Abstract
We report nine severe neonatal infections caused by a new variant of echovirus 11. All were male, eight were twins. At illness onset, they were 3–5 days-old and had severe sepsis and liver failure. This new variant, detected in France since April 2022, is still circulating and has caused more fatal neonatal enterovirus infections in 2022 and 2023 (8/496; 1.6%, seven associated with echovirus 11) compared with 2016 to 2021 (7/1,774; 0.4%). National and international alerts are warranted.
Keywords: Enterovirus, Echovirus-11, neonatal sepsis, liver failure
Enteroviruses (EV) are a common cause of neonatal infections with clinical manifestations ranging from asymptomatic to severe and sometimes fatal disease [1–3]. Between July 2022 and April 2023, nine cases of severe neonatal infection with a liver failure were reported in France. Seven of these children died. All were associated with a new variant of echovirus 11 (E-11). We describe the clinical and virological characteristics of this upsurge of highly severe neonatal EV infections.
Description of the cases
Between July 2022 and April 2023, nine children from three French metropolitan regions were hospitalised in paediatric intensive care units for suspected neonatal sepsis. A description of each case is available in Supplement 1. All patients were male. Only one was born at full term and was from a singleton pregnancy. The other eight patients were from twin pregnancies. They were born between 31 weeks and 5 days and 36 weeks and 3 days of gestation and were initially hospitalised in neonatology departments before developing the first symptoms (Table 1).
Table 1. Clinical characteristics of enterovirus-associated severe neonatal infections of male neonates with acute liver failure and multivisceral failure, France, 2022–2023 (n = 9).
| Patient number | Gestation length (weeks + days) | Date of birth | Mode of delivery | Birth weight (g) | Age at first symptoms (days) | Specimen type | Age at diagnosis of EV infection (days) | Maternal EV infection | Maternal EV infection (days from delivery) | Treatment | Dialysis | Age at death (days) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 39 + 0 | Jan 2023 | Vaginal | 3,480 | 5 | CSF | 5 | Yes | 0 | Pocapavir D7-D21 IVIg D5 | Yes | 34 |
| 2 | 35 + 4 | Dec 2022 | Caesarian | 2,400 | 5 | Blood | 5 | Yes | -3b | IVIg D7, D8 | Yes | 19 |
| 3 | 2,650 | Blood | IVIg D7, D8 | Yes | 40 | |||||||
| 4 | 31 + 5 | Oct 2022 | Vaginal | 1,805 | 5 | Blood | 6 | NA | NA | No | No | 7 |
| 5 | 2,320 | Post-mortem biopsies | No | No | 6 | |||||||
| 6 | 36 + 3 | Jul 2022 | Vaginal | 2,600 | 3 | CSF | 3 | Yes | -3b | IVIg D4 | No | 5 |
| 7 | 2,860 | CSF | IVIg D3, D4 | No | 5 | |||||||
| 8 | 34 + 0 | Mar 2023 | Vaginal | 2,375 | 4 | CSF | 8 | Yes | -1b | Pocapavir D8-D23 IVIg D18, 19 | No | NA |
| 9 | 1,970 | CSF | Pocapavir D8-D23 IVIg D18, 19 | No | NA |
CSF: cerebrospinal fluid; IVIg: polyvalent intravenous immunoglobulins; NA: not applicable.
a From a singleton pregnancy.
b Days before delivery.
In all patients, the first symptoms appeared between the third and fifth day of life. The initial symptoms were fever and apnoea, which were rapidly followed by signs of septic shock requiring active resuscitation including administration of amines, with the exception of Patients 8 and 9, who responded to vascular filling alone. All cases developed acute hepatocellular failure with severe cytolysis and disseminated intravascular coagulation as early as the first day of hospitalisation. Blood coagulation factor V, II, VII, X, Quick factor, and fibrinogen were undetectable in all patients. The patients had hyperammonaemia: five were treated with sodium benzoate and three with haemodialysis and one died before treatment. All patients had thrombocytopaenia, and seven patients required transfusions of platelets, plasma, and tranexamic acid to control the haemorrhagic syndrome. All cases had acute renal failure at the onset of the symptoms, one had myocarditis, three were diagnosed with meningoencephalitis and two had enterocolitis. Transfontanellar ultrasound was performed for all patients: five of them had normal findings, two had multiple hyperechogenic lesions in the white matter, confirmed by cerebral magnetic resonance imaging (MRI), and two had bilateral intraventricular haemorrhage, grade III.
