twelve fish farms in three districts of Punjab in Pakistan. E. tarda was recovered from 147 fish showing symptoms of exophthalmia, hemorrhages, skin depigmentation, ascites, and bacteria-filled nodules in enlarged liver and kidney. Antimicrobial susceptibility testing proved chloramphenicol, ciprofloxacin, and streptomycin effective, but amoxicillin, erythromycin, and flumequine ineffective in controlling edwardsiellosis. Maximum occurrence of qnrA, blaTEM, and sul3 genes of E. tarda was detected in 45% in the liver, 58%, and 42% respectively in the intestine; 46.5%, 67.2%, and 55.9% respectively in O. niloticus; 24%, 36%, and 23% respectively in summer with respect to fish organs, species, and season, respectively. Motility, H2S, indole, methyl red, and glucose tests gave positive results. Overall, E. tarda infected 27.2% of fish, which ultimately caused 7.69% mortality. The Chi-squared test of independence showed a significant difference in the occurrence of ABR genes of E. tarda with respect to sampling sites. In conclusion, the misuse of antibacterial agents has led to the emergence of ABR genes in E. tarda, which in association with high temperatures cause multiple abnormalities in infected fish and ultimately resulting in massive mortality.
Keywords: Oreochromis niloticus, antimicrobial susceptibility, Kirby-Bauer disk diffusion method, Mueller-Hinton agar (MHA), abnormalities, exophthalmia
Introduction
The rapid expansion of the global aquaculture industry has provided high-quality food products, more job opportunities, and economic benefits [1]. Meanwhile, aquatic organisms are an excellent source of life-sustaining proteins and nutrients [2, 3]. Fish and its various related products help to overcome food insecurity amidst a rapidly increasing human population. For this reason, fish has also become a crucial part of a sustainable diet from a future perspective [4]. Recent fish consumption worldwide is 20.5 kg per capita [5]. Tilapia is considered an economically and commercially important fish due to its high consumption rate, fast growth performance, efficient feed conversion ratio (FCR), high resistance against disease, and easy breeding nature [6]. These factors have positioned tilapia as the cheapest source of animal protein worldwide [7]. However, the culture of tilapia has also contributed to the emergence of antibiotic-resistant (ABR) bacteria [8]. At present, fish diseases caused by pathogenic bacteria pose a significant threat to aquaculture economics [9].
The close interaction between naturally resistant bacteria of terrestrial and aquatic environments contributes to the rapid transfer of ABR genes to pathogenic bacteria in fish [10]. So, fish is considered a vehicle for the dissemination of ABR bacteria and ABR genes [11]. Fish farmers employ multiple antibiotics to overcome fish mortality caused by ABR bacteria [12]. Their repeated and useless application increases antimicrobial-resistant (AMR) bacteria and their AMR genes in aquaculture [13]. ABR bacteria are an emerging and significant challenge to public health [14] as they are able to utilize mechanisms of genetic strategy by which they become resistant to the effects of antibiotics [15].
Edwardsiellosis is an enteric systemic disease in fish characterized by hernia, ascites, exophthalmia, hemorrhages, and severe lesions of the internal organs [16]. Edwardsiellosis outbreaks have caused significant losses at farms of various fish species in aquaculture [17]. Its causative agent, Edwardsiella tarda, is a gram-negative bacterium, a facultatively intracellular pathogen, and one of the emerging top causative pathogens [18, 19] of systemic hemorrhagic septicemic infection in fish, namely edwardsiellosis and hemorrhagic septicemia [16, 20, 21]. E. tarda has caused mass mortality in a wide variety of wild and cultured fish species of both marine and freshwater environments worldwide [22], especially in Oreochromis niloticus, Cyprinus carpio, Labeo rohita, Clarias gariepinus, Seriola quinqueradiata, Anguilla japonica, and Pagrus major [20, 23, 24]. The huge economic loss caused by E. tarda in commercially important fish species is due to its high resistance against multiple antibiotics [25, 26] as well as transmission of resistance from antibiotic-resistant E. tarda strains to non-resistant E. tarda strains [24, 27]. These ABR strains have antimicrobial resistance and virulence genes which cause pathogenicity [25, 21] and severe outbreaks, especially at tilapia and catfish farms [19, 28]. E. tarda is an opportunistic pathogen and infects fish under environmental stress, low water quality, high temperature and organic content, and stocking density [26, 29]. E. tarda poses high zoonotic risks [17] and infects a broad range of host animal species [20], including birds, reptiles, amphibians, mammals, and even humans [30-32].
In the current study, using conventional PCR, we sought to determine antibiotic susceptibility and check the occurrence of ABR genes in E. tarda detected in tilapia in the fish farms of Punjab, Pakistan.
Materials and Methods
Sample Collection, Clinical, and Postmortem Examination
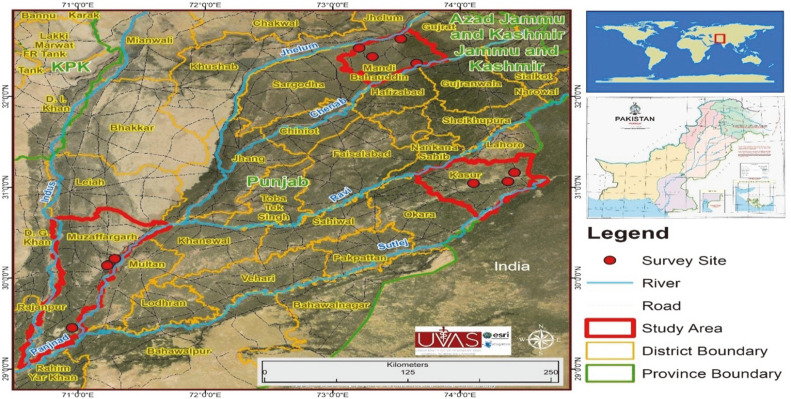
We randomly collected a total of 540 samples of tilapia fish species, viz., Oreochromis niloticus, O. aureus, and O. Mossambicus from selected fish farms of the districts of Kasur, Muzaffargarh, and Mandi-Bahauddin of Punjab in Pakistan. A GIS map showing the sampling sites (fish farms) in three districts is shown in Fig. 1. Ice-treated fish samples were transported to the laboratory of the Department of Fisheries and Aquaculture, University of Veterinary and Animal Sciences, Lahore, Pakistan. Suspected fish samples were examined for external and internal abnormalities during clinical and postmortem examination [33].
Fig. 1. GIS map shows sampling sites (fish farms) of three selected districts of Punjab.
Isolation, Phenotypic Characterization and Biochemical Identification of E. tarda
Suspected fish samples were disinfected with 70% ethanol and swabs from isolated organs (kidney, gills, liver, spleen, stomach, intestine, heart, and tail fins) were collected and inoculated onto trypticase soy agar (TSA, Oxoid, England) media plates and incubated at 37°C overnight. A single colony from freshly prepared culture was inoculated onto brain heart infusion agar (BHIA LAB, UK) media plates to obtain a pure culture of E. tarda, which was then incubated at 37°C for 24 h [34]. The pure culture of E. tarda isolates was stored at -20°C. A single colony from a freshly prepared E. tarda culture was subjected to phenotypic characterization and biochemical identification [35].
Isolation of DNA
DNA was extracted using a Genomic DNA Purification Kit (Thermo Scientific, GeneJET, USA) and quantified spectrophotometrically (Nanodrop, USA). DNA samples were evaluated by gel electrophoresis on 1% agarose gel stained with ethidium bromide (C21H20BrN3) and utilizing a standard-sized molecular marker [1Kb DNA Ladder RTU (Ready-to-Use) GeneDireX, Taiwan]. Isolated DNA was stored at -20°C for further use.
