Abstract
The Creb-Regulated Transcriptional Coactivator (Crtc) family of transcriptional coregulators drive Creb1-mediated transcription effects on metabolism in many tissues, but the in vivo effects of Crtc2/Creb1 transcription on skeletal muscle metabolism are not known. Skeletal muscle-specific overexpression of Crtc2 (Crtc2 mice) induced greater mitochondrial activity, metabolic flux capacity for both carbohydrates and fats, improved glucose tolerance and insulin sensitivity, and increased oxidative capacity, supported by upregulation of key metabolic genes. Crtc2 overexpression led to greater weight loss during alternate day fasting (ADF), selective loss of fat rather than lean mass, maintenance of higher energy expenditure during the fast and reduced binge-eating during the feeding period. ADF downregulated most of the mitochondrial electron transport genes, and other regulators of mitochondrial function, that were substantially reversed by Crtc2-driven transcription. Glucocorticoids acted with AMPK to drive atrophy and mitophagy, which was reversed by Crtc2/Creb1 signaling. Crtc2/Creb1-mediated signaling coordinates metabolic adaptations in skeletal muscle that explain how Crtc2/Creb1 regulates metabolism and weight loss.
Keywords: Creb1, Crtc2, intermittent fasting, skeletal muscle
1 |. INTRODUCTION
Long-term dieting to lose weight is usually not effective as most of the weight is regained through physiologic adaptations that increase hunger and slow metabolic rate.1 Weight loss reduces the metabolic rate due to a number of changes that include a disproportional decreased in circulating leptin (in proportion to the amount of fat lost) and decreased thyroid hormone and sympathetic nervous system (SNS) activity.2–4 People who maintain long-term weight loss almost invariably exercise,5 and both preclinical and clinical data suggest that this is through combating weight-loss induced adaptations, including raising metabolic rate,6–8 decreased energy efficiency,9 and increased energy expenditure from the increased physical activity.10 Exercise during dieting in formerly obese rats also ameliorated the effects of relapse including binge eating, weight gain and preferential carbohydrate usage and fat disposition,6,11,12 through unknown mechanisms.13
During stress such as strenuous exercise, Creb1-regulated transcription is stimulated by GPCRs, like the β-adrenergic receptors (βAR),14–17 which activate adenylyl cyclases to convert ATP to the second messenger cAMP, which in turn liberates the catalytic subunit of protein kinase A (PKA).18 Crtc1–3 family of transcriptional activators is highly conserved and functions as the primary coactivators for Creb1, playing a crucial role in cAMP transcriptional responses. Crtc1–3 activity is regulated via phosphorylation events. They are fully activated through synergistic dephosphorylation by phosphatases downstream of PKA and calcium flux in polarizable tissues including the skeletal muscle, and are rendered cytoplasmic and inactive by phosphorylation from salt inducible kinases SIK1 and Sik2,14,19 in the hippocampus,20 cortex,21 islet cells,19 and the renal proximal tubule.22 In skeletal muscle, Crtc/Creb1 is activated by exercise downstream of the β-adrenergic receptor and calcium flux, 14 stimulating anabolic transcriptional programs,23 including induction of PGC-1α14,24 and in vitro enhancement of oxidative capacity.24 While Crtc/Creb1 coordinates metabolic transcriptional programs in other tissues such as adipocytes, liver, and pancreas, its role in skeletal muscle metabolism has not been fully studied in vivo.
To investigate the effects of Crtc2/Creb1 activation in adult skeletal muscle, we generated doxycycline (dox)-inducible transgenic mice that show skeletal muscle-specific overexpression of Crtc2 (FLAG-Crtc2 S171A/S275A/K628R mutant that prevents cytoplasmic localization) under the control of a tetracycline response element.14 Induction of Crtc2 improved whole animal and skeletal muscle metabolism, enabling maintenance of higher energy expenditure during fasting and improved weight loss, and countered the effects of glucocorticoids that drive catabolic effects of fasting and weight loss in skeletal muscle.
2 |. MATERIALS AND METHODS
2.1 |. Statistical analyses
Changes in body weight and composition were analyzed with two-way analysis of variance. For VO2, respiratory quotient, and ambulation data, the 12-h libitum and fasted periods were analyzed separately with two-way ANOVA, and the refeeding data analyzed with Student’s t-test. For the energy consumption data, the fasted and re-fed data were analyzed together with two-way ANOVA. Gene expression data were analyzed with one-way ANOVA, with Dunnett’s test for multiple comparisons of each gene to its corresponding control. Glycemia and insulin changes over time were analyzed with two-way ANOVA, and the AUC assessed with Student’s t-test. The behavioral data for meal consumption were analyzed with Student’s t-tests.
2.2 |. Crtc2 transgenic mice and animal care
All animal procedures were approved by the Scripps Research Institute IACUC. We chose to study Crtc2 because its role in metabolism was better established at the time than Crtc3, the other isoform expressed in skeletal muscle25–27 and because the constitutively nuclear triple mutant Crtc2 was well characterized.19,26 TRE-Crtc2 tm mice were generated by cloning FLAG-Crtc2 (S171A, S275A, K628R) into pTRE-Tight. Oocyte injections were conducted into C57BL/6 mice at the University of Cincinnati. Double transgenic mice (DTg) were generated by crossing C57BL/6-Tg(TRE-Crtc2tm)1Mdc/j mice with 1256[3Emut] MCK-rtTA transgenic mice, which expresses the reverse tetracycline transactivator (rtTA) regulated by the 1256[3Emut] MCK promoter specifically in skeletal muscle,28 and we did not detect expression in heart.14 Wild type and TRE littermates were used as controls and treated with dox in the same manner as DTg mice. High-fat diet (Research Diet D12450J) contained 60 kcal% fat and matched control low-fat diet (D12492) contained 10kCal% fat and 7% sucrose.
2.3 |. Glucose and insulin tolerance tests
Glucose tolerance testing was performed after an overnight 12-h fast. Blood glucose was quantified at 15, 30, 60, 90, and 120 min post-i.p. administration of 20% D-glucose (1 g/kg fasted body weight) with an Accu-check glucose monitor as described.29 Insulin tolerance testing was performed on 6-h–fasted mice injected i.p. with human insulin (1.5 U/kg fasted body weight). Blood glucose levels were determined immediately before, and 15 and 30 min after, injection.
2.4 |. In vivo electroporation
Plasmid DNA (50 μg) was transfected into mouse tibialis anterior (TA) muscles by electroporation as described previously.14 One hour before electroporation mice were anesthetized, and a small incision was made through the skin covering the TA muscle and then injected with 30 Al of 0.5 U/Al hyaluronidase and then injected with plasmid DNA Crtc2 and GFP expression plasmids were purified from DH5a Escherichia coli cultures using an EndoFree plasmid kit (Qiagen, Valencia, CA, USA), and resuspended in 71 mM sterile PBS. After the injections, electric pulses were administered by using non-i nvasive platinum-coated tweezertrodes. Eight 100-ms square-wave electric pulses at a frequency of 1 Hz and at 200-m s intervals were delivered with an ECM 830 electroporation unit (BTX-Harvard Apparatus, Holliston, MA, USA) with a field strength of 40 V/cm. After the electroporation procedure, the incision was closed with Vetbond surgical glue.
2.5 |. Indirect calorimetry and energy expenditure
Mice were individually housed in a sixteen chamber Oxymax comprehensive lab animal monitoring system (CLAMS; Columbus Instruments, Columbus OH) at 26°C. Animals were allowed to acclimate for 1 day followed by VO2 and VCO2 were measured at 13 min intervals over a continuous duration of 60 h Energy expenditure (EE) was calculated using the Lusk equation, kcal/h = (3.815 + 1.232 × RQ) × VO2 in l/h, where RQ is the ratio of VCO2 to VO2. Opto-V arimetrix-3 sensor system in the x-, y-, and z-axes of each chamber recorded sterotype, ambulatory and z-axis activity. Consecutive adjacent infrared beam breaks in either the x- or y-axes were scored as ambulatory activity.
2.6 |. Body composition
Body composition was determined using time domain nuclear magnetic resonance (TD-NMR) in a Bruker minispec.
2.7 |. Automated food intake monitoring
Feeding behavior was assessed using a BioDAQ episodic Food Intake Monitor (BioDAQ, Research Diets, Inc) as described.30 Feeding parameters were calculated with BioDAQ Monitoring Software 2.2.02. Feeding bouts were defined as a change in stable weight of food. Meals consisted of two or more bouts within 5 s with a minimum meal amount of 0.02 g. Clusters of bouts occurring greater than 600 s apart were considered separate meals. Water or water containing dox was provided ad libitum from regular water bottles. Mice habituated to the new environment within 1 day and showed normal food intake and regular body weight gain.
2.8 |. mRNA-seq and bioinformatics
Total RNA was extracted from tissues using TRIzol™ reagent (Invitrogen), and cleaned up using the RNeasy kit with on-column DNA digestion (QIAGEN). Three mRNA-seq libraries per condition were prepared using the NEBNext Ultra II directional RNA library Prep kit for Illumina (New England Biolabs), and read in paired-end 40-c ycle sequencing runs on the NextSeq 500 system (Illumina). Sequences were aligned to the Ensembl Mus musculus GRCm38.83.chr.gtf genome (mm10) assembly using HISAT2 v2.0.5 (PMID: 25751142), sorted and indexed using SAMtools v1.731 and quantified for differential expression analysis using Subread v1.6.1 featureCounts.32 Differential expression analysis was performed using DESeq2 v1.18.1.33,34 Enriched canonical pathways were identified using the Ingenuity pathways analysis software (QIAGEN). Gene Ontology (GO) analysis excluding inferred electronic annotations, was performed using the Mouse Genome Informatics GO Browser and GO Term Mapper (http://www.informatics.jax.org). Genes encoding mouse nuclear hormone receptors were obtained from the Nuclear Receptor Signaling Atlas (NURSA).35 Genes encoding mitochondrial proteins were obtained from the mouse Mitocarta2.0.36,37 Genes encoding transcriptional coregulators were obtained from the EpiFactors database.38 Glucocorticoid-responsive genes were obtained by combining previously published datasets39 reanalyzed with GEO2R, with other dexamethasone-regulated genes that we identified in C2C12 myotubes, GEO accession no. GSE149 453. Heatmaps were prepared using Pheatmap v1.0.8, RColorBrewer v1.1–2, and other software packages available in R. Other data visualizations were performed using GraphPad Prism7.
2.9 |. Glucose uptake
Insulin-mediated 2-deoxy-D-[14C] glucose uptake was determined by incubating primary myotubes with PBS or PBS containing 100 nM insulin for 30 min followed by the addition of 0.1 mM of cold 2-deoxy-D-glucose and 0.1 μCi/well of 2-deoxy-[14C] D-glucose for 5 min prior to solubilization. Nonspecific deoxyglucose uptake was measured in the presence of 20 μM cytochalasin B and subtracted from the total glucose uptake.
2.10 |. Primary myoblast isolation and culture
Primary myoblasts were isolated from P1 to P3 (1–3 day old) C57BL/6J pups as described40 and cultured on collagen-coated plates in media consisting in 1:1 F-10/DMEM supplemented with 20% FCS and 25 μg/ml bFGF. Subconfluent myoblasts were differentiated into multinucleated myotubes using a 1:1 F-10/5 mM glucose DMEM supplemented with 4% heat-inactivated horse serum (Hyclone) for up to five days. For steroid-free culture conditions, cells were grown in charcoal:dextran-stripped FBS (Gemini Bio-products, cat no. 100–119).
