Abstract
Tuberculosis and sarcoidosis are inflammatory diseases characterized by granulomas that may occur in any organ but are often found in the lung. The panoply of classical HLA alleles associated with occurrence and/or severity of both diseases varies considerably across studies. This heterogeneity of results, due to variation in factors like ancestry and disease sub-phenotype, as well as the use of simple modeling strategies to elucidate likely complex relationships, has made conclusions about underlying commonalities difficult. Here we perform HLA association analyses in individuals of African ancestry, using a greater resolution to include sub-phenotypes of disease and employing more comprehensive analytical techniques. Using a novel application of nearest-neighbor feature selection to score allelic importance, we investigated HLA allele association with Mycobacterium tuberculosis exposure outcomes in the first analysis of both latent Mycobacterium tuberculosis infection and active disease compared to those who, despite long term exposure to active index cases, have neither positive diagnostic tests nor display clinical symptoms. We also compared persistent to resolved sarcoidosis. This led to identification of novel HLA associations and evidence of main effects and interaction effects. We found strikingly similar main effects and interaction effects at HLA-DRB1, -DQB1, and -DPB1 in those resistant to tuberculosis (either latent or active) and persistent sarcoidosis.
Keywords: HLA, Mycobacterium tuberculosis, sarcoidosis, nearest-neighbor feature selection, epistasis, resistance to infection
Introduction
Active tuberculosis disease is consistently among the most deadly infectious diseases each year and incidence of disease in African countries is among the highest worldwide (Global tuberculosis report, 2020). Tuberculosis disease is caused by Mycobacterium tuberculosis infection and is characterized by caseating granulomas. Although most instances of Mycobacterium tuberculosis exposure result in latent Mycobacterium tuberculosis infection or clinically active disease, some individuals, called ‘Resisters’, maintain negative diagnostic tests for Mycobacterium tuberculosis infection over many years of follow up despite long-term exposure to index cases with active disease (Stein et al., 2019, Stein et al., 2018). Sarcoidosis, also a granulomatous disease, is characterized by non-caseating granulomas and thought to be the host immune response to unknown antigen(s) (Iannuzzi, 2007) in individuals with a genetic predisposition. Phenotypes in sarcoidosis are also highly heterogeneous, ranging from complete resolution, with or without medical intervention, to persistent and even fibrotic disease (Baughman et al., 2009). In the United States, incidence of sarcoidosis is the highest (Rybicki et al., 1997, Mirsaeidi et al., 2015) and the clinical manifestations are more severe (Israel et al., 1986, Mirsaeidi et al., 2015) in African Americans as compared to other ancestral groups. Myriad infectious agents have been examined for a potential role in causing sarcoidosis (Esteves et al., 2016, Gupta et al., 2007, Zhou et al., 2013), with Mycobacterium tuberculosis being one of the most extensively studied, however, the etiology of sarcoidosis remains elusive. Regardless, by studying them jointly, much can be learned about both diseases due to their highly similar clinical manifestations.
Variants within the human leukocyte antigen (HLA) genomic region are associated with both active tuberculosis disease (Tong et al., 2015, Yang et al., 2016) and sarcoidosis (Fingerlin et al., 2015). The class II HLA loci: DRB1, DQB1, and DPB1, are of particular concern due to their central role in antigen recognition and presentation to helper T cells via antigen presenting cells, which is the initial mechanism leading to granuloma formation and persistence that characterizes both tuberculosis disease and sarcoidosis (Grunewald et al., 2019). A recent review summarizes and compares HLA associations in the two diseases (Malkova et al., 2020), making two broad conclusions: (1) DRB1*03, *07, and *15 predispose for sarcoidosis and protect against tuberculosis and (2) DRB1*04 predisposes for tuberculosis disease and protects against sarcoidosis. These comparisons are representative of one major limitation; to date, there are no comparisons of HLA associations in Mycobacterium tuberculosis exposure outcomes and sarcoidosis that include sub-phenotypes of disease, such as latent Mycobacterium tuberculosis infection or persistent sarcoidosis. Further, no studies report HLA associations with ‘Resisters’ (Stein et al., 2019, Stein et al., 2018). Discovering associations of class II HLA loci to all outcomes related to Mycobacterium tuberculosis exposure and sarcoidosis, could provide insight into potential mechanisms for long-term resistance against conversion to positive diagnostic tests.
Finally, the few existing HLA association studies in individuals of African ancestry indicate considerable deviation from the effects in other groups, the majority of which were identified in individuals of European or Asian populations (Fingerlin et al., 2015, Tong et al., 2015, Yang et al., 2016, Malkova et al., 2020). The present study probed for HLA associations with outcomes of Mycobacterium tuberculosis exposure in individuals from a densely populated region of Kampala, Uganda (Stein et al., 2019, Stein et al., 2018, McHenry et al., 2021), whereas associations with sarcoidosis outcomes were assessed in Americans of African ancestry (Baughman et al., 2001, ACCESS Research Group 1999, Rybicki et al., 2007). Uganda is a high-burden country for tuberculosis disease, with a recently estimated incidence of 196 per 100,000 (Global tuberculosis report, 2020); this is almost surely higher in the populous region of Kampala, with a recent estimate of 740 per 100,000 (Stein et al., 2018). In the United States, incidence of sarcoidosis in African Americans is recently estimated at 17.8 per 100,000; this incidence is more than double that of European Americans (8.1 per 100,000), more than four times that of Hispanics (4.3 per 100,000), and more than five times that of Asians (3.2 per 100,000) (Baughman et al., 2016).
Using a method called Nearest-neighbor Projected-Distance Regression (NPDR) that is designed for detecting complex associations in high-dimensional bioinformatic data (Le et al., 2020), we examined high-resolution HLA-DRB1, -DQB1, and -DPB1 alleles across distinct sub-phenotypes of Mycobacterium tuberculosis exposure and sarcoidosis in individuals of African ancestry, seeking to identify novel main effect and epistatic associations both unique to and common across diseases. These findings can help guide mechanistic hypotheses explaining the highly variable responses to these two clinically similar but etiologically distinct granulomatous disorders.
Materials and methods
Subjects and phenotype definitions.
Our Mycobacterium tuberculosis (Mtb) exposure outcome cohort derived from individuals participating in a household-contact study in an urban region of Kampala, Uganda (Stein et al., 2019, Stein et al., 2018, McHenry et al., 2021). Individuals living with a confirmed, culture positive, clinically active tuberculosis case at enrollment were screened with tuberculin skin tests (TSTs) performed every 3 months for the first 2 years to identify individuals with latent Mtb infection and ‘Resisters’ (Stein et al., 2018). Latent Mtb infection was defined as individuals who never converted to active disease in spite of positive tuberculin skin tests, while Resisters were individuals who consistently tested negative by both TSTs and interferon-gamma release assays (IGRA) over an average of 11 years (Stein et al., 2019, Stein et al., 2018). The current study focuses on three sub-phenotypes of Mtb exposure, including active tuberculosis disease (n=202), latent Mtb infection (n=228), and Resisters (n=87). Approximately 64% of the individuals were vaccinated with the bacille Calmette-Guérin (BCG) vaccine, and proportions of vaccinated individuals did not differ between groups (Stein et al., 2019, Stein et al., 2018). Our sarcoidosis data derive from family and case-control studies of African American individuals with sarcoidosis (n=664), confirmed by presence of granulomas in biopsy tissue and absence of mycobacterial or fungal infection. The healthy controls were family members and unrelated individuals matched to the cases for age, sex, and race (n=518) (Baughman et al., 2001, ACCESS Research Group 1999, Rybicki et al., 2007). Persistent (n=343) or resolved (n=134) sarcoidosis status was determined by radiographic evidence of continued lung involvement at 2-year follow-up.
