Abstract
The biological function of the novel zinc-finger SWIM domain-containing protein family (ZSWIM) during embryonic development remains elusive. Here, we conducted a genome-wide analysis to explore the evolutionary processes of the ZSWIM gene family members in mice, Xenopus tropicalis, zebrafish, and humans. We identified nine putative ZSWIM genes in the human and mouse genome, eight in the Xenopus genome, and five in the zebrafish genome. Based on multiple sequence alignment, three members, ZSWIM5, ZSWIM6, and ZSWIM8, demonstrated the highest homology across all four species. Using available RNA sequencing (RNA-seq) data, ZSWIM genes were found to be widely expressed across different tissues, with distinct tissue-specific properties. To identify the functions of the ZSWIM protein family during embryogenesis, we examined temporal and spatial expression patterns of zswim family genes in Xenopus embryos. Quantitative real-time polymerase chain reaction (qRT-PCR) revealed that each member had a distinct expression profile. Whole-mount in situ hybridization showed that both zswim1 and zswim3 were maternally expressed genes; zswim5 and zswim6 were expressed throughout embryogenesis and displayed dynamic expression in the brain, eyes, somite, and bronchial arch at the late tailbud stages; zswim7 was detected in the eye area; zswim8 showed a dynamic expression pattern during the tailbud stages, with expression detected in the brain, eyes, and somite; zswim9 was faintly expressed throughout embryonic development. This study provides a foundation for future research to delineate the functions of ZSWIM gene members.
Keywords: ZSWIM gene family, Xenopus, Gene expression, Phylogenetic analysis, Bioinformatics
INTRODUCTION
The zinc finger is a short finger-like conformation formed by the coordination of structural motifs in a protein with zinc ions (Zn2+) (Klug & Rhodes, 1987). First discovered in the African clawed frog (Xenopus laevis), zinc finger proteins (ZNFs) have since been identified in a variety of organisms (Klug, 2010; Klug & Rhodes, 1987) and are implicated in various biological processes, such as transcriptional regulation (Droll et al., 2013), gene expression (Nunez et al., 2011), cell migration (Zhang et al., 2012), cell differentiation (Powell et al., 2019), and embryonic development (Close et al., 2002). The classification of ZNFs is based on the structure of the zinc-finger domain. To date, the HUGO Gene Nomenclature Committee has approved 30 types of ZNFs, including the zinc-finger C2H2-type, RING-type, PHD-type, LIM-type, C3H-type, C2HC-type, ZZ-type, and SWIM-type (Gray et al., 2015). The zinc-finger SWIM-type is a novel and functionally uncharacterized zinc-finger domain with a CxCxnCxH molecular signature. The SWIM domain is evolutionarily conserved across many organisms, from archaea to mammals (Makarova et al., 2002), and has been identified in various prokaryotic and eukaryotic proteins, including bacterial ATPases in the SWI2/SNF2 (SWItch Sucrose Non-fermentable) family (Flaus et al., 2006), plant MuDR transposases (Benito & Walbot, 1997; Hershberger et al., 1995), and vertebrate MEK kinase-1 (Hagemann & Blank, 2001). First identified in SWI2/SNF2 and MuDR proteins, leading to the nomenclature SWIM, the domain is predicted to have DNA-binding and protein-protein interaction functions (Makarova et al., 2002). However, the functions of the novel zinc-finger SWIM domain-containing protein family (ZSWIM) remain poorly known, despite its identification in many organisms. Several studies have suggested that members of this protein family are involved in regulating various important biological processes. For example, ZSWIM1 is considered a biomarker in the differentiation of the T helper cells (Ko et al., 2014). ZSWIM2 is reported to regulate MEX E3 ubiquitin ligase activity in the testis (Nishito et al., 2006). ZSWIM4 is involved in the ubiquitination pathway (Forde et al., 2012). ZSWIM5 participates in the inhibition of the malignant progression of non-small-cell lung cancer (Xu et al., 2018). A point mutation in ZSWIM6 is associated with acromelic frontonasal dysostosis, causing multiple malformations of the skull, brain, and limbs (Twigg et al., 2016). ZSWIM8 is implicated in microRNA degradation by reducing the stability of Argonaute proteins (Shi et al., 2020). While these results strongly emphasize the biological importance of the ZSWIM protein family, its functions in embryonic development remain elusive.
Here, we present the first detailed and comprehensive analysis of the ZSWIM gene family in four well-characterized organisms, including humans, mice, X. tropicalis, and zebrafish. We first performed genome-wide identification of the ZSWIM gene family using various publicly available databases across the four organisms. Based on phylogenetic analysis, we examined the evolutionary relationships among all identified members of the ZSWIM gene family. In addition, we used various bioinformatics tools to gain a better understanding of the ZSWIM gene family, performing amino acid sequence alignment to identify protein sequence similarity, motif structure analysis to identify conserved motifs, gene structure analysis to determine intron/exon organization, gene expression analysis using RNA sequencing (RNA-seq) datasets to identify expression pattern among various tissues, Gene Ontology (GO) to predict their function, and gene interaction analysis to predict interacting partners. To gain a better understanding of how ZSWIM proteins play a role in embryogenesis, we examined the temporal and spatial expression patterns of ZSWIM family members using Xenopus embryos. Results showed that each member of this family exhibited a specific expression pattern. Notably, zswim5, zswim6, and zswim8 displayed distinct dynamic expression patterns. This study provides detailed information about members of the ZSWIM gene family and facilitates their further functional characterization.
MATERIALS AND METHODS
Identification of ZSWIM genes and SWIM domain
We searched various databases to identify ZSWIM family members, including the NCBI database (https://www.ncbi.nlm.nih.gov) for humans, MGI database (http://www.informatics.jax.org) for mice, Xenbase database (https://www.xenbase.org/entry/) for Xenopus, and ZFIN database (https://zfin.org) for zebrafish. The presence of the ZSWIM-type domain in all identified peptide sequences was further confirmed using the Pfam database (https://pfam.xfam.org). In addition, the physical and chemical properties of the ZSWIM domain-containing proteins were determined using the ExPASy program (https://web.expasy.org/protparam/), including molecular weight (MW), sequence length of amino acids (aa), and isoelectric point (pI) (Supplementary Table S2).
Phylogenetic, syntenic, and multiple sequence alignment analyses
A phylogenetic tree was constructed using the protein sequences of all identified members across the four organisms using MEGA v11 with the neighbor-joining (NJ) method and bootstrapping for 1 000 replicates (Tamura et al., 2021). Syntenic analysis was performed using the Circoletto web tool (http://tools.bat.infspire.org/circoletto/) with FASTA sequences in each block for all selected organisms. Multiple sequence alignment was carried out using Jalview software with default parameters. Heatmaps representing the percentage of similarity in protein sequences were generated using phyton.
Gene structure and conserved motif analyses
The exon-intron organization of ZSWIM genes was examined using the gene structure display server (GSDS) (http://gsds.cbi.pku.edu.cn/, 2.0) by aligning complementary DNA (cDNA) sequences with corresponding genomic sequences. The online MEME (Multiple EM for Motif Elicitation) suite was used to identify unknown conserved motifs (http://meme-suite.org/tools/meme) with default parameters (Bailey et al., 2015).
