Abstract
To prioritize circulating metabolites that likely play causal roles in the pathogenesis of multiple sclerosis (MS). Two-sample Mendelian randomization analysis was performed to estimate the causal effects of 571 circulating metabolites on the risk of MS. Genetic instruments for circulating metabolites were obtained from three previous genome-wide association studies (GWAS) of the blood metabolome (N = 7,824; 24,925; and 115,078; respectively), while genetic associations with MS were from a large GWAS by the International Multiple Sclerosis Genetics Consortium (14,802 cases and 26,703 control). The primary analysis was performed with the multiplicative random-effect inverse variance-weighted method, while multiple sensitivity analyses were conducted with the weighted median, weighted mode, MR-Egger, and MRPRESSO. A total of 29 metabolites had suggestive evidence of causal associations with MS. Genetically instrumented levels of serine (OR = 1.56, 95% CI = 1.25 – 1.95), lysine (OR = 1.18, 95% CI = 1.01 – 1.38), acetone (OR = 2.45, 95% CI = 1.02 – 5.90), and acetoacetate (OR = 2.47, 95% CI = 1.14 – 5.34) were associated with a higher MS risk. Total cholesterol and phospholipids in large very-low-density lipoprotein were associated with a lower MS risk (OR = 0.83, 95% CI = 0.69 – 1.00; OR = 0.80, 95% CI = 0.68 – 0.95), but risk-increasing associations (OR = 1.20, 95% CI = 1.04 – 1.40; OR = 1.13, 95% CI = 1.00 – 1.28) were observed for the same two lipids in very large high-density lipoprotein. Our metabolome-wide Mendelian randomization study prioritized a list of circulating metabolites, such as serine, lysine, acetone, acetoacetate, and lipids, that likely have causal associations with MS.
Keywords: Mendelian randomization, Metabolome, Metabolites, Multiple sclerosis
1. Introduction
Multiple sclerosis (MS) is an autoimmune disease in the central nervous system, characterized by neuroinflammation, demyelination, and neurodegeneration. While the exact causes of MS are still unknown, some lifestyle and environmental risk factors have been relatively well-established, such as female sex, smoking, Epstein–Barr virus (EBV) infection, low vitamin levels, and obesity (Olsson et al. , 2017). Metabolomics is a powerful approach to identifying metabolites that differentiate MS patients from healthy controls, revealing diagnostic or prognostic biomarkers, potential therapeutic targets, and insights into the pathogenesis (Bhargava and Anthony, 2020, Zahoor et al. , 2021). Metabolites in various metabolic pathways have been implicated in MS, such as higher plasma levels of acetoacetate, acetone, and 3-hydroxybutyrate in energy metabolism (Cocco et al. , 2016), higher circulating levels of gamma-glutamyl amino acids and lysine in amino acid metabolism (Bhargava et al. , 2017, Moussallieh et al. , 2014), elevated serum levels of uridine in nucleotide metabolism (Lazzarino et al. , 2017), and altered circulating profiles of lipids and lipoproteins in lipid metabolism (Lorincz et al. , 2022). Of note, lipoproteins are soluble complexes of proteins and lipids, with a hydrophilic membrane of phospholipids, free cholesterol, and apolipoproteins surrounding a hydrophobic core of cholesteryl esters and triglycerides. Based on their size, constituent lipids and apolipoproteins, lipoproteins can be divided into seven classes, chylomicrons, chylomicron remnants, very low-density lipoproteins (VLDL), VLDL remnants, low-density lipoproteins (LDL), high-density lipoproteins (HDL), and lipoprotein (a). These lipoprotein classes could be further divided into subclasses based on their size and density (Feingold, 2022). A metabolomics study, comparing relapsing-remitting MS patients (RRMS) to age- and sex-matched healthy volunteers, found that cholesterol, phospholipids, and triglycerides are elevated in the larger subclasses of VLDL and HDL (Gafson et al. , 2018). However, the causality of these lifestyle, environmental, or metabolomic risk factors is hard to establish due to the inherent limitations of observational associations, especially in case-control studies, such as reverse causation and residual confounding (Olsson, Barcellos, 2017).
Mendelian randomization (MR) is a genetic epidemiology method that leverages genetic effects to enable the inference of causality between an exposure and an outcome. It selects genetic variants with known effects on the exposure of interest. The random allocation of the two alleles at a genetic variant across generations mimics the random assignment of placebo or treatment to participants in a randomized controlled trial (Davies et al. , 2018). MR has been applied to MS, providing evidence for causal roles of high BMI (Harroud et al. , 2021a, Harroud et al. , 2021c, Jacobs et al. , 2020, Vandebergh et al. , 2022), increased interleukin-6 signaling (Vandebergh, Becelaere, 2022), and low vitamin D (Harroud, Manousaki, 2021a, Jacobs, Noyce, 2020). On the other hand, MR did not support the causal roles of uric acid (Niu et al. , 2020), leptin (Harroud, Manousaki, 2021a), adiponectin (Harroud, Manousaki, 2021a), and depression (Binzer et al. , 2021, Harroud et al. , 2021b). A systemic MR study of 65 possible risk factors for MS revealed robust evidence of causality for four of them, high childhood and adult BMI, low vitamin D, and low physical activity. It also found suggestive evidence for type 2 diabetes, waist circumference, body fat percentage, age of puberty, and high-density lipoprotein cholesterol (HDL-C) (Yuan et al. , 2021). Although large-scale MR analysis is a powerful approach to prioritizing causal risk factors for MS, no such study has been applied to all metabolites in a metabolome. Taking advantage of the recent large genome-wide association studies (GWAS) of human blood metabolites measured by metabolomics platforms (Kettunen et al. , 2016, Richardson et al. , 2022, Shin et al. , 2014), we performed a metabolome-wide MR study to prioritize causal circulating metabolites for MS.
