Abstract
Although circular RNAs (circRNAs) are involved in cell proliferation, differentiation, apoptosis, and invasion, the underlying regulatory mechanisms of circRNAs in thyroid cancer have not been fully elucidated. This article aimed to study the role of circRNA regulated by N6‐methyladenosine modification in papillary thyroid cancer (PTC). Quantitative real‐time PCR, western blotting, and immunohistochemistry were used to investigate the expressions of circRNA nuclear receptor‐interacting protein 1 (circNRIP1) in PTC tissues and adjacent noncancerous thyroid tissues. In vitro and in vivo assays were carried out to assess the effects of circNRIP1 on PTC glycolysis and growth. The N6‐methyladenosine mechanisms of circNRIP1 were evaluated by methylated RNA immunoprecipitation sequencing, luciferase reporter gene, and RNA stability assays. Results showed that circNRIP1 levels were significantly upregulated in PTC tissues. Furthermore, elevated circNRIP1 levels in PTC patients were correlated with high tumor lymph node metastasis stage and larger tumor sizes. Functionally, circNRIP1 significantly promoted glycolysis, PTC cell proliferation in vitro, and tumorigenesis in vivo. Mechanistically, circNRIP1 acted as a sponge for microRNA (miR)‐541‐5p and miR‐3064‐5p and jointly upregulated pyruvate kinase M2 (PKM2) expression. Knockdown of m6A demethylase α‐ketoglutarate‐dependent dioxygenase alkB homolog 5 (ALKBH5) significantly enhanced circNRIP1 m6A modification and upregulated its expression. These results show that ALKBH5 knockdown upregulates circNRIP1, thus promoting glycolysis in PTC cells. Therefore, circNRIP1 can be a prognostic biomarker and therapeutic target for PTC by acting as a sponge for oncogenic miR‐541‐5p and miR‐3064‐5p to upregulate PKM2 expression.
Keywords: circNRIP1, glycolysis, m6A, PKM2, PTC
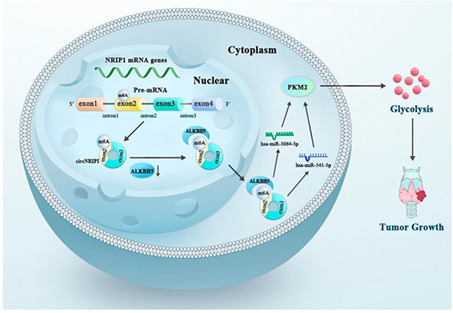
ALKBH5 regulated PKM2 by modifying circNRIP1 via competing for sponge actions of miR‐541‐5p and miR‐3064‐5p, thereby affecting the glycolytic functions of PTC cells
Abbreviations
- 18F‐FDG
18F‐fluoro‐2‐deoxyglucose
- ALKBH5
α‐ketoglutarate‐dependent dioxygenase alkB homolog 5
- ceRNA
competing endogenous RNA
- circRNA
circular RNA
- ECAR
extra cellular acidification rate
- FCCP
trifluoromethoxy carbonylcyanide phenylhydrazone
- FTO
fat‐mass and obesity‐associated protein
- GEO
Gene Expression Omnibus
- m6A
N6‐methyladenosine
- MeRIP
methylated RNA immunoprecipitation
- METTL
methyltransferase‐like
- miR
microRNA
- miRNA
microRNA
- Mut
mutant
- NC
non control
- ncRNA
noncoding RNA
- NRIP1
nuclear receptor‐interacting protein 1
- OCR
oxygen consumption rate
- OD
optical density
- PK
pyruvate kinase
- PKM2
pyruvate kinase M2
- PTC
papillary thyroid carcinoma
- qRT‐PCR
quantitative real‐time PCR
- SUVmax
maximum standardized uptake value
- WTAP
Wilms tumor 1 associated protein
- YTHDF
YTH domain family protein
1. INTRODUCTION
Thyroid cancer is the most common endocrine‐related malignancy, of which papillary thyroid cancer is the most prevalent subtype, accounting for approximately 90% of all thyroid cancer cases. 1 , 2 The prevalence of thyroid cancer has gradually increased over the past two decades, doubling yearly. 3 Surgical resection combined with radioactive iodine therapy and thyroid‐stimulating hormone suppression therapy is recommended for PTC treatment. 4 Although posttreatment prognostic outcomes are good, posttreatment recurrence and metastasis occur in 10%–15% of PTC patients. 5 The overall survival rate of PTC patients with local recurrence is 70%–85%. 6 However, some advanced, invasive, and locally recurrent or metastatic thyroid cancers do not respond to the above treatments and are often incurable. 7 , 8 Patients with such poor prognostic outcomes have a 10‐year survival rate of 40%. 9 Therefore, it is important to elucidate the molecular mechanisms involved in PTC development and establish prognostic factors for PTC therapy.
Circular RNAs were originally thought to be caused by splicing errors during RNA splicing and had no biological functions. 10 However, various circRNAs have become increasingly common in mammalian cells due to advances in RNA sequencing technology and bioinformatics. Circular RNAs are major transcripts for multiple human cell types, 11 implying that they are stable, conserved, and nonrandom products of RNA splicing. 12 Circular RNAs are involved in cell proliferation, differentiation, apoptosis, and invasion during tumor progression. 13 , 14 In addition, circRNAs may be associated with various miRNAs. For example, hsa_circRNA_104348 facilitate proliferation, migration, and invasion of hepatocellular carcinoma by sponging miR‐187‐3p 15 while hsa_circRNA_002178 act as a ceRNA to promote lung adenocarcinoma progression. 16 Also, hsa_circRNA_0088036 acts as a ceRNA to promote bladder cancer progression by sponging miR‐140‐3p. 17 However, the potential regulatory mechanisms of circRNA actions in PTC have not been fully elucidated.
Tumor cells are characterized by abnormal glucose metabolism. The efficiency of aerobic glycolysis in tumor cells is low, even under sufficient oxygen levels. Cancer cells generate energy through glycolysis, a process referred to as the “Warburg effect”. 18 , 19 , 20 Therefore, abnormal expression of some key glycolysis‐related genes, such as hexokinase 2 (HK2), PKM2, and glucose transporter genes, in various tumors promotes cancer cell proliferation and invasion. 21 , 22 Pyruvate kinase (PK) is the last rate‐limiting enzyme in glycolysis. Four PK isozymes have been identified (M, K, L, and R types). Pyruvate kinase M2 is aberrantly expressed in tumor cells. 23 Therefore, it is necessary to elucidate the mechanisms involved in PKM2 functions and dysregulation in thyroid cancer for informed PTC therapy. 24 , 25 Traditionally, epigenetic regulation refers to DNA‐ or histone‐associated chemical modifications that regulate gene expressions independent of changes in genomic sequences. 26 m6A was first reported in the 1970s and is the most prevalent internal chemical modification associated with eukaryotic mRNAs and ncRNAs. 27 , 28 m6A modification affects many steps in mRNA metabolism, including RNA processing, nuclear export, translation, degradation, and RNA–protein interactions. 29 , 30 Furthermore, m6A modification is a reversible chemical process that is dynamically regulated by balanced activities of N6‐methyladenosine (m6A). m6A modification regulates cancer progression through regulated mechanism of circRNAs. 31 , 32 , 33 , 34 For example, N6‐methyladenosine‐modified circCPSF6 drives malignancy in hepatocellular carcinoma, 35 and m6A‐mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. 36 Furthermore, METTL14‐mediated m6A modification of circORC5 suppresses gastric cancer progression by regulating the miR‐30c‐2‐3p/AKT1S1 axis. 37 However, the role of m6A modification in PTC and its potential regulatory mechanisms for circRNAs are unknown.
In this study, circNRIP1 expression was stably downregulated in PTC tissues and cell lines to assess the potential relationships between circNRIP1 levels and clinicopathological features of PTC. CircNRIP1 enhanced glycolysis in PTC cells by upregulating PKM2 levels and sponging miR‐541‐3p and miR‐3064‐5p. However, these effects were inhibited by ALKBH5, a demethylase. This study elucidates the mechanisms through which m6A modifications regulate PTC glycolysis and ceRNAs.
2. MATERIALS AND METHODS
2.1. Human samples and cell lines
A total of 102 pairs of PTC tissues and adjacent noncancerous tissues were obtained from patients undergoing surgery at the First Hospital of China Medical University between 2019 and 2020. None of the patients had received preoperative local or systemic therapy. Formalin‐fixed and paraffin‐embedded tissue samples were subjected to immunohistochemistry analysis, while fresh frozen tissue samples were used for qRT‐PCR and western blotting. Samples were independently confirmed by two histopathologists. The Research Ethics Committee of the First Hospital of China Medical University approved this study, which was carried out in accordance with the 1964 Declaration of Helsinki guidelines and its subsequent amendments. All study participants provided written informed consent. The human thyroid follicular epithelial cell line (Nthy‐ori3‐1) and PTC cell lines (TPC1, K1, IHH4, and BCPAP) were used in this study. The sources and culture methods for these cell lines are described in Table S1.
2.2. Cell transfection and lentiviral infections
Circular RNA NRIP1, ALKBH5, WTAP, YTHDF1, FTO, METTL3, METTL14, and NC siRNAs were obtained from GenePharma and their sequences are shown in Table S2. The inhibitors and mimics of miRNAs are shown in Table S3. Lipofectamine 3000 (Invitrogen) was used for transfections, following the manufacturer's instructions. A recombinant lentivirus containing sh‐circNRIP1 or sh‐ALKBH5 was established to stably knockdown circNRIP1 or ALKBH5. A nontargeting shRNA was used as the negative control. Circular RNA NRIP1 or ALKBH5 were overexpressed using recombinant lentiviruses containing complete coding sequences of these genes.