Molecular Detection of ABR Genes in E. tarda by PCR
E. tarda was detected by amplification of ABR genes (qnrA, blaTEM, and sul3) and gyrB gene by PCR (Bio-Rad, USA) using E. tarda-specific primers (Macrogen, Korea). A total of 25 μl of PCR reaction solution containing 1 μl of template DNA, 1 μl forward primer, 1 μl reverse primer, 10 μl injection water, and 12 μl GoTaq Green Master Mix (Promega, USA) were used to detect the ABR genes of E. tarda under amplification conditions for PCR as mentioned in Table 1. Amplified PCR products were analyzed on 1% agarose gel stained with ethidium bromide (C21H20BrN3) and utilizing a standard-sized molecular marker (1Kb DNA Ladder RTU, GeneDireX). PCR products revealing the thickest bands were sequenced by Sanger’s method at BGI Hong Kong Co. Ltd., China.
Table 1.
Primer sequences and conditions for amplification of 16S rRNA and antibiotic resistance genes of E. tarda by PCR.
| Target Gene | Primer sequence (5`-3`) | Amplified segment (bp) | Initial denaturation | Final denaturation | Annealing | Initial extension | Final extension | Reference |
|---|---|---|---|---|---|---|---|---|
| blaTEM | F-CATTTCCGTGTCGCCCTTATTC R-CGTTCATCCATAGTTGCCTGAC |
800 | 95°C For 2 min | 95°C For 30 s 35 cycles | 52°C For 30 s 35 cycles | 72°C For 1min 35 cycles | 72°C For 7 min | [39] |
| qnrA | F-ATTTCTCACGCCAGGATTTG R-GATCGGCAAAGGTTAGGTCA |
516 | 95°C For 10 min | 95°C For 1min 35 cycles | 58.5°C For 1min 35 cycles | 72°C For 1min 35 cycles | 72°C For 10 min | [40] |
| sul3 | F-AGATGTGATTGATTTGGGAGC R-TAGTTGTTTCTGGATTAGAGCCT |
443 | 93°C For 3 min | 93°C For 30s 35 cycles | 54.2°C For 30 s 35 cycles | 72°C For 1min 35 cycles | 72°C For 7min | [39] |
| 16SrRNA gene | F-AGAGTTTGATCCTGGCTCAG R-GGTTACCTTGTTACGACTT |
1503 | 94°C For 5 min | 90°C For 30 s 30 cycles | 52°C For 1min 30 cycles | 72°C For 1min 30 cycles | 72°C for 8 min | [41] |
| gyrB | F-GCATGGAGACCTTCAGCAAT R-GCGGAGATTTTGCTCTTCTT |
414 | 94°C For 1 min | 94°C For 1min 32 cycles | 55°C For 1min 32 cycles | 72°C For 1min 32 cycles | 72°C For 10 min | [66] |
Amplification, Sequencing, and Phylogenetic Tree Analysis of 16SrRNA Gene of E. tarda
One microliter of template DNA was added into a total of 25 μl reaction solution for PCR containing two primers of 16S rRNA gene; 1 μl forward primer (27F): AGAGTTTGATCCTGGCTCAG, 1 μl reverse primer (1492R): GGTTACCTTGTTACGACTT, 10 μl injection water and 12 μl GoTaq Green Master Mix (Promega, USA), under amplification conditions for PCR as mentioned in Table 1. PCR products were electrophoresed in 1%agarose gel stained with ethidium bromide (C21H20BrN3) and utilizing a standard-sized molecular marker (1Kb DNA Ladder RTU, GeneDireX). PCR products revealing the thickest bands were sequenced by Sanger’s method at BGI Hong Kong Co. Ltd., China. The obtained sequences were analyzed and compared for taxonomic identification using NCBI BLAST and submitted to the GenBank database. The phylogenetic relationship of E. tarda was checked by phylogenetic tree analysis of 16SrRNA gene of E. tarda by the bootstrap method using MEGA 11.0 (Molecular Evolutionary Genetic Analysis) with 1,000 bootstrap replications [36].
Antimicrobial Susceptibility Testing
The Kirby-Bauer disk diffusion method was applied to determine the antibiotic susceptibility pattern using Mueller-Hinton agar (MHA) plates [37]. A total of 14 antibiotic discs were applied on MHA plates with a suspension of E. tarda colonies containing 1.5 × 108 CFU/ml. Thereafter, 6 mm discs with discrete doses (5–30 μg) of antibiotics (Oxoid, UK), as mentioned in Table 8, were applied onto the MHA plate surfaces and incubated at 37°C for 24 h. The inhibition zones were measured to classify bacteria as resistant, moderately susceptible, and susceptible [38].
Table 8.
Results of antimicrobial susceptibility of E. tarda isolates and MIC values.
| Antibiotics (conc.) | Disc code | Concentration (μg) | Sensitive | Intermediate | Resistant | MIC50 (μg /ml) | MIC90 (μg /ml) |
|---|---|---|---|---|---|---|---|
| Amoxicillin | AMX | 10 | 0 | 0 | 100% | >64 | >128 |
| Ampicillin | AMP | 10 | 0 | 50% | 50% | <16 | <32 |
| Cefotaxime | CTX | 30 | 0 | 50% | 50% | >8 | >16 |
| Chloramphenicol | C | 30 | 100% | 0 | 0 | 0.5 | 1 |
| Ciprofloxacin | CIP | 5 | 100% | 0 | 0 | 2 | 4 |
| Doxycycline | DO | 5 | 0 | 50% | 50% | 8 | 16 |
| Erythromycin | E | 15 | 0 | 0 | 100% | >32 | >64 |
| Flumequine | FLU | 30 | 0 | 0 | 100% | >64 | >128 |
| Gentamicin | GM | 10 | 100% | 0 | 0 | 0.5 | 1 |
| Neomycin | N | 30 | 0 | 50% | 50% | >16 | <32 |
| Norfloxacin | NOR | 10 | 100% | 0 | 0 | <2 | <4 |
| Streptomycin | S | 10 | 100% | 0 | 0 | 1 | 2 |
| Sulfamethoxazole | SXT | 25 | 100% | 0 | 0 | 2 | 4 |
| Tetracycline | TE | 30 | 0 | 50% | 50% | 8 | 16 |
Histopathological Characterization of Edwardsiellosis
Tissues specimens were collected from liver, stomach, and small intestine of infected fish. These collected tissue specimens were examined for histopathological changes due to edwardsiellosis and were treated by 10% neutral buffered formalin solution for one day to fix them. The fixed tissue specimens were submitted to the laboratory of the Department of Pathology, University of Veterinary and Animal Sciences, Lahore, Pakistan for histopathological examination.
Statistical Analysis
Descriptive statistics such as proportions and percentage (%) were applied to summarize the data of prevalence. Chi-squared test of independence was applied to compare the occurrence of ABR genes of E. tarda with respect to fish species, organs of isolation, seasons, sampling sites, and fish sex using Statistical Package for Social Sciences (SPSS) version 21.0 software (IBM, USA).
Results
Isolation and Characterization of E. tarda
Swabs were collected from the organs of 540 fish samples and E. tarda was isolated by direct plating on TSA plates and BHIA plates. Quinolone resistance gene (qnrA), beta-lactam resistance gene (blaTEM), and sulphonamide resistance gene (sul3) of E. tarda were detected in 540 samples of three tilapia species (O. niloticus, O mossambicus and O. aureus) sampled from April to December. Colonies from a pure culture of E. tarda isolates showed phenotypic characteristics as opaque, large, irregular, translucent, and whitish-colored colonies on BHIA media plates while showing circular and round with grayish white-colored colonies on TSA media plates. Microscopic examination of E. tarda colonies revealed E. tarda as a motile and rod-shaped bacterium. Production of O2 caused bubble formation in the catalase test; red color appeared in Gram-staining, methyl red, and indole production test; diffused and hazy growths spread throughout the medium in the motility test, while black and purple-black color appeared in the H2S production and amylase tests, respectively. In addition, yellow color developed in the glucose fermentation test while no color change appeared in the arginine dihydrolase, cytochrome oxidase, citrate utilization, lactose fermentation, and Voges-Proskauer tests.
