Abstract
Research on the relationship between the microbiome and cancer has been controversial for centuries. Recent works have discovered that the intratumor microbiome is an important component of the tumor microenvironment (TME). Intratumor bacteria, the most studied intratumor microbiome, are mainly localized in tumor cells and immune cells. As the largest bacterial reservoir in human body, the gut microbiome may be one of the sources of the intratumor microbiome in gastrointestinal malignancies. An increasing number of studies have shown that the gut and intratumor microbiome play an important role in regulating the immune tone of tumors. Moreover, it has been recently proposed that the gut and intratumor microbiome can influence tumor progression by modulating host metabolism and the immune and immune tone of the TME, which is defined as the immuno-oncology-microbiome (IOM) axis. The proposal of the IOM axis provides a new target for the tumor microbiome and tumor immunity. This review aims to reveal the mechanism and progress of the gut and intratumor microbiome in gastrointestinal malignancies such as esophageal cancer, gastric cancer, liver cancer, colorectal cancer and pancreatic cancer by exploring the IOM axis. Providing new insights into the research related to gastrointestinal malignancies.
Keywords: Gut microbiome, Intratumor microbiome, Gastrointestinal malignancy, Tumor microenvironment, Immunity, Therapy
Core Tip: The gut and intratumor microbiome can influence tumor progression by modulating host metabolism and the immune and immune tone of the tumor microenvironment, which is defined as the immuno-oncology-microbiome (IOM) axis. The proposed the IOM axis provides a new target for tumor microbiome and tumor immunity. Current studies have shown that immunotherapy with fecal microbiota transplantation or microbial metabolism have certain effects. This review aims to explore the mechanism of the IOM axis of gastrointestinal malignancies, to reveal the mechanism and progress of gut and intratumor microbiome in gastrointestinal malignancies. Providing new insights into the research related to gastrointestinal malignancies.
INTRODUCTION
According to statistics, gastrointestinal tumors were one of the leading causes of death in the United States in 2020[1]. Improving the survival rate of patients with gastrointestinal tumors has always been an urgent task. In recent years, with the deepening of research on tumor immunity, various immune checkpoint inhibitors (ICIs) have shown great effects in clinical practice[2,3]. Among gastrointestinal tumors, there appears to be heterogeneity in the effect of immunotherapy across different tumor types[4,5]. Pembrolizumab improves the prognosis of patients with esophageal cancer (EC)[4]. However, the effect of niraparib plus nivolumab in pancreatic cancer was not satisfactory[5]. Recent studies have described that the gut microbiome can reprogram tumor microenvironment (TME) immunity by participating in innate and/or adaptive immunity[6]. Regulation of the microbiome can enhance the effectiveness of immunotherapy[7]. Of all tumors, gastrointestinal malignancies have received the most attention due to their large number of microbial residues in the gut[8]. Using microorganisms as one of the targets of immunotherapeutic effects seems to be an effective measure.
The human microbiome inhabits every surface and cavity of the body[9], including bacteria, archaea, eukaryotes, and viruses that colonize humans[10]. The microbiome affects overall immune function through many different mechanisms, resulting in a broad response from resistance to immune activation[10]. The gut microbiome has long been recognized as playing a major role in human health and disease[6], influencing host metabolism and shaping the immune system and disease conditions, including cancer[11]. The gut microbiota plays a key role in shaping the immune system[12]. The human gut microbiota can influence the development and progression of gastrointestinal tumors by disrupting DNA, activating oncogenic signaling pathways, producing protumor metabolites, and suppressing antitumor immune responses[13,14]. In recent years, with increasing research on the relationship between tumors and the microbiome, tumor tissues that were previously considered sterile are rich in microorganisms. After statistical analysis, the bacterial composition in tumor tissue is approximately 0.68%, equivalent to approximately 105 to 106 bacteria per 1 cm3 of tumor tissue[15,16]. Although the abundance of microorganisms is relatively lower for tumor genomes, this intratumor microbiome is a potentially important player in tumor progression[15]. The intratumor microbiome is mainly intracellular and present in cancer cells and immune cells[17]. The gut and intratumor microbiome can influence tumor progression by modulating host metabolism and the immune and immune tone of the TME, and these immune-mediated interactions and collective feedback loops have been defined as the he immuno-oncology-microbiome (IOM) axis[15]. The proposal of the IOM axis provides a new target for the tumor microbiome and tumor immunity. This review aims to reveal the mechanism and progress of the gut and intratumor microbiome and gastrointestinal malignancies by exploring the mechanism of the IOM axis in gastrointestinal malignancies. Providing new insights into the research related to gastrointestinal malignancies.
COLORECTAL CANCER
Colorectal cancer (CRC) is one of the most common cancers in humans, with a leading incidence rate[18]. The initiation of CRC is a heterogeneous process that is influenced by the environment, microbial exposure, and host immunity[9]. Interactions between CRC and the microbiome have been revealed in numerous studies, with increasing evidence highlighting the critical role of the TME in the initiation and progression of carcinogenesis. In this microenvironment, multiple relationships between tumor development, immune responses, and the microbiome have been identified. All stages of CRC are accompanied by an immune response[19]. The regulation of the tumor immune response by the microbiome plays an important role in the pathogenesis of CRC (Figure 1).
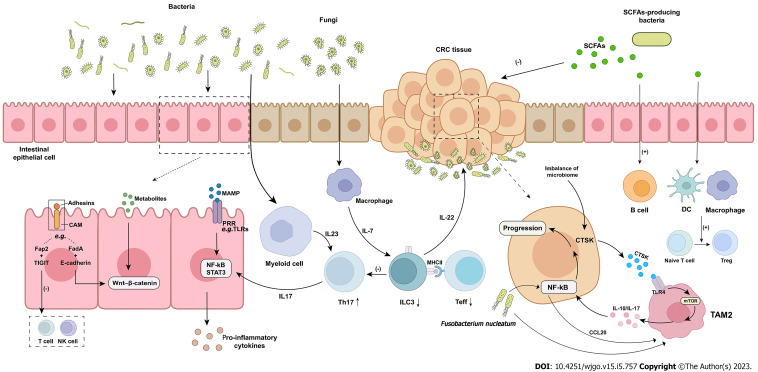
Figure 1.
The role of microbiome in the occurrence and development of colorectal cancer. Microbiome influences the occurrence and development of colorectal cancer through a variety of mechanisms. Microbiome and their metabolites can induce tumorigenesis through direct mutagenesis of intestinal epithelial cells or activation of intracellular carcinogenic signals. Bacterial metabolites can also trigger the release of pro-inflammatory signals, which further promote tumorigenesis. Pathogenic bacteria or their products activate tumor-associated myeloid cells and induce tumor-promoting inflammation. Symbiotic fungi activate the production of interleukin (IL)-7 in macrophages and induce the production of IL-22 in ILC3, leading to tumor progression. ILC3s inhibited Th17 cells and intestinal inflammation. ILC3s decreased significantly and Th17 increased in colorectal cancer. The dialogue between ILC3s, effector T cell and major histocompatibility complex class II is disrupted by colorectal cancer and intestinal inflammation, promoting the progress of colorectal cancer. Fusobacterium nucleatum upregulates the expression of chemokine (C-C motif) ligand 20 through nuclear factor-κB and induces M2 macrophages to polarize and promote tumor metastasis. Cathepsin K mediates tumor invasion and metastasis. Short chain fatty acids directly inhibits tumor cell growth and induces host macrophage, T and B cell responses to protect colitis-induced colorectal cancer. CRC: Colorectal cancer; CAM: Cell adhesion molecule; FadA: Fusobacterium adhesin A; TIGIT: T-cell immunoglobulin and ITIM domain; MAMP: Microbe-associated molecular pattern; PRR: Pattern recognition receptor; NF-κB: Nuclear factor-κB; STAT3: Signal transducer and activator of transcription 3; MHCII: Major histocompatibility complex class II; Teff: Effector T cell; CTSK: Cathepsin K; TLR4: Toll-like receptor 4; TAM: Tumor-associated macrophages; SCFA: Short chain fatty acid; DC: Dendritic cell; IL: Interleukin; NK: Natural killer; CCL20: Chemokine (C-C motif) ligand 20.
Tumor-infiltrating lymphocytes (TILs) are beneficial to the survival of human CRC[8,19]. One of the features of CRC is a strong imbalance of T cells[9,20]. One of the features of CRC is a strong imbalance of T cells[21]. Tosolini et al[22] analyzed T helper (Th) cell subsets in CRC and found that the expression of immune markers was different in adjacent mucosa and tumor tissue, suggesting that specific Th cell subsets were recruited at the tumor site. Experiments by Cremonesi et al[20] showed that infiltration of different T-cell subsets in CRC correlated with the expression of well-defined chemokine genes such as chemokine (C-C motif) ligand (CCL)3, CCL4, and CCL20. Exposure of tumor cells to gut bacteria induced upregulation of most chemokine genes by flow cytometry[20]. Upregulation of chemokines leads to higher T-cell recruitment into tumor xenografts. Therefore, whether CRC cells are exposed to intestinal bacteria and the degree of exposure may be one of the factors that affect the abundance of TILs. However, Fusobacterium nucleatum (F. nucleatum)[23-25], which is abundantly enriched within CRC tumors, can interact with the immune cell inhibitory receptor T-cell immunoglobulin (Ig) and ITIM domain expressed by TILs through the adhesin Fap2, inhibiting the activity of tumor-infiltrating T cells and protecting tumor cells from immune cell attack[26]. The effect of F. nucleatum on T cells may not be limited to this. In vitro experiments have shown that F. nucleatum inhibitory protein can block human T cells in the G1 phase of the cell cycle to prevent their proliferation[27]. F. nucleatum can also use the trimeric autotransporter adhesin CbpF on its surface, inhibiting T-cell function by activating the inhibitory receptor carcinoembryonic antigen cell adhesion molecule 1[28].
Th17 cells were found to be elevated to promote tumorigenesis[29]. Microbial metabolites penetrate tumors, activate tumor-associated bone marrow cells, and mediate interleukin (IL)-23 secretion. In turn, the IL-23-driven Th17 response can promote tumor growth. Enterotoxigenic Bacteroides fragilis, on the other hand, increases IL-17 expression by triggering a Th17-type inflammatory response, which shifts the colonic epithelium from an inflammatory to an oncogenic state[30]. The mechanism of bacterial induction of carcinogenesis can be explained by the hypothesis of the “bacterial driver–passenger model”, in which “driver bacteria” of CRC promote tumorigenesis by inducing a sustained Th17 type inflammatory response, which is subsequently replaced by opportunistic “passenger bacteria” within the tumor, disrupting local innate immunity and ultimately leading to cancer progression[30]. Group 3 innate lymphoid cells (ILC3s) are the innate counterpart of Th17 cells, which modulate adaptive Th17 cell responses and act with Th17 cells against extracellular microorganisms[31]. ILCs are lymphocytes that do not express multiple antigen receptor types expressed on T and B cells[31] and play a key role in regulating host-microorganism interactions on the intestinal mucosal barrier surface[21,32]. ILC3s are abundant in mucosal sites and are involved in the innate immune response to extracellular bacteria and the suppression of gut commensal bacteria[31], and intestinal T cells control microbiota composition and intestinal immune response[33]. One study found that CRC is characterized by a significant decrease in ILC3s, accompanied by an increase in Th17 cells, suggesting that the progression of CRC is associated with impaired dialog between gut innate and adaptive immunity[21]. Whereas ILC3s and effector T cells interact via major histocompatibility complex class II (MHCII), this experiment found that deletion of ILC3-specific MHCII in mice lead to increased CRC invasiveness and susceptibility. Thus, the disruption of the interaction between MHCII+ ILC3s, effector T cells, and microbiota may be a mechanism to increase CRC invasiveness. In addition, gut commensal fungi, such as Candida albicans (C. albicans), promoted CRC tumorigenesis in animal experiments[34]. Commensal fungi activate glycolysis and IL-7 production in macrophages, while IL-7 induces IL-22 production in ILC3s, leading to tumor progression. However, other studies suggest that ILC3s in the TME may have both pro- and antitumor functions, depending on the cytokine types in the microenvironment[35,36].