Bacteriological and virological investigations
Bacterial blood cultures were negative in seven patients within 48 h of the onset of the symptoms. Blood culture was positive for Escherichia coli in one neonate and for Staphylococcus epidermidis in another. The EV genome was detected in all available specimens, including blood, cerebrospinal fluid, throat, nasopharyngeal or rectal swabs, stool, post-mortem biopsies or dried blood spots (Table 2). We identified E-11 as the causative agent in all cases either by next generation sequencing of (sub)-complete genomes using direct RNA sequencing or amplicon sequencing adapted from the previously described method in Duval et al. [4]), or by Sanger sequencing of the 1D gene coding the VP1 capsid protein [5,6].
Table 2. Virological data on the echovirus-11-infected severe cases and their mothers, France, 2022–2023.
| Patient | Sample type | Age of the child at sampling (days) | Sequence | Accession number |
|---|---|---|---|---|
| 1 | Dried blood spot | 3 | Not typeable | NA |
| CSF | 5 | Complete 1DVP1 | OQ927993 | |
| Stool | 6 | Complete genome | OQ927998 | |
| Plasma | 6 | Complete genome | OQ923264 | |
| Mother 1 | Serum | 0 | Complete 1DVP1 | OQ927994 |
| Milk | 9 | Partial 1DVP1 | Not deposited | |
| 2 | Dried blood spot | 3 | Partial 1DVP1 | Not deposited |
| Plasma | 5 | Complete genome | OQ927999 | |
| 3 | Dried blood spot | 3 | Partial 1DVP1 | Not deposited |
| Plasma | 5 | Complete genome | OQ928000 | |
| Mother 2–3 | Serum | D-3a | Complete 1DVP1 | OQ927995 |
| 4 | Dried blood spot | 3 | Partial 1DVP1 | Not deposited |
| Psoas muscle biopsy | 6 | Complete 1DVP1 | OQ927996 | |
| Liver biopsy | 6 | Complete genome | OQ928001 | |
| Lung biopsy | 6 | Complete genome | OQ928002 | |
| 5 | Dried blood spot | 2 | Partial 1DVP1 | Not deposited |
| Blood | 6 | Complete genome | OQ927567 | |
| 6 | CSF | 3 | Complete genome | OQ928003 |
| 7 | CSF | 3 | Complete genome | OQ928004 |
| Mother 6–7 | Serum | 1 | Complete 1DVP1 | OQ927997 |
| 8 | Dried blood spot | 3 | Partial 1DVP1 | Not deposited |
| NP swab | 8 | Partial 1DVP1 | Not deposited | |
| Throat swab | 8 | Complete genome | OQ969164 | |
| Rectal swab | 8 | Complete genome | Not deposited (identical to OQ969164) | |
| Plasma | 10 | Complete 1DVP1 | Not deposited | |
| 9 | Dried blood spot | 3 | Partial 1DVP1 | Not deposited |
| NP swab | 8 | Partial 1DVP1 | Not deposited | |
| Throat swab | 8 | Complete genome | OQ969165 | |
| Rectal swab | 8 | Complete genome | Not deposited (identical to OQ969165) | |
| Plasma | 10 | Complete 1DVP1 | Not deposited | |
| Mother 8–9 | Serum | D-1a | Complete 1DVP1 | OQ971926 |
CSF: cerebrospinal fluid; D: delivery; ND: not determined; NP: nasopharyngeal; VP1: capsid protein; 1D: gene coding for VP1 capsid protein.
a Samples collected before delivery.
Maternal blood samples collected during the peripartum period were available for four of the five mothers with gastrointestinal symptoms or fever during the 3 days before or at delivery. All were EV-positive with subsequent identification of an E-11.
All patients received empiric antibiotic therapy at the onset of clinical symptoms. Treatment for EV infection included polyvalent intravenous immunoglobulins (IVIg) in seven patients and pocapavir for 14 days in three patients (Table 1). Seven of the nine patients died: four without a dialysis between 5 and 7 days of life and three between 19 and 40 days of life. A pair of twins (Patients 8 and 9) survived with no evidence of sequelae at the age corrected to term birth.
