Abstract
We aimed to investigate potential roles of LRRC75A‐AS1 delivered by M2 macrophage exosomes in inducing cervical cancer progression. We demonstrated LRRC75A‐AS1 was highly expressed in exosomes from M2 macrophages which could be absorbed by Hela cells. M2 macrophage‐derived exosomes promoted Hela cell proliferation, migration, invasion, and EMT process by delivering LRRC75A‐AS1. LRRC75A‐AS1 directly targeted and suppressed miR‐429 in Hela cells. The regulation of cell functions by exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages was abrogated by miR‐429 mimics. miR‐429 directly targeted and repressed SIX1 expression. SIX1 overexpression alleviated the modulation of cellular functions and STAT3/MMP‐9 signaling by miR‐429 mimics. Also, miR‐429 overexpression or SIX1 silence repressed tumor formation and metastasis in nude mice, which was mitigated by exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages. In conclusion, LRRC75A‐AS1 delivered by M2 macrophage exosomes repressed miR‐429 to elevate SIX1 expression and promote cervical cancer progression through activating the STAT3/MMP‐9 axis.
Keywords: cervical cancer, exosomes, LRRC75A‐AS1, M2 macrophages, miR‐429, SIX1
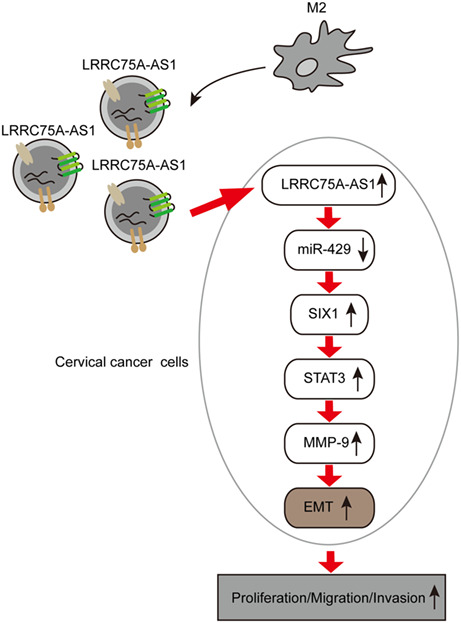
We aimed to investigate potential roles of LRRC75A‐AS1 delivered by M2 macrophage exosomes in inducing epithelial to mesenchymal transition (EMT) and cervical cancer progression.
Abbreviations
- Arg‐1
arginase‐1
- CRKL
Crk‐like adapter protein
- EMT
epithelial to mesenchymal transition
- HPV
human papilloma virus
- LRRC75A‐AS1
the antisense of leucine‐rich repeat‐containing protein 75A
- MMP‐9
matrix metalloproteinase 9
- NTA
nanoparticle tracking analysis
- PMA
phorbol 12‐myristate 13‐acetate
- SIX1
sine oculis homebox 1
- SPF
specific pathogen‐free
- STAT3
signal transducer and activator of transcription 3
- TEM
transmission electron microscopy
- TGF‐β1
transforming growth factor β1
- ZEB1
zinc finger E‐box binding homeobox 1
1. INTRODUCTION
Cervical cancer is a major malignant cancer in females worldwide, which is estimated to cause more than 260, 000 deaths each year. 1 , 2 , 3 The majority of cervical cancer cases are mainly attributed to the persistent infection with human papilloma virus (HPV), as well as other risk factors such as smoking and sexual activities. 2 , 4 , 5 The application of early screening and HPV vaccines has greatly reduced the incidence and mortality of cervical cancer. 1 , 3 However, its incidence still remains high, wherein 15% of cervical cancer patients are usually diagnosed at advanced stages with lymphatic or hematogenous metastasis. 6 Even worse, metastatic patients are commonly not suitable for the common treatments. 6 Therefore, it is of great clinical significance to elucidate the causes of cervical cancer metastasis. The epithelial to mesenchymal transition (EMT) is critical to metastasis of cervical cancer, 7 but its regulatory mechanisms underlying cervical cancer pathogenesis deserve further elucidation.
Exosomes and their noncoding RNA cargos have been regarded as key players in cervical cancer metastasis. 8 , 9 Recent research showed that exosomes from M2 macrophages substantially regulated cancer initiation, metastasis, and chemotherapy resistance. 10 , 11 , 12 , 13 For instance, the migration of gastric cancer cells was reported to be driven by M2 macrophage‐derived exosomes via delivery of the apolipoprotein E protein. 13 However, little is known about the roles of macrophage‐derived exosomes in cervical cancer growth and metastasis. Furthermore, the antisense of leucine‐rich repeat‐containing protein 75A (LRRC75A‐AS1) was characterized as one key lncRNA widely associated with colorectal carcinoma, glioblastoma, and breast cancer. 14 , 15 , 16 , 17 Bioinformatics data from Kaplan–Meier Plotter indicated that high LRRC75A‐AS1 expression was associated with poor survival rate of cervical cancer patients. Our preliminary experiments indicated LRRC75A‐AS1 was highly expressed in cervical cancer tissues and exosomes from M2 macrophages, but its potential roles delivered by M2 macrophage‐derived exosomes in cervical cancer remain largely unknown.
miR‐429 suppressed the invasion and migration of cervical cancer cells through targeting zinc finger E‐box binding homeobox 1 (ZEB1) and Crk‐like adapter protein (CRKL) to inhibit the EMT process. 18 Our bioinformatics analysis (Starbase) showed that LRRC75A‐AS1 might also target miR‐429, and their association with cervical cancer development deserves further investigation. On the other hand, sine oculis homebox 1 (SIX1) is a homeodomain protein of the SIX family that regulates the EMT process during invasion and metastasis, 19 , 20 , 21 , 22 during which SIX1 activated the signal transducer and activator of transcription 3 (STAT3) to promote matrix metalloproteinase 9 (MMP‐9) activity. 22 , 23 In cervical cancer development, high expression of SIX1 also contributed to cell proliferation, EMT, and lymphangiogenesis. 24 , 25 , 26 Moreover, the expression and role of SIX1 in cervical cancer cells were suppressed by miRNAs such as miR‐362 and miR‐23b. 27 , 28 However, whether miR‐429 could also target SIX1 in cervical cancer is still unaddressed.
In this study, we tested our hypothesis that lncRNA LRRC75A‐AS1 in M2 macrophage‐derived exosomes might sponge miR‐429 to upregulate SIX1 expression and activate the STAT3/MMP‐9 signaling axis, thus activating the EMT process and cervical cancer growth and metastasis.
2. MATERIALS AND METHODS
2.1. Patients and clinical tissues
Totally 30 cervical cancer patients who were diagnosed and underwent surgery at the Hunan Cancer Hospital were enrolled in this study. All experimental operations were approved in advance by the Medical Ethical Committee of the Hunan Cancer Hospital. Written informed consent was signed by all cervical cancer patients. Cervical tissues were collected during surgery and immediately stored in liquid nitrogen for following assays. Detailed clinicopathological characteristics of cervical cancer patients are listed in Table 1.
TABLE 1.
Clinicopathological significance of LRRC75A‐AS1 expression in cervical cancer patients.
| Clinicopathological characteristics | Number of cases | LRRC75A‐AS1 expression | p‐value | |
|---|---|---|---|---|
| High | Low | |||
| Age (year) | ||||
| <50 | 13 | 5 | 8 | 0.4621 |
| ≥50 | 17 | 10 | 7 | |
| Stages | ||||
| I–II | 12 | 2 | 10 | 0.0078 |
| III–IV | 18 | 13 | 5 | |
| Lymph node metastasis | ||||
| No | 16 | 4 | 12 | 0.0092 |
| Yes | 14 | 11 | 3 | |
| Differentiation degree | ||||
| Weak–moderate | 15 | 8 | 7 | 1.0000 |
| Poor | 15 | 7 | 8 | |
| Tumor size | ||||
| ≥6 cm | 20 | 11 | 9 | 0.6999 |
| <6 cm | 10 | 4 | 6 | |
2.2. Bioinformatic analysis
The correlation between LRRC75A‐AS1/miR‐429 expression and overall survival time of cervical cancer patients on a large scale was analyzed using the Kaplan–Meier Plotter platform (http://kmplot.com/analysis/). 29 SIX1 expression in large‐scale cervical cancer population was analyzed by searching TCGA using the UALCAN website (http://ualcan.path.uab.edu/index.html). 30 The interaction between LRRC75A‐AS1 and miRNAs was predicted via Starbase. 31 The association of SIX1 mRNA with miR‐429 was predicted with TargetScan (http://www.targetscan.org/vert_71/). 32
2.3. M2 macrophage polarization induction
The induction of M2 polarization of macrophages was performed as previously described. 10 , 33 Briefly, the differentiation of THP‐1 cells into macrophages was induced by treating with phorbol 12‐myristate 13‐acetate (PMA; #ab120297; Abcam; 320 nM) at 37°C for 6 h, which were then incubated with PMA, IL‐4 (#ab155733; Abcam; 20 ng/mL), and IL‐13 (#ab270079; Abcam; 20 ng/mL) for 18 h at 37°C. The identity of M2 macrophages was confirmed by detecting the expression of M2 macrophage biomarkers including CD206, CD163, Arginase‐1 (Arg‐1), transforming growth factor β1 (TGF‐β1), and IL‐10 through flow cytometry and quantitative RT‐PCR.