Macrophages play a vital role in the maintenance of the innate immune response[37]. Tumor-associated macrophages (TAMs) are the main component of immune cells in the TME. On the one hand, M1 TAMs are induced by cytokines such as tumor necrosis factor (TNF)-α, secrete IL-6 and IL-23, participate in the polarized Th1 response, and exert antitumor immunity[37,38]. Akkermansia muciniphila (A. muciniphila) is a gut probiotic. Compared with the control group, the levels of M1-like TAMs were increased in A. Muciniphila-treated ApcMin/+ mice, and M1-like TAM-related cytokines, such as IL-23, TNF-α, and IL-27, were significantly induced in CRC[39]. It was also shown that A. Muciniphila promotes the enrichment of M1-like macrophages in a nucleotide-binding and oligomerization domain-like receptor thermal protein domain associated protein 3 (NLRP3)-dependent manner in vivo and in vitro, acting as a suppressor of CRC proliferation. NLRP3 activation and macrophage phenotypic polarization may be induced by toll-like receptor (TLR) 2. On the other hand, M2 is the main phenotype of TAMs. M2 TAMs are induced by IL-4, IL-10, and IL-13, secrete anti-inflammatory cytokines such as IL-10 and IL-1β, and participate in the polarized Th2 response, while activated Th2 cells produce lymphocytes producing IL-4 and IL-13, enhancing the expression of epidermal growth factor in TAMs and promoting the occurrence and development of tumors[37,38]. Gut dysbacteriosis results in increased expression of cathepsin K (CTSK) in colon cancer cells, and CTSK binds to TLR4 to stimulate M2 polarization of TAMs via the mechanistic target of rapamycin-dependent pathway[40]. At the same time, CTSK can stimulate M2 TAMs to secrete cytokines, including IL-10 and IL-17, which in turn mediate CRC cell invasion and metastasis through the nuclear factor kappa B (NF-κB) pathway. In addition to dysbacteriosis, F. nucleatum can also promote macrophage infiltration through CCL20 activation while inducing polarization of M2 macrophages to promote CRC metastasis[41].
TAMs can also promote the proliferation and invasion of tumor cells by interacting with tumor cells through microbiota-derived exosomes[37]. Microbiota-derived exosomes have the potential to activate macrophages. The extracellular vesicles released from bacteria are named outer membrane vesicles, and the coculture of outer membrane vesicles and macrophages leads to a large production of type M1 and M2 cytokines and chemokines. TLRs are an important component of the host defense mediated by the innate immune system[39] and are also involved in tumorigenesis[40]. Colon epithelial cells can sense the gut microbiome through pattern recognition receptors, including TLRs[42]. Thus, bacterial-induced chemokine gene expression is most likely initiated when TLRs trigger tumor cells[20].
In addition to the microbiome itself, microbiota-derived metabolites are also significant factors in the regulation of TME formation[43]. It is an important mediator of host-microbiome interactions[44]. Short-chain fatty acids (SCFAs), beneficial metabolites of the gut microbiome, are fermented from dietary fiber, including acetic acid, propionic acid, and butyrate[43-46]. SCFAs regulate intestinal motility and energy metabolism by secreting peptides YY and glucagon-like peptide 1 from intestinal endocrine L cells, directly inhibiting tumor cell growth and inducing host macrophage, T-cell and B-cell responses to protect against colitis-induced CRC[45].
Of these, butyrate is the most relevant bacterial metabolite of SCFAs[47] and has important immunomodulatory functions that can mediate the switching of pro-inflammatory cytokine expression by inhibiting histone deacetylases[48]. The significant reduction in butyrate-producing bacteria in the gut microbiome of patients with CRC may constitute a major structural imbalance in the gut microbiome of patients with CRC[49]. Thus, SCFAs play an important role in regulating host energy metabolism and the immune system[50]. Studies have shown that supplementation with probiotics that produce SCFAs can inhibit the development of intestinal tumors[51,52]. However, under certain conditions, butyrate metabolized by microorganisms may have the opposite effect. Several studies have shown that at lower concentrations, butyrate may stimulate the proliferation of colonic epithelial cells and thus promote CRC[53,54]. This discrepancy in the effects of butyrate on CRC is known as the “butyrate paradox”[53,54]. Therefore, increasing metabolites, such as SCFAs, has the potential to be used as an adjuvant treatment and preventive measure for CRC. However, it is necessary to further explore the action threshold of butyrate to avoid the opposite effect. Another gut bacterial metabolite, conjugated linoleic acid (CLA), is one of the most important fatty acids in the gut and is mainly produced by Roseburia species[55]. The reduction in Roseburia species was closely associated with the occurrence of CRC[49]. CLA also inhibits TNF-α expression and induces the immunomodulatory cytokine transforming growth factor β1 (TGF-β1) to participate in and regulate the pathways of apoptosis and immune response, reducing the risk of CRC[56]. Thus, the antiproliferative and anti-inflammatory capacity of CLA on colon cells may play an important role in CLA’s protection of the host against CRC[57].
GASTRIC CANCER
Gastric cancer (GC) is the fifth most common cancer in the world and the third most common cause of cancer death[58]. The host microbiome is closely associated with the occurrence and development of GC. The gastric microbiome affects inflammation and immunity at the local mucosal level and systemically. Disorders in the close interaction between the gastric microbiome and the immune system in the TME may contribute to GC development by triggering a tumor-promoting immune response[59] (Figure 2).
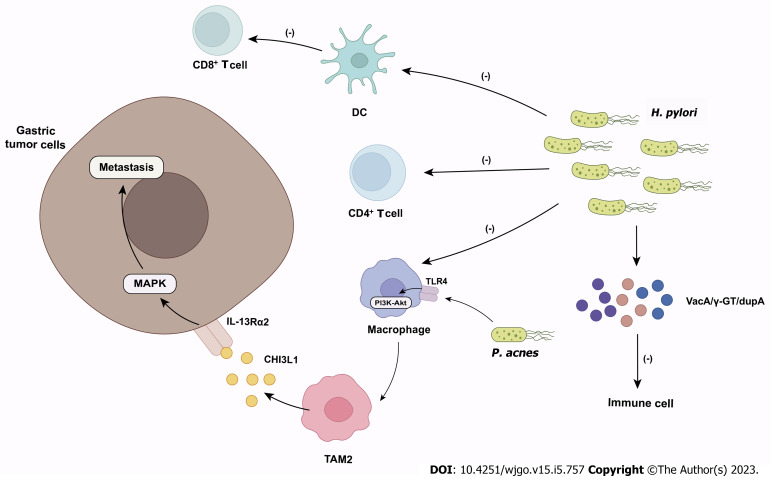
Figure 2.
The role of microbiome in gastric cancer. Helicobacter pylori inhibits the effector functions of CD4+ T cells, dendritic cells and macrophages and suppresses antitumor CD8+ T cell responses by altering the cross-presentation activity of dendritic cells. Helicobacter pylori also produce virulence factors (e.g., VacA, γ-GT, dupA) to impair immune cell activity. Propionibacterium acnes promote M2 polarization of macrophages via toll-like receptor 4/PI3K/Akt signaling. M2 macrophages secrete chitinase 3-like protein 1 that binds specifically to the interleukin-13 receptor α2 chai of tumor cells, triggering the mitogen-activated protein kinase signaling pathway and promoting gastric cancer metastasis. DC: Dendritic cell; TLR4: Toll-like receptor 4; TAM: Tumor-associated macrophages; CHI3L1: Chitinase 3-like protein 1; IL-13Rα2: Interleukin-13 receptor α2 chain; MAPK: Mitogen-activated protein kinase; H. pylori: Helicobacter pylori; P. acnes: Propionibacterium acnes.
Helicobacter pylori (H. pylori) infection is a risk factor for GC[60]. H. pylori colonizes the gastric mucosa of up to 50% of the population, manipulating host tissue to establish and maintain an immunosuppressive environment for chronic infection[61]. H. pylori inhibits the effector functions of CD4+ T cells, dendritic cells (DCs), and macrophages by altering the cross-presentation activity of DCs to suppress antitumor CD8+ T-cell responses, thereby suppressing the innate and adaptive immune responses of the infected host[61,62]. During GC formation, H. pylori may actively participate in GC by altering gastric mucosal immunity, particularly regulatory T (Treg)/Th17 imbalance[61]. H. pylori can also produce virulence factors (e.g., VacA, γ-GT, DupA) to impair immune cell activity[63]. For example, VacA affects the inflammatory response of the host primarily by inhibiting the proliferation and effector functions of T cells[63], but also induces the proinflammatory effects of T cells by activating nuclear factor kappa B and leading to the upregulation of IL-8[64]. γ-GT induces T-cell cycle block by disrupting RAS signaling and inhibits T-cell proliferation[65]. DupA increases the expression of proinflammatory cytokines (e.g., IL-12 and IL-23) via monocytes[66]. However, only H. pylori colonization in the stomach is not sufficient to induce gastric carcinogenesis[61]. In the past, the human stomach was thought to be the only habitat for H. pylori and was considered unsuitable for microbial habitation. However, recently, various studies have demonstrated that different gastric environments result in different microbial ecosystems within the stomach, and changes in specific microbial species may make the gastric microbiome more carcinogenic[67]. Bacteria other than H. pylori were previously thought to be unrelated to the development of GC[60]. Lofgren et al[68] demonstrated that the presence of a complex microbiome promoted the development of H. pylori-induced GC. However, the specific mechanism of the intragastric microbiome interaction with H. pylori on immunity remains unclear.