Expression profiles of ZSWIM gene family by RNA-seq analysis
For humans, ZSWIM gene family expression data in 27 organs/tissues, including the adrenal gland, appendix, bone marrow, brain, colon, duodenum, endometrium, esophagus, fat, gall bladder, heart, kidney, liver, lung, lymph node, ovary, pancreas, placenta, prostate, salivary gland, skin, small intestine, spleen, stomach, testis, thyroid, and urinary bladder, were obtained from publicly available transcriptome datasets in the NCBI database (Fagerberg et al., 2014). Reads per kilobase million (RPKM) values were used to generate expression levels with the online Heatmapper tool (http://heatmapper.ca/expression/) by selecting the distance function (Euclidean) and hierarchical clustering (average) method. For Xenopus, zswim gene family expression data in five major organs/tissues, including the brain, heart, kidney, liver, and skeletal muscles, were obtained from publicly available transcriptome datasets in the Xenbase database (http://www.xenbase.org/entry/). Transcripts per million (TPM) values were used to generate the graph for gene expression analysis.
GO enrichment and protein-protein interaction (PPI) network analysis
GO analysis was performed to analyze the biological processes associated with the human ZSWIM protein family using the publicly available online tool Enrichr (https://maayanlab.cloud/Enrichr/). PPI network analysis of the ZSWIM protein in humans was carried out using the online STRING database (https://string-db.org).
RNA extraction and purification from Xenopus embryos
Total RNA was extracted from different embryonic stages, including egg (stage 1), early cleavage (stage 2), blastula (stage 6), gastrula (stage 10), neurula (stage 13 and stage 18), early tailbud (stage 23 and stage 28), and late tailbud (stage 35) using TRIzol reagent (Invitrogen, 15596-026, USA) following the manufacturer’s instructions. RNA was then purified using an RNA Clean-up Kit (Favorgen, FAATR001-1, Taiwan, China). RNA concentration was measured using a NanoDrop spectrophotometer (Thermo, ND-2000, USA), with samples stored at −80 °C until use.
Reverse transcription and quantitative real-time polymerase chain reaction (qRT-PCR)
For each embryonic developmental stage, 1 μg of total RNA was reverse transcribed to cDNA using Superscript III Reverse Transcriptase (Invitrogen, 18080-044, USA). The obtained cDNA samples were then analyzed by qRT-PCR using SYBR Green PCR Master Mix (Takara Bio, China) on an ABI Quant Studio7 Flex Real-Time PCR System (USA). The primers were designed using Primer v3 software (https://primer3.ut.ee). Ornithine decarboxylase (odc) was used as an internal control. Relative expression was calculated using the 2-ΔΔCT method (Livak & Schmittgen, 2001).
Embryo collection by in vitro fertilization for whole-mount in situ hybridization
Wild-type female Xenopus laevis frogs were injected with 500 IU of human chorionic gonadotropin (HCG) the night before embryo collection. The injected frogs were then kept in a separate water tank overnight. The next day, testes were dissected from a mature male frog, with small pieces then blended with 1× MMR (Marc’s modified ringer’s solution) to expel sperm and mixed with eggs in a 10 cm petri dish. After 3–5 min, 0.2× MMR (20 mmol/L NaCl, 0.4 mmol/L KCl, 0.2 mmol/L MgSO4, 0.4 mV CaCl2, 1 mmol/L HEPES, 0.02 mmol/L EDTA, pH 7.8) was added to the dish and incubated for 30–40 min at room temperature. The eggs rotated after successful fertilization, resulting in the animal hemisphere facing upward. The embryos were collected at the desired stages and fixed in HEMFA buffer (0.1 mol/L HEPES, 2 mmol/L EGTA, 1 mmol/L MgSO4, 4% formaldehyde) for 1 h, then stored in 100% ethanol at −20 °C. The embryos were cultured in 0.2× MMR at room temperature.
Probes synthesis, whole-mount in situ hybridization, and vibratome sectioning
All members of the zswim gene family (zswim1, zswim3, zswim4, zswim5, zswim6, zswim7, zswim8, and zswim9) were subcloned into the pBluescript II KS (+) vector. Following confirmation by sequencing, the plasmids were linearized and purified using a PCR purification kit (Favorgen, FAGCK001-1, Taiwan, China). Antisense probes were synthesized using DIG RNA Labeling Mix (Roche, 11277073910, Germany), T3 RNA polymerase (Promega, P2083, USA), and linearized plasmid as templates. Standard protocols for whole-mount in situ hybridization were followed, as described previously (Harland, 1991). To further delineate expression pattern details of the zswim gene family, embryos were sectioned using a vibratome after whole-mount in situ hybridization. First, embryos were fixed in a solution (containing 5 g/L gelatin, 380 g/L chick egg albumin, and 200 g/L sucrose in 0.1 mol/L phosphate buffer, pH 7.4) with 0.2 mL of 25% (v/v) glutaraldehyde. Sectioning was then carried out at a thickness of 50 mm using a vibratome (Leica VT1200 S, Leica Biosystems, Germany), followed by coverslip mounting with Mowiol (Merck KGaA, Germany). Images were captured using a stereomicroscope (Olympus SZX16, Japan).
RESULTS
Identification, physiochemical properties, and phylogenetic analysis of ZSWIM family members
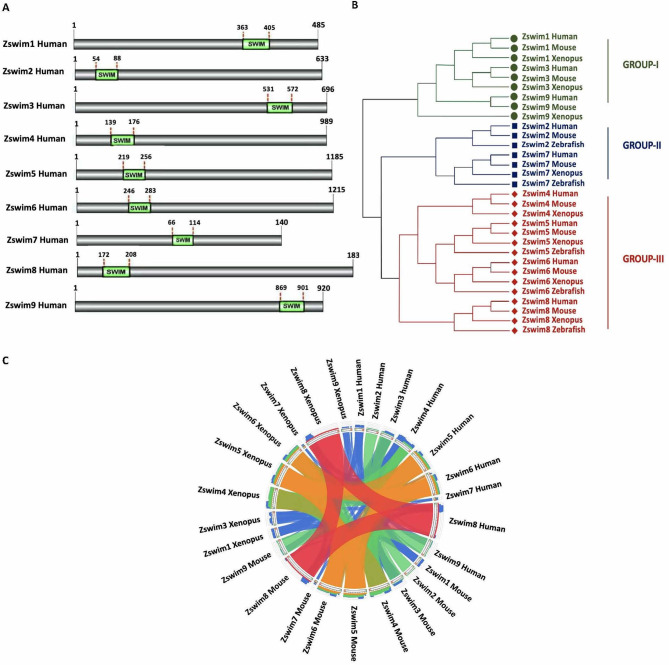
We identified nine putative ZSWIM genes in the human and mouse genomes (ZSWIM1 to ZSWIM9), eight genes in the Xenopus genome (zswim1, zswim3, zswim4, zswim5, zswim6 zswim7, zswim8, and zswim9), and five genes in the zebrafish genome (zswim2, zswim5, zswim6, zswim7, and zswim8) (Supplementary Table S1). All ZSWIM members identified in the four organisms contained the conserved SWIM domain at a specific position (Figure 1A; Supplementary Figure S1A–C). Furthermore, we identified various biochemical properties, such as molecular weight (MW), sequence length of amino acids (aa), isoelectric point (pI) using the expasy tool (https://www.expasy.org) (Supplementary Table S2). Each member exhibited distinct physical and chemical characteristics, suggesting that each ZSWIM protein exerts a distinct biological function. Furthermore, we performed phylogenetic analysis to explore the evolutionary relationships among ZSWIM family members across all four organisms. The amino acid sequences of all identified ZSWIM family members were used to construct the evolutionary tree with the NJ method (Figure 1B). Based on their phylogenetic distribution, the ZSWIM family members were categorized into three major groups (Groups I, II, and III). Group I consisted of ZSWIM1, ZSWIM3, and ZSWIM9; Group II consisted of ZSWIM2 and ZSWIM5; and Group III consisted of ZSWIM4, ZSWIM5, ZSWIM6, and ZSWIM8. Classification showed that the genes in each group were more closely related than the genes in other groups. To further identify the evolutionary process of gene duplication in the ZSWIM gene family, we conducted a comparative analysis of all ZSWIM genes across all four species. Results confirmed the phylogenetic tree pattern, with ZSWIM5, ZSWIM6, and ZSWIM8 conserved across all four species (Figure 1C).