2. Methods
2.1. Data sources
Genetic associations with circulating metabolites were obtained from three GWAS of human blood metabolome. Their summary statistics were compiled and made available through the MRC IEU OpenGWAS project (Elsworth et al. , 2020, Hemani et al. , 2018), and the three GWAS were labeled as met-a (Shin, Fauman, 2014), met-c (Kettunen, Demirkan, 2016), and met-d (Richardson, Leyden, 2022). All three GWAS were performed in participants of European ancestry. The met-a study covers 452 metabolites and 7,824 participants, the met-c study 123 metabolites and up to 24,925 participants, and the met-d study 249 metabolites and 115,078 individuals. For met-a, which performed GWAS on raw phenotypes, we rescaled SNP effect sizes to one standard deviation (SD) of the circulating metabolite level (Shin, Fauman, 2014). The effect sizes in met-c and met-d were already standardized to SD because of the inverse rank-based normal transformation of phenotypic values before GWAS (Kettunen, Demirkan, 2016, Richardson, Leyden, 2022). Genetic associations with MS in Europeans were obtained from the discovery GWAS by the International Multiple Sclerosis Genetics Consortium (14,802 cases and 26,703 control). We obtained access to the summary statistics on the designated website (https://nettskjema.no/a/imsgc-data-access#/). We further confirmed that the same summary statistics were available on the OpenGWAS project with a dataset ID of ieu-b-18.
2.2. Selection of instrumental variables
Two significance thresholds (P < 5 × 10−8 and P < 1 × 10−6) were used to select single nucleotide polymorphisms (SNPs) as instrumental variables (IVs). The genome-wide significance threshold of P < 5 × 10−8 is commonly used for the selection of genetic instruments to fulfill the relevance assumption of MR. We additionally used the suggestive significance threshold of P < 1 × 10−6 to include more metabolites in the analysis, which would otherwise be excluded due to the lack of genetic instruments under the stringent genome-wide significance cutoff. For other metabolites, the usage of the less stringent significance threshold increases the number of genetic instruments and offers an opportunity to assess the robustness of the MR estimates using different sets of genetic instruments. However, we would like to emphasize that these two sets of genetic instruments do not represent independent replications due to their overlaps. We used linkage disequilibrium (LD) clumping (r2 < 0.001 within a 10 Mb window) to identify independent SNPs. For exposure-associated SNPs not present in the MS GWAS dataset, we searched for proxy SNPs in high LD (r2 ≥ 0.8). A threshold of F-statistics greater than 10 indicates strong instruments (Pierce et al. , 2011). The effects of IVs on exposure and outcome were harmonized to rule out strand mismatches and ensure alignment of effect sizes. All IV selection, clumping, and harmonization were implemented in R v4.2.1 using the TwoSampleMR package (v0.5.6) (Hemani, Zheng, 2018).
2.3. Statistical analyses
We performed two-sample MR analysis only for metabolites that have at least three independent genetic instruments in order to apply statistical testing of and correction for potential pleiotropy. The primary analysis utilized the multiplicative random-effect inverse variance-weighted (IVW) method, which used a meta-analysis approach to combine Wald estimates for each SNP and obtain an overall estimate of the effect of each metabolite on MS (Burgess et al. , 2013). The Cochran’s Q test was used to determine the homogeneity within the causal estimates of different SNPs (Greco et al. , 2015). Sensitivity analyses were performed with MR-Egger (Bowden et al. , 2015), weighted median (WME) (Bowden et al. , 2016), and weighted mode (WMO) methods (Hartwig et al. , 2017). The MR-Egger provides robust effect estimates in the presence of balanced pleiotropy. The WME method provides reliable estimates when at least 50% of the weight comes from valid IVs. The WMO method reports the effect estimate supported by the largest number of genetic instruments. The MR-Egger intercept test was applied to evaluate the presence of unbalanced horizontal pleiotropy (Bowden, Davey Smith, 2015, Burgess and Thompson, 2017). Moreover, we applied the MR-PRESSO method for detecting overall horizontal pleiotropy (i.e., the global test), identifying specific outliers (i.e., the outlier test), and re-calculating effect estimates after outlier removal (Verbanck et al. , 2018). Scatter plots, forest plots, and leave-oneout plots were generated to visualize the relationships and the impacts of individual genetic instruments. These sensitivity analyses aimed to ensure robustness and validity of the findings while accounting for potential biases due to pleiotropy. In addition, we applied the MR Steiger method to infer the direction of causality (Hemani et al. , 2017). Candidate metabolites were defined into two groups, consistent and suggestive. The consistent group includes metabolites that have nominally significant (P < 0.05) and directionally consistent MR IVW estimates under both p-value cutoffs (P < 5 × 10−8 and P < 1 × 10−6) for genetic instruments. The suggestive group includes metabolites that have nominally significant MR IVW estimates under either p-value cutoff. Note that some metabolites only have genetic instruments under the less stringent cutoff of P < 1 × 10−6. All analyses were conducted in R v4.2.1 using MendelianRandomization (v0.6.0, IVW, MR-Egger, WME, and WMO analyses) (Broadbent et al. , 2020), TwoSampleMR (v0.5.6, MR Steiger analysis, scatter plots, forest plots, and leave-one-out plots) (Hemani, Zheng, 2018), and MRPRESSO (v1.0, MR-PRESSO analysis) (Verbanck, Chen, 2018).
2.4. Standard protocol approvals, registrations, and patient consent
This study was conducted with previously published summary-level data. No individual-level data were used.
2.5. Data availability
All GWAS summary statistics can be accessed through the OpenGWAS project (Elsworth, Lyon, 2020, Hemani, Zheng, 2018). The MR analysis scripts can be found at https://github.com/yitangsun/metabolome-wide-MR-for-MS. The key MR results have been replicated by another author, and the scripts can be found here https://github.com/angelage678/Mendelian-Randomization---multiple-sclerosis-and-metabolites.