The microbial diversity and abundance in GC tumors were higher than those in nontumor tissues[67,69]. The intratumoral microbiome influences GC by modulating immune responses in the TME. A recent study by Peng et al[59] found that the infiltration of CD8+ tissue-resident memory T cells (TRM cells) in the TME of patients with GC is negatively correlated with the abundance of Methylobacterium in gastric tumors. The population of memory T cells in TILs, known as TRM cells, is derived from T cells that enter tissues during the primary response (such as in response to viral invasion) and plays a role in tumor immune surveillance[70]. CD8+ TRM cells produce and release various cytokines, such as interferon-γ (IFN-γ), which promote the activation of other immune cells and play an important role in antitumor immunity[71]. Intratumor Methylobacterium may promote the development of GC by inhibiting the immune infiltration of CD8+ TRM cells in the TME, leading to poor prognosis[59]. Furthermore, the abundance of Methylobacterium was also found to be significantly negatively correlated with TGF-β and IL-2[59]. TGF-β induces the expression of CD103, which plays an essential role in the permanent residence of TRM cells in epithelial tissues[71]. This may provide a new target for the development of immunotherapy for GC.
TAMs, mainly M2 TAMs (i.e., polarized M2 macrophages), are important in the progression of GC[72]. Recent experiments have shown that Propionibacterium acnes (P. acnes) is significantly increased in GC tissues, especially in H. pylori-negative tissue, and promotes M2 polarization of macrophages via TLR4/PI3K/Akt signaling when comparing microbial communities in GC tissues and adjacent normal tissues[73]. M2 macrophages secrete chitinase 3-like protein 1, which binds specifically to the interleukin-13 receptor α2 chain of cancer cells and triggers the mitogen-activated protein kinase signaling pathway to promote regional or distal metastasis of GC[72]. The abundance of P. acnes is positively correlated with M2 macrophages in GC tissues[73]. Therefore, P. acnes may be one of the possible factors for GC progression beyond H. pylori. Recent reports have shown a sustained increase in the abundance of lactic acid bacteria (LAB) in GC patients. LAB can increase exogenous lactate production[74]. Lactate produced by the glycolytic pathway leads to the formation of an acidic TME[75], which is involved in cancer progression. Lactate mediates M2-like polarization of TAMs and increases vascular endothelial growth factor and arginase 1 expression in these cells, thereby facilitating immune escape[74]. Lactate also inhibits the function and survival of T cells and natural killer (NK) cells and increases the number of myeloid-derived suppressor cells, which further inhibits the cytotoxicity of NK cells. Because LAB can produce large amounts of lactate in a short period of time[76], it is possible that microbial lactate can shape the TME like host lactate, altering the immune response[74].
In addition to bacteria, intratumoral fungi can also affect GC progression through the IOM axis. Yang et al[77] analyzed the gastric fungal microbiome in patients with GC and healthy individuals by high-throughput sequencing and noted that the ecological dysbiosis of the gastric fungal microbiota may be related to the occurrence of GC. They also measured the mRNA levels of cytokines and chemokines in tumor and normal tissues. The results showed that the levels of pro-inflammatory cytokines and chemokines such as CXCL9, CXCL10, CXCL11, and TNF-α were significantly increased, while anti-inflammatory cytokines and chemokines such as CCL17, IL-4, IL-6, and IL-8 were significantly decreased in the GC group. It is suggested that fungi that promote the production of proinflammatory cytokines and chemokines may be involved in promoting the tumor immune response, while fungi that promote the production of anti-inflammatory cytokines and chemokines may enhance the anti-inflammatory response in GC. IL-10 is highly expressed in various types of cancer[78,79], and can downregulate the inflammatory cytokines IL-6 and IL-8[77]. Therefore, the decreased levels of IL-6 and IL-8 in the GC group may be due to the increase in IL-10 levels in the local area of the tumor. IL-10 is released by TAMs, and IL-10+ TAMs infiltrate the tumor, which drives immune escape from the TME characterized by Treg-cell infiltration and CD8+ T-cell dysfunction[80]. C. albicans is the most studied fungal species in GC. C. albicans was found to be a fungal marker of GC[81]. In vitro, the mannose protein of C. albicans could promote tumor adhesion and liver metastasis[82]. Similar to CRC, fungi may have a complex role in the Th17 cell family. On the one hand, Th17 cells produce IL-17 to initiate downstream immunity against C. albicans. On the other hand, other cytokines, such as IL-23, produced by Th17 cells can promote tumorigenesis and growth. In addition, Th-17 can promote the progression of GC through an indirect mechanism. IL-17 secreted by Th17 cells can antagonize IL-12. IL-12 induces the production of IFN-γ and promotes the infiltration of cytotoxic T cells, which plays a critical role in the Th1-type antitumor immune response[83,84]. The promotion of cancer progression by IL-17 is also associated with neutrophil recruitment[85]. Additionally, the complement receptor 3-related protein (CR3-RP) of C. albicans has antigenic and structural similarity to CR3. CR3 is involved in leukocyte adhesion to endothelial cells and the subsequent extravasation process. Thus, antibodies against C. albicans CR3-RP may cross-react with leukocyte CR3 and disrupt the host’s antitumor defense[85].
In general, bacteria and fungi in the stomach, which are considered unsuitable for microbial habitation, have an important role in the regulation of GC progression. They can interact with immune cells in the body or the TME, affecting the progression of GC. Moreover, the role of fungi in GC may be underestimated. However, it is not clear whether fungi exist in tumor tissue as viable or partial components.
EC
EC is one of the most invasive malignant diseases[86]. Unlike other luminal organs of the digestive system, the esophagus does not retain food contents. It was first thought that the esophagus was aseptic, but after the study of traditional bacterial culture methods, it was found that the esophagus contained a small number of microorganisms swallowed from the oropharynx or excreted from the stomach through gastroesophageal reflux[87,88]. With the progress of culture technology, increasing evidence shows that the normal esophagus has a unique, stable, resident microbiome[87,88]. The distal esophageal microbiome was divided into two groups: Type I and type II[89]. Among them, the type I microbiome, dominated by Streptococcus species, seems to be the main component of the normal esophageal microbiome[89,90]. However, a study found that the microbiome of the normal esophagus is not the same as that of the esophagus with inflammation, Barrett’s esophagus (BE), and EC[87]. Type II microbiome, dominated by gram-negative bacteria, is usually associated with an abnormal esophagus[89].
Gastroesophageal reflux disease (GERD) and BE are significant risk factors for esophageal adenocarcinoma (EAC)[91-93]. Gram-negative bacteria that predominate in GERD and BE produce specific components, such as lipopolysaccharide (LPS)[87,88]. LPS directly or indirectly stimulates TLR4[94] in epithelial cells or inflammatory cells, leading to NF-кB activation[87] promote the expression of proinflammatory cytokines and sustain the innate immune response in the esophagus[88,94]. LPS binds plasma-derived LPS-binding protein and transmits to membrane-bound CD14 in monocytes, interacting with CD14[95]. CD14 plays a key role in LPS-mediated signal transduction by enhancing leukocyte adhesion, activation, and cytokine production. LPS stimulation of monocytes or epithelial cells leads to the activation of TLR4 and downstream NF-κB pathways, which triggers an inflammatory response[94]. LPS may also indirectly activate the NF-κB pathway of epithelial cells by triggering the production of inflammatory cytokines such as TNF-α, IL-1, and IL-6 by monocytes and macrophages[94,95].
In addition, Campylobacter was experimentally demonstrated to be significantly increased in GERD and BE[96]. Cytokines related to carcinogenesis, such as IL-18, were more highly expressed in the tissues colonized by Campylobacter[87]. Studies have shown that there is a very close relationship between esophageal Campylobacter colonization and IL-18 epithelial cell production[97]. IL-18 is produced by gastrointestinal epithelial cells, including squamous esophagus cells, which can stimulate both congenital and adaptive responses (Th1 and Th2) and induce NK cell activity and apoptosis[98]. Another study found that Campylobacter infection induced the secretion of IL-8 and TNF-α[99]. However, the increase in IL-8 secretion was not associated with the secretion of TNF-α stimulated by Campylobacter. It is not clear whether IL-8 can play a role in the initiation and maintenance of malignant transformation from GERD and BE to EAC. However, Campylobacter species is associated with a range of gastrointestinal diseases and may play a role in the progression of EAC similar to H. pylori in GC[87,100].
Moreover, the microbiome can also directly promote the occurrence of EAC by stimulating the human systemic immune system. For example, Lopetuso et al[90] identified Leptotrichia as the main taxon distinguishing EAC by LefSe analysis. Leptotrichia can promote the release of proinflammatory cytokines, such as IL-6 and IL-8. In turn, IL-6 and IL-8 can attract granulocytes and lymphocytes, thus inducing the host cellular immune response[101]. Serum antibodies against Leptotrichia are very common and belong to the IgG and IgM classes[102]. It is speculated that the immune activity caused by Leptotrichia may promote the occurrence of esophageal tumors[90].
Although there are few definitive conclusions about the impact of the microbiome on EAC, the change in the microbiome in the esophagus induces GERD and BE by activating the innate immune system and makes them progress to EAC, which seems to be a very important link in the pathogenesis of EAC. Compared with EAC, the characteristics of the microbiome in esophageal squamous cell carcinoma (ESCC) are not very clear[103]. However, there is growing evidence showing that the microbiome plays an important role in the occurrence and development of ESCC[87]. In a study by NHANES III, an increase in serum IgG of Porphyromonas gingivalis (P. gingivalis) was associated with increased mortality from oral and gastrointestinal cancers[104]. The results of Gao et al[105] showed that P. gingivalis infected the cancerous and adjacent esophageal mucosa of patients with ESCC. The titers of IgG and IgA against P. gingivalis in patients with ESCC were significantly higher than those in patients with esophagitis and healthy controls, which provided direct evidence for the involvement of P. gingivalis in the pathogenesis of ESCC. However, the specific mechanism by which P. gingivalis is involved in the progression of ESCC is unknown. Although the mechanism by which P. gingivalis affects the progression of ESCC through the IOM axis is still unclear, P. gingivalis can promote the infiltration of tumor-associated neutrophil 2 in tumor tissues in pancreatic cancer[106].
The poor prognosis of ESCC is also related to the presence of F. nucleatum[107]. In the relationship between F. nucleatum and cancer, especially CRC, F. nucleatum is considered to selectively amplify myeloid-derived immune cells to regulate the tumor immune microenvironment[23,25]. The immune response mediated by myeloid cells may provide the driving force for inflammation, genotoxicity, and epigenetic changes that lead to cancer[23]. In addition, F. nucleatum may promote tumor invasiveness by activating chemokines, such as CCL20, in EC tissue[87]. CCL20 plays a critical role in the migration of Treg lymphocytes. The specific receptor of CCL20 is CCR6, which is highly expressed in immune cells (e.g., Treg and Th17) and epithelial tumors[108]. Treg cells promote migration to tumor tissue in a CCR6-dependent manner in response to CCL20[109], and the concentration of CCL20 in the tumor is positively correlated with the number of tumor-infiltrating Tregs[108,110].
