Abstract
Despite the urgent need for conservation consideration, strategic action plans for the preservation of the Asian honeybee, Apis cerana Fabricius, 1793, remain lacking. Both the convergent and divergent adaptations of this widespread insect have led to confusing phenotypical traits and inconsistent infraspecific taxonomy. Unclear subspecies boundaries pose a significant challenge to honeybee conservation efforts, as it is difficult to effectively prioritize conservation targets without a clear understanding of subspecies identities. Here, we investigated genome variations in 362 worker bees representing almost all populations of mainland A. cerana to understand how evolution has shaped its population structure. Whole-genome single nucleotide polymorphisms (SNPs) based on nuclear sequences revealed eight putative subspecies, with all seven peripheral subspecies exhibiting mutually exclusive monophyly and distinct genetic divergence from the widespread central subspecies. Our results demonstrated that most classic morphological traits, including body size, were related to the climatic variables of the local habitats and did not reflect the true evolutionary history of the organism. Thus, such morphological traits were not suitable for subspecific delineation. Conversely, wing vein characters showed relative independence to the environment and supported the subspecies boundaries inferred from nuclear genomes. Mitochondrial phylogeny further indicated that the present subspecies structure was a result of multiple waves of population divergence from a common ancestor. Based on our findings, we propose that criteria for subspecies delineation should be based on evolutionary independence, trait distinction, and geographic isolation. We formally defined and described eight subspecies of mainland A. cerana. Elucidation of the evolutionary history and subspecies boundaries enables a customized conservation strategy for both widespread and endemic honeybee conservation units, guiding colony introduction and breeding.
Keywords: Apis cerana, Integrative taxonomy, Species concept, Pollinator insect, Centrifugal diversification, Morphology, Genomics
INTRODUCTION
The Asian honeybee (Apis cerana) is an important pollinator native to much of Asia (Figure 1A), providing crucial pollination services for agricultural production, forestry, biodiversity, and natural resource maintenance, supporting more than half of the human population (Abrol, 2013). However, the species faces numerous threats, including the application of pesticides, floral resource shortages, and habitat destruction and fragmentation (Abrol, 2013). In addition to these common challenges shared by most pollinators, the early introduction and extensive importation of the western honeybee (A. mellifera Linnaeus 1758) has resulted in direct colony replacements by beekeepers across a large range of the native bees’ habitat and introduced new pathogens such as the sacbrood virus (SBV) (Vung et al., 2018) and A. mellifera filamentous virus (AmFV) (Hou et al., 2016). Consequently, A. cerana populations have experienced a significant reduction in both distribution and size over the past century (Yang, 2009).
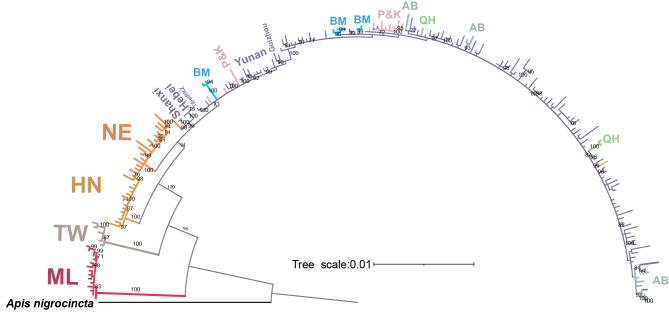
Figure 1.
Population structure, phylogenetic relationships, and genetic divergence of mainland Asian honeybee A. cerana
A: Native ranges of eight subspecies constituting the mainland lineage of A. cerana (indicated by dash lines), and Sundaland, Palawan, and Luzon-Mindanao lineages. Note, cavity-nesting bees of India (yellow) and Philippines (beige) have recently been recognized as outgroups to the common ancestor of Sundaland and mainland lineages (Lo et al., 2010). Dots represent sampling sites. B: Population structure based on nuclear SNPs and NetView. C: Phylogenetic tree based on nuclear SNPs, showing phylogenetic monophyly of seven peripheral descendant subspecies and Malaysian out-group. D: FST matrix constructed from estimates of pairwise FST between all eight subspecies of mainland Asian honeybee and Malaysian out-group. Codes for subspecies and geographic populations are: A. c. cerana (central group, CT), A. c. japonica (Northeast group, NE), A. c. guidensis (Qinghai group, QH), A. c. abansis (Aba group, AB), A. c. skorikovi (Bomi group, BM), A. c. kashmirensis (Pakistan and Kashmir group, K&P), A. c. hainana (Hainan group, HN), A. c. taiwanensis (Taiwan group, TW), and Malaysian out-group (ML).
Although the need for immediate conservation action for this species is widely acknowledged, strategic plans are rarely discussed. Prioritizing conservation efforts for species with restricted distribution (including most endangered species) may not present a challenge, but the same cannot be said for taxa with large geographic ranges (Phillimore & Owens, 2006). Notably, regarding native bee populations, cross-population breeding and bee farming practices are obvious issues (Wang et al., 2021). In China, the importation of exotic colonies from remote provinces to local villages to promote regional economies (Wang et al., 2021; Zhao, 2018) presents significant difficulties for assessing the potential impacts of human interference on local honeybees, given the lack of understanding of genealogy and fitness variations in different environments. Conservation actions require a clear definition of target species or subspecies and the differentiation of evolutionary factors from contemporary human disturbance. Although both may influence populations, the latter can be immediately ameliorated by implementing proper actions (Gutiérrez & Helgen, 2013; Morrison III et al., 2009; Requier et al., 2019).
The establishment of proper conservation units is an essential prerequisite for successful conservation. However, the infraspecific taxonomy of A. cerana remains enigmatic. Over the past two centuries, approximately 20 equivocal “subspecies” (Chen, 1993; Engel, 1999; Ilyasov et al., 2018, 2019; Lo et al., 2010; Ma & Huang, 1981; Maa, 1953; Radoszkowski, 1887; Smith & Hagen, 1996; Yang et al., 1986; Zhuang, 1989), 31 putative biometric groups (Hepburn et al., 2001b), and various ecotypes (National Animal Genetic Resources Committee, 2011) and morphoclusters (Radloff et al., 2010) have been proposed. In particular, taxonomists hold divergent views at the subspecies level, ranging from the recognition of eight subspecies (Engel, 1999) to the identification of only six morphoclusters across Asia (Radloff et al., 2010). Recently, based on whole-genome single nucleotide polymorphisms (SNPs) of representative populations, we revealed the presence of seven genetically distinctive groups within mainland A. cerana (the only group distributed across tropical and temperate regions), showing genomic divergence at the subspecies level (Ji et al., 2020). The reasons for this long-lasting debate are multifaceted (Engel, 1999; Hepburn et al., 2001b; Peng et al., 1989; Radloff et al., 2010), but include discrepancies in sampling strategy, conflicting results produced by varied characteristics (e.g., morphology and genetics), and innate challenges in achieving a uniform subspecies concept (De Queiroz, 2007, 2020).