Occurrence of ABR Genes in E. tarda
ABR genes of E. tarda (qnrA, blaTEM, and sul3) were amplified using E. tarda-specific primers and their amplification by PCR revealed bands at 801, 654, and 444, respectively. Maximum occurrence of qnrA was recorded in 45% of liver, while that of blaTEM and sul3 was found in 58% and 42% respectively in intestine, with respect to fish organs. Similarly, maximum occurrence of qnrA, blaTEM and sul3 was recorded in 46.5%, 67.2%, and 55.9% respectively in O. niloticus with respect to fish species, while 24%, 36%, and 23% were recorded respectively in summer with respect to season. Meanwhile, 32.3%, 56.2%, and 18.7% were recorded respectively in female fish samples with respect to fish sex, and 36%, 52%, and 26% was recorded respectively at 37°C and pH 6.67 (Tables 3-7). Chi-squared test of independence showed a significant difference (p < 0.05) in the occurrence of ABR genes of E. tarda, with respect to sampling sites, while insignificant (p > 0.05) differences were shown with respect to fish species, sex, and organs of isolation and season (Table 8). Accession numbers allotted by the NCBI GenBank database against all detected genes are given in Table 9.
Table 3.
Occurrence of 16S rRNA and antibiotic resistance genes of E. tarda with respect to fish species.
| Fish species | qnrA gene | blaTEM gene | sul3 gene | 16SrRNA gene |
|---|---|---|---|---|
| Oreochromis niloticus | 139 (46.5%) | 201 (67.2%) | 170 (56.9%) | 183 (61.2%) |
| O. Mossambicus | 51 (36.9%) | 75 (54.3%) | 63 (45.6%) | 77 (55.8%) |
| O. aureus | 46 (44.6%) | 57 (55.3%) | 34 (33.3%) | 40 (38.8%) |
Table 4.
Occurrence of 16S rRNA and antibiotic resistance genes of E. tarda with respect to season.
| Season | qnrA gene | blaTEM gene | sul3 gene | 16SrRNA gene |
|---|---|---|---|---|
| Summer | 36 (24%) | 54 (36%) | 34 (23%) | 193 (64.3%) |
| Winter | 4 (8%) | 5 (10%) | 3 (6%) | 21 (7.0%) |
| Spring | 10 (20%) | 11 (22%) | 9 (18%) | 50 (16.7%) |
| Autumn | 8 (16%) | 8 (16%) | 5 (10%) | 36 (12.0%) |
Table 5.
Occurrence of 16S rRNA and antibiotic resistance genes of E. tarda with respect to fish sex.
| Sex | qnrA gene | blaTEM gene | sul3 gene | 16SrRNA gene |
|---|---|---|---|---|
| Male | 52 (28.6%) | 84 (46.1%) | 32 (17.6%) | 187 (62.3%) |
| Female | 62 (32.3%) | 108 (56.2%) | 36 (18.7%) | 113 (37.7%) |
Table 6.
Occurrence of 16S rRNA and antibiotic resistance genes of E. tarda with respect to water temperature (oC) and pH.
| Month | Temperature (°C) | pH | qnrA gene | blaTEM gene | sul3 gene | 16SrRNA gene |
|---|---|---|---|---|---|---|
| Apr-20 | 25.58°C | 7.92 | 2 (8%) | 3 (12%) | 1 (4%) | 11 (27.5%) |
| May-20 | 26.5°C | 7.68 | 4 (16%) | 6 (24%) | 3 (12%) | 23 (57.5%) |
| Jun-20 | 28.91°C | 7.42 | 11 (22%) | 14 (28%) | 9 (18%) | 48 (80.0%) |
| Jul-20 | 37°C | 6.67 | 18 (36%) | 26 (52%) | 13 (26%) | 73 (81.1%) |
| Aug-20 | 36.17°C | 6.8 | 14 (28%) | 18 (36%) | 12 (24%) | 67 (74.4%) |
| Sep-20 | 28.75°C | 7.87 | 5 (20%) | 6 (24%) | 4 (16%) | 36 (40.0%) |
| Oct-20 | 26.83°C | 7.95 | 3 (12%) | 4 (16%) | 2 (8%) | 20 (33.3%) |
| Nov-20 | 24.75°C | 8.27 | 2 (8%) | 3 (12%) | 1 (4%) | 14 (40.0%) |
| Dec-20 | 19.92°C | 8.4 | 1 (4%) | 2 (8%) | 0 (0%) | 8 (22.8%) |
Table 7.
Results of statistical analysis; chi-squared test of independence showing X2-value and p-value with respect to selected parameters.
| Parameters | X2-value | p-value |
|---|---|---|
| Sampling sites | 294 | 0.00*** |
| Fish Farms | 7.364 | 0.998ns |
| Season | 0.233 | 1.00ns |
| Organs | 3.262 | 0.999ns |
| Sex | 0.844 | 0.656ns |
*** = Shows significant result, ns = non-significant result
Table 9.
Accession numbers of 16S rRNA and antibiotic resistance genes of E. tarda.
| Gene | District | Accession number |
|---|---|---|
| blaTEM | Kasur | OP919343 |
| Mandi Bahauddin | OP919344 | |
| Muzaffargarh | OP919345 | |
| qnrA | Kasur | OP901500 |
| Mandi Bahauddin | OP901501 | |
| Muzaffargarh | OP901502 | |
| sul3 | Kasur | OP919347 |
| Mandi Bahauddin | OP919348 | |
| Muzaffargarh | OP919349 | |
| 16SrRNA | Kasur | ON508800 |
| Mandi Bahauddin | ON415282 | |
| Muzaffargarh | ON524384 | |
| gyrB | Kasur | OQ326581 |
| Mandi Bahauddin | OQ326582 | |
| Muzaffargarh | OQ326583 |
Overall Prevalence of E. tarda and Fish Mortality
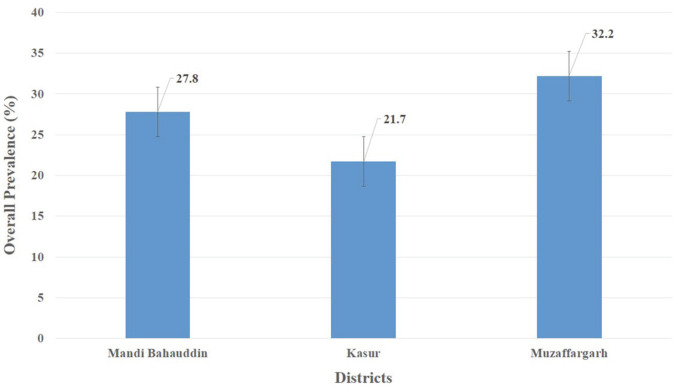
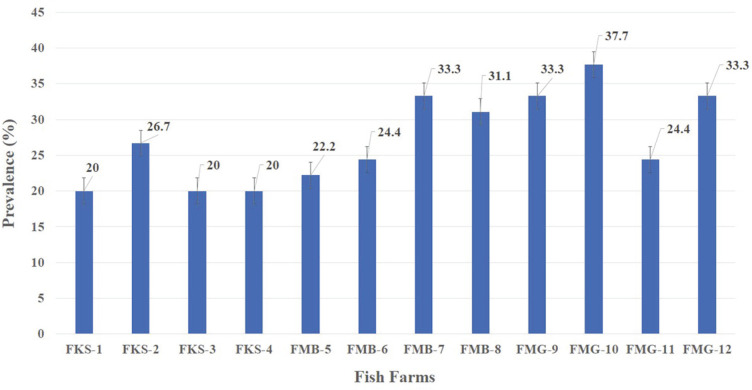
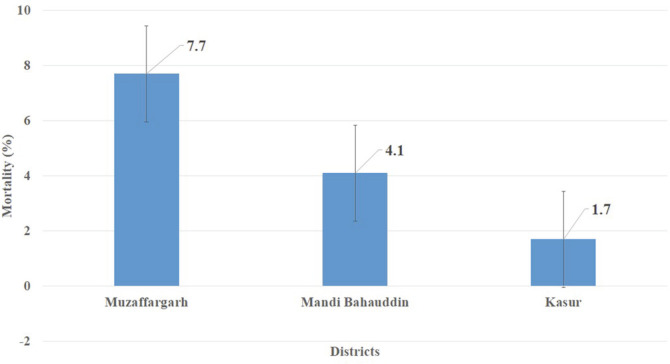
Overall, E. tarda infected 27.2% of fish (Fig. 2), but a maximum prevalence of 37.8% was recorded at a fish farm in Muzaffargarh Province designated as FMG-10, and minimum prevalence of 20% was seen at fish farms of Kasur Province, designated as FKS-1, FKS-3, and FKS-4 (Fig. 3). E. tarda caused 7.69% overall mortality (Fig. 4).