PANCREATIC CANCER
The central position of the pancreas in the abdominal cavity and the surrounding blood vessels and lymphatic vessels promote local and distant tumor spread. The TME of pancreatic cancer is characterized by a high matrix level and low immune activity[111]. This has also become one of the reasons for the poor efficacy of ICIs in pancreatic cancer. Improving this immunosuppressive microenvironment has become an urgent problem to be solved. In recent years, with the deepening of the study of the microbiome and the application of high-throughput sequencing technology[112], it has been found that the gut and intratumor microbiome have an important effect on pancreatic cancer[113-115]. This is especially true in immune-related research. The role of the IOM axis in pancreatic cancer has a dual nature. Most studies have reported the cancer-promoting effect of fungi and bacteria through the IOM axis in pancreatic ductal adenocarcinoma (PDAC).
TAMs have important potential in activating tumor-specific CD8+ T cells, while the important role of the microbiome has long been neglected in research on TAMs in the TME, as well as in pancreatic cancer[116-119]. Pushalkar et al[120] reported that ablation of the gut flora of mice with pancreatic cancer resulted in a decrease in immunosuppressive CD206+ M2-like TAMs with a concomitant increase in M1-like TAMs and increased expression of MHCII, CD86, TNF-α, IL-12, and IL-6. Moreover, like other gastrointestinal tumors, microbial ablation increased TLR expression in macrophages. Thus, the effect of the microbiome on PDAC through the IOM axis may be partly caused by acting on TLRs on macrophages. At the same time, their research also reported that the cell-free extract of the gut microbiome from pancreatic cancer mice can also promote the transformation of TAMs to an immunosuppressive phenotype. This suggests that the cellular components or metabolites of the microbiome located in the gut or tumor may play an important role in TAM-related reactions. Indole, a metabolite of the gut microbiome, such as Lactobacillus murinus, was found to activate the aryl hydrocarbon receptor (AhR) on TAMs. AhR activation leads to the expression of arginase 1 and IL-10 in TAMs and suppresses the expression of IFN-γ in CD8+ T cells. Another metabolite of the gut microbiome, trimethylamine N-oxide (TMAO), is different from indole in its production, which is completely dependent on gut bacteria[121]. Choline is the main source of circulating TMAO. However, unlike the effect of indole, choline-supplemented mice had increased TMAO levels, accompanied by a significant reduction in PDAC burden[122]. Flow cytometry showed that the expression of MHCI and MHCII on TAMs was significantly increased, accompanied by a significant increase in activated CD8+ and CD4+ T cells. Further research found that TMAO directly changed TAMs into a phenotype that could support the T-cell response and reduce the burden of PDAC. However, the role of CD8+ T cells mediated by the microbiome in the gut/tumor seems not limited to the stimulation of gut microbiome metabolites. Riquelme et al[12] found that patients with long-term survival had higher intratumor microbiome diversity than those with short-term survival. Moreover, PDAC tumor volume was reduced in mice that were gavaged with feces from long-term survival patients, and this effect could be eliminated by the depletion of CD8+ T cells. Although the causal relationship between the enrichment of microbial communities and tumors is still unclear, it suggests that the construction of microbial communities in the gut/tumors may be more effective than studying the role of a single strain.
The role of IL-33 and ILC2s in pancreatic cancer is highly controversial. KRAS mutant PDAC cells can secrete IL-33 through the KRAS-MEK-ERK pathway[123]. Intratumor fungi and their cell-free extracts can activate dectin-1 on PDAC. The activation of dectin-1 promotes the secretion of IL-33 by PDAC cells through the Src-Syk-CARD9 pathway. This shows the important role of intratumoral fungi in mediating the secretion of PDAC cells. However, the source of IL-33 may not be limited to PDAC cells. Another study showed a different result. Sun et al[124] conducted immunohistochemistry on 20 human PDAC tissues and found that pericytes and cancer-associated fibroblasts were the major cell sources of IL-33 production in PDAC tissues and promoted PDAC metastasis through the IL-33-ST2-CXCL3-CXCR2 axis, although both studies found that the expression of IL-33 in PDAC was increased. The increase in IL-33 in the TME of PDAC promotes the infiltration of TH2 cells and ILCs in PDAC through the IL-33/ST2 axis[123]. These immune cells promote the tumorigenic process through their cytokine networks, leading to PDAC progression[125-127]. In addition to IL-33, the role of its downstream ILC2s is also controversial. The role of bipartisan politicians of ILC2s is also reflected in PDAC. ILC2s can also promote the effective infiltration of immune cells. ILC2s can inhibit the progression of PDAC tumors through the ILC2-CD103+ DC-CD8+ T axis[128]. The level of immunity in PDAC appears to affect the effect of ILCs.
THE MICROBIOME AND CANCER THERAPY
Antibiotics
In a retrospective clinical study, increased overall survival (OS) and progression-free survival (PFS) in patients with metastatic PDAC were associated with antibiotic use[129]. Several preclinical studies have found that ablation of the gut and intratumor microbiome with antibiotics in a PDAC in situ mouse model can prevent tumor progression[12,120,130]. In the abnormal esophagus, the use of selective antibiotics or probiotics to reverse the type II microbiome to a type I microbiome in the esophagus can reduce the risk of esophageal carcinogenesis[94]. However, the effect caused by this antibiotic is related to the type of tumor and the type of antibiotics. Treatment of mice carrying colon cancer xenografts with the antibiotic metronidazole was found to reduce cancer cell proliferation and tumor growth[131]. However, treatment with metronidazole in pancreatic cancer significantly increased the tumor load[122]. In pancreatic cancer, quinolone therapy is linked to the improvement of OS. Postoperative quinolone therapy may prolong the survival time of preoperative treatment and resection of pancreatic cancer[132]. The use of antibiotics can inhibit or kill the pathogenic microbiome in the host. However, frequent use of broad-spectrum antibiotics may interfere with the gut microbiome, leading to ecological disorders and even cancer development[133].
Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) is a method that transplants the entire gut microbiota from a healthy donor into the patient’s gut to correct gut microbial abnormalities and reconstruct the structure and function of normal gut microbiota[134]. Metagenomic analysis showed that FMT significantly increased the abundance of potentially beneficial species[135]. The microbiome remodeling induced by FMT may be related to an improved tumor immune microenvironment. Riquelme et al[12] concluded that compared with short-term PDAC survivors or healthy controls, PDAC tumor-bearing mice transplanted with the fecal microbiome from long-term survivors of PDAC had antitumor immunity. The bacteria found in the tumor transferred from the intestinal tract to the pancreas, affecting the composition of the tumor microbiome and antitumor immunity in the pancreas. Rosshart et al[136] reported that FMT enhanced host resistance to mutagen/inflammation-induced colorectal tumorigenesis. The beneficial role of FMT in the treatment of diseases has been confirmed, but it has not been widely studied in gastrointestinal tumors.
Probiotics
Probiotics can alter the composition of the gut microbiome and have been shown to inhibit tumor development by downregulating the levels of LPS, inflammatory factors, and chemokines[137]. Various studies have found that supplementation with probiotics that produce SCFAs can inhibit the development of tumors[51,52]. For example, both Lactobacillus coryniformis MXJ32 and Lacticaseibacillus rhamnosus LS8 can reduce intestinal inflammation by downregulating the expression of inflammatory cytokines (e.g., TNF-α, IL-1β, IL-6, IL-γ, and IL-17a) and chemokines (e.g., CXCL1, CXCL2, CXCL3, CXCL5, and CCL7) and effectively improve colitis-associated CRC[138,139]. However, the effects of these two probiotics on the mechanism of regulating the gut microbiome and specific immune response are not clear. In the study of Heydari et al[140], which uses an animal colon cancer model, treatment with probiotics suppressed the increase in miRNA expression and decreased the level of oncogenes, and such treatment was considered beneficial for colon cancer treatment. Lactobacillus brevis SBL8803-derived polyphosphate leads to apoptosis of CRC cells by activating extracellular signal-regulated kinase signal transduction. Heptelidic acid, a metabolite of the probiotic Aspergillus oryzae, passes through the gut mucosa to reach the pancreas and induces apoptosis in pancreatic cancer cells by activating the p38MAPK signaling pathway[141]. Although probiotic supplements may alter the structure of the microbiome and regulate inflammation to prevent cancer[142], probiotics may adversely affect the reconstruction of gut mucosal host-microbiome ecosystems after antibiotic treatment[143]. Therefore, probiotic therapy may be a promising intervention method[138], but many problems urgently require further research.
Immunotherapy
The use of ICIs has made remarkable progress in the treatment of many cancers, among which the most widely used ICIs are monoclonal antibodies targeting programmed cell death protein 1 (PD-1) and its ligand PD-L1[144]. Although immunotherapy based on anti-PD-1 has a limited response in CRC patients, a growing body of research supports the important role of the gut microbiome in the immune system. The gut microbiome seems to influence the expression of PD-1/PD-L1 indirectly through systemic or locally mediated immune function, thus affecting the efficacy of anti-PD-1 and anti-PD-L1 therapy[134]. The mechanisms by which the gut microbiome improves the efficacy of anti-PD-1 are as follows[145-148]: (1) An increase in beneficial bacteria; (2) Enhancement of the function of DCs; (3) An increase in antitumor CD8+ T-cell activity; and (4) Promotion of T-cell tumor infiltration. Several studies have reported that oral combinations of specific symbiotic bacteria and anti-PD-1/PD-L1 antibodies almost eliminated tumor growth[145,149]. In a mouse cancer model, oral live Lactobacillus rhamnosus GG enhanced the antitumor activity of anti-PD-1 immunotherapy by increasing tumor-infiltrating DCs and T cells and significantly inhibited tumor growth[146].
Antibiotics and immunotherapy
There is growing evidence that the gut microbiome can influence immunotherapy responses in patients treated with ICIs[61,62,134]. Preclinical experiments in mice showed that the use of antibiotics could decrease the efficacy of ICIs[144]. In a meta-analysis enrolling 2740 cancer patients, antibiotic use significantly reduced OS and PFS in patients treated with ICIs[150]. But, In a study on pancreatic cancer, researchers discovered that by using broad-spectrum antibiotics to eliminate gut microbiota, they could trigger immunogenic reprogramming within the TME. This made treatment with ICIs more effective by increasing the expression of PD-1[120]. Thus, whether the use of antibiotics can improve the efficacy of ICIs is controversial. At present, some meta-analyses suggest that antibiotic administration may be associated with poor prognosis of tumor patients receiving ICIs[151,152]. However, these studies focused on lung cancer, renal cell carcinoma, urothelial carcinoma, and melanoma[151]. Research on gastrointestinal malignancies is still insufficient. When CRC cells were implanted into germ-free or specific pathogen-free mice, broad-spectrum antibiotics reduced their ICI efficacy[153]. In a mouse model, H. pylori infection partially blocked the activity of ICIs and reduced the effect of tumor immunotherapy[61]. However, eradication of H. pylori infection through antibiotic therapy did not restore the decreased response of H. pylori-induced cancer to immunotherapy. Therefore, the administration of antibiotics to cure H. pylori infection is not a good choice to improve the efficacy of cancer immunotherapy. Han et al[154] recently demonstrated that antibiotic-induced microbiome disorders enhanced the antitumor efficacy of γ-δ T cells during immunotherapy in a mouse model of hepatocellular carcinoma. γ-δ T cells can generate immune responses to a wide range of antigens. They are believed to serve as a bridge between innate and adaptive immune responses[7]. γ-δ T cells can also infiltrate GC, pancreatic cancer, and colon cancer[155-157]. Thus, the effect of antibiotics on ICIs in patients with gastrointestinal malignancies still needs to be further studied.
