Highlights
-
•
Coagulase-negative staphylococci (CoNS), particularly S. chromogenes are the most predominant organisms associated with bovine sub-clinical mastitis (SCM).
-
•
Multidrug resistance isolates were identified in all five Staphylococcus species. The detection of chromosome-mediated mecA gene and the virulence gene within the CoNS could be a potential challenge for the clinical management of SCM.
-
•
‘Use of antimicrobials’ and ‘History of previous clinical mastitis’ were identified as significant risk factors for methicillin-resistant staphylococci in SCM-affected cows.
Keywords: SCM, Staphylococcus spp., mecA gene, Virulence genes, Risk factors
Abstract
This study was conducted to investigate the diversity and antimicrobial resistance profiling of Staphylococcus species causing sub-clinical mastitis (SCM) in dairy herds in Bangladesh as well as putative risk factors associated with the infections. Individual quarter milk samples were collected from a total of 284 lactating cows from 30 dairy farms were screened by means of California mastitis test; 178 (62.7%) of them had at least of quarter affected by SCM. After conventional microbiological isolation procedures, PCR tests were used for Staphylococcus species identification and detection of antimicrobial resistance and virulence genes. S. chromogenes (65.7%) was the most predominant species followed by, S. epidermidis (20.2%), S. haemolyticus (19.1%), S. aureus (15.7%), and S. sciuri (5.6%). High levels of antimicrobial resistance to ampicillin and amoxicillin/clavulanic acid were observed in S. aureus (82.1% and 75%) and S. sciuri (80% and 70%), while resistance to cefepime was markedly higher in S. chromogenes (95.7%), S. haemolyticus (94.1%), and S. epidermidis (97.2%). Multidrug resistance isolates were identified in all five species. The mecA gene was detected in S. aureus (32.1%) and S. chromogenes (5.98%). In addition, 20% S. sciuri and 17.7% S. haemolyticus carried the cytotoxin (pvl) gene, while 14.3% S. aureus harbored the toxic shock syndrome toxin (tst) gene. Multivariable logistic regression analysis identified “Old aged” (OR [CI]: 3.5 [1–12.4]); “Early stage of lactation” (OR [CI]: 3.4 [1.2–9.7]) and, “Firm udder condition” (OR [CI]: 4.2 [1.2–14.6]) as risk factors associated with SCM caused by S. aureus, S. chromogenes, and S. haemolyticus, respectively. Moreover, “Use of antimicrobials” (OR [CI]: 10.4 [3.4–32.1] and “History of previous clinical mastitis” (OR [CI]: 4.9 [1.2–19.7] for the carriage of methicillin-resistant Staphylococcus spp.
1. Introduction
Sub-clinical mastitis (SCM) is the most frequent diseases in dairy farming across the world causing serious economic burden (Hoque et al., 2018). Although there is usually a significant decrease in milk production due to damage of the mammary gland, there are no clinical signs of mastitis compromising diagnosis (Simojoki et al., 2009 and De Buck et al., 2021). Although staphylococci are considered as major mastitis-causing agents responsible for clinical and SCM (De Visscher et al., 2017), it is worth nothing that the contagious Staphylococcus aureus and others species of coagulase-negative staphylococci (CoNS) such as Staphylococcus haemolyticus, Staphylococcus chromogenes, Staphylococcus epidermidis, Staphylococcus sciuri, and Staphylococcus simulans, are frequently isolated from bovine SCM cases (De Visscher et al., 2017; De Buck et al., 2021). Nonetheless, differentiation between species of mastitis-associated CoNS by conventional microbiological methods is laborious and time consuming (Capurro et al., 2009) due to the close phenotypic resemblance within the CoNS species. CoNS species can colonize the mammary glands tissue for prolonged periods of time or even throughout the lactation period being continuously shed into milk (Taponen et al., 2007; Taponen & Pyörälä, 2009). Furthermore, CoNS species can be highly transmittable (Ster et al., 2013and Schabauer et al., 2018) and rapidly spread within the dairy farm through the milkers as well as by milking vehicles like milking machine or instruments. In addition, most of the organisms encode important virulence factors, as well as multiple antimicrobial resistance genes (Hosseinzadeh et al., 2014; Frey et al., 2013, Zhou et al., 2017 and González-Martín et al., 2020) and cause persistent infection in mammary gland (Cervinkova et al., 2013 and Mahato et, al.,2017). Some CoNS strains exhibit poor response to a wide range of antimicrobials leading to low cure and high culling rates in the dairy farms (Hoque et al., 2018 and Schnitt et al., 2021). Consequently, the emergence and spread of antimicrobial resistant CoNS species, particularly multidrug resistant (MDR) strains is of utmost concern for both mastitis treatment, (Dabele et al., 2021) and public health (Dhaouadi et al., 2020). Therefore, the rapid identification of Staphylococcus species associated with udder colonization in dairy herds is key importance for the control of the infection (Piessens et al., 2011). In this regard, rapid DNA-based molecular diagnostic techniques can be highly reliable, accurate, and reproducible (Zadoks & Watts, 2009) towards the identification of staphylococci at the species level as well as virulence and antimicrobial resistance determinants. Although, there are several studies on the damaging effect and pathogenic significance of S. aureus (Hoque etal., 2018; Priyantha et al., 2021 and Rana et al., 2022a), but there is few information on the species-specific significance, virulence and antimicrobial resistance profile of CoNS associated with bovine SCM in developing region including Bangladesh. Therefore, the aim of the present study was to characterize S. aureus and CoNS and identify risk factors associated with their occurrence in dairy herds in Bangladesh.
2. Materials and methods
2.1. Ethical approval
Both field (SCM screening and sample collection) and laboratory procedures of the current study were performed with the permission of the Chattogram Veterinary and Animal Sciences University's (CVASU) Ethical Approval Committee [Approval no. CVASU/Dir (R&E) EC/2020/165 (1)].
2.2. Study design
Across-sectional study was conducted in the Cumilla district of Bangladesh (N 23.4607°, E 91.1809°) from January 2020 to September 2022. The majority of them used crossbred cows (Holstein-Friesian × Indigenous), Jerseys, and non-descriptive breeds. Only dairy farms having more than 10 lactating cows and a minimum average milk production of 50 liters per day were selected for this study.
2.3. Data collection
Pre-tested questionnaires (supplementary file-1) were used for the collection of all farm and animal related data including husbandry management, udder health information, all hygienic practices and antimicrobials used for the treatment of bovine mastitis. Finally, individual data from a SCM positive cow were gathered to determine further association of various risk factors with identified Staphylococcus spp and antimicrobial resistance.
2.4. Screening and diagnosis of SCM
California Mastitis Test (CMT) was performed for all the lactating cows on each dairy farm for the initial screening of SCM. The test and interpretation were performed according to the protocol described by Abebe et al. (2016). In brief, 2 mL of milk from each quarter was collected in CMT paddles, and an equal volume of CMT reagent was mixed for 60 s in a gentle circular motion. The CMT results were scored and interpreted as 0 (negative [healthy quarter], somatic cell count [SCC] ≤100,000 cells/ml milk), 1+ (weak positive, SCC >100,000–500,000 cells/ml milk), 2+ (distinct positive, SCC >500,000–1000,000 cells/ml milk), and 3+; (strong positive, SCC ≥1000,000 cells/ml milk). Finally, udder quarters showing a CMT score of 1+ were considered as positive for SCM. Furthermore, if any of the tested cows showed mixed CMT results (different scores) within the same udder from different quarters, we considered the maximum CMT score for grading SCM in cows.
2.5. Sample collection and transportation
After confirmation of SCM, approximately 10 mL of milk was collected from each CMT positive quarter by following the procedures of the National Mastitis Council (NMC), 2004. Before milk collection the teat of the infected quarter was cleaned using 70% ethanol, then fore milk has been discarded. After that, the samples were taken aseptically into the individual sterile plastic tubes for microbiological analysis. Finally, all milk samples were transferred under refrigeration to the Department of Microbiology and Veterinary Public Health, CVASU. Samplings were performed weekly by a trained veterinarian.