3. Results
Our workflow is summarized in Fig. 1. Three GWAS of human blood metabolome were included in our analysis, each with 452, 123, and 249 metabolites, respectively. Metabolites with less than three genetic instruments were excluded from our analysis. With a significance cutoff of P < 5 × 10−8 for the selection of genetic instruments, we obtained MR results for a total of 404 metabolites (Supplementary Table 1). When we relaxed the significance cutoff to P < 1 × 10−6, an additional 167 metabolites were included for MR analysis, reaching a total of 571 metabolites (Supplementary Table 2). For all the 404 metabolites included in analyses with both significance cutoffs, the effect estimates are highly concordant (Supplementary Fig. 1). A total of 29 metabolites were identified as potential causal for MS. Six metabolites have nominally significant and directionally consistent effect estimates with both significance cutoffs. Another 23 metabolites have nominally significant signals with one cutoff. Fourteen of these 23 only have genetic instruments under the significance threshold of P < 1 × 10−6. For the other nine that have genetic instruments under both significance thresholds, the MR effect estimates are directionally consistent and close to each other (Fig. 2). Sensitivity analyses with MR-Egger, WME, WMO, and MR-PRESSO revealed directionally consistent effect estimates, although not always reaching nominal significance. The MR Steiger test further supports the causal direction from the metabolite to MS, instead of the reverse (Supplementary Tables 1 and 2).
Fig. 1.
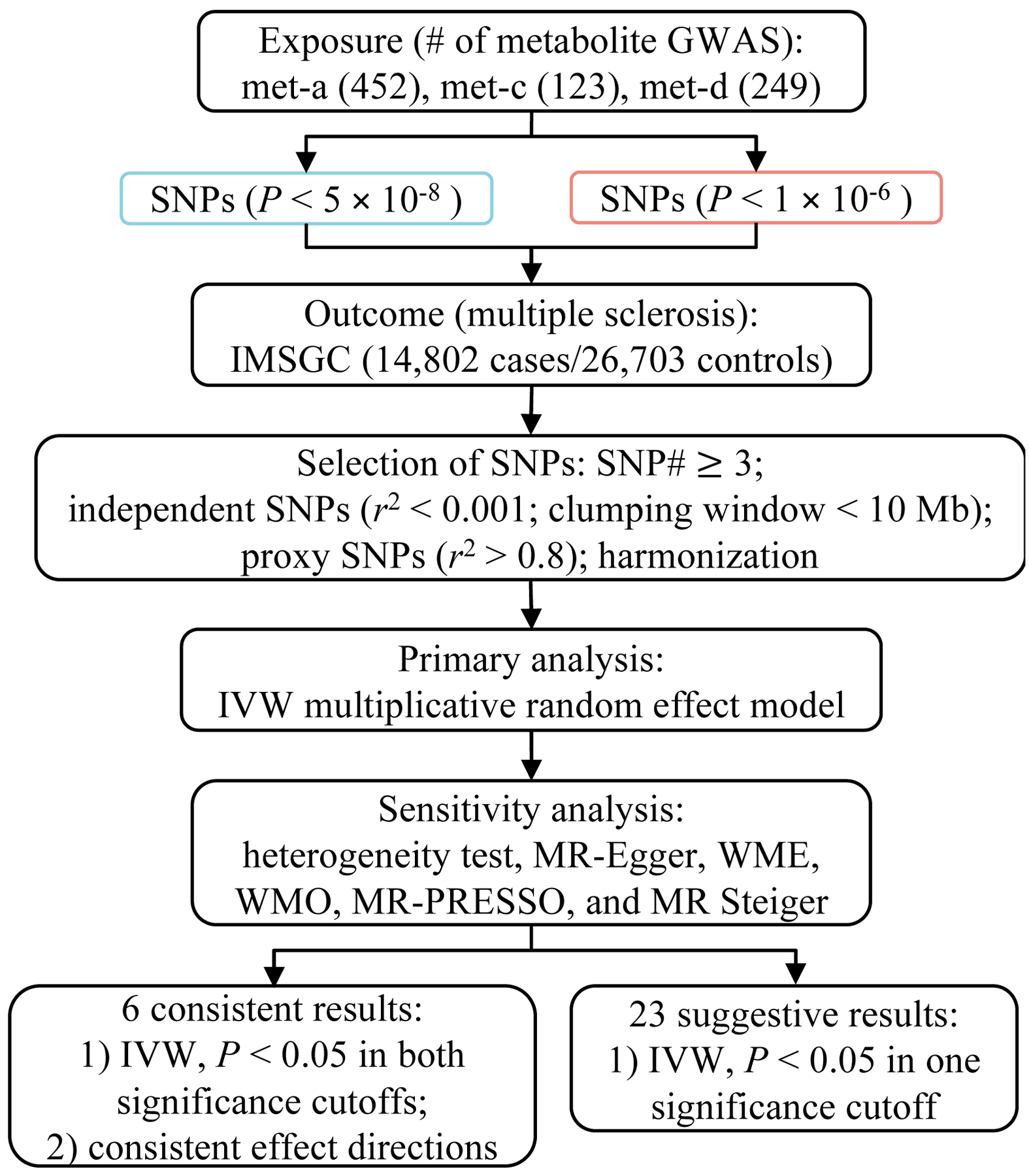
Flowchart of the MR study.
MR: Mendelian randomization; GWAS: genome-wide association studies; SNPs: single nucleotide polymorphisms; IMSGC: International Multiple Sclerosis Genetics Consortium; IVW: inverse variance-weighted; WME: weighted median; WMO: weighted mode.
Fig. 2.