FMT and immunotherapy
FMT is a potential way to improve the efficacy of anti-PD-1 therapy[158]. Huang et al[158] found that compared with colon cancer-bearing mice treated with anti-PD-1 or FMT alone, FMT combined with anti-PD-1 showed higher survival and tumor control. The enhancement of anti-PD-1 therapy induced by FMT may be mediated by changes in the microbial genome and blood metabolism. Through metagenomic analysis, FMT altered the composition of the gut microbiome. The relative abundance of Bacteroides species and Parabacteroides species increased significantly. Metabonomic analysis of mouse plasma showed that after FMT, several metabolites, including taxic acid and aspirin, may promote the response to anti-PD-1 therapy through their immunomodulatory function. Accordingly, the composition and function of the gut microbiome may be able to influence the ICI response in cancer[7]. However, FMT did not improve the response to immunotherapy in cancer patients infected with H. pylori[62]. In the absence of ICIs, modulation of the gut microbiome with bacteria or FMT has a limited impact on the antitumor immune response or tumor growth[134]. Therefore, FMT may serve as an important therapeutic modality to assist patients treated with ICIs to enhance systemic and antitumor immunity in cancer patients.
Microbial metabolites and immunotherapy
Intestinal epithelial cells are closely related to the immune system. Bacterial metabolites, such as SCFAs, occur in the immune response and are strongly associated with innate immunity and antibody production[159]. The results from a cohort study showed that high levels of fecal or plasma SCFAs were associated with PD-1 treatment response and longer PFS[160]. In mice humanized with gut microbiota from patients, butyrate promoted T-cell infiltration in the TME, thus improving the efficacy of anti-PD-1 monoclonal antibodies[147]. T Tryptophan is an essential amino acid for the human body. Indole, a metabolite of tryptophan, is a biologically active compound that plays an important role in tumor and immune regulation[161]. Indole drives AhR on TAMs and suppresses antitumor immunity. Macrophage AhR is the central driver of TAM function in PDAC. In patients with PDAC, high expression of AhR is associated with rapid disease progression and mortality. TMAO, a metabolite of natural microorganisms, suppresses the immunostimulatory phenotype of macrophages, promotes the activity of effector T cells, and enhances antitumor immunity against PDAC[122]. The combination of TMAO and anti-PD1 in a PDAC mouse model significantly reduced the tumor burden and improved the survival rate compared to TMAO or ICIs alone. Therefore, the immunomodulatory mechanism associated with microbial metabolites may become a new direction to improve PD-1 efficacy in cancer patients[160].
CONCLUSION
In recent years, some progress has been made in the study of microorganisms and tumors. Although the role of the IOM axis in GC and EC needs to be further clarified, it plays an important role in the occurrence and development of gastrointestinal malignancies. Macrophages may be a key component in linking the microbiome and immunity, which has been reflected in the variety of tumors mentioned above. The microbiome may influence tumor immune responses through TLRs on macrophages. ILCs also play a vital role in the host microbiome and, together with T cells, regulate the IOM axis. The IOM axis provides a new direction for the treatment of gastrointestinal malignancies. An increasing number of studies have shown the role of the microbiome in immunotherapy. For example, specific antibiotic use may prevent tumor progression, whereas the combination of antibiotics with ICIs may reduce the efficacy of ICIs. In contrast, FMT has been found to improve the efficacy of immunotherapy. However, the effect of the microbiome on immunotherapy is still controversial, and the mechanism of action is still elusive and needs to be widely validated by more preclinical models and clinical trials.
Footnotes
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: December 26, 2022
First decision: March 13, 2023
Article in press: April 14, 2023
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ekine-Afolabi B, United Kingdom; Paparoupa M, Germany S-Editor: Wang JJ L-Editor: A P-Editor: Li X
Contributor Information
Quan Lin, Department of Surgery, Wenzhou Central Hospital, The Dingli Clinical Institute of Wenzhou Medical University, Wenzhou 325000, Zhejiang Province, China.
Shi-Wei Guan, Department of Surgery, Wenzhou Central Hospital, The Dingli Clinical Institute of Wenzhou Medical University, Wenzhou 325000, Zhejiang Province, China.
Hai-Bo Yu, Department of Surgery, Wenzhou Central Hospital, The Dingli Clinical Institute of Wenzhou Medical University, Wenzhou 325000, Zhejiang Province, China. yuhaibozjwz@163.com.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Bagchi S, Yuan R, Engleman EG. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu Rev Pathol. 2021;16:223–249. doi: 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 3.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Diéras V, Hegg R, Im SA, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA IMpassion130 Trial Investigators. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 4.Kojima T, Shah MA, Muro K, Francois E, Adenis A, Hsu CH, Doi T, Moriwaki T, Kim SB, Lee SH, Bennouna J, Kato K, Shen L, Enzinger P, Qin SK, Ferreira P, Chen J, Girotto G, de la Fouchardiere C, Senellart H, Al-Rajabi R, Lordick F, Wang R, Suryawanshi S, Bhagia P, Kang SP, Metges JP KEYNOTE-181 Investigators. Randomized Phase III KEYNOTE-181 Study of Pembrolizumab Versus Chemotherapy in Advanced Esophageal Cancer. J Clin Oncol. 2020;38:4138–4148. doi: 10.1200/JCO.20.01888. [DOI] [PubMed] [Google Scholar]
- 5.Reiss KA, Mick R, Teitelbaum U, O'Hara M, Schneider C, Massa R, Karasic T, Tondon R, Onyiah C, Gosselin MK, Donze A, Domchek SM, Vonderheide RH. Niraparib plus nivolumab or niraparib plus ipilimumab in patients with platinum-sensitive advanced pancreatic cancer: a randomised, phase 1b/2 trial. Lancet Oncol. 2022;23:1009–1020. doi: 10.1016/S1470-2045(22)00369-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, Li Q. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. 2022;15:47. doi: 10.1186/s13045-022-01273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain T, Sharma P, Are AC, Vickers SM, Dudeja V. New Insights Into the Cancer-Microbiome-Immune Axis: Decrypting a Decade of Discoveries. Front Immunol. 2021;12:622064. doi: 10.3389/fimmu.2021.622064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557–566. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 12.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, Scheet P, Xu H, Hanash SM, Feng L, Burks JK, Do KA, Peterson CB, Nejman D, Tzeng CD, Kim MP, Sears CL, Ajami N, Petrosino J, Wood LD, Maitra A, Straussman R, Katz M, White JR, Jenq R, Wargo J, McAllister F. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178:795–806.e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021;371 doi: 10.1126/science.abc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sepich-Poore GD, Guccione C, Laplane L, Pradeu T, Curtius K, Knight R. Cancer's second genome: Microbial cancer diagnostics and redefining clonal evolution as a multispecies process: Humans and their tumors are not aseptic, and the multispecies nature of cancer modulates clinical care and clonal evolution: Humans and their tumors are not aseptic, and the multispecies nature of cancer modulates clinical care and clonal evolution. Bioessays. 2022;44:e2100252. doi: 10.1002/bies.202100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, Meltser A, Douglas GM, Kamer I, Gopalakrishnan V, Dadosh T, Levin-Zaidman S, Avnet S, Atlan T, Cooper ZA, Arora R, Cogdill AP, Khan MAW, Ologun G, Bussi Y, Weinberger A, Lotan-Pompan M, Golani O, Perry G, Rokah M, Bahar-Shany K, Rozeman EA, Blank CU, Ronai A, Shaoul R, Amit A, Dorfman T, Kremer R, Cohen ZR, Harnof S, Siegal T, Yehuda-Shnaidman E, Gal-Yam EN, Shapira H, Baldini N, Langille MGI, Ben-Nun A, Kaufman B, Nissan A, Golan T, Dadiani M, Levanon K, Bar J, Yust-Katz S, Barshack I, Peeper DS, Raz DJ, Segal E, Wargo JA, Sandbank J, Shental N, Straussman R. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Li H, Xie H, Wu X, Lan P. The malignant role of exosomes in the communication among colorectal cancer cell, macrophage and microbiome. Carcinogenesis. 2019;40:601–610. doi: 10.1093/carcin/bgy138. [DOI] [PubMed] [Google Scholar]
- 20.Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, Trella E, Galati-Fournier V, Oertli D, Däster SR, Droeser RA, Weixler B, Bolli M, Rosso R, Nitsche U, Khanna N, Egli A, Keck S, Slotta-Huspenina J, Terracciano LM, Zajac P, Spagnoli GC, Eppenberger-Castori S, Janssen KP, Borsig L, Iezzi G. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut. 2018;67:1984–1994. doi: 10.1136/gutjnl-2016-313498. [DOI] [PubMed] [Google Scholar]
- 21.Goc J, Lv M, Bessman NJ, Flamar AL, Sahota S, Suzuki H, Teng F, Putzel GG JRI Live Cell Bank, Eberl G, Withers DR, Arthur JC, Shah MA, Sonnenberg GF. Dysregulation of ILC3s unleashes progression and immunotherapy resistance in colon cancer. Cell. 2021;184:5015–5030.e16. doi: 10.1016/j.cell.2021.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F, Galon J. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 23.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenker BJ, Datar S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun. 1995;63:4830–4836. doi: 10.1128/iai.63.12.4830-4836.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galaski J, Shhadeh A, Umaña A, Yoo CC, Arpinati L, Isaacson B, Berhani O, Singer BB, Slade DJ, Bachrach G, Mandelboim O. Fusobacterium nucleatum CbpF Mediates Inhibition of T Cell Function Through CEACAM1 Activation. Front Cell Infect Microbiol. 2021;11:692544. doi: 10.3389/fcimb.2021.692544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, Datz C, Feng Y, Fearon ER, Oukka M, Tessarollo L, Coppola V, Yarovinsky F, Cheroutre H, Eckmann L, Trinchieri G, Karin M. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 31.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, Powrie F, Spits H. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 32.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, Wherry EJ, Koni PA, Bushman FD, Elson CO, Eberl G, Artis D, Sonnenberg GF. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, Wieduwild E, Putoczki T, Mondot S, Lantz O, Demon D, Papenfuss AT, Smyth GK, Lamkanfi M, Carotta S, Renauld JC, Shi W, Carpentier S, Soos T, Arendt C, Ugolini S, Huntington ND, Belz GT, Vivier E. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol. 2016;17:179–186. doi: 10.1038/ni.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Shi T, Lu X, Xu Z, Qu J, Zhang Z, Shi G, Shen S, Hou Y, Chen Y, Wang T. Fungal-induced glycolysis in macrophages promotes colon cancer by enhancing innate lymphoid cell secretion of IL-22. EMBO J. 2021;40:e105320. doi: 10.15252/embj.2020105320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillerey C. Roles of cytotoxic and helper innate lymphoid cells in cancer. Mamm Genome. 2018;29:777–789. doi: 10.1007/s00335-018-9781-4. [DOI] [PubMed] [Google Scholar]