An empty lentivirus (vector) was used as the negative control. Lentiviral vectors were constructed by Obio Technologies. Infected cells were selected using puromycin. Quantitative real‐time PCR and immunoblotting were carried out to assess the transfection and infection efficiencies.
2.3. Total RNA extraction and qRT‐PCR
Using RNAiso Plus (Takara), total RNA was extracted from frozen tissue samples and cell samples. PrimeScriptTM RT Master Mix was used for reverse transcription. SYBR Premix Ex Taq II (Takara) was used for qRT‐PCR on a LightCycler 480 system (Roche). Table S4 shows the primer sequences that were utilized in the current study. The relative expression levels were calculated using the 2‐ △△ Ct technique. The internal control was GAPDH.
2.4. Seahorse metabolic analysis
The Seahorse XF Glycolytic Stress Test Kit and the Seahorse XF Cell Aqueous Stress Test Kit (Agilent Technologies) were used to determine the ECAR and OCR, respectively. Transfected PTC cells were seeded in 96‐well plates and cultured in the Seahorse machine overnight. Glucose, oligomycin, and 2‐deoxyglucose were then sequentially added to the corresponding wells on a sensor cartridge for ECAR measurements, while oligomycin, FCCP, antimycin A, and rotenone were sequentially added to the corresponding wells for OCR assay. The prepared cell plates were analyzed using the Seahorse XF analyzer, while the Seahorse wave software was used for data analysis.
2.5. Glucose uptake and lactate production
Glucose uptake was determined using a glucose colorimetric assay kit (Biovision), following the manufacturer's instructions. Briefly, the transfected PTC cells were seeded in 6‐well plates and incubated for 48 h. Specifically, the glucose enzyme mixture oxidizes glucose to form a product (OD value 570 nm) that reacts with the dye, resulting in color generation. Glucose levels in cell culture medium were evaluated, after which cells in each well were counted to normalize glucose levels. Glucose uptake was indirectly determined by measuring the amounts of glucose remaining in cell culture medium. Lactate levels were determined using the lactate colorimetric assay kit (Biovision), following the manufacturer's protocol. Lactate reacts with the enzyme mixture to produce a product that interacts with the lactate probe to generate a color (OD value 570 nm). Lactate production in the cell culture medium was detected and cells in each well were counted to normalize lactate concentrations.
2.6. Methylated RNA immunoprecipitation assays
The RNAiso Plus (Takara) was used for total RNA extraction, after which DNase was added to remove DNA. The m6A RNA enrichment kit (EpigenTek) was used to detect MeRIP levels, following the manufacturer's instructions. The m6A‐containing target fragment was pulled down using a bead‐bound m6A capture Ab, then the RNA sequence containing both ends of the m6A region was cleaved using a lyase cocktail. Enriched RNA was released, purified, and eluted. Quantitative real‐time PCR was carried out after MeRIP to quantify changes in target gene m6A methylation.
2.7. Nude mouse xenograft models and 18F‐FDG PET imaging
Five‐week‐old female athymic BALB/c nude mice from Beijing HFK Biosciences Co. Ltd were used to construct xenograft tumor models. The transfected TPC1 cells were injected into the subcutaneous tissues of mice. The mice were killed after 4 weeks to excise and measure the tumors. Tumor volumes were determined as follows: tumor volume (mm3) = 1/2 × long diameter × short diameter2. The day before they were killed, the mice were fasted for 8 h, anesthetized using 1% sodium pentobarbital, then injected with approximately 200–300 μCi 18F‐FDG into the lateral tail vein. The mice were then kept in cages at room temperature for 1 h. The mice were placed in the prone position on the examination bed for micro‐PET and micro‐computed tomography imaging. The 18F‐FDG uptake was quantified by plotting regions of interest and SUVmax values using the MetisViewer software. The use of animals was approved by the Institutional Animal Care and Use Committee of China Medical University.
2.8. Statistical analysis
The SPSS 26.0 (IBM) and GraphPad Prism 8.3.0 software were used for data analyses. Data are shown as mean ± SD for n = 3. Student's t‐test and ANOVA were used for between‐ and among‐group comparisons of means, respectively. The Wilcoxon signed‐rank test was used to assess differences in relative expression levels of circNRIP1, hsa‐miR‐541‐5p, hsa‐miR‐3064‐5p, PKM2, and ALKBH5 in PTC tissues and adjacent noncancerous tissues. Correlations between circNRIP1 levels and clinicopathological characteristics of patients were determined using the χ2‐test. p ≤ 0.05 was the threshold for statistical significance.
3. RESULTS
3.1. Elevated circNRIP1 levels are correlated with higher TNM stage and larger tumor sizes in PTC patients
The differential expressions of circRNAs in PTC tissues and normal thyroid tissues were determined using the GEO database to investigate the roles of circRNAs in PTC. Compared with normal tissues, expressions of 14 circRNAs in two GEO datasets (GSE93522 and GSE171011) were significantly different (Figure 1A). Heat map analysis revealed that hsa‐circ‐0004771 (circNRIP1) had the largest expression difference (Fold change = 3.13) p < 0.01 (Figure 1B). Furthermore, qRT‐PCR analysis showed that circNRIP1 levels were higher in 102 PTC tissues than in normal tissues. Expression levels in paired adjacent noncancerous tissues are shown in Figures 1C and S1A,B. Elevated circNRIP1 levels in PTC patients were correlated with higher TNM stage (p = 0.006) and larger tumor size (p = 0.029) (Table 1). Quantitative real‐time PCR analysis revealed that circNRIP1 levels were significantly high in patients with higher TNM stages and larger tumor volumes (Figure S1C,D). Consistent with PTC tissue sample data, circNRIP1 levels were significantly higher in PTC cell lines (TPC1, BCPAP, K1, and IHH4) than in the normal thyroid cell line, Nthy‐ori‐3‐1 (Figure 1D). Circular RNA NRIP1 in TPC1 cells was characterized by qRT‐PCR, Sanger sequencing, and RNase R treatment. Quantitative real‐time PCR analysis was undertaken using different primers. The sequenced PCR products corresponded to 5′ exon 3 to 3′ exon 2 (Figure 1E). Resistance experiments to RNase R exon nuclease digestion confirmed that this RNA species has a circular RNA structure (Figure 1F). Quantitative real‐time PCR analysis revealed that the half‐life of circNRIP1 was over 24 h after treatment with actinomycin D (transcription inhibitor), while the half‐life of the related linear transcript was approximately 4 h (Figure 1G), indicating that circNRIP1 was more stable in PTC cells. Nucleocytoplasmic cell isolation and FISH analysis revealed that circNRIP1 was mainly localized in the cytoplasm of TPC1, BCPAP, and Nthy‐ori3‐1 cells (Figures 1H,I and S1E,F). These results suggest that circNRIP1 is a circRNA stably expressed in PTC cytoplasm.
FIGURE 1.
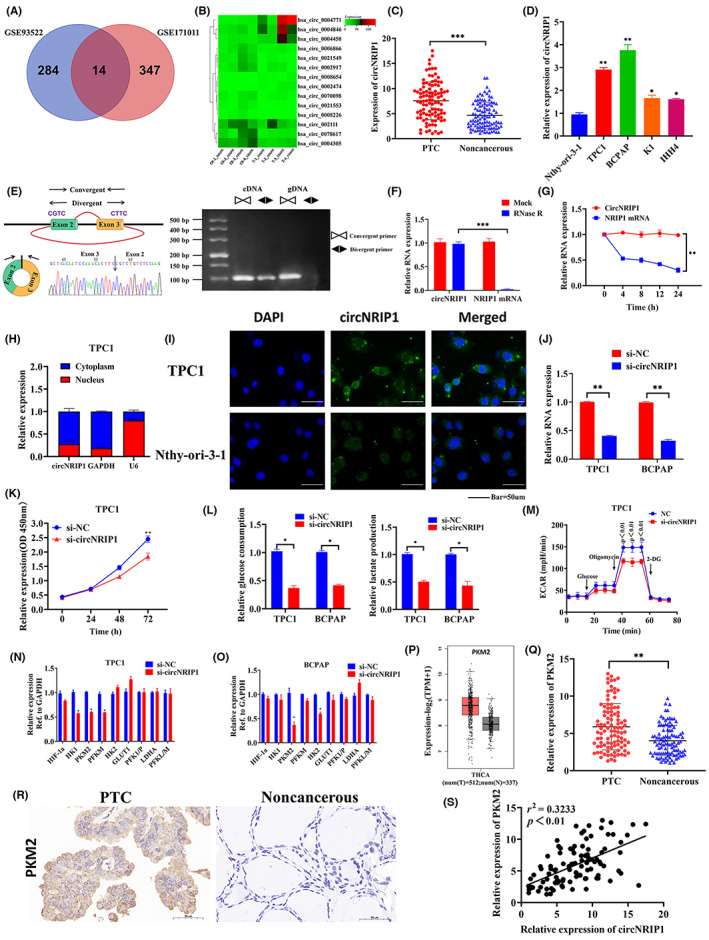
Circular RNA nuclear receptor‐interacting protein 1 (circNRIP1) upregulation in papillary thyroid cancer (PTC): characterization of circNRIP1 in PTC; circNRIP1 promotes PKM2 expressions in PTC cells. (A,B) circNRIP1 was selected using (A) the Gene Expression Omnibus database (GSE93522 and GSE171011) and (B) the bioinformatics software. (C) Relative expression of circNRIP1 in 102 pairs of PTC tissues and adjacent noncancerous tissues. (D) Relative expression of circNRIP1 in PTC cells and normal thyroid cells determined by quantitative real‐time PCR (qRT‐PCR). (E) Genomic locus of circNRIP1. Left, expression of circNRIP1 assessed by qRT‐PCR followed by Sanger sequencing. Arrows represent divergent primers binding the genome region of circNRIP1. Right, qRT‐PCR products with divergent primers showing circNRIP1 circularization. gDNA, genomic DNA. (F) qRT‐PCR showing the expression of circNRIP1 and NRIP1 mRNA after treatment with RNase R in TPC1 cells. (G) qRT‐PCR showing the expression of circNRIP1 and NRIP1 mRNAs after treatment with actinomycin D at indicated time points in TPC1 cells. (H) Nuclear and cytoplasmic RNA fractions isolated from TPC1 cells. CircNRIP1 was mainly located in the cytoplasm; GAPDH and U6 were used as controls. (I) FISH verification showing that circNRIP1 is mainly localized in the cytoplasm. Scale bar, 50 μm. (J) Quantitative PCR showing that si‐circNRIP1 can significantly inhibit circNRIP1 expression in TPC1 and BCPAP cells. (K) CCK‐8 assay showing the proliferative abilities of TPC1 cells after circNRIP1 knockdown. (L) Glucose uptake and lactate production determined after transfection with si‐NC (non‐silencing contron) and si‐circNRIP1 in TPC1 and BCPAP cells. (M) Extra cellular acidification rate (ECAR) determined by Seahorse metabolic analysis after transfection with si‐NC and si‐circNRIP1 in TPC1 cells. (N,O) Relative expression of glycolysis‐related genes after circNRIP1 downregulation detected by qRT‐PCR in (N) TPC1 and (O) BCPAP cells. (P) pyruvate kinase M2 (PKM2) expression in PTC and normal tissues from The Cancer Genome Atlas database. (Q) Relative expression of PKM2 in 102 pairs of PTC tissues and adjacent noncancerous tissues. (R) Representative PKM2 immunohistochemical staining of PTC tissues, compared with paired normal thyroid tissues. Scale bar, 50 um. (S) Correlations between circNRIP1 and PKM2 levels in PTC tissues analyzed by Pearson's correlation analysis. *p < 0.05, **p < 0.01, ***p < 0.001
TABLE 1.