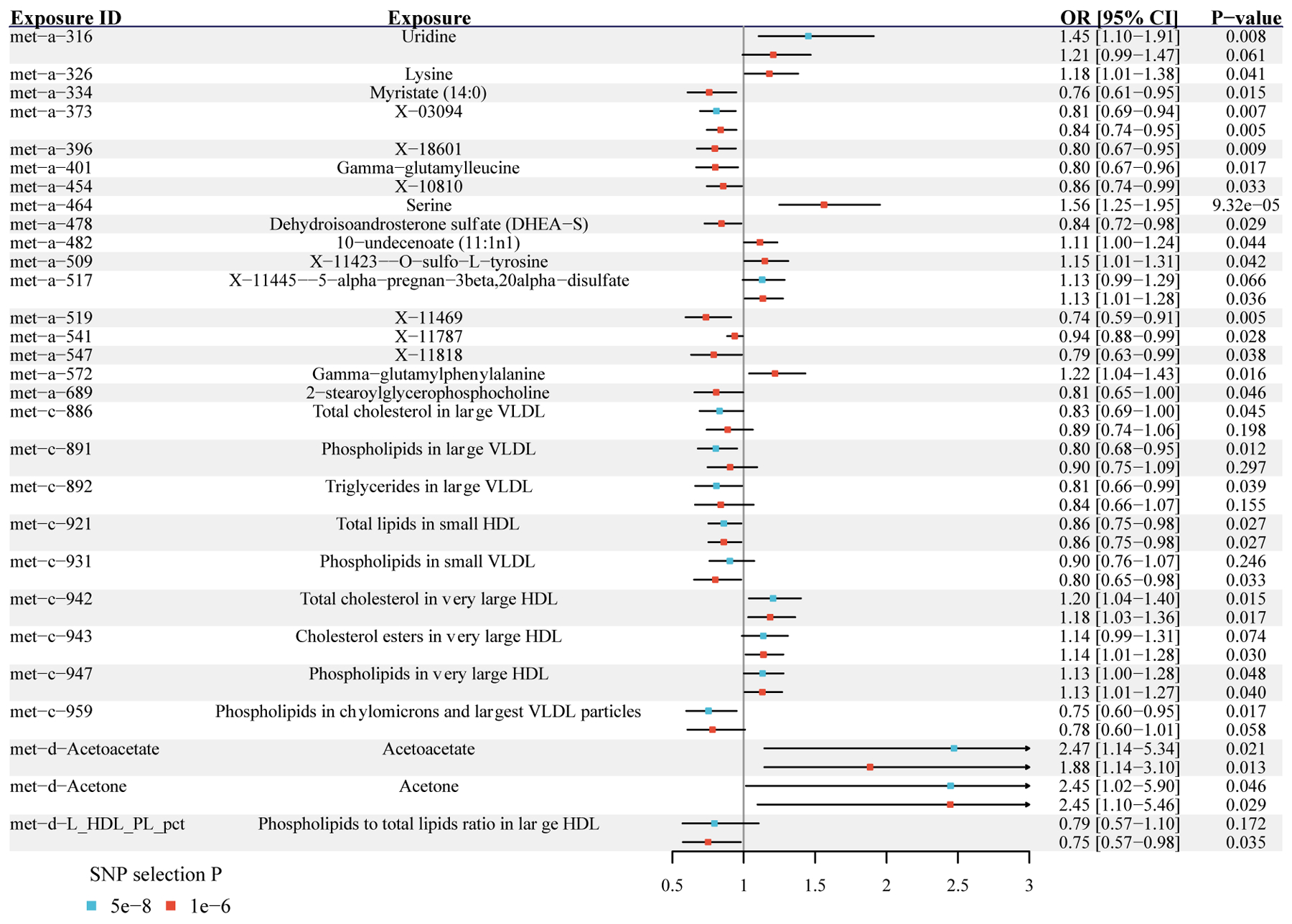
Metabolites with significant MR estimated effects on the risk of MS.
Odds ratios and 95% confidence intervals are scaled to per genetically predicted 1 SD increase in circulating metabolite levels. MR: Mendelian randomization; MS: multiple sclerosis; OR: odds ratios; 95% CI: 95% confidence intervals; SD: standard deviation; SNP: single nucleotide polymorphism.
Among 29 potential causal metabolites, ten are lipids in specific lipoprotein subclasses. The genetically predicted circulating levels of total cholesterol (OR = 0.83, 95% CI = 0.69 – 1.00, P = 0.045), phospholipids (OR = 0.80, 95% CI = 0.68 – 0.95, P = 0.012), and triglycerides (OR = 0.81, 95% CI = 0.66 – 0.99, P = 0.039) in large VLDL are associated with a lower risk of MS. Phospholipids in small VLDL (OR = 0.80, 95% CI = 0.60 – 0.95, P = 0.017) and in chylomicrons and the largest VLDL particles (OR = 0.75, 95% CI = 0.65 – 0.98, P = 0.033) are both negatively associated with the MS risk. In contrast, the genetically predicted circulating levels of total cholesterol (OR = 1.20, 95% CI = 1.04 – 1.40, P = 0.015), phospholipids (OR = 1.13, 95% CI = 1.00 – 1.28, P = 0.048), and cholesterol esters (OR = 1.14, 95% CI = 1.01 – 1.28, P = 0.030) in very large HDL are positively associated with the risk of MS.
Five of the 29 metabolites are amino acids. The genetically predicted circulating levels of serine (Fig. 3A and 3D, OR = 1.56, 95% CI = 1.25 – 1.95, P = 9.32 × 10−5), lysine (OR = 1.18, 95% CI = 1.01 – 1.38, P = 0.041) and O-sulfo-L-tyrosine (OR = 1.15, 95% CI = 1.01 – 1.31, P = 0.042) are all associated with a higher risk of MS. The additional leave-one-out analysis for serine demonstrated that the causal estimate was not driven by any single SNP (Fig. 3G). The other two are gamma-glutamyl amino acids, and they have opposite associations. Gamma-glutamyl leucine is negatively (OR = 0.80, 95% CI = 0.67 – 0.96, P = 0.017), while gamma-glutamylphenylalanine is positively (OR = 1.22, 95% CI = 1.04 – 1.43, P = 0.016) associated with the MS risk.