- 36.Wu J, Cheng H, Wang H, Zang G, Qi L, Lv X, Liu C, Zhu S, Zhang M, Cui J, Ueno H, Liu YJ, Suo J, Chen J. Correlation Between Immune Lymphoid Cells and Plasmacytoid Dendritic Cells in Human Colon Cancer. Front Immunol. 2021;12:601611. doi: 10.3389/fimmu.2021.601611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Tian T, Zhang J. Tumor-Associated Macrophages (TAMs) in Colorectal Cancer (CRC): From Mechanism to Therapy and Prognosis. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22168470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Italiani P, Boraschi D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front Immunol. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan L, Xu C, Ge Q, Lin Y, Wong CC, Qi Y, Ye B, Lian Q, Zhuo W, Si J, Chen S, Wang L. A. Muciniphila Suppresses Colorectal Tumorigenesis by Inducing TLR2/NLRP3-Mediated M1-Like TAMs. Cancer Immunol Res. 2021;9:1111–1124. doi: 10.1158/2326-6066.CIR-20-1019. [DOI] [PubMed] [Google Scholar]
- 40.Li R, Zhou R, Wang H, Li W, Pan M, Yao X, Zhan W, Yang S, Xu L, Ding Y, Zhao L. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019;26:2447–2463. doi: 10.1038/s41418-019-0312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, Chen Z, Wang L, Long Y, Hou T, Si J, Chen S. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13:1980347. doi: 10.1080/19490976.2021.1980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Liu B, Wei Y, Kuang DM. Influence of gut and intratumoral microbiota on the immune microenvironment and anti-cancer therapy. Pharmacol Res. 2021;174:105966. doi: 10.1016/j.phrs.2021.105966. [DOI] [PubMed] [Google Scholar]
- 44.Gasaly N, de Vos P, Hermoso MA. Impact of Bacterial Metabolites on Gut Barrier Function and Host Immunity: A Focus on Bacterial Metabolism and Its Relevance for Intestinal Inflammation. Front Immunol. 2021;12:658354. doi: 10.3389/fimmu.2021.658354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 46.Makki K, Deehan EC, Walter J, Bäckhed F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe. 2018;23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16:605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 48.Khan I, Huang G, Li XA, Liao W, Leong WK, Xia W, Bian X, Wu J, Hsiao WLW. Mushroom polysaccharides and jiaogulan saponins exert cancer preventive effects by shaping the gut microbiota and microenvironment in Apc(Min/+) mice. Pharmacol Res. 2019;148:104448. doi: 10.1016/j.phrs.2019.104448. [DOI] [PubMed] [Google Scholar]
- 49.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- 51.Hou H, Chen D, Zhang K, Zhang W, Liu T, Wang S, Dai X, Wang B, Zhong W, Cao H. Gut microbiota-derived short-chain fatty acids and colorectal cancer: Ready for clinical translation? Cancer Lett. 2022;526:225–235. doi: 10.1016/j.canlet.2021.11.027. [DOI] [PubMed] [Google Scholar]
- 52.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 53.Bultman SJ, Jobin C. Microbial-derived butyrate: an oncometabolite or tumor-suppressive metabolite? Cell Host Microbe. 2014;16:143–145. doi: 10.1016/j.chom.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Surendra A, Kumar S, Green B, Geddes K, Pezo RC, Navarre WW, Milosevic M, Wilson BC, Girardin SE, Wolever TMS, Edelmann W, Guttman DS, Philpott DJ, Martin A. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 55.Devillard E, McIntosh FM, Duncan SH, Wallace RJ. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J Bacteriol. 2007;189:2566–2570. doi: 10.1128/JB.01359-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans NP, Misyak SA, Schmelz EM, Guri AJ, Hontecillas R, Bassaganya-Riera J. Conjugated linoleic acid ameliorates inflammation-induced colorectal cancer in mice through activation of PPARgamma. J Nutr. 2010;140:515–521. doi: 10.3945/jn.109.115642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bassaganya-Riera J, Reynolds K, Martino-Catt S, Cui Y, Hennighausen L, Gonzalez F, Rohrer J, Benninghoff AU, Hontecillas R. Activation of PPAR gamma and delta by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology. 2004;127:777–791. doi: 10.1053/j.gastro.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 58.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 59.Peng R, Liu S, You W, Huang Y, Hu C, Gao Y, Jia X, Li G, Xu Z, Chen Y. Gastric Microbiome Alterations Are Associated with Decreased CD8+ Tissue-Resident Memory T Cells in the Tumor Microenvironment of Gastric Cancer. Cancer Immunol Res. 2022;10:1224–1240. doi: 10.1158/2326-6066.CIR-22-0107. [DOI] [PubMed] [Google Scholar]
- 60.Artola-Borán M, Fallegger A, Priola M, Jeske R, Waterboer T, Dohlman AB, Shen X, Wild S, He J, Levesque MP, Yousefi S, Simon HU, Cheng PF, Müller A. Mycobacterial infection aggravates Helicobacter pylori-induced gastric preneoplastic pathology by redirection of de novo induced Treg cells. Cell Rep. 2022;38:110359. doi: 10.1016/j.celrep.2022.110359. [DOI] [PubMed] [Google Scholar]
- 61.Oster P, Vaillant L, Riva E, McMillan B, Begka C, Truntzer C, Richard C, Leblond MM, Messaoudene M, Machremi E, Limagne E, Ghiringhelli F, Routy B, Verdeil G, Velin D. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut. 2022;71:457–466. doi: 10.1136/gutjnl-2020-323392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oster P, Vaillant L, McMillan B, Velin D. The Efficacy of Cancer Immunotherapies Is Compromised by Helicobacter pylori Infection. Front Immunol. 2022;13:899161. doi: 10.3389/fimmu.2022.899161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalali B, Mejías-Luque R, Javaheri A, Gerhard M. H. pylori virulence factors: influence on immune system and pathology. Mediators Inflamm. 2014;2014:426309. doi: 10.1155/2014/426309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takeshima E, Tomimori K, Takamatsu R, Ishikawa C, Kinjo F, Hirayama T, Fujita J, Mori N. Helicobacter pylori VacA activates NF-kappaB in T cells via the classical but not alternative pathway. Helicobacter. 2009;14:271–279. doi: 10.1111/j.1523-5378.2009.00683.x. [DOI] [PubMed] [Google Scholar]
- 65.Schmees C, Prinz C, Treptau T, Rad R, Hengst L, Voland P, Bauer S, Brenner L, Schmid RM, Gerhard M. Inhibition of T-cell proliferation by Helicobacter pylori gamma-glutamyl transpeptidase. Gastroenterology. 2007;132:1820–1833. doi: 10.1053/j.gastro.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 66.Hussein NR, Argent RH, Marx CK, Patel SR, Robinson K, Atherton JC. Helicobacter pylori dupA is polymorphic, and its active form induces proinflammatory cytokine secretion by mononuclear cells. J Infect Dis. 2010;202:261–269. doi: 10.1086/653587. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336–348. doi: 10.1016/j.ebiom.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, Wang TC, Fox JG. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dai D, Yang Y, Yu J, Dang T, Qin W, Teng L, Ye J, Jiang H. Interactions between gastric microbiota and metabolites in gastric cancer. Cell Death Dis. 2021;12:1104. doi: 10.1038/s41419-021-04396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang N, Bevan MJ. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39:687–696. doi: 10.1016/j.immuni.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amsen D, van Gisbergen KPJM, Hombrink P, van Lier RAW. Tissue-resident memory T cells at the center of immunity to solid tumors. Nat Immunol. 2018;19:538–546. doi: 10.1038/s41590-018-0114-2. [DOI] [PubMed] [Google Scholar]
- 72.Chen Y, Zhang S, Wang Q, Zhang X. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol. 2017;10:36. doi: 10.1186/s13045-017-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Q, Wu W, Gong D, Shang R, Wang J, Yu H. Propionibacterium acnes overabundance in gastric cancer promote M2 polarization of macrophages via a TLR4/PI3K/Akt signaling. Gastric Cancer. 2021;24:1242–1253. doi: 10.1007/s10120-021-01202-8. [DOI] [PubMed] [Google Scholar]
- 74.Vinasco K, Mitchell HM, Kaakoush NO, Castaño-Rodríguez N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer. 2019;1872:188309. doi: 10.1016/j.bbcan.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Zhang L, Li S. Lactic acid promotes macrophage polarization through MCT-HIF1α signaling in gastric cancer. Exp Cell Res. 2020;388:111846. doi: 10.1016/j.yexcr.2020.111846. [DOI] [PubMed] [Google Scholar]
- 76.Neal-McKinney JM, Lu X, Duong T, Larson CL, Call DR, Shah DH, Konkel ME. Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS One. 2012;7:e43928. doi: 10.1371/journal.pone.0043928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang P, Zhang X, Xu R, Adeel K, Lu X, Chen M, Shen H, Li Z, Xu Z. Fungal Microbiota Dysbiosis and Ecological Alterations in Gastric Cancer. Front Microbiol. 2022;13:889694. doi: 10.3389/fmicb.2022.889694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fortis C, Foppoli M, Gianotti L, Galli L, Citterio G, Consogno G, Gentilini O, Braga M. Increased interleukin-10 serum levels in patients with solid tumours. Cancer Lett. 1996;104:1–5. doi: 10.1016/0304-3835(96)04213-9. [DOI] [PubMed] [Google Scholar]
- 79.Ahmad N, Ammar A, Storr SJ, Green AR, Rakha E, Ellis IO, Martin SG. IL-6 and IL-10 are associated with good prognosis in early stage invasive breast cancer patients. Cancer Immunol Immunother. 2018;67:537–549. doi: 10.1007/s00262-017-2106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang H, Li R, Cao Y, Gu Y, Lin C, Liu X, Lv K, He X, Fang H, Jin K, Fei Y, Chen Y, Wang J, Liu H, Li H, Zhang H, He H, Zhang W. Poor Clinical Outcomes and Immunoevasive Contexture in Intratumoral IL-10-Producing Macrophages Enriched Gastric Cancer Patients. Ann Surg. 2022;275:e626–e635. doi: 10.1097/SLA.0000000000004037. [DOI] [PubMed] [Google Scholar]