2.6. Isolation and identification of Staphylococcus spp
Microbiological analysis was performed within 12 h of sampling. Initially, all milk samples were directly incubated at 37°C for 8 h which enhanced the growth rate of mastitic organisms. Then, 20 μL of milk was streaked on 5% bovine blood agar (Oxoid Ltd., Cambridge, UK) and the agar plates were incubated at 37°C for 48 h. All agar plates were examined in every 24 h to check the optimum growth of desired organisms. Suspicious staphylococci colonies were initially identified based on appearance (round, smooth, raised, pigmented or non-pigmented, yellow or creamy white, presence or absence of hemolysis) and Gram staining. In addition, selected staphylococci colonies were further subcultured on Mannitol Salt Agar (MSA) (Oxoid Ltd., Basingstoke, UK), and incubated at 37 °C for 48 h. Subsequently, a catalase test was performed to differentiate the catalase-positive Staphylococcus spp. from the catalase-negative Streptococcus spp. and tube coagulase test was performed to distinguish coagulase-positive staphylococci (CoPS) from coagulase-negative staphylococci (CoNS) or non-aureus staphylococci (NAS) (Quinn et al., 1998).
2.7. Extraction of bacterial chromosomal DNA and multiplex PCR program
In the current study, we used a multiplex- polymerase chain reaction (m-PCR) assay for the species-specific confirmation of investigated S. aureus, S. haemolyticus, S. chromogenes, S. epidermidis, S. sciuri, and S. simulans. The chromosomal DNA of Staphylococcus spp. was extracted from freshly grown cultures by the crude boiling lysis method described by Hoque et al. (2018) with some modifications. All staphylococcal genomic DNA were investigated for the confirmation of species level by m-PCR targeting different species specific oligonucleotide primers (Table 1) as described previously by Shome et al. (2011). A total of 50 μL of PCR reaction mixture was thermally cycled once for denaturation at 94°C for 5 min, followed by 30 times at 94°C for 30 s; 60°C for 30 s,72°C for 45 s and then once for final extension at 72°C for 10 min. Bacterial species was confirmed based on PCR-amplified product size, and the amplicons were measured using a 1 kb plus DNA ladder on a 1.5% agarose gel (Promega Corporation, Madison, WI, USA) containing ethidium bromide. Previously confirmed S. aureus was used as positive control, while S. pseudintermedius (Rana et al., 2020b) was used as negative control for every m-PCR reaction. Finally, all genomic DNA and PCR confirmed pure isolates were cultured on brain heart infusion broth (BHIB) (Oxoid Ltd., Basingstoke, UK) and stocked at –80°C for further analysis.
Table 1.
Primer sequences used for the amplification of different Staphylococcus spp.
| Organism | Gene primer | Primer designation | Primer sequence 5´−3´ | Amplicon size (bp) | References |
|---|---|---|---|---|---|
| S. aureus | 23S rRNA | SAS2F | AGCGAGTCTGAATAGGGCGTTT | 894 | Shome et al., 2011 |
| SAS2R | CCCATCACAGCTCAGCCTTAAC | ||||
| S. haemolyticus | sodA | SHS1F | CAAATTAAATTCTGCAGTTGAGG | 214 | |
| SHS1R | AGAGCCCCATTGTTCTTTGA | ||||
| S. chromogenes | sodA | SCHS1F | GCGTACCAGAAGATAAACAAACTC | 222 | |
| SCHS1R | CATTATTTACAACGAGCCATGC | ||||
| S. epidermidis | rdr | SERF | AAGAGCGTGGAGAAAAGTATCAAG | 130 | |
| SERR | TCGATACCATCAAAAAGTTGG | ||||
| S. sciuri | gap | SSCGF | GATTCCGCGTAAACGGTAGAG | 306 | |
| SSCGR | CATCATTTAATACTTTAGCCATTG | ||||
| S. simulans | gap | SSMF | AGCTTCGTTTACTTCTTCGATTGT | 472 | |
| SSMR | AAAAGCACAAGCTCACATTGAC |
2.8. Antimicrobial resistance profile of diverse Staphylococcus spp
After PCR confirmation, individual staphylococcal isolates were subjected to susceptibility testing by means of standard disk diffusion method using eleven different antimicrobials (Oxoid Ltd., Basingstoke, UK) from eight different antimicrobial classes: amoxycillin/clavulanic acid (AMC) 30 µg, ampicillin (AMP) 10 μg, ceftriaxone (CRO) 30 µg, cefoxitin (FOX) 10 µg, cefepime (FEP) 30 µg, erythromycin (E) 15 µg, gentamicin (CN) 10 µg, tetracycline (TE) 10 µg, ciprofloxacin (CIP) 5 µg, trimethoprim/sulfamethoxazole (SXT) 25 µg and imipenem (IMP) 10 µg. The zone of inhibition of individual disks was measured and interpreted as susceptible (S), intermediate (I), or resistant (R) by following the guidelines of Clinical and Laboratory Standards Institute (CLSI, 2018) for veterinary pathogens. Any bacterial isolates exhibiting resistance against ≥3 different classes of antimicrobial was classified as MDR (Magiorakos et al., 2012).
2.9. Detection of resistance genes
Any staphylococcal isolates showing resistance to cefoxitin was further screened for the detection of the mecA gene by PCR, as described earlier by Larsen et al., 2008 (Table 2). A previously confirmed MRSA strain (Rana et al., 2020b) and nuclease-free water were used as positive and negative controls, respectively in PCR reactions.
Table 2.
Primer sequences used for nuc, mecA and virulence genes amplification.
| Organism | Gene Primer | Primer designation | Primer sequence 5´−3´ | Amplicon size (bp) | References |
|---|---|---|---|---|---|
| Staphylococcus spp. | nuc | nucF | TCGCTTGCTATGATTGTGG | 359 | Sasaki et al., 2010 |
| nucR | GCCAATGTTCTACCATAGC | ||||
| mecA | MecF | TCCAGATTACAACTTCACCAGG | 162 | Larsen et al., 2008 | |
| MecR | CCACTTCATATCTTGTAACG | ||||
| sea | SF | GGTTATCAATGTGCGGGTGG | 102 | Zhou et al., 2017 | |
| SR | CGGCACTTTTTTCTCTTCGG | ||||
| seb | SF | GTATGGTGGTGTAACTGAGC | 164 | ||
| SR | CCAAATAGTGACGAGTTAGG | ||||
| sec | SF | AGATGAAGTAGTTGATGTGTATGG | 451 | ||
| SR | CACACTTTTAGAATCAACCG | ||||
| tst | SF | ACCCCTGTTCCCTTATCATC | 326 | ||
| SR | TTTTCAGTATTTGTAACGCC | ||||
| eta | SF | ATATCAACGTGAGGGCTCTAGTAC | 1155 | ||
| SR | ATGCAGTCAGCTTCTTACTGCTA | ||||
| etb | SF | CACACATTACGGATAATGCAAG | 604 | ||
| SR | TCAACCGAATAGAGTGAACTTATCT | ||||
| pvl | SF | GTGCCAGACAATGAATTACCC | 255 | ||
| SR | TTCATGAGTTTTCCAGTTCACTTC |
2.10. Detection of virulence genes
All PCR confirmed Staphylococcus species were further screened for the detection of the following virulence genes: enterotoxins (sea, seb and sec), cytotoxin (pvl), exfoliative toxins (eta and etb) and a toxic shock syndrome toxin (tst) previously described by Zhou et al., 2017 (Table 2). In addition, all S. aureus isolates displaying positive results for the tube coagulase test were further confirmed for the presence of the nuc gene (Sasaki et al., 2010).
2.11. Data management and analysis
All the data obtained from the dairy farm during sample collection and laboratory analyses was entered into spreadsheets using commercial software (Microsoft Excel 2010). The descriptive statistics were performed to calculate the prevalence of the current study. The prevalence of SCM was calculated by dividing the number of SCM affected cows (numerator) by the total number of tested cows (the denominator). All putative risk factors for SCM were analyzed considering six outcomes in mastitic milk: the presence of S. aureus, S. haemolyticus, S. chromogenes, S. epidermidis, S. sciuri, and methicillin-resistant Staphylococcus spp. However, none of the milk samples tested positive for S. simulans. Shortly, univariable logistic regression analysis was performed to identify possible risk factors for the six mentioned outcomes. Any variables showing a p value of ≤0.20 were selected for the multivariable logistic regression model. Stepwise forward selection approach was followed to construct the absolute model. Variables having p value of ≤0.05 were considered significant for the final model. All statistical analysis was performed using STATA® 13.0 software.