Fig. 3.
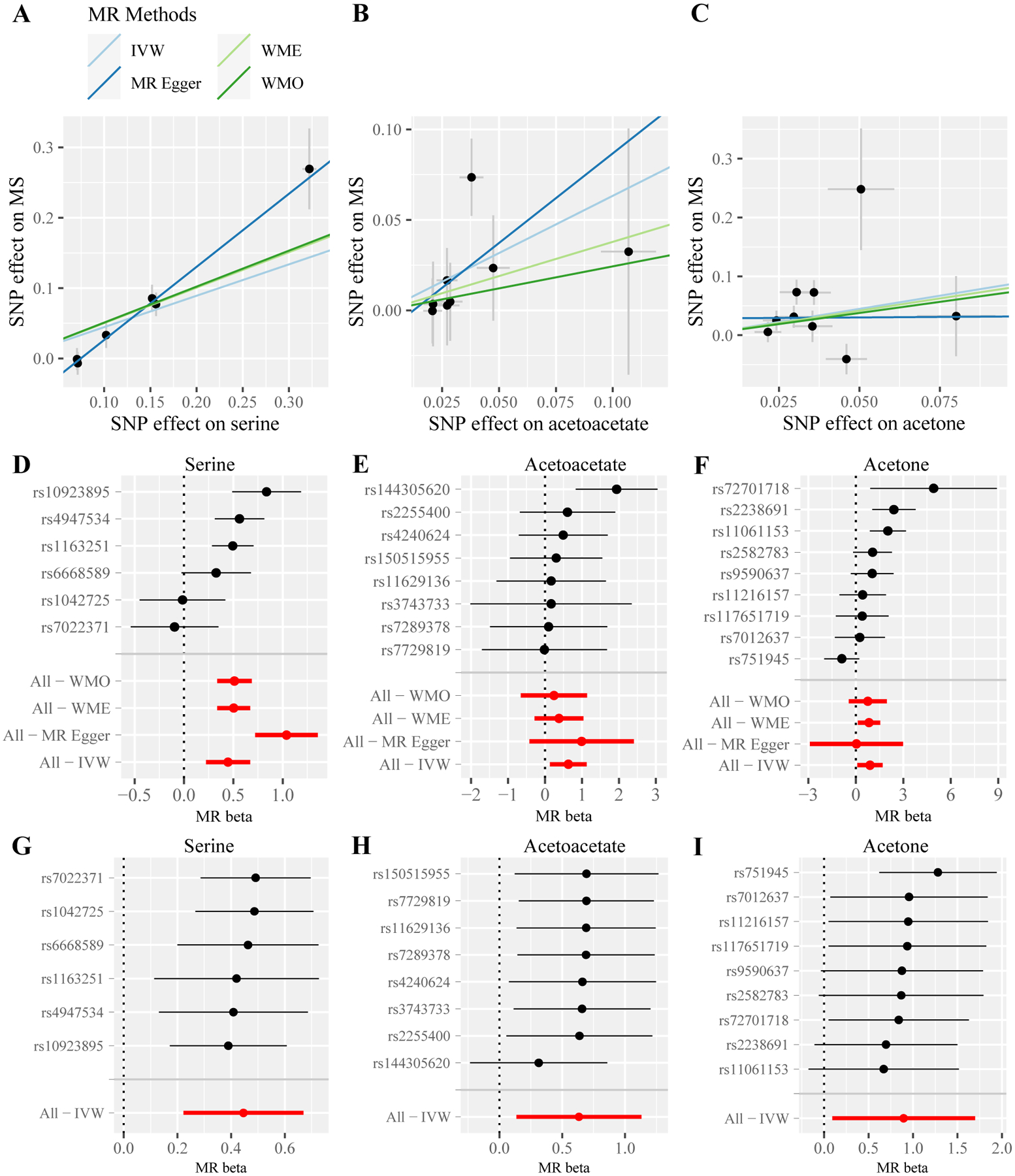
MR estimated effects of three metabolites on MS. Scatter plots for serine (A), acetoacetate (B), and acetone (C) illustrate the individual SNP effects on the metabolite and MS and the estimated linear causal relationship between the metabolite and MS by applying four MR methods. Forest plots for serine (D), acetoacetate (E), and acetone (F) show the causal effect estimates based on individual SNPs and based on all SNPs using four MR methods. Leave-one-out plots for serine (G), acetoacetate (H), and acetone (I) evaluate whether any SNP is driving the causal effect. MR: Mendelian randomization; MS: multiple sclerosis; IVW: inverse variance-weighted; WME: weighted median; WMO: weighted mode; SNP: single nucleotide polymorphism.
Two of the six consistently significant metabolites are acetoacetate (OR = 2.47, 95% CI = 1.14 – 5.34, P = 0.021) and acetone (OR = 2.45, 95% CI = 1.02 – 5.90, P = 0.046), both of which are associated with a higher MS risk (Fig. 3). Visual inspection of the leave-one-out plots suggested the potential presence of outliers of IVs for acetoacetate and acetone (Fig. 3H and 3I). However, further MR-PRESSO analysis did not find any significant outliers for acetoacetate (global test P > 0.05), while the causal estimate of acetone remained significant after removing the outlier SNPs (OR = 3.60, 95% CI = 1.86 – 6.97, P = 0.007). Another notable metabolite is uridine, which is positively associated with the MS risk (OR = 1.45, 95% CI = 1.10 – 1.91, P = 0.008).