- 81.Zhong M, Xiong Y, Zhao J, Gao Z, Ma J, Wu Z, Song Y, Hong X. Candida albicans disorder is associated with gastric carcinogenesis. Theranostics. 2021;11:4945–4956. doi: 10.7150/thno.55209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramirez-Garcia A, Arteta B, Abad-Diaz-de-Cerio A, Pellon A, Antoran A, Marquez J, Rementeria A, Hernando FL. Candida albicans increases tumor cell adhesion to endothelial cells in vitro: intraspecific differences and importance of the mannose receptor. PLoS One. 2013;8:e53584. doi: 10.1371/journal.pone.0053584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 84.Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 85.Ramirez-Garcia A, Rementeria A, Aguirre-Urizar JM, Moragues MD, Antoran A, Pellon A, Abad-Diaz-de-Cerio A, Hernando FL. Candida albicans and cancer: Can this yeast induce cancer development or progression? Crit Rev Microbiol. 2016;42:181–193. doi: 10.3109/1040841X.2014.913004. [DOI] [PubMed] [Google Scholar]
- 86.Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390:2383–2396. doi: 10.1016/S0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 87.Baba Y, Iwatsuki M, Yoshida N, Watanabe M, Baba H. Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann Gastroenterol Surg. 2017;1:99–104. doi: 10.1002/ags3.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neto AG, Whitaker A, Pei Z. Microbiome and potential targets for chemoprevention of esophageal adenocarcinoma. Semin Oncol. 2016;43:86–96. doi: 10.1053/j.seminoncol.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588–597. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lopetuso LR, Severgnini M, Pecere S, Ponziani FR, Boskoski I, Larghi A, Quaranta G, Masucci L, Ianiro G, Camboni T, Gasbarrini A, Costamagna G, Consolandi C, Cammarota G. Esophageal microbiome signature in patients with Barrett's esophagus and esophageal adenocarcinoma. PLoS One. 2020;15:e0231789. doi: 10.1371/journal.pone.0231789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maret-Ouda J, Markar SR, Lagergren J. Gastroesophageal Reflux Disease: A Review. JAMA. 2020;324:2536–2547. doi: 10.1001/jama.2020.21360. [DOI] [PubMed] [Google Scholar]
- 92.Katzka DA, Kahrilas PJ. Advances in the diagnosis and management of gastroesophageal reflux disease. BMJ. 2020;371:m3786. doi: 10.1136/bmj.m3786. [DOI] [PubMed] [Google Scholar]
- 93.Spechler SJ, Souza RF. Barrett's esophagus. N Engl J Med. 2014;371:836–845. doi: 10.1056/NEJMra1314704. [DOI] [PubMed] [Google Scholar]
- 94.Yang L, Francois F, Pei Z. Molecular pathways: pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clin Cancer Res. 2012;18:2138–2144. doi: 10.1158/1078-0432.CCR-11-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang Q, Akashi S, Miyake K, Petty HR. Lipopolysaccharide induces physical proximity between CD14 and toll-like receptor 4 (TLR4) prior to nuclear translocation of NF-kappa B. J Immunol. 2000;165:3541–3544. doi: 10.4049/jimmunol.165.7.3541. [DOI] [PubMed] [Google Scholar]
- 96.Macfarlane S, Furrie E, Macfarlane GT, Dillon JF. Microbial colonization of the upper gastrointestinal tract in patients with Barrett's esophagus. Clin Infect Dis. 2007;45:29–38. doi: 10.1086/518578. [DOI] [PubMed] [Google Scholar]
- 97.Blackett KL, Siddhi SS, Cleary S, Steed H, Miller MH, Macfarlane S, Macfarlane GT, Dillon JF. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther. 2013;37:1084–1092. doi: 10.1111/apt.12317. [DOI] [PubMed] [Google Scholar]
- 98.Yasuda K, Nakanishi K, Tsutsui H. Interleukin-18 in Health and Disease. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20030649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng J, Meng J, Zhao S, Singh R, Song W. Campylobacter-induced interleukin-8 secretion in polarized human intestinal epithelial cells requires Campylobacter-secreted cytolethal distending toxin- and Toll-like receptor-mediated activation of NF-kappaB. Infect Immun. 2008;76:4498–4508. doi: 10.1128/IAI.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Man SM. The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol. 2011;8:669–685. doi: 10.1038/nrgastro.2011.191. [DOI] [PubMed] [Google Scholar]
- 101.Eribe ERK, Olsen I. Leptotrichia species in human infections II. J Oral Microbiol. 2017;9:1368848. doi: 10.1080/20002297.2017.1368848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morgenstein AA, Citron DM, Orisek B, Finegold SM. Serious infection with Leptotrichia buccalis. Report of a case and review of the literature. Am J Med. 1980;69:782–785. doi: 10.1016/0002-9343(80)90452-0. [DOI] [PubMed] [Google Scholar]
- 103.Di Pilato V, Freschi G, Ringressi MN, Pallecchi L, Rossolini GM, Bechi P. The esophageal microbiota in health and disease. Ann N Y Acad Sci. 2016;1381:21–33. doi: 10.1111/nyas.13127. [DOI] [PubMed] [Google Scholar]
- 104.Gao S, Liu Y, Duan X, Liu K, Mohammed M, Gu Z, Ren J, Yakoumatos L, Yuan X, Lu L, Liang S, Li J, Scott DA, Lamont RJ, Zhou F, Wang H. Porphyromonas gingivalis infection exacerbates oesophageal cancer and promotes resistance to neoadjuvant chemotherapy. Br J Cancer. 2021;125:433–444. doi: 10.1038/s41416-021-01419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao SG, Yang JQ, Ma ZK, Yuan X, Zhao C, Wang GC, Wei H, Feng XS, Qi YJ. Preoperative serum immunoglobulin G and A antibodies to Porphyromonas gingivalis are potential serum biomarkers for the diagnosis and prognosis of esophageal squamous cell carcinoma. BMC Cancer. 2018;18:17. doi: 10.1186/s12885-017-3905-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, Yuan W, Ma J. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes. 2022;14:2073785. doi: 10.1080/19490976.2022.2073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y, Yoshida N, Watanabe M, Baba H. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res. 2016;22:5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 108.Kadomoto S, Izumi K, Mizokami A. The CCL20-CCR6 Axis in Cancer Progression. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21155186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, Craig S, Watowich SS, Jetten AM, Tian Q, Dong C. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008;181:8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen KJ, Lin SZ, Zhou L, Xie HY, Zhou WH, Taki-Eldin A, Zheng SS. Selective recruitment of regulatory T cell through CCR6-CCL20 in hepatocellular carcinoma fosters tumor progression and predicts poor prognosis. PLoS One. 2011;6:e24671. doi: 10.1371/journal.pone.0024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hessmann E, Buchholz SM, Demir IE, Singh SK, Gress TM, Ellenrieder V, Neesse A. Microenvironmental Determinants of Pancreatic Cancer. Physiol Rev. 2020;100:1707–1751. doi: 10.1152/physrev.00042.2019. [DOI] [PubMed] [Google Scholar]
- 112.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, Thaiss CA, Reuben A, Livny J, Avraham R, Frederick DT, Ligorio M, Chatman K, Johnston SE, Mosher CM, Brandis A, Fuks G, Gurbatri C, Gopalakrishnan V, Kim M, Hurd MW, Katz M, Fleming J, Maitra A, Smith DA, Skalak M, Bu J, Michaud M, Trauger SA, Barshack I, Golan T, Sandbank J, Flaherty KT, Mandinova A, Garrett WS, Thayer SP, Ferrone CR, Huttenhower C, Bhatia SN, Gevers D, Wargo JA, Golub TR, Straussman R. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thomas RM, Gharaibeh RZ, Gauthier J, Beveridge M, Pope JL, Guijarro MV, Yu Q, He Z, Ohland C, Newsome R, Trevino J, Hughes SJ, Reinhard M, Winglee K, Fodor AA, Zajac-Kaye M, Jobin C. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis. 2018;39:1068–1078. doi: 10.1093/carcin/bgy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, Yang G, You L, Yang J, Feng M, Qiu J, Zhao F, Liu Y, Cao Z, Zheng L, Zhang T, Zhao Y. Role of the microbiome in occurrence, development and treatment of pancreatic cancer. Mol Cancer. 2019;18:173. doi: 10.1186/s12943-019-1103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ghaddar B, Biswas A, Harris C, Omary MB, Carpizo DR, Blaser MJ, De S. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell. 2022;40:1240–1253.e5. doi: 10.1016/j.ccell.2022.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, McCullagh J, Sansom SN, Arancibia-Cárcamo CV, Uhlig HH, Powrie F. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity. 2019;50:432–445.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol. 2015;33:445–474. doi: 10.1146/annurev-immunol-032414-112043. [DOI] [PubMed] [Google Scholar]
- 118.Lankadasari MB, Mukhopadhyay P, Mohammed S, Harikumar KB. TAMing pancreatic cancer: combat with a double edged sword. Mol Cancer. 2019;18:48. doi: 10.1186/s12943-019-0966-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lecoultre M, Dutoit V, Walker PR. Phagocytic function of tumor-associated macrophages as a key determinant of tumor progression control: a review. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, Werba G, Zhang K, Guo Y, Li Q, Akkad N, Lall S, Wadowski B, Gutierrez J, Kochen Rossi JA, Herzog JW, Diskin B, Torres-Hernandez A, Leinwand J, Wang W, Taunk PS, Savadkar S, Janal M, Saxena A, Li X, Cohen D, Sartor RB, Saxena D, Miller G. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018;8:403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.al-Waiz M, Mikov M, Mitchell SC, Smith RL. The exogenous origin of trimethylamine in the mouse. Metabolism. 1992;41:135–136. doi: 10.1016/0026-0495(92)90140-6. [DOI] [PubMed] [Google Scholar]
- 122.Mirji G, Worth A, Bhat SA, El Sayed M, Kannan T, Goldman AR, Tang HY, Liu Q, Auslander N, Dang CV, Abdel-Mohsen M, Kossenkov A, Stanger BZ, Shinde RS. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci Immunol. 2022;7:eabn0704. doi: 10.1126/sciimmunol.abn0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, Zhang Y, Gomez EC, Morreale B, Senchanthisai S, Li J, Turowski SG, Sexton S, Sait SJ, Singh PK, Wang J, Maitra A, Kalinski P, DePinho RA, Wang H, Liao W, Abrams SI, Segal BH, Dey P. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. 2022;40:153–167.e11. doi: 10.1016/j.ccell.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun X, He X, Zhang Y, Hosaka K, Andersson P, Wu J, Jing X, Du Q, Hui X, Ding B, Guo Z, Hong A, Liu X, Wang Y, Ji Q, Beyaert R, Yang Y, Li Q, Cao Y. Inflammatory cell-derived CXCL3 promotes pancreatic cancer metastasis through a novel myofibroblast-hijacked cancer escape mechanism. Gut. 2022;71:129–147. doi: 10.1136/gutjnl-2020-322744. [DOI] [PubMed] [Google Scholar]