Molecular characterisation of a new E-11 variant
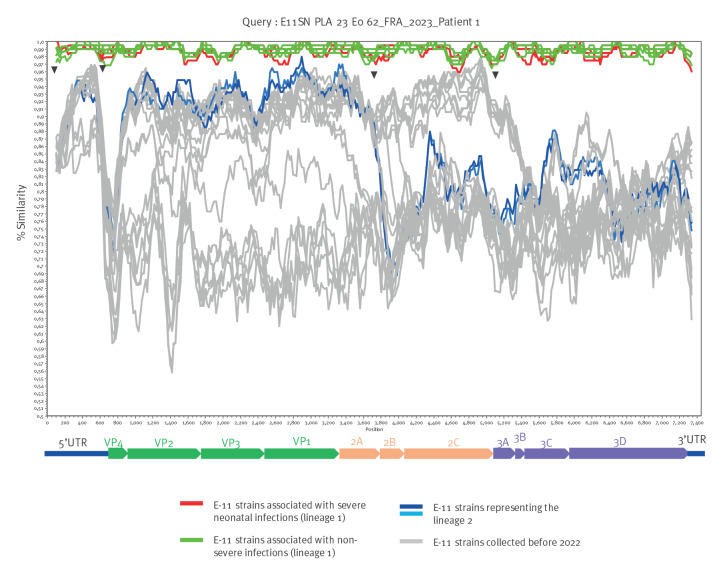
Phylogenetic analyses were performed on complete 1DVP1 E-11 sequences determined from specimens collected in France between 2010 and 2023 from patients (n = 104) in the five hospitals from where the severe neonatal cases were reported, as well as sequences from other countries for which complete genome sequences were available from GenBank (n = 37). E-11 strains detected in 2022 and 2023 fell into two separate lineages, distinct from the E-11 strains isolated before 2022 (Figure 1). The lineage 1 included all E-11 sequences associated with severe neonatal infections as well as sequences from non-severe neonatal and non-neonatal infections. The remaining French sequences from 2022 were grouped in lineage 2 and were related to strains isolated in China in 2018 and 2019. Similarity plot analysis of the E-11 complete genome sequences suggested a recombinant origin for both lineages. The lineage 1 representing genomes appears to be a mosaic defined by distinct similarity patterns characterised by sharp decrease in nt similarity within the 5’-untranslated region and in the 3A coding region, possibly arising through recombination (Figure 2) compared with E-11 strains collected before 2022. They also differed from lineage 2 complete genomes whose similarity plots are characterised by a recombination signal in the 2A coding region.
Figure 1.
Phylogenetic tree of echovirus 11 complete 1DVP1 sequences from neonatal and non-neonatal infections in France and other countries, 2010–2023 (n = 142)
MEGA: molecular evolutionary genetics analysis.
The phylogenetic tree was constructed by the neighbour joining method and evaluated with 1,000 bootstrap pseudoreplicates, using MEGA6. Only bootstrap values > 70% are indicated. Genetic distances were calculated with Tamura-Nei’s model of evolution and branch length is drawn to the indicated scale (proportion of nt substitution per site). The strains collected in patients with severe neonatal infection are labelled with a filled triangle. Strains collected in France (n = 104) between 2010 and 2023, accession numbers OQ927567, OQ923264, OQ927993–927997, OQ927998–928004, OQ969158-OQ969177, OQ971926-OQ971949; OR029978–030028) and in other countries (n = 37) (selected sequences among complete E-11 genomes available in GenBank, as of 28 April 2023) are labelled with a filled circle or empty circle, respectively. Year of isolation is colour coded. For clarity, the taxon names are not indicated in the tree. See Supplement 2 for taxon names. The French sequences from 2010 to 2023 were detected from samples from hospitalised patients in Orléans, Lyon, Clermont-Ferrand, Rouen, Cochin-Port Royal (Paris) and Necker-Enfants Malades (Paris) Hospitals, where the severe cases were reported.