Correlation between circular RNA nuclear receptor‐interacting protein 1 (circNRIP1) expression and clinicopathological features in papillary thyroid cancer tissues (n = 102)
| Characteristic | n | circNRIP1 | p value | |
|---|---|---|---|---|
| High expression(%) | Low expression(%) | |||
| Gender | ||||
| Male | 39 | 18(46.2) | 21(53.8) | 0.541 |
| Female | 63 | 33(52.4) | 30(47.6) | |
| Age, years | ||||
| <55 | 60 | 33(55.0) | 27(45.0) | 0.227 |
| ≥55 | 42 | 18(42.9) | 24(57.1) | |
| Tumor size, cm | ||||
| <2 | 55 | 22(40.0) | 33(60.0) | 0.029* |
| ≥2 | 47 | 29(61.7) | 18(38.3) | |
| Extrathyroidal extension | ||||
| Yes | 35 | 20(57.1) | 15(42.9) | 0.297 |
| No | 67 | 31(46.3) | 36(53.7) | |
| LNM | ||||
| Yes | 64 | 36(56.2) | 28(43.8) | 0.101 |
| No | 38 | 15(39.5) | 23(60.5) | |
| Multifocality | ||||
| Yes | 32 | 17(53.1) | 15(46.9) | 0.669 |
| No | 70 | 34(48.6) | 36(51.4) | |
| TNM staging | ||||
| I‐II | 86 | 38(44.2) | 48(55.8) | 0.006* |
| III–IV | 16 | 13(81.2) | 3(18.8) | |
| Hashimoto thyroiditis | ||||
| Yes | 27 | 13(48.1) | 14(51.9) | 0.822 |
| No | 75 | 38(50.7) | 37(49.3) | |
Abbreviation: LNM, lymph node metastasis.
Statistically significant (p < 0.05).
3.2. Circular RNA NRIP1 promotes glycolysis in PTC cells
The oncogenic phenotypes in TPC1 and BCPAP cells were characterized by circNRIP1 silencing to assess the function of circNRIP1. Quantitative PCR results showed that si‐circNRIP1 significantly inhibited circNRIP1 expression in TPC1 and BCPAP cells (Figure 1J). The CCK‐8 assays revealed that circNRIP1 knockdown in TPC1 and BCPAP cells inhibited proliferation (Figures 1K and S1G). Extracellular glucose and lactate level analyses were carried out to elucidate the roles of circNRIP1 on cell proliferation. Suppression of circNRIP1 markedly inhibited glucose uptake and lactate production in PTC cells (Figure 1L). The OCR and ECAR of cells were assessed using Seahorse metabolic assays after circNRIP1 suppression. The ECAR value was significantly reduced in TPC1 and BCPAP cells, implying the inhibition of glycolysis in PTC. These findings indicate that circNRIP1 suppression decreases glycolysis levels in post‐PTC cells (Figures 1M and S1H). The OCR value was significantly increased, indicating the inhibition of PTC mitochondrial oxidative phosphorylation and that circNRIP1 promotes glycolysis (Figure S1I,J). Circular RNA NRIP1 was knocked down to investigate the regulatory mechanisms of circNRIP1 in glycolysis. The levels of key glycolysis‐related enzymes were also evaluated. Quantitative real‐time PCR analysis showed that circNRIP1 silencing suppressed PKM2 levels in TPC1 and BCPAP cell lines (Figure 1N,O). Analysis of The Cancer Genome Atlas dataset revealed that PKM2, a key enzyme in glycolysis, was significantly upregulated in PTC tissues, compared to normal tissues (Figure 1P). Relative expressions of PKM2 in 102 pairs of PTC tissues and matched adjacent noncancerous tissues were verified using qRT‐PCR. Pyruvate kinase M2 levels were higher in PTC tissues than in adjacent noncancerous tissues (Figure 1Q). Furthermore, immunohistochemistry revealed that PKM2 protein levels were significantly upregulated in PTC tissues and were mainly localized in the cytoplasm of PTC cells (Figure 1R). Spearman's correlation analysis revealed that circNRIP1 was positively correlated with PKM2 levels in PTC tissues (Figure 1S).
3.3. Circular RNA NRIP1 promotes glycolysis by regulating PKM2 levels through competitively binding to miR‐541‐5p and miR‐3064‐5p
Subcellular localization of circRNAs is closely associated with their biological functions and potential molecular roles. The ceRNA theory is an important circRNA regulatory mechanism stating that circRNAs highly expressed in cytoplasm can compete with miRNAs to bind and regulate downstream target genes. Therefore, circNRIP1 can regulate PTC cell functions through a ceRNA mechanism. Herein, miRWalk (http://mirwalk.umm.uni‐heidelberg.de/) and ENCORI (http://starbase.sysu.edu.cn/) databases revealed 12 miRNAs that could bind circNRIP1 and PKM2 by complementary base pairing (Figure 2A). Expression of candidate miRNAs after circNRIP1 downregulation in TPC1 and BCPAP cells was evaluated by qRT‐PCR (Figure 2B,C). Among the candidate miRNAs, miR‐541‐5p, miR‐3064‐5p, and miR‐3140‐5p may be involved in the regulation of PKM2 expression. The candidate miRNAs were overexpressed to confirm the hypothesis, and PKM2 levels in PTC cells were also assessed. Western blotting showed that only miR‐541‐5p and miR‐3064‐5p overexpression suppressed PKM2 (Figure 2D), suggesting that PKM2 might be regulated by miR‐541‐5p and miR‐3064‐5p. Quantitative real‐time PCR revealed that miR‐541‐5p and miR‐3064‐5p levels were suppressed in 98 PTC tissues compared with paired adjacent noncancerous tissues (Figure 2E,F). Both miR‐541‐5p and miR‐3064‐5p were negatively correlated with circNRIP1 and PKM2 levels in PTC tissues (Figure 2G,H). The PTC cells with suppressed miR‐541‐5p and miR‐3064‐5p were constructed to verify the effects of miR‐541‐5p and miR‐3064‐5p on cell proliferation and glycolysis. Western blotting revealed that miR‐541‐5p and miR‐3064‐5p knockdown promoted PKM2 expression (Figure 2I); CCK‐8 analysis showed that miR‐541‐5p and miR‐3064‐5p knockdown promoted PTC cell proliferation (Figures 2J and S2A). Extracellular glucose and lactate analyses showed that both glucose uptake and lactate production were significantly elevated in PTC cells when miR‐541‐5p and miR‐3064‐5p were downregulated (Figures 2K and S2B). Seahorse XF metabolic assays were used to evaluate OCR and ECAR values of TPC1 cells after inhibition of miR‐541‐5p and miR‐3064‐5p. The ECAR values of PTC cells were significantly elevated (Figures 2L and S2C), while OCR values were significantly suppressed (Figure S2D,E). These findings imply that miR‐541‐5p and miR‐3064‐5p knockdown enhance glycolysis in PTC cells.
FIGURE 2.