3. Results
3.1. Prevalence of SCM
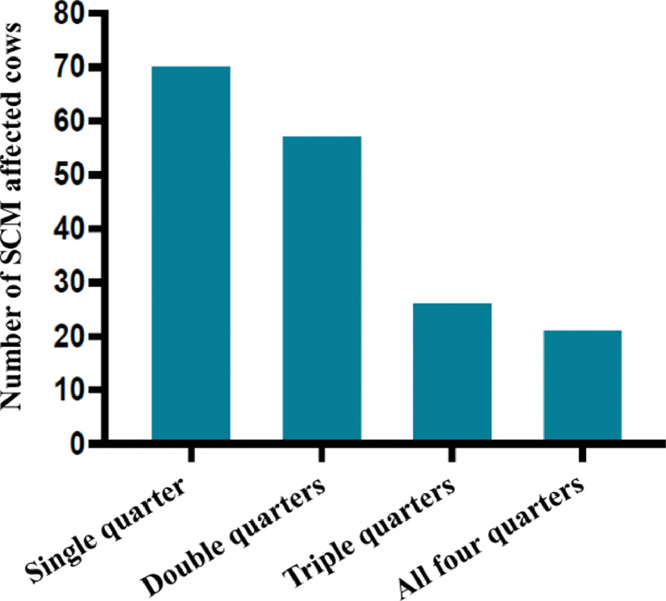
A total of 284 lactating cows (1136 quarters) were screened for SCM on 30 selected dairy farms. Out of them, 178 [62.7%, 95% Confidence Interval (CI): 56.9 to 68.1] cows were positive for the CMT and confirmed as having SCM (Table 3). Among the SCM affected cows, 71 (39.9%, 95% CI: 33 to 47.2) cases were associated with a single quarter, 58 (32.6%, 95% CI: 26.1 to 39.8) with two quarters, 27 (15.2%, 95% CI: 10.6 to 21.2) with three quarters, and 22 (12.4%, 95% CI: 8.2 to 18) with four quarters (Fig. 1). Moreover, a total of 356 (31.3%, 95% CI: 28.7 to 34.1) quarters were found positive for SCM through the CMT test. All the dairy farms (100%, 95% CI: 86.5 to 100) were positive for SCM (Fig. 2).
Table 3.
Prevalence of different Staphylococcus spp. associated with SCM affected lactating cows.
| Total number of dairy farm | Total number of lactating cows | SCM affected cows | PCR confirmed bacterial species | Prevalence |
|---|---|---|---|---|
| 30 | 284 | 178 (62.7%) | S. aureus | 28 (15.7%) |
| S. chromogenes | 117 (65.7%) | |||
| S. haemolyticus | 34 (19.1%) | |||
| S. epidermidis | 36 (20.2%) | |||
| S. sciuri | 10 (5.6%) |
Fig. 1.
Frequency of SCM in different quarters of CMT- positive cows.
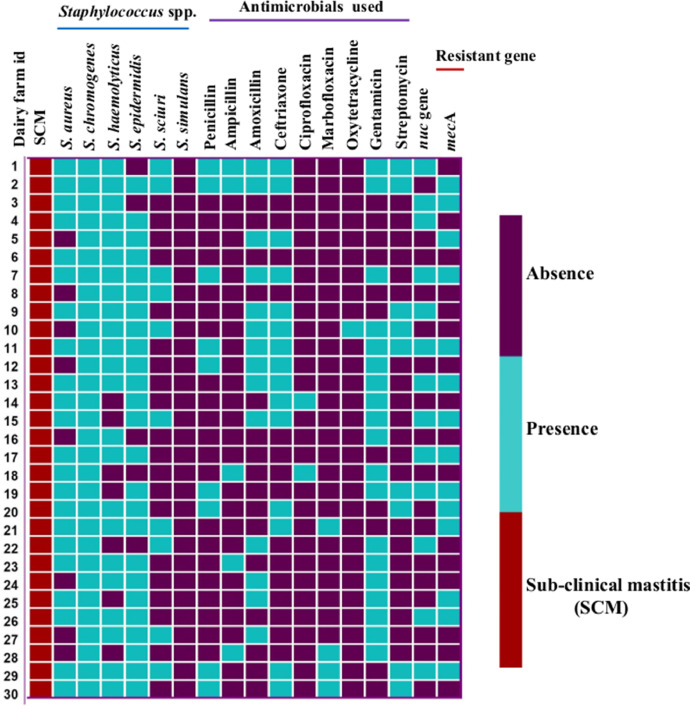
Fig. 2.
The heat map represents the distribution of diverse Staphylococcus spp. isolated from SCM-affected cows and different classes of antimicrobials used in dairy farms, as well as the presence of the nuc gene and the methicillin-resistant gene (mecA). Each row represents an individual dairy farm.
3.2. Detection rate of SCM based on CMT score
Considering all 284 lactating cows, 106 (37.3%, 95% CI: 31.9 to 43.1) had negative CMT results. Moreover, out of 178 SCM positive cows, 97 (55%, 95% CI: 47.2 to 61.6) were diagnosed as weak positive (1+). In addition, 71 (40%, 95% CI: 33 to 47.2) and 10 (6%, 95% CI: 3 to 10.2) cows were identified as distinct (2+) and strong positive (3+) for SCM, respectively.
3.3. Frequency of different Staphylococcus spp. on SCM affected cows
The frequency calculation was performed based on the different PCR confirmed Staphylococcus spp. out of 356 cultured milk samples (Fig. 3a). Among Staphylococcus spp., S. chromogenes was the most frequent pathogen in SCM-affected cows (117/178; 65.7%, 95% CI: 58.5 to 72.3) at the cow level (Table 3). S. epidermidis, S. haemolyticus, and S. aureus were identified in 36 (20.2%, 95% CI: 15 to 26.8), 34 (19.1%, 95% CI: 14 to 25.5) and 28 (15.7%, 95% CI: 12 to 21.8) cows, respectively (Table 3). On the contrary, S. sciuri was detected in ten cows only (5.6%, 95% CI: 3 to 11) and none of the mastitic milk samples were positive for S. simulans.
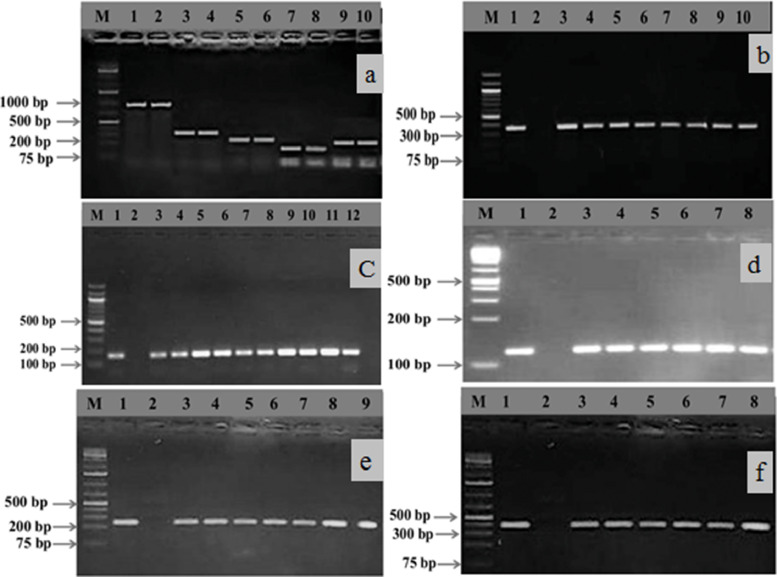
Fig. 3.