2.4. M2 macrophage detection by flow cytometry
CD206 and CD163 expression on the surface of M2 macrophages or tumor tissues was analyzed by flow cytometry as previously described. 12 Briefly, tissues were digested with trypsin for release of macrophages. Subsequently, macrophages isolated from the tissues or induced from THP‐1 cells were washed with PBS two times, incubated with primary antibodies targeting CD11b (labeled with FITC, #301404; Biolegend), CD206 (labeled with APC, #321110; Biolegend) or CD163 (labeled with PE, #333606; Biolegend) for 45 min at 4°C, washed again with PBS, and analyzed in flow cytometer (BD Biosciences).
2.5. Exosome isolation and characterization
Exosomes from M2 macrophages were extracted using the ExoQuick® ULTRA EV Isolation System Kit (#EQULTRA‐20A‐1; System Biosciences) following the manufacturer's instructions. Briefly, culture medium was collected when cell confluence was between 80% and 90% by centrifuge. The supernatant containing exosomes was incubated with 100 μL ExoQuick solution on ice for 30 min with frequent inverting or flicking. After centrifugation at 3200g for 12 min at 4°C, the pellets containing exosomes were resuspended in 200 μL buffer B, mixed with 200 uL buffer A, and loaded into a purification column. Following mixing for 12 min, exosomes were collected by centrifugation at 900g for 30 s. The morphology and diameter of exosomes were then observed by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) as previously described. 11 , 12
2.6. Exosome uptake analysis
The uptake of M2 macrophage‐derived exosomes by Hela cells was validated as previously described. 12 Briefly, the isolated exosomes were first labeled with PKH26 using the PKH26 Red Fluorescent Cell Linker Kit (#PKH26PCL; Sigma‐Aldrich) strictly following the manufacturer's instruction. Excess fluorescent labelling dyes were then removed by passing through the MW3000 exosome spin columns (Thermo Fishier Scientific). Subsequently, the fluorescent‐labeled exosomes were then incubated with Hela cells for 24 h at 37°C, counter‐stained with DAPI, and finally observed under laser confocal microscopy (Zeiss). The emergence of red fluorescence in Hela cells indicated the uptake of exosomes.
2.7. In vivo tumorigenesis
The in vivo tumorigenesis of cervical cancer was evaluated through the xenograft model as previously introduced with minor modification. 11 Briefly, female BALB/c nude mice at the age of 6 weeks were purchased from the SLAC Laboratory Animal Center and maintained at 24°C–26°C under specific pathogen‐free (SPF)‐grade condition with humidity of 40%–60%. All experiments using mice were approved by the Animal Care and Use Committee of the Hunan Cancer Hospital.
For assessment of the pathogenic roles of LRRC75A‐AS1, totally 50 nude mice were randomly divided into control, OE‐NC‐Exos, OE‐LRRC75A‐AS1‐Exos, shNC‐Exos, and shLRRC75A‐AS1‐Exos groups (n = 10 per group). All mice were subcutaneously or intravenously injected with approximately 2 × 106 or 1 × 106 (for intravenous injection) Hela cells resuspended in PBS and sustained in normal condition for induction of tumor cells‐based xenografts. In the exosomes‐treated groups, mice were treated with 1 × 1012 exosome particles (suspended in 100 μL PBS) isolated from M2 macrophages twice a week through tail vein injection. Mice of the control group were subjected to injection of the same volume of PBS. All mice were sacrificed 30 days after cell injection, and tumor sizes and weights were recorded to evaluate Hela cell tumorigenesis. For metastasis model, lung tissues were collected for the histological evaluation and accounting of lung nodule numbers.
For evaluation of the pathogenic roles of miR‐429 and SIX1, another group of 60 nude mice were randomly divided into the following groups (n = 10): mimics NC, miR‐429 mimics, miR‐429 mimics+OE‐LRRC75A‐AS1‐Exos, sh‐NC, sh‐SIX1, and sh‐SIX1 + OE‐LRRC75A‐AS1‐Exos. Mice were also subcutaneously or intravenously subjected to injection of Hela cells transfected with miR‐429 mimics or sh‐SIX1 followed by treatment with exosomes from M2 macrophages overexpressing LRRC75A‐AS1 as introduced above.
More detailed methods are shown in Appendix S1
3. RESULTS
3.1. High expression of LRRC75A‐AS1 detected in exosomes from M2 macrophages
We first induced the differentiation of THP‐1 cells into M2 macrophages, and found the expression levels of CD206, CD163, Arg‐1, IL‐10, and TGF‐β1 in M2 macrophages were all greatly elevated compared with the control group (Figure 1A,B). These results validated the successful induction of M2 macrophages. Next, we isolated exosomes from the culture medium of M2 macrophages, which exhibited the regular elliptical shape in our TEM observation (Figure 1C). Both the TEM and NTA results showed that the diameters of the majority of these exosomes were about 100–150 nm (Figure 1C,D). Also, exosome markers CD63, TSG101, and CD9 were significantly expressed in M2 macrophage‐derived exosomes (Figure 1E). More importantly, these M2 macrophage‐derived exosomes labeled with PKH26 could effectively be absorbed into Hela cells (Figure 1F). Interestingly, LRRC75A‐AS1 expression in M2 macrophage‐derived exosomes was remarkably higher than that from THP‐1 cells (Figure 1G).
FIGURE 1.
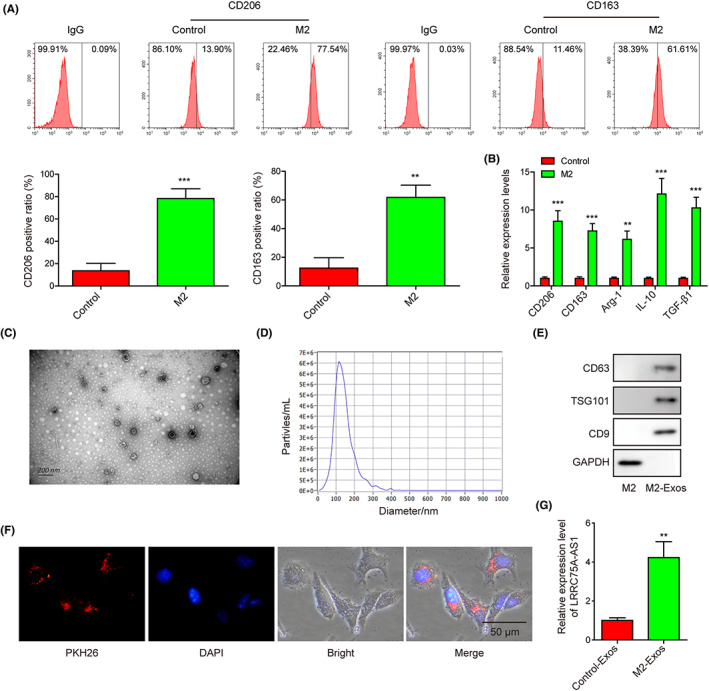
High expression of LRRC75A‐AS1 is detected in exosomes from M2 macrophages. (A) Significant increases of CD206 and C163 expression in M2 macrophages were evaluated by flow cytometry. (B) Great increases of CD206, CD163, Arg‐1, IL‐10, and TGF‐β1 gene expression in M2 macrophages were measured by qRT‐PCR assay. (C) The morphological features of M2 macrophage‐derived exosomes observed through transmission electron microscopy. (D) Diameter distribution of M2 macrophage‐derived exosomes detected by nanoparticle tracking analysis. (E) The expression levels of CD63, TSG101, and CD9 were detected in the exosomes isolated from the M2 macrophages. (F) The uptake of M2 macrophage‐derived exosomes by the Hela cells. (G) Significantly higher LRRC75A‐AS1 expression in M2 macrophage‐derived exosomes detected by qRT‐PCR. **p < 0.01 and ***p < 0.001.