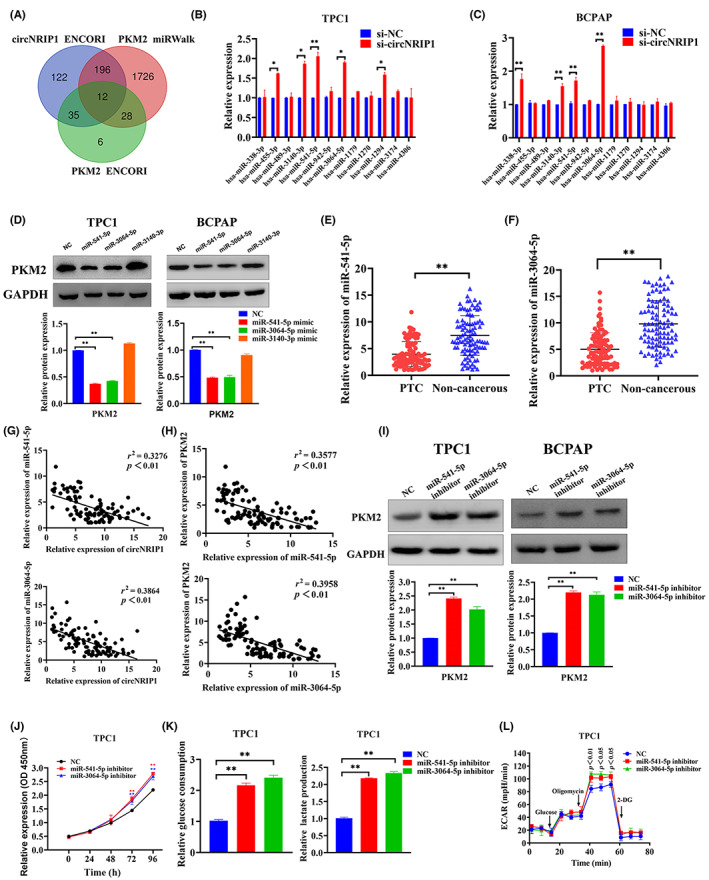
MicroRNA (miR)‐541‐5p and miR‐3064‐5p targeting inhibited pyruvate kinase M2 (PKM2) expression in papillary thyroid cancer (PTC) cells. Knockdown of miR‐541‐5p and miR‐3064‐5p promoted PTC cell proliferation and glycolysis. (A) miRNAs potentially targeting circular RNA nuclear receptor‐interacting protein 1 (circNRIP1) and PKM2 predicted using bioinformatics software. (B,C) Quantitative real‐time PCR (qRT‐PCR) showing relative expressions of predicted miRNAs after si‐NC (non‐silencing control) and si‐circNRIP1 transfection in (B) TPC1 and (C) BCPAP cells. (D) Western blot detection of glycolysis‐associated proteins (PKM2) after miR‐541‐5p, miR‐3064‐5p, and miR‐3140‐3p mimic transfection in PTC cells. (E,F) qRT‐PCR showing relative expressions of miR‐541‐5p and miR‐3064‐5p in 98 pairs of PTC tissues and adjacent noncancerous tissues. (G) Correlations between circNRIP1 and miR‐541‐5p and miR‐3064‐5p in PTC tissues determined by Pearson's correlation. (H) Correlations between miR‐541‐5p, miR‐3064‐5p, and PKM2 in PTC tissues analyzed by Pearson's correlation. (I) PKM2 levels determined by western blotting. (J) TPC1 cell proliferation evaluated by the CCK‐8 assay. (K) Glucose uptake and lactate production after transfection with NC, miR‐541‐5p inhibitor, and miR‐3064‐5p inhibitor in TPC1 cells. (L) Extracellular acidification rate (ECAR) determined by Seahorse metabolic analysis after transfections with NC, miR‐541‐5p inhibitor, and miR‐3064‐5p inhibitor in TPC1 cells.*p < 0.05, **p < 0.01
3.4. Circular RNA NRIP1 regulates PKM2 expression by directly binding miR‐541‐5p and miR‐3064‐5p
In this study, we also determined whether circNRIP1 can function as a ceRNA by directly sponging miR‐541‐5p and miR‐3064‐5p. Bioinformatic predictions indicated that miR‐541‐5p and miR‐3064‐5p could bind circNRIP1 and directly target the 3′‐UTR of PKM2. Dual‐luciferase reporter gene assays showed that miR‐541‐5p upregulation significantly reduced the luciferase activities of PTC cells cotransfected with circNRIP1‐WT, whereas luciferase activities of cells with circNRIP1‐Mut1 and upregulated miR‐541‐5p did not change after the cotransfection (Figure 3A). Cotransfection of the miR‐3064‐5p mimic and circNRIP1‐WT significantly reduced luciferase activities, whereas cotransfections of the miR‐3064‐5p mimic and circNRIP1‐Mut2 did not alter luciferase activities (Figure 3B), indicating that circNRIP1 directly binds miR‐541‐5p and miR‐3064‐5p. A dual‐luciferase reporter system was then used to determine whether PKM2 is a direct target of miR‐541‐5p and miR‐3064‐5p. Compared with the NC group, overexpressed miR‐541‐5p significantly inhibited PKM2‐3′‐UTR‐WT luciferase activities, while it did not affect PKM2‐3′‐UTR‐Mut activities (Figure 3C). Overexpressed miR‐3064‐5p significantly inhibited PKM2‐3′‐UTR‐WT luciferase activities, but did not affect PKM2‐3′‐UTR‐Mut activities (Figure 3D). Therefore, these results indicate that miR‐541‐5p and miR‐3064‐5p specifically bound PKM2‐3′‐UTR in PTC cells. Another dual‐luciferase reporter assay was undertaken to confirm whether circNRIP1 can regulate PKM2 by interacting with miR‐541‐5p and miR‐3064‐5p. Circular RNA NRIP1‐WT significantly increased the luciferase activities of WT PKM2, whereas circNRIP1‐Mut did not affect the luciferase activities of WT PKM2 (Figure 3E,F). These results indicate that circNRIP1 regulates PKM2 expression as a ceRNA by sponging miR‐541‐5p and miR‐3064‐5p.
FIGURE 3.
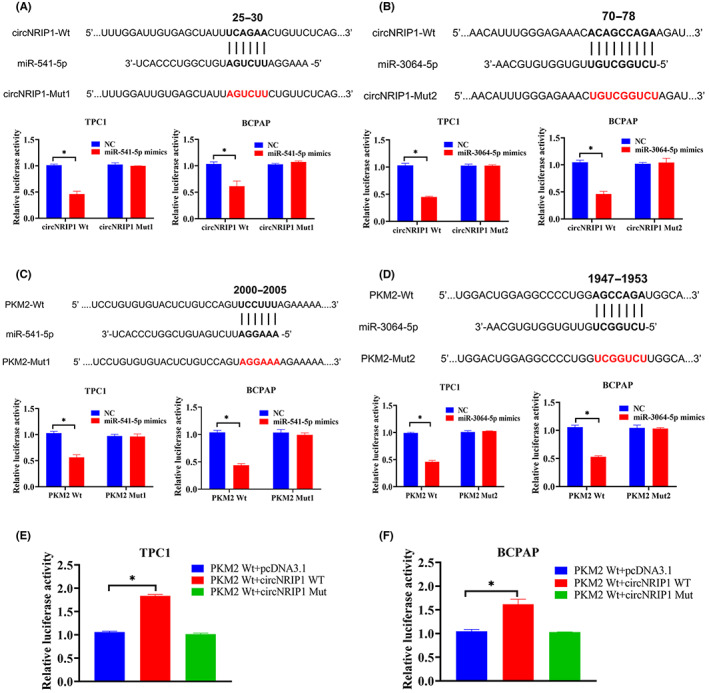
Circular RNA nuclear receptor‐interacting protein 1 (circNRIP1) acts as a competing endogenous RNA and regulates pyruvate kinase M2 (PKM2) expression by binding microRNA (miR)‐541‐5p and miR‐3064‐5p. (A,B) Predicted (A) miR‐541‐5p and (B) miR‐3064‐5p binding sites in circNRIP1 (circNRIP1‐WT) and designed mutant sequences (circNRIP1‐Mut1, circNRIP1‐Mut2). TPC1 and BCPAP cells were transfected with circNRIP1‐WT, circNRIP1‐Mut1, circNRIP1‐Mut2, and indicated miRNAs, after which the luciferase reporter assay was carried out. (C,D) Predicted (C) miR‐541‐5p and (D) miR‐3064‐5p binding sites in PKM2 3′‐UTR (PKM2 3′‐UTR‐WT) and designed mutant sequences (PKM2 3′‐UTR‐Mut1, PKM2 3′‐UTR‐Mut2). TPC1 and BCPAP cells were transfected with PKM2 3′‐UTR‐WT, PKM2 3′‐UTR‐Mut1, PKM2 3′‐UTR‐Mut2, and the indicated miRNAs, after which the luciferase reporter assay was carried out. (E,F) Luciferase reporter assay of (E) TPC1 and (F) BCPAP cells cotransfected with PKM2 3′‐UTR‐WT, pcDNA3.1, circNRIP1‐WT, and circNRIP1‐Mut1 vectors. *p < 0.05
3.5. Overexpressed miR‐541‐5p and miR‐3064‐5p inhibit circNRIP1‐induced glycolysis and proliferation of PTC cells
Rescue assays showed that miR‐541‐5p and miR‐3064‐5p restored the effects of circNRIP1 on PTC cell proliferation and glycolysis, and western blotting revealed that miR‐541‐5p and miR‐3064‐5p mimics significantly reversed the glycolysis of circNRIP1 and inhibited PKM2 expression (Figure 4A,B). The CCK‐8 assay showed that miR‐541‐5p and miR‐3064‐5p attenuated the promotion effects of circNRIP1 on TPC1 and BCPAP cell proliferation (Figures 4C,D and S2H,I). Extracellular glucose and lactate level assays showed that miR‐541‐5p and miR‐3064‐5p antagonized the promoting effects of circNRIP1 in TPC1 and BCPAP cells, thus inducing glucose uptake and lactate production (Figure 4E–H). Moreover, Seahorse XF metabolic assays showed that miR‐541‐5p and miR‐3064‐5p attenuated the glycolytic effects of circNRIP1 in PTC cells (Figures 4I,J and S2J,K).