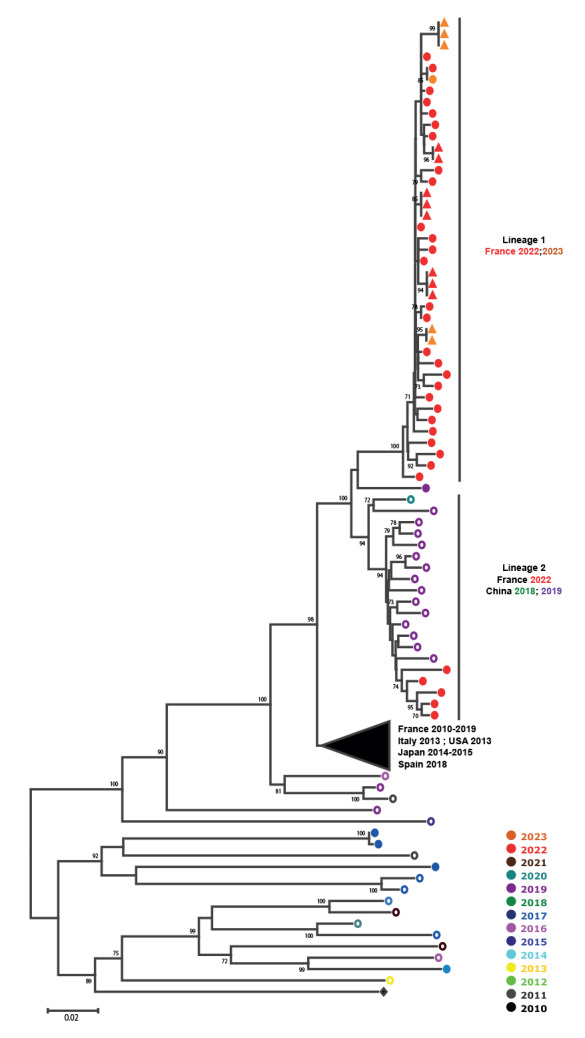
Figure 2.
Analysis of nt similarity of echovirus 11 complete genome sequences from neonatal and non-neonatal infections, France and other countries, 2010–2023 (n = 38)
UTR: Untranslated region.
Complete genome sequences of the French E-11 strains of 2022–2023 (n = 26) were compared with each other and with other E-11 strains (11 from other countries and the prototype strain X.80059) using SimPlot 3.5.1. A schematic diagram of the enterovirus genome is shown at the bottom of the panel. Recent E-11 genomes, including those of the strains associated with severe neonatal infection or non-severe infection within the lineage 1 and the other French strains from 2022 to 2023 (lineage 2) are represented in red, green and blue, respectively. The genomes of strains collected before 2022 are represented in grey. Potential recombination breakpoints are indicated by black arrows.
National surveillance data
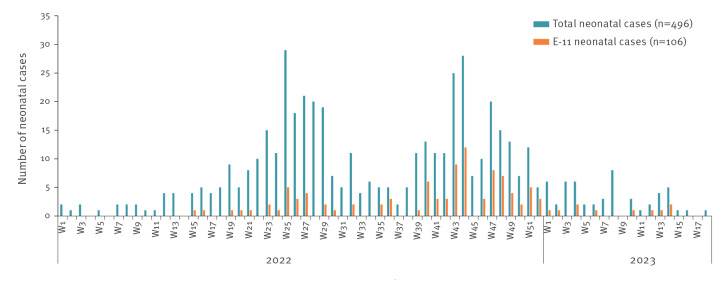
The hospital-based EV surveillance in France is a voluntary system involving 36 virology or microbiology laboratories, covering approximatively 100 hospitals [7] distributed in all French regions, except Corsica, coordinated by the National Public Health Agency, Santé publique France, and the two National Reference Laboratories (NRLs). Participants report to the NRLs the number and type of samples analysed for EV and the relevant epidemiological and standardised clinical data of the EV-positive cases [8]. EV-positive samples are sent to the NRL laboratories for genotyping, whenever possible. Between January 2022 and April 2023, 2,026 EV cases were reported, among which neonatal cases represented 24.5% of the cases. Echovirus-11 was the most predominant EV type identified among neonates (31.1%, 106 of 341) and non-neonates (15.3%, 158 of 1,035). The epidemic curve (Figure 3) shows that E-11 has been continuously detected since April 2022.