Fig. 2. Overall prevalence (%) of E. tarda at selected districts.
Fig. 3. Overall prevalence (%) of E. tarda at selected fish farms of three districts (FKS: fish farms of district Kasur, FMB: Fish farms of Mandi Bahauddin, FMG: Fish farms of Muzaffargarh).
Fig. 4. Mortality (%) in tilapia fish of all selected fish farms of three districts. Fig.
Antimicrobial Susceptibility Test
This test was performed on 147 isolates of E. tarda. Chloramphenicol, ciprofloxacin, gentamicin, norfloxacin, streptomycin, and sulfamethoxazole were found 100% effective in controlling edwardsiellosis, but amoxicillin, erythromycin, flumequine, and neomycin were found ineffective to control E. tarda infection, and all E. tarda isolates showed 100% resistance. Whereas, all E. tarda isolates showed intermediate resistance (50%) against ampicillin, cefotaxime, tetracycline, and doxycycline (Table 8).
Clinical and Post-Mortem Examination
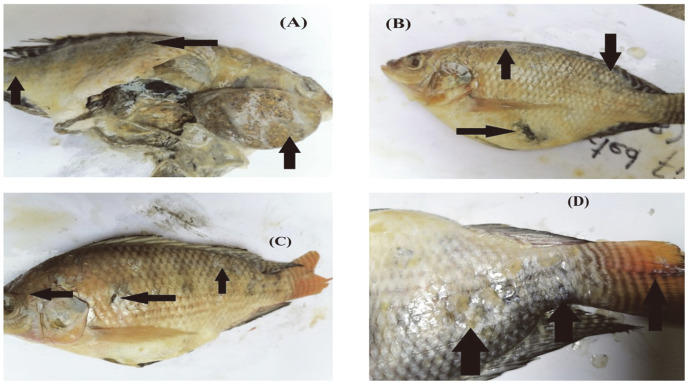
Clinical and post-mortem examination revealed E. tarda infection, or edwardsiellosis, in 147 fish samples (27.2% prevalence). Infected fish showed a variety of external and internal abnormalities, such as hemorrhages, exophthalmia, dark spots, and skin lesions. Congested gills, vent, and fins, hemorrhaged and protruded anus, hernia, and swollen abdomen filled with ascitic fluid (ascites) were also observed in infected tilapia. White, bacteria-filled nodules were observed in the enlarged liver, intestine, gills, spleen and kidney (Fig. 5).
Fig. 5. Clinical and post-mortem abnormalities.
(A) enlarged liver with black dots and white bacteria filled nodules, (B) skin depigmentation, exophthalmia, and swollen abdomen, (C) hemorrhages, (D) scale loss and lesions on head, vent and tail.
Phylogenetic Tree Analysis
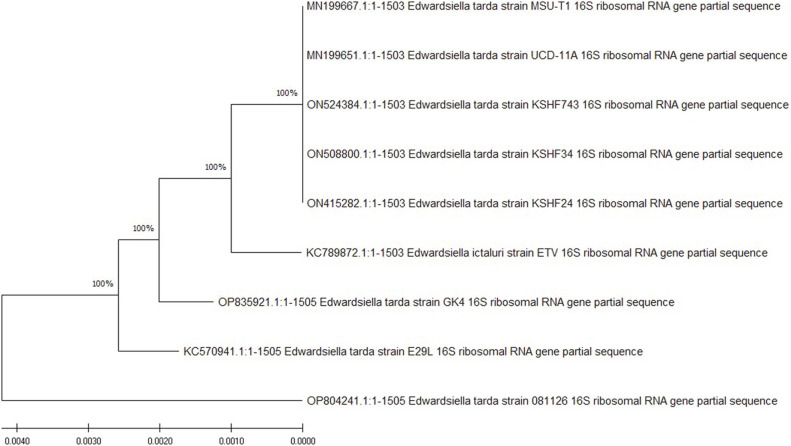
The phylogenetic tree revealed that our isolated E. tarda strains, such as KSHF743 (ON524384), KSHF34 (ON508800), and KSHF24 (ON415282) shared 100% similarity and also 100% with previously isolated E. tarda strain UCD-11A (MN199651, USA), MSU-T1 (MN199667, USA), GK4 (OP835921, India), E29L (KC570941, China), and 081126 (OP804241, China). Phylogenetic tree analysis of the 16S rRNA gene of E. tarda is shown in Fig. 6.
Fig. 6. Phylogenetic tree of 16S rRNA gene of E. tarda by using the Neighbour-joining bootstrap method with 1000 bootstrap replicates.