Figure 1.
Identification of ZSWIM family members
A: Identification of human ZSWIM family members and precise position of SWIM domain in each member. Identified SWIM domain is shown in green, and its position is labeled at the top of each domain. B: Phylogenetic relationships among human, mouse, Xenopus, and zebrafish ZSWIM proteins. Phylogenetic tree was constructed using the NJ method in MEGA v11. The three groups are indicated by different colors and designated as groups I–III. C: Syntenic analysis of ZSWIM family members in humans, mice, Xenopus, and zebrafish. In the circle, ribbons represent four quartiles of the maximum score, represented by semitransparent blue, green, orange, and red.
Amino acid sequence alignment and interaction analysis
To identify the conserved regions across protein sequences of all four species, we aligned the protein sequences of all ZSWIM members and identified conserved regions. To further determine which ZSWIM family members share the highest degree of homology across all four species, we aligned the protein sequences of each member, i.e., ZSWIM1 to ZSWIM9, separately. Interestingly, three members of the ZSWIM family, i.e., ZSWIM5, ZSWIM6, and ZSWIM8, shared the highest homology across all four species (Supplementary Figure S2A). To identify the interacting partners of the ZSWIM family members, we performed network analysis using the STRING database (https://string-db.org). Interestingly, ZSWIM5, ZSWIM6, and ZSWIM8 interacted with each other (Supplementary Figure S2B). Additionally, we calculated the percentage of similarity among all members, and found that ZSWIM5, ZSWIM6, and ZSWIM8 shared the highest percentage of similarity (Supplementary Figure S3A–C). Conversely, other members, e.g., ZSWIM1, ZSWIM2, ZSWIM3, ZSWIM7, and ZSWIM9, did not show high similarity in their amino acid sequences (data not shown).
Gene structure and conserved motif analysis of ZSWIM family members
To further explore the structural diversity of identified ZSWIM genes, their exon-intron organization was also analyzed. Gene structure diversity is the main source of evolution of multigene families (Cao et al., 2016; Mercereau-Puijalon et al., 2002; Pellicer et al., 2018). Analysis revealed that all identified members of the ZSWIM gene family exhibited a similar pattern of exon-intron organization across the four studied organisms, e.g., ZSWIM1 was intronless in all organisms. Similarly, other members exhibited almost identical exon-intron patterns (Supplementary Figure S4A–D).
To predict the function of each family member based on their motif sequences, we conducted a BLAST search of each protein family member using the MEME suite. Our analysis identified various conserved and different motifs in each member among the four organisms, suggesting that each member has a unique biological value. Remarkably, ZSWIM2 not only contained the ZSWIM type domain but also other types of zinc-finger domains, including the PHD-type zinc finger, ubiquitin-binding zinc finger, ZZ-type zinc finger, and Ring-type zinc finger (Supplementary Figure S5C). These findings suggest that the ZSWIM2 protein may be involved in a variety of critical biological processes, such as transcriptional regulation, signal transduction, and ubiquitin-mediated protein degradation. Furthermore, we obtained the sequence logos for each identified motif of ZSWIM2 (Supplementary Figure S5D). Conversely, while various known conserved motifs were identified in other members, each member contained a motif distinct from the others (Supplementary Figure S5A, B, E–R).
Furthermore, we identified conserved motifs in the ZSWIM protein family using the MEME suite (https://meme-suite.org/meme/tools/meme). Interestingly, four members of this family, i.e., ZSWIM4, ZSWIM5, ZSWIM6, and ZSWIM8, exhibited a variety of common motifs in their protein structures (Supplementary Figure S5S). The sequence logos of each motif were also obtained using the MEME suite (Supplementary Figure S5T).
Expression and GO analyses of ZSWIM genes
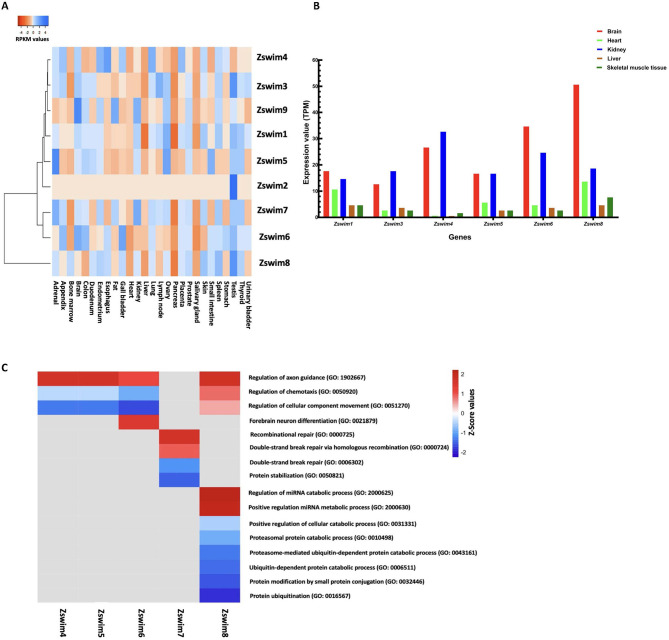
To delineate ZSWIM expression in different organs, all identified human ZSWIM genes were investigated using available RNA-seq data of 27 major human organs/tissues (adrenal gland, appendix, bone marrow, brain, colon, duodenum, endometrium, esophagus, fat, gall bladder, heart, kidney, liver, lung, lymph node, ovary, pancreas, placenta, prostate, salivary gland, skin, small intestine, spleen, stomach, testis, thyroid, and urinary bladder). Overall, ZSWIM gene expression was tissue-specific in the 27 tissues, indicating that each family member may play a specific and essential role in that tissue. For example, ZSWIM1 was highly expressed in the ovary (RPKM: 3.001) and testis (RPKM: 2.745). ZSWIM2 was only expressed in the testis (RPKM: 5.29), suggesting an essential role in spermatogenesis. ZSWIM3 was expressed in the lymph node (RPKM: 1.762) and testis (RPKM: 2.241), while ZSWIM4 was expressed in the esophagus (RPKM: 2.253) and lungs (RPKM: 1.963). Furthermore, ZSWIM5 was expressed in the adrenal gland (RPKM: 5.603), ovary (RPKM: 4.245), and small intestine (RPKM: 3.621), while ZSWIM6 was expressed in the gall bladder (RPKM: 6.767) and brain (RPKM: 6.391), ZSWIM7 was expressed in adrenal gland (RPKM: 6.129) and kidney (RPKM: 6.183), ZSWIM8 was expressed in the testis (RPKM: 21.564) and spleen (RPKM: 16.023), and ZSWIM9 was expressed in the brain (RPKM: 2.958), fat (RPKM: 2.308), and kidney (RPKM: 2.198) (Figure 2A).