Figure 3.
Distribution of neonatal cases by week, France, January 2022–April 2023 (n = 496)
In 2022–23, 28 (5.6%) of the 496 neonatal cases presented with severe EV infection (defined as at least one organ failure requiring intensive care support). We identified E-11 in 14 of the 26 cases with a known type, among which nine had severe sepsis with liver failure and five had apnoea requiring high-flow oxygen therapy. By comparison, from 2016 to 2021, 62 (3.5%) severe neonatal infections were reported, and E-11 was identified in 6.2% of the cases with a known type (3/48). Moreover, a higher mortality was observed in 2022 and 2023 among the EV neonatal cases: eight (1.6%) of the 496 children died, all but one associated with E-11 compared with seven (0.4%) of 1,774 in 2016–2021, none associated with E-11.
Clinical presentation of other E-11 infections in children did not seem to have changed during this period. Finally, male children were overrepresented among the severe E-11 neonatal cases (12/14) and among the severe EV neonatal cases (20/28), whereas the proportion of males among all EV neonatal cases was 56.3% (276/490).
Discussion
We report a cluster of severe neonatal cases with liver failure and a high mortality rate associated with a new variant of E-11, leading to an alert in the European Union Early Warning and Response System on 4 May 2023, and inclusion in the ECDC Communicable Disease Threat Report [9]. Enterovirus infections in neonates may be associated with severe clinical illness and mortality depending on (i) the infecting EV type, of which the group B coxsackieviruses and E-11 are the most commonly EVs associated with neonatal sepsis [10] and (ii) the clinical infection phenotype, where the mortality rate is the highest for severe hepatitis or myocarditis manifestations [11].
Fulminant hepatitis associated with E-11 has already been described, but the high fatality rate within a nine-month period and the high proportion of twin cases were striking in this cluster. All patients presented with one or more well-known risk factors for severe neonatal EV infection, which are maternal evidence of a recent EV infection (four of the five mothers in this report), prematurity and onset of illness within the first few days of life [1,11].
The emergence of a new E-11 variant of recombinant origin in 2022, after a period of a low-level circulation of this type since 2008 may have led to a higher proportion of women susceptible to E-11 infection and subsequently to the higher proportion of E-11 associated neonatal infections observed in 2022–2023. Recombination, which frequently occurs in enteroviruses, is considered a factor driving viral emergence. Whether the genetic changes of the predominant circulating E-11 lineage have influenced the pathogenicity needs further investigation. Host genetic factors affecting immunity might also have influenced the clinical severity observed in the nine cases (by comparison with other patients infected with the same E-11 variant during their first week of life) as previously described in EV rhombencephalitis cases [12]. High prevalence of boys in severe neonatal EV infection might speak for a predisposition associated with X-chromosome.
Therapeutic options for neonatal EV infections are limited and include intravenous immunoglobulins or investigational/compassionated specific antiviral therapy consisting of pleconaril or pocapavir [13–15]. We used pocapavir for three patients, of whom two survived, but the efficacy of the treatment cannot be concluded from our data.
Conclusion
This report suggests that a new variant of E-11 is currently circulating and associated with a high risk of severe neonatal infection and death at least in France. Clinicians should be aware of potential involvement of EV in severe clinical presentations in neonates, as they are at the frontline to detect such cases. Enterovirus surveillance in France, as in most European country, is a voluntary system reporting EV-positive cases which could lead to an underestimation of the number of severe cases if EV genome detection is not considered. As a reminder, symptomatic neonatal EV disease initially presents as a neonatal sepsis, which is clinically indistinguishable from bacterial or herpes simplex virus infections. Neonates with an unexplained sepsis who present with signs of myocarditis or liver failure with cytolysis should be rapidly evaluated for EV infection, especially if the mother has had acute symptoms of gastroenteritis in the days before birth. Blood, as well as respiratory, cerebrospinal fluid and stool samples should be collected for initial testing and further sequencing. Therapeutic options as IVIg or pocapavir should be considered.
Ethical statement
Ethical approval was not needed for this retrospective study because the study was part of routine management and treatments for children. Nevertheless, parents of children have been informed and consented to this report.