FIGURE 4.
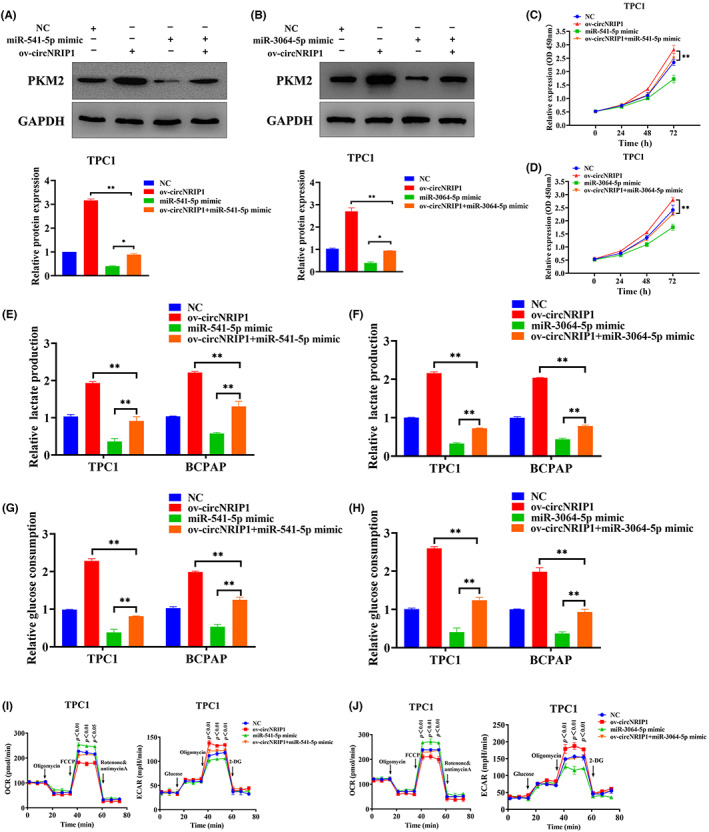
MicroRNA (miR)‐541‐5p and miR‐3064‐5p overexpression partially impair circular RNA nuclear receptor‐interacting protein 1 (circNRIP1)‐induced promotion of malignant behaviors in papillary thyroid cancer (PTC) cells. (A,B) Pyruvate kinase M2 (PKM2) levels in TPC1 cells analyzed by Western blotting. (C,D) TPC1 cell proliferation analyzed by the CCK‐8 assay. (E–H) Glucose uptake and lactate production in PTC cells analyzed using the Glucose Colorimetric Assay Kit. (I,J) Oxygen consumption rate (OCR) and extra cellular acidification rate (ECAR) analyzed by Seahorse metabolic in TPC1 cells. *p < 0.05, **p < 0.01. NC, non control; OD, optical density; ov‐circNRIP1, overexpression‐circNRIP1.
3.6. Circular RNA NRIP1 promotes in vivo tumor growth
A control vector (pcDNA3.1), a circNRIP1 overexpression vector, a mixed vector of circNRIP1 overexpression and miR‐541‐5p mimic, and a mixed vector of circNRIP1 overexpression and miR‐3064‐5p mimic were stably transfected to establish whether circNRIP1 affects tumor growth in vivo. Stained TPC1 cells were also injected into the back of mice. The growth of subcutaneous tumors was observed after every 3 days from the 10th day after injection. Long and short tumor diameters were measured using a vernier caliper to calculate tumor volumes. The circNRIP1 overexpressed ov‐circNRIP1 group showed larger xenograft tumor volumes than pcDNA3.1, ov‐circNRIP1 + miR‐541‐5p mimic, and ov‐circNRIP1 + miR‐3064‐5p mimic groups (Figure 5A). Also, overexpression of miR‐541‐5p and miR‐3064‐5 suppressed the tumor volumes and weights (Figure 5B,C). Overexpression of circNRIP1 in the resected tumor masses was associated with increased circNRIP1 and PKM2 levels as well as suppressed miR‐541‐5p and miR‐3064‐5p levels (Figure 5D). Histochemical detection revealed that PKM2 protein levels were significantly upregulated in the circNRIP1 overexpression group, consistent with in vitro findings. However, miR‐541‐5p and miR‐3064‐5 overexpression downregulated PKM2 protein levels (Figure 5E). In vivo changes in glycolysis were measured using 18F‐FDG‐PET to determine glucose uptake in mice tumors. Glucose uptake in xenograft mice tumor models were significantly increased after circNRIP1 overexpression, compared with control group. However, miR‐541‐5p and miR‐3064‐5p antagonized this effect (Figure 5F).
FIGURE 5.
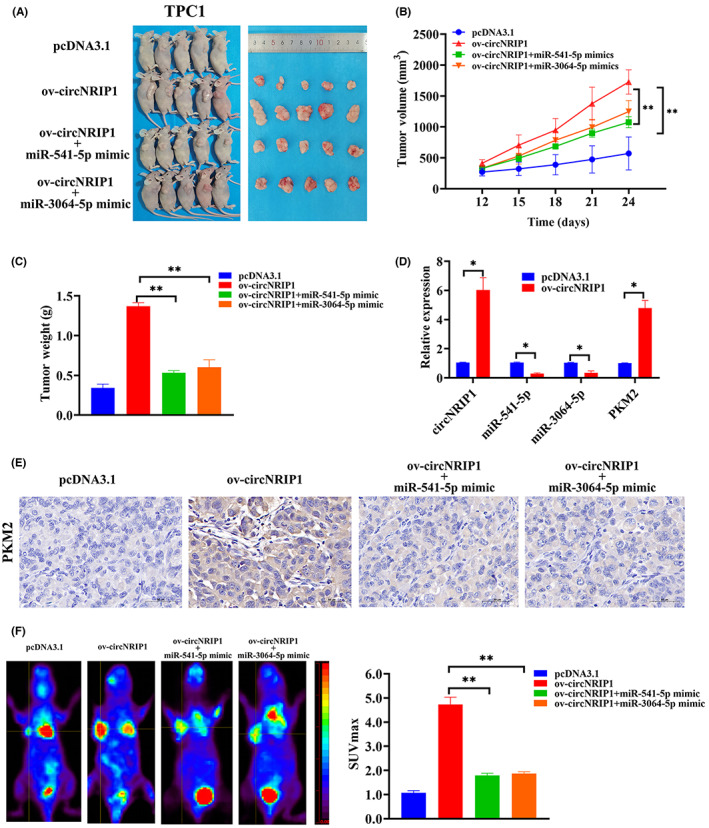
Circular RNA nuclear receptor‐interacting protein 1 (circNRIP1) promotes tumor growth in vivo. (A) Subcutaneous implant mouse model established after inoculation with pcDNA3.1, overexpression‐circNRIP1 (ov‐circNRIP1), ov‐circNRIP1 + mir‐541‐5p mimic, and ov‐circNRIP1 + mir‐3064‐5p mimic TPC1 cells (n = 5). (B) Tumor volumes and (C) tumor weights. (D) Quantitative real‐time PCR showing relative expressions of circNRIP1, miR‐541‐5p, miR‐3064‐5p, and pyruvate kinase M2 (PKM2) in pcDNA3.1 and ov‐circNRIP1 groups. (E) Tumor sections stained with H&E. PKM2 levels were evaluated by immunohistochemistry. (F) Representative images of18F‐fluoro‐2‐deoxyglucose (18F‐FDG) uptake in pcDNA3.1, ov‐circNRIP1, ov‐circNRIP1 + mir‐541‐5p mimic, and ov‐circNRIP1 + mir‐3064‐5p mimic xenograft mouse models. Red circles indicate tumor glucose uptake. Maximum uptake values (SUVmax) for xenografts were determined by 18F‐FDG PET. *p < 0.05, **p < 0.01
3.7. ALKBH5 inhibits PTC development by suppressing circNRIP1
m6A is crucial in RNA methylation modification of circRNAs. Herein, the potential roles of m6A in thyroid cancer were valuated using bioinformatics analyses. Results showed that the mRNA levels of methylases (METTL3, METTL14, and WTAP), demethylases (FTO and ALKBH5), and methylation recognition proteins were significantly different between PTC and normal tissues (Figure S3A,B). The major m6A methyltransferases were disrupted in TPC1 as well as BCPAP cells and expressions of circNRIP1 were detected. Only ALKBH5 knockdown affected circNRIP1 expression in both cell lines (Figure 6A,B). The tissue sample size was increased to 102 pairs to establish the effect of ALKBH5 downregulation in PTC tissues. The qRT‐PCR analysis showed that ALKBH5 level was lower in PTC tissues than in paired adjacent noncancerous tissues (Figure 6C). Furthermore, ALKBH5 levels were negatively correlated with circNRIP1 (Figure 6D). Moreover, the actinomycin D assays showed that the stability of circNRIP1 was decreased with increasing ALKBH5 overexpression (Figure 6E). Knockdown of ALKBH5 promoted TPC1 and BCPAP cell proliferation (Figures 6F and S4A) and significantly increased glucose uptake as well as lactate production in both cells (Figure 6G,H). Similarly, OCR and ECAR values of PTC cells were assessed after ALKBH5 suppression by hippocampal metabolic analysis experiments. The ECAR values were significantly increased in TPC1 and BCPAP cells (Figures 6I and S4B), whereas OCR values were significantly decreased (Figure S4C,D). These findings indicate that ALKBH5 suppression promotes PTC mitochondrial oxidative phosphorylation and inhibits glycolysis. Furthermore, m6A‐modified fragments were enriched using m6A Abs through the MeRIP assay, after which qRT‐PCR assays were carried out using primers containing the m6A‐modified sequence of circNRIP1, as predicted by the methylated MeRIP sequencing data. Agarose electrophoresis showed that circNRIP1 contained m6A‐related enzyme enrichment sites. The MeRIP‐qPCR experiments indicated that m6A enrichment levels of circNRIP1 were significantly increased after ALKBH5 suppression (Figures 6J and S4E). A luciferase reporter gene containing WT and Mut circNRIP1 was then constructed to assess the effects of m6A modification on circNRIP1 levels. The overexpression of ALKBH5 decreased the luciferase activities of WT and the A in the m6A modification site of circNRIP1 was mutated to C, whereas Mut circNRIP1 did not change (Figures 6K and S4F). Moreover, ALKBH5 interference significantly increased luciferase activities of WT. However, luciferase activities did not change in Mut circNRIP1 (Figure S4G,H), implying that ALKBH5 affects circNRIP1 expression through m6A modifications.