2.11 |. Adenoviruses
Control GFP-expressing and Crtc2-expressing adenoviruses have been described (Wu et al.24; Koo et al.25). For all experiments, myotubes were infected after 72 h of differentiation with 4 × 108 viral particles per ml per well for 24–48 h.
2.12 |. Immunoblotting analysis
Immunoblot analysis of in vitro differentiated myotubes was performed as described.14 Whole cell extracts were prepared in HEPES whole cell lysis buffer (20 mM HEPES, pH 7.4, 1% TX-100, 1× phosphoSTOP complete inhibition cocktail tablets [Roche], and 1X Complete-EDTA Free protease inhibitor cocktail [Roche]) and the supernatant was collected after centrifugation. The protein concentration was determined using the protein assay reagent (Bio-Rad). About 60–80 μg of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes for immunoblotting. Images were quantified with ImageJ.
Analysis of frozen TA muscle was performed as described.41 Frozen TA muscles were homogenized in sucrose lysis buffer (50 mM Tris pH 7.5, 250 mM sucrose, 1 mM EDTA, 1 mM EGTA, 1% Triton X 100, and 50 mM NaF). The supernatant was collected following centrifugation at 8000 g for 10 min and protein concentrations were determined using the 660 nm-protein assay (Thermo Fisher Scientific, Waltham, MA). Twelve micrograms of protein were subjected to SDS-P AGE on 4%–20% Criterion TGX stain-free gels (Bio-Rad, Hercules, CA) and transferred to a polyvinylidene diflouride membranes (PVDF, Millipore, Burlington, MA) for immunoblotting.41 Image acquisition and band quantification were performed using the Azure C400 System (Azure Biosystems, Dublin, CA,) and Image Laboratory, v 6.0.1 (Bio-Rad), respectively.
Antibodies used: Nur77/Nr4a1 #2518 Novus Biological. CRTC2 #A300–638A Bethyl Laboratories. Irs2 (L1326) #3089S Cell Signaling Technology. Tubulin #T9026 Sigma-Aldrich. pAKT(Thr308) (244f1)Rb and pAKT(Ser473) (971s) Cell Signaling Technology. PGC-1α ab54481 Abcam. Puromycin Cat# MABE343 EMD-Millipore, S6Kp (Thr389) Cat# 9205 Cell Signaling Technology, S6K Cat# 9202 Cell Signaling Technology. S6p (ser 240/244) Cat# 5364 Cell Signaling Technology. S6 Cat# 2217 Cell Signaling Technology. 4E-BP1-p (Thr37/46) Cat# 2855 Cell Signaling Technology. 4E-BP1 Cat# 9644 Cell Signaling Technology.
2.13 |. Quantitative RT-PCR
Total RNA from tibialis anterior muscle or cells was harvested using Rneasy RNA Kit (Qiagen) and cDNA prepared using Transcriptor High Fidelity cDNA Synthesis Kit (Roche). Relative abundance of cDNAs was determined a Roche LightCycler 480, and the resulting data were normalized to Rpl-23 ribosomal RNA (IDT) as described.42 The primer pairs are as follows: Nr4a1 (5′-ttctgctcaggcctggtact′, 5′-gattctgcagctcttccacc-3′); Nr4a3 (5′-tcagcctttttggagctgtt′, 5′-tgaagtcgatgcaggacaag-3′); Irs2 (5′-acaacctatcgtggcacctc-3′, 5′-gacggtggtggtagaggaaa-3′); Idh3a (5′-gtgacaagaggttttgctggt-3′, 5′-tgaaatttctgggccaattc-3′); Cytc (5′-gatgccaacaagaacaaaggt-3′, 5′-tgggattttccaaatactc cat-3′); Glut4 (5′-tgtggctgtgccatcttg-3′, 5′-cagggccaatctcaa agaag-3′).
2.14 |. Mitochondrial DNA analysis
Total DNA was extracted using the GenElute™ mammalian genomic DNA miniprep kit (Sigma-Aldrich). Triplicate real-time PCR reactions were performed using 50 ng of total DNA, CoxI (5′-agcattcccacgaataaataaca-3′, 5′-agcattcccacgaataaataacat-3′) or CoxII (5′-tttcaacttggcttac aagacg-3′, 5′-tttcaacttggcttacaagacg-3′) mitochondrial DNA (mt-DNA) primers, and a control genomic DNA primer, Gapdh (5′-gaaaaggagattgctacg-3′, 5′-gcaagaggctaggggc-3′).
2.15 |. Histology
Dissected muscle tissue was mounted in OCT medium (TissueTek) and froze in liquid N2–cooled isopentane. Tissue sections were stained for succinate dehydrogenase activity using the published methods.43 Gastrocnemius cryosections Taken directly from −20C were placed in coplin jar containing SDH solution and incubated for 20 min to Stop enzymatic reaction by the slides were dipped once quickly in ddH2O and Air-dry for ~10 min in the dark, then slides were mounted and the nitroblue-diformozan precipitate analyzed. Regions of high or low SDH activity were identified by gross anatomy.
2.16 |. Fatty acid metabolism
Fatty acid oxidation was assayed by incubating (9,10(n)-3H) palmitic acid (60 Ci/mmol) bound to fatty-acid free albumin (final concentration:100 μM, palmitate:albumin 2:1) and 1 mM carnitine with primary myocytes for 2 h. Tritiated water released was collected and quantitated.
2.17 |. High-content imaging and analysis
Primary or C2C12 myotubes in black 96-or 384-well tissue culture plates with clear bases (Greiner Bio-One, North America, Inc) were stained for 15 min with 200 nM MitoTracker® Orange CM-H2TMRos dye (Invitrogen™ by ThermoFisher Scientific). The myotubes were rinsed with steroid-free DMEM to remove un-incorporated dye, and then incubated in the differentiation media for 45 min to reach peak fluorescence. Myotubes were fixed in 4% formaldehyde for 20 min, stained with 300 nM DAPI for 5 min, permeabilized in PBS containing 0.1% Triton X-100 for 20 min, blocked for 1 h with 1× TBS containing 0.1% Tween-20 (TBS-T) and 2.5% normal goat serum, and incubated at 4°C overnight with an AlexaFluor® 488-conjugated anti-skeletal muscle myosin (F59) antibody (Santa Cruz Biotechnology, Inc cat no. sc-32732 AF488). The next day, the myotubes were washed 4 times with TBS-T to remove unbound antibodies and rinsed twice with PBS. The stained myotubes were imaged at 10–20× magnification on the IN Cell Analyzer 6000 platform (GE Healthcare). For each treatment condition, a stack of 24–27 images containing an average of 50–100 myotubes per image were analyzed using the IN Cell Developer Toolbox image analysis software, with a customized segmentation protocol for myotubes. The average (mean) diameter and mitochondrial potential (i.e., MitoTracker staining density × area) of myotubes in these images were then calculated.
2.18 |. Muscle protein synthesis
Changes in muscle protein synthesis (MPS) were assessed in transfected Crtc2 TA muscles under Fed and ADF conditions using the SUnSET method by measuring the incorporation of exogenous puromycin into nascent peptides as described previously.41,44 Puromycin is a tyrosyl tRNA analogue that results in the premature release of translation products—puromycin conjugates. These conjugates can be subsequently measured in protein lysates through western blotting using an antibody against puromycin. Puromycin (EMD Millipore, Billerica, MA; Cat. No. 540222) was dissolved in sterile saline and delivered (0.02 μmol/g body weight by intraperitoneal injection) 30 min before muscle collection.
2.19 |. Citrate synthase activity assay
Frozen transfected TA muscles under fed and ADF conditions were powdered and 10 mg of tissue was homogenized in RIPA buffer containing protease and phosphatase inhibitors (Thermo Fisher scientific, Cat no. A32959). Samples were then centrifuged at 12 000 g for 15 min at 4°C and the supernatant collected. For the citrate synthase assay, 8 μg of protein was added to a reaction buffer (36.5 mM KH2PO4, 83.5 mM K2HPO4, 10 mM EDTA) containing a citrate synthase cocktail (0.1 mM 5,5′-Dithoibis-2[2-nitrobenzoic acid] (Millipore Sigma, Cat no. D2818200); 0.1 mM Acetyl-SCoA (Millipore Sigma, Cat no. A2056) and 0.1% Triton X-100) in a 96-well plate. Immediately prior to the assay plate being measured, 10 mM Oxalocetic Acid solution (Millipore Sigma, Cat no. O4126) was added to each well. The 96-well plate was measured on a SpectraMax M2 plate reader (Molecular Devices, San Jose, CA) at an absorbance of 412 nm. Samples were analyzed in duplicate. Data are presented as units per milligram.
3 |. RESULTS
3.1 |. Crtc2 stimulates mitochondrial activity and improves oxidative capacity
To further understand the roles of Crtc2 in skeletal muscle, we compared aged Crtc2 (rtTA/Crtc2tm) mice and WT (rtTA/WT) mice in the C57BL/6 background. After one week of dox treatment, Crtc2 mice showed increased mitochondrial succinate dehydrogenase activity in the gastrocnemius muscle, reflecting greater respiratory complex II activity (Figure 1A, Supporting Information Figure S1A,B). Myotubes overexpressing Crtc2 or Crtc3 oxidized more palmitate than control myotubes (Figure 1B), as previously reported.24 Crtc2 mice displayed increased expression of Cytochrome C (Cycs), Ppargc1a/PGC-1α,14,24 and Nr4a3/NOR-1 nuclear receptor (Figure 1C) which induces mitochondrial biogenesis and increases oxidative capacity,45,46 and is induced by high-intensity exercise.14 These mitochondria-related genes were also upregulated in primary myotubes overexpressing Crtc2, indicating a skeletal muscle-intrinsic effect of Crtc2 that mimics some of the effects of exercise adaptation (Figure 1D). We also observed a doubling of mt-DNA after in vivo electroporation of Crtc2 into skeletal muscle that was not significant, and a statistically significant increase in mitochondrial displacement loop (D-loop) region, containing third strands of DNA that are the sites of replication (Figure 1E). These data support a model where Crtc2/PGC-1α transcriptional control programs mediate exercise-induced mitochondrial biogenesis and oxidative capacity.
FIGURE 1.