3.2. LRRC75A‐AS1 in M2 macrophage‐derived exosomes induces Hela cell functions and EMT process through STAT3/MMP‐9 signaling
We subsequently found that LRRC75A‐AS1 expression level in Hela cells was greatly decreased compared with the control cell line Ect1/E6E7 or M2 macrophages (Figure 2A,B). Silence and overexpression of LRRC75A‐AS1 in M2 macrophages or its exosomes were achieved (Figure 2C,D). We also confirmed that LRRC75A‐AS1 expression in Hela cells was greatly reduced or elevated by treatment with exosomes from M2 macrophages transfecting LRRC75A‐AS1 silence or LRRC75A‐AS1 overexpression vector (Figure 2E). Moreover, the capabilities of M2 macrophage‐derived exosomes in promoting Hela cell proliferation, migration, and invasion were impaired by LRRC75A‐AS1 silence and enhanced by LRRC75A‐AS1 overexpression in M2 macrophages (Figure 2F–H). In consistence, the STAT3 phosphorylation, MMP‐9, N‐cadherin, and slug protein levels in Hela cells by M2 macrophage‐derived exosomes were abrogated by LRRC75A‐AS1 silence and enhanced by LRRC75A‐AS1 overexpression (Figure 2I). Also, E‐cadherin protein abundances in Hela cells showed opposite effects (Figure 2I). These results showed that LRRC75A‐AS1 in M2 macrophage exosomes promoted the proliferation, migration, invasion, and EMT in Hela cells through STAT3/MMP‐9 signaling.
FIGURE 2.
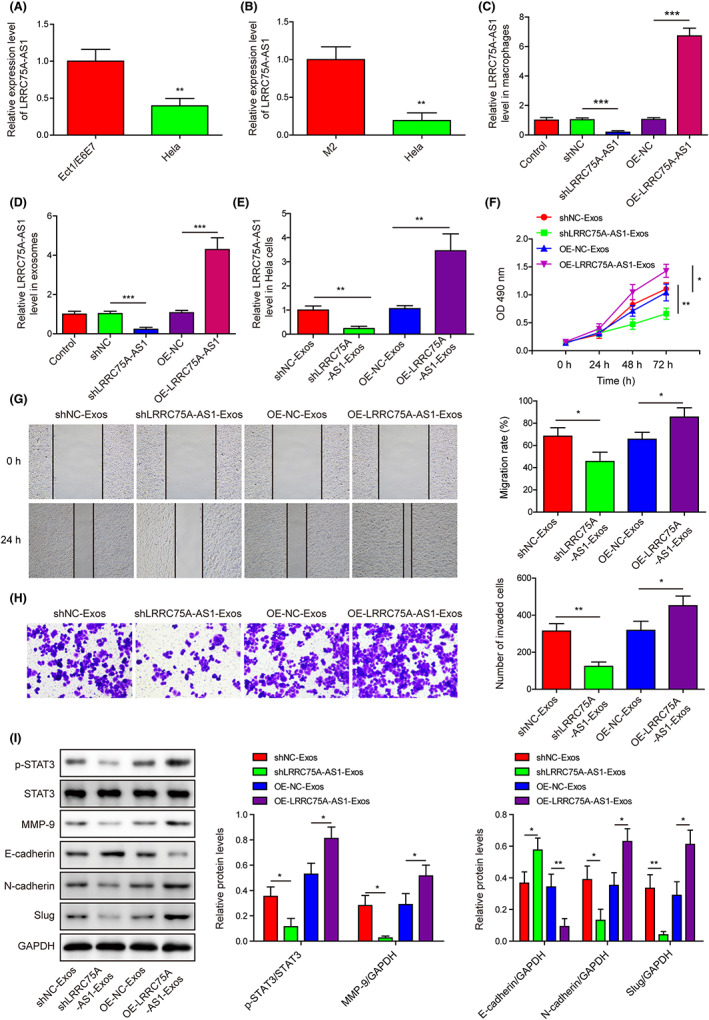
LRRC75A‐AS1 in M2 macrophage‐derived exosomes induces Hela cell proliferation, migration, invasion, and epithelial to mesenchymal transition (EMT) process through STAT3/MMP‐9 signaling. (A) LRRC75A‐AS1 expression in Hela and Ect1/E6E7 cell lines was quantitated by qRT‐PCR. (B) Differential expression of LRRC75A‐AS1 between M2 macrophages and Hela cells tested by qRT‐PCR. (C) LRRC75A‐AS1 expression in M2 macrophages with LRRC75A‐AS1 knockdown or overexpression. (D) LRRC75A‐AS1 expression in exosomes from M2 macrophages with LRRC75A‐AS1 knockdown or overexpression. (E) LRRC75A‐AS1 expression in Hela cells treated by exosomes isolated from M2 macrophages with LRRC75A‐AS1 knockdown or overexpression. (F) The proliferation of Hela cells treated with exosomes secreted by M2 macrophages with LRRC75A‐AS1 knockdown or overexpression. (G) The migration of Hela cells treated with exosomes secreted by the M2 macrophages with LRRC75A‐AS1 knockdown or overexpression. (H) The invasion of Hela cells treated with exosomes secreted by the M2 macrophages with LRRC75A‐AS1 knockdown or overexpression. (I) The protein levels of p‐STAT3, STAT3, MMP‐9, E‐cadherin, N‐cadherin, and slug in Hela cells treated with exosomes isolated from M2 macrophages with LRRC75A‐AS1 knockdown or overexpression. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.3. LRRC75A‐AS1 is highly expressed in cervical cancer tissues and negatively regulates miR‐429 expression
Kaplan–Meier Plotter database showed that higher expression of LRRC75A‐AS1 in cervical cancer tissues was significantly correlated with poor survival of cervical cancer patients (Figure 3A). Also, we showed the LRRC75A‐AS1 expression in the cervical cancer tissues was much higher than in adjacent normal cervical tissues (Figure 3B). According to the median expression of LRRC75A‐AS1, high expression of LRRC75A‐AS1 in our clinical samples was positively associated with tumor stages and lymph node metastasis (Table 1). Interestingly, the ratio of M2 macrophages (CD163+CD206+) in cervical cancer tissues collected from 15 patients was significantly higher than in corresponding adjacent normal cervical tissues (Figure 3C). Also, the CD163 protein level was also upregulated in the abovementioned cervical cancer tissues (Figure 3D). Importantly, LRRC75A‐AS1 was predicted to directly target miR‐429 (Starbase database) (Figure 3E). We found the lower expression of miR‐429 in cervical cancer tissues was significantly correlated with poor survival of cervical cancer patients via the Kaplan–Meier Plotter platform (Figure 3F). Moreover, miR‐429 expression levels in cervical cancer tissues were much lower than in adjacent cervical tissues (Figure 3G). Furthermore, LRRC75A‐AS1 and miR‐429 expression in cervical cancer tissues exhibited significantly negative correlation (Figure 3H). To validate this, we showed the colocalization of LRRC75A‐AS1 and miR‐429 in Hela cell cytosols (Figure 3I). We also found that LRRC75A‐AS1 precipitated with Ago2 protein was greatly increased by transfecting Hela cells with miR‐429 mimics (Figure 3J). In addition, miR‐429 mimics greatly reduced the luciferase activity in the Hela cells expressing wild‐type LRRC75A‐AS1, but not in those expressing the mutant LRRC75A‐AS1 (Figure 3K). Consistently, miR‐429 expression in Hela cells was remarkably promoted by LRRC75A‐AS1 knockdown and greatly repressed by LRRC75A‐AS1 overexpression (Figure 3L). These results showed that LRRC75A‐AS1 could directly target and inhibit miR‐429 expression in Hela cells.
FIGURE 3.