4. Discussion
Our metabolome-wide MR study, the first of its kind for MS, prioritized a list of 29 circulating metabolites that likely have causal associations with the risk of MS. Our results highlighted metabolites in lipid metabolism (e.g., cholesterol and phospholipids in large VLDL and very large HDL), amino acid metabolism (e.g., serine and lysine), and energy metabolism (e.g., acetoacetate and acetone).
Altered lipid metabolism is well-known in MS patients (Lorincz, Jury, 2022). When comparing RRMS patients to age- and sex-matched controls, a metabolomics study found that cholesterol, phospholipids, and triglycerides are elevated in the larger subclasses of VLDL and HDL (Gafson, Thorne, 2018). Two previous MR studies examined the causal roles of HDL-C, low-density lipoprotein cholesterol (LDL-C), and triglycerides in MS. Using GWAS of blood lipids that are independent of our GWAS of metabolomics, they found that HDL-C is positively associated with the MS risk, while no significant effects were found for LDL-C and triglycerides (Almramhi et al. , 2022, Yuan, Xiong, 2021). Our results consistently revealed that total cholesterol, cholesterol esters, and phospholipids in very large HDL are associated with a higher MS risk. Also, we did not find significant effects of lipids in LDL. Our study showed that lipids in large VLDL are associated with a lower MS risk. It is important to note that the previously observed elevated levels of lipids in the larger subclasses of VLDL in RRMS patients may be confounded by reserve causation, as lipid levels may respond to the progression of MS. Our study highlighted the importance of examining the role of lipoprotein subclasses in MS.
Altered circulating levels of amino acids and gamma-glutamyl amino acids have been observed in MS patients (Bhargava and Anthony, 2020, Zahoor, Rui, 2021). A higher serum level of lysine was observed in MS patients when compared to healthy controls (Moussallieh, Elbayed, 2014), and also in MS patients who are in relapse in comparison to those that are a few months after the last relapse (Yeo et al. , 2021). The pattern for serine is more complex. A lower plasma concentration of serine was observed in RRMS patients (Sylvestre et al. , 2020), but a higher serum serine level was found in secondary progressive MS patients (Rzepinski et al. , 2022). On the other hand, in the experimental allergic encephalomyelitis rat model of MS, elevated levels of both lysine and serine were observed in the spinal cord and the brain (Battini et al. , 2018). Our MR analysis indicates that individuals with genetic capacities for higher lysine and serine are more likely to develop MS. Interestingly, it has been shown that EBV infection, a known risk factor for MS (Olsson, Barcellos, 2017), upregulates the import and biosynthesis of serine in B cells (Wang et al. , 2019). Moreover, serine is a precursor for phosphatidylserine and sphingomyelin, both of which are key lipids in myelin and implicated in the demyelination process of MS (Beyer et al. , 2018, Ho et al. , 2012). Therefore, our observations of serine and lipids may be related. As for the two gamma-glutamyl amino acids, both gamma-glutamylleucine and gamma-glutamylphenylalanine were observed to be higher in MS patients than in healthy controls, although only gamma-glutamylleucine reached statistical significance. But both of them were significantly elevated in MS patients receiving vitamin D supplementation (Bhargava, Fitzgerald, 2017). Our MR analysis suggests that gamma-glutamylleucine increases, while gamma-glutamylphenylalanine decreases, the risk of MS. Our results call for future studies into the effects of these amino acids before the onset of MS.
Disrupted nucleotide metabolism and energy metabolism are commonly observed in MS (Bhargava and Anthony, 2020, Zahoor, Rui, 2021). Consistent with our MR result that individuals with a higher genetic capacity for uridine have a higher MS risk, it has been observed that the serum uridine level is higher in MS patients (Lazzarino, Amorini, 2017). Similarly, previous case-control studies observed that MS patients have elevated levels of acetoacetate and acetone in the plasma and the cerebrospinal fluid (Cocco, Murgia, 2016, Kim et al. , 2017). Our MR analysis supports the causal roles of these metabolites in the development of MS. Notably, the higher circulating levels of ketone bodies (i.e., acetoacetate and acetone) may reflect a protective shift in energy metabolism in MS patients, and ketogenic diets have shown suggestive benefits for MS patients (Lin et al. , 2022). It is of great interest to investigate the roles of ketone bodies in the development of MS, in addition to its treatment.
The present study has multiple strengths. First, the two-sample MR study design mitigates biases from residual confounding and reverse causation in observational association studies. Second, we examined an extensive list of metabolites to systematically investigate their causal roles in MR risk. Third, two thresholds (P < 5 × 10−8 and P < 1 × 10−6) were applied to select genetic instruments, and results are consistent across the two analyses. All metabolites examined have strong genetic instruments (all F-statistics >10), mitigating possible biases from weak instruments. Fourth, we applied six MR methods to assess the robustness of causal associations and effect directions, including IVW with a multiplicative random-effects model, MR-Egger, WME, WMO, MR-PRESSO, and MR Steiger. Fifth, most of our identified metabolites have been previously associated with MS status or severity in traditional epidemiological studies. One (i.e., HDL-C) has been found in previous MR studies, while the other metabolites are novel findings from our study.
Nonetheless, several limitations should be considered when interpreting our results. First, our study could not completely rule out the possible presence of horizontal pleiotropy, although we performed comprehensive MR analyses to confirm consistent causal estimations. Second, some metabolites in the original metabolomics data were excluded from our analysis due to their lack of three or more genetic instruments. Third, some metabolites were present in two metabolomics GWAS, mainly met-c and met-d, but they were only significant in one MR analysis. The different cohort characteristics, study designs, and sample sizes may be underlying these differences. However, we did find that the MR estimates between the two metabolomics GWAS are highly correlated (Supplementary Fig. 1). Fourth, our study could not differentiate between MS subtypes. It is of great interest to perform a similar analysis for MS subtypes in the future when large GWAS of these subtypes become available. Fifth, the current MR methods assume a linear relationship between the exposure and the outcome, which may not be the case for some metabolite and MS. Sixth, the MR estimates reflect the lifelong effects of an exposure and provide no information about the critical window of the exposure action. Last, our study was restricted to individuals of European descent to reduce possible bias from population stratification, but it limits the generalizability of our results to other populations.