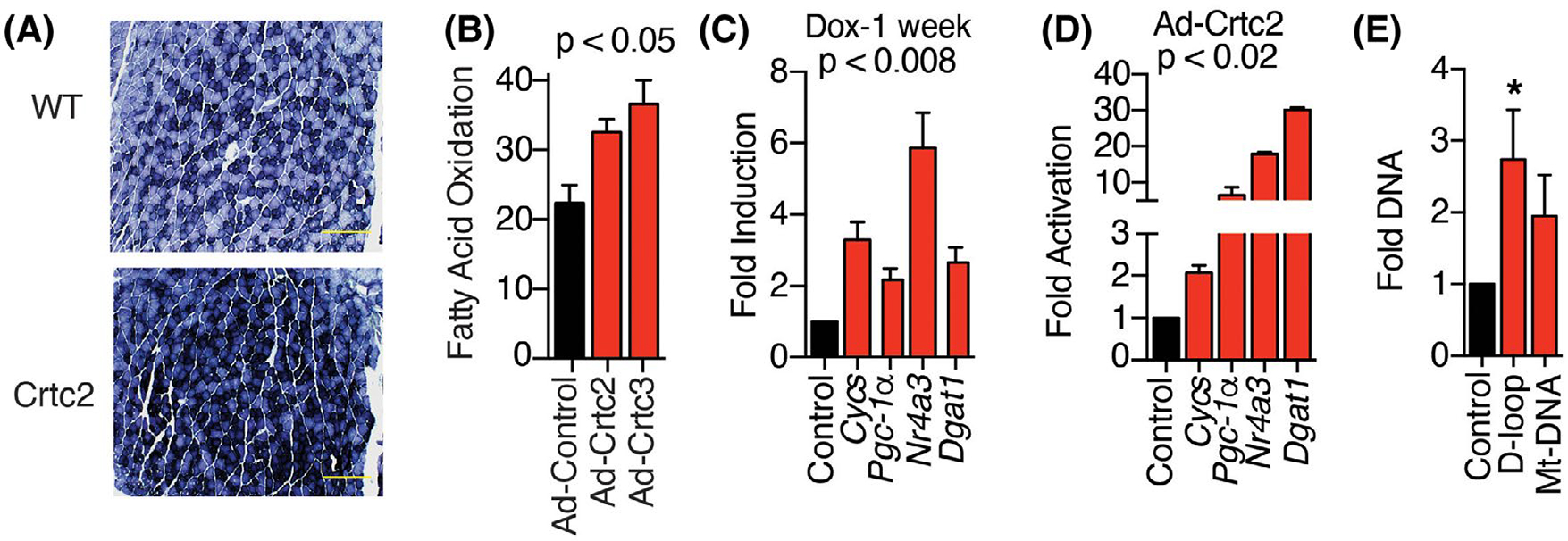
Crtc2 expression in skeletal muscle enhances mitochondrial biogenesis. (A) Histological analysis of succinate dehydrogenase in myofibers from gastrocnemius muscle sections after dox treatment. Scale bar = 500 μm. (B) Oxidation of 3H-Palmitic acid in the supernatant of primary myocytes transduced with control, Crtc2, or Crtc3-expressing adenovirus. (C) Gene induction calculated relative to WT mice from isolated tibialis anterior muscles. N = 8 mice per group. (D) Gene expression in primary myocytes transduced with adenovirus expressing Crtc2 or GFP control 72-h post-transduction. (E) Crtc2 expression plasmid was electroporated into the tibialis anterior muscle and GFP plasmid in the contralateral leg. After 7 days, DNA was extracted for assessment of mitochondrial DNA (mt-DNA) for Crtc2/GFP control. N = 5 mice per group. *p < .05. Data are shown as mean ± SEM
3.2 |. Crtc2 improves glucose tolerance and carbohydrate metabolism
To probe for Crtc2-dependent changes in carbohydrate metabolism, we performed glucose tolerance tests comparing Crtc2 and WT mice. After one week of dox treatment, the Crtc2 mice displayed significantly improved glucose disposal, including a dramatic improvement in their ability to reduce glycemia (Figure 2A, Supporting Information Figure S2), and a lower insulin excursion (Figure 2B). Crtc2 overexpression also improved insulin tolerance (Figure 2C). In primary myotubes, expression of Crtc2 was sufficient to significantly increase the uptake of 2-Deoxy-D-glucose (Figure 2D).
FIGURE 2.
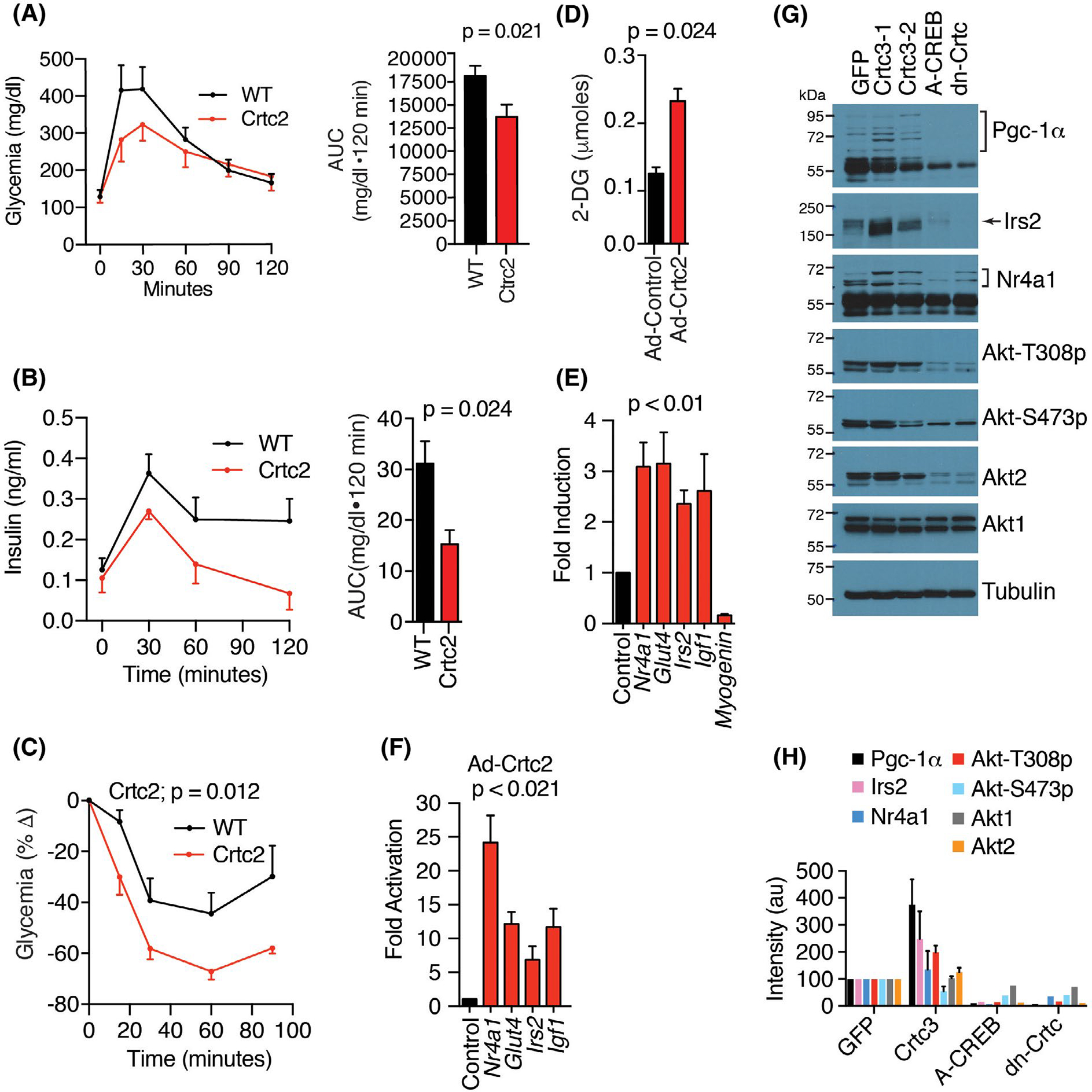
Crtc2 transgenic mice maintain enhanced glucose disposal and insulin tolerance. (A) Glucose tolerance test (GTT). Right panel, area under the curve. N = 10 mice per group. (B) Plasma insulin levels during GTT described in panel B. (C) Insulin tolerance tests. N = 10 mice per group. (D) Insulin-mediated uptake of 2-deoxy-D-glucose in primary myocytes transduced with adenovirus expressing Crtc2 or GFP control. N = 10 per group. Student’s t-test. (E) Crtc2 mice were treated +/− dox for 1 week and the gene induction compared by qPCR relative to the no dox groups. N = 10 mice per group. (F) Gene expression in primary myocytes transduced with adenovirus expressing Crtc2 or GFP control were compared by qPCR. N = 4 per group. (G) Myocytes stably expressing GFP, Crtc3 (lanes 2 and 3), dominant-negative Creb (A-CREB), or dominant-negative Crtc2 (dn-Crtc) were compared by western blot. See also Supporting Information Figure S3. (H) Quantitation of western blots shown in panel G. Arbitrary units (au). Data are mean ± SEM
Crtc2 mice and Crtc2/3-overexpressing myotubes upregulated a number of carbohydrate metabolism and signaling genes at both the RNA and protein levels (Figure 2E–H, Supporting Information Figure S3), including the glucose transporter, Glut4, whose insulin-dependent translocation to the membrane drives glucose uptake into skeletal muscle, accounting for 80% of glucose disposal. The Nr4a1 gene encodes Nur77, a nuclear receptor family transcription factor that regulates transcriptional programs for carbohydrate metabolism, lipolysis, and mitochondrial biogenesis,47 and is upregulated by high intensity exercise or adrenergic signaling.14 Irs2, Akt1, and Akt2 encode core components of the insulin receptor signaling pathway that can coordinate anabolic signaling and improve carbohydrate metabolism. We also identified a significant downregulation of myogenin by Crtc2 (Figure 2E), a primary driver of muscle atrophy during fasting. To validate on-target mechanism of action, we generated stable cell lines overexpressing Crtc3, dominant-negative A-Creb, or dominant-negative Crtc2 expressing only the Creb-binding domain.19 These results demonstrate that A-Creb and dn-Crtc2 blocked basal expression of PGC-1α, Irs2, and Akt2, and prevented phosphorylation of Akt1/2 at Thr308 (Figure 2G,H). These experiments demonstrate that activation of skeletal muscle Crtc2/Creb1 transcriptional programs recapitulates beneficial metabolic responses to exercise.
3.3 |. Crtc2 overexpression in skeletal muscle facilitates weight loss
We noticed during the GTTs that the Crtc2 mice tend to lose more weight after the overnight fast, so we tested them in the context of a common dieting scheme of intermittent fasting. We studied aged Crtc2 or WT mice, which in the C57BL/6 background naturally gain fat mass as they age (18 weeks, 30–39 g, Figure 3A). During dox treatment, Crtc2 mice and control WT mice gained weight similarly (Supporting Information Figure S4A). When subject to alternate day fasting (ADF), three separate cohorts of Crtc2 mice displayed significantly greater and sustained weight loss than WT mice, with Crtc2 mice losing about twice as much weight in each case, 9%–14% of body weight (Figure 3B, Supporting Information Figure S4B).
FIGURE 3.