Histopathological Characterization of Edwardsiellosis
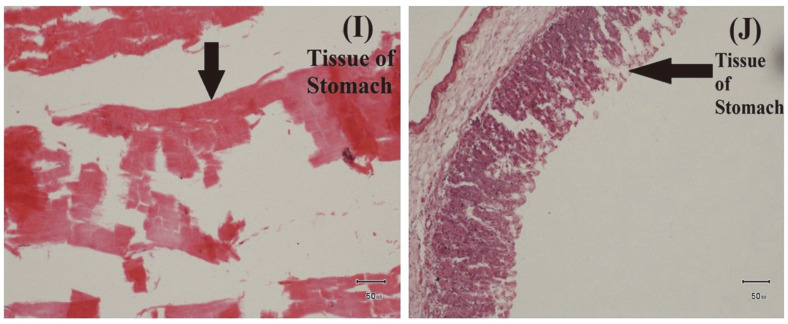
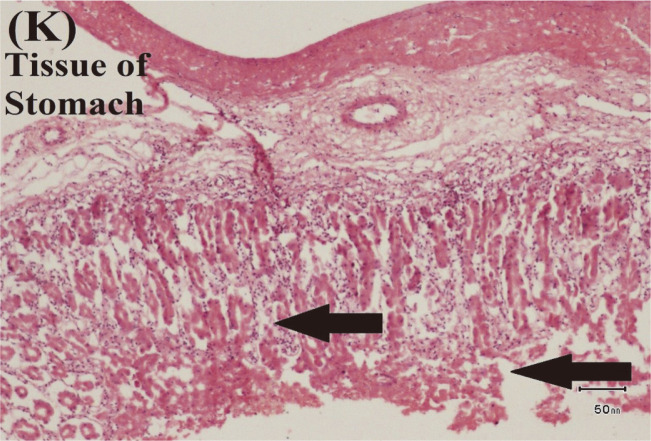
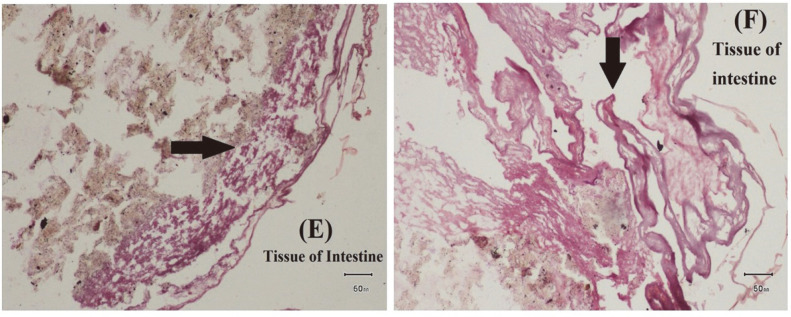
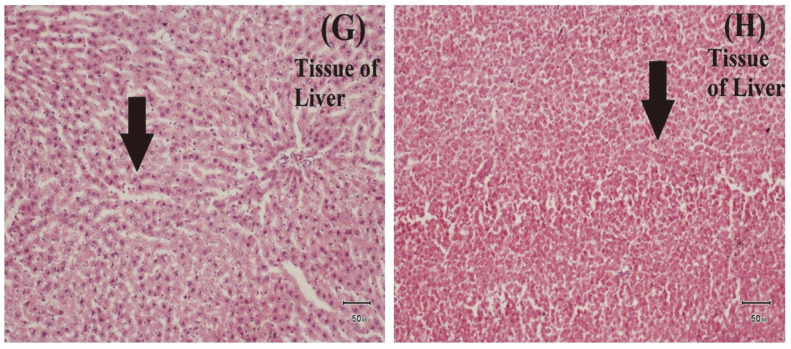
E. tarda causes severe changes in tissues of edwardsiellosis-infected fish samples. Sloughing and necrosis of gastric epithelial cells were observed in the stomach tissues of infected fish. Some mononuclear inflammatory cells and artefactual changes were also observed in tissues of the infected stomach (Figs. 9 and 10). In addition, artifactual changes and cellular swelling were observed in the infected intestine (Fig. 7). Mild cellular swelling was seen in many hepatocytes of the infected liver (Fig. 8).
Fig. 9. Examination of Histopathological Impact of E. tarda.
(I) Sloughing and necrosis of gastric epithelial cells, (J) Some mononuclear inflammatory cells in tissue of stomach.
Fig. 10. Examination of Histopathological Impact of E. tarda.
(K) Artefactual changes in tissue of stomach.
Fig. 7. Examination of Histopathological Impact of E. tarda.
(E) Mild cellular swelling in tissue of intestine (F) Artefactual changes in tissue of intestine.
Fig. 8. Examination of Histopathological Impact of E. tarda.
(G) and (H) Cellular swelling in many hepatocytes of tissue of liver.
Discussion
Edwardsiellosis has been reported in a wide variety of fish species worldwide and caused massive mortality in farmed and wild fish under stress [43]. Various antibiotics are being applied as one of the basic measures to control bacterial infections in aquaculture [44]. However, useless and uncontrolled application of antibiotics has enabled harmful bacteria to develop ABR genes while also giving rise to multiple antibiotic-resistant strains of pathogenic bacteria having ABR genes [45]. Such bacteria have caused severe outbreaks and high mortality at fish farms, ultimately resulting in huge economic losses for fish farmers [42]. Hence, routine antimicrobial susceptibility testing is vital to select suitable antibiotics to control the problem [45].
The acquisition of resistance against multiple antibiotics and the emergence of ABR genes in E. tarda can result in genetic mutations that may alter the target site of antibiotics or provide alternative pathways for survival against antibiotics with efficacy against gram-positive bacteria. Regarding the antibiotic susceptibility testing, retrieved E. tarda isolates revealed a significant difference in their sensitivity to different applied antibiotics. In the present study, chloramphenicol, ciprofloxacin, gentamicin, norfloxacin, streptomycin, and sulfamethoxazole were found 100% effective in controlling edwardsiellosis. However, in prior studies, E. tarda was observed sensitive to chloramphenicol, gentamicin, norfloxacin, streptomycin [26, 46], and ciprofloxacin [47]. Similarly, in the current study, amoxicillin, erythromycin, and flumequine were found 100% ineffective to control E. tarda while all E. tarda isolates showed intermediate resistance (50%) against tetracycline, ampicillin, cefotaxime, and doxycycline. Whereas, a prior study found tetracycline ineffective to control edwardsiellosis [46]. However, previous studies observed E. tarda isolates resistant to ampicillin [48, 49]. E. tarda isolates were resistant to tetracycline, erythromycin, and streptomycin [50], which may be due less to the application of these antibiotics than the increase in fish farming, high stocking density, application of animal waste as fertilizer, naturally antibiotic-resistant aquatic bacteria, and also the transfer of ABR genes from livestock and humans to fish [10, 51]. Recently, in another study, all E. tarda isolates were found susceptible against tetracycline, chloramphenicol and ciprofloxacin, and resistant to erythromycin, ampicillin, cefotaxime, and sulfamethoxazole, with intermediate resistance results against streptomycin, and gentamycin [28]. E. tarda had qnrA and sul3 genes but still E. tarda was found sensitive against ciprofloxacin and sulfamethoxazole, which may be due to the presence of other ABR genes, multiple resistance mechanisms, mutations, and high exposure to these antibiotics. Previously, E. tarda isolates were found resistant to amoxicillin and norfloxacin [26, 47, 52]. Similarly, neomycin was found effective in controlling edwardsiellosis [24, 52], but in our study, all E. tarda isolates showed intermediate resistance against neomycin. These differences in susceptibility might be due to different frequencies and quantities of applied antibiotics and also to varying E. tarda strains.
The existence of ABR genes, whether plasmid or chromosome, can be demonstrated using plasmid curing studies [21]. Recently, in a previous study, beta-lactam resistance gene blaTEM was detected in E. tarda and amplified at 60°C and sul3 at 53°C [27], while blaTEM gene was amplified at 52°C and sul3 gene at 54°C in the current study. This difference of 8°C and 1°C respectively might be due to variations in E. tarda strains with different antibiotic resistance profile, variation in application of beta-lactams and sulphonamides, and also different geographical distribution. Our recorded occurrence of β-lactam resistance gene (blaTEM), was nearly in accordance with findings of [53]. High occurrence may be due to the reason of their chromosomal-mediated, intrinsic resistance and fast transmission into the next pathogenic bacterial generations [54]. In the current study, 16S rRNA was amplified at 1,503 bp at 52°C for 1 min while previously, 16S rRNA bands were detected at 56°C for 30 s [25], at 1,465 bp size [55]. Currently, our isolated E. tarda strains revealed 100% similarity with E. tarda strains isolated in China, India, and the USA. Recently, in a previous study, similarity of 16S rRNA gene was recorded at 75%, 55% and 48% similarity [21], while 97% was reported by [27]. In the current study, E. tarda isolates showed positive results in motility, methyl red, H2S and indole production tests, while negative results were shown in gram-staining, citrate, lactose, amylase and urease tests. Similar results of biochemical identification of E. tarda were reported in previous studies [57, 58]. With respect to phenotypic characterization, the retrieved E. tarda isolates showed specific biochemical profiles, culture conditions, and phenotypic characteristics of E. tarda [59, 60]. Both biochemical and phenotypic characteristics are key factors for differentiation between E. tarda and other members of Enterobacteriaceae [61]. However, various studies showed variations in biochemical and phenotypic characteristics of E. tarda isolates [62]. These variations were attributed to the presence of plasmid that controls the metabolism in bacteria or to their ability to get energy for growth utilizing only citrate [63].