Geographic coverage of population sampling can impact the representativeness of genealogy and the subsequent interpretation of results. Broad sampling of a wide-ranging species is inevitably challenging, and key lineages many remain unsampled. For instance, in various studies conducted in the Far East, the inferences of A. cerana colonization routes between mainland China and Japan varied with the inclusion of new samples (Ilyasov et al., 2018, 2019). Incomplete sampling seems to have at least partially contributed to previous designations of invalid subspecies delineations. For example, three A. cerana subspecies and one endemic type identified in local studies in the Far East are now hypothesized to belong to the same subspecies (Ilyasov et al., 2018, 2019; Ji et al., 2020; Liu et al., 2022; National Animal Genetic Resources Committee, 2011).
The continued emergence of new characters and evolution of new methodologies have added yet another level of complexity to subspecies delineation. In particular, the application of novel molecular techniques has cast doubt upon prior studies. For example, recent genomic analyses have largely redefined the distribution of the Aba race of A. cerana in montane regions (Chen et al., 2018a; Ji et al., 2020). Once thought to span a large transition zone from the Sichuan basin to the Qinghai-Xizang (Tibet) Plateau (QTP) (central and northern Sichuan, eastern Qinghai, and southern Gansu) (Peng et al., 1989; National Animal Genetic Resources Committee, 2011; Yang et al., 1995), it is now thought to be restricted to the upper reaches of the Dadu River. This substantial difference in range could strongly influence the efficacy of conservation strategies, as an action plan based on the larger hypothesized distribution range may result in no management efforts in the smaller area where these honeybees actually inhabit. However, the lack of criteria for judging which set of characters (e.g., morphology vs. genetics) is more reliable remains a dilemma.
The inconsistent application of subspecies definitions also represents a substantial conceptual challenge. Understanding species boundaries is essential for answering questions related to evolution, adaptation, conservation, and ecology (Christmas et al., 2021; De Queiroz, 2007; Gutiérrez & Helgen, 2013; Hillis, 2019; Morrison III et al., 2009). However, subspecies are relatively less regulated and are not explicitly covered by the International Code of Zoological Nomenclature. Despite considerable species definitions (Groves et al., 2017), multiple lines of evidences have been applied to infer evolutionary or reproductive isolation and subsequent completion of genetic sorting between sister taxon pairs that share a common ancestor. However, a central challenge is the need to determine isolation, which is difficult to implement for all but the best-known species (Malinsky et al., 2015). Because subspecies reflect an intermediate evolutionary phase of a focal species, they are transient units on the verge of diverging into new incipient species, or alternatively, being replaced by other subspecies, younger descendants, or their hybrids (De Queiroz, 2020). Subsequently, given the lower levels of divergence expected among subspecies, the integration of multiple forms of data (e.g., molecular, morphology) is expected to produce the best results (Guillot et al., 2012).
In accordance with the evolutionary model established in our previous work (Ji et al., 2020), we herein formally describe eight mainland A. cerana subspecies. Compared with Ji et al. (2020), we focused on taxonomic delineations and descriptions of the proposed subspecies, with the designation of type specimens for each newly described subspecies and for all previously defined subspecies lacking types. Our streamlined pipeline integrated genetic divergence, phylogenetic relationships, and morphology to test the hypothesis that genetically and morphologically distinct population groups revealed within mainland A. cerana are valid subspecies. We demonstrated that multiple independent divergence events may have shaped the current population structure, resulting in a total of eight subspecies, three of which are described for the first time. In addition to clarifying conservation units, we also explored the unique threats faced by these subspecies, highlighting their evolutionary histories and genetic features, and providing important information for strategizing and enacting novel conservation priorities.
MATERIALS AND METHODS
Genome sequencing
Whole-genome sequences of 306 worker bee samples were obtained from our previous study (Ji et al., 2020, NCBI archive PRJNA592293). Additionally, 56 worker bees collected from Pakistan, Kashmir, Myanmar, Malaysia, and China (Supplementary Table S1) were newly sequenced in this study. The combined data covered all eight provisional subspecies of the mainland Asian honeybee (with A. cerana indica recently recognized as an outgroup to the common ancestor of the Sundaland and mainland lineages) (Lo et al., 2010). The 56 new samples were collected on flowers, and the head and thorax were then processed for DNA extraction using phenol chloroform. Genomic DNA was sequenced on the BGISEQ-500 platform at the BGI, China (3.5 to 5 Gb per sample). The newly sequenced data were deposited in the Sequence Read Archive database (PRJNA870246).
We applied Fastp v0.13.1 (Chen et al., 2018b) to remove low-quality reads with the parameters “-q 10 -n 10 -u 40.”. The clean reads of each sample were mapped to the genome assembly of A. cerana (Ref ID: ACSNU-2.0, GCF_001442555.1) using BWA ALN (Li & Durbin, 2009) with polymerase chain reaction (PCR) duplications being masked using SamBamba (Tarasov et al., 2015). We then applied the Genome Analysis Toolkit v4.0.4 (McKenna et al., 2010) for short variant identification and retained biallelic SNP sites that met the following criteria: (1) average depth >1/3 and <2× mean depth of the whole dataset; (2) quality score>20; (3) genotype quality>30; (4) minor allele frequency>0.01; (5) proportion of missing genotypes<50%. Genome heterozygosity and allelic missing rate were estimated for each individual using VCFtools (Danecek et al., 2011).
Population structure and phylogeny
We inferred kinship relationships between individual pairs using VCFtools (Danecek et al., 2011) following Manichaikul et al. (2010) and only retained unrelated individuals for downstream analyses. Individuals with >10% missing data were also removed. Finally, a total of 273 worker bees (75.6%) were retained for subsequent analyses (Supplementary Table S1). We applied a balanced minimum evolution method implemented in Fastme (Lefort et al., 2015) for phylogenetic tree construction based on a whole-genome identical-by-state (IBS) matrix calculated using Plink v1.9 (Purcell et al., 2007). Pairwise phylogenetic distances (PD) between different individuals were calculated using an in-house Perl script. The IBS matrix was also used for genetic structure estimation with a mutual k-nearest neighbor graph method implemented in NetView (Neuditschko et al., 2012).
We performed admixture analyses of bee populations from Hainan (HN), Bomi (BM), Northeast (NE), Qinghai (QH), Taiwan (TW), and Pakistan & Kashmir (KP). For each bee population, we created a dataset that respectively included: (1) all individuals from the focal locality; (2) individuals from Malaysia as an outgroup; (3) individuals from adjacent regions that belong to the central population according to Ji et al. (2020) (Supplementary Table S1).
We estimated the admixture level for each group using NGSAdmix (Skotte et al., 2013) based on the genotype likelihood calculated using ANGSD (Korneliussen et al., 2014) with the following parameters: base quality≥20, mapping quality≥20, minor allele frequency (MAF)≥0.05, SNP P-value≥1e-6, and informative individuals≥10. For admixture analysis, we applied a predefined K value of 3 and obtained a convergent estimation when the three highest likelihood values fell within the range of five likelihood units.