FIGURE 6.
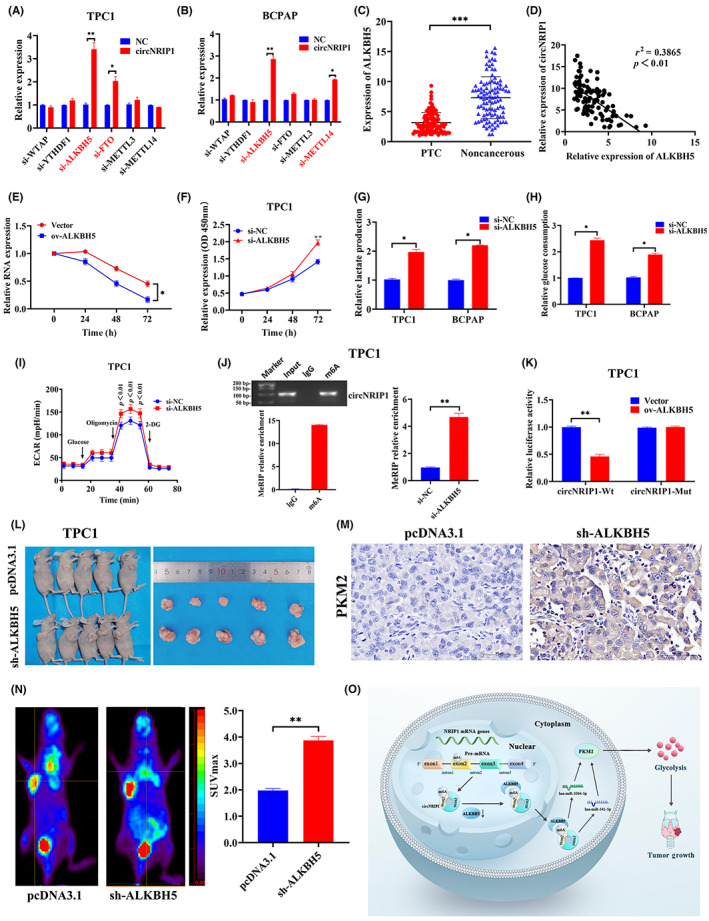
α‐Ketoglutarate‐dependent dioxygenase alkB homolog 5 (ALKBH5) inhibits circular RNA nuclear receptor‐interacting protein 1 (circNRIP1) expression in papillary thyroid cancer (PTC) cells. ALKBH5 is negatively correlated with circNRIP1 and regulates PTC proliferation by modulating glycolysis. (A,B) Quantitative real‐time PCR (qRT‐PCR) showing relative expressions of predicted circNRIP1 after downregulation of m6A‐related genes in (A) TPC1 and (B) BCPAP cells. (C) qRT‐PCR showing relative expression of ALKBH5 in 102 pairs of PTC tissues and adjacent noncancerous tissues. (D) Correlations between ALKBH5 and circNRIP1 in PTC tissues analyzed by Pearson's correlation analyses. (E) Actinomycin D assays showing the expression and stability of circNRIP1 after ALKBH5 overexpression. (F) TPC1 cell proliferation evaluated using CCK‐8 assay. (G) Lactate production and (H) glucose uptake by TPC1 and BCPAP cells after transfections with non‐silencing control (si‐NC) and si‐ALKBH5. (I) Extra cellular acidification rate (ECAR) in TPC1 cells determined by Seahorse metabolic analysis after transfections with si‐NC and si‐ALKBH5. (J) Left, direct binding between m6A Ab and circNRIP1 validated by agarose electrophoresis and methylated RNA immunoprecipitation (MeRIP) assays in TPC1 cells. Right, relative enrichment of the m6A Ab and circNRIP1 determined by MeRIP‐qPCR after ALKBH5 knockdown in TPC1 cells. (K) Relative luciferase activities of circNRIP1‐WT and circNRIP1‐Mut after ALKBH5 overexpression in PTC cells. (L) Subcutaneous implant mouse model established after inoculation with TPC1 cells transfected with pcDNA3.1 or sh‐ALKBH5 vectors (n = 5). (M) Tumor sections stained with H&E. Pyruvate kinase M2 (PKM2) was assessed by immunohistochemistry. (N) Representative images of 18F‐fluoro‐2‐deoxyglucose (18F‐FDG) uptake by micro‐PET imaging in pcDNA3.1 and sh‐ALKBH5 xenograft mouse models. Red circles indicate tumor glucose uptake. Maximum uptake values (SUVmax) for xenografts were determined by 18F‐FDG PET. (O) Mechanistic diagram showing the relationships among circNRIP1, m6A modification, PTC tumor growth, and glycolysis metabolism. *p < 0.05, **p < 0.01, ***p < 0.001
3.8. ALKBH5 promotes in vivo tumor growth
TPC1 cells stably transfected with ALKBH5 KO vector and control vector (pcDNA3.1) were injected into the backs of mice to assess whether ALKBH5 affects tumor growth in vivo. The growth of subcutaneous tumors was observed every 3 days from the 10th day after injection. Long and short diameters of tumors were measured using a vernier caliper, and tumor volumes were calculated. The xenograft tumors were larger in the sh‐ALKBH5 group than in the NC vector (Figure 6L). Compared with the NC vehicle group, tumor volumes and weights were significantly increased in the ALKBH5 KO group (Figure S4L,J). PKM2 protein levels were upregulated in the sh‐ALKBH5 group, consistent with in vitro findings (Figure 6M). Furthermore, ALKBH5 knockdown was associated with low ALKBH5 levels and increased circNRIP1 levels (Figure S4K). The SUVmax value of the tumor was significantly higher in the sh‐ALKBH5 group than in the pcDNA3.1 group (Figure 6N), indicating that ALKBH5 knockdown promotes glucose uptake in PTC xenografts. ALKBH5 regulated PKM2 by modifying circNRIP1 through competing for sponge actions of miR‐541‐5p and miR‐3064‐5p, thereby affecting the glycolytic functions of PTC cells (Figure 6O).
4. DISCUSSION
Circular RNAs are a novel type of ncRNAs with covalently closed loop structures. 38 Circular RNAs are generated by exon skipping or back splicing without 5′–3′ polarity or polyadenylation tail. 12 , 39 Several exonic and intronic circRNAs have been identified in eukaryotes in the past 20 years, 40 indicating that circRNAs are not byproducts of aberrant splicing, but have multiple potential biological functions. Furthermore, circRNAs are more stable and durable compared with linear RNAs because they lack free ends for RNase‐mediated degradation. 41 In addition, circRNAs promote tumor progression. 42 , 43 , 44 In this study, circNRIP1, whose host gene is NRIP1, was upregulated in both PTC tissues and cell lines, indicating that it might be involved in PTC occurrence and development. Functionally, circNRIP1 promotes cervical cancer cell migration and invasion by sponging miR‐629‐3p and regulating the PTP4A1/ERK1/2 pathway, 45 esophageal squamous cell migration and invasion through the miR‐339‐5p/CDC25A axis, progression of squamous cell carcinoma, 46 or by targeting the ZEB2 signaling pathway to activate miR‐653, thus inhibiting breast cancer cell proliferation while inducing apoptosis. 47 In this study, circNRIP1 levels were significantly elevated in thyroid cancer tissues compared with normal noncancerous tissues. Levels of circNRIP1 were also upregulated in 102 PTC tissues compared with adjacent noncancerous tissues. Elevated circNRIP1 levels were associated with poorer prognostic outcomes, including higher TNM stages and larger tumor volumes, suggesting that circNRIP1 promotes PTC progression. Therefore, circNRIP1 could be involved in malignant progression of PTC.