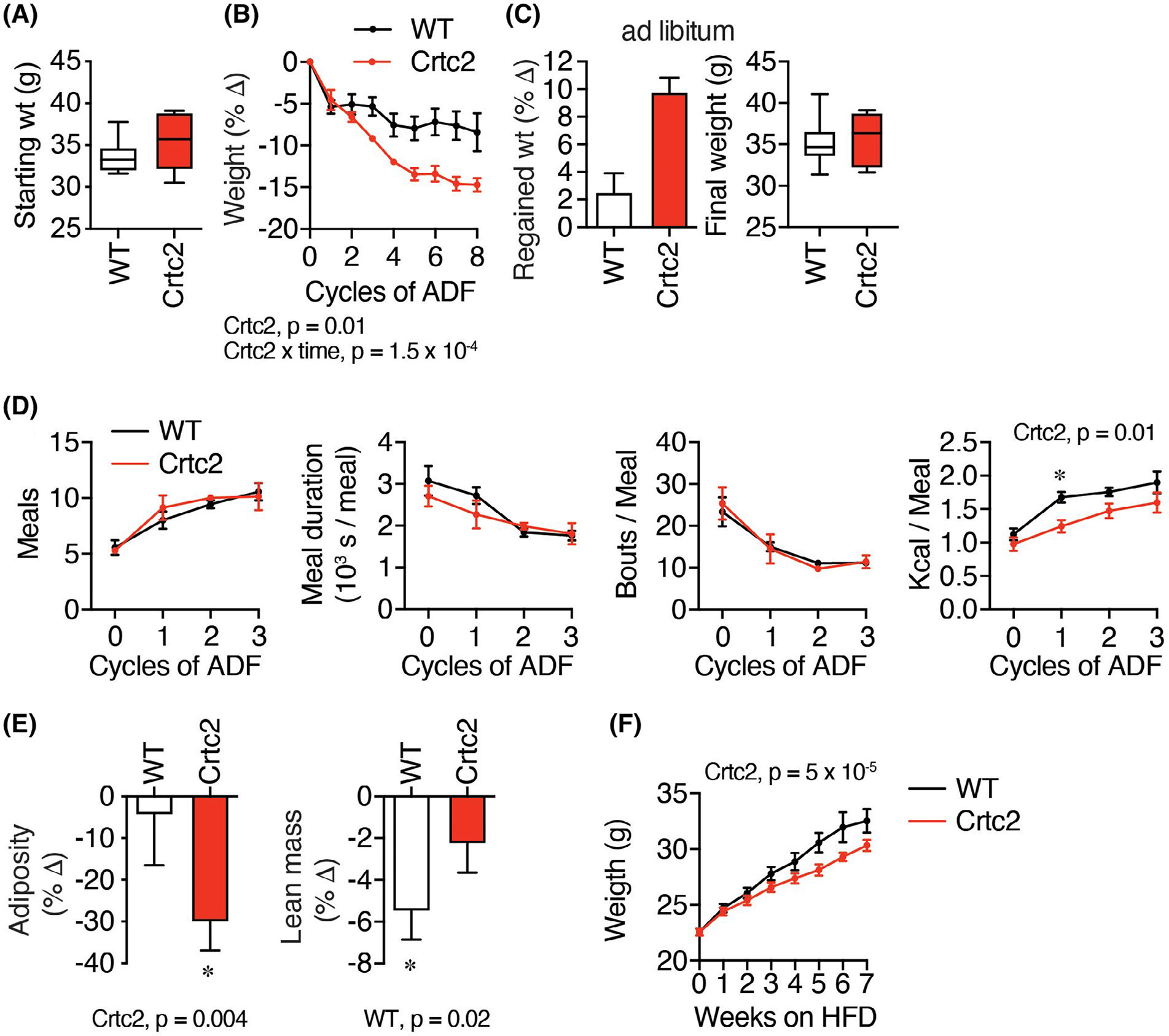
Crtc2/Creb1 signaling enables greater loss of weight and adiposity during alternate day fasting (ADF). (A) 18-week-old C57BL/6 male control (WT) and Crtc2 mice were treated with dox for 35 days and weighed. Data are shown as box-and-whisker plot, indicating the middle quartiles and range. N = 6 (WT) or 5 (Crtc2) mice per group. (B–E) Mice from (A) were subject to alternate day fasting (ADF). (B) Changes in the body weight during ADF. (C) Weight regain after the ADF treatment and 3 weeks of ad libitum feeding. (D) Behaviors were assayed with Biodak monitoring system, including number of meals, bouts per meal, meal duration, and kilocalories consumed over the 12-h re-feeding period. *significant in Sidak’s test for planned comparisons. (E) Adiposity and lean mass determined by whole-body NMR measurements taken before and after 8 cycles of ADF. Data analyzed with 1-way ANOVA. (F) 8-week-old C57BL/6 male control and Crtc2 mice were treated with dox and placed on a high-fat diet (HFD). N = 9 (WT) or 7 (Crtc2) mice per group. Time course data was analyzed by two-way ANOVA. Data are shown as mean ± SEM
After 3 weeks of ad libitum refeeding, the Crtc2 mice re-gained more weight to catch up with the WT mice (Figure 3C), which may be due to the anabolic effects of higher insulin sensitivity (Figure 2). We found that locomotion was similarly reduced in both groups during the refeeding period (Supporting Information Figure S4C), the number of meals doubled in both groups, while meal duration and number of bouts per meal declined similarly (Figure 3D). We observed a reduction in kilocalories consumed in the Crtc2 relative to the WT group, which was significant only during the second ADF cycle, when the WT mice increased consumption at a faster rate than the Crtc2 mice (Figure 3D), similar to the effect of exercise on reducing binge eating following relapse from dieting.6,11,12 Control mice lost more lean mass, while Crtc2 mice conserved lean mass and burned the excess fat accumulated during aging (Figure 3E), similar to the effects of exercise.48 To verify that Crtc2 helps maintain lower body weight in a different model, we placed mice on a high-fat diet, and found that Crtc2 mice also maintained lower body weight than WT mice in this context (Figure 3F). To test for pre-fasting changes in the central control of feeding and energy expenditure, we completed a leptin infusion study, as leptin secretion from adipocytes both suppresses hunger and stimulates the SNS. The WT and Crtc2 mice displayed identical patterns of weight loss following leptin infusion, and there were also no differences in plasma levels of feeding hormones, cholesterol, or triglycerides (Supporting Information Figure S4D–F). Thus, the activation of Crtc2/Creb1 signaling in skeletal muscle significantly modulated body composition and weight loss, mimicking the effects of exercise training including preferential loss of fat mass.
3.4 |. Crtc2 maintains energy expenditure during fasting
Lowered metabolic rate is one of the major defenses against fasting-induced weight loss. As expected, ad libitum oxygen consumption cycled with activity in the mice, which increased at night and decreased during the day (Figure 4A, Supporting Information Figure S5A). Fasting reduced oxygen consumption at every cycle, and this effect was more pronounced during the day as the fast was extended (Figure 4A). However, Crtc2 mice consistently showed higher oxygen consumption, independent of fasting (Figure 4A).
FIGURE 4.
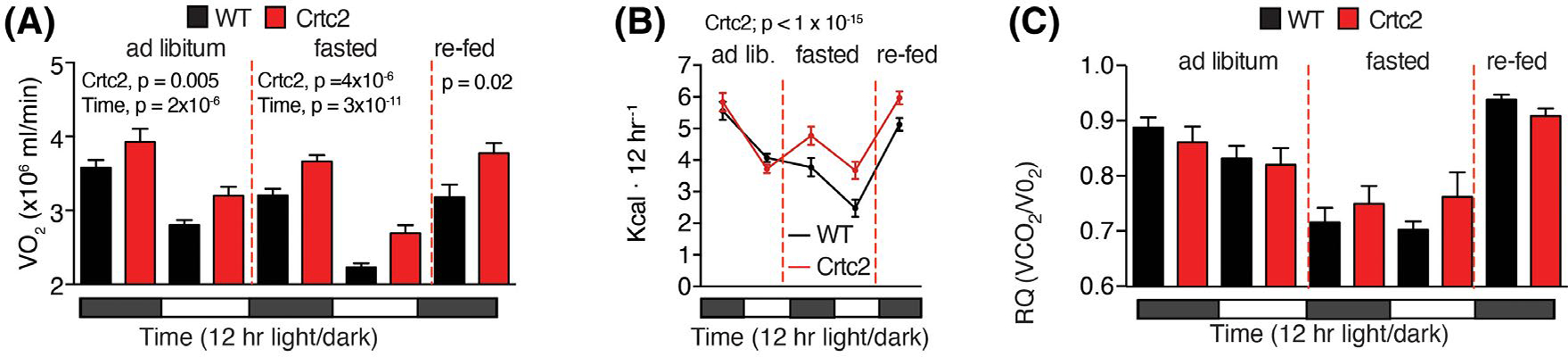
Whole-body energy expenditure is reduced by weight loss and fasting and is reversed by Crtc2 expression. (A) VO2 and VCO2 were measured continuously (Figure S2A) for 72 h. in a CLAMS animal monitoring system. Shown are the 12-h averages. N = 8 mice per group. (B) Total energy expenditure was calculated from VO2 and VCO2 values using the Lusk equation. (C) The respiratory quotient (RQ) was determined during the CLAMS experiment. Data are shown as mean ± SEM and were analyzed by two-way ANOVA for the different feeding conditions separately
When fed ad libitum, Crtc2 and control mice display similar rates of energy expenditure (EE) (Figure 4B). We calculated EE from indirect calorimetry measurements of VO2 and VCO2 and performed ANCOVA analysis using body weight and NMR data as covariates to adjust the EE to total body weight and body composition (Supporting Information Figure S5B). While fasting, the EE of WT mice dropped significantly. In contrast, during fasting EE in Crtc2 mice did not drop below the ad libitum levels (Figure 4B). Respiratory quotient was also reduced during fasting, indicating greater utilization of fat for fuel, but the differences between WT and Crtc2 mice were not significant (Figure 4C, Supporting Information Figure S5C). Maintenance of a higher energy expenditure during fasting thus links Crtc2 activity to increased weight loss during fasting.
3.5 |. The transcriptional response to fasting and weight loss and its regulation by Crtc2
To understand the skeletal muscle intrinsic effects of Crtc2 on dieting and weight loss, we performed transcriptional profiling of mouse tibialis anterior (TA) muscles electroporated with control and Crtc2 expression plasmids in contralateral legs. One cohort of electroporated mice was subject to 3 cycles of ADF, while the other cohort was fed ad libitum. TA muscle tissues were collected from both cohorts at the end of the third 24-h fast. The electroporation increased Crtc2 mRNA levels by ~2.5-fold across groups (Figure 5A). In response to ADF, we identified 1880 differentially expressed genes (DEGs) in the control-transduced muscle and 3197 DEGs in Crtc2-transduced muscle, with 1380 genes in common between the two groups (Figure 5B). Analysis of canonical signaling pathways and gene ontology (GO) annotations revealed that ADF modulated the transcriptional programs for lipid and carbohydrate metabolism, fatty acid beta-oxidation, and insulin signaling (Figure 5C–E, annotated gene lists are in Supporting Information Dataset 1). There were also >200 DEGs regulated by ADF that encode transcription factors and transcriptional coregulators, and expression of >50 transcription factors, including nuclear receptors, and coregulator genes were altered by Crtc2 over-expression (Figure 5C–E), demonstrating the complexity of the transcriptional response to intermittent fasting in skeletal muscle. ADF also regulated other transcriptional programs that control protein balance, including autophagy, ubiquitin-dependent proteasomal degradation, and RNA splicing (Figure 5C–E).
FIGURE 5.