Genotyping.
Genome-wide genotyping for the Mtb exposure outcome cohort is reported in a GWA study (McHenry et al., 2021). Briefly, active cases from the original 2-year study (Stein et al., 2018) and those with latent Mtb infection and Resisters in the long-term follow up study (Stein et al., 2019) were typed from a combination of the Illumina MEGAEX and Illumina Omni5 chips. Genome-wide genotyping for our sarcoidosis outcome data is reported in another GWA study (Adrianto et al., 2012). Genotyping for sarcoidosis cases and healthy controls was performed using the Illumina HumanOmni1-Quad chip (Adrianto et al., 2012).
Identification of HLA alleles.
In our Mtb exposure dataset, HLA-DRB1, -DQB1, and -DPB1 alleles were identified via deep sequencing for 37 individuals (Wang et al., 2012), while those for the remaining 728 individuals were imputed with HLA Genotype Imputation with Attribute Bagging (HIBAG) (Zheng et al., 2014) from genome-wide SNP data mentioned above. In our sarcoidosis dataset, HLA-DRB1, -DQB1, and -DPB1 alleles were identified via PCR amplification and hybridization with sequence specific oligonucleotide (PCR-SSO) probes in 325 individuals who also had genome-wide genotyping (ACCESS Research Group 1999, Rossman et al., 2003). These 325 individuals were used as reference for imputing alleles in the remaining 857 individuals with HIBAG using the genome-wide SNP data mentioned above. Accuracy of the HIBAG method specific to our sarcoidosis dataset has previously been reported (Levin et al., 2015, Levin et al., 2014). For our Mtb exposure dataset, we implemented HIBAG using pre-built classifiers with published parameter estimates, trained on samples of African ancestry (Zheng et al., 2014), to predict HLA-types from genome-wide genotypes (McHenry et al., 2021, Stein et al., 2019, Stein et al., 2018). Prior to imputation, we filtered out all SNPs with missing call rate in excess of 0.01 and those with minor allele frequency less than 0.05.
HLA allelic importance.
To identify the HLA-DRB1, -DQB1, and -DPB1 alleles that best delineate disease groups, we applied a machine learning algorithm that scores the importance of a predictor variable in the presence of covariates (Nearest-neighbor Projected-Distance Regression, NPDR) (Le et al., 2020). Importance scores were adjusted for background relatedness and imputation uncertainty. For background relatedness adjustment, we used the first 10 principal components of the genetic relationship matrix for each cohort. We adjusted for imputation uncertainty using HIBAG posterior probabilities, assigning all unimputed HLA alleles a probability of 1. HLA alleles with counts less than 5 in both phenotype groups for a given outcome were excluded prior to determining importance scores. NPDR p-values were adjusted for multiple testing by Bonferroni correction.
Nearest-neighbor feature selection is usually applied with only one neighborhood method (Urbanowicz et al., 2018), however using simulated data it has been shown that the ability of a nearest-neighbor feature selection method to detect main effects and/or interaction effects changes as a function of neighborhood size (Dawkins et al., 2021, McKinney et al., 2013, Urbanowicz et al., 2018). Here NPDR importance scores were calculated using three different neighborhood methods: (1) an adaptive-radius method (Urbanowicz et al., 2018) that allows each instance (or sample) to have its own neighborhood radius, (2) a fixed-k method (Dawkins et al., 2021, Le et al., 2020) that takes into account moments of the sample distance distribution, and (3) a variable-wise optimized fixed-k method (McKinney et al., 2013) that allows each allele to have its own optimized neighborhood size (or fixed-k). Only those alleles with statistically significant adjusted NPDR p-values for all three methods (termed here as “consensus positives”) were considered relevant for classifying a particular outcome. The top 15 associations, ranked according to NPDR consensus importance, are described below and the remainder are included in supplementary materials (Figure S1).
Simulated data.
We used simulated case-control data to compare the performance of our NPDR consensus method to each of the three input methods, as well as univariate logistic regression (Figure S2). Main effect and interaction effect simulations were generated using the createSimulation() function from a software package called private Evaporative Cooling (privateEC) (Le et al., 2017). Main effect simulations were based on an approach that uses a linear model to generate data characteristic of bioinformatic applications (Leek and Storey, 2007). Interaction effect simulations were based on a method that generates differential correlation from a random network with Erdős-Rényi degree distribution (Lareau et al., 2015). We generated 30 replicate simulations for assessing algorithm performance in detecting main effects and interaction effects, respectively. Each simulated dataset had m = 50 samples and p = 1000 features with 10% functional (i.e., relevant to the outcome). For main effect simulations, we set the effect size parameter (bias) of createSimulation() to 0.8 which corresponds to statistical power of about 40% (Le et al., 2017). For interaction effects, bias was set to 0.4 and is inversely proportional to the level of correlation between functional features (i.e., features involved in interaction effects relevant to the outcome) in the ‘control’ group. In the ‘case’ group, correlation between functional features is disrupted through random permutation (Lareau et al., 2015).
The Area Under the Precision-Recall Curve (AUPRC), precision, and recall were chosen as performance metrics for comparing different feature selection algorithms. In order to calculate each performance metric, we defined “positives” as features that were functionally related (i.e. relevant) to the binary outcome. Thus, a true positive (TP) is a relevant feature that was detected by a given feature selection algorithm. AUPRC was derived by calculated precision and recall across a grid of increasing feature selection thresholds. We also calculated precision and recall using a p-value threshold of nominal significance (P < 0.05) for each feature selection algorithm. We found that NPDR consensus positives had higher precision overall than NPDR positives among the three individual neighborhood methods, respectively (Figure S2 A). We also compared performance between our NPDR consensus approach and univariate logistic regression with respect to detecting simulated main effects and interaction effects (Figure S2 B). While univariate logistic regression had an advantage in detecting main effects, it completely failed in detecting interaction effects (Figure S2 B). To the contrary, our NPDR consensus approach allowed for detection of both main effects and interaction effects, significantly improving overall detection of functional features (Figure S2 B).
Regression analysis.
