Abstract
The plant-specific RNA Polymerase V (Pol V) plays a key role in gene silencing, but its role in repair of double stranded DNA breaks is unclear. Excision of the transposable element mPing creates double stranded breaks that are repaired by NHEJ. We measured mPing excision site repair in multiple DNA methylation mutants including pol V using an mPing : GFP reporter. Two independent mutant alleles of pol V showed less GFP expression, indicating that the Pol V protein plays a role in excision site repair. Sequence analysis of the pol V excision sites indicated an elevated rate of large deletions consistent with less efficient repair. These results clarify the role of Pol V, but not other RNA-directed DNA methylation proteins (Pol IV) or maintenance DNA methylation pathways ( MET1 ), in the repair of double-strand DNA breaks.
Figure 1. mPing Excision Site Analysis .
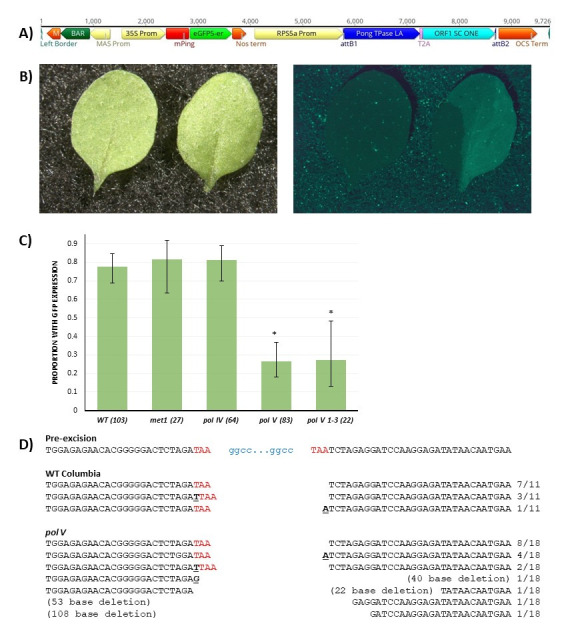
A. Diagram of pEarleyGate100RMoA mPing T-DNA that was used in Arabidopsis transformation. Promoters are shown in yellow; terminators are shown in orange. B. White light (left) and blue light (right) images of WT leaves with (right leaf) and without (left leaf) GFP expression induced by mPing excision. C. Graph of the proportion of Arabidopsis plants with GFP expression for each line tested. Axis labels are genotype (number of plants). Error bars indicate the standard deviation among each sample. * Indicates significantly different from WT, met1 , and pol IV . D. Sequencing results for cloned excision site PCR products from WT and pol V plants. Pre-excision shows the location of the mPing element (blue) before transposition. The target site duplication bases are shown in red; insertions are underlined. Numbers on the right indicate the number of each sequence obtained/total number of sequences for that genotype.
Description
Transposable elements are mobile sequences of DNA that are found in virtually all eukaryotic organisms and make up a sizeable percentage of the genome in some species, especially plants (Fedoroff 2012, Stitzer et al. 2021) . DNA transposable elements are excised and inserted by transposase proteins that cleave at the end of the elements, producing a double stranded DNA break (DSB) that must be repaired (Yuan and Wessler 2011, Hickman and Dyda 2016) . These DSBs are often repaired by the non-homologous end-joining (NHEJ) pathway, which often results in mutations at the excision site (Plasterk et al. 1999) . Previous studies indicate that small RNAs play a role in DSB repair in eukaryotes by potentially recruiting DNA repair enzymes or directly functioning as a repair template (Yang and Qi 2015, Bader et al. 2020, Franklin and Steele 2021) . Plants generate noncoding RNAs as part of the RNA-directed DNA Methylation (RdDM) pathway designed to silence repetitive sequences, especially transposable elements (Matzke and Mosher 2014) . Small interfering RNAs are produced by RNA Polymerase IV (Pol IV) and the plant specific RNA Polymerase V (Pol V) generates nascent scaffolding transcripts (Herr et al. 2005, Onodera et al. 2005, Lahmy et al. 2009) . Arabidopsis plants missing either of these RNA polymerases or the Dicer-like proteins that process the resulting RNAs were shown to exhibit reduced ability to repair a 35S:GU-US reporter gene cleaved between the repeated region with a restriction enzyme (Wei et al. 2012) . The repair of this reporter by homologous recombination was associated with the production of diRNAs (double-stranded break induced small RNAs) with homology to the sequences flanking the DSB (Wei et al. 2012) . A similar study showed that cleavage of the GU-US reporter by CRISPR also resulted in homologous 21nt small RNAs when it was highly expressed, but they were not detected from cleavage of a promoter-less GU-US construct or endogenous sequences (Miki et al. 2017) . In contradiction to the prior study, they didn’t detect a decrease in homologous recombination-mediated repair of the GU-US reporter in Dicer-like mutants (Miki et al. 2017) . Thus, there are still many unanswered questions about the role of RNA in DSB repair in plants.