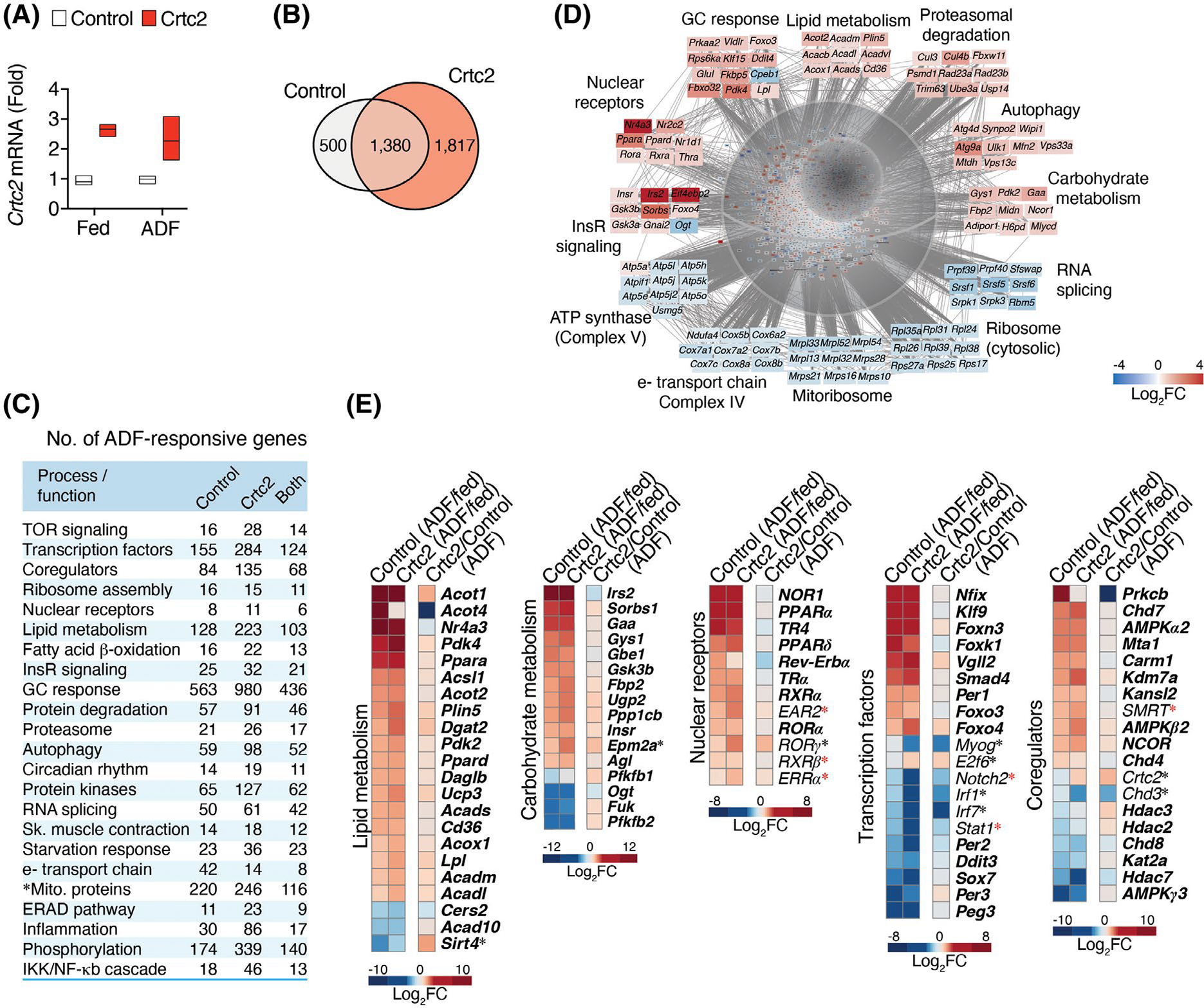
Transcriptional control of fasting and weight loss and its regulation by Crtc2. (A) Mice tibialis anterior (TA) muscle were transduced with GFP control or Crtc2 expression vector. Crtc2 mRNA levels in the TA of mice fed ad libitum (Fed), or subject to three cycles of ADF (Fast) were compared by qPCR. N = 3 mice per group. (B) Venn diagram showing the numbers of DEGs identified by mRNA-seq comparing the effects of ADF in control versus Crtc2-transduced TA muscles. (C) Biological processes and functions regulated by ADF and Crtc2. Gene ontology (GO) analysis suggests that ADF regulates several processes/functions in a Crtc2-sensitive manner. The numbers of DEGs involved in each process are shown. See Supporting Information Dataset S1 for a complete list of represented GO annotations. (D) Examples of ADF-regulated transcriptional programs and mRNAs in control TA muscles. (E) Gene expression profiles in control and Crtc2-transduced TA muscles of mice subjected to ADF relative to the ad libitum fed mice (columns 1–2) , and effect of Crtc2 transduction relative to control during ADF (column 3). ADF-regulated genes in control muscle appear in bold. *Crtc2-regulated genes in mice subjected to ADF. *ADF-regulated genes in Crtc2-transduced muscle but not control. FC, fold change
We also considered that Crtc2 might be regulating secreted proteins that modulate the response to dieting, using the Metazoan Secretome Knowledgebase.49 Among 2500 genes encoding likely secreted proteins, 171 DEGs were regulated by Crtc2 during ADF (Secretome, Supporting Information Dataset 1). Using the Reactome. org peer-reviewed pathway database, we detected significant enrichment of pathways regulating extracellular matrix, inter-cell signaling, and immune function, among others (Supporting Information Table S1), but no changes in myokines that would drive weight loss. However, Crtc2 signaling in skeletal muscle is expected to affect other tissues via changes in skeletal muscle metabolic flux capacity (Figures 1 and 2).
3.6 |. Crtc2 regulation of mTOR signaling and protein synthesis
In mice electroporated with Crtc2 versus GFP control in the contralateral TA muscle, ADF significantly reduced TA weight, but in both the fed and ADF groups Crtc2 increased weight compared to the GFP control muscle (Figure 6A). To further understand the effects of ADF and Crtc2 overexpression on protein synthesis, we treated mice with puromycin for an in vivo SuNSET assay, where immunoblotting for newly incorporated puromycin into growing polypeptide chains allows analysis of new protein synthesis. Crtc2 significantly increased protein synthesis, which planned comparisons showed were significant in the ad libitum fed cohort (Figure 6B).
FIGURE 6.
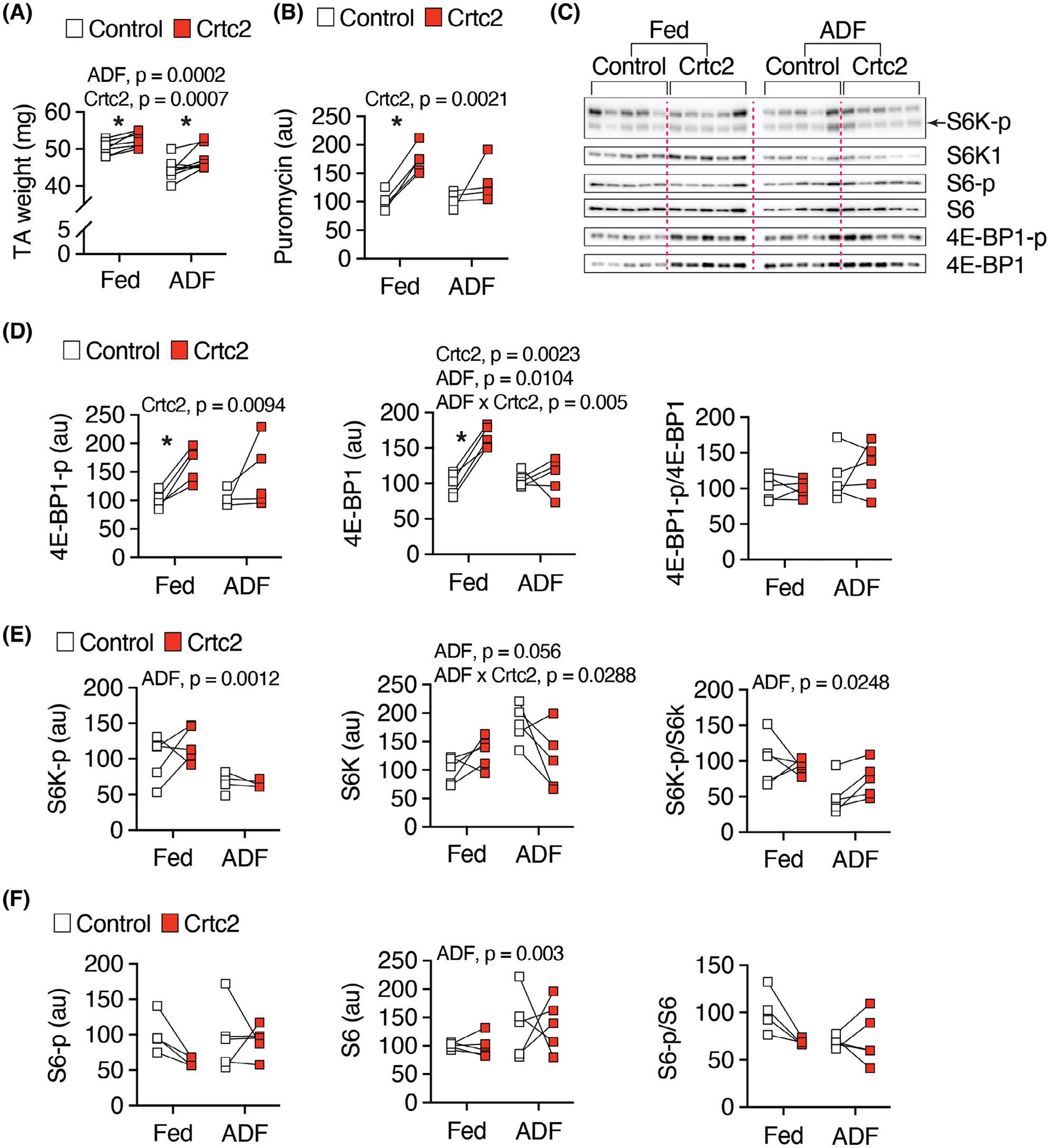
Protein synthesis and mTOR signaling. (A) Mice tibialis anterior (TA) muscle were transduced with GFP control or Crtc2 expression vector. Crtc2 mRNA levels in the TA of mice fed ad libitum (Fed), or subject to three cycles of ADF (Fast) were isolated and weighted. N = 8 mice per group. (B) Quantitation of western blot (Supporting Information Figure S6B) for SUnSET protein synthesis assay. Mice electroporated and fed as in (A) were fasted overnight and treated with puromycin for 30 min and then the TA muscles collected for western blot. N = 4 mice per group. (C) TA muscle were isolated from mice electroporated and fed as in (A) for western blot analyses. N = 5 per group. (D–F) Quantitation of (C)
Crtc2 and ADF regulated the expression of genes in the mTOR signaling pathway that controls protein synthesis (Supporting Information Figure S6A, Supporting Information Dataset S1). Surprisingly, Crtc2 stimulated the expression of the protein synthesis inhibitor gene Eif4ebp1 during ADF (Supporting Information Figure S6B). Upon activation, mTOR phosphorylates the Eif4ebp1 gene product 4E-BP1 thereby releasing Eif4e from inhibiting the translation initiation factor, Eif4e to stimulate protein synthesis. Western blotting showed that the increase in 4E-BP-1 expression was accompanied by a stimulation of phosphorylation of 4E-BP-1 by Crtc2, preferentially in the ad libitum fed cohort (Figure 6C,D). As expected, the downstream marker of mTOR activity, phosphorylation of S6K, was inhibited by ADF, while we did not observe changes in an additional marker, phosphorylation of S6 (Figure 6E,F). Thus, Crtc2 induced anabolic effects on protein synthesis supporting retention of muscle mass during the fast (Figures 3B and 6A).