Mitochondrial phylogeny
From the genome sequences of each individual honeybee, we obtained a consensus sequence for the mitochondrial genome using the doFasta function imbedded in ANGSD (Korneliussen et al., 2014), requiring a depth ≥5X. We then obtained all 13 protein coding genes (PCGs) for each sample based on the reference annotation information (NC_014295.1). In addition, the mitochondrial genome of A. nigrocincta (NC_038114.1) was applied to root the tree. After sequence alignment of the 13 PSGs using MAFFT (Nakamura et al., 2018), we concatenated the alignments to generate a “supergene” and inferred a phylogenetic tree using IQTREE (Minh et al., 2020) with the best-fit substitution models estimated by ModelFinder (Kalyaanamoorthy et al., 2017). To present a simplified view for better illustration, we included only a single representative for each unique haplotype from the same sampling location.
Morphological analyses
The honeybee worker specimens used for morphological analyses were obtained from two sources: (1) A total of 119 specimens collected from seven peripheral provisional subspecies population throughout China, India, and Japan from 2010 to 2021. (2) A total of 207 colonies from 101 localities, covering much of the distribution of the central population, recorded in the Kunming Honeybee Database (Tan et al., 2006). The geographical origins of all samples representing 234 colonies from 115 localities are listed in Supplementary Table S2.
Morphological measurements
The morphological traits described in Ruttner (1988), as well as two additional effective characters (cubital index and hamuli number), were employed to characterize A. cerana. Overall, the 35 morphometric characters included 12 relevant to size, 16 relevant to forewing, five relevant to color, one relevant to hair, and one relevant to hamuli (Tan et al., 2008).
Morphometric analysis
We first assessed differences among different datasets, i.e., legacy data obtained from Tan et al. (2006) and the Morphological Bank and new data obtained from the current study. We then performed independent principal component analysis (PCA) for size-, wing-, and color-related traits to remove traits showing apparent divergences across data sources or outliner samples from each data source. Subsequently, we used the filtered trait set to conduct cluster analysis using samples from Hainan Island and Northeast China/Korea/Japan populations, for which overlapping sampling is found between legacy and new data. We divided the focal populations into eight provisional subspecies, with the central subspecies (i.e., A. cerana cerana) further divided into three sub-clusters (Northern cerana, Indo Chinese cerana and Himalayan cerana), based on combined genomic (Ji et al., 2020) and morphological evidence (Radloff et al., 2010). Mean values and standard deviations were computed for each character for all colonies using the morphometric Image Analysis System 11 program. The “princomp” function in R was used for PCA to identify possible morphoclusters. Morphometric similarities were investigated using hierarchical cluster analyses based on location means of Chinese populations and group means of adjacent country populations.
Bioclimatic analysis
We extracted bioclimatic variables for all sampling sites using DIVA-GIS (www.diva-gis.org). We performed PCA using the “princomp” function in R to obtain the first two principal components. Correlations between Dim 1–5 scores of PCA for morphological characteristics and bioclimatic variables, including altitude, latitude, longitude, and Bio1–19, were calculated using the R package “corrplot”. *: P<0.05; **: P<0.01; ***: P<0.001.
Geometric morphometrics
A total of 221 forewings from A. cerana worker bees collected from 24 sites (see Supplementary Table S3) were used for geometric morphometric analysis. All wing specimens were photographed using a Nikon D700 camera (Japan). These images were then used in subsequent morphological analyses. A total of 20 forewing landmarks were identified following our previous work (Ji et al., 2020). All landmarks were digitized using tpsDig v2.0 and tpsUtil v1.40 (http://life.bio.sunysb.edu/morph/). We divided the 24 sampling sites into eight groups following our subspecies definitions. Size-corrected shape was used to conduct canonical variable analysis (CVA) in Morphoj v1.06 to determine the degree of morphological discrimination among subspecies. Mahalanobis distances (MDs) among groups and P-values from permutation tests (10 000 iterations) were calculated during CVA. The thin plate spline (TPS) method in Morphoj v1.06 (Klingenberg, 2011) was used to identify morphological differences in wing shape between each pair of subspecies. We estimated morphological divergences between peripheral and central subspecies using MD. We calculated the correlation efficiency between fixation index (FST) and MD among groups to evaluate the level of coherence between genetic and morphological divergences. All calculations were performed using SPSS v20.0.
RESULTS
Provisional subspecies suggested by nuclear genetic divergence, phylogenetic relationships, and evolutionary history
Phylogenetic relationships based on nuclear genomes (nrDNA) showed that, in congruence with our previous findings (Ji et al., 2020), the six peripheral population groups (i.e., Hainan, Taiwan, Northeast, Bomi, Aba, and Qinghai) and the out-group (Malaysia) (Figure 1A) were genetically distinct from the central group, each forming a monophyletic lineage (Figure 1B). In addition, the newly sampled population from Pakistan and Kashmir (P&K) formed a new monophyletic group, albeit at a relatively lower divergence level from the central group compared to other peripheral groups (Figure 1A–C; Supplementary Figure S1). Thus, all genetic divergence values between the central and peripheral groups (FST=0.135±0.06, Figure 1D) were comparable to subspecies variations found in A. mellifera (FST=0.163±0.073 (Dogantzis et al., 2021)). With reference to historical taxonomic studies on relevant geographic populations of mainland A. cerana, we defined these genetically distinct groups as provisional subspecies: A. cerana cerana (central group), A. c. japonica (Northeast group), A. c. guidensis (Qinghai group), A. c. abansis (Aba group), A. c. skorikovi (Bomi group), A. c. kashmirensis (Pakistan and Kashmir group), A. c. hainana (Hainan group), and A. c. taiwanensis (Taiwan group). The taxonomic considerations will be discussed in the subsequent subspecies section.
Although our results suggested clear groupings, genetic introgression has occurred frequently between the proposed peripheral and central subspecies. For example, individuals from the Liaoning population showed a much closer phylogenetic relationship to the central subspecies compared to the remaining Northeast samples (Supplementary Figure S1), with admixture analysis suggesting it may be a hybridized population between the central and Northeast subspecies (Supplementary Figure S2). Similarly, all but the Kashmir population contained hybridized individuals, suggesting recent independent genetic introgressions between peripheral and central (ancestral) subspecies (Supplementary Figures S1–S3). Although no obvious hybridization was detected in the Kashmir population, this could be attributed to insufficient sampling in intervening areas where interbreeding is more likely.
Mitochondrial phylogeny indicates hidden history of multiple radiation waves
Unlike nuclear phylogeny, where the aforementioned genetic subspecies all split from the common central subspecies (Figure 1C), mitochondrial genomes revealed a temporal order among subspecies divisions (Figure 2; Supplementary Figure S4). These differences in phylogenetic structure are likely due to variations in evolutionary rates between nuclear and mitochondrial genomes, as mitochondrial genes are known to complete lineage sorting more rapidly (Barr et al., 2005; Li et al., 2017). Alternatively, genetic drift associated with recent population bottlenecks may have led to a similar pattern (Larson et al., 2012). Within the mainland lineage, A. c. taiwanensis (TW) and A. c. hainana (HN) diverged consecutively, following the split of the mainland lineage from the ML outgroup (Figure 2). Each of these early divergent subspecies formed a separate monophyletic group in the mitochondrial phylogeny, mirroring the nuclear genome results (Supplementary Figure S5). Following these early divisions, three clades formed a trichotomy in the mitochondrial phylogeny – one representing the A. c. japonica (NE) subspecies; one including populations from Hebei, Shanxi, and Liaoning; and one including the four remaining peripheral subspecies, A. c. kashmirensis (KP), A. c. skorikovi (BM), A. c. abansis (AB), and A. c. guidensis (QH), and geographic populations of A. c. cerana (CT). The Liaoning samples from the second clade showed mitochondrial introgression between A. c. japonica and the northern China populations (A. c. cerana) (Figure 2), in congruence with findings based on nuclear genomes, thus confirming hybridization. The third clade contained multiple peripheral subspecies and geographic populations of A. c. cerana, none of which showed mutual monophyly, reflecting recent divergence history and ongoing hybridization (Supplementary Figure S6). A more detailed tree structure is shown in Supplementary Figure S4. Overall, the mitochondrial phylogeny indicated that the current population structure of mainland A. cerana resulted from multiple waves of divergence across evolutionary history.