Gel electrophoresis image (3a) of PCR products of Staphylococcus spp. showing specific amplified bands on 1.5% agarose gel, where lanes 1 and 2 = S. aureus (894 bp), 3 and 4 = S. sciuri (306 bp), 5 and 6 = S. chromogenes (222 bp), 7 and 8 = S. epidermidis (130 bp), and 9 and 10 = S. haemolyticus (214 bp). Image-3b and 3c display the specific amplification of the nuc (359 bp) and mecA (162 bp) genes, respectively. In addition, images 3d, 3e, and 3f exhibit the amplification of virulence genes like sea (102 bp), pvl (255 bp), and tst (326 bp), respectively. In all gel electrophoresis systems, we used a 1 kb plus DNA marker except for images 3b and 3c, where we used 100 bp DNA markers for confirmation of amplification of specific genes. Moreover, lanes 1 and 2 were used as positive and negative controls for the 3b, 3c, 3d, 3e, and 3f images, respectively.
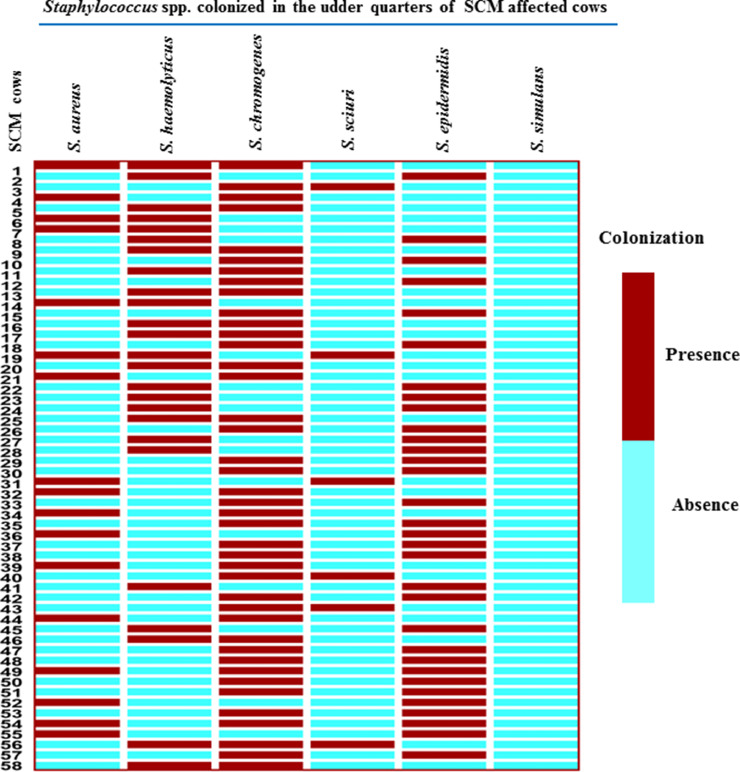
3.4. Mixed infection of diverse Staphylococcus spp. on udder of SCM affected cows
Across the 178 SCM-affected cows, 58 (32.6%) were co-infected by Staphylococcus spp. (Fig. 3). Among them, 4 cows harbored three different Staphylococcus species in same udder quarters, while 54 SCM- cows showed quarters with various two Staphylococcus species combinations (Fig. 3).
3.5. Antimicrobial use on the dairy herds for SCM treatment
From the antimicrobial therapeutic history of dairy farms, 19 (63.3%) and 9 (30%) herds used gentamicin and streptomycin (aminoglycosides) respectively for treatment of SCM. Furthermore, 15 (50%), 14 (47%), and 9 (30%) farms used β-lactam antibiotics such as ceftriaxone, amoxicillin, and penicillin (Fig. 2). Oxytetracycline 1 (3.3%) was the least used drug for the treatment of bovine SCM (Fig. 2).
3.6. Antimicrobial resistance profiles of Staphylococcus spp
Antimicrobial susceptibility profiles of Staphylococcus spp. are shown in Table-4. Overall, S. aureus isolates exhibited the highest resistance levels against ampicillin (82. 1%) and amoxycillin/clavulanic acid (75%) (Table 4). In addition, most of the S. chromogenes isolates were susceptible to majority of the tested antimicrobials, and the highest resistance levels were observed against cefepime, tetracycline, which were 95.7% and 54.7%, respectively. On the other hand, all S. haemolyticus were susceptible to ampicillin, amoxycillin/clavulanic acid, imipenem, and gentamicin (Table-4). Cefepime (97.2%), erythromicin (80.6%) and cefoxitin (77.8%) exhibit resistant against S. epidermidis, while 70% of S. sciuri isolates were susceptible to ceftriaxone and ciprofloxacin. All isolates were susceptible to imipenem (Table 4).
Table 4.
Antimicrobial sensitivity profiles of different Staphylococcus spp. causing SCM in lactating cows.
| Organism | Antimicrobial susceptibility | AM | AMC | CRO | FOX | FEP | IMP | CIP | E | CN | SXT | TE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus (28) | Sensitive (%) | 17.9 | 25 | 67.9 | 71.4 | 46.4 | 100 | 64.3 | 39.3 | 42.9 | 35.7 | 53.6 |
| Resistant (%) | 82.1 | 75 | 32.1 | 28.6 | 53.6 | 0 | 35.7 | 60.7 | 57.1 | 64.3 | 46.4 | |
| S. chromogenes (117) | Sensitive (%) | 58.1 | 71.8 | 63.3 | 73.5 | 4.3 | 100 | 67.5 | 66.7 | 76.1 | 74.4 | 45.3 |
| Resistant (%) | 41.9 | 28.2 | 36.8 | 26.5 | 95.7 | 0 | 32.5 | 33.3 | 23.9 | 25.6 | 54.7 | |
| S. haemolyticus (34) | Sensitive (%) | 100 | 100 | 64.7 | 73.5 | 5.9 | 100 | 91.2 | 61.8 | 100 | 76.5 | 88.2 |
| Resistant (%) | 0 | 0 | 35.3 | 26.5 | 94.1 | 0 | 8.8 | 38.2 | 0 | 26 | 11.8 | |
| S. epidermidis (36) | Sensitive (%) | 36.1 | 80.6 | 33.3 | 22.2 | 2.8 | 100 | 94.4 | 19.4 | 86.1 | 36.1 | 33.3 |
| Resistant (%) | 63.9 | 19.4 | 66.7 | 77.8 | 97.2 | 0 | 5.6 | 80.6 | 13.9 | 63.9 | 66.7 | |
| S. sciuri (10) | Sensitive (%) | 20 | 30 | 70 | 60 | 40 | 100 | 70 | 30 | 60 | 30 | 40 |
| Resistant (%) | 80 | 70 | 30 | 40 | 60 | 0 | 30 | 70 | 40 | 70 | 60 |
AM = ampicillin, AMC = amoxycillin/clavulanic acid, CRO = ceftriaxone, FOX = cefoxitin, FEP = cefepime, IPM = imipenem, CIP = ciprofloxacin, E = erythromicin, CN = gentamicin, SXT = trimethoprim/sulfamethoxazole and TE = tetracycline.
3.7. Detection rate of MDR in Staphylococcus spp
MDR isolates were identified among all five Staphylococcus spp. Among all MDR isolates, 19/28 (67.9%) S. aureus and 68/117 (58.1%) S. chromogenes showed resistance up to seven classes of antimicrobials. A lower frequency of MDR isolates was detected among S. epidermidis (16/ 36; 44.4%), S. haemolyticus (12/34; 35.3%) and S. sciuri (3/10; 30%).
3.8. Distribution of resistance genes among diverse Staphylococcus spp
The mecA gene conferring methicillin resistance was screened in all cefoxitin resistant staphylococci isolates. It was detected in 9 (32.1%) S. aureus (Fig. 3c) that were classified as methicillin-resistant S. aureus (MRSA). Isolates belonging to S. chromogenes 7 (6%), S. epidermidis 2 (5.6%), and S. sciuri 1 (10%), also carried the mecA gene (Table 5), and therefore were designated as MRSC, MRSE, and MRSS.
Table 5.