3.7 |. Crtc2 attenuates fasting-induced effects on mitochondria
In addition to lower metabolic rate, fasting, and weight loss render humans and other mammals more energetically efficient, such that the same amount of work is performed with less ATP production, suggesting a remodeling of glycolytic or oxidative capacity.3,50 ADF modulated the expression of >200 genes whose proteins localize in mitochondria according to MitoCarta36 (Figure 7A). ADF downregulated most of the genes that encode the electron transport chain (complexes I–IV), the ATP synthase (complex V), the mitoribosome, and the cytosolic ribosome (Figure 7A–C, Supporting Information Figure S7A), but significantly upregulated genes such as Sdha, Uqcrc1, and Atp5a1 (Figure 6B, Supporting Information Figure S7A). While we observed that Crtc2 upregulated mitochondrial complex II activity (Figure 1A, Supporting Information Figure S1), we did not observe changes in citrate synthase activity in the electroporation experiment, although ADF modestly stimulated mRNA expression (Supporting Information Figure S7B).
FIGURE 7.
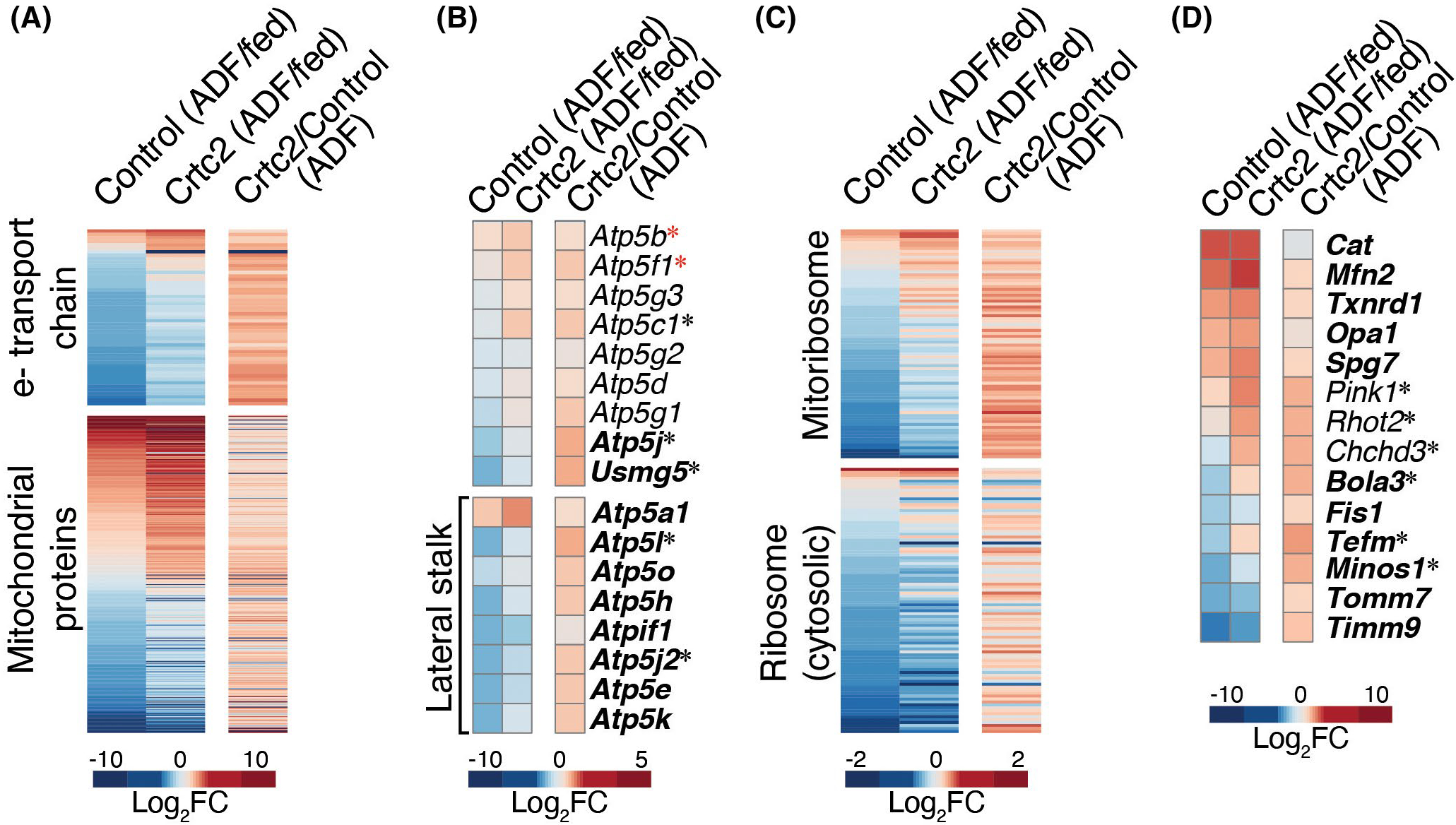
Alternate day fasting and Crtc2 regulation of mitochondrial gene expression. (A–D) Gene expression profiles in control and Crtc2-transduced TA muscles of mice subjected to ADF relative to the ad libitum fed mice (columns 1–2) , and effect of Crtc2 transduction relative to control during ADF (column 3). The profiles of all genes encoding (A) mitochondrial proteins, including subunits of electron transport chain (ETC) complexes, (B) subunits of the mitochondrial ATPase (complex V), (C) ribosomal subunits, and (D) highlighted mitochondrial proteins, are shown. ADF-regulated genes in control muscle appear in bold. *Crtc2-regulated genes in mice subjected to ADF. *ADF-regulated genes in Crtc2-transduced muscle but not control. FC, fold change
We also observed a significant ADF-induced expression of genes encoding ROS scavenger proteins such as thioredoxin (Txnrd1) and catalase (Cat), and genes regulating mitochondrial stability, Tomm7, Timm9, and the AAA protease Spg,7 known to be active during the removal of damaged mitochondrial proteins (Figure 7D). Among the ATP synthase genes, there was a more pronounced loss of gene expression in the lateral stalk genes (Figure 7B), which may decrease the conductance of mitochondrial permeability transition pore (mPTP) and thus protect mitochondria from detrimental swelling and oxidative injury.51 ADF also induced increases in mitochondrial fusion (Mfn2 and Opa1), and decreased fission (Fis) gene expression, consistent with the protective effects of fusion during nutrient deprivation.52 These adaptations could be advantageous for weight loss as they prepare the mitochondria for the reduced metabolic rate and increased reliance on lipid metabolism during fasting, without causing permanent damage to the mitochondrial pool within the cell.
In mice subjected to ADF, Crtc2 transduction altered expression of 80 genes whose proteins localize in the mitochondria. Crtc2 expression strongly attenuated the ADF-dependent repression of electron transport chain genes and the mitoribosome (Figure 7A–C, Supporting Information Figure S7A), and generally upregulated the other genes encoding proteins that localize to mitochondria. In addition, Crtc2 enhanced expression of ribosomal genes that facilitate mitoribosome assembly, and genes such as Spg7, Pink1, RhoT2, and Chchd3 that control the quality of mitochondrial proteins, but blocked ADF-dependent downregulation of Minos1, a central component of the mitochondrial inner membrane organizing system (Figure 7D). Crtc2 also upregulated the expression of Tefm, the mitochondrial transcriptional elongation factor, and Bola3, which participates in the electron transport chain assembly.53 These data suggest potential mechanisms to explain how physical activity—through Crtc2/Creb1-regulated transcriptional programs—can help maintain a higher metabolic rate and a lower body weight by upregulating expression of mitochondrial respiratory chain subunits and modulating the expression of other mitochondria-localized gene products, consistent with the well-known beneficial effects of exercise on mitochondrial biogenesis and energy expenditure.
3.8 |. Crtc2 reverses catabolic effects of glucocorticoids on skeletal muscle
To further understand the signaling pathways that are regulated by fasting and Crtc2, we noted that glucocorticoids (GCs) are one of the primary drivers of systemic catabolism during fasting. In response to ADF and Crtc2 overexpression, we observed over a thousand DEGs that were annotated as “glucocorticoid response” genes (Figure 5C). In the context of an overnight fast, treatment with dexamethasone (Dex) more than doubled the loss of lean mass (Figure 8A). Using high-content imaging, we observed that Dex reduced the diameter and mitochondrial potential of nutrient-deprived primary mouse skeletal myotubes (Figure 8B). GCs can also activate the master energy sensor, AMP-activated protein kinase (AMPK),54 which can act as a glucocorticoid receptor (GR) coactivator.55 AMPK is activated by a lower ATP/ADP ratio during fasting to inhibit anabolism and stimulate catabolic processes such as autophagy and mitophagy. In primary myotubes, we observed a Dex-dependent activating phosphorylation of AMPK that increased over time, consistent with an underlying transcriptional mechanism (Figure 8C). Activation of AMPK with both Dex and the AMP analog, AICAR, had an additive detrimental effect on myotube diameter (Figure 8D), demonstrating that GR collaborates with AMPK to drive skeletal muscle atrophy. Primary myotubes transduced with dominant-negative Creb (A-Creb) were significantly smaller than control, suggesting that Crtc2 may be acting through Creb1 to regulate anabolic effects. Myotubes transduced with Crtc2 were larger than control and reversed the catabolic effects of Dex (Figure 8E), indicating that GR and Crtc2 drive opposing effects on skeletal muscle.
FIGURE 8.