Figure 2.
Expression profiles of ZSWIM genes in human organs and GO analysis
A: Heatmap showing expression profiles of ZSWIM genes in twenty-seven major human organs/tissues. Different colors represent RPKM values, as indicated in the bar at the top of the figure. B: Graphical representation of zswim gene expression profiles in five major organs/tissues in Xenopus. C: Heatmap shows GO terms associated with various biological processes involving ZSWIM family members.
Similarly, we analyzed the expression of all identified zswim genes in Xenopus using publicly available RNA-seq data obtained from the Xenbase database (http://www.xenbase.org/entry/). The zswim genes were mainly expressed in five major organs, including the brain, heart, kidney, liver, and skeletal muscles. Remarkably, zswim6 and zswim8 were highly expressed in the brain, while zswim4 was highly expressed in the kidney and brain. Other members, such as zswim1, zswim3, zswim5, and zswim7, were moderately expressed in all other organs, including the brain, heart, kidney, liver, and skeletal muscles (Figure 2B). These findings indicate that zswim6 and zswim8 may perform key roles in the regulation of brain development in Xenopus.
Next, we performed GO analysis of zswim family members to reveal their involvement in critical biological processes. Results showed that zswim4 and zswim5 were involved in the regulation of axon guidance (GO: 1902667), zswim6 was involved in forebrain neuron differentiation (GO: 0021879), zswim7 was involved in recombinational repair (GO: 0000725), and zswim8 was involved in the regulation of miRNA catabolic process (GO: 2000625) and positive regulation of miRNA metabolic process (GO: 2000630) (Figure 2C).
Temporal expression pattern analysis of ZSWIM family members in Xenopus
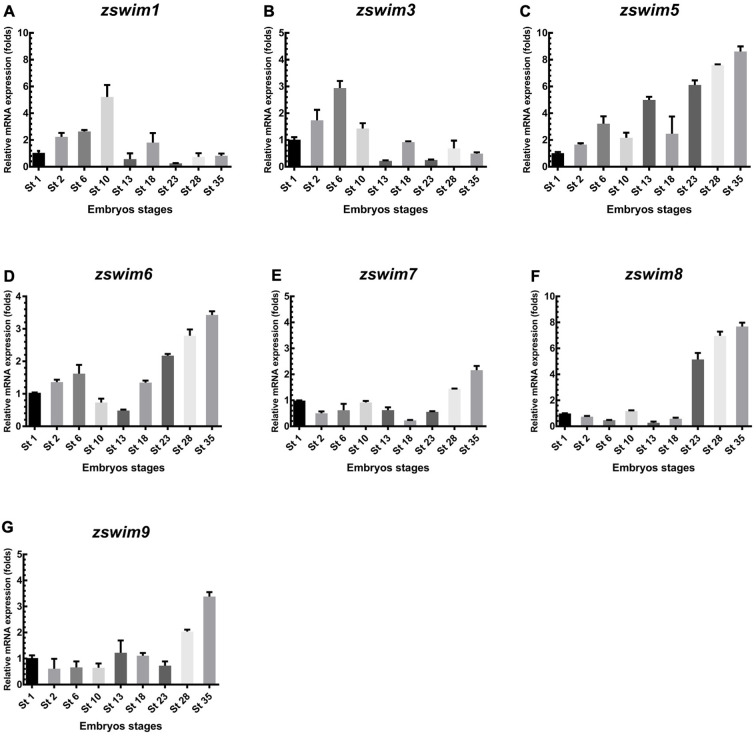
To examine whether zswim genes are expressed during embryonic development, we carried out temporal and spatial expression analysis in X. laevis embryos. To analyze the temporal expression pattern of zswim genes during embryonic development, total RNA was isolated at different stages of development and analyzed by qRT-PCR. Based on qRT-PCR analysis, zswim1 and zswim3 showed maternal expression patterns and were increased during the early cleavage (stage 2) to blastula stages (stage 6). The increase in zswim1 continued to stage 10, then sharply declined during the gastrulation, neurulation, and late tailbud stages (Figure 3A). For zswim3, expression peaked at stage 6 and then declined (Figure 3B). These findings suggest that zswim1 and zswim3 play important roles before gastrulation. The expression patterns of zswim5 and zswim6 were similar. Both were detected before gastrulation, then increased continually at a high level during the early and late tailbud stages (stages 23 to 35) (Figure 3C, D). These findings suggest that zswim5 and zswim6 exert functions during the neurula and tailbud stages. Results further showed that zswim7 expression remained low throughout embryogenesis (Figure 3E). In addition, zswim8 expression remained at a low level before the tailbud stages, then increased to a high expression level during the early and late tailbud stages (stages 23, 28, and 35) (Figure 3F). Likewise, zswim9 was moderately expressed from the early cleavage (stage 2) to early tailbud stage (stage 23), but showed the highest expression at the late tailbud stage (stage 35) (Figure 3G).
Figure 3.
qRT-PCR analysis of zswim gene expression in Xenopus embryos
A–G: mRNA expression level of Xenopus zswim genes was measured at different embryonic stages, including in fertilized egg (stage 1), cleavage (stage 2), blastula (stage 6), gastrula (stage 10), neurula (stages 13, 18), early tailbud (stages 23, 28), and late tailbud stages (stage 35). ODC served as the internal standard control.