mPing is an active miniature inverted repeat transposable element from rice (Jiang et al. 2003, Kikuchi et al. 2003, Nakazaki et al. 2003) that can be induced to transpose in Arabidopsis by expression of the ORF1 and Transposase (TPase) proteins from the related Ping or Pong elements (Yang et al. 2007) . Transposition frequency is measured using a mPing :GFP reporter ( Fig. 1A ) in which green fluorescent protein (GFP) is only expressed after mPing excision and the repair of the excision site (Yang et al. 2007) . Unlike the 35S:GU-US experiments described above, excision of mPing leaves behind compatible ends that are usually repaired by NHEJ, resulting in precise restoration to the state that would have been present before mPing insertion (Yang et al. 2007, Gilbert et al. 2015) . The purpose of these experiments was to test if the Pol IV ( AT1G63020 ) and Pol V ( AT2G40030 ) proteins are required for mPing excision site repair in Arabidopsis . As mutations of these genes result in alteration of DNA methylation, we also tested DNA repair in plants missing the main maintenance of DNA methylation gene, MET1 ( AT5G49160 ).
GFP expression was measured in met1, pol IV, pol V, pol V 1-3 , and Columbia (WT) Arabidopsis transformed with the pEarleyGate100RMOA mPing T-DNA ( Fig. 1A ). The pol V line used is a null allele while the pol V 1-3 line is a single amino acid mutant that produces Pol V protein that is defective for RNA synthesis (Kanno et al. 2005, Lahmy et al. 2009) . We observed that ~80% of WT, met1, and pol IV had GFP expression, while the pol V and pol V 1-3 lines had significantly lower percentage of plants with GFP expression [*p≤0.000137] ( Fig. 1B ). This indicates that in the absence of functional Pol V, either mPing excision frequency is reduced or the excision sites are not being repaired precisely by the Pol V mutants. mPing excision was detected in WT and pol V mutants using non-quantitative PCR with primers that flank the mPing excision site, suggesting that Pol V is not required for transposition. Sequence analysis of these excision site amplicons showed that 100% of the excision site repair in WT plants resulted in either precise repair or a 1 base insertion ( Fig. 1C ). In contrast, 22% of the excision sites in pol V plants had large deletions (>20 bp), with one deleting part of the promoter ( Fig. 1C ).
Together these results support a model in which RNAs produced by the Pol V protein play a role in NHEJ repair of the mPing excision sites. The finding that pol V and pol V 1-3 mutants both exhibited the same decreased GFP expression indicates that Pol V produced transcripts and not simply the presence of the protein is required. In addition, excision site sequencing evidence suggest that abnormal repair is the cause of decreased GFP expression. The large deletions left behind in some pol V plants is consistent with stalled or slow NHEJ repair in which extra bases were excised from the ends. Our sequence analysis is also likely an underrepresentation of the quality of the excision site repair in pol V plants because PCR amplicons were only produced when the repair retained both primer binding sites.