Our metabolome-wide MR study prioritized metabolites, such as lysine, serine, acetone, acetoacetate, and various lipids, in the lipid, amino acids and energy metabolism that likely play causal roles in the development of MS. They may serve as diagnostic biomarkers to identify individuals at high risk for early prevention. Future studies on these metabolites will further our understanding of the MS pathogenesis and evaluate the efficacy of these metabolites as therapeutic targets.
Supplementary Material
Supplementary Fig. 1. Consistency of MR estimates for the same metabolites between two significant cutoffs, P < 5 × 10−8 and P < 1 × 10−6 (A, B), and between two metabolomics GWAS, met-c and met-d (C, D). Only point estimates from the multiplicative random effect IVW method were shown in A and C. The blue line is the linear regression line. The corresponding 95% confidence intervals were shown in B and D. The red dashed line indicates y = x.
Supplementary Table 1. All Mendelian randomization results using the instrumental variables selection threshold (P < 5 × 10−8). b: causal effect size; se: standard error; pval: p-value; IVW_MRE: inverse-variance weighted random-effects model; IVW_FE: inverse-variance weighted fixed-effects model; Egger: MR-Egger; Het: heterogeneity; W_Med: weighted median; W_Mod: weighted mode; nsnps: number of SNPs retained for this analysis; SD: standard deviation of metabolites.
Supplementary Table 2. All Mendelian randomization results using the instrumental variables selection threshold (P < 1 × 10−6). b: causal effect size; se: standard error; pval: p-value; IVW_MRE: inverse-variance weighted random-effects model; IVW_FE: inverse-variance weighted fixed-effects model; Egger: MR-Egger; Het: heterogeneity; W_Med: weighted median; W_Mod: weighted mode; nsnps: number of SNPs retained for this analysis; SD: standard deviation of metabolites.
Highlights.
Mendelian randomization analysis leverages genetic variants associated with a biomarker to infer possible causal effects of the biomarker on a disease.
We performed the first metabolome-wide Mendelian randomization analysis to prioritize potential causal circulating metabolites for multiple sclerosis.
Twenty-nine metabolites, including serine, lysine, acetone, acetoacetate, and lipids, likely have causal roles in multiple sclerosis.
Acknowledgments
We thank the IMSGC and the MRC IEU OpenGWAS for access to the GWAS summary statistics.
Funding
This work was funded by the University of Georgia Research Foundation and by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM143060.
Footnotes
Declaration of Competing Interest
No competing interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almramhi MM, Finan C, Storm CS, Schmidt AF, Kia DA, Coneys R, et al. Exploring the Role of Plasma Lipids and Statins Interventions on Multiple Sclerosis Risk and Severity: A Mendelian Randomization Study. medRxiv. 2022:2022.08.01.22277781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battini S, Bund C, Moussallieh FM, Cicek AE, De Seze J, Namer IJ. Metabolomics approaches in experimental allergic encephalomyelitis. J Neuroimmunol. 2018;314:94–100. [DOI] [PubMed] [Google Scholar]
- Beyer BA, Fang M, Sadrian B, Montenegro-Burke JR, Plaisted WC, Kok BPC, et al. Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat Chem Biol. 2018;14:22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava P, Anthony DC. Metabolomics in multiple sclerosis disease course and progression. Mult Scler. 2020;26:591–8. [DOI] [PubMed] [Google Scholar]
- Bhargava P, Fitzgerald KC, Calabresi PA, Mowry EM. Metabolic alterations in multiple sclerosis and the impact of vitamin D supplementation. JCI Insight. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzer S, Jiang X, Hillert J, Manouchehrinia A. Depression and multiple sclerosis: A bidirectional Mendelian randomisation study. Mult Scler. 2021;27:1799–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent JR, Foley CN, Grant AJ, Mason AM, Staley JR, Burgess S. MendelianRandomization v0.5.0: updates to an R package for performing Mendelian randomization analyses using summarized data. Wellcome Open Res. 2020;5:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MREgger method. Eur J Epidemiol. 2017;32:377–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco E, Murgia F, Lorefice L, Barberini L, Poddighe S, Frau J, et al. (1)H-NMR analysis provides a metabolomic profile of patients with multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2016;3:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsworth B, Lyon M, Alexander T, Liu Y, Matthews P, Hallett J, et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv. 2020:2020.08.10.244293. [Google Scholar]
- Feingold KR. Lipid and Lipoprotein Metabolism. Endocrinology and Metabolism Clinics of North America. 2022;51:437–58. [DOI] [PubMed] [Google Scholar]
- Gafson AR, Thorne T, McKechnie CIJ, Jimenez B, Nicholas R, Matthews PM. Lipoprotein markers associated with disability from multiple sclerosis. Sci Rep. 2018;8:17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34:2926–40. [DOI] [PubMed] [Google Scholar]
- Harroud A, Manousaki D, Butler-Laporte G, Mitchell RE, Davey Smith G, Richards JB, et al. The relative contributions of obesity, vitamin D, leptin, and adiponectin to multiple sclerosis risk: A Mendelian randomization mediation analysis. Mult Scler. 2021a;27:1994–2000. [DOI] [PubMed] [Google Scholar]
- Harroud A, Marrie RA, Fitzgerald KC, Salter A, Lu Y, Patel M, et al. Mendelian randomization provides no evidence for a causal role in the bidirectional relationship between depression and multiple sclerosis. Mult Scler. 2021b;27:2077–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harroud A, Mitchell RE, Richardson TG, Morris JA, Forgetta V, Davey Smith G, et al. Childhood obesity and multiple sclerosis: A Mendelian randomization study. Mult Scler. 2021c;27:2150–8. [DOI] [PubMed] [Google Scholar]
- Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PP, Kanter JL, Johnson AM, Srinagesh HK, Chang EJ, Purdy TM, et al. Identification of naturally occurring fatty acids of the myelin sheath that resolve neuroinflammation. Sci Transl Med. 2012;4:137ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs BM, Noyce AJ, Giovannoni G, Dobson R. BMI and low vitamin D are causal factors for multiple sclerosis: A Mendelian Randomization study. Neurol Neuroimmunol Neuroinflamm. 2020;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen J, Demirkan A, Wurtz P, Draisma HH, Haller T, Rawal R, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Jeong IH, Hyun JS, Kong BS, Kim HJ, Park SJ. Metabolomic profiling of CSF in multiple sclerosis and neuromyelitis optica spectrum disorder by nuclear magnetic resonance. PLoS One. 2017;12:e0181758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarino G, Amorini AM, Petzold A, Gasperini C, Ruggieri S, Quartuccio ME, et al. Serum Compounds of Energy Metabolism Impairment Are Related to Disability, Disease Course and Neuroimaging in Multiple Sclerosis. Mol Neurobiol. 2017;54:7520–33. [DOI] [PubMed] [Google Scholar]
- Lin WS, Lin SJ, Liao PY, Suresh D, Hsu TR, Wang PY. Role of Ketogenic Diets in Multiple Sclerosis and Related Animal Models: An Updated Review. Adv Nutr. 2022;13:2002–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz B, Jury EC, Vrablik M, Ramanathan M, Uher T. The role of cholesterol metabolism in multiple sclerosis: From molecular pathophysiology to radiological and clinical disease activity. Autoimmun Rev. 2022;21:103088. [DOI] [PubMed] [Google Scholar]
- Moussallieh FM, Elbayed K, Chanson JB, Rudolf G, Piotto M, De Seze J, et al. Serum analysis by 1H nuclear magnetic resonance spectroscopy: a new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult Scler. 2014;20:558–65. [DOI] [PubMed] [Google Scholar]
- Niu PP, Song B, Wang X, Xu YM. Serum Uric Acid Level and Multiple Sclerosis: A Mendelian Randomization Study. Front Genet. 2020;11:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson T, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13:25–36. [DOI] [PubMed] [Google Scholar]
- Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson TG, Leyden GM, Wang Q, Bell JA, Elsworth B, Davey Smith G, et al. Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol. 2022;20:e3001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzepinski L, Koslinski P, Gackowski M, Koba M, Maciejek Z. Amino Acid Levels as Potential Biomarkers of Multiple Sclerosis in Elderly Patients: Preliminary Report. J Clin Neurol. 2022;18:529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre DA, Slupsky CM, Aviv RI, Swardfager W, Taha AY. Untargeted metabolomic analysis of plasma from relapsing-remitting multiple sclerosis patients reveals changes in metabolites associated with structural changes in brain. Brain Res. 2020;1732:146589. [DOI] [PubMed] [Google Scholar]
- Vandebergh M, Becelaere S, Group CIW, Dubois B, Goris A. Body Mass Index, Interleukin-6 Signaling and Multiple Sclerosis: A Mendelian Randomization Study. Front Immunol. 2022;13:834644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Shen H, Nobre L, Ersing I, Paulo JA, Trudeau S, et al. Epstein-Barr-Virus-Induced One-Carbon Metabolism Drives B Cell Transformation. Cell Metab. 2019;30:539–55 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo T, Probert F, Sealey M, Saldana L, Geraldes R, Hoeckner S, et al. Objective biomarkers for clinical relapse in multiple sclerosis: a metabolomics approach. Brain Commun. 2021;3:fcab240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Xiong Y, Larsson SC. An atlas on risk factors for multiple sclerosis: a Mendelian randomization study. J Neurol. 2021;268:114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahoor I, Rui B, Khan J, Datta I, Giri S. An emerging potential of metabolomics in multiple sclerosis: a comprehensive overview. Cell Mol Life Sci. 2021;78:3181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Consistency of MR estimates for the same metabolites between two significant cutoffs, P < 5 × 10−8 and P < 1 × 10−6 (A, B), and between two metabolomics GWAS, met-c and met-d (C, D). Only point estimates from the multiplicative random effect IVW method were shown in A and C. The blue line is the linear regression line. The corresponding 95% confidence intervals were shown in B and D. The red dashed line indicates y = x.
Supplementary Table 1. All Mendelian randomization results using the instrumental variables selection threshold (P < 5 × 10−8). b: causal effect size; se: standard error; pval: p-value; IVW_MRE: inverse-variance weighted random-effects model; IVW_FE: inverse-variance weighted fixed-effects model; Egger: MR-Egger; Het: heterogeneity; W_Med: weighted median; W_Mod: weighted mode; nsnps: number of SNPs retained for this analysis; SD: standard deviation of metabolites.
Supplementary Table 2. All Mendelian randomization results using the instrumental variables selection threshold (P < 1 × 10−6). b: causal effect size; se: standard error; pval: p-value; IVW_MRE: inverse-variance weighted random-effects model; IVW_FE: inverse-variance weighted fixed-effects model; Egger: MR-Egger; Het: heterogeneity; W_Med: weighted median; W_Mod: weighted mode; nsnps: number of SNPs retained for this analysis; SD: standard deviation of metabolites.
Data Availability Statement
All GWAS summary statistics can be accessed through the OpenGWAS project (Elsworth, Lyon, 2020, Hemani, Zheng, 2018). The MR analysis scripts can be found at https://github.com/yitangsun/metabolome-wide-MR-for-MS. The key MR results have been replicated by another author, and the scripts can be found here https://github.com/angelage678/Mendelian-Randomization---multiple-sclerosis-and-metabolites.