Funding statement
No funding was required for this study. The Centre National de Référence des Enterovirus-Parechovirus is supported by an annual grant from the French national public health network (Santé publique France).
Data availability
Data of this study is available in the manuscript and in the Supplements.
Acknowledgements
We thank parents of the children who gave their consent for this publication, and the caregivers who looked after the children.
We thank Dr Robert Ratiney (Necker-Enfants malades Hospital) and Jeffrey Hincks (President at ViroDefense Inc) to allow us to obtain pocapavir to treat some of the children.
We thank Adeline Duard and Laetitia Planche from Clermont-Ferrand NRL for excellent technical assistance for EV genome detection and genotyping. We warmly thank Delphine Falcon, Emmanuelle Groscarret and Laure Zanghellini from Lyon NRL for EV detection and routine typing and Thibault Corsin and Quentin Semanas for complete genome NGS sequencing.
We are grateful to all members of the EV surveillance network in Amiens (Dr Marie Louchet Ducoroy), Angers (Dr Caroline Lefeuvre, Pr Alexandra Ducancelle), Bayonne (Drs David Leyssene and Anne-Christine Jaouen), Besançon (Pr Quentin Lepiller), Bordeaux (Prs Marie-Edith Lafon and Sonia Burrel), Bourgoin-Jallieu (Drs Charlotte Tellini), Brest (Dr Léa Pilorgé and Pr Christopher Payan), Caen (Dr Cécile Schanen, Pr Astrid Vabret), Dijon (Dr Katia Balay, Pr Alexis de Rougemont), Frejus (Dr Gillon), Grenoble (Pr Sylvie Larrat), Lille (Dr Mouna Lazrek, Pr Didier Hober), Limoges (Prs Sylvie Rogez and Sophie Alain), Mantes-La-Jolie (Dr Emeline Riverain), Marseille (Drs Antoine Nougairède and Laetitia Ninove), Montpellier (Dr Vincent Foulongne, Pr Philippe Van de Perre), Nancy (Dr Véronique Vénard, Pr Evelyne Schvoerer), Nantes (Dr Marianne Coste-Burel), Nice (Dr Gonfrier), Orléans (Dr Clémence Guillaume and Jerôme Guinard), Paris – Cochin (Dr Anne-Sophie L’honneur, Pr Véronique Avettand Fenoël), Paris – Necker (Drs Hanène Abid, Marianne Burgard, and Marianne Leruez-Ville), Paris – Trousseau (Drs Kenda Saloum and Aurélie Schnuriger), Poitiers (Dr Anne Bourgoin, Pr Nicolas Lévêque), Reims (Pr Laurent Andreoletti), Rennes (Dr Gisèle Lagathu, Pr Vincent Thibault), Roanne (Drs Jean-Benjamin Murat), Rouen (Dr Véronique Lémée, Pr Jean-Christophe Plantier), St Etienne (Dr Sylvie Pillet, Pr Thomas Bourlet), Strasbourg (Dr Floriane Gallais, Pr Samira Fafi-Kremer), Suresnes (Dr Eric Farfour), Toulouse (Drs Jean-Michel Mansuy and Pauline Trémeaux, Pr Jacques Izopet), Toulon-CHI (Drs Anne-Lise Toyer and Cécile Poggi), Tours (Dr Karl Stefic, Pr Catherine Gaudy), Versailles (Dr Stéphanie Marque-Juillet), Villefranche (Dr Marine Jourdain).
Supplementary Data
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: MG, DP, AB, MB1, BK, MK, LR, YV, SR, PHJ, ID, FL, AL and MA took care of the children/mothers during illness. MG collected clinical and biological data. ASLH, VL, CG and MLV analysed the clinical specimens and collected virological data of the children in each hospital. AM, MJ, CH and IS were involved in the analysis of the epidemiological and virological data of enterovirus infections reported by the EV surveillance network. AM, IS and MB2 were involved in the sequence analyses. MA coordinated the study group on severe neonatal enterovirus infections. MG, AM, CH, IS and MA wrote the first draft of the manuscript. All authors were involved in the study group on severe neonatal enterovirus infections, contributed to the manuscript and approved the final version.