Spatial expression pattern analysis of zswim family members
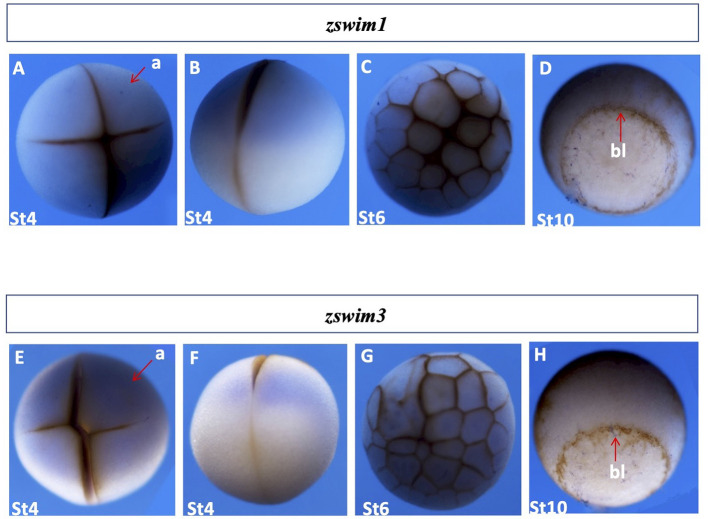
zswim1 and zswim3: We next cloned the coding sequences of zswim family members from X. laevis using qRT-PCR. Spatial expression of zswim family members during embryonic development was analyzed by whole-mount in situ hybridization. Results revealed that both zswim1 and zswim3 were maternally expressed, and their signals were mainly detected in the animal region during the early cleavage (stage 4) and early blastula stages (stage 6) (Figure 4A–C, E–G). Furthermore, only weak signals were detected during the early gastrula stage (stage 10), exclusively from the yolk plug (Figure 4D, H). These results indicate that zswim1 is a maternally expressed gene and may function during oocyte maturation and before mid-blastula transition (MBT).
Figure 4.
Spatial gene expression patterns of zswim1 and zswim3
Spatial expression levels of zswim1 and zswim3 were analyzed by whole-mount in situ hybridization. Embryonic stages are shown at the bottom. Detailed explanations of expression patterns are described in the Results section. A, E: Stage 3, animal view. B, F: Stage 3, lateral view. C, G: Early blastula stage embryo (stage 6), animal view. D, H: Gastrula stage embryo (stage 11), blastopore lip indicated by the red arrow. a, animal; bl, blastopore lip; v, vegetal.
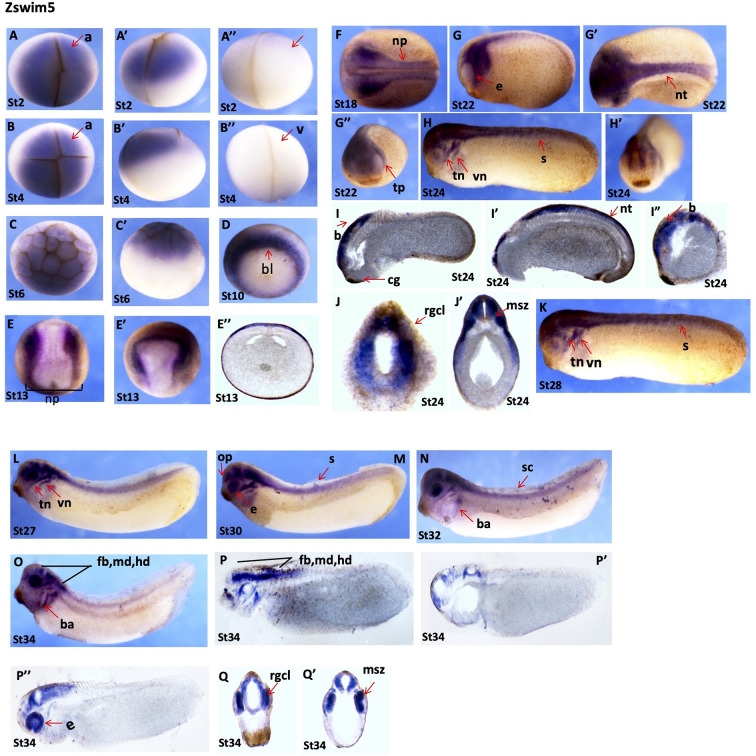
zswim5: The zswim5 gene showed a highly discrete and dynamic spatial expression pattern during embryonic development, with detection during all developmental stages. Whole-mount in situ hybridization revealed that zswim5 was maternally expressed in the animal region during the early cleavage (stages 2 and 3) and early blastula stages (stage 6) (Figure 5A–C’). At the onset of gastrulation (stage 11), zswim5 signals were highly restricted around the yolk plug, with higher expression in the dorsal blastopore lip (Figure 5D). At the early neurula stage (stage 14), strong zswim5 signals were detected at both edges of the neural plate and were highly expressed in the neural crest region (Figure 5E, E’). Transverse sectioning at stage 13 showed zswim5 expression in the dorsal ectoderm region (Figure 5E’’). At the late neurula stage (stage 18), strong signals were detected in the neural plate and neural crest region (Figure 5F). During the early tailbud stages (stages 22 and 24), zswim5 expression was mainly detected in the neural tube and various regions of the head, including the eye, trigeminal placode, trigeminal nerve, and vestibulocochlear nerve (Figure 5G–H’). Longitudinal sections at stage 24 revealed clear and dominant expression in the head region and neural tube (Figure 5I–I”), while transverse sections at the same stage showed expression in the retinal ganglion cell layer and marginal and subventricular zone (Figure 5J, J’). In addition, in late tailbud stage embryos (stages 27 to 34), strong and highly dynamic signals of zswim5 were detected in the branchial arch, eyes, spinal cord, pineal gland, olfactory placode, and various regions of the brain, including the forebrain, midbrain, and hindbrain (Figure 5K–O). Longitudinal sections of tailbud embryos (stage 34) revealed zswim5 expression in the eyes and various regions of the brain, including forebrain, midbrain, and hindbrain (Figure 5P–P”). Transverse sections at stage 34 showed expression in the retinal ganglion cell layer and marginal and subventricular zone (Figure 5Q, Q’).
Figure 5.
Dynamic expression of zswim5 throughout embryogenesis
A–A”: Stage 2 embryo, animal view (A), lateral view (A’), vegetal view (A”). B–B”: Stage 3 embryo, animal view (B), lateral view (B’), vegetal view (B”). C, C’: Early blastula stage (stage 6), animal view (C), lateral view (C’). D: Gastrula stage (stage 10), blastopore lip is indicated by red arrow. E, E’: Early neurula stage (stage 14), dorsal view (E), lateral view (E’). E”: Transverse section of stage 14 embryo. F: Late neural stage (stage 18), dorsal view. G–G”: Neural stage (stage 22), lateral, dorsal, and anterior views. H, H’: Early tailbud stage (stage 24), lateral view (H), anterior view (H’). I–I”: Longitudinal sections from stage 24 embryo. J, J’: Transverse sections from stage 24 embryos. K–O: Tailbud stages (stages 28 to 34), lateral view. P–P’’: Longitudinal section from late tailbud stage (stage 34). Q, Q’: Transverse section from late tailbud stage (stage 34). a, animal; b, brain; bl, blastopore lip; cg, cement gland; e, eye; fb, forebrain; md, midbrain; hb, hindbrain; msz, marginal and subventricular zone; np, neural plate; op, olfactory placode; pg, pineal gland; rgcl, retinal ganglion cell layer; s, somites; tn, trigeminal nerve; tp, trigeminal placode; vn, vestibulocochlear nerve.