Compared with normal cells, cancer cells absorb large amounts of glucose even under aerobic conditions to produce lactate, a phenomenon known as “aerobic glycolysis” or the Warburg effect. 48 Glycolysis has been widely used as a marker of tumor progression because large amounts of lipids, proteins, and nucleotides are produced during glucose metabolism, accelerating cancer cell proliferation and division. 49 , 50
Glycolysis or oxidative phosphorylation can be controlled by the regulation of glycolytic flux through glycolytic enzymes. Among the several key enzymes in glycolysis, PK catalyzes the final glycolysis reaction by transferring high‐energy phosphates from phosphoenolpyruvate to ADP for ATP and pyruvate generation. Enzyme PK has four isoforms, with PKM2 being the predominant type in cancer cell proliferation. 51 Overexpression of PKM2 increases the rate of glycolysis, and thus most glucose is converted to lactate, and ATP is rapidly produced. 52 Recent findings indicated that PKM2 can be regulated by circRNA to promote tumor glycolysis and accelerate tumor progression. For instance, exosome‐delivered hsa_circ_0005963 promotes glycolysis through the miR‐122‐PKM2 axis, thus inducing chemoresistance in colorectal cancer. 53 Circular RNA MAT2B promotes glycolysis and malignancy in hepatocellular carcinoma under hypoxic stress through the miR‐338‐3p/PKM2 axis. 24 Furthermore, circATP2B1 promotes aerobic glycolysis in gastric cancer cells by regulating PKM2. 54 However, it is unknown whether circNRIP1 can regulate PKM2.
In this study, circNRIP1 promoted PTC cell proliferation and glycolysis. Furthermore, PKM2 was significantly suppressed in the si‐circNRIP1 group in both TPC1 and BCPAP cell lines. Tissue expression assays showed that downregulated PKM2 in PTC tissues was positively correlated with circNRIP1, indicating that circNRIP1 might be involved in tumor progression by regulating PKM2.
Additionally, circRNAs can regulate gene expression by acting as ceRNAs. 55 , 56 Recent findings indicated that hsa_circRNA_102002 facilitates metastasis of PTC by regulating the miR‐488‐3p/HAS2 axis, 57 and hsa_circRNA_104565 promotes cell proliferation in PTC by sponging miR‐134. 58 Also, the hsa_circ_0088494/miR‐876‐3p/CTNNB1/CCND1 axis participates in carcinogenesis and progression of PTC. 59 Furthermore, circITCH suppresses PTC progression through the miR‐22‐3p/CBL/β‐catenin pathway. 60 However, the relationship between circNRIP1 and glycolysis of PTC is unknown. In this study, circNRIP1 was found to be mainly located in the cytoplasm of PTC cells. Therefore, circNRIP1 might also function as a ceRNA and regulate PKM2 expression by sponging miRNAs in PTC. Bioinformatics analysis and target prediction tools identified miRNAs that could target PKM2 and the circNRIP1 binding sites.
Initially, 12 miRNAs were predicted to interact with circNRIP1 and PKM2. However, qRT‐PCR found that miR‐541‐5p, miR‐3064‐5p, and miR‐3140‐3p could be candidate miRNAs. Western blotting indicated that only miR‐541‐5p and miR‐3064‐5p could downregulate PKM2 levels. MicroRNA‐541‐5p is involved in the progression of hepatocellular carcinoma and intrahepatic cholangiocarcinoma, 61 , 62 whereas miR‐3064‐5p is involved in the development of colorectal cancer and cervical cancer. 63 , 64 Therefore, miR‐541‐5p and miR‐3064‐5p might also be involved in thyroid cancer progression, and PKM2 is a common target gene for both miR‐541‐5p and miR‐3064‐5p.
In this study, the expression of miR‐541‐5p and miR‐3064‐5p was significantly lower in 98 pairs of PTC tissues than in normal tissues. Furthermore, circNRIP1 levels were negatively correlated with miR‐541‐5p and miR‐3064‐5p levels. Functional assays showed that miR‐541‐5p and miR‐3064‐5p can promote PTC cell proliferation and glycolysis. In addition, an in vivo and in vitro rescue strategy was used to confirm whether both miR‐541‐5p and miR‐3064‐5p can antagonize the proliferative and glycolytic effects of circNRIP1 on PTC cells. Dual‐luciferase reporter assays were also undertaken to confirm whether circNRIP1 contains binding sites for miR‐541‐5p and miR‐3064‐5p. Furthermore, miR‐541‐5p and miR‐3064‐5p were found to be important miRNAs that bind circNRIP1 and PKM2 3′‐UTR. These findings reveal that circNRIP1 promotes glycolysis in PTC cells through the miR‐541‐5p/PKM2 and miR‐3064‐5p/PKM2 axes.
m6A is a new epigenetic regulatory layer that regulates cell growth, differentiation, and self‐renewal by controlling RNA splicing, translation, and stability. 65 , 66 , 67 m6A is also an abundant cotranscriptional modifier of mRNA 68 , 69 and is involved in many aspects of posttranscriptional mRNA metabolism. 70 , 71 , 72 Recent studies have indicated that m6A is involved in modification of ncRNAs, including circRNAs. 73 For instance, m6A modification of circHPS5 promotes hepatocellular carcinoma progression through HMGA2 expression. 74 m6A‐modified circRNA‐SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β‐catenin signaling. 75 m6A modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through the caspase1 pathway. 76 However, the relationship between circRNAs and m6A modification in PTC is unknown.
These results indicate that circNRIP1 can promote thyroid cancer progression and possesses an m6A site. Therefore, m6A regulates the biological behavior of PTC cells by rationally inducing circNRIP1 demethylation. To the best of our knowledge, this is the first study to demonstrate that circNRIP1 is a direct downstream target of ALKBH5‐mediated m6A modification, revealing the mechanisms by which ALKBH5 can manipulate circNRIP1 and regulate cell proliferation, as well as glycolytic capacity. Among the many key m6A genes found in TPC1 and BCPAP cells, only ALKBH5 was downregulated in PTC, while circNRIP1 was significantly changed compared with the normal thyroid tissue. ALKBH5 also inhibited PTC cell proliferation and glycolysis, suggesting its inhibitory role in PTC tumorigenesis. In addition, ALKBH5 levels were negatively correlated with circNRIP1. The MeRIP assays showed that circNRIP1 has an m6A site, whereas luciferase assays indicated that ALKBH5 can modify circNRIP1 expression, thereby affecting tumor growth and glycolytic functions of PTC.
In conclusion, downregulation of ALKBH5 activates circNRIP1‐mediated glycolytic functions of PTC cells by sponging oncogenic miR‐541‐5p and miR‐3064‐5p as well as by upregulating PKM2 levels. Therefore, circNRIP1 is a promising biomarker and therapeutic target for PTC.
AUTHOR CONTRIBUTIONS
XJ, HZ, and WS designed this study, performed the statistical analysis, and drafted the manuscript. XJ, WS, JH, WD, and CL carried out the experiments. JH and CL analyzed the data. XJ and WS provided clinical samples as well as clinical information, and JH and CL supervised the study. All authors read and gave final approval of the manuscript.
FUNDING INFORMATION
National Natural Science Foundation of China (81902726), the Natural Science Foundation of Education Bureau of Liaoning Province (QNZR2020002), Natural Science Foundation of Education Bureau of Liaoning Province (QNZR2020009), the Natural Science Foundation of Liaoning Province (grant numbers 2020‐MS‐143, 2020‐MS‐186, and 2021‐MS‐193), and the Science and Technology Project of Shenyang City (21–173–9‐31).
ETHICS STATEMENT
Approval of the research protocol by an institutional review board: The research was approved by the Ethics Committee of the First Hospital of China Medical University.
Informed consent: N/A.
Registry and registration no. of the study/trial: N/A.
Animal studies: All animal studies were conducted in accordance with the principles and procedures outlined in the guidelines of the Institutional Animal Care and Use Committee (IACUC) of China Medical University. (IACUC approval number: KT2020136).
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Table S1.
Table S2‐S4.
ACKNOWLEDGMENTS
We thank the Home for Researchers editorial team for language editing service and the Department of Endocrinology and Metabolism, Institute of Endocrinology, NHC Key Laboratory of Diagnosis and Treatment of Thyroid Diseases, The First Affiliated Hospital of China Medical University, Shenyang, China.
Ji X, Lv C, Huang J, Dong W, Sun W, Zhang H. ALKBH5‐induced circular RNA NRIP1 promotes glycolysis in thyroid cancer cells by targeting PKM2 . Cancer Sci. 2023;114:2318‐2334. doi: 10.1111/cas.15772
Contributor Information
Wei Sun, Email: sun19890208@126.com.
Hao Zhang, Email: haozhang@cmu.edu.cn.