References
- 1. Abzug MJ. Presentation, diagnosis, and management of enterovirus infections in neonates. Paediatr Drugs. 2004;6(1):1-10. 10.2165/00148581-200406010-00001 [DOI] [PubMed] [Google Scholar]
- 2. Sandoni M, Ciardo L, Tamburini C, Boncompagni A, Rossi C, Guidotti I, et al. Enteroviral Infections in the first three months of life. Pathogens. 2022;11(1):60. 10.3390/pathogens11010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chuang YY, Huang YC. Enteroviral infection in neonates. J Microbiol Immunol Infect. 2019;52(6):851-7. 10.1016/j.jmii.2019.08.018 [DOI] [PubMed] [Google Scholar]
- 4. Duval M, Mirand A, Lesens O, Bay JO, Caillaud D, Gallot D, et al. Retrospective study of the upsurge of enterovirus D68 clade D1 among adults (2014-2018). Viruses. 2021;13(8):1607. 10.3390/v13081607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mirand A, le Sage FV, Pereira B, Cohen R, Levy C, Archimbaud C, et al. Ambulatory pediatric surveillance of hand, foot and mouth disease as signal of an outbreak of coxsackievirus A6 infections, France, 2014-2015. Emerg Infect Dis. 2016;22(11):1884-93. 10.3201/eid2211.160590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44(8):2698-704. 10.1128/JCM.00542-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuffenecker I, Mirand A, Josset L, Henquell C, Hecquet D, Pilorgé L, et al. Epidemiological and clinical characteristics of patients infected with enterovirus D68, France, July to December 2014. Euro Surveill. 2016;21(19):30226. 10.2807/1560-7917.ES.2016.21.19.30226 [DOI] [PubMed] [Google Scholar]
- 8.French Reference Centre for enteroviruses and parechovirus. CNR enterovirus. Clermont-Ferrand-Lyon: France. [Accessed: 6 May 2023]. Available from: http://cnr.chu-clermontferrand.fr/CNR
- 9.European Centre for Disease Prevention and Control (ECDC). Weekly bulletin. Communicable Disease Threats Report. Week 18, 30 April–6 May 2023. Stockholm: ECDC; 2023. Available from: https://www.ecdc.europa.eu/en/publications-data/communicable-disease-threats-report-30-april-6-may-2023-week-18
- 10. Khetsuriani N, Lamonte A, Oberste MS, Pallansch M. Neonatal enterovirus infections reported to the national enterovirus surveillance system in the United States, 1983-2003. Pediatr Infect Dis J. 2006;25(10):889-93. 10.1097/01.inf.0000237798.07462.32 [DOI] [PubMed] [Google Scholar]
- 11. Zhang M, Wang H, Tang J, He Y, Xiong T, Li W, et al. Clinical characteristics of severe neonatal enterovirus infection: a systematic review. BMC Pediatr. 2021;21(1):127. 10.1186/s12887-021-02599-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J, Jing H, Martin-Nalda A, Bastard P, Rivière JG, Liu Z, et al. Inborn errors of TLR3- or MDA5-dependent type I IFN immunity in children with enterovirus rhombencephalitis. J Exp Med. 2021;218(12):e20211349. 10.1084/jem.20211349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yen MH, Huang YC, Chen MC, Liu CC, Chiu NC, Lien R, et al. Effect of intravenous immunoglobulin for neonates with severe enteroviral infections with emphasis on the timing of administration. J Clin Virol. 2015;64:92-6. 10.1016/j.jcv.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 14. Abzug MJ, Michaels MG, Wald E, Jacobs RF, Romero JR, Sánchez PJ, et al. A Randomized, double-blind, placebo-controlled trial of pleconaril for the treatment of neonates with enterovirus sepsis. J Pediatric Infect Dis Soc. 2016;5(1):53-62. 10.1093/jpids/piv015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Torres-Torres S, Myers AL, Klatte JM, Rhoden EE, Oberste MS, Collett MS, et al. First use of investigational antiviral drug pocapavir (v-073) for treating neonatal enteroviral sepsis. Pediatr Infect Dis J. 2015;34(1):52-4. 10.1097/INF.0000000000000497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.