Only those outcomes having at least one allele with statistically significant NPDR importance were included in follow up regression analysis. We used a generalized linear model to determine odds ratios for each HLA allele for which NPDR importance was statistically significant (Tables S1 – S2). For each HLA allele detected by NPDR in at least one outcome of Mtb exposure or sarcoidosis, respectively, we calculated odds ratios for all possible two-way interactions for which there was sufficient data, excluding combinations of alleles that were observed in fewer than 5 individuals (Tables S3 – S4). We report only nominally significant (P < 0.05) interactions involving at least one allele detected by NPDR, possibly including alleles for which NPDR importance was not statistically significant; for visualization purposes only, odds ratios were calculated in this latter scenario. Each model included the first 10 principal components of the genetic relationship matrix and HIBAG posterior probabilities for the corresponding HLA loci. We used a dominant model assumption for all main effect and interaction effect odds ratios. Testing for the presence of interaction was done using a likelihood ratio test. We also calculated the standardized difference of proportions for each HLA allele in order to illustrate NPDR allelic importance and the corresponding unadjusted directionality in disease risk. We visualized main effects and interaction effects in a regression-based Genetic Association Interaction Network (reGAIN) (Lareau et al., 2015, McKinney et al., 2009, Pandey et al., 2012), showing pairwise interactions reaching nominal significance and corresponding main effects in the absence of any interaction term (Figures S3 – S7).
Results
In order to determine which HLA alleles contributed most significantly to outcomes in Mtb exposure and sarcoidosis, we used NPDR to calculate allelic importance scores (Figure 1A – E). Larger importance scores correspond to stronger relationships between alleles and the log-odds of two individuals having a different phenotype. Thus, alleles with a higher rank of importance can be thought of as being better for classification of a given outcome, or sub-phenotype comparison, regardless of directionality of disease risk. Main effects (Tables S1 – S2) and interaction effects (Tables S3 – S4) were determined to annotate allelic importance with directionality for odds of disease.
Figure 1. HLA allelic importance in Mtb exposure and sarcoidosis outcomes.
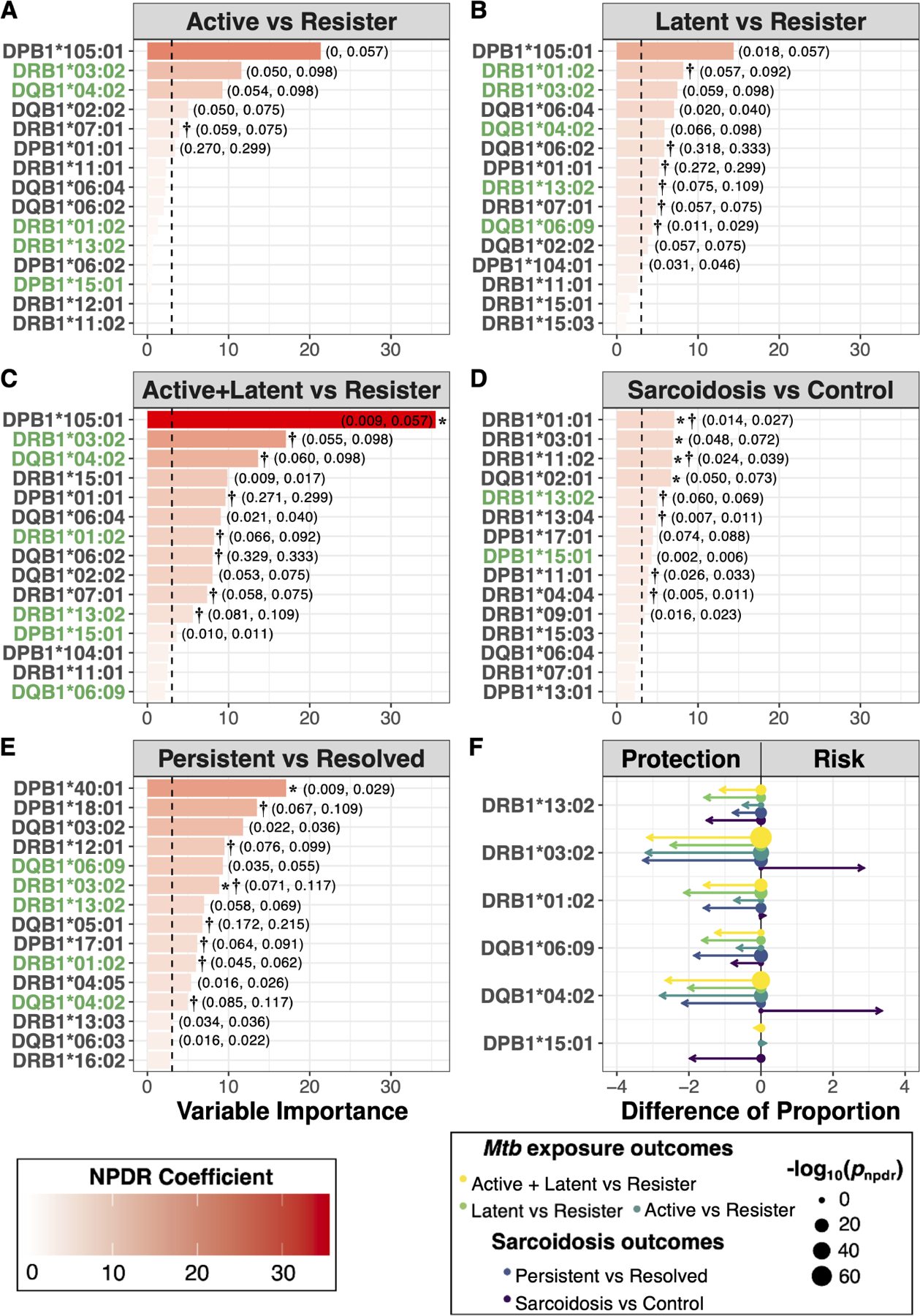
(A) – (E) Top 15 HLA alleles, ranked in decreasing magnitude of NPDR standardized regression coefficient – i.e. variable importance, are shown for each outcome. Mtb exposure outcomes: (A) Active (n=202) vs Resister (n=87), (B) Latent (n=228) vs Resister (n=87), (C) Active + Latent (n=430) vs Resister (n=87). Sarcoidosis outcomes: (D) Sarcoidosis (n=664) vs Control (n=518) and (E) Persistent (n=343) vs Resolved (n=134). Vertical-dashed line is the standard normal quantile corresponding to the NPDR p-value significance threshold for each respective outcome (A) – (E). HLA alleles with significant adjusted NPDR p-values in at least one Mtb exposure and sarcoidosis outcome are highlighted in green (A) – (E). For these overlapping HLA alleles, the standardized difference of proportion and −log10 NPDR p-value are shown (F). Arrow length is proportional to the magnitude difference in allele proportions between phenotype groups. Arrows pointing left correspond to alleles more common in the “control” group, while arrows pointing right correspond to alleles more common in the “case” group. Point sizes are proportional to the −log10 NPDR p-value. Alleles with nominally significant (P < 0.05) regression coefficients are labeled: main effects (*) and interaction effects (†). Allele frequencies are provided in parentheses for significant alleles (case frequency, control frequency).
HLA-DRB1, -DQB1, and -DPB1 associations in Mtb exposure outcomes.