Because the mPing excision site used in this study is directly after the 35S promoter, it is likely to be highly expressed similar to the 35S:GU-US reporter (Wei et al. 2012, Miki et al. 2017) . The 35S promoter has been shown to be a target for silencing mechanisms, including the Pol IV and Pol V mediated RdDM pathways (Mlotshwa et al. 2010, Li et al. 2012) . The observation that excision site repair was not significantly altered in other RdDM related mutants also provides important information. The fact that we did not see a difference in GFP expression in the pol IV knock-out tells us that the process of RdDM is not directly acting on NHEJ. Similarly, we can conclude that CG maintenance methylation is not responsible for DNA repair because the met1 mutant had no change in GFP expression. Additional experiments will be needed to determine if the Dicer-like proteins play a role in the repair of the mPing excision site. It would also be interesting to test the repair of transposon excision sites that are not highly expressed or potentially targeted by RdDM pathways.
Methods
Arabidopsis Lines
The KS019 ( met1 allele “ddm2-1” ) line was previously backcrossed multiple times into wild-type Columbia (Kankel et al. 2003) . Salk_083051 ( pol IV ), Salk_017795 ( pol V ), and CS69544 ( pol V 1-3 ) were obtained from the Arabidopsis Biological Resource Center and genotyped by PCR and restriction digest analysis to confirm homozygosity. The genotype of CS69544 ( pol V 1-3 ) was confirmed by sequencing the PCR amplicon from the following primers: AT2G40030 1317 For – 5’ CCCTCTGATGTGTAGCCCCCTCAG and AT2G40030 1627 Rev – 5’ GGAAAACTGTCCAAGCTGGGCCAG. Young plants were grown at 23°C in 10 hours of light per day, mature plants were grown at 25°C with constant light.
Construct development and plant transformation
The 35S promoter in pEarleyGate 100 (Earley et al. 2006) between the Xho I and Stu I sites was replaced with the RPS5a promoter and then the Pong TPase L418A, L420A/T2A/ORF1SC1 ONE construct was gateway cloned into the plasmid. The mPing :e GFP reporter from pBIN (Yang et al. 2007) was cloned into the Bgl II site by ligation. The sequence verified construct was transferred to GV3101 Agrobacterium and Arabidopsis transformation was performed by floral dip (Clough and Bent 1998) . Transgenic seedlings were selected by spraying daily with a 1:500 dilution of Liberty herbicide before being transplanted to new flats.
GFP screening
Fluorescent microscopy was performed on 3–4-week-old seedlings using a Leicia M165 FC dissecting microscope equipped with a pE-300 lite LED light source and an ET GFP - M205FA/M165FC filter. Plants were visualized by two individuals and classified as having GFP expression in a binary (+/-) process. Fluorescence that results from transposition is usually localized to sectors or large spots on the leaves (Yang et al. 2007) , allowing us to differentiate from other fluorescent sources.
DNA Analysis
Arabidopsis DNA was extracted using the CTAB method. Briefly, a young leaf was ground in 600 μl of CTAB buffer (1% CTAB, 0.1 M Tris pH 8, 0.02 M EDTA, 5 mM ascorbic acid, 0.02% PVP-40, 4 mM DIECA) and incubated at 65°C for 30 minutes before adding 400 μl of chloroform. After centrifugation, the supernatant was transferred to a new tube and precipitated with 300 μl isopropanol, washed with 70% EtOH, dried, then resuspended in 50 μl of TE. PCR was performed using the pBIN Flank For – 5’ AGACGTTCCAACCACGTCTTCAAAGCAA and pBIN Flank Rev – 5’ CCTCTCCACTGACAGAAAATTTGTGCCCA primers with the following conditions: 30 cycles of 95°C for 30 sec, 58°C for 30 sec, and 72°C for 90 sec. The bands containing the excision sites were gel purified and cloned into pGEM® T Easy (Promega) before Sanger sequencing. The sequence results were aligned to the expected template for precise mPing excision site repair using Geneious Prime 2022.2.2 software (https://www.geneious.com).
Reagents
|
STRAIN |
GENOTYPE |
AVAILABLE FROM |
|
CS70000 |
Wild type Columbia |
ABRC |
|
KS019 |
met1 ( ddm2-1 allele) |
Keith Slotkin Laboratory |
|
Salk-083051 |
pol IV |
ABRC |
|
Salk-017795 |
pol V |
ABRC |
|
CS69544 |
pol V 1-3 |
ABRC |
|
PLASMID |
GENOTYPE |
DESCRIPTION |
|
pEarleyGate 100RMOA mPing |
Rps5a: Pong TPase L418A, L420A/T2A/ORF1SC1 ONE, 35S: mPing : eGFP |
Addgene #196137 |
Acknowledgments
Acknowledgments
Thanks to Priscilla Redd for assistance with a chi-square statistical analysis for production of the GFP expression graph.