Figure 2.
Phylogenetic relationships of subspecies of mainland A. cerana and Malaysian lineage based on mitochondrial genomes
Maximum-likelihood phylogeny was rooted using A. nigrocincta. Internodes with support values >70 are marked with numbers. Color codes follow those of Figure 1.
Subspecies delineation is supported by morphological characters
We compared morphological traits among the three different datasets (Supplementary Figure S7A–D) and excluded all five color-related traits from subsequent analyses due to obvious divergences across datasets (Supplementary Figure S7C). In addition, 23 outliner samples were identified in the size- and wing-related traits and were also excluded from subsequent analyses (Supplementary Figure S7E, F). The 30 remaining traits were employed to cluster samples from the HN and NE populations, where new samples nested readily within subspecies boundaries delineated by legacy samples, showing consistency in subspecies delineation in the different morphological datasets (Supplementary Figure S7G). Thus, we employed the 30 morphological characters from 211 colonies for downstream analyses. Morphological differentiations among A. cerana lineages and provisional subspecies of the mainland lineage were correlated with climatic characters in the local habitats. Based on hierarchical cluster analysis and PCA, the mainland Asian honeybee could be divided into two large morphological clusters, corresponding to tropical and temperate groups. The tropical bees (Indo Chinese cerana, A. c. hainana, and A. c. taiwanensis) formed a compact morphological cluster distinctively separate from those of mainland China, Korea, Japan, and Pakistan & Kashmir (Figures 3A, B; Supplementary Figure S8A). Notably, the Malaysian out-group, the sister lineage to all mainland A. cerana, was also clustered with the tropical mainland populations based on hierarchical cluster analysis. PCA showed that PC1 was mostly influenced by body size characters (Figure 3B; Supplementary Figure S8B), explaining 39.6% of the differentiation between the two morphological clusters. The size division between tropical and temperate bees was further supported by analysis using wing length characters only (Supplementary Figure S8C).
Figure 3.
Morphological differentiation among provisional subspecies of mainland Asian honeybee
A: Hierarchical cluster analysis using ward linkage method with location means of morphometric characters. B: Principal component plot of mainland Asian honeybee based on 30 morphometric characters. C: Correlations among factor analysis Dim 1–5 scores and environmental variables. *: P<0.05; **: P<0.01; ***: P<0.001. Dim 1–5 represents the first five factors in PCA for traditional morphological data. Bio1–19 represents 19 bioclimatic variables (specific meanings can be found at https://www.worldclim.com/bioclim). D: Scatterplot of CV1 vs. CV2 for wing shape among eight subspecies of mainland A. cerana and Malaysian lineage. Ellipses indicate 95% confidence intervals. Color codes in A, B, and D follow that of Figure 1. Circle, square, and triangle in purply blue color represent Northern, Himalayan, and Indo-Chinese A. c. cerana, respectively.
Similarly, within both tropical and temperate groups, morphological affinity among provisional subspecies and geographic populations was also correlated with local climatic parameters, where peripheral subspecies inhabiting similar habitats showed apparent morphological convergence (Supplementary Figure S8D). Further analyses of the correlations between Dim 1–5 scores and environmental factors indicated that Dim 1 (related characters, e.g., hair length and appendage size; Supplementary Figure S8B) was highly correlated with temperature-related environmental factors (Figure 3C; Supplementary Figure S8B). These results suggest that conventional morphological characteristics were largely influenced by geography and microclimate.
Our results also showed that wing vein angles exhibited relatively weak associations with environmental factors (Dim 2 and Dim 3 in Figure 3C, Supplementary Figure S8B). Thus, wing morphological geometrics were applied to validate morphological differences among the eight provisional subspecies defined by nuclear genomic divergences and evolutionary history. All eight provisional subspecies and the Malaysian out-group showed consistent morphological clustering based on wing geometric morphology (Figure 3D), and P-values for MD among groups were less than 0.001 (Supplementary Table S4), supporting the subspecies delineations. We discovered a significant correlation between FST (Figure 1D) and MD (Supplementary Table S4) (R=0.44, P=0.008), indicating a consistent relationship between genetic and morphological differentiations among groups (Supplementary Figure S8E). In addition, pairwise MDs between peripheral and central subspecies (4.21±0.75) were significantly lower than those between peripheral subspecies (5.49±0.73) (P<0.001, two-tailed Mann-Whitney test). The same pattern was observed for FST estimations (0.135±0.06 vs. 0.28±0.08, P<0.001, two-tailed Mann-Whitney test). These findings indicate that the central subspecies was the closest lineage to each of the seven peripheral subspecies, as supported by both molecular- and morphology-based evidence. The wing shape groups with positive and negative loadings on CV1 and CV2 are shown on Supplementary Figure S8F.
Subspecies descriptions and taxonomic annotations on mainland A. cerana
Based on convergent evidence from morphology, nuclear genetic divergence, evolutionary history, and phylogenetic relationships, we herein formally define eight subspecies for the mainland A. cerana lineage: i.e., central subspecies A. c. cerana and seven peripheral subspecies, A. c. taiwanensis, A. c. hainana, A. c. kashmirensis, A. c. skorikovi, A. c. abansis, A. c. guidensis, and A. c. japonica. A revision of the subspecies taxonomy of mainland A. cerana, including descriptions of three newly defined subspecies, is provided in Supplementary Text. The holotypes of A. c. taiwanensis, A. c. guidensis, and A. c. kashmirensis and neotypes of A. c. hainana and A. c. abansis, as well as locations of four highland subspecies, are shown in Supplementary Figures S9–11. Morphometric character measurements of worker bees of the eight A. cerana subspecies are provided in Supplementary Table S5.