There were several alleles important for distinguishing those with active disease (Figure 1A), latent Mtb infection (Figure 1B), and the combined group of active disease or latent Mtb infection (Figure 1C) from Resisters. We found no alleles distinguishing those with active disease from those with latent Mtb infection, thus this contrast was excluded from further regression analysis. When comparing the combined group of active disease or latent Mtb infection to Resisters, two alleles were associated with decreased risk: DQB1*02:02 and DPB1*105:01 (Table S1). Most of the HLA alleles with significant importance were involved in pairwise interactions (Table S3), not main effects. DPB1*01:01 was ranked among the highest in terms of importance for Mtb exposure outcomes (Figure 1A – C), and coupled with DPB1*04:01, had the strongest interaction associated with increased risk when comparing those with latent Mtb infection to Resisters (Table S3). This same effect was observed when comparing the combined group of active disease and latent Mtb infection to Resisters (Table S3). DRB1*03:02 was also among those with the highest importance scores, and, coupled with DQB1*02:01, had the strongest interaction associated with decreased risk when comparing the combined group of active disease and latent Mtb infection to Resisters (Table S3). There were several other nominally significant (P < 0.05) interactions, predominantly associated with decreased risk (Table S3).
HLA-DRB1, -DQB1, and -DPB1 associations in sarcoidosis outcomes.
We confirmed several previously reported associations for our sarcoidosis cohort (Levin et al., 2015, Iannuzzi et al., 2003). These included an association with increased risk of sarcoidosis: DRB1*03:02 (Table S2). Also among these were associations with decreased risk of sarcoidosis onset: DRB1*01:01, DRB1*03:01, DRB1*09:01, DRB1*13:04, and DQB1*02:01 (Table S2). Finally, we confirmed an association with decreased risk of developing persistent disease for DRB1*03:02 (Table S2). In addition to confirming known associations, we found several novel associations with sarcoidosis outcomes for this cohort (Figure 1D – E). Among these, DRB1*11:02 was associated with decreased risk of sarcoidosis onset, while DPB1*40:01 was associated with decreased risk for persistent sarcoidosis (Table S2). Most of the HLA alleles with significant importance scores were involved in pairwise interactions (Table S4). DRB1*04:04 was ranked among the highest in terms of importance for the comparison of sarcoidosis cases with healthy controls; in combination with DPB1*03:01, it had one of the strongest interaction effects associated with increased risk of sarcoidosis onset (Table S4). Other highly ranked alleles for the comparison of sarcoidosis cases and healthy controls had nominally significant (P < 0.05) interactions, predominantly associated with increased risk of onset (Table S4). DRB1*12:01 and DPB1*17:01 were ranked among the highest in terms of importance for the comparison of persistent and resolved sarcoidosis; interactions between DRB1*12:01 and DRB1*11:01 and between DPB1*17:01 and DRB1*03:01 had the strongest associations with decreased risk for persistent disease (Table S4). DRB1*01:02 was also ranked among the highest for the comparison of persistent and resolved sarcoidosis, and coupled with DPB1*01:01, had the strongest interaction associated with increased risk for persistent disease (Table S4). There were several other nominally significant interactions (P < 0.05) involving highly ranked alleles for the comparison of persistent and resolved sarcoidosis as well (Table S4).
Shared HLA importance and directionality between Resister phenotype of Mtb exposure and resolved sarcoidosis.
Alleles that were important for distinguishing those with active tuberculosis disease or latent Mtb infection from Resisters (Figure 1A – C), tended to also be important for distinguishing persistent from resolved sarcoidosis (Figure 1E); there was less overlap with the comparison of sarcoidosis cases versus controls (Figure 1D). Overall, there were six alleles of overlapping importance between Mtb exposure and sarcoidosis outcomes (Figure 1F): DRB1*01:02, DRB1*03:02, DRB1*13:02, DQB1*04:02, DQB1*06:09, and DPB1*15:01. With one exception, these alleles were found in higher proportion among Resisters and those with resolved sarcoidosis, relative to their counterpart comparison phenotypes. On the contrary, two of these resistance-associated alleles were more frequent in sarcoidosis cases than healthy controls: DRB1*03:02 and DQB1*04:02; this was also true after covariate adjustment (Table S2).
Interaction networks reveal high-order similarities between Resister phenotype of Mtb exposure and resolved sarcoidosis.
As mentioned above, there were many nominally significant interaction effects involving alleles with significant importance scores for outcomes of Mtb exposure (Table S3) and sarcoidosis (Table S4). In order to make a comprehensive, high-order comparison of the interactions between different outcomes, we created regression-based Genetic Association Interaction Networks (reGAIN) for each outcome of Mtb exposure (Figures S3 – S5) and sarcoidosis (Figures S6 – S7). Each node of a given network represents a single HLA allele and the edges (or connections) between nodes represent interactions, which we only allowed if the interaction was at least nominally (P < 0.05) significant. By comparing reGAINs involving HLA alleles with significant importance scores, we found many similar interaction subnetworks between outcomes of Mtb exposure and sarcoidosis (Figure 2). Four subnetworks were highly concordant for two outcomes in particular: (1) persistent versus resolved sarcoidosis and (2) the combined active tuberculosis disease or latent Mtb infection versus Resister. In all but the comparison of active tuberculosis disease versus Resister, we found interactions between DPB1*02:01 and DQB1*06, primarily DQB1*06:09. Finally, we found that an interaction between DPB1*01:01 and DQB1*03 was shared in the comparison of sarcoidosis case versus control, as well as comparing active tuberculosis disease or latent Mtb infection to Resister.
Figure 2. Interaction subnetwork similarities for Mtb exposure and sarcoidosis outcomes.