Severe outbreaks and massive mortality due to edwardsiellosis have been reported in farmed and wild fish under stress worldwide [43]. Our study concluded overall 7.69% mortality with 5.35% in O. niloticus, 4.52% in O. mossambicus and 3.47% in O. aureus. In prior studies, 40% mortality was recorded in O. niloticus of fish farms of Dakahlia Governorate, Egypt [19] while 60% was reported in infected fish of farms in Ernakulam, Kerala [21]. Similarly, 40% mortality was reported in autumn, 63.6% in winter, and 69.9% in spring [34]. This difference in fish mortality was due to variations in temperature, stocking density, water quality parameters and different geographical locality.
E. tarda has been associated with severe fish diseases, viz., hemorrhagic septicemia and lesions of skin and internal organs, thereby leading to massive mortality in various fish species [56]. In the current study, the results of clinical and post-mortem examination were in agreement with [20]. Moreover, the current post-mortem findings were highly compatible with those reported by [57], who observed muscular hemorrhage, ascitic fluid and congestion with small, white bacteria-filled nodules in enlarged spleen, kidney, and liver infected O. niloticus. Similarly, severe lesions were reported in the infected tilapia [64]. These variations in symptoms could be related to the pathogen’s ability to invade the fish immune status, severity of infection, site of sample collection, and other environmental conditions. Pathogenic bacteria cause infection in fish through oral and gill routes. In prior studies, histopathological changes were observed after experimental infection; however, the current study observed the impact of E. tarda infection in naturally infected fish tissues [26, 47, 65]. Moreover, in the current study many hepatocytes of infected liver showed cellular swelling while multifocal areas of necrosis were observed in infected liver [21]. Artifactual changes were also observed in intestine but lymphocytic enteritis was observed in intestine of infected fish [17].
The overuse of antibiotics in fish farms leads to the spread of antibiotic resistance and emergence of ABR genes, makes it more challenging to treat infections, especially edwardsiellosis in fish, which ultimately enhances the rate of pathogenicity of E. tarda and making it a major potential threat for tilapia culture. The antibiotic resistance pattern causes multiple abnormalities and severe infections in infected fish resulting in major outbreaks and ultimately causing massive mortality and huge economic loss for fish farmers. To mitigate this, antibiotics must be used responsibly and alternative strategies for disease control must be implemented.
Table 2.
Occurrence of 16S rRNA and antibiotic resistance genes of E. tarda with respect to fish organs.
| Organ | qnrA gene | blaTEM gene | sul3 gene | 16SrRNA gene |
|---|---|---|---|---|
| Liver | 135 (45%) | 157 (52%) | 122 (41%) | 300 (100%) |
| Kidney | 123 (41%) | 143 (48%) | 114 (38%) | 284 (95%) |
| Spleen | 118 (39%) | 132 (44%) | 98 (33%) | 293 (98%) |
| Intestine | 133 (44%) | 174 (58%) | 126 (42%) | 297 (99%) |
| Stomach | 67 (22%) | 109 (36%) | 42 (14%) | 277 (92%) |
| Gills | 41 (14%) | 83 (28%) | 37 (12%) | 189 (63%) |
| Tail fins | 18 (6%) | 26 (9%) | 12 (4%) | 106 (35%) |
| Heart | 26 (9%) | 41 (14%) | 20 (7%) | 148 (49%) |
| Overall | 83 (27.5%) | 108 (36.04%) | 71 (23.8%) | 237 (78.9%) |
Acknowledgments
Mr. Ghulam Muhayyodin (Department of Wildlife and Ecology, University of Veterinary and Animal Sciences, Lahore, Pakistan) helped in designing a GIS map of selected fish farms of three districts. Mr. Haroon Aslam (Aqua Feed, Muzaffargarh) cooperated and assisted in visiting and sampling from fish farms of Muzaffargarh and Mandi Bahauddin.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
References
- 1.Manage PM. Heavy use of antibiotics in aquaculture: emerging human and animal health problems.A review. Sri Lanka J. Aquat. Sci. 2018;23:13–27. doi: 10.4038/sljas.v23i1.7543. [DOI] [Google Scholar]
- 2.Troell M, Kautsky N, Beveridge M, Henriksson P, Primavera J, Ronnback P, et al. In reference module in life sciences. Aquac. 2017 doi: 10.1016/B978-0-12-809633-8.02007-0. https://doi.org/10.1016/B978-0-12-809633-8.02007-0. [DOI] [Google Scholar]
- 3.FAO, author. Food and Agriculture Organization of the United Nations. The State of World Fisheries and Aquaculture; Rome: 2016. [Google Scholar]
- 4.Froehlich HE, Runge CA, Gentry RR, Gaines SD, Halpern BS. Comparative terrestrial feed and land use of an aquaculturedominant world. Proc. Natl. Acad. Sci. USA. 2018;115:5295–5300. doi: 10.1073/pnas.1801692115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FAO, author. Sustainability in action. The state of world fisheries and aquaculture series. Food and Agriculture Organization of the United Nations; 2020. [Google Scholar]
- 6.Sousa SMN, Freccia A, dos Santos LD, Meurer F, Tessaro L, Bombardelli RA. Growth of Nile tilapia post-larvae from broodstock fed diet with different levels of digestible protein and digestible energy. R. Bras. Zootec. 2013;42:535–540. doi: 10.1590/S1516-35982013000800001. [DOI] [Google Scholar]
- 7.Amal MN, Koh CB, Nurliyana M, Suhaiba M, Nor-Amalina Z, Santha S, et al. A case of natural co-infection of tilapia lake virus and Aeromonas veronii in a Malaysian red hybrid tilapia (Oreochromis niloticus × O. mossambicus) farm experiencing high mortality. Aquac. 2018;485:12–16. doi: 10.1016/j.aquaculture.2017.11.019. [DOI] [Google Scholar]
- 8.Watts EMJ, Schreier JH, Lanska L, Hale SM. The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Mar. Drugs. 2017;15:158. doi: 10.3390/md15060158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seth M, Chandrasekaran N, Mukherjee A, Thomas J. Pathogenicity of Edwardsiella tarda in Oreochromis mossambicus and treatment by Tamarindus indica seed extract. Aquac Inter. 2021;29:1829–1841. doi: 10.1007/s10499-021-00719-0. [DOI] [Google Scholar]
- 10.Cantas L, Shah SQ, Cavaco LM, Manaia CM, Walsh F, Popowska M, et al. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front. Microbiol. 2013;4:96. doi: 10.3389/fmicb.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marti E, Variatza E, Balcazar JL. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014;22:36–41. doi: 10.1016/j.tim.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Schar D, Klein EY, Laxminarayan R, Gilbert M, Van Boeckel TP. Global trends in antimicrobial use in aquaculture. Sci. Rep. 2020;10:21878. doi: 10.1038/s41598-020-78849-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabello FC, Godfrey HP, Buschmann AH, Dolz HJ. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016;16:127–33. doi: 10.1016/S1473-3099(16)00100-6. [DOI] [PubMed] [Google Scholar]