- 125.Mahajan UM, Langhoff E, Goni E, Costello E, Greenhalf W, Halloran C, Ormanns S, Kruger S, Boeck S, Ribback S, Beyer G, Dombroswki F, Weiss FU, Neoptolemos JP, Werner J, D'Haese JG, Bazhin A, Peterhansl J, Pichlmeier S, Büchler MW, Kleeff J, Ganeh P, Sendler M, Palmer DH, Kohlmann T, Rad R, Regel I, Lerch MM, Mayerle J. Immune Cell and Stromal Signature Associated With Progression-Free Survival of Patients With Resected Pancreatic Ductal Adenocarcinoma. Gastroenterology. 2018;155:1625–1639.e2. doi: 10.1053/j.gastro.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 126.Liu X, Xu J, Zhang B, Liu J, Liang C, Meng Q, Hua J, Yu X, Shi S. The reciprocal regulation between host tissue and immune cells in pancreatic ductal adenocarcinoma: new insights and therapeutic implications. Mol Cancer. 2019;18:184. doi: 10.1186/s12943-019-1117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dey P, Li J, Zhang J, Chaurasiya S, Strom A, Wang H, Liao WT, Cavallaro F, Denz P, Bernard V, Yen EY, Genovese G, Gulhati P, Liu J, Chakravarti D, Deng P, Zhang T, Carbone F, Chang Q, Ying H, Shang X, Spring DJ, Ghosh B, Putluri N, Maitra A, Wang YA, DePinho RA. Oncogenic KRAS-Driven Metabolic Reprogramming in Pancreatic Cancer Cells Utilizes Cytokines from the Tumor Microenvironment. Cancer Discov. 2020;10:608–625. doi: 10.1158/2159-8290.CD-19-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moral JA, Leung J, Rojas LA, Ruan J, Zhao J, Sethna Z, Ramnarain A, Gasmi B, Gururajan M, Redmond D, Askan G, Bhanot U, Elyada E, Park Y, Tuveson DA, Gönen M, Leach SD, Wolchok JD, DeMatteo RP, Merghoub T, Balachandran VP. ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity. Nature. 2020;579:130–135. doi: 10.1038/s41586-020-2015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mohindroo C, Hasanov M, Rogers JE, Dong W, Prakash LR, Baydogan S, Mizrahi JD, Overman MJ, Varadhachary GR, Wolff RA, Javle MM, Fogelman DR, Lotze MT, Kim MP, Katz MHG, Pant S, Tzeng CD, McAllister F. Antibiotic use influences outcomes in advanced pancreatic adenocarcinoma patients. Cancer Med. 2021;10:5041–5050. doi: 10.1002/cam4.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, Guo Y, Saxena A, Vardhan M, Diskin B, Wang W, Leinwand J, Kurz E, Kochen Rossi JA, Hundeyin M, Zambrinis C, Li X, Saxena D, Miller G. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574:264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weniger M, Hank T, Qadan M, Ciprani D, Michelakos T, Niess H, Heiliger C, Ilmer M, D'Haese JG, Ferrone CR, Warshaw AL, Lillemoe KD, Werner J, Liss A, Fernández-Del Castillo C. Influence of Klebsiella pneumoniae and quinolone treatment on prognosis in patients with pancreatic cancer. Br J Surg. 2021;108:709–716. doi: 10.1002/bjs.12003. [DOI] [PubMed] [Google Scholar]
- 133.Xu H, Cao C, Ren Y, Weng S, Liu L, Guo C, Wang L, Han X, Ren J, Liu Z. Antitumor effects of fecal microbiota transplantation: Implications for microbiome modulation in cancer treatment. Front Immunol. 2022;13:949490. doi: 10.3389/fimmu.2022.949490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rezasoltani S, Yadegar A, Asadzadeh Aghdaei H, Reza Zali M. Modulatory effects of gut microbiome in cancer immunotherapy: A novel paradigm for blockade of immune checkpoint inhibitors. Cancer Med. 2021;10:1141–1154. doi: 10.1002/cam4.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mao J, Wang D, Long J, Yang X, Lin J, Song Y, Xie F, Xun Z, Wang Y, Li Y, Sun H, Xue J, Zuo B, Zhang J, Bian J, Zhang T, Zhang L, Sang X, Zhao H. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J Immunother Cancer. 2021;9 doi: 10.1136/jitc-2021-003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mima K, Nakagawa S, Sawayama H, Ishimoto T, Imai K, Iwatsuki M, Hashimoto D, Baba Y, Yamashita YI, Yoshida N, Chikamoto A, Baba H. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 2017;402:9–15. doi: 10.1016/j.canlet.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 138.Wang T, Zhang L, Wang P, Liu Y, Wang G, Shan Y, Yi Y, Zhou Y, Liu B, Wang X, Lü X. Lactobacillus coryniformis MXJ32 administration ameliorates azoxymethane/dextran sulfate sodium-induced colitis-associated colorectal cancer via reshaping intestinal microenvironment and alleviating inflammatory response. Eur J Nutr. 2022;61:85–99. doi: 10.1007/s00394-021-02627-8. [DOI] [PubMed] [Google Scholar]
- 139.Wang T, Zheng J, Dong S, Ismael M, Shan Y, Wang X, Lü X. Lacticaseibacillus rhamnosus LS8 Ameliorates Azoxymethane/Dextran Sulfate Sodium-Induced Colitis-Associated Tumorigenesis in Mice via Regulating Gut Microbiota and Inhibiting Inflammation. Probiotics Antimicrob Proteins. 2022;14:947–959. doi: 10.1007/s12602-022-09967-9. [DOI] [PubMed] [Google Scholar]
- 140.Heydari Z, Rahaie M, Alizadeh AM, Agah S, Khalighfard S, Bahmani S. Effects of Lactobacillus acidophilus and Bifidobacterium bifidum Probiotics on the Expression of MicroRNAs 135b, 26b, 18a and 155, and Their Involving Genes in Mice Colon Cancer. Probiotics Antimicrob Proteins. 2019;11:1155–1162. doi: 10.1007/s12602-018-9478-8. [DOI] [PubMed] [Google Scholar]
- 141.Konishi H, Isozaki S, Kashima S, Moriichi K, Ichikawa S, Yamamoto K, Yamamura C, Ando K, Ueno N, Akutsu H, Ogawa N, Fujiya M. Probiotic Aspergillus oryzae produces anti-tumor mediator and exerts anti-tumor effects in pancreatic cancer through the p38 MAPK signaling pathway. Sci Rep. 2021;11:11070. doi: 10.1038/s41598-021-90707-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mendes MCS, Paulino DS, Brambilla SR, Camargo JA, Persinoti GF, Carvalheira JBC. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J Gastroenterol. 2018;24:1995–2008. doi: 10.3748/wjg.v24.i18.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Ben-Zeev Brik R, Federici S, Horn M, Cohen Y, Moor AE, Zeevi D, Korem T, Kotler E, Harmelin A, Itzkovitz S, Maharshak N, Shibolet O, Pevsner-Fischer M, Shapiro H, Sharon I, Halpern Z, Segal E, Elinav E. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell. 2018;174:1406–1423.e16. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 144.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 145.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Si W, Liang H, Bugno J, Xu Q, Ding X, Yang K, Fu Y, Weichselbaum RR, Zhao X, Wang L. Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade. Gut. 2022;71:521–533. doi: 10.1136/gutjnl-2020-323426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang SL, Mao YQ, Zhang ZY, Li ZM, Kong CY, Chen HL, Cai PR, Han B, Ye T, Wang LS. Pectin supplement significantly enhanced the anti-PD-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics. 2021;11:4155–4170. doi: 10.7150/thno.54476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and Anticancer Immunosurveillance. Cell. 2016;165:276–287. doi: 10.1016/j.cell.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 149.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huang XZ, Gao P, Song YX, Xu Y, Sun JX, Chen XW, Zhao JH, Wang ZN. Antibiotic use and the efficacy of immune checkpoint inhibitors in cancer patients: a pooled analysis of 2740 cancer patients. Oncoimmunology. 2019;8:e1665973. doi: 10.1080/2162402X.2019.1665973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wilson BE, Routy B, Nagrial A, Chin VT. The effect of antibiotics on clinical outcomes in immune-checkpoint blockade: a systematic review and meta-analysis of observational studies. Cancer Immunol Immunother. 2020;69:343–354. doi: 10.1007/s00262-019-02453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu Q, Liu J, Wu S, Xie X. The impact of antibiotics on efficacy of immune checkpoint inhibitors in malignancies: A study based on 44 cohorts. Int Immunopharmacol. 2021;92:107303. doi: 10.1016/j.intimp.2020.107303. [DOI] [PubMed] [Google Scholar]
- 153.Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, Lewis IA, Geuking MB, McCoy KD. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 154.Han J, Zhang S, Xu Y, Pang Y, Zhang X, Hu Y, Chen H, Chen W, Zhang J, He W. Beneficial Effect of Antibiotics and Microbial Metabolites on Expanded Vδ2Vγ9 T Cells in Hepatocellular Carcinoma Immunotherapy. Front Immunol. 2020;11:1380. doi: 10.3389/fimmu.2020.01380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang L, Chen C, Chai D, Li C, Guan Y, Liu L, Kuang T, Deng W, Wang W. The association between antibiotic use and outcomes of HCC patients treated with immune checkpoint inhibitors. Front Immunol. 2022;13:956533. doi: 10.3389/fimmu.2022.956533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, Wang Z, Wang C, Zhang Z, Xia W, Chen Z, Wang K, Zhang T, Xu J, Han Y, Wu X, Wang J, Gong W, Zheng S, Qiu F, Yan J, Huang J. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity. 2014;40:785–800. doi: 10.1016/j.immuni.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Daley D, Zambirinis CP, Seifert L, Akkad N, Mohan N, Werba G, Barilla R, Torres-Hernandez A, Hundeyin M, Mani VRK, Avanzi A, Tippens D, Narayanan R, Jang JE, Newman E, Pillarisetty VG, Dustin ML, Bar-Sagi D, Hajdu C, Miller G. γδ T Cells Support Pancreatic Oncogenesis by Restraining αβ T Cell Activation. Cell. 2016;166:1485–1499.e15. doi: 10.1016/j.cell.2016.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Huang J, Zheng X, Kang W, Hao H, Mao Y, Zhang H, Chen Y, Tan Y, He Y, Zhao W, Yin Y. Metagenomic and metabolomic analyses reveal synergistic effects of fecal microbiota transplantation and anti-PD-1 therapy on treating colorectal cancer. Front Immunol. 2022;13:874922. doi: 10.3389/fimmu.2022.874922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koustas E, Trifylli EM, Sarantis P, Papadopoulos N, Aloizos G, Tsagarakis A, Damaskos C, Garmpis N, Garmpi A, Papavassiliou AG, Karamouzis MV. Implication of gut microbiome in immunotherapy for colorectal cancer. World J Gastrointest Oncol. 2022;14:1665–1674. doi: 10.4251/wjgo.v14.i9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nomura M, Nagatomo R, Doi K, Shimizu J, Baba K, Saito T, Matsumoto S, Inoue K, Muto M. Association of Short-Chain Fatty Acids in the Gut Microbiome With Clinical Response to Treatment With Nivolumab or Pembrolizumab in Patients With Solid Cancer Tumors. JAMA Netw Open. 2020;3:e202895. doi: 10.1001/jamanetworkopen.2020.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, Jiang W, Cai S, Zhao P, Song R, Li P, Qin N, Fang W. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7:193. doi: 10.1186/s40425-019-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]