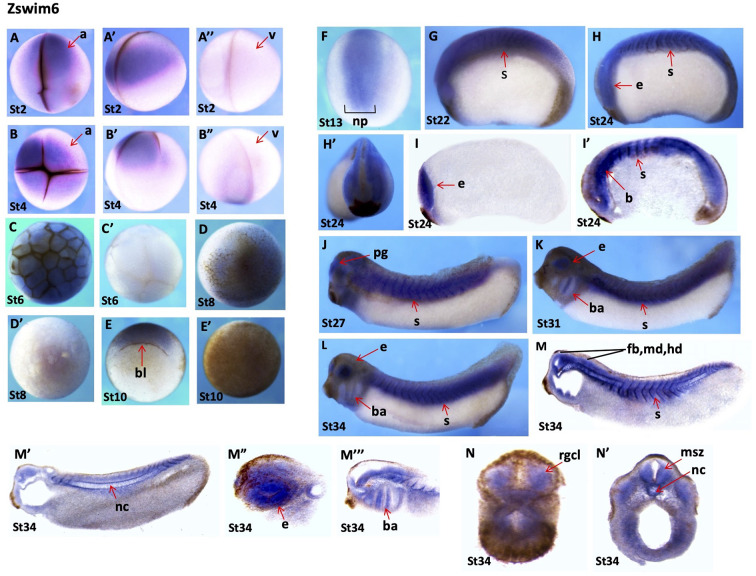
zswim6: The zswim6 gene also showed very dynamic spatial expression patterns at all stages of embryonic development. Notably, zswim6 expression was detected in the animal region during the early cleavage (stages 2 and 3) and blastula stages (stages 6 and 8), suggesting that zswim6 was maternally expressed (Figure 6A–D’). However, faint signals were detected in the dorsal blastopore lip region at the early gastrula stage (stage 10) (Figure 6E). No signals were found at the animal pole of the embryo (Figure 6E’). At the early neurula stage (stage 13), zswim6 expression was detected in the neural plate, predominantly at the middle region of the primary neurons (Figure 6F). At the early tailbud stages (stages 22, 24, and 27), its expression became evident at the neural plate, developing somites, pineal gland, and trigeminal placode (Figure 6G–H’, J). Longitudinal sections of embryos at the early tailbud stage (stage 24) showed clear expression in the eyes, somites, and brain region (Figure 6I, I’). Obvious and dominant expression during the late tailbud stages (stages 27 to 34) was detected in eyes, branchial arches, and somites (Figure 6K, L). Longitudinal sections of embryos at the tailbud stage (stage 34) showed highly specific and dynamic expression patterns in various regions of the brain, including the forebrain, midbrain, hindbrain, eyes, branchial arch, somites, and notochord (Figure 6M–M”’). Transverse sections of embryos at the same stage showed expression in the marginal and subventricular zone, notochord, and retinal ganglion cell layer (Figure 6N, N’).
Figure 6.
Spatial gene expression patterns of zswim6s
A–A”: Stage 2 embryo, animal view (A), lateral view (A’), vegetal view (A”). B–B”: Stage 3 embryo, animal view (B), lateral view (B’), and vegetal view (B”). C, C’: Early blastula stage (stage 6), animal view (C), vegetal view (C’). D, D’: Embryo at late blastula stage, animal view (D), vegetal view (D’). E: Embryo at gastrula stage (stage 10), blastopore lip is indicated by red arrow. F: Animal view. Embryo at neurula stage (stage 13), dorsal view. G: Early tailbud stage (stage 22). H, H’: Stage 24, lateral view (H), anterior view (H’). I, I’: Longitudinal sections of embryo at stage 24. J–L: Late tailbud stages (stages 27 to 34), lateral view. M–M”’: Tailbud stage (stage 34), longitudinal section. N, N’: Late tailbud stage (stage 34), transverse section. a, animal; b, brain; bl, blastopore lip; br, branchial arch; e, eye; msz, marginal and subventricular zone; nc, notochord; np, neural plate; pg, pineal gland; rgcl, retinal ganglion cell layer; s, somites; v, vegetal.
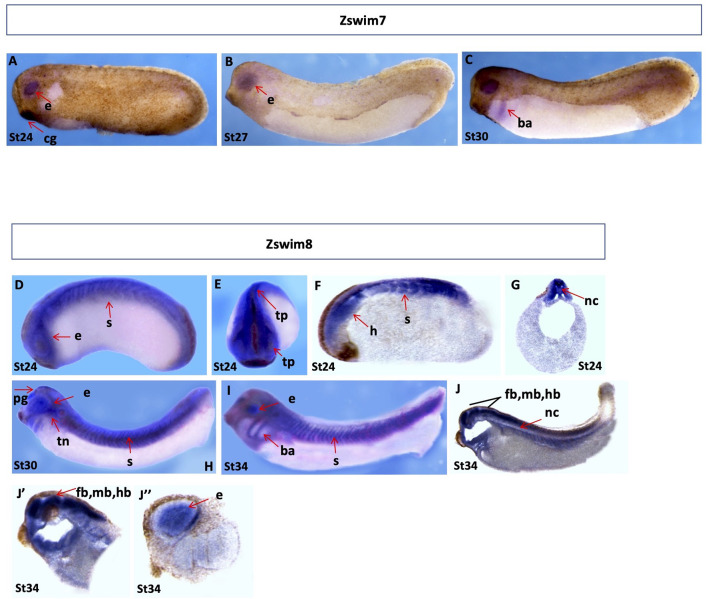
zswim7 and zswim8: Expression of zswim7 was not evident in embryos prior to stage 23 (results not shown). However, during the early and late tailbud stages (stages 24 and 30), faint signals were detected in the eyes (Figure 7A–C).
Figure 7.
Spatial gene expression patterns of zswim7 and zswim8
Spatial expression levels of zswim7 and zswim8 were analyzed by whole-mount in situ hybridization. Embryonic stages are indicated at the bottom of each image. A–C: Spatial expression of zswim7. Tailbud stages (stages 24 to 30), lateral view. D, E: Spatial expression of zswim8. D–G: Embryos at early tailbud stage (stage 24), lateral view (D), anterior view (E), longitudinal section (F), and transverse section (G). H, I: Embryos at tailbud stages (H, stage 30 and I, stage 34), lateral view. J–J’’: Longitudinal section of embryos at late tailbud (stage 34). b, brain; ba, branchial arch; e, eye; fb, fore brain; hb, hind brain; mb, mid brain; nc, notochord; pg, pineal gland; s, somites; tn, trigeminal nerves.