REFERENCES
- 1. Zhu X, Yao J, Tian W. Microarray technology to investigate genes associated with papillary thyroid carcinoma. Mol Med Rep. 2015;11(5):3729‐3733. [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 4. Brose MS, Cabanillas ME, Cohen EE, et al. Vemurafenib in patients with BRAF(V600E)‐positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: a non‐randomised, multicentre, open‐label, phase 2 trial. Lancet Oncol. 2016;17(9):1272‐1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mazzaferri EL, Kloos RT. Clinical review 128: current approaches to primary therapy for papillary and follicular thyroid cancer. J Clin Endocrinol Metab. 2001;86(4):1447‐1463. [DOI] [PubMed] [Google Scholar]
- 6. Shaha AR. Recurrent differentiated thyroid cancer. Endocr Pract. 2012;18(4):600‐603. [DOI] [PubMed] [Google Scholar]
- 7. Haddad RI, Nasr C, Bischoff L, et al. NCCN guidelines insights: thyroid carcinoma, version 2.2018. J Natl Compr Canc Netw. 2018;16(12):1429‐1440. [DOI] [PubMed] [Google Scholar]
- 8. Llamas‐Olier AE, Cuéllar DI, Buitrago G. Intermediate‐risk papillary thyroid cancer: risk factors for early recurrence in patients with excellent response to initial therapy. Thyroid. 2018;28(10):1311‐1317. [DOI] [PubMed] [Google Scholar]
- 9. Fröhlich E, Wahl R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat Rev. 2014;40(5):665‐674. [DOI] [PubMed] [Google Scholar]
- 10. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis‐splicing yields circular RNA molecules. FASEB J. 1993;7(1):155‐160. [DOI] [PubMed] [Google Scholar]
- 11. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLOS One. 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L, Zhou Q, Qiu Q, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR‐9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. 2019;18(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guanqun H, Min L, Haiyan L, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR‐187‐3p/RTKN2 axis and activating Wnt/β‐catenin pathway. Cell Death Dis. 2020;11(12):1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Zhao X, Wang Y, et al. circRNA‐002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang J, Qi M, Fei X, Wang X, Wang K. Hsa_circRNA_0088036 acts as a ceRNA to promote bladder cancer progression by sponging miR‐140‐3p. Cell Death Dis. 2022;13(4):322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stacpoole PW. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) Axis in cancer. J Natl Cancer Inst. 2017;109(11):djx071. [DOI] [PubMed] [Google Scholar]
- 19. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309‐314. [DOI] [PubMed] [Google Scholar]
- 20. Warburg O, Wind F, Negelein E. The metabolism of tumors IN the body. J Gen Physiol. 1927;8(6):519‐530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102(13):932‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teoh ST, Lunt SY. Metabolism in cancer metastasis: bioenergetics, biosynthesis, and beyond. Wiley Interdiscip Rev Syst Biol Med. 2018;10(2):e1406. [DOI] [PubMed] [Google Scholar]
- 23. Li Q, Chen P, Zeng Z, et al. Yeast two‐hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37(9):12503‐12512. [DOI] [PubMed] [Google Scholar]
- 24. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR‐338‐3p/PKM2 Axis under hypoxic stress. Hepatology. 2019;70(4):1298‐1316. [DOI] [PubMed] [Google Scholar]
- 25. Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt‐dependent regulation of the E‐cadherin repressor snail. J Biol Chem. 2005;280(12):11740‐11748. [DOI] [PubMed] [Google Scholar]
- 26. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465‐476. [DOI] [PubMed] [Google Scholar]
- 27. Beemon K, Keith J. Localization of N6‐methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977;113(1):165‐179. [DOI] [PubMed] [Google Scholar]
- 28. Chen‐Kiang S, Nevins JR, Darnell JJ. N‐6‐methyl‐adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J Mol Biol. 1979;135(3):733‐752. [DOI] [PubMed] [Google Scholar]
- 29. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15(5):293‐306. [DOI] [PubMed] [Google Scholar]
- 30. Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m(6)a modification. Trends Genet. 2020;36(3):177‐188. [DOI] [PubMed] [Google Scholar]
- 31. Tang B, Yang Y, Kang M, et al. M(6)a demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF‐1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yue B, Song C, Yang L, et al. METTL3‐mediated N6‐methyladenosine modification is critical for epithelial‐mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen M, Wei L, Law CT, et al. RNA N6‐methyladenosine methyltransferase‐like 3 promotes liver cancer progression through YTHDF2‐dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254‐2270. [DOI] [PubMed] [Google Scholar]
- 34. Cai X, Wang X, Cao C, et al. HBXIP‐elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let‐7g. Cancer Lett. 2018;415:11‐19. [DOI] [PubMed] [Google Scholar]
- 35. Chen Y, Ling Z, Cai X, et al. Activation of YAP1 by N6‐Methyladenosine‐modified circCPSF6 drives malignancy in hepatocellular carcinoma. Cancer Res. 2022;82(4):599‐614. [DOI] [PubMed] [Google Scholar]
- 36. Du A, Li S, Zhou Y, et al. M6A‐mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. 2022;21(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan HN, Chen ZY, Chen XY, et al. METTL14‐mediated m(6)a modification of circORC5 suppresses gastric cancer progression by regulating miR‐30c‐2‐3p/AKT1S1 axis. Mol Cancer. 2022;21(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ashwal‐Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell. 2014;56(1):55‐66. [DOI] [PubMed] [Google Scholar]
- 39. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell‐type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333‐338. [DOI] [PubMed] [Google Scholar]
- 41. Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang F, Fang E, Mei H, et al. Cis‐acting circ‐CTNNB1 promotes β‐catenin signaling and cancer progression via DDX3‐mediated transactivation of YY1. Cancer Res. 2019;79(3):557‐571. [DOI] [PubMed] [Google Scholar]
- 43. Yang Z, Qu CB, Zhang Y, et al. Dysregulation of p53‐RBM25‐mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L‐miR‐193a‐5p‐Pcdha pathway. Oncogene. 2019;38(14):2516‐2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang R, Zhang S, Chen X, et al. EIF4A3‐induced circular RNA MMP9 (circMMP9) acts as a sponge of miR‐124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Ma N, Zhang Y, et al. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR‐629‐3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis. 2020;11(5):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang E, Fu J, Yu Q, et al. CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR‐339‐5p/CDC25A axis. EPIGENOMICS‐UK. 2020;12(7):587‐603. [DOI] [PubMed] [Google Scholar]
- 47. Xie R, Tang J, Zhu X, Jiang H. Silencing of hsa_circ_0004771 inhibits proliferation and induces apoptosis in breast cancer through activation of miR‐653 by targeting ZEB2 signaling pathway. Biosci Rep. 2019;39(5):BSR20181919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891‐899. [DOI] [PubMed] [Google Scholar]
- 50. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646‐674. [DOI] [PubMed] [Google Scholar]
- 51. Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356(2 Pt A):184‐191. [DOI] [PubMed] [Google Scholar]
- 52. Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. 2012;37(8):309‐316. [DOI] [PubMed] [Google Scholar]
- 53. Wang X, Zhang H, Yang H, et al. Exosome‐delivered circRNA promotes glycolysis to induce chemoresistance through the miR‐122‐PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao X, Tian Z, Liu L. circATP2B1 promotes aerobic glycolysis in gastric cancer cells through regulation of the miR‐326 gene cluster. Front Oncol. 2021;11:628624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27(3):471‐481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351‐379. [DOI] [PubMed] [Google Scholar]
- 57. Zhang W, Liu T, Li T, Zhao X. Hsa_circRNA_102002 facilitates metastasis of papillary thyroid cancer through regulating miR‐488‐3p/HAS2 axis. Cancer Gene Ther. 2021;28(3–4):279‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gong J, Kong X, Qi J, Lu J, Yuan S, Wu M. CircRNA_104565 promoted cell proliferation in papillary thyroid carcinoma by sponging miR‐134. Int J Gen Med. 2021;14:179‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lou W, Ding B, Wang J, Xu Y. The involvement of the hsa_circ_0088494‐miR‐876‐3p‐CTNNB1/CCND1 Axis in carcinogenesis and progression of papillary thyroid carcinoma. Front Cell Dev Biol. 2020;8:605940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang M, Chen B, Ru Z, Cong L. CircRNA circ‐ITCH suppresses papillary thyroid cancer progression through miR‐22‐3p/CBL/β‐catenin pathway. Biochem Biophys Res Commun. 2018;504(1):283‐288. [DOI] [PubMed] [Google Scholar]
- 61. Li D, Zhang J, Yang J, et al. CircMTO1 suppresses hepatocellular carcinoma progression via the miR‐541‐5p/ZIC1 axis by regulating Wnt/β‐catenin signaling pathway and epithelial‐to‐mesenchymal transition. Cell Death Dis. 2021;13(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tang J, Wang R, Tang R, Gu P, Han J, Huang W. CircRTN4IP1 regulates the malignant progression of intrahepatic cholangiocarcinoma by sponging miR‐541‐5p to induce HIF1A production. Pathol Res Pract. 2022;230:153732. [DOI] [PubMed] [Google Scholar]
- 63. Wei M, Chen Y, Du W. LncRNA LINC00858 enhances cervical cancer cell growth through miR‐3064‐5p/ VMA21 axis. Cancer Biomark. 2021;32(4):479‐489. [DOI] [PubMed] [Google Scholar]
- 64. Luo Z, Hao S, Yuan J, et al. Long non‐coding RNA LINC00958 promotes colorectal cancer progression by enhancing the expression of LEM domain containing 1 via microRNA miR‐3064‐5p. Bioengineered. 2021;12(1):8100‐8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dai D, Wang H, Zhu L, Jin H, Wang X. N6‐methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Geula S, Moshitch‐Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002‐1006. [DOI] [PubMed] [Google Scholar]
- 67. Du Y, Hou G, Zhang H, et al. SUMOylation of the m6A‐RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46(10):5195‐5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352(6292):1408‐1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127‐150. [DOI] [PubMed] [Google Scholar]
- 70. Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)a reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61(4):507‐519. [DOI] [PubMed] [Google Scholar]
- 71. Wang X, Lu Z, Gomez A, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang X, Zhao BS, Roundtree IA, et al. N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhou C, Molinie B, Daneshvar K, et al. Genome‐wide maps of m6A circRNAs identify widespread and cell‐type‐specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20(9):2262‐2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rong D, Wu F, Lu C, et al. m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids. 2021;26:637‐648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu J, Wan Z, Tang M, et al. N(6)‐methyladenosine‐modified CircRNA‐SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β‐catenin signaling. Mol Cancer. 2020;19(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu P, Fang X, Liu Y, et al. N6‐methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12(4):298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Table S1.
Table S2‐S4.