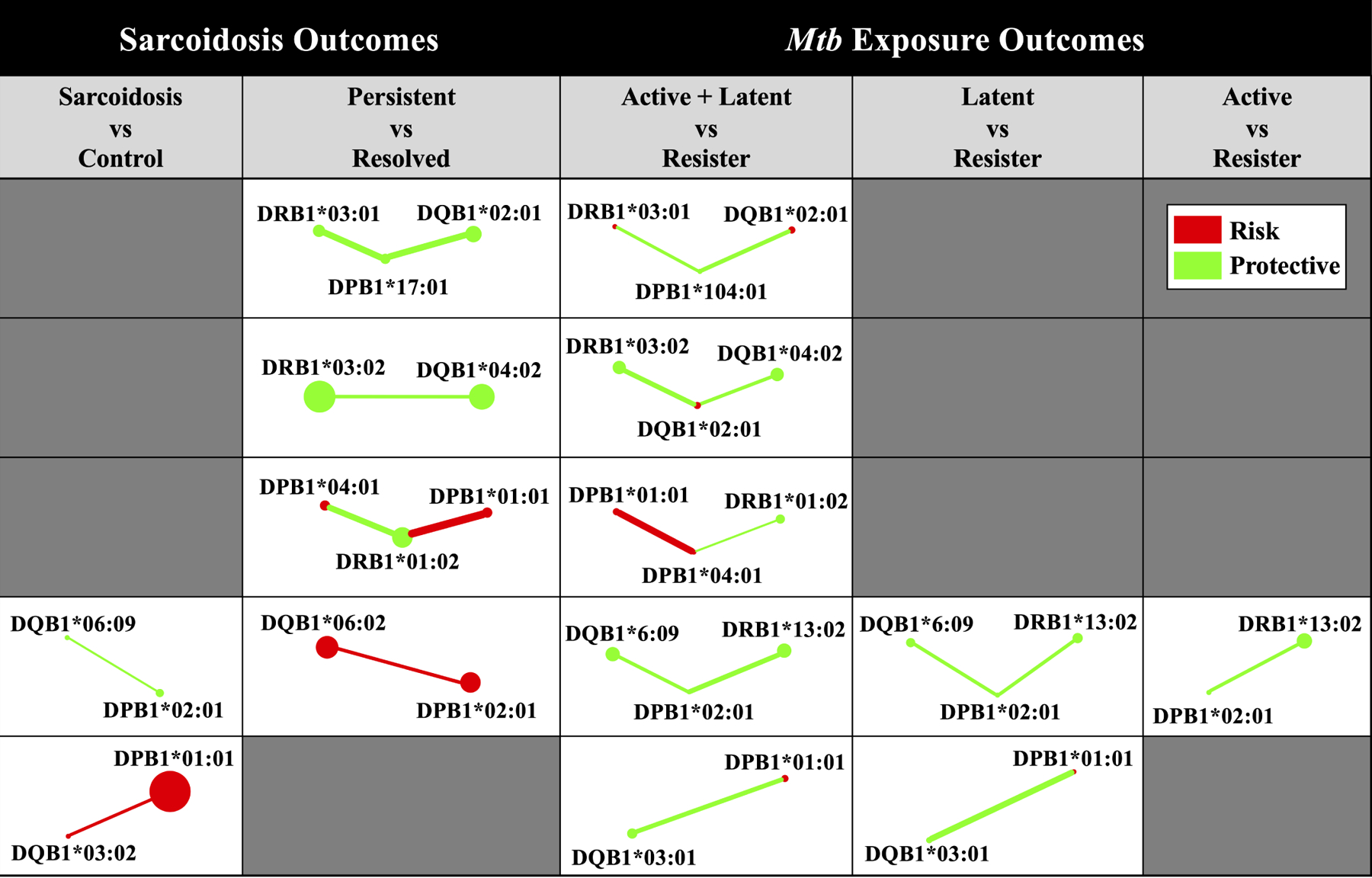
Subnetworks present were extracted from reGAINs for each Mtb exposure (Supplementary Figures S3 – S5) and sarcoidosis (Supplementary Figures S6 – S7) outcome. Each subnetwork includes only those interactions that were nominally significant (P < 0.05) and involving alleles detected by NPDR (Supplementary Tables S3 – S4). Node color corresponds to the adjusted odds ratio in the absence of non-additive effects, indicating whether the allelic odds ratio was protective (green, 0 < OR < 1) or predisposing (red, OR > 1) for a given disease sub-phenotype. Edge color corresponds to the interaction effect odds ratio for a given pair of adjacent alleles, similarly indicating whether the interaction effect odds ratio was protective (green, 0 < OR < 1) or predisposing (red, OR > 1) for a given disease sub-phenotype. Node size and edge width not drawn to scale.
Discussion
In this study, we compared HLA associations with outcomes of Mtb exposure and sarcoidosis in individuals of African ancestry. We conducted the first HLA association analysis to include individuals with consistently negative diagnostic tests for Mtb infection, despite long-term exposure to index cases with active disease. This led to the discovery of several HLA-DRB1, -DQB1, and -DPB1 alleles distinguishing this resistant phenotype from both active disease and latent Mtb infection. We also found novel allelic associations in HLA-DRB1, -DQB1, and -DPB1 distinguishing sarcoidosis cases from healthy controls, as well as persistent from resolved sarcoidosis. Although a subset of the associations detected with nearest-neighbor feature selection could be attributed to main effects, most were explained only by pairwise interactions. Comparing HLA associations between outcomes of Mtb exposure and sarcoidosis, there was striking similarity; HLA alleles important for distinguishing sub-phenotypes of Mtb exposure from resisters tend to also distinguish persistent and resolved sarcoidosis. These similarities extended beyond just allelic importance. We observed parallels in directionality of disease risk, both for main effects and interaction effects, revealing a common interaction network architecture for these two granulomatous disorders.
One of the more striking findings of our study is that we found no association with HLA-DRB1, -DQB1, or -DPB1 when comparing individuals with latent Mtb infection to those with active disease. The comparison of active disease and latent Mtb infection is relatively rare (Oliveira-Cortez et al., 2016); control groups are often heterogeneous and consisting of individuals with positive and negative tuberculin skin tests, respectively (Magira et al., 2012, Duarte et al., 2011), which are known (along with interferon-γ release assays) to poorly distinguish between latent Mtb infection and active disease (Abubakar et al., 2018). While no specific associations were found in our study, other HLA-dependent genetic or immunological mechanisms may be at play. For example, expression of HLA-DR on Mtb-specific IFN-γ+TNF-α+ cells is reportedly higher in individuals with active disease compared to those with latent Mtb infection (Luo et al., 2021). Future work examining activated cellular immune responses to Mtb or cell-type specific expression may gain additional mechanistic insight into the role of specific HLA genotypes.
Interestingly, most of the HLA associations that were common to Mtb exposure outcomes and sarcoidosis tended to be those that were previously reported. For example, DRB1*03:01 and *03:02 as well as DQB1*02:01 are strongly associated with sarcoidosis across multiple ancestries (Fingerlin et al., 2015, Levin et al., 2015) and DQB1*04:02 has been shown to be protective against active tuberculosis across multiple ancestries (Oliveira-Cortez et al., 2016). However, it is important to note that our use of a more inclusive approach allowed for the identification of HLA alleles involved in pairwise interactions, which are important in delineating outcomes of Mtb exposure and sarcoidosis and would not have been found using traditional approaches. For example, DQB1*04:02 has not previously been associated with sarcoidosis, but here, we show it significantly interacts with DRB1*03:02 to protect against persistent sarcoidosis. Evidence for more complex effects involving DRB1*03:02 and DQB1*04:02 also exists outside the context of sarcoidosis, with the DRB1*03:02-DQA1*04:01-DQB1*04:02 haplotype reported to be highly protective against Type 1 diabetes in African Americans (Howson et al., 2013). Similarly, DQB1*04:02 was shown here to interact with DQB1*02:01 and associate with resistance against active tuberculosis, but to predispose to disease in combination with DRB1*11:01. Finally, we found DPB1*02:01 and *04:01 involved in nearly identical interaction subnetworks for Mtb exposure resisters and resolved sarcoidosis. These HLA alleles have strong effects in chronic beryllium disease (McCanlies et al., 2003), a differential diagnosis of sarcoidosis.
Finally, while mechanistic studies of these HLA alleles are outside of the scope of this paper, it is important to note that in vitro and in vivo effects may not be tied directly to the role of HLA-DRB1, -DQB1, or -DPB1 in antigen presentation. Previous studies have suggested both a unique innate and adaptive immune response to Mtb in Resisters (Seshadri et al., 2017) via peripheral monocytes differentially activating pathways controlled by histone deacetylase. This pathway, along with IFN-γ signaling, is known to be aberrant in sarcoidosis, via dysregulation of the enzyme 1α-hydroxylase and the systemic excess of 1,25(OH)2D3 and hypercalcemia seen in sarcoidosis (Overbergh et al., 2006). In addition, a previous study of this exact Mtb exposure cohort found a distinct immunological profile of Resisters characterized by IgM, class-switched IgG antibody responses, and non-IFN-γ T cell responses to the Mtb-specific proteins ESAT6 and CFP10, confirming exposure without pathology (Lu et al., 2019). Th17 cells from sarcoidosis patients also show reduced IFN-γ responses to ESAT6 (Richmond et al., 2013). These studies suggest resistance to persistent forms of sarcoidosis or Mtb infection may be controlled by multiple shared pathways, including IFN-γ responses.