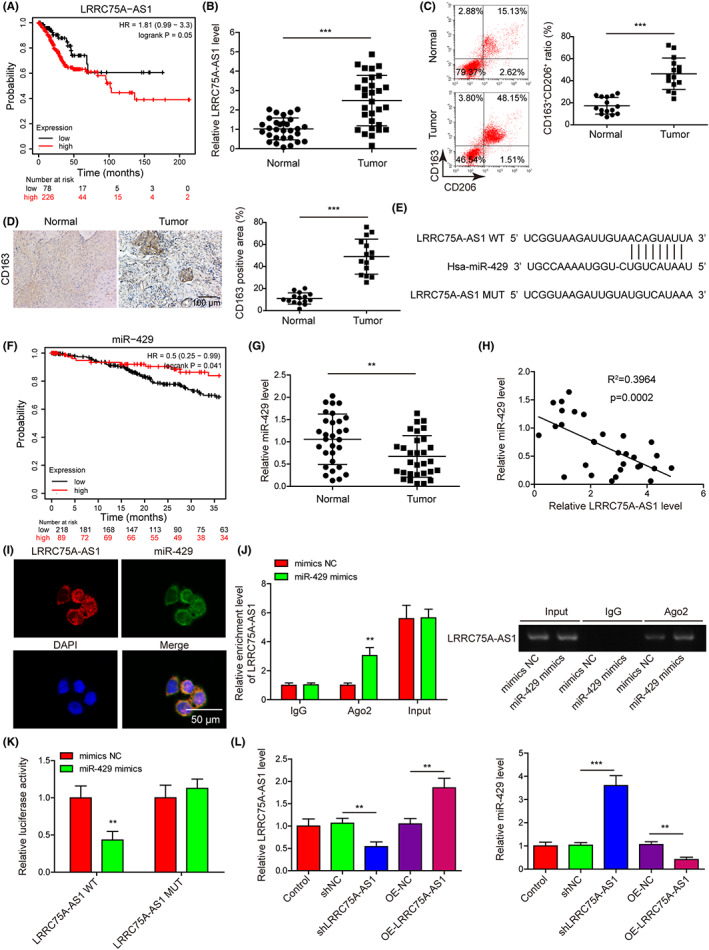
LRRC75A‐AS1 is highly expressed in cervical cancer tissues and negatively regulates miR‐429 expression. (A) The correlation of LRRC75A‐AS1 expression with the overall survival of cervical cancer patients by Kaplan–Meier Plotter platform. (B) The expression levels of LRRC75A‐AS1 in cervical cancer tissues compared with the adjacent normal cervical tissues were detected by qRT‐PCR (n = 30). (C) The ratio of M2 macrophages (CD206+CD163+) in normal and cervical cancer tissues was assessed by flow cytometry. (D) The protein level of CD163 in normal and cervical cancer tissues by immunohistochemistry assay. (E) Bioinformatic prediction of the direct targeting of miR‐429 by LRRC75A‐AS1. (F) The correlation of miR‐429 expression with the survival of cervical cancer patients by Kaplan–Meier Plotter platform. (G) Lowered expression of miR‐429 in cervical cancer tissues compared with the normal cervical tissues shown by qRT‐PCR (n = 30). (H) The negative correlation of LRRC75A‐AS1 and miR‐429 expression in cervical cancer tissues (n = 30). (I) The colocalization of LRRC75A‐AS1 and miR‐429 in the cytosol of Hela cells by FISH method. (J, K) Direct interaction between LRRC75A‐AS1 and miR‐429 in Hela cells was validated through the RIP (J) and dual luciferase reporter (K) assays. RIP electrophoretic graphs are shown in the right. (L) LRRC75A‐AS1 and miR‐429 expression in Hela cells by silence or overexpression of LRRC75A‐AS1. **p < 0.01 and ***p < 0.001.
3.4. LRRC75A‐AS1 in M2 macrophage‐derived exosomes represses miR‐429 to promote Hela cell proliferation, migration, invasion, and EMT
We then isolated exosomes from M2 macrophages with LRRC75A‐AS1 overexpression, which were used to incubate with the Hela cells transfected with miR‐429 mimics. We found that LRRC75A‐AS1 expression in Hela cells was substantially increased by exosomes from normal M2 macrophages, and exosomes from M2 macrophages with LRRC75A‐AS1 overexpression induced greater expression of LRRC75A‐AS1, which was not affected by miR‐429 mimics (Figure 4A). The miR‐429 expression in Hela cells was effectively suppressed by exosomes secreted by normal M2 macrophages and M2 macrophages with LRRC75A‐AS1 overexpression, which was significantly abrogated by miR‐429 mimics (Figure 4A). Also, Hela cell proliferation, migration, and invasion were effectively enhanced by exosomes secreted by M2 macrophages with LRRC75A‐AS1 overexpression, which were also abrogated by miR‐429 mimics (Figure 4B‐D). Moreover, STAT3 phosphorylation, MMP‐9, N‐cadherin, and slug protein levels in Hela cells were remarkably upregulated, while E‐cadherin protein was substantially downregulated by exosomes from M2 macrophages with LRRC75A‐AS1 overexpression (Figure 4E). In addition, miR‐429 mimics transfection in Hela cells significantly mitigated the above alterations. These assays indicated that the regulation of Hela cell functions and EMT by LRRC75A‐AS1 in M2 macrophage‐derived exosomes was mediated by its inhibition on miR‐429 expression.
FIGURE 4.
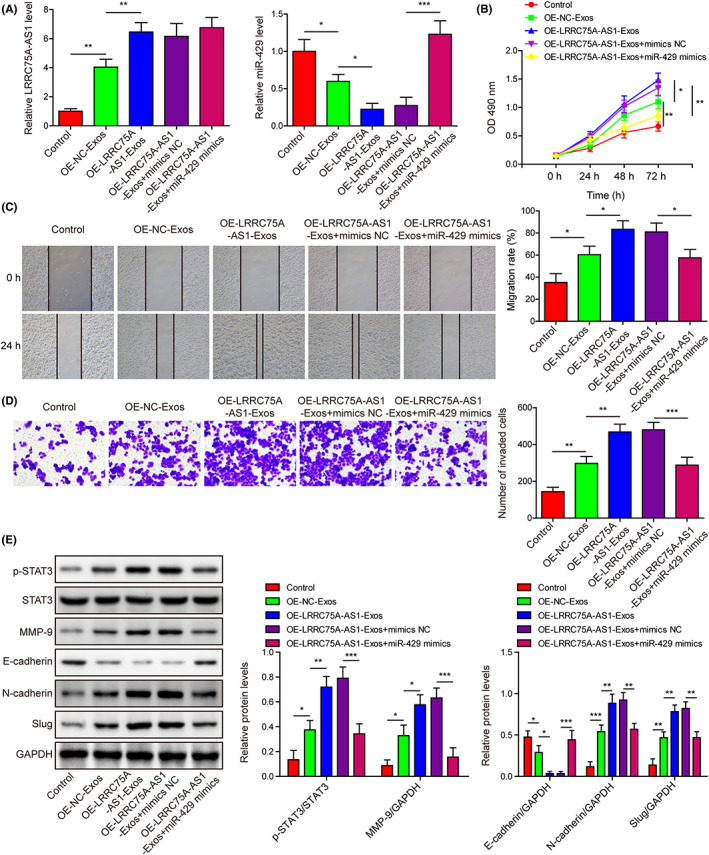
LRRC75A‐AS1 in M2 macrophage‐derived exosomes represses miR‐429 expression to promote Hela cell proliferation, migration, invasion, and epithelial to mesenchymal transition (EMT) process. (A) Expressional changes of LRRC75A‐AS1 and miR‐429 in Hela cells treated with miR‐429 mimics and exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages. (B) Influences of exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages on the proliferation of Hela cells transfected with miR‐429 mimics. The CCK‐8 assay was done to analyze Hela cell proliferation. (C, D) The migration and invasion ability of Hela cells transfected with miR‐429 mimics and exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages. The wound healing assay (C) and Transwell system (D) were used to detect Hela cell migration and invasion, respectively. (E) The protein levels of p‐STAT3, STAT3, MMP‐9, E‐cadherin, N‐cadherin, and slug in the Hela cells transfected with miR‐429 mimics following treatment with exosomes secreted by LRRC75A‐AS1‐overexpressing M2 macrophages. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.5. miR‐429 targets and negatively regulates SIX1 expression in cervical cancer cells
Through bioinformatic prediction (Targetscan), miR‐429 might directly interact with the 3’‐UTR region of the SIX1 gene (Figure 5A). By searching the UALCAN database, we found that SIX1 expression levels in cervical cancer tissues were much higher than in normal cervical tissues (Figure 5B). SIX1 expression in cancerous tissues was greatly elevated compared with adjacent cervical tissues (Figure 5C). The miR‐429 and SIX1 expression in cervical cancer tissues demonstrated significantly negative correlation (Figure 5D). The dual luciferase reporter assay showed great decrease of luciferase activities in Hela cells expressing the wild type of SIX1 by miR‐429 mimics, other than in those expressing the mutant SIX1 (Figure 5E). miR‐429 expression was elevated or knocked down in Hela cells by miR‐429 mimics or inhibitor, respectively (Figure 5F). The SIX1 mRNA and protein levels in Hela cells were significantly repressed by miR‐429 mimics, but greatly elevated by miR‐429 inhibitor (Figure 5G,H). These results proved that miR‐429 repressed SIX1 expression in Hela cells through directly binding with its 3’‐UTR region.