DISCUSSION
Subspecies of mainland A. cerana
In the present study, we analyzed 362 A. cerana specimens, representing almost all distinct groups of mainland A. cerana known to date. Our results demonstrated that all previously identified genetically distinct groups (i.e., six peripheral groups – NE, QH, AB, BM, HN, and TW) and the newly added KP group each formed a monophyletic group based on nuclear SNP phylogeny. In addition, all peripheral groups showed notable genetic divergences from the central group, while the latter formed a paraphyly intermingled with all peripheral groups (Figure 1C). In this new analysis, we only retained non-kin individuals to better characterize intra- and inter-population genetic features. With this stringent sampling scheme, genetic divergence between populations was expected to decrease, explaining the slight reduction in FST between the QH and CT groups compared with our previous work (0.09 and 0.10 in current study and Ji et al. (2020), respectively). Even so, the genetic divergence values detected between pairs of mainland A. cerana subspecies (FST range 0.07–0.26, between each peripheral subspecies and central subspecies, Figure 1D) were on par with those of A. mellifera subspecies (0.038–0.263) (Dogantzis et al., 2021). In addition, both phylogenetic distances from the current study (Supplementary Figure S1) and FST estimates from our previous work (Ji et al., 2020, Supplementary Figure S6B) revealed limited differentiation within the central group, even among distantly located geographic populations. Therefore, we regarded all central group populations as a single subspecies. Based on phylogenetic monophyly and divergence values, we propose that these genetically distinct groups are putative subspecies. The taxonomic boundaries were further supported by morphometrics using forewing characters, where genomic and morphological results showed consistent differentiations among the proposed subspecies (Figure 3D).
Mitochondrial phylogeny further indicated that the current population structure of mainland A. cerana was the result of multiple waves of divergence. This newly discovered temporal sequence is an important addition to the centrifugal diversification model (Ji et al., 2020). Island subspecies represented by A. c. taiwanensis and A. c. hainana were among the first diverging sublineages (Figure 2). The relatively long divergence time (inferred by basal phylogenetic positions) and ocean isolation may explain the genetic distinctiveness of the island subspecies. Alternatively, the observed genetic distinctiveness and basal phylogenetic placement may have resulted from recent genetic drift following population bottlenecks (Larson et al., 2012). Distinguishing between these two evolutionary models requires further investigation, with direct evidence from historical honeybee samples. The A. c. japonica subspecies was likely the first continentally distributed group to separate from the common ancestor (Figure 2). It remains unclear how this subspecies maintained genetic isolation from current A. c. cerana in the absence of obvious geographic barriers. Despite the presence of hybrid individuals identified in the Liaoning region, genetic introgression from the central subspecies was not prevalent in A. c. japonica. Genomic incompatibility (e.g., structural variations) and differing fitness (e.g., cold hardiness (Liu et al., 2022)) are among the possible mechanisms remaining to be tested. Finally, the most recent range expansion(s) led to parallel invasions of the central ancestor into the peripheral valleys of the QTP (Figure 2). Formed by continuous mountain formation events in the Himalayans, these deep valleys are characterized by abrupt vertical transitions in vegetation, with the tree line confined to mid-hill elevations (Wang et al., 2022). As a cavity-nesting honeybee, A. cerana relies on tree hollows to build nests. Therefore, the barren ridges defining these valleys may have effectively served as terrestrial barriers, preventing local honeybee populations from cross-valley migration and direct interbreeding. Compared to the three species that diverged earlier, the four QTP valley subspecies (A. c. guidensis, A. c. abansis, A. c. skorikovi, and A. c. kashmirensis) showed elevated levels of admixture with A. c. cerana. Both the relatively young divergence time and lack of absolute geographic isolation (lasting secondary contact with A. c. cerana at the valley opening) are likely reasons for their relatively low genetic divergences from the central subspecies (Figure 1D), whereas ongoing hybridization and retention of ancestral polymorphisms likely explain the paraphylies based on mitochondrial phylogeny (Figure 2). Individuals from QH (A. c. guidensis), AB (A. c. abansis), and BM (A. c. skorikovi) contained mitochondrial haplotypes derived from A. c. cerana (Supplementary Figure S6) and the QH and AB populations exhibited elevated genome-wide heterozygosity rates (Supplementary Figure S3), indicating hybridization with A. c. cerana. Conversely, individuals from BM (A. c. skorikovi) demonstrated no clear signs of mitochondrial introgression (Supplementary Figure S6, this study), but exhibited a lower genome-wide heterozygosity rate (Supplementary Figure S3), suggesting that retention of ancestral polymorphism is the most likely evolutionary cause of mitonuclear discordance, although additional sampling is needed for confirmation.
Although the divergence order of these young subspecies remains unknown, their evolutionary independence not only helps clarify their genetic boundaries but also the constraining factors that determine their distributional ranges. For the QTP valley subspecies, understanding how valley isolation has shaped their formation provides critical clues for drawing fine distributional borders. Additionally, a global understanding of the phylogenetic relationship based on comprehensive sampling is essential for revealing the genealogy of local populations. For example, in a recent study, the close phylogenetic relationship between A. cerana samples collected in the plains region of northern Pakistan was interpreted as the result of human introduction (Tan et al., 2021). However, our new sampling scheme covering major geographic populations of mainland A. cerana indicated an alternative hypothesis, namely, that the Pakistani populations were part of the widely distributed A. c. cerana. Further sampling of A. cerana populations in South Asian countries will better clarify the boundaries among A. c. kashmirensis, A. c. cerana, A. c. skorikovi, and A. c. indica. In addition, given the wide distribution range of A. cerana in Asia, we anticipate that new subspecies may be discovered in future research, especially in regions not included in the current study, such as deep QTP valleys, central and southern India, and Sri Lanka (Oldroyd et al., 2006; Shanas et al., 2022).
Integrative taxonomy of honeybee subspecies
Subspecies are taxonomic assignments used to classify evolutionary units showing evolutionary independence and genetic/morphological distinctiveness from other such units (Braby et al., 2012). Our study showed that the A. cerana subspecies resulted from multiple divergence events, representing transient evolutionary units along the temporal spectrum of speciation. Thus, all characters used to delineate subspecies are dependent on the divergence time of the focal population from its ancestor and subsequent interactions (e.g., hybridization) with other evolutionary units. However, it remains challenging, and in many cases impossible, to identify a fixed level of variation for any given character, including morphology and genetics.
This issue became evident when quantifying genetic divergence and completeness of lineage sorting among the A. cerana subspecies. Notably, the early diverging subspecies (A. c. taiwanensis, A. c. hainana, and A. c. japonica) exhibited larger genetic divergences and exclusive monophylies in both the nuclear and mitochondrial trees. In contrast, the younger QTP valley subspecies experienced more intensive and recent introgression events from A. c. cerana, resulting in lower genetic divergences and paraphylies in the mitochondrial tree (Figures 1, 2). Therefore, for these later diverging subspecies, subspecies delineation based on genetic divergence or monophyly alone will require arbitrary thresholds. Moreover, the level of genomic variation observed in the honeybee populations resulted from multiple factors, including genomic divergence due to historical division and isolation, natural selection, ongoing introgression, and anthropogenic influences. Clarification of the relative contributions of these factors is necessary to understand the extent to which observed genetic features are a consequence of evolutionary isolation.
Morphological subspecies delimitation faces similar challenges. Our study demonstrated that honeybee morphology, including characters typically employed in differentiating subspecies (e.g., body size), was strongly related to environmental features, in congruence with findings from regional studies (Tan et al., 2003). Such a pattern has created major challenges in the application of morphological characters in defining honeybee subspecies. This issue is particularly common for species with large geographic ranges, such as A. cerana, where geographic populations inhabit and have adapted to highly heterogenous environments (Hepburn et al., 2001a; Tan et al., 2006). In some cases, a wide-ranging lineage may retain relatively distinctive characteristics despite experiencing continuous within-group genetic exchange, such as in A. c. cerana. On the other hand, distantly related (and genetically distinct) populations living under similar climatic conditions may show convergent traits. For example, several independent montane subspecies of A. cerana share a darkened body coloration and enlarged body size, whereas tropical populations are almost all lighter in color and smaller in size. In these cases, morphological characters alone would incorrectly group distant lineages as the same or close related subspecies. Thus, morphological traits influenced by the environment should not be used in subspecies delineation, as these traits cannot correctly trace underlying evolutionary history.