The main objective of this study was to compare class II HLA associations with outcomes of Mtb exposure and sarcoidosis, including sub-phenotypes, in order to learn more about both diseases. We showed that by using a tool free of model specification, we could confirm previously reported HLA associations, including DRB1*01:01, *03:01, *03:02, *09:01, *13:04, and DQB1*02:01, and identify novel associations. The overlapping genetic associations between resistant phenotypes of Mtb exposure and sarcoidosis suggest commonalities in immune-mediated resistance to both diseases and suggest etiological and mechanistic insight to be gained by their joint examination, as well as the importance of examining both single-allele and more complex associations.
Supplementary Material
Acknowledgements
We thank our patients and referring physicians as well as our lab and technical staff: Sarah Cioli, Judy Harris, Cherilyn Pritchett-Frazee, Sharon Johnson, Kiely Grundahl, and Stuart Glenn for technical support. We also thank Dr. Marcelo A. Fernández-Viña at the Stanford Blood Center for collaboration in HLA typing. We want to acknowledge the contributions made by senior physicians, medical officers, health visitors, laboratory and data personnel: Drs. Alphonse Okwera, W. Henry Boom, Mary Nsereko, and Moses Joloba, Ms. Dorcas Lamunu, LaShaunda Malone, Deborah Nsamba, Annet Kawuma, Saidah Menya, Joan Nassuna, Joy Beseke, Michael Odie, Henry Kawoya, Shannon Pavsek, Dr. E. Chandler Church, Anna Duewiger, Keith Chervenak, and Bonnie Thiel. This study would not be possible without the generous participation of the Ugandan patients and families. Lastly, we would like to acknowledge the invaluable contributions to early data analyses by Dr. Robert Igo, Jr, who passed away prior to the preparation of this manuscript.
Sources of Support:
This work was supported by the Foundation for Sarcoidosis research (Chicago, IL), the National Institutes of Health [R01HL113326-05, P30 GM110766-01, R61/R33AI38272, U54GM104938-06, 5T32AI007633-19], and the Bill and Melinda Gates Foundation [OPP1151836].
Footnotes
Disclosures: The research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- ACCESS Research Group 1999. Design of a case-control etiologic study of sarcoidosis (ACCESS). ACCESS Research Group. Journal of Clinical Epidemiology, 52, 1173–1186. [DOI] [PubMed] [Google Scholar]
- ABUBAKAR I, DROBNIEWSKI F, SOUTHERN J, SITCH AJ, JACKSON C, LIPMAN M, DEEKS JJ, GRIFFITHS C, BOTHAMLEY G, LYNN W, BURGESS H, MANN B, IMRAN A, SRIDHAR S, TSOU CY, NIKOLAYEVSKYY V, REES-ROBERTS M, WHITWORTH H, KON OM, HALDAR P, KUNST H, ANDERSON S, HAYWARD A, WATSON JM, MILBURN H, LALVANI A & TEAM PS 2018. Prognostic value of interferon-gamma release assays and tuberculin skin test in predicting the development of active tuberculosis (UK PREDICT TB): a prospective cohort study. Lancet Infect Dis, 18, 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ADRIANTO I, LIN CP, HALE JJ, LEVIN AM, DATTA I, PARKER R, ADLER A, KELLY JA, KAUFMAN KM, LESSARD CJ, MOSER KL, KIMBERLY RP, HARLEY JB, IANNUZZI MC, RYBICKI BA & MONTGOMERY CG 2012. Genome-wide association study of African and European Americans implicates multiple shared and ethnic loci in sarcoidosis susceptibility. PLoS ONE, 7, e43907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMIRZARGAR A, YALDA A, HAJABOLBAGHI M, KHOSRAVI F, JABBARI H, REZAEI N, NIKNAM MH, ANSARI B, MORADI B & NIKBIN B 2004. The association of HLA-DRB, DQA1, DQB1 alleles and haplotype frequency in Iranian patients with pulmonary tuberculosis. Int J Tuberc Lung Dis, 8, 1017–1021. [PubMed] [Google Scholar]
- ARCHAKOVA LI 2008. Improving therapy based on the study of immunogenetic factors in the formation of pulmonary tuberculosis. Rus Immune J, 11, 188–189. [Google Scholar]
- BAUGHMAN RP, FIELD S, COSTABEL U, CRYSTAL RG, CULVER DA, DRENT M, JUDSON MA & WOLFF G 2016. Sarcoidosis in America. Analysis Based on Health Care Use. Annals of the American Thoracic Society, 13. [DOI] [PubMed] [Google Scholar]
- BAUGHMAN RP, NAGAI S, BALTER M, COSTABEL U, DRENT M, DU BOIS R, GRUTTERS JC, JUDSON MA, LAMBIRI I, MULLER-QUERNHEIM J, PRASSE A, RIZZATO G, ROTTOLI P, SPAGNOLO P & TEIRSTEIN A 2009. Defining the Clinical Outcome Status (COS) in Sarcoidosis: Results of WASOG Task Force. Sarcoidosis Vasc Diffuse Lung Dis, 28, 56–64. [PubMed] [Google Scholar]
- BAUGHMAN RP, TEIRSTEIN AS, JUDSON MA, ROSSMAN MD, YEAGER HJ, BRESNITZ EA, DEPALO L, HUNNINGHAKE G, IANNUZZI MC, J. C. J, MCLENNON G, MOLLER DR, NEWMAN LS, RABIN DL, ROSE C, RYBICKI B, WEINBERGER SE, TERRIN ML, KNATTERUD GL, CHERNIAK R & GROUP, C. C. E. S. O. S. A. R. 2001. Clinical characteristics of patients in a case control study of sarcoidosis. Am J Respir Crit Care Med, 164, 1885–1889. [DOI] [PubMed] [Google Scholar]
- DAWKINS BA, LE TT & MCKINNEY BA 2021. Theoretical properties of distance distributions and novel metrics for nearest-neighbor feature selection. PLoS ONE, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUARTE R, CARVALHO C, PEREIRA C, BETTENCOURT A, CARVALHO A, VILLAR M, DOMINGOS A, BARROS H, MARQUES J, COSTA PP, MENDONÇA D & MARTINS B 2011. HLA class II alleles as markers of tuberculosis susceptibility and resistance. Rev Port Pneumol, 17, 15–19. [DOI] [PubMed] [Google Scholar]
- ESTEVES T, APARICIO G & GARCIA-PATOS V 2016. Is there any association between Sarcoidosis and infectious agents?: a systematic review and meta-analysis. BMC Pulmonary Medicine, 16, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FINGERLIN TE, HAMZEH N & MAIER LA 2015. Genetics of Sarcoidosis. Clinics in Chest Medicine, 36, 569–584. [DOI] [PubMed] [Google Scholar]