FIGURE 5.
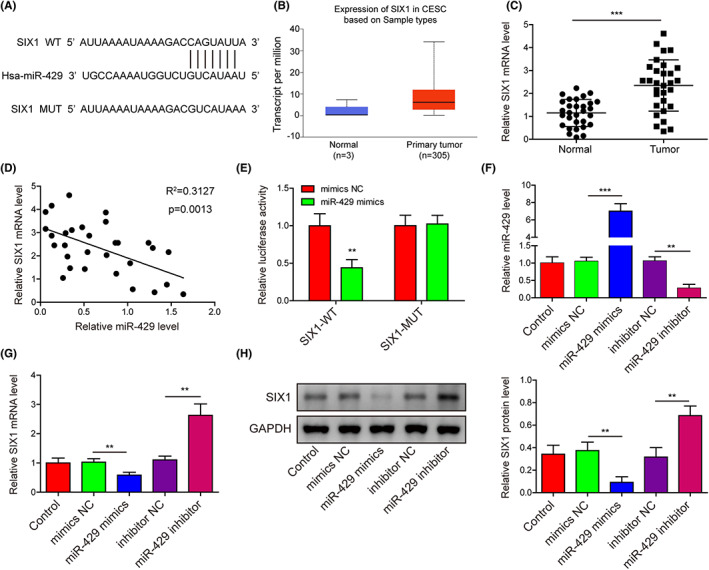
miR‐429 targets and negatively regulates SIX1 expression in cervical cancer cells. (A) Bioinformatic prediction (Targetscan) of the interaction between miR‐429 and the 3’‐UTR region of the SIX1 gene. (B) Relative expression of the SIX1 gene in cervical cancer tissues shown by the UALCAN database. (C) The elevation of SIX1 expression in cervical cancer tissues surgically collected from cervical cancer patients (n = 30). SIX1 mRNA levels were detected using qRT‐PCR. (D) Negative correlation of SIX1 and miR‐429 in cervical cancer tissues (n = 30). (E) Validation of the association of miR‐429 with the 3’‐UTR region of SIX1 in Hela cells. The dual luciferase reporter assay was performed to verify the miR‐429/SIX1 interaction. (F–G) Relative expression levels of miR‐429 (F) and SIX1 (G) in the Hela cells transfected with miR‐429 mimics or inhibitor. SIX1 mRNA and miR‐429 were determined by qRT‐PCR method. (H) Alteration of SIX1 protein level in Hela cells caused by miR‐429 mimics or inhibitor. The abundance of SIX1 protein was detected with Western blotting assay. **p < 0.01 and ***p < 0.001.
3.6. miR‐429 represses SIX1 expression to modulate Hela cell functions and EMT process through STAT3/MMP‐9 signaling
The miR‐429 expression in Hela cells was markedly elevated by miR‐429 mimics, which was not affected by SIX1 overexpression (Figure 6A). However, SIX1 mRNA and protein levels in Hela cells were greatly reduced by miR‐429 mimics, which were then remarkably alleviated by SIX1 overexpression (Figure 6B,C). Hela cell proliferation rates were greatly repressed by miR‐429 mimics, but SIX1 overexpression effectively abrogated the inhibition of cell proliferation (Figure 6D). On the contrary, miR‐429 mimics significantly accelerated Hela cell apoptosis, which was also significantly mitigated by SIX1 overexpression (Figure 6E). Hela cell migration and invasion exhibited similar alteration as cell proliferation (Figure 6F,G). Furthermore, STAT3 phosphorylation, MMP‐9, N‐cadherin and slug protein levels in Hela cells were substantially reduced by miR‐429 mimics, which, however, caused great increase in the E‐cadherin protein level (Figure 6H). These alterations of EMT biomarker proteins and STAT3/MMP‐9 signaling in Hela cells by miR‐429 mimics were all significantly abrogated by SIX1 overexpression (Figure 6H). These observations demonstrated that miR‐429 repressed the proliferation, migration, invasion, and EMT but promoted apoptosis in Hela cells through suppressing SIX1 expression.
FIGURE 6.
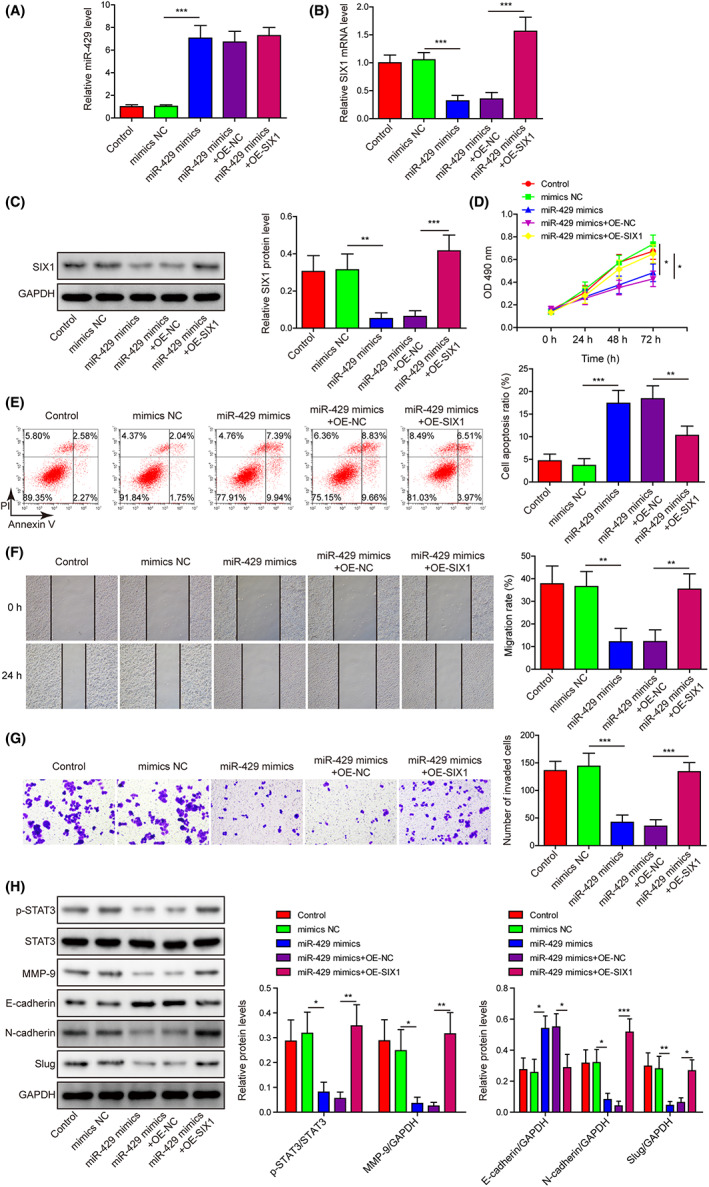
miR‐429 represses SIX1 expression to modulate Hela cell proliferation, apoptosis, migration, invasion, and epithelial to mesenchymal transition (EMT) process through STAT3/MMP‐9 signaling. (A, B) Relative expression levels of miR‐429 (A) and SIX1 mRNA (B) in Hela cells regulated by miR‐429 mimics and SIX1 overexpression by qRT‐PCR method. (C) Alteration of SIX1 protein level in Hela cells induced by miR‐429 mimics and SIX1 overexpression by Western blotting. (D) Regulation of Hela cell proliferation by miR‐429 mimics and SIX1 overexpression by the CCK‐8 method. (E) The percentages of apoptotic Hela cells by flow cytometry after being transfected with miR‐429 mimics and SIX1‐overexpressing plasmid. (F, G) The influences of miR‐429 mimics and SIX1 overexpression on the migration and invasion of Hela cells by the wound‐healing assay (F) and Transwell system (G), respectively. (H) The effects of miR‐429 mimics and SIX1 overexpression on the abundances of p‐STAT3, STAT3, MMP‐9, E‐cadherin, N‐cadherin, and slug proteins in Hela cells by using Western blotting assay. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.7. LRRC75A‐AS1 in M2 macrophage‐derived exosomes promotes Hela cell tumorigenesis and metastasis in nude mice
Exosomes from the OE‐NC‐Exos and shNC‐Exos groups increased the tumor volumes and weights in nude mice, and the volumes and weights of tumors were further promoted by exosomes derived from LRRC75A‐AS1‐overexpressing M2 macrophages, but effectively reduced by exosomes derived from LRRC75A‐AS1‐silencing M2 macrophages (Figure 7A–C). In tumor tissues of nude mice, SIX1, N‐cadherin, and LRRC75A‐AS1 expressions were promoted by exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages, but inhibited by exosomes from LRRC75A‐AS1‐silencing M2 macrophages (Figure 7D,E). Completely opposite alterations of miR‐429 levels were observed in the above tumor tissues (Figure 7E). Furthermore, STAT3 phosphorylation and MMP‐9 expression in the above tumor tissues were also positively regulated by exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages and repressed by exosomes from LRRC75A‐AS1‐silencing M2 macrophages (Figure 7F).