- 14.Woo SJ, Kim MS, Jeong MG, Do MY, Hwang SD, Kim WJ. Establishment of epidemiological cut-off values and the distribution of resistance genes in Aeromonas hydrophila and Aeromonas veronii isolated from aquatic animals. Antibiot. 2022;11:343. doi: 10.3390/antibiotics11030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munita JM, Arias CA. Mechanisms of antibiotic resistance. In: Kudva IT, editor. Virulence mechanisms of bacterial pathogens. Hoboken, NJ: John Wiley and Sons; 2016. pp. 481–511. [Google Scholar]
- 16.Rodrigues MV, Falcone-Dias MF, Francisco CJ, David GS, Da Silva RI, Junior JPA. Occurrence of Edwardsiella tarda in Nile tilapia Oreochromis niloticus from Brazilian Aquaculture Edwardsiella tarda in Nile tilapia. Acta. Sci. Microbiol. 2019;2:13–19. [Google Scholar]
- 17.Davies MY, de Oliveira MG, Cunha PVM, Franco SL, Santos SSL, Moreno ZL, et al. Edwardsiella tarda outbreak affecting fishes and aquatic birds in Brazil. Vet. Q. 2018;38:99–105. doi: 10.1080/01652176.2018.1540070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du C, Huo X, Gu H, Wu D, Hu Y. Acid resistance system CadBA is implicated in acid tolerance and biofilm formation and is identified as a new virulence factor of Edwardsiella tarda. Vet. Res. 2021;52:117. doi: 10.1186/s13567-021-00987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Algammal AM, Mabrok M, Ezzat M, Alfifi KJ, Esawy AM, Elmasry N, et al. Prevalence, antimicrobial resistance (AMR) pattern, virulence determinant and AMR genes of emerging multi-drug resistant Edwardsiella tarda in Nile tilapia and African catfish. Aquac. 2022;548:737643. doi: 10.1016/j.aquaculture.2021.737643. [DOI] [Google Scholar]
- 20.Butar-Butar OD, Suryanto D, Ilyas S. Detection of Edwardsiella tarda infection of catfish (Clarias gariepinus) in Central Tapanuli Regency, North Sumatra, Indonesia. IOSR J. Agric. Vet. Sci. 2020;13:6–13. [Google Scholar]
- 21.Preena PG, Dharmaratnam A, Swaminathan RT. A peek into mass mortality caused by antimicrobial resistant Edwardsiella tarda in goldfish, Carassius auratus in Kerala. Biologia. 2022;77:1161–1171. doi: 10.1007/s11756-022-01007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh WT, Jun JW, Kim HJ, Giri SS, Yun S, Kim SG, et al. Characterization and pathological analysis of a virulent Edwardsiella anguillarum strain isolated from Nile tilapia (Oreochromis niloticus) in Korea. Front. Vet. Sci. 2020;7:14. doi: 10.3389/fvets.2020.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charles OS, Olabisi IO, Olufemi OI, Bolarinwa AO. Detection and antibiogram of Edwardsiella tarda from Oreochromis niloticus (Tilapia fish) obtained from selected farms in Ibadan, Nigeria. J. Food Safe Hyg. 2020;6:38–46. doi: 10.18502/jfsh.v6i1.6024. [DOI] [Google Scholar]
- 24.Kumar P, Adikesavalu H, Abraham TJ. Prevalence of Edwardsiella tarda in commercially important finfish and shellfish of Bihar and West Bengal, India. J. Coast. Life Med. 2016;4:30–35. doi: 10.12980/jclm.4.2016apjtd-2014-0184. [DOI] [Google Scholar]
- 25.Nantongo M, Mkupasi EM, Byarugaba DK, Wamala SP, Mdegela RH, Walakira JK. Molecular characterization and antibiotic susceptibility of Edwardsiella tarda isolated from farmed Nile tilapia and African catfish from Wakiso, Uganda. Uganda. J. Agri. Sci. 2019;19:51–64. doi: 10.4314/ujas.v19i1.5. [DOI] [Google Scholar]
- 26.Nagy EA, Fadel, Al-Moghny FA, Ibrahim MS. Isolation, identification and pathogenicity characterization of Edwardsiella tarda isolated from Oreochromis niloticus fish farms in Kafr-Elshiekh, Egypt. Alex. J. Vet. Sci. 2018;57:171–179. doi: 10.5455/ajvs.294237. [DOI] [Google Scholar]
- 27.Niu G, Wongsathein D, Boonyayatra S, Khattiya K. Occurrence of multiple antibiotic resistance and genotypic characterization in Edwardsiella tarda isolated from cage‐cultured hybrid red tilapia (Oreochromis sp.) in the Ping River, Northern Thailand. Aquac. Res. 2019;50:3643–3652. doi: 10.1111/are.14322. [DOI] [Google Scholar]
- 28.Wimalasena SHMP, Pathirana HNKS, De Silva BCJ, Hossain S, Sugaya E, Nakai T, et al. Antibiotic resistance and virulenceassociated gene profiles of Edwardsiella tarda isolated from cultured fish in japan. Turk. J. Fish Aquat. Sci. 2018;19:141–148. doi: 10.4194/1303-2712-v19_2_06. [DOI] [Google Scholar]
- 29.Park SB, Aoki T, Jung TS. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet. Res. 2012;43:67. doi: 10.1186/1297-9716-43-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou JH, Hu YH, Zhang M, Sun L. Identification and characterization of the AcrR/AcrAB system of a pathogenic Edwardsiella tarda strain. J. Gen. Appl. Microbiol. 2009;55:191–199. doi: 10.2323/jgam.55.191. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Sun B, Ning X, Jiang S, Sun L. A comparative analysis of Edwardsiella tarda induced transcriptome profiles in RAW264.7 cells reveals new insights into the strategy of bacterial immune evasion. Int. J. Mol. Sci. 2019;20:5724. doi: 10.3390/ijms20225724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamuro T, Fukuhara A, Kang J, Takamatsu J. A case of necrotizing fasciitis following Edwardsiella tarda septicemia with gastroenteritis. J. Infect. Chemother. 2019 doi: 10.1016/j.jiac.2019.05.017. S1341-321X. 19. 30149-30147. [DOI] [PubMed] [Google Scholar]
- 33.Noga EJ. Fish disease diagnosis and treatment (Iowa: Iowa State University Press) 2010 doi: 10.1002/9781118786758. [DOI] [Google Scholar]
- 34.Muratori MCS, Martins NE, Peixoto MTD, Oliveira AL, Ribeiro LP, Costal APR, et al. Mortalidade por "septicemia dos peixes tropicais" em tilapias criadas em consorciacao com suínos. Arq. Bras. Med. Vet. Zootec. 2001;53:658–662. doi: 10.1590/S0102-09352001000600007. [DOI] [Google Scholar]
- 35.Austin B, Austin DA. Bacterial Fish Pathogens. Springer International Publishing; 2016. Miscellaneous Pathogens; pp. 603–642. [Google Scholar]
- 36.Shah D, Shiringi S, Besser T, Call D. Molecular detection of food borne pathogens, Boca Raton: CRC Press, In Liu. (Edition) Taylor and Francis group; Florida, USA: 2009. pp. 369–389. [Google Scholar]
- 37.Tenover FC. Encyclopedia of microbiology. In Schaechter M. (ed.), Antibiotic susceptibility testing, fourth ed. Academic Press, Cambridge. 2014;2009:67–77. doi: 10.1016/B978-012373944-5.00239-X. [DOI] [Google Scholar]
- 38.Weinstein MP, Lewis JS. The clinical and laboratory standards institute subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes. J. Clin. Microbiol. 2020;58:e01864–19. doi: 10.1128/JCM.01864-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge M, Jin X, Zhang Y, Fang H, Chen C. Detection of drug resistance and four drug resistance genes in bacterial pathogen Edwardsiella tarda (in Chinese) Fish. Sci. 2015;34:300–304. [Google Scholar]