- GLOBAL TUBERCULOSIS REPORT, W. 2020. World Health Organization. Global tuberculosis report 2020. Geneva, Switzerland: World Health Organization. [Google Scholar]
- GRUNEWALD J, GRUTTERS JC, ARKEMA EV, SAKETKOO LA, MOLLER DR & MÜLLER-QUERNHEIM J 2019. Sarcoidosis. Nature Reviews Disease Primers, 5, 45. [DOI] [PubMed] [Google Scholar]
- GUPTA D, AGARWAL R, AGGARWAL AN & JINDAL SK 2007. Molecular evidence for the role of mycobacteria in sarcoidosis: a meta-analysis. Eur Respir J, 30, 508–16. [DOI] [PubMed] [Google Scholar]
- HOWSON JMM, ROY MS, ZEITELS L, STEVENS H & TODD JA 2013. HLA class II gene associations in African American Type 1 diabetes reveal a protective HLA-DRB1*03 haplotype. Diabetic Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IANNUZZI MC 2007. Genetics of sarcoidosis. Seminars in Respiratory and Critical Care Medicine, 28, 015–021. [DOI] [PubMed] [Google Scholar]
- IANNUZZI MC, MALIARIK MJ, POISSON LM & RYBICKI BA 2003. Sarcoidosis susceptibility and resistance HLA-DQB1 alleles in African Americans. Am J Respir Crit Care Med, 167, 1225–1231. [DOI] [PubMed] [Google Scholar]
- ISRAEL HL, KARLIN P, MENDUKE H & DELISSER OG 1986. Factors Affecting Outcome of Sarcoidosis. Annals of the New York Academy of Sciences, 465, 609–618. [DOI] [PubMed] [Google Scholar]
- LAREAU CA, WHITE BC, OBERG AL & MCKINNEY BA 2015. Differential co-expression network centrality and machine learning feature selection for identifying susceptibility hubs in networks with scale-free structure. BioData Mining, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LE TT, DAWKINS BA & MCKINNEY BA 2020. Nearest-neighbor Projected-Distance Regression (NPDR) for detecting network interactions with adjustments for multiple tests and confounding. Bioinformatics, 36, 2770–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LE TT, SIMMONS KW, MISAKI M, BODURKA J, WHITE BC, SAVITZ J & MCKINNEY BA 2017. Differential privacy-based evaporative cooling feature selection and classification with relief-F and random forests. Bioinformatics, 33, 2906–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEEK JT & STOREY JD 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet, 3, 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVIN AM, ADRIANTO I, DATTA I, IANNUZZI MC, TRUDEAU S, LI J, DRAKE WP, MONTGOMERY CG & RYBICKI BA 2015. Association of HLA-DRB1 with Sarcoidosis Susceptiblity and Progression in African Americans. Am J Respir Cell Mol Biol, 53, 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVIN AM, ADRIANTO I, DATTA I, IANNUZZI MC, TRUDEAU S, MCKEIGUE P, MONTGOMERY CG & RYBICKI BA 2014. Performance of HLA allele prediction methods in African Americans for class II genes HLA-DRB1, -DQB1, and -DPB1. BMC Genetics, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU LL, SMITH MT, YU KKQ, LUEDEMANN C, SUSCOVICH TJ, GRACE PS, CAIN A, YU WH, MCKITRICK TR, LAUFFENBURGER D, CUMMINGS RD, MAYANJA-KIZZA H, HAWN TR, BOOM WH, STEIN CM, FORTUNE SM, SESHADRI C & ALTER G 2019. IFN-g-independent immune markers of Mycobacterium tuberculosis exposure. Nat Med, 25, 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUO Y, XUE Y, TANG G, LIN Q, SONG H, LIU W, YIN B, HUANG J, WEI W, MAO L, WANG F & SUN Z 2021. Combination of HLA-DR on Mycobacterium tuberculosis-Specific Cells and Tuberculosis Antigen/Phytohemagglutinin Ratio for Discriminating Active Tuberculosis From Latent Tuberculosis Infection. Frontiers in Immunology, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAGIRA EE, PAPASTERIADES C, KANTERAKIS S, TOUBIS M, ROUSSOS C & MONOS DS 2012. HLA-A and HLA-DRB1 amino acid polymorphisms are asscociated with susceptibility and protection to pulmonary tuberculosis in a Greek population. Hum Immunol, 73, 641–646. [DOI] [PubMed] [Google Scholar]
- MALKOVA A, STARSHINOVA A, ZINCHENKO Y, BASANTSOVA N, MAYEVSKAYA V, YABLONSKIY P & SHOENFELD Y 2020. The opposite effect of human leukocyte antigen genotypes in sarcoidosis and tuberculosis: a narrative review of the literature. ERJ Open Res, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCANLIES EC, KREISS K, ANDREW M & WESTON A 2003. HLA-DPB1 and Chronic Beryllium Disease: A HuGE Review. American Journal of Epidemiology, 157, 388–398. [DOI] [PubMed] [Google Scholar]
- MCHENRY ML, BENCHEK P, MALONE L, NSEREKO M, MAYANJA-KIZZA H, BOOM WH, WILLIAMS SM, HAWN TR & STEIN C 2021. Resistance to TST/IGRA Conversion in Uganda: Heritability and Genome-Wide Association Study. [DOI] [PMC free article] [PubMed]
- MCKINNEY BA, CROWE JR JE, GUO J & TIAN D 2009. Capturing the Spectrum of Interaction Effects in Genetic Association Studies by Simulated Evaporative Cooling Network Analysis. PLoS Genet, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCKINNEY BA, WHITE BC, GRILL DE, LI PW, KENNEDY RB, POLAND GA & OBERG AL 2013. ReliefSeq: A Gene-Wise Adaptive-K Nearest-neighbor Feature Selection Tool for Finding Gene-Gene Interactions and Main Effects in mRNA-Seq Gene Expression Data. PLoS ONE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIRSAEIDI M, MACHADO RF, SCHRAUFNAGEL D, SWEISS NJ & BAUGHMAN RP 2015. Racial difference in sarcoidosis mortality in the United States. Chest, 147, 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORAIS A, LIMA B, PEIXOTO MJ, ALVES H, MARQUES A & DELGADO L 2012. BTNL2 gene polymorphism associations with susceptibility and phenotype expression in sarcoidosis. Respiratory Medicine, 106, 1771–1777. [DOI] [PubMed] [Google Scholar]