FIGURE 7.
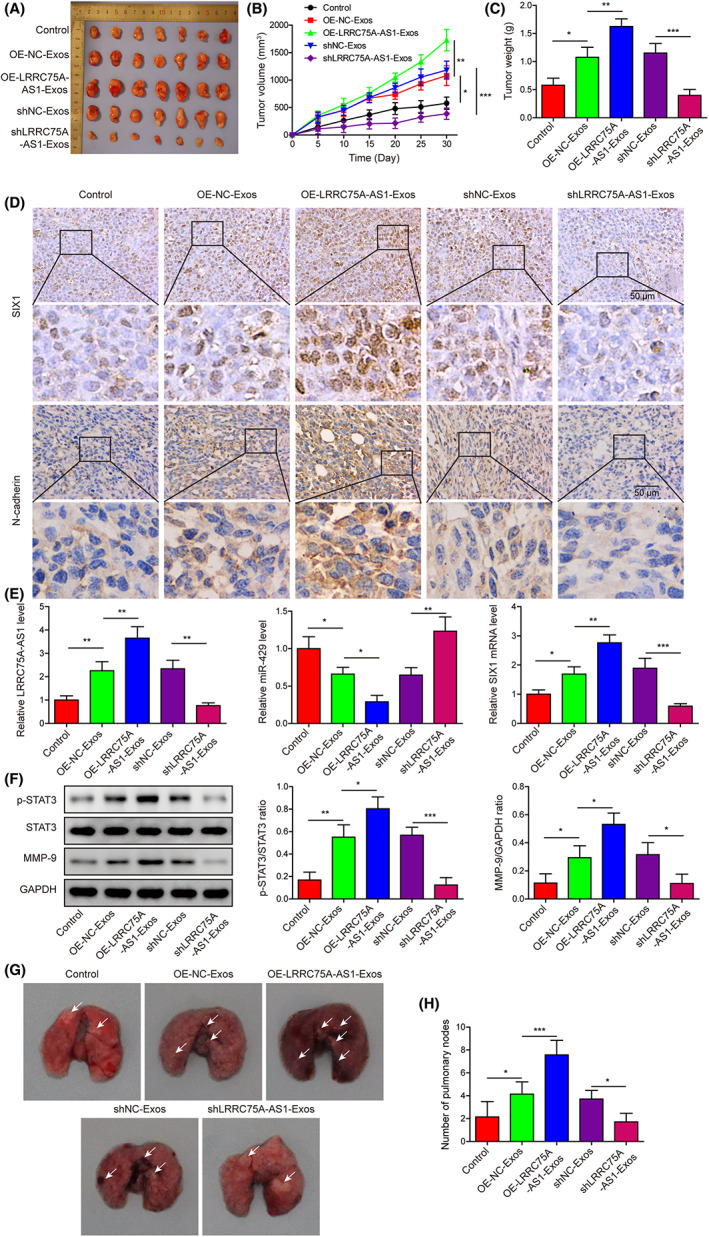
LRRC75A‐AS1 in M2 macrophage‐derived exosomes promotes Hela cell tumorigenesis and metastasis in nude mice. (A) Representative images of nude mice harboring tumors formed from Hela cells after treatment with exosomes from M2 macrophages with LRRC75A‐AS1 overexpression or knockdown. (B, C) The volumes (B) and weights (C) of tumors derived from Hela cells after treatment with exosomes from M2 macrophages with LRRC75A‐AS1 overexpression or knockdown. (D) Regulation of SIX1 and N‐cadherin protein levels in tumor tissues by exosomes from M2 macrophages with LRRC75A‐AS1 overexpression or knockdown. (E) Modulation of LRRC75A‐AS1, miR‐429, and SIX1 mRNA levels in tumor tissues by exosomes from M2 macrophages with LRRC75A‐AS1 overexpression or knockdown. (F) Alterations of STAT3 protein phosphorylation and MMP‐9 protein expression in Hela cells‐induced tumors caused by exosomes secreted by M2 macrophages with LRRC75A‐AS1 overexpression or knockdown. (G) Histological evaluation of mice lung tissues in the Hela metastasis model treated with exosomes from LRRC75A‐AS1‐overexpressing or ‐silenced M2 macrophages. (H) The numbers of lung nodules formed in the Hela metastasis model following treatment with exosomes secreted by LRRC75A‐AS1‐overexpressing or ‐silenced M2 macrophages. *p < 0.05, **p < 0.01, and ***p < 0.001.
In addition, the metastasis potential of Hela cells was then evaluated by histological evaluation and lung nodule numbers (Figure 7G,H). Compared with the corresponding NC groups, the numbers of lung nodules formed in nude mice were remarkably increased by treatment with exosomes purified from M2 macrophages overexpressing LRRC75A‐AS1, but significantly decreased by treatment of exosomes from LRRC75A‐AS1‐silenced M2 macrophages (Figure 7G,H). These observations proved that LRRC75A‐AS1 from M2 macrophage‐derived exosomes could effectively promote both the tumorigenesis and metastasis potential of Hela cells in vivo.
3.8. LRRC75A‐AS1 in M2 macrophage‐derived exosomes induces Hela cell tumorigenesis by regulating the miR‐429/SIX1 axis in vivo
Compared with the corresponding NC groups, the transfection with miR‐429 mimics or sh‐SIX1 in Hela cells significantly reduced the weights and volumes of tumors formed in nude mice, which were then partially abrogated by treatment with exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages (Figure 8A–C). Also, the expression of N‐cadherin in tumor tissues in nude mice was greatly reduced by transfection with miR‐429 mimics or sh‐SIX1 in Hela cells, which were also partially recovered by exosomes purified from M2 macrophages with LRRC75A‐AS1 overexpression (Figure 8D). In addition, SIX1, MMP‐9, and STAT3 phosphorylation in tumor tissues in nude mice were significantly inhibited by the transfection of Hela cells with miR‐429 mimics or sh‐SIX1, which were also substantially mitigated by the LRRC75A‐AS1‐overexpressing M2 macrophage exosomes (Figure 8E). miR‐429 mimics or sh‐SIX1 transfection in Hela cells greatly decreased the number of lung nodules in nude mice, which was remarkably alleviated by exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages (Figure 8F,G). These in vivo results verified that miR‐429 overexpression and SIX1 knockdown could suppress the tumorigenesis and metastasis of Hela cells, and exosomal LRRC75A‐AS1 promoted Hela cell tumorigenesis and metastasis by regulating miR‐429/SIX1 axis.
FIGURE 8.