The highest expression of zswim8 was detected from the early to late tailbud stages (stages 24 to 34). At the early tailbud stage (stage 24), strong signals were detected in the neural tube, trigeminal placode, eyes, and somites (Figure 7D, E). Longitudinal and transverse sections of embryos at stage 24 further confirmed dominant expression near the head region, somites, and notochord (Figure 7F, G). In addition, during the late tailbud stages (stages 30 and 34), highly dynamic expression was detected in the somites, eyes, brachial arches, pineal gland, and trigeminal nerve (Figure 7H, I). Longitudinal and transverse sections of embryos at stage 34 revealed expression in the eye, various regions of the brain, including the forebrain, midbrain, and hindbrain, and notochord (Figure 7J–J”).
zswim9: The zswim9 gene was faintly expressed at all stages of embryonic development, from the early cleavage to tailbud stages (stages 2 to 34). During the early cleavage (stages 2 and 4) and blastula stages (stages 6 and 8), expression was detected in the animal pole (Supplementary Figure S6A–D’). At the onset and during the early gastrula stage (stages 10 and 11), signals were detected around the blastopore lip (Supplementary Figure S6E, F). From the early to late neural stages (stages 13 to 20), faint expression was detected in the neural plate and trigeminal placode (Supplementary Figure S6G–H’’). At the early tailbud stage (stage 24), faint signals were detected in the eyes, developing somite, and neural plate (Supplementary Figure S6I, I’). Longitudinal and transverse sections of stage 24 embryos showed expression in the neural tube and head region (Supplementary Figure S6J, J’). During the late tailbud stages, faint expression was detected in the somite, eyes, branchial arches, and brain (Supplementary Figure S6K–O). In addition, longitudinal sections at the late tailbud stages showed expression in the branchial arches, brain, eyes, and spinal cord (Supplementary Figure S6P, R, R’). Transverse sections at the same stage showed expression in the retinal ganglion cell layer and foregut (Supplementary Figure S6Q–Q’’’).
DISCUSSION
The first ZNFs were discovered in the late 1980s (Miller et al., 1985). Proteins from this novel class are known to bind with specific DNA sequences (Vrana et al., 1988), and are involved in many important cellular processes, including cell migration, DNA repair, and transcriptional regulation (Cassandri et al., 2017). Around 3% of all human gene products contain one or more zinc-binding domains, which are structurally stabilized into a folded assembly by zinc ions (Klug, 2010). ZNFs are classified based on their domain structure, with each domain directly implicated in the regulation of various biological processes (Cassandri et al., 2017). For instance, C2H2-type ZNFs are mostly transcription factors and play significant roles in regulating gene expression, e.g., ZNF217 contains multiple C2H2 domains involved in repressing target gene expression (Nunez et al., 2011). RING-type ZNFs consist of various important E3-ubiquitin ligases, including Mouse Double Minute 2 (MDM2), which mediates ubiquitination of p53 (Linke et al., 2008). PHD-type ZNFs are mainly involved in the transcriptional regulation (Aasland et al., 1995). LIM-type ZNFs mediate target gene expression and cytoskeletal functions (Kadrmas & Beckerle, 2004). While the novel SWIM-type ZNFs are functionally uncharacterized, previous research has reported that the SWIM domain of mouse mitogen-activated protein kinase kinase 1 (MEKK1) is involved in protein-protein interactions and functions as a novel substrate receptor for c-Jun ubiquitylation (Rieger et al., 2012). Moreover, the SWIM domain of the Sws1 protein is involved in ATPase activity (Martín et al., 2006), further demonstrating the biological significance of the SWIM domain.
In this study, we conducted a genome-wide analysis of ZSWIM family members in four organisms, i.e., mice, Xenopus, zebrafish, and humans. We first identified total members of the ZSWIM family across all four organisms by searching various publicly available databases. In total, we identified nine ZSWIM genes in humans and mice (ZSWIM1–ZSWIM9), eight genes in Xenopus (zswim1, zswim3, zswim4, zswim5, zswim6, zswim7, zswim8, and zswim9), and five genes in zebrafish (zswim2, zswim5, zswim6, zswim7, and zswim8). We further confirmed the presence of the SWIM domain in all identified members using the Pfam database (Figure 1A; Supplementary Figure S1A–C) and identified various biochemical properties of each identified member (Supplementary Table S1). We constructed a phylogenetic tree using the NJ method to explore the evolutionary relationship among all identified ZSWIM members. Based on phylogenetic analysis, all ZSWIM family members were divided into three groups: Group I contained ZSWIM1, ZSWIM3, and ZSWIM9; Group II contained ZSWIM2 and ZSWIM5; and Group III contained ZSWIM4, ZSWIM5, ZSWIM6, and ZSWIM8 (Figure 1B). Moreover, syntenic analysis confirmed the phylogenetic pattern, suggesting that ZSWIM4, ZSWIM5, ZSWIM6, and ZSWIM8 were more evolutionarily conserved across the four species (Figure 1C). Next, we performed various bioinformatics analyses to explore the functions of the ZSWIM family members. Gene structure analysis showed that all identified members exhibited similar exon-intron organization (Supplementary Figure S4A–D). Multiple sequence alignment revealed that ZSWIM5, ZSWIM6, and ZSWIM8 shared the highest degree of similarity in protein structure across all four species (Supplementary Figure S2A). Motif sequence analysis indicated that ZSWIM4, ZSWIM5, ZSWIM6, and ZSWIM8 contained several conserved unknown motifs in their protein structure across all four species (Supplementary Figure S5S).
We also utilized publicly available RNA-seq data and examined tissue-specific expression of ZSWIM genes in various major human and Xenopus organs/tissues. Results revealed that each member showed the highest expression in specific organs/tissues, e.g., ZSWIM2 was only expressed in the human testis (Figure 2A). Xenopus also showed organ/tissue-specific expression patterns of zswim genes, e.g., zswim6 and zswim8 were highly expressed in the brain and zswim4 was highly expressed in the kidney and brain (Figure 2B). Additionally, we performed network analysis to identify the interacting partners of the ZSWIM family members. Interestingly, ZSWIM5, ZSWIM6, and ZSWIM8 were predicted to interact with each other (Supplementary Figure S2B).
Xenopus is considered an excellent model organism in developmental biology due to its easy breeding and maintenance (Dawid & Sargent, 1988). Therefore, we selected Xenopus to determine the contribution of each member to embryonic development. We first analyzed the temporal expression patterns of ZSWIM family members using qRT-PCR. Our results revealed that zswim1 and zswim3 were highly expressed from stages 2 to 6, with expression then declining at the onset of the gastrula stage. Both zswim5 and zswim6 showed similar expression patterns, with high expression in the neural and tailbud stages, gradually increasing from the neurula to tailbud stage. The zswim7 gene showed low expression throughout embryogenesis, while zswim8 and zswim9 were highly expressed during the late tailbud stages (Figure 3A–G). We also analyzed the spatial expression patterns using whole-mount in situ hybridization (Harland, 1991). Results showed that zswim1 and zswim3 were maternally expressed, suggesting essential roles during early embryonic development. The zswim5 and zswim6 genes exhibited highly discrete and dynamic expression patterns throughout embryogenesis, with both expressed in various major regions of the brain, including the forebrain, midbrain, and hindbrain. The zswim7 gene showed very faint expression in the brain region at the tailbud stage. Interestingly, zswim7 was expressed explicitly in the eye region. The zswim8 gene was expressed during the late tailbud stages, while zswim9 was faintly expressed throughout embryogenesis.