Distribution of antimicrobial resistance and virulence genes among different Staphylococcus spp. isolates from SCM affected lactating cows.
| Organism | Gene | Number of isolates | Detection rate (%) |
|---|---|---|---|
| S. aureus (28) | mecA | 9 | 32.1 |
| nuc | 13 | 46.4 | |
| sea | 5 | 17.9 | |
| seb | 2 | 7.1 | |
| sec | 0 | 0 | |
| pvl | 3 | 10.7 | |
| eta | 2 | 7.1 | |
| etb | 0 | 0 | |
| tst | 4 | 14.3 | |
| S. chromogenes (117) | mecA | 7 | 6 |
| sea | 2 | 1.7 | |
| S. haemolyticus (34) | pvl | 6 | 17.7 |
| eta | 2 | 5.9 | |
| tst | 3 | 8.8 | |
| S. epidermidis (36) | mecA | 2 | 5.6 |
| S. sciuri (10) | mecA | 1 | 10 |
| pvl | 2 | 20 |
3.9. Distribution of virulence genes among diverse Staphylococcus spp
The enterotoxin-encoding genes sea was detected in S. aureus (17.9%) and S. chromogenes (1.7%), (Fig. 3d), while the seb gene was only detected for S. aureus (7.1%) (Table 5). The cytotoxin (pvl) gene was detected in S. sciuri (20%) and S. haemolyticus (17.7%) (Fig. 3e). The toxic shock syndrome toxin-encoding gene (tst) was detected in S. aureus (14.3%) (Fig. 3f).The exfoliative toxins-encoding gene (eta) was detected in S. aureus (7.1%) and S. haemolyticus (5.9%). None of the isolates carried sec or etb genes (Table 5).
3.10. Risk factors associated with Staphylococcus spp. carriage in SCM- affected lactating cows
The univariable logistic regression analysis identified five potential different risk factors (p ≤ 0.20) associated with the carriage of S. aureus and S. chromogenes in SCM affected cows (Supplementary Tables 1 and 2). In the subsequent multivariable logistic regression analysis, two different factors were found to be significant risk factors associated with the carriage of both S. aureus and S. chromogenes. The significantly associated risk factors were "Old aged" (OR: 3.5, 95% CI: 1–12.4, p ≤ 0.05) and "Front left side quarter" (OR: 2.5, 95% CI: 1.1–5.7, p ≤ 0.03) for S. aureus (Table 6). In case of S. chromogenes, two factors, "Early stage of lactation" (OR: 3.4, 95% CI: 1.2–9.7, p ≤ 0.02) and "Hind left side quarter" (OR: 2.1, 95% CI: 1.1–4.0, p ≤ 0.02), were significantly associated with SCM (Table 6). Six potential risk factors (p ≤ 0.20) were particularly associated with the presence of S. haemolyticus (supplementary Table 3). However, "Firm udder condition" (OR: 4.2, 95% CI: 1.2–14.6, p ≤ 0.02) and "Front right side quarter" (OR: 2.8, 95% CI: 1.1–7, p ≤ 0.03) were significantly associated with presence of S. haemolyticus in SCM affected cows. A total of four and two different factors (p ≤ 0.20) were initially associated with S. epidermidis and S. sciuri, respectively (supplementary Table 4 and 5). In multivariable logistic regression model, none of the factors were significantly associated with SCM in cows. Fig. 4
Table 6.
Multivariable logistic regression model for assessing the risk factors independently associated with the S. aureus, S. haemolyticus, S. chromogenes and methicillin-resistant (MR) Staphylococcus spp. from SCM affected lactating cows.
| Outcome variable | Explanatory variable | Description | OR (95% CI) | p-value |
|---|---|---|---|---|
| S. aureus | Age | Adult | 1 | Reference |
| Young adult | 1.8 (0.7–4.9) | 0.21 | ||
| Old | 3.5 (1–12.4) | 0.05* | ||
| Front left side quarter | Yes | 2.5 (1.1–5.7) | 0.03* | |
| No | 1 | Reference | ||
| S. haemolyticus | Udder condition | Firm | 4.2 (1.2–14.6) | 0.02* |
| Normal | 1 | Reference | ||
| Front right side quarter | Yes | 2.8 (1.1–7) | 0.03* | |
| No | 1 | Reference | ||
| S. chromogenes | Stage of lactation | Early | 3.4 (1.2–9.7) | 0.02* |
| Middle | 1.1 (0.5–2.4) | 0.78 | ||
| Last | 1 | Reference | ||
| Hind left side quarter | Yes | 2.1 (1.1–4.0) | 0.02* | |
| No | 1 | Reference | ||
| MR Staphylococcus spp. | Use of antimicrobials | Yes | 10.4 (3.4–32.1) | 0.00* |
| No | 1 | Reference | ||
| History of previous clinical mastitis | Yes | 4.9 (1.2–19.7) | 0.02* | |
| No | 1 | Reference |
Abbreviation: OR= odds ratio; CI=confidence interval; significance *(≤0.05).
Fig. 4.
The heat map depicts the mixed colonization of multiple Staphylococcus spp. on udder quarter of SCM affected cows.
3.11. Risk factors associated with carriage of methicillin-resistant Staphylococcus spp. in SCM affected lactating cows
Twelve out of the thirty factors were primarily identified in the univariable analysis as the potential risk factors for the carriage of methicillin-resistant Staphylococcus spp. in SCM affected lactating cows (supplementary Table 6). Two variables “Use of antimicrobials” (OR: 10.4, 95% CI: 3.4–32.1, p ≤ 0.00) and “History of previous clinical mastitis” (OR: 4.9, 95% CI: 1.2–19.7, p ≤ 0.02) were found significant in the final model (Table 6).
4. Discussion
Staphylococcal mastitis is an ongoing challenge for successful management and operation of dairy industry throughout the world, including Bangladesh. As per our knowledge, this is the first comprehensive molecular study for identification of diverse CoNS species which enabled us to representing the species-specific prevalence of SCM in affected cows in Bangladesh context. This study estimated overall prevalence of SCM on different dairy farms as 62.7% at cow level and 31.3% at quarter level. This result is higher than the previous findings of, Ramírez et al., 2014; Rana et al., 2022a and Zeryehun & Abera, 2017 in Colombia, Bangladesh, and Eastern Ethiopia, which were 37.2%, 38.4%, and 51.8%, respectively. The variation in prevalence may have occurred due to individual cow and farm factors like breed, age, stages of lactation, parity, udder immunity, farm ecology, management, hygiene, and milking practices in dairy farms (Rana et al., 2022a; Ashraf et al., 2018).
Both CoPS and CoNS are the most prevalent pathogens involved in bovine SCM (Taponen & Pyörälä, 2009; Pyörälä & Taponen, 2009; Schabauer et al., 2018; Rana et al., 2022a), even if the species-specific bacterial prevalence and distribution mightily vary among different dairy herds. The isolation rate of S. aureus in the current study is in line with previous studies by Kasa et al., 2020, and Rana et al., 2022a. However, the secretion of the coagulase enzyme is one of the key virulence strategies of S. aureus that encourages the initiation of mammary gland damage and the pathogenesis of bovine mastitis (Bonar et al., 2018). Several studies have described that the presence of S. aureus is significantly involved in clinical and SCM and causes pronounced inflammatory responses in cow udder (De Buck et al., 2021).
In addition, we revealed that S. chromogenes (65.73%) S. epidermidis (20.2%) and S. haemolyticus (19.1%) were the most prevalent CoNS species isolated from SCM affected cows. The isolation rate of these CoNS species was agreed upon in the previous study of the USA, Canada, and Argentina, which was reported by Hasan et al., 2022; Jenkins et al., 2019; De Visscher et al., 2016; Raspanti et al., 2016. As bovine skin and farm bedding materials are the fundamental sources of S. chromogenes, S. epidermidis, and S. haemolyticus (Taponen & Pyörälä, 2009; Schabauer et al., 2018) such reports give evidence to the origin of bovine mastitis. The internal udder environment or intrinsic factors of the mammary gland also favor the colonization of diverse groups of organisms, especially bacterial species (Rana et al., 2020a). Moreover, the CoNS species, especially S. chromogenes, S. haemolyticus, and S. sciuri, are responsible for persistent SCM and markedly increased somatic cell count (SCC) in lactating cows without visible clinical signs (De Buck et al., 2021; Simojoki et al., 2009). These observations were kept up in our study, and the majority of CoNS isolates have been recovered from CMT-positive milk from cows with usual udder conditions. As a result, the pathogenic potentialities of CNS species, ranging from commensal organisms to actual mastitis pathogens, remain uncertain (Isaac et al., 2017; Pyörälä & Taponen, 2009). The early detection of bovine SCM and its causal agent is crucial for making treatment and management decisions (Schabauer et al., 2018). Moreover, the exact identification and recording of bacterial species involved in mastitis is inevitable to ameliorate dairy herd health.