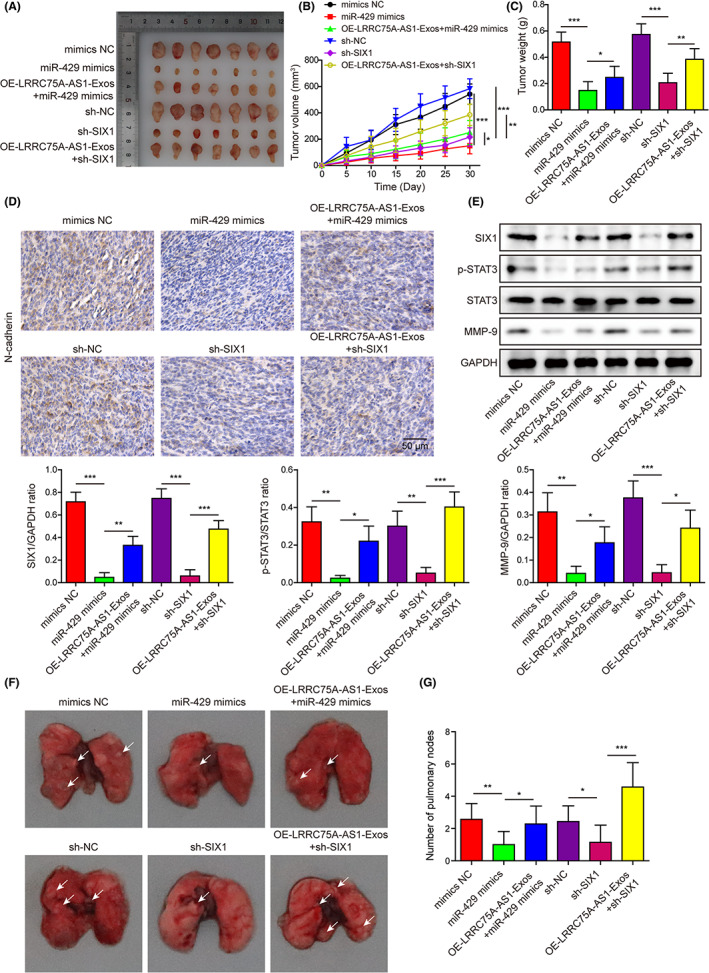
LRRC75A‐AS1 in M2 macrophage‐derived exosomes induces Hela cell tumorigenesis by regulating the miR‐429/SIX1 axis in vivo. (A) The tumors formed in nude mice injected with Hela cells transfected with the miR‐429 mimics or sh‐SIX1 and treated with OE‐LRRC75A‐AS1‐overexpressing M2 macrophage exosomes. (B, C) Statistics of volumes and weights of tumors developed from Hela cells in (A). The tumor volumes (B) and weights (C) were quantitated 30 days after Hela cell injection. (D) Expressional alteration of N‐cadherin protein in tumors formed in nude mice injected with Hela cells transfected with miR‐429 mimics or sh‐SIX1 and treated with exosomes secreted by LRRC75A‐AS1‐overexpressing M2 macrophages. N‐cadherin protein levels in tumor tissues were tested by immunohistochemistry. (E) The changes of SIX1, phosphorylated STAT3, and MMP‐9 proteins in tumors developed from Hela cells transfected with miR‐429 mimics or sh‐SIX1 and treated with exosomes secreted by LRRC75A‐AS1‐overexpressing M2 macrophages. (F, G) Mice metastasis model of Hela cells transfected with miR‐429 mimics or sh‐SIX1, combined with exosomes from LRRC75A‐AS1‐overexpressing M2 macrophages. The metastasis of modified Hela cells was evaluated in lung tissues (F) and lung nodule numbers (G). *p < 0.05, **p < 0.01, and ***p < 0.001.
4. DISCUSSION
The progression of EMT during cervical cancer was known to be regulated by a number of inducers and inhibitors, 7 but the epigenetic mechanism mediating EMT in cervical cancer still remains poorly understood. In this study, we comprehensively investigated the pathogenic roles of LRRC75A‐AS1 in cervical cancer progression. We proved that exosomes secreted by M2 macrophages promoted the proliferation, migration, invasion, and EMT of cervical cancer through delivery of LRRC75A‐AS1, which was mediated by miR‐429, SIX1, and the downstream STAT3/MMP‐9 pathway.
Exosomes have been regarded as essential signaling platforms in cancer development. 34 , 35 For instance, recent report showed that the exosomes derived from M2 macrophages effectively promoted the migration and invasion of colon cancer cells. 10 The development of pancreatic ductal adenocarcinoma could also be promoted by M2 macrophage‐derived exosomes. 11 However, the roles of M2 macrophage‐derived exosomes in cervical cancer progression remain largely unexplored. The lncRNA LRRC75A‐AS1 was involved in development of several cancers such as colorectal carcinoma and glioblastoma, 14 , 15 but not studied in cervical cancer. Also, the potential delivery of LRRC75A‐AS1 by exosomes has not been previously reported. In the present study, we found that the expression level of LRRC75A‐AS1 in Hela cells was decreased compared with normal cervical epithelial cells Ect1/E6E7 and M2 macrophages; thus, we provided the first evidence that LRRC75A‐AS1 was delivered by exosomes from M2 macrophages to Hela cells. Also, we proved firstly that exosomal LRRC75A‐AS1 promoted proliferation, migration, invasion, and EMT of cervical cancer cells. The pathogenic roles of LRRC75A‐AS1 delivered by M2 macrophage exosomes in cervical cancer were finally validated by our tumorigenesis in vivo model.
The interaction of lncRNAs with miRNAs in regulating gene expression is a key epigenetic mechanism driving cancer pathogenesis. For instance, the prognosis and chemotherapy resistance of gastric cancer were reported to be regulated by the association of the lncRNA EIF3J‐DT with the miRNA MIR188‐3p, mediated by their modulation of the autophagy process. 36 Previous reports showed that miR‐429 acted as an important tumor suppressor in cervical cancer, 18 , 37 and its expression in cervical cancer cells could be negatively modulated by MALAT1, 38 but its potential targeting by LRRC75A‐AS1 was not previously addressed. We proved here for first time that LRRC75A‐AS1 could directly sponge miR‐429 in Hela cells, and miR‐429 expression in Hela cells could be repressed by LRRC75A‐AS1 delivered by M2 macrophage exosomes, which contributed to cervical cancer development. Recent research demonstrated that miR‐429 also performed regulatory roles in other human cancers such as ovarian cancer and glioma. 39 , 40 Specifically, the inhibition of miR‐429 was proved to promote ovarian cancer development and drug resistance via modulation of the expression of ZEB1. 40
SIX1 contributed to EMT progression in various cancer cells through activating the STAT3 signaling pathway and MMP‐9 activity. 22 , 23 The proliferation rate of thyroid carcinoma cells could be enhanced by SIX1‐activated STAT3 signaling, through elevating EYA1 gene expression at the post‐transcriptional level. 23 More importantly, the expression of SIX1 in cervical cancer cells was known to be negatively regulated by multiple miRNAs like miR‐23b and miR‐362, 27 , 28 but the potential roles of miR‐429 in regulating SIX1 expression was not validated before. In this study, we provided the first evidence of the direct association of miR‐429 with SIX1 gene, as well as their roles in cervical cancer pathogenesis. Above results persuasively proved the roles of SIX1 in cervical cancer development by miR‐429 and LRRC75A‐AS1 in M2 macrophage exosomes. Furthermore, SIX1 was known to regulate EMT by activating STAT3 and MMP‐9 expression, 22 , 23 which was also confirmed in our in vivo metastasis study.
In summary, we revealed in this study that LRRC75A‐AS1, which was delivered by the exosomes secreted by M2 macrophages, negatively regulated miR‐429 expression to elevate SIX1 expression and promote cervical cancer progression through activating STAT3/MMP‐9 signaling. These investigations provided new insights into molecular mechanisms underlying cervical cancer development.
AUTHOR CONTRIBUTIONS
HYS: conceptualization, funding acquisition. XYC: writing—original draft. CXS: data curation, resources. ZPY: methodology, formal analysis. HCL: investigation, software, visualization. FW: project administration, supervision, validation, writing—review & editing. All authors have read and approved the final version of this manuscript to be published.
FUNDING INFORMATION
This work was supported by the funding project of Turpan Science and Technology Bureau (2019BJ001), National Cancer Center Climbing Fund (NCC2018B66), and Key R&D Project of Scientific Research Climbing Plan of Hunan Cancer Hospital (YF2020003).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
All experimental operations using human cervical cancer tissues were approved in advance by the Medical Ethical Committee of the Hunan Cancer Hospital.