- 40.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. Qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agent Chemother. 2006;50:2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiao M, Ying GG, Singer AC, Zhu YG. Review of antibiotic resistance in China and its environment. Environ. Int. 2018;110:160–72. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 43.Wamala SP, Mugimba KK, Mutoloki S, Evensen O, Mdegela R, Byarugaba DK, et al. Occurrence and antibiotic susceptibility of fish bacteria isolated from Oreochromis niloticus (Nile tilapia) and Clarias gariepinus (African catfish) in Uganda. Fish. Aquat. Sci. 2018;21:1–10. doi: 10.1186/s41240-017-0080-x. [DOI] [Google Scholar]
- 44.Lo DY, Lee YJ, Wang JH, Kuo HC. Antimicrobial susceptibility and genetic characterization of oxytetracycline resistant Edwardsiella tarda isolated from diseased eels. Vet. Record. 2014;175:203. doi: 10.1136/vr.101580. [DOI] [PubMed] [Google Scholar]
- 45.Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA. Prevalence, antimicrobial resistance profiles, virulence and enterotoxin-determinant genes of MRSA isolated from subclinical bovine mastitis samples in Egypt. Pathogen. 2020;9:362. doi: 10.3390/pathogens9050362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali MH, Chowdhury FS, Ashrafuzzaman M, Nayem MA, Chowdhury, Haque MRU, et al. Identification, pathogenicity, antibiotic and herbal sensitivity of Edwardsiella tarda causing fish disease in Bangladesh. Cur. Res. Micro. Biotech. 2014;2:292–297. [Google Scholar]
- 47.Noor El Deen AIE, El-Gohary MS, Abdou MS, Adel El-Gamal M. Molecular characterization of Edwardsiella tarda bacteria causing severe mortalities in cultured Oreochromis niloticus fish with treatment trials. Inter. J. Cur. Res. 2017;9:50962–50969. [Google Scholar]
- 48.Lee SW, Wendy W. Antibiotic and heavy metal resistance of Aeromonas hydrophila and Edwardsiella tarda isolated from red hybrid tilapia (Oreochromis spp.) coinfected with motile Aeromonas septicemia and edwardsiellosis. Vet. World. 2017;10:803–807. doi: 10.14202/vetworld.2017.803-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogbonne FC, Ukazu ER, Egbe FC. Antibiotics resistance pattern and plasmid profiling of Edwardsiella tarda isolated from Heterobranchus longifilis. J. Biosci. Med. 2018;6:95–105. doi: 10.4236/jbm.2018.64008. [DOI] [Google Scholar]
- 50.Newaj‐Fyzul A, Mutani A, Ramsubhag A, Adesiyun A. Prevalence of bacterial pathogens and their anti‐microbial resistance in tilapia and their pond water in Trinidad. Zoonos. Public Health. 2008;55:206–213. doi: 10.1111/j.1863-2378.2007.01098.x. [DOI] [PubMed] [Google Scholar]
- 51.Penders J, Stobberingh EE. Antibiotic resistance of motile aeromonads in indoor catfish and eel farms in the southern part of The Netherlands. Int. J. Antimicrob. Agents. 2008;31:261–265. doi: 10.1016/j.ijantimicag.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Mahmoud E, El-Tarabili RM, Esawy AM, Elmasry N. Antibiotic resistance and antibiotic resistance genes among Edwardsiella tarda isolated from fish. Suez Canal. Vet. Med. J. SCVMJ. 2021;26:171–188. doi: 10.21608/scvmj.2021.184978. [DOI] [Google Scholar]
- 53.Sedek A, El Tawab AA, El-Hofy F, El-Gohary M. Antibiotic resistance genes of Edwardsiella tarda isolated from Oreochromis niloticus and Claris gariepinus. Benha Vet. Med. J. 2020;38:131–135. doi: 10.21608/bvmj.2020.29323.1205. [DOI] [Google Scholar]
- 54.Kummerer K. Antibiotics in the aquatic environment-a review-part I. Chemosphere. 2009;75:417–434. doi: 10.1016/j.chemosphere.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 55.Katharios P, Kokkari C, Dourala N, Smyrli M. First report of Edwardsiellosis in cage-cultured sharp snout sea bream, Diplodus puntazzo from the Mediterranean. Vet. Res. 2015;11:155–161. doi: 10.1186/s12917-015-0482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubey S, Maiti B, Kim SH, Sivadasan SM, Kannimuthu D, Pandey PK, et al. Genotypic and phenotypic characterization of Edwardsiella isolates from different fish species and geographical areas in Asia: implications for vaccine development. Fish Dis. 2019;42:835–850. doi: 10.1111/jfd.12984. [DOI] [PubMed] [Google Scholar]
- 57.El-Seedy FR, Radwan IA, Abd El-Galil MA, Sayed HH. Phenotypic and Genotypic characterization of Edwardsiella tarda isolated from Oreochromis niloticus and Clarias gariepinus at Sohag Governorate. J. Americ. Sci. 2015;11:68–75. [Google Scholar]
- 58.Moustafa EM, Omar AA, Abdo WS. Insight into the virulence-related genes of Edwardsiella tarda isolated from cultured freshwater fish in Egypt. World Vet. J. 2016;6:101–109. doi: 10.5455/wvj.20160874. [DOI] [Google Scholar]
- 59.Enany M, AL-Gammal A, Hanora A, Shagar G, El Shaffy N. Sidr honey inhibitory effect on virulence genes of MRSA strains from animal and human origin. Suez Canal Vet. Med. J. 2018a;20:23–30. doi: 10.21608/scvmj.2015.64562. [DOI] [Google Scholar]
- 60.Enany M, Shalaby A, El Deen AN, Wahdan A, El Kattawy Z. Characterization of Edwardsiella species isolated from fish by using genomic DNA fingerprinting technique. Suez Canal Vet. Med. J. 2018b;23:215–223. doi: 10.21608/scvmj.2018.60542. [DOI] [Google Scholar]
- 61.Sakazaki R. Edwardsiella. Wiley; 2015. pp. 1–12. [Google Scholar]
- 62.Kim KI, Kang JY, Park JY, Joh SJ, Lee HS, Kwon YK. Phenotypic traits, virulence-associated gene profile and genetic relatedness of Edwardsiella tarda isolates from Japanese eel Anguilla japonica in Korea. Lett. Appl. Microbiol. 2014;58:168–176. doi: 10.1111/lam.12172. [DOI] [PubMed] [Google Scholar]
- 63.Panangala VS, Shoemaker CA, McNulty ST, Arias CR, Klesius PH. Intra- and interspecific phenotypic characteristics of fishpathogenic Edwardsiella ictaluri and E. tarda. Aquac. Res. 2006;37:49–60. doi: 10.1111/j.1365-2109.2005.01394.x. [DOI] [Google Scholar]
- 64.Iregui CA, Guarin M, Tibata VM, Ferguson HW. Novel brain lesions caused by Edwardsiella tarda in a red tilapia (Oreochromis spp.) J. Vet. Diagn. Investig. 2012;24:446–449. doi: 10.1177/1040638711435232. [DOI] [PubMed] [Google Scholar]
- 65.Devi TB, Abraham TJ, Kamilya D. Susceptibility and pathological consequences of catla, Catla catla (Hamilton) experimentally infected with Edwardsiella tarda. Arch. Pol. Fish. 2016;24:209–217. doi: 10.1515/aopf-2016-0018. [DOI] [Google Scholar]
- 66.Huong NTT, Thuy HL, Gallardo WG, Thanh HN. Bacterial population in intensive tilapia (Oreochromis niloticus) culture pond sediment in Hai Duong province, Vietnam. Int. J. Fish. 2014;6:133–139. [Google Scholar]