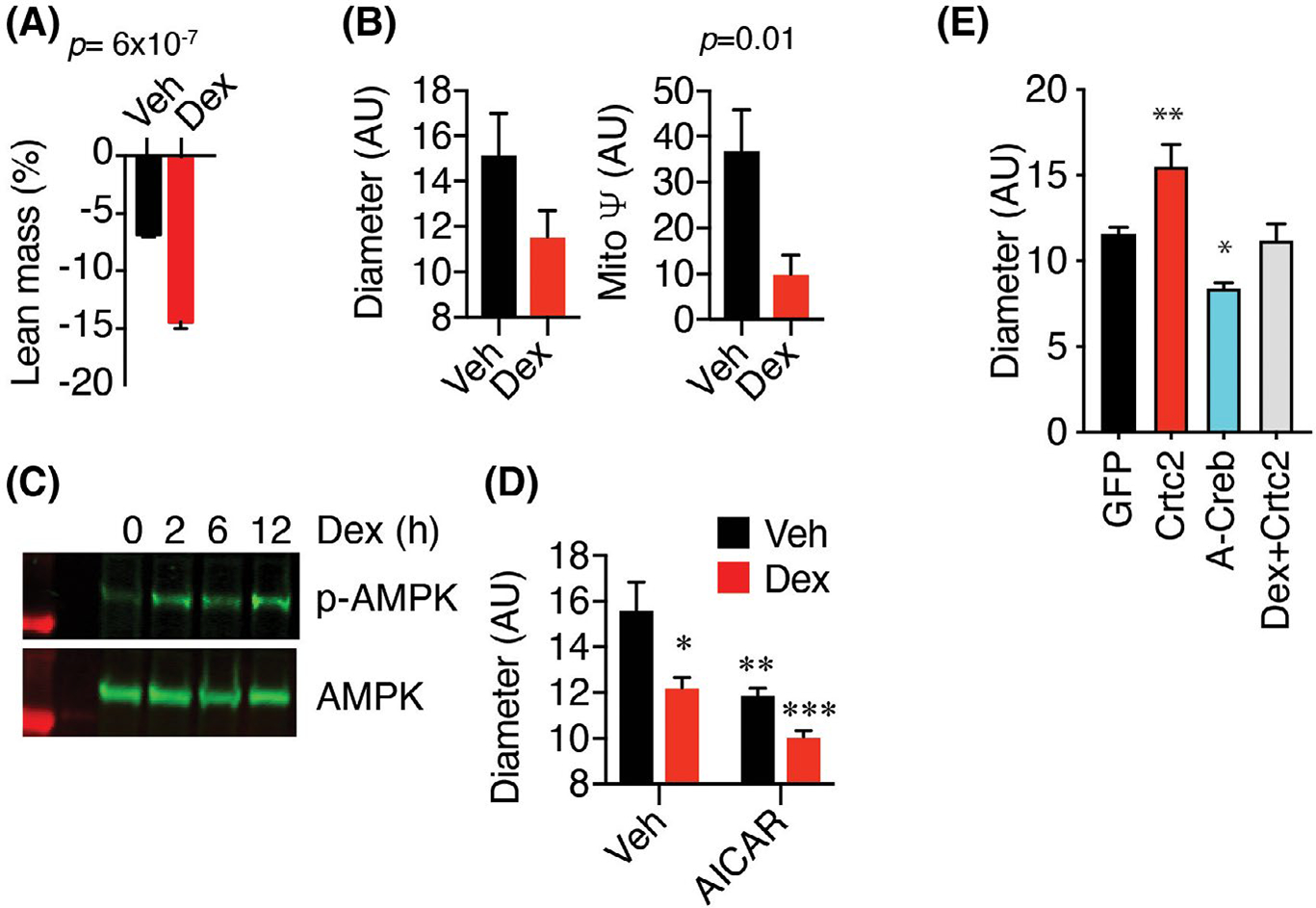
Glucocorticoids and AMPK regulate skeletal muscle atrophy and mitophagy. (A) Effects of Dex on lean body mass. Mice injected with vehicle or 10 mg/kg Dex were fasted overnight. Lean mass was measured by whole-body NMR before and after treatment. N = 5 mice per group. (B) Diameter and mitochondrial potential of steroid-deprived primary myotubes treated for 2 days with vehicle or 100 nM Dex were compared by confocal high-content imaging. N = 24–27 images × 50–100 myotubes per image. (C) Steroid-deprived C2C12 myotubes were stimulated with 100 nM Dex for 0–12 h. Levels of phosphorylated and total AMPK in whole lysates were compared by western blot. (D) Diameter of steroid deprived C2C12 myotubes treated for 48 h with vehicle, 100 nM Dex, or 1 μM AICAR were compared by confocal high content imaging. N = 24–27 images × 50–100 myotubes per image. (E) C2C12 myocytes were transduced with adenovirus GFP, Crtc2, or A-Creb genes and analyzed by confocal high content imaging. N = 24–27 images × 50–100 myotubes per image
4 |. DISCUSSION
During exercise, the SNS through actions of catecholamines is thought to control blood supply(cardiodynamics) and the catabolic processes necessary to regulate immediate energy during an intense activity, such as lipolysis and glycogen brakedown. This work demonstrates that the transcriptional arm the SNS in skeletal muscle, Crtc2/Creb1, mediates metabolic rewiring having most of the hallmarks described as part of exercise adaptation response, including mitochondrial biogenesis and increased respiratory capacity, increased lipid flux, improved insulin sensitivity and glucose utilization (Figures 1 and 2). The combination of b-AR and Ca2+ signaling are required to maximally stimulate nuclear translocation of Crtc2 in skeletal muscle,14 providing a mechanism through which the physiological signal cardiometabolic stress mediated by SNS/Crtc2/Creb1 signaling can synergize with the stimulus from the contracting muscle to mediate adaptive responses specifically in the working muscle. Here, overexpression of Crtc2 circumvented the neuronal activation of Creb1 (Supporting Information Figure S8). Combined with our earlier work showing increased exercise performance, myotube diameter, and storage of IMTG and glycogen14 in the Crtc2 mice, this work suggests a revision of the role of the SNS in exercise physiology as coordinating both catabolic processes during the stressor and subsequent anabolic and metabolic adaptations.
Metabolic effects of Crtc2 were supported by upregulation of mitochondrial biogenesis master regulatory genes, Nr4a1, Nr4a3, Essrra/ERRα, and Ppargc1a/PGC-1α, genes whose proteins localize to mitochondria, and key regulators of carbohydrate and fat metabolism (Figures 1, 2, 5, 7). Crtc3 is also expressed in skeletal muscle and induces PGC-1α and mitochondrial biolgenesis.24 While Crtc2 and Crtc3 act similarly in many tissues, there are differences in activation pathways that may provide integration of exercise-specific signals.56,57
We present a model of exercise-induced adaptations in skeletal muscle that shows how genes such as PGC-1α can respond to SNS signals, but also other inputs such as Ca++-induced activation of calmodulin, or AMPK activation (Supporting Information Figure S8). The SNS is activated logarithmically by exercise intensity and linearly with time, highlighting that different forms of exercise will activate different response pathways. Overexpression of PGC-1α increases mitochondrial density and exercise performance, but without the increase in metabolic rate that can occur from exercise training,58–61 highlighting the requirement for additional exercised-induced signals. Basal sympathetic tone also regulates fiber composition and exercise capacity, as β-AR KO led to a switch from glycolytic to oxidative fibers, accompanied by greater running capacity. However, these mice experienced a greater decline in performance and increased muscle atrophy following myocardial infarction,62 highlighting the complexity of skeletal muscle adaptations. These observations may explain the variable role of the SNS in activating PGC-1α and regulating exercise responses that may be specific to certain forms of exercise.63–65
Defense of body weight during fasting is through lower metabolic rate and increased hunger. The metabolic adaptations in muscle enabled Crtc2 to reset to a lower body weight, consistent with the effects of rigorous exercise on maintaining weight loss (reviewed in Ref. [5]). In humans, successful weight loss maintainers showed higher overall activity and physical energy expenditure compared to weight matched controls.10 The Crtc2 induced changes in metabolism enabled the maintenance of higher energy expenditure during fasting, accompanied by a maintenance of fat free mass (FFM) and selective loss of adiposity.
ADF-induced binge eating was attenuated in the Crtc2 mice. We didn’t observe differences in satiety/hunger hormones or myokine gene expression and the Crtc2 mice showed no differences in feeding in response to leptin infusion (Supporting Information Figure S4E). However, exercise alters the response to binge eating, enabling greater utilization of fats and reduced adiposity.66 In the context of weight reduced formerly obese rats allowed to relapse, exercise reduced food intake, lowered adiposity, and favored fat oxidization instead of storage,6,11,12 as we observed here in Crtc2 mice. The effects of skeletal muscle on satiety are poorly studied and of unknown mechanisms,13 but the Crtc2 may provide a useful genetic model to study muscle-brain crosstalk.
Reduced energy expenditure and increased hunger during fasting are partly mediated by a lowering of the satiety hormone leptin.2–4 Leptin is secreted basally from adipose tissue in proportion to fat mass, so it is chronically lower during fasting, which lowers sympathetic tone and energy expenditure, reduces thyroid hormone levels, and stimulates the HPA axis67 (Supporting Information Figure S8). We showed that GCs catabolic effects on mitochondria and muscle mass during fasting included activation of AMPK and were reversed by Crtc2. Crtc2 also stimulated new protein synthesis, supported by activation of mTOR signaling. We previously demonstrated that Creb1 and GR work together to coordinate gluconeogenesis transcriptional programs in the liver, where GR doubles the number of Creb1 binding sites and increases binding of Creb1.68 This work suggests that the effects of exercise on fasting may also involve a coordination of Creb1 and GR signaling as master transcriptional regulators of metabolism, in conjunction with other stress-response signals. It is noteworthy that HPA and AMPK activation occur during both exercise and fasting, but those activities have opposing effects on mitochondria. We suggest a model where GR and AMPK signaling induce mitophagy in both settings, while the mitochondrial remodeling is guided by other catabolic or anabolic signaling, such as increased expression of PGC-1α (Supporting Information Figure S8A). This work adds to a body of evidence on Crtc2 and Creb1 as global regulators of metabolism in the liver, pancreas, adipose tissue, and other endocrine tissues by coordinating with other tissue- and stressor-specific transcriptional signaling programs.
Supplementary Material
ACKNOWLEDGMENTS
JCN was supported by the Frenchman’s Creek Women for Cancer Research. The authors thank Ana Lira (University of Iowa) for technical assistance on this project.
Funding information
Frenchman’s Creek Women for Cancer Research
Abbreviations:
- ADF
alternate day fasting
- AICAR
5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside
- ANOVA
analysis of variance
- cAMP
cyclic ATP monophosphate
- CLAMS
comprehensive lab animal monitoring system
- Dex
dexamethasone
- Dox
doxycycline
- GC
glucocorticoid
- GPCR
G-protein coupled receptor
- GTT
glucose tolerance test
- ITT
insulin tolerance test
- SDH
succinate dehydrogenase
- SNS
sympathetic nervous system
- TA
tibialis anterior
- βAR
β-adrenergic receptor
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
DATA AVAILABILITY STATEMENT
The raw and processed RNA-seq dataset available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/geo/and can be retrieved with the accession number GSE14 9150, which will be released upon publication of this manuscript. The processed RNA-seq dataset is also provided in SI Appendix, Dataset S1. Relevant protocols and raw data not detailed in the Methods or Supplemental Information are available upon request.