Approval of the research protocol by an Institutional Reviewer Board.
Informed Consent: Before the surgical operation, written informed consent was signed by all cervical cancer patients.
Registry and the Registration No: N/A.
Animal Studies: All experiments using mice were approved in advance by the Animal Care and Use Committee of the Hunan Cancer Hospital.
CONSENT FOR PUBLICATION
The informed consent was obtained from study participants.
Supporting information
Appendix S1.
ACKNOWLEDGEMENTS
The authors have nothing to report.
Sui H‐Y, Cui X‐Y, Shi C‐X, Yan Z‐P, Li H‐C, Wang F. LRRC75A‐AS1 delivered by M2 macrophage exosomes promotes cervical cancer progression via enhancing SIX1 expression. Cancer Sci. 2023;114:2634‐2649. doi: 10.1111/cas.15780
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Hillemanns P, Soergel P, Hertel H, Jentschke M. Epidemiology and early detection of cervical cancer. Oncol Res Treat. 2016;39:501‐506. [DOI] [PubMed] [Google Scholar]
- 2. Johnson CA, James D, Marzan A, Armaos M. Cervical cancer: an overview of pathophysiology and management. Semin Oncol Nurs. 2019;35:166‐174. [DOI] [PubMed] [Google Scholar]
- 3. Small W Jr, Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer. 2017;123:2404‐2412. [DOI] [PubMed] [Google Scholar]
- 4. Aballea S, Beck E, Cheng X, et al. Risk factors for cervical cancer in women in China: a meta‐model. Womens Health (Lond). 2020;16:1745506520940875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brito‐Marcelino A, Duarte‐Tavares RJ, Marcelino KB, Silva‐Neto JA. Cervical cancer related to occupational risk factors: review. Rev Bras Med Trab. 2020;18:103‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li H, Wu X, Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. J Gynecol Oncol. 2016;27:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett. 2015;356:321‐331. [DOI] [PubMed] [Google Scholar]
- 8. Sadri Nahand J, Moghoofei M, Salmaninejad A, et al. Pathogenic role of exosomes and microRNAs in HPV‐mediated inflammation and cervical cancer: a review. Int J Cancer. 2020;146:305‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J, Liu SC, Luo XH, et al. Exosomal long noncoding RNAs are differentially expressed in the cervicovaginal lavage samples of cervical cancer patients. J Clin Lab Anal. 2016;30:1116‐1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lan J, Sun L, Xu F, et al. M2 macrophage‐derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79:146‐158. [DOI] [PubMed] [Google Scholar]
- 11. Yin Z, Ma T, Huang B, et al. Macrophage‐derived exosomal microRNA‐501‐3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3‐mediated TGF‐beta signaling pathway. J Exp Clin Cancer Res. 2019;38:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng P, Chen L, Yuan X, et al. Exosomal transfer of tumor‐associated macrophage‐derived miR‐21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng P, Luo Q, Wang W, et al. Tumor‐associated macrophages‐derived exosomes promote the migration of gastric cancer cells by transfer of functional apolipoprotein E. Cell Death Dis. 2018;9:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J, Lan J, Ye Z, et al. Long noncoding RNA LRRC75A‐AS1 inhibits cell proliferation and migration in colorectal carcinoma. Exp Biol Med (Maywood). 2019;244:1137‐1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han L, Li Z, Jiang Y, Jiang Z, Tang L. SNHG29 regulates miR‐223‐3p/CTNND1 axis to promote glioblastoma progression via Wnt/beta‐catenin signaling pathway. Cancer Cell Int. 2019;19:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li S, Wu D, Jia H, Zhang Z. Long non‐coding RNA LRRC75A‐AS1 facilitates triple negative breast cancer cell proliferation and invasion via functioning as a ceRNA to modulate BAALC. Cell Death Dis. 2020;11:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X, Wang H, Zhang R, Li D, Gao MQ. LRRC75A antisense lncRNA1 knockout attenuates inflammatory responses of bovine mammary epithelial cells. Int J Biol Sci. 2020;16:251‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Dong X, Hu B, Wang XJ, Wang Q, Wang WL. The effects of Micro‐429 on inhibition of cervical cancer cells through targeting ZEB1 and CRKL. Biomed Pharmacother. 2016;80:311‐321. [DOI] [PubMed] [Google Scholar]
- 19. Kahlert C, Lerbs T, Pecqueux M, et al. Overexpression of SIX1 is an independent prognostic marker in stage I‐III colorectal cancer. Int J Cancer. 2015;137:2104‐2113. [DOI] [PubMed] [Google Scholar]
- 20. Lerbs T, Bisht S, Scholch S, et al. Inhibition of Six1 affects tumour invasion and the expression of cancer stem cell markers in pancreatic cancer. BMC Cancer. 2017;17:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ono H, Imoto I, Kozaki K, et al. SIX1 promotes epithelial‐mesenchymal transition in colorectal cancer through ZEB1 activation. Oncogene. 2012;31:4923‐4934. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y, Wang S, Liu Z, Yang L, Liu J, Xiu M. Increased Six1 expression in macrophages promotes hepatocellular carcinoma growth and invasion by regulating MMP‐9. J Cell Mol Med. 2019;23:4523‐4533. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Kong D, Li A, Liu Y, et al. SIX1 activates STAT3 signaling to promote the proliferation of thyroid carcinoma via EYA1. Front Oncol. 2019;9:1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu D, Li L, Zhang XX, et al. SIX1 promotes tumor lymphangiogenesis by coordinating TGFbeta signals that increase expression of VEGF‐C. Cancer Res. 2014;74:5597‐5607. [DOI] [PubMed] [Google Scholar]
- 25. Liu D, Zhang XX, Xi BX, et al. Sine oculis homeobox homolog 1 promotes DNA replication and cell proliferation in cervical cancer. Int J Oncol. 2014;45:1232‐1240. [DOI] [PubMed] [Google Scholar]
- 26. Sun SH, Liu D, Deng YT, et al. SIX1 coordinates with TGFbeta signals to induce epithelial‐mesenchymal transition in cervical cancer. Oncol Lett. 2016;12:1271‐1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li YM, Li XJ, Yang HL, Zhang YB, Li JC. MicroRNA‐23b suppresses cervical cancer biological progression by directly targeting six1 and affecting epithelial‐to‐mesenchymal transition and AKT/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:4688‐4697. [DOI] [PubMed] [Google Scholar]
- 28. Shi C, Zhang Z. MicroRNA‐362 is downregulated in cervical cancer and inhibits cell proliferation, migration and invasion by directly targeting SIX1. Oncol Rep. 2017;37:501‐509. [DOI] [PubMed] [Google Scholar]
- 29. Lanczky A, Gyorffy B. Web‐based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23:e27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA‐ceRNA, miRNA‐ncRNA and protein‐RNA interaction networks from large‐scale CLIP‐seq data. Nucleic Acids Res. 2014;42:D92‐D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15‐20. [DOI] [PubMed] [Google Scholar]
- 33. Liu Z, Kuang W, Zhou Q, Zhang Y. TGF‐beta1 secreted by M2 phenotype macrophages enhances the stemness and migration of glioma cells via the SMAD2/3 signalling pathway. Int J Mol Med. 2018;42:3395‐3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13‐27. [DOI] [PubMed] [Google Scholar]
- 35. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208‐1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo Y, Zheng S, Wu Q, et al. Long noncoding RNA (lncRNA) EIF3J‐DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy. 2021;17:4083‐4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan JY, Fan YJ, Wang XL, et al. miR‐429 is involved in regulation of NF‐kappaBactivity by targeting IKKbeta and suppresses oncogenic activity in cervical cancer cells. FEBS Lett. 2017;591:118‐128. [DOI] [PubMed] [Google Scholar]
- 38. Shen F, Zheng H, Zhou L, Li W, Xu X. Overexpression of MALAT1 contributes to cervical cancer progression by acting as a sponge of miR‐429. J Cell Physiol. 2019;234:11219‐11226. [DOI] [PubMed] [Google Scholar]
- 39. Cheng Z, Li Z, Ma K, et al. Long non‐coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR‐429. J Cancer. 2017;8:4106‐4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zou J, Liu L, Wang Q, et al. Downregulation of miR‐429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. Am J Transl Res. 2017;9:1357‐1368. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1.
Data Availability Statement
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