The delineation of species and subspecies is integral to conservation because they are the units most used for conservation management. However, taxonomic research is always evolving, based on continuous effort in formulating working hypotheses and ever-improving species/subspecies definitions (Orr et al., 2021). This process is built upon many pioneering studies, with the goal of understanding the taxonomic boundaries and evolutionary mechanisms that have led to the biodiversity we see today. How we do this has immense ramifications for conservation and science. Therefore, we propose that multiple lines of evidence, including, but not limited to, morphology, genetics, biology, and ecology, should be employed to determine subspecies boundaries. This practice should abide by the general rules of taxonomy (e.g., establishment and curation of type specimens) and data sharing in public domains to better enable subsequent studies to leverage and synthesize data.
To ensure best practices and facilitate replicability and comparability, we propose that honeybee subspecies must meet the following criteria: (1) Different subspecies should have undergone independent evolutionary histories supported by integrative evidence of monophyly, preferably including molecular data. Given the difference in inheritance patterns between nuclear and mitochondrial genes, discordance in phylogenetic history is frequently revealed by nuclear and mitochondrial data (Toews & Brelsford, 2012), especially in young and radiating species (Shaw, 2002). Here, we used nuclear genome sequences as the primary data to better reconstruct the evolutionary history of A. cerana populations. Additionally, we employed mitochondrial data to specify evolutionary mechanisms causing genetic admixture among populations, such as hybridization and retention of ancestral polymorphism (Garnery et al., 1992; Hausdorf et al., 2011; Rodríguez et al., 2010); (2) Each subspecies should be confined to a distribution that is largely disjunct from other such lineages; and (3) Subspecies should be diagnosable based on a suite of combined characteristics, as well as molecular, morphological, and other types of evidence.
Conservation of mainland A. cerana
From an evolutionary perspective, delineating honeybee subspecies necessitates determining the relative contributions of evolutionary and contemporary events to the current status of a focal population. Such an endeavor perfectly aligns with conservation initiatives, where the challenges posed to the studied organism lie in both historical and ongoing processes.
We identified seven peripheral subspecies within the mainland A. cerana lineage, each with an independent history and constrained to their current ranges by various geographical and biological features. Compared with the widely distributed A. c. cerana subspecies, these isolated subspecies exhibited distinct genetic differentiation and restricted habitat ranges, indicating that each should be treated as an independent conservation unit. Clarification of subspecies boundaries is crucial in prioritizing conservation efforts. Once these target units are defined, further dissection of potential historical and contemporary threats can help to identify conservation priorities, in line with the concept of evolutionarily significant units (ESUs) (Ryder, 1986), which have been extensively discussed in the context of conservation biology (Casacci et al., 2014; De Guia & Saitoh, 2007; Dogantzis & Zayed, 2019; Requier et al., 2019).
Conservation strategies for peripheral subspecies should aim to maintain their integrity, in line with their parallel evolutionary history and isolated distributions. For example, A. c. taiwanensis and A. c. hainana likely formed via geographical isolation. On concern, elevated hybridization with A. c. cerana has been found in the Hainan subspecies, in concordance with extensive colony introductions from the Guangxi and Guangdong provinces in the 1990s (Zhao et al., 2017). This has had a significant influence on the current genetic structure of the Hainan subspecies, highlighting the need for immediate intervention and prohibition of colony introductions from the mainland to prevent further genetic degradation.
Conversely, geographic populations belonging to the central subspecies A. c. cerana showed much more homogenized genetic structure due to long-term introgression. Major geographic barriers, including the Qinling Mountains, which define the western boundary between the Palearctic and Oriental Realms, showed limited effects in preventing introgression among geographic populations within A. c. cerana. Therefore, a primary conservation goal for this subspecies should include prevention of population compartmentalization caused by anthropogenic activities, such as urbanization, habitat fragmentation, uniform crop plantation, and replacement with introduced honeybee species such as A. mellifera (Abrol, 2013; Yang, 2009). Furthermore, the knowledge that honeybee subspecies have independently evolved and adapted to their current habitats (Ji et al., 2020) warrants increased caution in honeybee cross-breeding. For example, queens of montane (e.g., A. c. guidensis and A. c. abansis) and northern subspecies (A. c. japonica) and races are often introduced to queenless tropical colonies to improve honey production (Wang et al., 2021). Hybridized progeny often show rapid trait regression, leading to the persistent importation of even more exogenous queens. Hybridization regression is unsurprising considering that local subspecies are adapted to specific tropical or montane environments. However, to what extent these anthropogenic activities are changing the genetic structure of the target population remains largely untested. Unlike in the western honeybee A. mellifera, most A. cerana breeding practices have been conducted within the native range of the species. Understanding how these activities may alter the genetics and fitness of endemic honeybees is crucial for their future survival. The clear delineation of conservation targets provides a pathway to understand ongoing impacts and track long-term influences, enabling more effective management and the preservation of these unique honeybees.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Xin Z. and S.L. designed the study. L.Q. performed morphometrics analysis. S.L. conducted population genomics and mitochondrial analyses. J.D. assisted with sequencing data analysis. S.H.P. and A.M. acquired Indian specimens. Q.N., T.J., X.L., and Xin Z. organized sampling. X.L. provided samples from Northeast China and Japan. M.O., C.Z., X Zhang., Xuguo Z., and S. Luo assisted in manuscript writing. K.T. provided morphological data for a series of geographic populations. Xin Z., S.L., and L.Q. wrote the first drafts, and all authors contributed to and proof-read the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We gratefully acknowledge sample contributions from many collaborators, including Hu Li, Syed I.A. Shah, Qingtao Wu, Zeqing Niu, Dan Zhang, Jinglin Gao, Yihai Zhong, Jiahui Hu, Zhenhua Chen, and Jialong Huang, among many others. We thank Guanhuang Yang, Chao Chen, and Wei Shi from the Institute of Apicultural Research of the Chinese Academy of Agricultural Sciences for help in tracing type specimens of A. c. hainana and A. c. abansis subspecies. S.H.P. acknowledges the National Biodiversity Authority (NBA) of the Government of India for providing approval to send honeybee samples to Xin Z. for non-commercial research purposes.
Funding Statement
The work was supported by the National Natural Science Foundation (NSF) of China (32270475), Program of Ministry of Science and Technology of China (2018FY100403), National Special Support Program for High-level Talents (Ten-Thousand Talents Program), and 2115 Talent Development Program of China Agricultural University through Xin Z. S.L. is supported by Funds for International Cooperation and Exchange of the National Natural Science Foundation of China (3211001043). Sample collection was also supported by the NSF of China (31470123) and Jilin Science and Technology Program (20030561) through X.L. S.H.P. is supported by the National Mission on Himalayan Studies (NMHS) - Almora, Ministry of Environment, Forest and Climate Change, Government of India, through grant GBPNI/NMHS-2017-18/MG-12
Contributor Information
Shanlin Liu, Email: shanlin.liu@cau.edu.cn.