In summary, we comprehensively analyzed the ZSWIM family members in four well-characterized organisms. Phylogenetic and syntenic analyses provided insight into the evolution of ZSWIM family members. Expression pattern analysis using Xenopus embryos clearly demonstrated the involvement of each member in embryonic development and confirmed their tissue-specific function. This study provides a basis for future functional study of the zswim genes.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
I.U.H. performed the experiments, analyzed the data, prepared the figures, and wrote the manuscript. H.M.R. conducted bioinformatics analysis. Z.L. and L.Z. provided valuable suggestions. M.K.S. and C.W. revised the manuscript, Z.R., X.Q., and D.C. provided analytical tools. H.Z. conceived, designed, and supervised this study, wrote the manuscript, and approved the final draft. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Key R&D Program of China, Synthetic Biology Research (2019YFA0904500), and Research Grants Council of Hong Kong (14119120, 14112618, and CRF C5033-19E to H.Z.). Additional support was provided by the Hong Kong Branch of CAS Center for Excellence in Animal Evolution and Genetics, Chinese University of Hong Kong
References
- Aasland R, Gibson TJ, Stewart AF The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends in Biochemical Sciences. 1995;20(2):56–59. doi: 10.1016/S0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, et al The MEME suite. Nucleic Acids Research. 2015;43(W1):W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito MI, Walbot V Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Molecular and Cellular Biology. 1997;17(9):5165–5175. doi: 10.1128/MCB.17.9.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YP, Han YH, Meng DD, et al Structural, evolutionary, and functional analysis of the class III peroxidase gene family in Chinese pear (Pyrus bretschneideri) Frontiers in Plant Science. 2016;7:1874. doi: 10.3389/fpls.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassandri M, Smirnov A, Novelli F, et al Zinc-finger proteins in health and disease. Cell Death Discovery. 2017;3(1):17071. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close R, Toro S, Martial JA, et al. 2002. Expression of the zinc finger Egr1 gene during zebrafish embryonic development. Mechanisms of Development, 118(1–2): 269–272.
- Dawid IB, Sargent TD Xenopus laevis in developmental and molecular biology. Science. 1988;240(4858):1443–1448. doi: 10.1126/science.3287620. [DOI] [PubMed] [Google Scholar]
- Droll D, Minia I, Fadda A, et al Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 2013;9(4):e1003286. doi: 10.1371/journal.ppat.1003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, et al Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DMA, Barton GJ, et al Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Research. 2006;34(10):2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde N, Duffy GB, McGettigan PA, et al Evidence for an early endometrial response to pregnancy in cattle: both dependent upon and independent of interferon tau. Physiological Genomics. 2012;44(16):799–810. doi: 10.1152/physiolgenomics.00067.2012. [DOI] [PubMed] [Google Scholar]
- Gray KA, Yates B, Seal RL, et al Genenames. org: the HGNC resources in 2015. Nucleic Acids Research. 2015;43(D1):D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann C, Blank JL The ups and downs of MEK kinase interactions. Cellular Signalling. 2001;13(12):863–875. doi: 10.1016/S0898-6568(01)00220-0. [DOI] [PubMed] [Google Scholar]
- Harland RM Appendix G: in situ hybridization: an improved whole-mount method for Xenopus embryos. Methods in Cell Biology. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hershberger RJ, Benito MI, Hardeman KJ, et al Characterization of the major transcripts encoded by the regulatory MuDR transposable element of maize. Genetics. 1995;140(3):1087–1098. doi: 10.1093/genetics/140.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC The LIM domain: from the cytoskeleton to the nucleus. Nature Reviews Molecular Cell Biology. 2004;5(11):920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Klug A The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annual Review of Biochemistry. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Klug A, Rhodes D Zinc fingers: a novel protein fold for nucleic acid recognition. Cold Spring Harbor Symposia on Quantitative Biology. 1987;52:473–482. doi: 10.1101/SQB.1987.052.01.054. [DOI] [PubMed] [Google Scholar]
- Ko KK, Powell MS, Hogarth PM ZSWIM1: a novel biomarker in T helper cell differentiation. Immunology Letters. 2014;160(2):133–138. doi: 10.1016/j.imlet.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, et al Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death & Differentiation. 2008;15(5):841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
-
Livak KJ, Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the
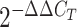 2-ddCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
2-ddCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar] - Makarova KS, Aravind L, Koonin EV SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends in Biochemical Sciences. 2002;27(8):384–386. doi: 10.1016/S0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- Martín V, Chahwan C, Gao H, et al Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. The EMBO Journal. 2006;25(11):2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercereau-Puijalon O, Barale JC, Bischoff E Three multigene families in Plasmodium parasites: facts and questions. International Journal for Parasitology. 2002;32(11):1323–1344. doi: 10.1016/S0020-7519(02)00111-X. [DOI] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. The EMBO Journal. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishito Y, Hasegawa M, Inohara N, et al MEX is a testis-specific E3 ubiquitin ligase that promotes death receptor-induced apoptosis. Biochemical Journal. 2006;396(3):411–417. doi: 10.1042/BJ20051814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez N, Clifton MMK, Funnell APW, et al The multi-zinc finger protein ZNF217 contacts DNA through a two-finger domain. Journal of Biological Chemistry. 2011;286(44):38190–38201. doi: 10.1074/jbc.M111.301234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Hidalgo O, Dodsworth S. et al Genome size diversity and its impact on the evolution of land plants. Genes. 2018;9(2):88. doi: 10.3390/genes9020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MD, Read KA, Sreekumar BK, et al Ikaros zinc finger transcription factors: regulators of cytokine signaling pathways and CD4+ T helper cell differentiation. Frontiers in Immunology. 2019;10:1299. doi: 10.3389/fimmu.2019.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger MA, Duellman T, Hooper C, et al The MEKK1 SWIM domain is a novel substrate receptor for c-Jun ubiquitylation. Biochemical Journal. 2012;445(3):431–439. doi: 10.1042/BJ20120406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CY, Kingston ER, Kleaveland B, et al The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science. 2020;370(6523):eabc9359. doi: 10.1126/science.abc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S MEGA11: molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SRF, Ousager LB, Miller KA, et al Acromelic frontonasal dysostosis and ZSWIM6 mutation: phenotypic spectrum and mosaicism. Clinical Genetics. 2016;90(3):270–275. doi: 10.1111/cge.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana KE, Churchill ME, Tullius TD, et al Mapping functional regions of transcription factor TFIIIA. Molecular and Cellular Biology. 1988;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Liu B, Ma YG, et al A novel SWIM domain protein ZSWIM5 inhibits the malignant progression of non-small-cell lung cancer. Cancer Management and Research. 2018;10:3245–3254. doi: 10.2147/CMAR.S174355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Jing Y, Qin Y, et al The zinc finger transcription factor ZKSCAN3 promotes prostate cancer cell migration. The International Journal of Biochemistry & Cell Biology. 2012;44(7):1166–1173. doi: 10.1016/j.biocel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.