REFERENCES
- 1.Melby CL, Paris HL, Foright RM, Peth J. Attenuating the biologic drive for weight regain following weight loss: must what goes down always go back up? Nutrients. 2017;9(5):468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arone LJ, Mackintosh R, Rosenbaum M, Leibel RL, Hirsch J. Autonomic nervous system activity in weight gain and weight loss. Am J Physiol. 1995;269:R222–R225. [DOI] [PubMed] [Google Scholar]
- 3.Rosenbaum M, Vandenborne K, Goldsmith R, et al. Effects of experimental weight perturbation on skeletal muscle work efficiency in human subjects. Am J Physiol Regul Integr Comp Physiol. 2003;285:R183–R192. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum M, Goldsmith R, Bloomfield D, et al. Low-dose leptin reverses skeletal muscle, autonomic, and neuroendocrine adaptations to maintenance of reduced weight. J Clin Invest. 2005;115:3579–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foright RM, Presby DM, Sherk VD, et al. Is regular exercise an effective strategy for weight loss maintenance? Physiol Behav. 2018;188:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacLean PS, Higgins JA, Wyatt HR, et al. Regular exercise attenuates the metabolic drive to regain weight after long-term weight loss. Am J Physiol Regul Integr Comp Physiol. 2009;297:R793–R802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turk Y, Theel W, Kasteleyn MJ, et al. High intensity training in obesity: a meta-analysis. Obes Sci Pract. 2017;3:258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mourier A, Gautier J-F, Kerviler ED, et al. Mobilization of visceral adipose tissue related to the improvement in insulin sensitivity in response to physical training in NIDDM. Effects of branched-chain amino acid supplements. Diabetes Care. 1997;20:385–391. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbaum M, Heaner M, Goldsmith RL, et al. Resistance training reduces skeletal muscle work efficiency in weight-reduced and non-weight-reduced subjects. Obesity. 2018;26:1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostendorf DM, Caldwell AE, Creasy SA, et al. Physical activity energy expenditure and total daily energy expenditure in successful weight loss maintainers. Obesity. 2019;27:496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steig AJ, Jackman MR, Giles ED, et al. Exercise reduces appetite and traffics excess nutrients away from energetically efficient pathways of lipid deposition during the early stages of weight regain. Am J Physiol Regul Integr Comp Physiol. 2011;301:R656–R667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins JA, Jackman MR, Brown IL, et al. Resistant starch and exercise independently attenuate weight regain on a high fat diet in a rat model of obesity. Nutr Metab. 2011;8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grannell A, De Vito G, Murphy JC, le Roux CW. The influence of skeletal muscle on appetite regulation. Expert Rev Endocrinol Metab. 2019;14:267–282. [DOI] [PubMed] [Google Scholar]
- 14.Bruno NE, Kelly KA, Hawkins R, et al. Creb coactivators direct anabolic responses and enhance performance of skeletal muscle. The EMBO J. 2014;33:1027–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popov DV, Makhnovskii PA, Shagimardanova EI, et al. Contractile activity-specific transcriptome response to acute endurance exercise and training in human skeletal muscle. Am J Physiol Endocrinol Metab. 2019;316:E605–E614. [DOI] [PubMed] [Google Scholar]
- 16.Egan B, Carson BP, Garcia-Roves PM, et al. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J Physiol. 2010;588:1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popov DV, Lysenko EA, Bokov RO, et al. Effect of aerobic training on baseline expression of signaling and respiratory proteins in human skeletal muscle. Physiol Rep. 2018;6:e13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 19.Screaton RA, Conkright MD, Katoh Y, et al. The CREB coactivator TORC2 functions as a calcium-and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. [DOI] [PubMed] [Google Scholar]
- 20.Ch’ng T, Uzgil B, Lin P, et al. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter PS, Martel MA, McMahon A, Kind PC, Hardingham GE. Pituitary adenylate cyclase-activating peptide induces long-lasting neur oprotection through the induction of activity-dependent signaling via the cyclic AMP response element-binding protein-regulated transcription co-activ ator 1. J Neurochem. 2011;118:365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taub M, Garimella S, Kim D, Rajkhowa T, Cutuli F. Renal proximal tubule Na, K-ATPase is controlled by CREB-regulated transcriptional coactivators as well as salt-inducible kinase 1. Cell Signal. 2015;27:2568–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berdeaux R, Hutchins C. Anabolic and pro-metabolic functions of CREB-CRTC in skeletal muscle: advantages and obstacles for type 2 diabetes and cancer cachexia. Front Endocrinol. 2019;10:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Z, Huang X, Feng Y, et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci U S A. 2006;103:14379–14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo S-H, Flechner L, Qi L, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. [DOI] [PubMed] [Google Scholar]
- 26.Dentin R, Liu YI, Koo S-H, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. [DOI] [PubMed] [Google Scholar]
- 27.Dentin R, Hedrick S, Xie J, Yates J 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. [DOI] [PubMed] [Google Scholar]
- 28.Grill MA, Bales MA, Fought AN, Rosburg KC, Munger SJ, Antin PB. Tetracycline-inducible system for regulation of skeletal muscle-specific gene expression in transgenic mice. Transgenic Res. 2003;12(1):33–43. [DOI] [PubMed] [Google Scholar]
- 29.Heikkinen S, Argmann CA, Champy MF, Auwerx J. Evaluation of glucose homeostasis. Curr Protoc Mol Biol. 2007;Chapter 29:Unit 29B.3. [DOI] [PubMed] [Google Scholar]
- 30.Stengel A, Goebel M, Wang L, et al. Activation of brain somatostatin 2 receptors stimulates feeding in mice: analysis of food intake microstructure. Physiol Behav. 2010;101(5):614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margolis RN, Evans RM, O’Malley BW, N. A. Consortium. The nuclear receptor signaling atlas: development of a functional atlas of nuclear receptors. Mol Endocrinol. 2005;19:2433–2436. [DOI] [PubMed] [Google Scholar]
- 36.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medvedeva YA, Lennartsson A, Ehsani R, et al. EpiFactors: a comprehensive database of human epigenetic factors and complexes. Database. 2015;2015:bav067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo T, Lew MJ, Mayba O, et al. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc Natl Acad Sci USA. 2012;109:11160–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Springer ML, Blau HM. High-efficiency retroviral infection of primary myoblasts. Somat Cell Mol Genet. 1997;23(3):203–209. [DOI] [PubMed] [Google Scholar]
- 41.Hughes DC, Turner DC, Baehr LM, et al. Knockdown of the E3 ubiquitin ligase UBR5 and its role in skeletal muscle anabolism. Am J Physiol Cell Physiol. 2021;320:C45–C56. [DOI] [PubMed] [Google Scholar]
- 42.Conkright MD, Canettieri G, Screaton R, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12(2):413–423. [DOI] [PubMed] [Google Scholar]
- 43.Blanco CE, Sieck GC, Edgerton VR. Quantitative histochemical determination of succinic dehydrogenase activity in skeletal muscle fibres. Histochem J. 1988;20:230–243. [DOI] [PubMed] [Google Scholar]
- 44.Goodman CA, Mabrey DM, Frey JW, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drake JC, Wilson RJ, Yan Z. Molecular mechanisms for mitochondrial adaptation to exercise training in skeletal muscle. FASEB J. 2016;30:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearen MA, Muscat GE. Minireview: nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maxwell MA, Cleasby ME, Harding A, Stark A, Cooney GJ, Muscat GEO. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J Biol Chem. 2005;280:12573–12584. [DOI] [PubMed] [Google Scholar]
- 48.Verheggen RJHM, Maessen MFH, Green DJ, Hermus ARMM, Hopman MTE, Thijssen DHT. A systematic review and meta-analysis on the effects of exercise training versus hypocaloric diet: distinct effects on body weight and visceral adipose tissue. Obes Rev. 2016;17:664–690. [DOI] [PubMed] [Google Scholar]
- 49.Meinken J, Walker G, Cooper CR, Min XJ. MetazSecKB: the human and animal secretome and subcellular proteome knowledgebase. Database. 2015;2015:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldsmith R, Joanisse DR, Gallagher D, et al. Effects of experimental weight perturbation on skeletal muscle work efficiency, fuel utilization, and biochemistry in human subjects. Am J Physiol Regul Integr Comp Physiol. 2010;298:R79–R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giorgio V, von Stockum S, Antoniel M, et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci U S A. 2013;110:5887–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108:10190–10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasta V, Suraci D, Gourdoupis S, Ciofi-Baffoni S, Banci L. A pathway for assembling [4Fe-4 S]2+ clusters in mitochondrial iron-sulfur pr otein biogenesis. FEBS J. 2020;287(11):2312–2327. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Peng Y, Wang X, et al. Mitochondrial dysfunction launches dexamethasone-induced skeletal muscle atrophy via AMPK/FOXO3 signaling. Mol Pharm. 2016;13:73–84. [DOI] [PubMed] [Google Scholar]
- 55.Ratman D, Mylka V, Bougarne N, et al. Chromatin recruitment of activated AMPK drives fasting response genes cocontrolled by GR and PPARalpha. Nucleic Acids Res. 2016;44:10539–10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Than TA, Lou H, Ji C, Win S, Kaplowitz N. Role of cAMP-responsive element-binding protein (CREB)-regulated transcription coactivator 3 (CRTC3) in the initiation of mitochondrial biogenesis and stress response in liver cells. J Biol Chem. 2011;286:22047–22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonntag T, Ostojić J, Vaughan JM, et al. Mitogenic signals stimulate the CREB coactivator CRTC3 through PP2A recruitment. iScience. 2019;11:134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi CS, Befroy DE, Codella R, et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci U S A. 2008;105:19926–19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tadaishi M, Miura S, Kai Y, Kano Y, Oishi Y, Ezaki O. Skeletal muscle-specific expression of PGC-1alpha-b, an exerciseresponsive isoform, increases exercise capacity and peak oxygen uptake. PLoS One. 2011;6:e28290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shook RP, Hand GA, Paluch AE, et al. Moderate cardiorespiratory fitness is positively associated with resting metabolic rate in young adults. Mayo Clin Proc. 2014;89:763–771. [DOI] [PubMed] [Google Scholar]
- 61.Gilliat-Wimberly M, Manore MM, Woolf K, Swan PD, Carroll SS. Effects of habitual physical activity on the resting metabolic rates and body compositions of women aged 35 to 50 years. J Am Diet Assoc. 2001;101:1181–1188. [DOI] [PubMed] [Google Scholar]
- 62.Voltarelli VA, Bechara LR, Bacurau AV, et al. Lack of β2-adrenoceptors aggravates heart failure-induced skeletal muscle myopathy in mice. J Cell Mol Med. 2014;18(6):1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miura S, Kawanaka K, Kai Y, et al. An increase in murine skeletal muscle peroxisome proliferator-activated receptorgamma coactivator-1alpha (PGC-1alpha) mRNA in response to exercise is mediated by beta-adrenergic receptor activation. Endocrinology. 2007;148:3441–3448. [DOI] [PubMed] [Google Scholar]
- 64.Brandt N, Nielsen L, Thiellesen Buch B, et al. Impact of β-adrenergic signaling in PGC-1α-mediated adaptations in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2018;314(1):E1–E20. [DOI] [PubMed] [Google Scholar]
- 65.Kim SH, Asaka M, Higashida K, Takahashi Y, Holloszy JO, Han DH. beta-Adrenergic stimulation does not activate p38 MAP kinase or induce PGC-1alpha in skeletal muscle. Am J Physiol Endocrinol Metab. 2013;304:E844–E852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levin BE, Dunn-Meynell AA. Chronic exercise lowers the defended body weight gain and adiposity in diet-induced obese rats. Am J Physiol Regul Integr Comp Physiol. 2004;286:R771–R778. [DOI] [PubMed] [Google Scholar]
- 67.Perry RJ, Resch JM, Douglass AM, et al. Leptin’s hunger-suppressing effects are mediated by the hypothalamicpituitary-adrenocortical axis in rodents. Proc Natl Acad Sci USA. 2019;116:13670–13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldstein I, Baek S, Presman DM, Paakinaho V, Swinstead EE, Hager GL. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 2017;27:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed RNA-seq dataset available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/geo/and can be retrieved with the accession number GSE14 9150, which will be released upon publication of this manuscript. The processed RNA-seq dataset is also provided in SI Appendix, Dataset S1. Relevant protocols and raw data not detailed in the Methods or Supplemental Information are available upon request.