Xin Zhou, Email: xinzhou@cau.edu.cn.
References
- Abrol DP. 2013. Asiatic Honeybee Apis cerana: Biodiversity Conservation and Agricultural Production. Dordrecht: Springer.
- Barr CM, Neiman M, Taylor DR Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytologist. 2005;168(1):39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- Braby MF, Eastwood R, Murray N The subspecies concept in butterflies: has its application in taxonomy and conservation biology outlived its usefulness? Biological Journal of the Linnean Society. 2012;106(4):699–716. doi: 10.1111/j.1095-8312.2012.01909.x. [DOI] [Google Scholar]
- Casacci LP, Barbero F, Balletto E The “Evolutionarily Significant Unit” concept and its applicability in biological conservation. Italian Journal of Zoology. 2014;81(2):182–193. doi: 10.1080/11250003.2013.870240. [DOI] [Google Scholar]
- Chen C, Wang HH, Liu ZG, et al Population genomics provide insights into the evolution and adaptation of the Eastern Honey Bee (Apis cerana) Molecular Biology and Evolution. 2018a;35(9):2260–2271. doi: 10.1093/molbev/msy130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Zhou YQ, Chen YR, et al fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018b;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC. 1993. Apiculture of China. Beijing: China Agricultural Publication. (in Chinese)
- Christmas MJ, Jones JC, Olsson A, et al Genetic barriers to historical gene flow between cryptic species of Alpine Bumblebees revealed by comparative population genomics. Molecular Biology and Evolution. 2021;38(8):3126–3143. doi: 10.1093/molbev/msab086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guia APO, Saitoh T The gap between the concept and definitions in the Evolutionarily Significant Unit: the need to integrate neutral genetic variation and adaptive variation. Ecological Research. 2007;22(4):604–612. doi: 10.1007/s11284-006-0059-z. [DOI] [Google Scholar]
- De Queiroz K Species concepts and species delimitation. Systematic Biology. 2007;56(6):879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- De Queiroz K An updated concept of subspecies resolves a dispute about the taxonomy of incompletely separated lineages. Herpetological Review. 2020;51(3):459–461. [Google Scholar]
- Dogantzis KA, Tiwari T, Conflitti IM, et al Thrice out of Asia and the adaptive radiation of the western honey bee. Science Advances. 2021;7(49):eabj2151. doi: 10.1126/sciadv.abj2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogantzis KA, Zayed A Recent advances in population and quantitative genomics of honey bees. Current Opinion in Insect Science. 2019;31:93–98. doi: 10.1016/j.cois.2018.11.010. [DOI] [PubMed] [Google Scholar]
- Engel MS The taxonomy of recent and fossil honey bees (Hymenoptera: Apidae; Apis) Journal of Hymenoptera Research. 1999;8(2):165–196. [Google Scholar]
- Garnery L, Cornuet JM, Solignac M Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Molecular Ecology. 1992;1(3):145–154. doi: 10.1111/j.1365-294X.1992.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Groves CP, Cotterill FPD, Gippoliti S, et al Species definitions and conservation: a review and case studies from African mammals. Conservation Genetics. 2017;18(6):1247–1256. doi: 10.1007/s10592-017-0976-0. [DOI] [Google Scholar]
- Guillot G, Renaud S, Ledevin R, et al A unifying model for the analysis of phenotypic, genetic, and geographic data. Systematic Biology. 2012;61(6):897–911. doi: 10.1093/sysbio/sys038. [DOI] [PubMed] [Google Scholar]
- Gutiérrez EE, Helgen KM Outdated taxonomy blocks conservation. Nature. 2013;495(7441):314. doi: 10.1038/495314e. [DOI] [PubMed] [Google Scholar]
- Hausdorf B, Wilkens H, Strecker U Population genetic patterns revealed by microsatellite data challenge the mitochondrial DNA based taxonomy of Astyanax in Mexico (Characidae, Teleostei) Molecular Phylogenetics and Evolution. 2011;60(1):89–97. doi: 10.1016/j.ympev.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Hepburn HR, Radloff SE, Verma S, et al Morphometric analysis of Apis cerana populations in the southern Himalayan region. Apidologie. 2001a;32(5):435–447. doi: 10.1051/apido:2001142. [DOI] [Google Scholar]
- Hepburn HR, Smith DR, Radloff SE, et al Infraspecific categories of Apis cerana: morphometric, allozymal and mtDNA diversity. Apidologie. 2001b;32(1):3–23. doi: 10.1051/apido:2001108. [DOI] [Google Scholar]
- Hillis DM Species delimitation in herpetology. Journal of Herpetology. 2019;53(1):3–12. doi: 10.1670/18-123. [DOI] [Google Scholar]
- Hou CS, Li BB, Luo YX, et al First detection of Apis mellifera filamentous virus in Apis cerana cerana in China. Journal of Invertebrate Pathology. 2016;138:112–115. doi: 10.1016/j.jip.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Ilyasov RA, Park J, Takahashi J, et al Phylogenetic uniqueness of Honeybee Apis cerana from the Korean peninsula inferred from the mitochondrial, nuclear, and morphological Data. Journal of Apicultural Science. 2018;62(2):189–214. doi: 10.2478/jas-2018-0018. [DOI] [Google Scholar]
- Ilyasov RA, Youn HG, Lee ML, et al Phylogenetic relationships of Russian Far-East Apis cerana with other north Asian populations. Journal of Apicultural Science. 2019;63(2):289–314. doi: 10.2478/jas-2019-0024. [DOI] [Google Scholar]
- Ji YK, Li XG, Ji T, et al Gene reuse facilitates rapid radiation and independent adaptation to diverse habitats in the Asian honeybee. Science Advances. 2020;6(51):eabd3590. doi: 10.1126/sciadv.abd3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP MORPHOJ: an integrated software package for geometric morphometrics. Molecular Ecology Resources. 2011;11(2):353–357. doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics. 2014;15(1):356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Karlsson EK, Perri A, et al Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(23):8878–8883. doi: 10.1073/pnas.1203005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V, Desper R, Gascuel O FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Molecular Biology and Evolution. 2015;32(10):2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Zhang R, Liu SL, et al The molecular evolutionary dynamics of oxidative phosphorylation (OXPHOS) genes in Hymenoptera. BMC Evolutionary Biology. 2017;17(1):269. doi: 10.1186/s12862-017-1111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NN, Liu HM, Ju Y, et al Geometric morphology and population genomics provide insights into the adaptive evolution of Apis cerana in Changbai Mountain. BMC Genomics. 2022;23(1):64. doi: 10.1186/s12864-022-08298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, Gloag RS, Anderson DL, et al A molecular phylogeny of the genus Apis suggests that the giant honey bee of the Philippines, A. breviligula Maa, and the plains honey bee of southern India, A. indica Fabricius, are valid species. Systematic Entomology. 2010;35(2):226–233. doi: 10.1111/j.1365-3113.2009.00504.x. [DOI] [Google Scholar]
- Ma DF, Huang WC Apiculture in the New China. Bee World. 1981;62(4):163–166. doi: 10.1080/0005772X.1981.11097840. [DOI] [Google Scholar]