- MOUTSIANAS L, JOSTINS L, BEECHAM AH, DILTHEY AT, XIFARA DK, BAN M, SHAH TS, PATSOPOULOS NA, ALFREDSSON L, ANDERSON CA, ATTFIELD KE, BARANZINI SE, BARRETT J, BINDER TMC, BOOTH D, BUCK D, CELIUS EG, COTSAPAS C, D’ALFONSO S, DENDROU CA, DONNELLY P, DUBOIS B, FONTAINE B, FUGGER L, GORIS A, GOURRAUD PA, GRAETZ C, HEMMER B, HILLERT J, INTERNATIONAL IBDGC, KOCKUM I, LESLIE S, LILL CM, MARTINELLI-BONESCHI F, OKSENBERG JR, OLSSON T, OTURAI A, SAARELA J, SONDERGAARD HB, SPURKLAND A, TAYLOR B, WINKELMANN J, ZIPP F, HAINES JL, PERICAK-VANCE MA, SPENCER CCA, STEWART G, HAFLER DA, IVINSON AJ, HARBO HF, HAUSER SL, DE JAGER PL, COMPSTON A, MCCAULEY JL, SAWCER S & MCVEAN G 2015. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat Genet, 47, 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLIVEIRA-CORTEZ A, MELO AC, CHAVES VE, CONDINO-NETO A & CAMARGOS P 2016. Do HLA class II genes protect againsts pulmonary tuberculosis? A systematic review and meta-analysis. Eur J Clin Microbiol Infect Dis, 35, 1567–1580. [DOI] [PubMed] [Google Scholar]
- OVERBERGH L, STOFFLES K, WAER M, VERSTUYF A, BOUILLON R & MATHIEU C 2006. Immune regulation of 25-hydroxyvitamin D-1alpha-hydroxylase in human monocytic THP1 cells: mechanisms of interferon-gamma-mediated induction. J Clin Endocrinal Metab, 91, 3566–3574. [DOI] [PubMed] [Google Scholar]
- PANDEY A, DAVIS NA, WHITE BC, PAJEWSKI NM, SAVITZ J, DREVETS WC & MCKINNEY BA 2012. Epistasis network centrality analysis yields pathway replication across two GWAS cohorts for bipolar disorder. Transl Psychiatry, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICHMOND BW, PLOETZE K, ISOM J, CHAMBERS-HARRIS I, BRAUN NA, TAYLOR T, ABRAHAM S, MAGETO Y, CULVER DA, OSWALD-RICHTER KA & DRAKE WP 2013. Sarcoidosis Th17 cells are ESAT-6 antigen specific but demonstrate reduced IFN-\gamma expression. J Clin Immunol, 33, 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSSMAN MD, THOMPSON B, FREDERICK M, MALIARIK M, IANNUZZI MC, RYBICKI BA, PANDEY JP, NEWMAN LS, MAGIRA E, BEZNIK-CIZMAN B, MONOS D & GROUP A 2003. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet, 73, 720–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RYBICKI BA, MAJOR M, POPOVICH JR J, MALIARIK MJ & IANNUZZI MC 1997. Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am J Epidemiol, 145, 234–241. [DOI] [PubMed] [Google Scholar]
- RYBICKI BA, SINHA R, IYENGAR S, GRAY-MCGUIRE C, ELSTON RC, IANNUZZI MC & CONSORTIUM SS 2007. Genetic linkage analysis of sarcoidosis phenotypes: the sarcoidosis genetic analysis (SAGA) study. Genes Immun, 8, 379–386. [DOI] [PubMed] [Google Scholar]
- SESHADRI C, SEDAGHAT N, CAMPO M, PETERSON G, WELLS GS, SHERMAN DR, STEIN CM, MAYANJA-KIZZA H, SHOJAIE A, BOOM WH, HAWN TR & (TBRU), T. R. U. 2017. Transcriptional networks are associated with resistance to Mycobacterium tuberculosis infection. PLoS ONE, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STARSHINOVA A, DOVGALYUK I, BERKOS A, OVCHINNIKOVA Y, BUBNOVA L & YABLONSKIY P 2018. The effect of human leukocyte Antigens-DRB1 alleles on development of different tuberculosis forms in children. Int J Mycobacteriol, 7, 117–121. [DOI] [PubMed] [Google Scholar]
- STEIN CM, NSEREKO M, MALONE LL, OKWARE B, KISINGO H, NALUKWAGO S, CHERVENAK K, MAYANJA-KIZZA H, HAWN TR & BOOM WH 2019. Long-term Stability of Resistance to Latent Mycobacterium tuberculosis Infection in Highly Exposed Tuberculosis Household Contacts in Kampala, Uganda. Clin Infect Dis, 68, 1705–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEIN CM, ZALWANGO S, MALONE LL, THIEL B, MUPERE E, NSEREKO M, OKWARE B, KISINGO H, LANCIONI CL, BARK CM, WHALEN CC, JOLOBA ML, BOOM WH & MAYANJA-KIZZA H 2018. Resistance and Susceptibility to Mycobacterium tuberculosis Infection and Disease in Tuberculosis Households in Kampala, Uganda. Am J Epidemiol, 187, 1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERÁN-ESCANDÓN D, TERÁN-ORTIZ L, CAMARENA-OLVERA A, GONZÁLEZ-AVILA G, VACA-MARÍN MA, GRANADOS J & SELMAN M 1999. Human leukocyte antigen-associated susceptibility to pulmonary tuberculosis: molecular analysis of class II alleles by DNA amplification and oligonucleotide hybridization in Mexican patients. Chest, 115, 428–433. [DOI] [PubMed] [Google Scholar]
- TONG X, CHEN L, LIU S, YAN Z, PENG S, ZHANG Y & FAN H 2015. Polymorphisms in HLA-DRB1 Gene and the Risk of Tuberculosis: A Meta-analysis of 31 Studies. Lung, 193, 309–318. [DOI] [PubMed] [Google Scholar]
- URBANOWICZ RJ, OLSON RS, SCHMITT P, MEEKER M & MOORE JH 2018. Benchmarking relief-based feature selection methods for bioinformatics data mining. Journal of Biomedical Informatics, 85, 168–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VEJBAESYA S, CHIERAKUL N, LUANGTRAKOOL K, SRINAK D & STEPHENS HAF 2002. Associations of HLA class II alleles with pulmonary tuberculosis in Thais. Eur J Immunogenet, 29, 431–434. [DOI] [PubMed] [Google Scholar]
- WANG C, KRISHNAKUMAR S, WILHELMY J, BABRZADEH F, STEPANYAN L, SE LF, LEVINSON D, FERNANDEZ-VIÑA MA, DAVIS RW, DAVIS MM & MINDRINOS M 2012. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc Natl Acad Sci USA, 109, 8676–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU F, ZHANG W, ZHANG L, WU J, LI C, MENG X, WANG X, HE P & ZHANG J 2013. NRAMP1, VDR, HLA-DRB1, and HLA-DQB1 gene polymorphisms in susceptibility to tuberculosis among the Chinese Kazakh population: a case-control study. Biomed Res Int. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG P-L, HE X-J, ZANG Q-J, LI H-P, WANG G-Y & QIN J 2016. Association of human leukocyte antigen DRB1 polymorphism and tuberculosis: a meta-analysis. Int J Tuberc Lung Dis, 20, 121–128. [DOI] [PubMed] [Google Scholar]
- ZHENG X, SHEN J, COX C, WAKEFIELD JC, EHM MG, NELSON MR & WEIR BS 2014. HIBAG-HLA Genotype imputation with attribute bagging. The Pharmacogenomics Journal, 14, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU Y, HU Y & LI H 2013. Role of Propionibacterium Acnes in Sarcoidosis: A Meta-analysis. Sarcoidosis Vasc Diffuse Lung Dis, 30, 262–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