We identified 32.6% of SCM cows carrying mixed Staphylococcus spp. infection with multiple combinations, which was strongly supported by Ethiopian and Estonian dairy herd findings and described by Kasa et al., 2020 and Kalmus et al., 2011. The possible reasons for this mixed colonization of bacterial species in udder might be occurred due to coexistence of mastitis pathogens together in farm environments (Kalmus et al., 2011). Additionally, the high dominance of mixed Staphylococcus spp. infection might occur due to fugitive transmission of these pathogens during hand milking of dairy cows.
In this study, S. aureus isolates displayed a high level of resistance against ampicillin (82.1%), amoxicillin and clavulanic acid (75%), which were the frequently used antimicrobials in dairy farms. This high level of resistance is in agreed with other previous reports by Priyantha et al., 2021; Rana et al., 2022a. Furthermore, the results showed that a significant number of CoNS species were resistant to various β-lactam antimicrobials. β-lactams (ampicillin, amoxicillin, ceftriaxone and cefoxitin) are the most used antimicrobials in the majority of dairy herds for the treatment of bovine clinical and SCM. This could be one of the major causes of β -lactam resistance in S. aureus and the CoNS due to repeated exposure and selective antimicrobial pressure (Christaki et al., 2020; Rana et al., 2022a). Furthermore, the production of β-lactamase has been reported as a resistance mechanism for Staphylococcus spp., which is resistant to a variety of antimicrobials (De Buck et al., 2021). In addition, the current study, all the Staphylococcus spp. was highly susceptible to imipenem (100%). This is highly expected as imipenem has not been used in the field of veterinary practices in the context of Bangladesh, and this antibiotic is totally reserved for human health.
In the present study, all of the S. aureus and CoNS isolates displayed MDR (i.e. resistance to ≥ 3 antimicrobial classes), and these findings are supported the previous several studies conducted in dairy farms for mastitic cows (Rana et al., 2022a, Dabele et al., 2021). Overexposure and repetitive use of similar groups of antimicrobials and antimicrobials having similar mode of action may induce the development of MDR (Rana et al., 2022b, Das et al., 2022). On the other hand, bacterial intrinsic factors, biofilm formation, and the encoding of multiple resistance genes were also reported for MDR expression (Christaki et al., 2020).
Our study detected 9 (32.1%) S. aureus and certain percentages of CoNS species that encode the mecA gene that displayed resistance against cefoxitin, ampicillin, amoxicillin/clavulanic acid, cefepime, and ceftriaxone antibiotics. However, due to the encoding of the mecA gene, which modifies penicillin binding protein (PBP) 2a on the bacterial cell wall, these broad-spectrum -lactam antibiotics become resistant to staphylococci. And these methicillin-resistant phenomena have already been reported in various farm animals (Monecke et al., 2013; Rana et al., 2020a; Schabauer et al., 2018). Specifically, the methicillin-resistant S. chromogenes, S. epidermidis, and S. sciuri were already reported in different countries like China (73.2%), Switzerland (62%), the Netherlands (14.1%), United Kingdom (4.1%), the USA (2.4%) and Canada (0.9%) (Schnitt et al., 2021) but these findings are noble for Bangladesh. Moreover, it is a matter of great concern that methicillin-resistant CoNS species are pivotal reservoirs of AMR genes, which create a clinical and management challenge for successful control of bovine mastitis in dairy farms.
We identified both S. aureus and CoNS species harbored a multiple number of virulence genes, including enterotoxins (sea, seb and sec), cytotoxin (pvl), exfoliative toxins (eta, etb) and a toxic shock syndrome toxin (tst). Interestingly, 17.9%, 14.3%, and 10.7% of S. aureus carried sea, tst, and pvl genes, respectively. These virulence determinants aligned with the previous reports of Mello et al., 2016, and Zhou et al., 2017. Moreover, alarming percentages of coagulase negative mastitis isolates like S. haemolyticus, S. chromogenes, and S. sciuri carried sea, pvl, eta and tst genes, and these findings are supported by the previous reports of González-Martín et al., 2020, Mahato et al., 2017. The presence of virulence genes and their known pathogenic potentialities are directly associated with bovine mastitis in farm environments. In addition, the information and evidence regarding the virulence factors of CoNS species and their gene regulations are much sparser in bovine mastitis.
The study also revealed that a significant statistical association between the prevalence of SCM and different risk factors like, age of cow, udder condition, stage of lactation, and position of the udder quarter. This finding is consistent with the findings of Lakew et al., 2019 who reported that the risk of SCM increases significantly with the advancement of lactation stage as well as the age of lactating cows. According to Radostitis et al. (2007) and Lakew et al. (2019) an older cow with more relaxed udder attachments (pendulous udder) and frequent floor touch of the cow's quarter allows contagious organisms to enter the cow's udder. In addition, previous history of clinical mastitis with prolonged mild infection dramatically increased SCC (McDougall et al., 2021) and initiated fibrosis of mammary gland tissue, changed the consistency of the udder to a firm texture, and, established SCM. Lactating cows' udder defense mechanism reduces the severity of the clinical nature of bovine mastitis (Lakew et al., 2019), which eventually conceals the clinical signs and cows perpetuate the disease's sub-clinical form.
We identified use of antimicrobials and history of previous clinical mastitis as the risk factors associated with higher prevalence of methicillin resistant staphylococci in SCM cows and the similar factors were obtained by Rana et al., 2022a and Mahato et al., 2017.Concurrent use of antimicrobials for the clinical management of mastitis case might be a potent reason behind this association. In addition, antimicrobial therapy and its selection pressure is a well-documented factor for methicillin resistance in staphylococci (Gnanamani et al., 2017). However, the imprudent use of antimicrobials in the treatment of methicillin resistance staphylococci should be unsanctioned and the absolute mastitis treatment regimen should be based on the antimicrobial susceptibility testing.
However, due to resource constraints, we were unable to perform detailed genotypic characterization of S. aureus and CoNS species involved in bovine SCM in the current study. Moreover, it would be an appropriate task to explore the sequence types of all staphylococcal isolates that are circulating in bovine mastitis in the context of Bangladesh. Future research should overcome these limitations.
5. Conclusion
The SCM in the dairy herds was highly prevalent, and the exalted burden of CoNS, markedly S. chromogenes, S. epidermidis, and S. haemolyticus were associated with the SCM. The alarming percentages of isolates found were not only MDR, but a significant number encoded the mecA gene, which is noted for methicillin resistance. In addition to S. aureus, a particular number of CoNS species harbor multiple virulence genes and assert their pathogenic potentialities. Moreover, old aged, early stage of lactation, and firm udder condition were identified as potential risk factors that amalgamated with SCM, while previous use of antimicrobials and history of previous clinical mastitis seem to be positively related with the prevalence of methicillin resistant Staphylococcus spp. in SCM cows. Therefore, intensive monitoring of the MDR pathogen in SCM is pivotal since the transmission of Staphylococcus spp. is dynamic and involves farm animals, workers, consumers, and veterinarians.
Ethical approval
The field task (SCM screening and sample collection) and entire laboratory procedures of the current study were performed with the permission of Chattogram Veterinary and Animal Sciences University's (CVASU) Ethical Approval Committee [Approval no. CVASU/Dir (R&E) EC/2020/165 (1)].
CRediT authorship contribution statement
Md Abul Fazal: Conceptualization, Data curation, Methodology, Project administration, Resources, Validation, Formal analysis, Visualization, Writing – review & editing. Eaftekhar Ahmed Rana: Conceptualization, Funding acquisition, Data curation, Investigation, Methodology, Project administration, Resources, Validation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. Sazeda Akter: Resources, Validation, Visualization, Writing – review & editing. Mohammad Abdul Alim: Visualization, Validation, Writing – review & editing. Himel Barua: Project administration, Supervision, Writing – review & editing. Abdul Ahad: Conceptualization, Funding acquisition, Project administration, Supervision.
Declaration of Competing Interest
We, the authors of this paper have no financial interest to declare or personal relations that could have influenced the findings presented in this manuscript.