Funding Statement
Funding for this project was provided by the National Science Foundation grant #1651666 and the University of South Carolina Aiken INBRE supported by a grant from the National Institutes of Health National Institute of General Medical Sciences (P20GM13499-20).
References
- Bader AS, Hawley BR, Wilczynska A, Bushell M. The roles of RNA in DNA double-strand break repair. Br J Cancer. 2020 Jan 2;122(5):613–623. doi: 10.1038/s41416-019-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998 Dec 1;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006 Feb 1;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012 Nov 9;338(6108):758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- Franklin A, Steele EJ. RNA-directed DNA repair and antibody somatic hypermutation. Trends Genet. 2021 Nov 2;38(5):426–436. doi: 10.1016/j.tig.2021.10.005. [DOI] [PubMed] [Google Scholar]
- Gilbert DM, Bridges MC, Strother AE, Burckhalter CE, Burnette JM 3rd, Hancock CN. Precise repair of mPing excision sites is facilitated by target site duplication derived microhomology. Mob DNA. 2015 Sep 7;6:15–15. doi: 10.1186/s13100-015-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005 Feb 3;308(5718):118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Dyda F. DNA Transposition at Work. Chem Rev. 2016 May 17;116(20):12758–12784. doi: 10.1021/acs.chemrev.6b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR. An active DNA transposon family in rice. Nature. 2003 Jan 9;421(6919):163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003 Mar 1;163(3):1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005 May 29;37(7):761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Terauchi K, Wada M, Hirano HY. The plant MITE mPing is mobilized in anther culture. Nature. 2003 Jan 9;421(6919):167–170. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci U S A. 2009 Jan 13;106(3):941–946. doi: 10.1073/pnas.0810310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Qian W, Zhao Y, Wang C, Shen J, Zhu JK, Gong Z. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci U S A. 2012 Jun 25;109(28):11425–11430. doi: 10.1073/pnas.1208557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014 May 8;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Miki D, Zhu P, Zhang W, Mao Y, Feng Z, Huang H, Zhang H, Li Y, Liu R, Zhang H, Qi Y, Zhu JK. Efficient Generation of diRNAs Requires Components in the Posttranscriptional Gene Silencing Pathway. Sci Rep. 2017 Mar 22;7(1):301–301. doi: 10.1038/s41598-017-00374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Gao Z, Mgutshini NL, Li J, Chen X, Bowman LH, Vance V. Transcriptional silencing induced by Arabidopsis T-DNA mutants is associated with 35S promoter siRNAs and requires genes involved in siRNA-mediated chromatin silencing. Plant J. 2010 Oct 8;64(4):699–704. doi: 10.1111/j.1365-313X.2010.04358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T. Mobilization of a transposon in the rice genome. Nature. 2003 Jan 9;421(6919):170–172. doi: 10.1038/nature01219. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005 Mar 11;120(5):613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Plasterk RH, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999 Aug 1;15(8):326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- Stitzer MC, Anderson SN, Springer NM, Ross-Ibarra J. The genomic ecosystem of transposable elements in maize. PLoS Genet. 2021 Oct 14;17(10):e1009768–e1009768. doi: 10.1371/journal.pgen.1009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012 Mar 22;149(1):101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Yang G, Zhang F, Hancock CN, Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007 Jun 19;104(26):10962–10967. doi: 10.1073/pnas.0702080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Qi Y. RNA-directed repair of DNA double-strand breaks. DNA Repair (Amst) 2015 May 1;32:82–85. doi: 10.1016/j.dnarep.2015.04.017. [DOI] [PubMed] [Google Scholar]
- Yuan YW, Wessler SR. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci U S A. 2011 Apr 25;108(19):7884–7889. doi: 10.1073/pnas.1104208108. [DOI] [PMC free article] [PubMed] [Google Scholar]