- Maa TC An inquiry into the systematics of the tribus Apidini or honeybees (Hym. ) Treubia. 1953;21(3):525–640. [Google Scholar]
- Malinsky M, Challis RJ, Tyers AM, et al Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350(6267):1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, et al Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison III WR, Lohr JL, Duchen P, et al The impact of taxonomic change on conservation: Does it kill, can it save, or is it just irrelevant? Biological Conservation. 2009;142(12):3201–3206. doi: 10.1016/j.biocon.2009.07.019. [DOI] [Google Scholar]
- Nakamura T, Yamada KD, Tomii K, et al Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34(14):2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Animal Genetic Resources Committee. 2011. Animal Genetic Resources in China: Bees. Beijing: China Agriculture Press. (in Chinese)
- Neuditschko M, Khatkar MS, Raadsma HW NetView: a high-definition network-visualization approach to detect fine-scale population structures from genome-wide patterns of variation. PLoS One. 2012;7(10):e48375. doi: 10.1371/journal.pone.0048375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd BP, Reddy MS, Chapman NC, et al Evidence for reproductive isolation between two colour morphs of cavity nesting honey bees (Apis) in south India. Insectes Sociaux. 2006;53(4):428–434. doi: 10.1007/s00040-005-0889-2. [DOI] [Google Scholar]
- Orr MC, Ferrari RR, Hughes AC, et al Taxonomy must engage with new technologies and evolve to face future challenges. Nature Ecology & Evolution. 2021;5(1):3–4. doi: 10.1038/s41559-020-01360-5. [DOI] [PubMed] [Google Scholar]
- Peng YS, Nasr ME, Locke SJ Geographical races of Apis cerana Fabricius in China and their distribution. Review of recent Chinese publications and a preliminary statistical analysis. Apidologie. 1989;20(1):9–20. doi: 10.1051/apido:19890102. [DOI] [Google Scholar]
- Phillimore AB, Owens IPF Are subspecies useful in evolutionary and conservation biology? Proceedings of the Royal Society B:Biological Sciences. 2006;273(1590):1049–1053. doi: 10.1098/rspb.2005.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff SE, Hepburn C, Hepburn HR, et al. 2010. Population structure and classification of Apis cerana. Apidologie, 41(6): 589–601.
- Radoszkowski O Hyménoptères de Korée. Horae Societatis Entomologicae Rossicae. 1887;21:428–436. [Google Scholar]
- Requier F, Garnery L, Kohl PL, et al The conservation of native Honey Bees is crucial. Trends in Ecolgy & Evolution. 2019;34(9):789–798. doi: 10.1016/j.tree.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Rodríguez F, Pérez T, Hammer SE, et al Integrating phylogeographic patterns of microsatellite and mtDNA divergence to infer the evolutionary history of chamois (genus Rupicapra) BMC Evolutionary Biology. 2010;10(1):222. doi: 10.1186/1471-2148-10-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttner F. 1988. Biogeography and Taxonomy of Honeybees. Berlin: Springer.
- Ryder OA Species conservation and systematics: the dilemma of subspecies. Trends in Ecology & Evolution. 1986;1(1):9–10. doi: 10.1016/0169-5347(86)90035-2. [DOI] [PubMed] [Google Scholar]
- Shanas S, Anju KG, Mashhoor K Identity of cavity nesting honey bees of the Indian subcontinent with description of a new species (Hymenoptera: Apidae: Apinae: Apini: Apis) Entomon. 2022;47(3):197–220. doi: 10.33307/entomon.v47i3.755. [DOI] [Google Scholar]
- Shaw KL Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: What mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(25):16122–16127. doi: 10.1073/pnas.242585899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotte L, Korneliussen TS, Albrechtsen A Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195(3):693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hagen RH The biogeography of Apis cerana as revealed by mitochondrial DNA sequence data. Journal of the Kansas Entomological Society. 1996;69(4):294–310. [Google Scholar]
- Tan HW, Naeem M, Ali H, et al Genome sequence of the Asian honeybee in Pakistan sheds light on its phylogenetic relationship with other honeybees. Insects. 2021;12(7):652. doi: 10.3390/insects12070652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Fuchs S, Koeniger N, et al Morphological characterization of Apis cerana in the Yunnan province of China. Apidologie. 2003;34(6):553–561. doi: 10.1051/apido:2003049. [DOI] [Google Scholar]
- Tan K, Hepburn HR, Radloff SE, et al Multivariate morphometric analysis of the Apis cerana of China. Apidologie. 2008;39(3):343–353. doi: 10.1051/apido:2008014. [DOI] [Google Scholar]
- Tan K, Meixner MD, Fuchs S, et al Geographic distribution of the eastern honeybee, Apis cerana (Hymenoptera: Apidae), across ecological zones in China: Morphological and molecular analyses. Systematics and Biodiversity. 2006;4(4):473–482. doi: 10.1017/S1477200006002015. [DOI] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, et al Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews DPL, Brelsford A The biogeography of mitochondrial and nuclear discordance in animals. Molecular Ecology. 2012;21(16):3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- Vung NN, Kim I, Lee MY, et al Controlling sacbrood virus disease in Apis cerana colonies with biological methods in Korea. Journal of Apiculture. 2018;33(4):283–295. doi: 10.17519/apiculture.2018.11.33.4.283. [DOI] [Google Scholar]
- Wang XY, Wang T, Xu JF, et al Enhanced habitat loss of the Himalayan endemic flora driven by warming-forced upslope tree expansion. Nature Ecology & Evolution. 2022;6(7):890–899. doi: 10.1038/s41559-022-01774-3. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen DH, Wang Q, et al Research on the development status and countermeasures of the bee seed industry in China. Journal of Bee. 2021;41(11):12–18. [Google Scholar]
- Yang GH The effect of Apis cerana cerana on forest ecosystem. Apiculture of China. 2009;60(4):5–7,10. [Google Scholar]
- Yang GH, Lin GL, Sun QH, et al The distribution, morphological variation and behavior of Chinese honeybee in northwestern plateau of Sichuan province. Scientia Agricultura Sinica. 1995;28(S1):202–206. [Google Scholar]
- Yang GH, Xu SY, Kuang BY, et al The distribution of the genus Apis cerana Fab. in China and some of its subspecies. Journal of Yunnan Agricultural University. 1986;1(1):89–92. [Google Scholar]
- Zhao HL Problems and countermeasures of bee cultivation in poor mountainous areas of Hainan. Anhui Agricultural Science Bulletin. 2018;24(12):10–12. [Google Scholar]
- Zhao WZ, Wang M, Liu YQ, et al Phylogeography of Apis cerana populations on Hainan island and southern mainland China revealed by microsatellite polymorphism and mitochondrial DNA. Apidologie. 2017;48(1):63–74. doi: 10.1007/s13592-016-0450-x. [DOI] [Google Scholar]
- Zhuang DA. 1989. New subspecies of Apis cerana. Southwest China Journal of Agricultural Sciences, 2(4): 61–65. (in Chinese)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.