Acknowledgments
Funding
The current study was financially supported by the Bangladesh Bureau of Educational Information and Statistics (BANBEIS), Ministry of Education, [grant numbers: LS20191030], The Ministry of Science and Technology [grant numbers: Reg.22 & 33], and Director Research and Extension, CVASU [grant numbers: SL-1], The People's Republic of Bangladesh.
Acknowledgments
The authors would like to thank the all dairy farm owners of Cumilla district for supporting the collection of the milk samples from SCM affected cows. We also would like to express our gratitude to the Department of Microbiology and Veterinary Public Health, CVASU, Chattogram, Bangladesh for their ongoing laboratory assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.vas.2023.100297.
Appendix. Supplementary materials
References
- Abebe R., Hatiya H., Abera M., Megersa B., Asmare K. Bovine mastitis: Prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC veterinary research. 2016;12(1):1–11. doi: 10.1186/s12917-016-0905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf A., Imran M., Yung-Fu C. Antimicrobial resistance of Escherichia coli isolates from mastitic milk and its possible relationship with resistance and virulence genes. Pakistan Journal of Zoology. 2018;50(4) doi: 10.17582/journal.pjz/2018.50.4.1435.1441. [DOI] [Google Scholar]
- Capurro A., Artursson K., Waller K.P., Bengtsson B., Ericsson-Unnerstad H., Aspán A. Comparison of a commercialized phenotyping system, antimicrobial susceptibility testing, and tuf gene sequence-based genotyping for species-level identification of coagulase-negative staphylococci isolated from cases of bovine mastitis. Veterinary microbiology. 2009;134(3–4):327–333. doi: 10.1016/j.vetmic.2008.08.028. [DOI] [PubMed] [Google Scholar]
- Cervinkova D., Vlkova H., Borodacova I., Makovcova J., Babak V., Lorencova A., et al. Prevalence of mastitis pathogens in milk from clinically healthy cows. Veterinarni medicina. 2013;58(11):567–575. [Google Scholar]
- Christaki E., Marcou M., Tofarides A. Antimicrobial resistance in bacteria: Mechanisms, evolution, and persistence. Journal of molecular evolution. 2020;88(1):26–40. doi: 10.1007/s00239-019-09914-3. [DOI] [PubMed] [Google Scholar]
- CLSI . Clinical and Laboratory Standards Institute; Wayne (PA): 2018. Performance standards for antimicrobial disk susceptibility tests. 13th ed. clsi standard M02. [Google Scholar]
- Dabele D.T., Borena B.M., Snr P.A., Gebremedhin E.Z., Marami L.M. Prevalence and risk factors of mastitis and isolation, identification and antibiogram of staphylococcus species from mastitis positive zebu cows in toke kutaye, cheliya, and dendi districts, west shewa zone, Oromia, Ethiopia. Infection and Drug Resistance. 2021;14:987. doi: 10.2147/IDR.S295257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T., Rana E.A., Dutta A., Bostami M.B., Rahman M., Deb P., et al. Antimicrobial resistance profiling and burden of resistance genes in zoonotic Salmonella isolated from broiler chicken. Veterinary Medicine and Science. 2022;8(1):237–244. doi: 10.1002/vms3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck J., Ha V., Naushad S., Nobrega D.B., Luby C., Middleton J.R., et al. Non-aureus Staphylococci and bovine udder health: Current understanding and knowledge gaps. Frontiers in veterinary science. 2021;8 doi: 10.3389/fvets.2021.658031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Visscher A., Piepers S., Haesebrouck F., De Vliegher S. Intramammary infection with coagulase-negative staphylococci at parturition: Species-specific prevalence, risk factors, and effect on udder health. Journal of Dairy Science. 2016;99(8):6457–6469. doi: 10.3168/jds.2015-10458. [DOI] [PubMed] [Google Scholar]
- De Visscher A., Piepers S., Haesebrouck F., Supré K., De Vliegher S. Coagulase-negative Staphylococcus species in bulk milk: Prevalence, distribution, and associated subgroup-and species-specific risk factors. Journal of Dairy Science. 2017;100(1):629–642. doi: 10.3168/jds.2016-11476. [DOI] [PubMed] [Google Scholar]
- Dhaouadi S., Soufi L., Campanile F., Dhaouadi F., Sociale M., Lazzaro L., et al. Prevalence of meticillin-resistant and-susceptible coagulase-negative staphylococci with the first detection of the mecC gene among cows, humans and manure in Tunisia. International journal of antimicrobial agents. 2020;55(1) doi: 10.1016/j.ijantimicag.2019.10.007. [DOI] [PubMed] [Google Scholar]
- Frey Y., Rodriguez J.P., Thomann A., Schwendener S., Perreten V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. Journal of dairy science. 2013;96(4):2247–2257. doi: 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- Gnanamani A., Hariharan P., Paul-Satyaseela M. Staphylococcus aureus: Overview of bacteriology, clinical diseases, epidemiology, antibiotic resistance and therapeutic approach. Frontiers in Staphylococcus aureus. 2017;4(28):10–5772. [Google Scholar]
- González-Martín M., Corbera J.A., Suárez-Bonnet A., Tejedor-Junco M.T. Virulence factors in coagulase-positive staphylococci of veterinary interest other than Staphylococcus aureus. Veterinary Quarterly. 2020;40(1):118–131. doi: 10.1080/01652176.2020.1748253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.S., Kober A.K.M.H., Rana E.A., Bari M.S. Association of udder lesions with subclinical mastitis in dairy cows of Chattogram, Bangladesh. Adv. Anim. Vet. Sci. 2022;10(2):226–235. [Google Scholar]
- Hoque M.N., Das Z.C., Rahman A.N.M.A., Haider M.G., Islam M.A. Molecular characterization of Staphylococcus aureus strains in bovine mastitis milk in Bangladesh. International journal of veterinary science and medicine. 2018;6(1):53–60. doi: 10.1016/j.ijvsm.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinzadeh S., Saei H.D. Staphylococcal species associated with bovine mastitis in the North West of Iran: Emerging of coagulase-negative staphylococci. International Journal of Veterinary Science and Medicine. 2014;2(1):27–34. doi: 10.1016/j.ijvsm.2014.02.001. [DOI] [Google Scholar]
- Isaac P., Bohl L.P., Breser M.L., Orellano M.S., Conesa A., Ferrero M.A., et al. Commensal coagulase-negative Staphylococcus from the udder of healthy cows inhibits biofilm formation of mastitis-related pathogens. Veterinary microbiology. 2017;207:259–266. doi: 10.1016/j.vetmic.2017.05.025. [DOI] [PubMed] [Google Scholar]
- Jenkins S.N., Okello E., Rossitto P.V., Lehenbauer T.W., Champagne J., Penedo M.C., Arruda A.G., Godden S., Rapnicki P., Gorden P.J., Timms L.L., Aly S.S. Molecular epidemiology of coagulase-negative Staphylococcus species isolated at different lactation stages from dairy cattle in the United States. PeerJ. 2019;7:e6749. doi: 10.7717/peerj.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmus P., Aasmäe B., Kärssin A., Orro T., Kask K. Udder pathogens and their resistance to antimicrobial agents in dairy cows in Estonia. Acta Veterinaria Scandinavica. 2011;53:1–7. doi: 10.1186/1751-0147-53-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasa G., Tegegne B., Tadesse B. Isolation and identification of major pathogenic bacteria from clinical mastitic cows in Asella Town, Ethiopia. Veterinary Medicine International. 2020;2020 doi: 10.1155/2020/6656755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakew B.T., Fayera T., Ali Y.M. Risk factors for bovine mastitis with the isolation and identification of Streptococcus agalactiae from farms in and around Haramaya district, eastern Ethiopia. Tropical animal health and production. 2019;51(6):1507–1513. doi: 10.1007/s11250-019-01838-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A.R., Stegger M., Sørum M. spa typing directly from a mecA, spa and pvl multiplex PCR assay—A cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clinical Microbiology and Infection. 2008;14(6):611–614. doi: 10.1111/j.1469-0691.2008.01995.x. [DOI] [PubMed] [Google Scholar]
- Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Mahato S., Mistry H.U., Chakraborty S., Sharma P., Saravanan R., Bhandari V. Identification of variable traits among the methicillin resistant and sensitive coagulase negative staphylococci in milk samples from mastitic cows in India. Frontiers in microbiology. 2017;8:1446. doi: 10.3389/fmicb.2017.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall S., Williamson J., Gohary K., Lacy-Hulbert J. Detecting intramammary infection at the end of lactation in dairy cows. Journal of Dairy Science. 2021;104(9):10232–10249. doi: 10.3168/jds.2020-20036. [DOI] [PubMed] [Google Scholar]
- Mello P.L., Moraes Riboli D.F., Pinheiro L., de Almeida Martins L., Vasconcelos Paiva Brito M.A., Ribeiro de Souza da Cunha M.D.L. Detection of enterotoxigenic potential and determination of clonal profile in Staphylococcus aureus and coagulase-negative staphylococci isolated from bovine subclinical mastitis in different Brazilian states. Toxins. 2016;8(4):104. doi: 10.3390/toxins8040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Gavier-Widen D., Mattsson R., Rangstrup-Christensen L., Lazaris A., Coleman D.C., et al. Detection of mecC-positive Staphylococcus aureus (CC130-MRSA-XI) in diseased European hedgehogs (Erinaceus europaeus) in Sweden. PloS one. 2013;8(6):e66166. doi: 10.1371/journal.pone.0066166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Mastitis Council (NMC) 4th ed. National Mastitis Council; Verona, WI: 2004. Microbiological procedures for use in the diagnosis of bovine udder infection and determination of milk quality. [Google Scholar]
- Piessens V., Van Coillie E., Verbist B., Supré K., Braem G., Van Nuffel A., et al. Distribution of coagulase-negative Staphylococcus species from milk and environment of dairy cows differs between herds. Journal of dairy science. 2011;94(6):2933–2944. doi: 10.3168/jds.2010-3956. [DOI] [PubMed] [Google Scholar]
- Priyantha M.A., Fernando P.S., De Alwis P.S. Emerging antimicrobial resistance in coagulase-positive Staphylococci and E. coli isolated from bovine clinical mastitis in Sri Lanka. Asian journal of animal and veterinary advances. 2021;28:29–35. [Google Scholar]
- Pyörälä S., Taponen S. Coagulase-negative staphylococci—Emerging mastitis pathogens. Veterinary microbiology. 2009;134(1–2):3–8. doi: 10.1016/j.vetmic.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Quinn P.J., Carter M.E., Markey B.K., Carter G.R. 2nd ed. Mosby publish ring; UK, London: 1998. Clinical veterinary microbiology. 1998. [Google Scholar]
- Radostits O.M., Gay C.C., Hinchcliff K.W., Constable P.D. Diseases associated with bacteria. Veterinary Medicine: A textbook of the diseases of cattle, horses, sheep, pigs, and goats. 2007;10:1007–1060. [Google Scholar]
- Ramírez N.F., Keefe G., Dohoo I., Sánchez J., Arroyave O., Cerón J., et al. Herd-and cow-level risk factors associated with subclinical mastitis in dairy farms from the High Plains of the northern Antioquia, Colombia. Journal of Dairy Science. 2014;97(7):4141–4150. doi: 10.3168/jds.2013-6815. [DOI] [PubMed] [Google Scholar]
- Rana E.A., Das T., Dutta A., Rahman M., Bostami M.B., Akter N., et al. Coagulase-positive methicillin-resistant Staphylococcus aureus circulating in clinical mastitic goats in Bangladesh. Veterinary World. 2020;13(7):1303. doi: 10.14202/vetworld.2020.1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana E.A., Fazal M.A., Alim M.A. Frequently used therapeutic antimicrobials and their resistance patterns on Staphylococcus aureus and Escherichia coli in mastitis affected lactating cows. International Journal of Veterinary Science and Medicine. 2022;10(1):1–10. doi: 10.1080/23144599.2022.2038494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana E.A., Islam M.Z., Das T., Dutta A., Ahad A., Biswas P.K., et al. Methicillin-resistant coagulase positive Staphylococcus aureus and Staphylococcus pseudintermedius circulating in dogs in Bangladesh. International Journal of Infectious Diseases. 2020;101:34–35. doi: 10.1016/ijid.2020.09.124. [DOI] [Google Scholar]
- Rana E.A., Islam M.Z., Das T., Dutta A., Ahad A., Biswas P.K., et al. Prevalence of coagulase-positive methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in dogs in Bangladesh. Veterinary medicine and science. 2022;8(2):498–508. doi: 10.1002/vms3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspanti C.G., Bonetto C.C., Vissio C., Pellegrino M.S., Reinoso E.B., Dieser S.A., et al. Prevalence and antibiotic susceptibility of coagulase-negative Staphylococcus species from bovine subclinical mastitis in dairy herds in the central region of Argentina. Revista Argentina de Microbiologia. 2016;48(1):50–56. doi: 10.1016/j.ram.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Tsubakishita S., Tanaka Y., Sakusabe A., Ohtsuka M., Hirotaki S., et al. Multiplex-PCR method for species identification of coagulase-positive staphylococci. Journal of clinical microbiology. 2010;48(3):765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabauer A., Pinior B., Gruber C.M., Firth C.L., Käsbohrer A., Wagner M., et al. The relationship between clinical signs and microbiological species, spa type, and antimicrobial resistance in bovine mastitis cases in Austria. Veterinary microbiology. 2018;227:52–60. doi: 10.1016/j.vetmic.2018.10.024. [DOI] [PubMed] [Google Scholar]
- Schnitt A., Lienen T., Wichmann-Schauer H., Tenhagen B.A. The occurrence of methicillin-resistant non-aureus staphylococci in samples from cows, young stock, and the environment on German dairy farms. Journal of Dairy Science. 2021;104(4):4604–4614. doi: 10.3168/jds.2020-19704. [DOI] [PubMed] [Google Scholar]
- Shome B.R., Das Mitra S., Bhuvana M., Krithiga N., Velu D., Shome R., et al. Multiplex PCR assay for species identification of bovine mastitis pathogens. Journal of applied microbiology. 2011;111(6):1349–1356. doi: 10.1111/j.1365-2672.2011.05169.x. [DOI] [PubMed] [Google Scholar]
- Simojoki H., Orro T., Taponen S., Pyörälä S. Host response in bovine mastitis experimentally induced with Staphylococcus chromogenes. Veterinary Microbiology. 2009;134(1–2):95–99. doi: 10.1016/j.vetmic.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Ster C., Allard M., Boulanger S., Boulet M.L., Mulhbacher J., Lafontaine D.A., et al. Experimental treatment of Staphylococcus aureus bovine intramammary infection using a guanine riboswitch ligand analog. Journal of dairy science. 2013;96(2):1000–1008. doi: 10.3168/jds.2012-5890. [DOI] [PubMed] [Google Scholar]
- Taponen S., Koort J., Björkroth J., Saloniemi H., Pyörälä S. Bovine intramammary infections caused by coagulase-negative staphylococci may persist throughout lactation according to amplified fragment length polymorphism-based analysis. Journal of Dairy Science. 2007;90(7):3301–3307. doi: 10.3168/jds.2006-860. [DOI] [PubMed] [Google Scholar]
- Taponen S., Pyörälä S. Coagulase-negative staphylococci as cause of bovine mastitis—Not so different from Staphylococcus aureus? Veterinary microbiology. 2009;134(1–2):29–36. doi: 10.1016/j.vetmic.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Zadoks R.N., Watts J.L. Species identification of coagulase-negative staphylococci: Genotyping is superior to phenotyping. Veterinary microbiology. 2009;134(1–2):20–28. doi: 10.1016/j.vetmic.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Zeryehun T., Abera G. Prevalence and bacterial isolates of mastitis in dairy farms in selected districts of Eastern Harrarghe zone, Eastern Ethiopia. Journal of veterinary medicine. 2017;2017 doi: 10.1155/2017/6498618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Zhang M., Li H., Yang H., Li X., Song X., Wang Z. Prevalence and molecular characterization of Staphylococcus aureus isolated from goats in Chongqing, China. BMC Veterinary Research. 2017;13:1–8. doi: 10.1186/s12917-017-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.